Abstract
Use of high-throughput, in vitro bioactivity data in setting a point-of-departure (POD) has the potential to accelerate the pace of human health safety evaluation by informing screening level assessments. The primary objective of this work was to compare PODs based on high-throughput predictions of bioactivity, exposure predictions, and traditional hazard information for 448 chemicals. PODs derived from new approach methodologies (NAMs) were obtained for this comparison using the 50th (PODNAM,50) and the 95th (PODNAM,95) percentile credible interval estimates for the steady-state plasma concentration used in in vitro to in vivo extrapolation of administered equivalent doses (AEDs). Of the 448 substances, 89% had a PODNAM,95 that was less than the traditional POD (PODtraditional) value. For the 48 substances for which PODtraditional < PODNAM,95, the PODNAM and PODtraditional were typically within a factor of 10 of each other, and there was an enrichment of chemical structural features associated with organophosphate and carbamate insecticides. When PODtraditional < PODNAM,95, it did not appear to result from an enrichment of PODtraditional based on a particular study type, (e.g. developmental, reproductive, chronic studies). Bioactivity:exposure ratios (BERs), useful for identification of substances with potential priority, demonstrated that high-throughput exposure predictions were greater than the PODNAM,95 for 11 substances. When compared to threshold of toxicological concern (TTC) values, the PODNAM,95 was greater than the corresponding TTC value 90% of the time. This work demonstrates the feasibility, and continuing challenges, of using in vitro bioactivity as a protective estimate of POD in screening level assessments via a case study.
Keywords: High-throughput screening; high-throughput toxicokinetics; threshold of toxicological concern (TTC), point of departure (POD); new approach methodologies
1. Introduction
The future of chemical risk assessment is moving towards high-throughput approaches that can provide preliminary estimates of hazard and exposure. The utilization and sharing of these new approaches and associated data internationally is imperative because each regulatory authority is addressing distinct but related challenges in chemical screening and evaluation. A commonality among these challenges is the need to prioritize chemicals for further evaluation and conduct screening-level assessments, as there are thousands of chemicals with potential human exposures but with minimal hazard information (Egeghy et al., 2012; Judson et al., 2009). New approach methodologies (NAMs) (ECHA, 2016; EPA, 2018a) include in vitro and in silico approaches for prediction of hazard and exposure, thereby enabling solutions to some of these regulatory challenges. NAMs for hazard evaluation can be used in high-throughput formats, and in some cases may identify chemical mechanisms of action. NAMs for exposure provide rapid estimates using limited information for more chemicals than lower-throughput models can achieve. The promise of NAMs is motivating regulatory authorities to define and adopt fit-for-purpose NAMs, and support efforts to reduce, refine, and replace resource-intensive vertebrate animal tests. International collaborative efforts that deepen understanding of NAMs and their application while preventing duplicative efforts have become a salient need.
There are several key regulatory drivers of international use of NAMs in toxicology applications. In the US, the amended Toxic Substances Control Act (TSCA) (Lautenberg, 2016) requires a risk-based screening process for prioritizing chemicals as high-priority substances for risk evaluation or low-priority sibstances for which risk evaluations are not warranted. The amended TSCA requires the U.S. Environmental Protection Agency (EPA) to develop a plan, “to promote the development and implementation of alternative test methods and strategies to reduce, refine, or replace vertebrate animal testing and provide information of equivalent or better scientific quality and relevance for assessing risks of injury to health or the environment” (EPA, 2018a). In the European Union (EU), the European Chemical Agency (ECHA) regulates chemical substances under the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) (Commission, 2007), that also promotes the use of NAMs as a means to increase the data availability for data poor substances (ECHA, 2016, 2017). Health Canada (HC) and Environment and Climate Change Canada (ECCC) are continuing work under the Chemicals Management Plan (CMP) to address human health and ecological concerns for approximately 4,300 prioritized substances on the Canadian Domestic Substances List (DSL) by the year 2020 (ECCC/HC, 2016a). High-throughput NAMs have been identified as a possible means to meet near-term timelines for screening assessments and to inform selection of future priorities as the program continues to evolve post-2020 (ECCC/HC, 2016b). These and other regulatory drivers underscore the need for an international discussion about how to apply NAMs in a transparent and effective way.
One aspect of prioritization and screening-level assessment strategies is a trade-off between speed and uncertainty. Though the specific implications of these strategies are likely to differ by regulatory authority, each of the aforementioned regulatory agencies are responsible for protecting human and environmental health and have traditionally relied on in vivo studies in efforts to achieve this mission. Despite increasing interest in prediction of human health hazard directly, rather than relying on animal models and associated extrapolation concerns, a conceptual bridge for toxicologists to understand how current practice could be augmented by incorporation of NAMs will enable greater discussion and progress. Thus, there is a clear need and opportunity to demonstrate how preliminary screening-level risk assessment using a NAM-based approach would perform when compared to traditional points of departure (PODs). Acknowledging and documenting the caveats and limitations of this comparison is central to building the confidence and insight needed to employ NAMs. Hence, the study documented here sought to use as many chemicals as possible to illustrate how the current state-of-the-science would support NAM-based screening-level risk assessment.
NAMs for exposure, bioactivity, and in vitro to in vivo extrapolation are available, for differing numbers of substances, to inform this exercise of demonstrating a risk-based screening-level assessment approach. High-throughput exposure predictions have been generated using a series of computational models under the ExpoCast project, the second version of which used a series of heuristics (Wambaugh et al., 2014) including chemical use type (Dionisio et al., 2015) and production volume to quantitatively predict exposure for thousands of chemicals. The US Environmental Protection Agency (EPA) Toxicity Forecaster (ToxCast) program (Kavlock et al., 2012) and the interagency Tox21 project (Thomas et al., 2018; Tice et al., 2013) provide publicly available, high-throughput in vitro bioactivity information for a diverse biological and chemical space. Additionally, for this case study, researchers at the Singapore Agency for Science, Technology, and Research (A*STAR) provided high-throughput phenotypic profiling information from three cell-based toxicity models for lung, kidney, and liver toxicity for a subset of substances (Lee et al., 2018; Su et al., 2016). Using high-throughput toxicokinetic (httk) information and reverse dosimetry, all of these bioactivity data, i.e. the micromolar concentration of a substance that altered an assay signal in vitro, were transformed into administered equivalent doses (AEDs) in milligram per kilogram bodyweight per day (mg/kg/day) units within a complex process referred to as in vitro to in vivo extrapolation (IVIVE). The reverse dosimetry component of IVIVE in this case relies on the assumption that a nominal in vitro assay concentration approximates an in vivo serum concentration using steady state kinetics, and then involves a toxicokinetic model to estimate the external exposures (in mg/kg/day units) that may have resulted in that concentration (Bell et al., 2017; Jamei et al., 2009; Sipes et al., 2017b; Wambaugh et al., 2018; Wetmore et al., 2014; Wetmore et al., 2015; Wetmore et al., 2013; Wetmore et al., 2012). A NAM-based POD, or PODNAM, can be selected from the range of AEDs. This PODNAM can be compared to exposure predictions to develop a bioactivity:exposure ratio (BER) to provide a risk-based context.
To understand the possible added benefit of in vitro bioactivity-derived PODNAM, a well-known, protective in silico approach that can be used in the absence of in vitro bioactivity information, the threshold of toxicological concern (TTC), was also included for comparison to PODNAM and exposure values. In addition, the PODNAM can be compared to traditional in vivo data for these chemicals, aggregated and summarized as the traditional POD (PODtraditional). Curation of these traditional data from in vivo toxicity testing has provided an important resource to evaluate whether PODNAM is protective relative to the PODtraditional (understanding that the PODtraditional itself is an approximation using animal models). Two examples of publicly-available curated traditional data include the Toxicity Reference Database (ToxRefDB) (Martin et al., 2009a; Martin et al., 2009b) and the Toxicity Value Database (ToxValDB) (Williams et al., 2017) the latter of which aggregates summary level information from over 40 sources, including ToxRefDB; EPA sources, such as the High Production Volume Information System (HPVIS), Integrated Risk Information System (IRIS) (https://www.epa.gov/iris), Provisional Peer-Reviewed Toxicity Values (PPRTVs) (https://hhpprtv.ornl.gov/), curated data from Office of Water (OW), Office of Land and Emergency Management (OLEM), and the Office of Pollution Prevention and Toxics (OPPT); other US state and federal sources, such as the Food and Drug Administration (FDA), U.S. Geological Survey (USGS), Department of Defense (DOD), Department of Energy (DOE), and California EPA (CalEPA); and international sources, such as ECHA via eChem Portal (https://www.echemportal.org/echemportal/index.action) and EFSA via the Chemical Hazards Database (https://www.efsa.europa.eu/en/data/chemical-hazards-data), the World Health Organization (WHO), the Cosmetics Ingredients Safety (COSMOS) database (https://cosmosdb.eu/cosmosdb.v2/accounts/login/?next=/cosmosdb.v2/), Health Canada, and the Hazard Evaluation Support System (HESS) (Williams, et al., 2017). The traditional data included for derivation of a PODtraditional included many study designs, including repeat-dose studies such as subacute, subchronic, chronic, reproductive, developmental and/or multi-generation reproduction studies, among others. Given that most of the in vitro bioactivity data measures disruption of a molecular target, pathway, or cellular function, rather than adversity at the tissue, organism, or population level as measured in in vivo toxicity studies, this work evaluates the hypothesis that the PODNAM would be protective relative to the PODtraditional across multiple study types and durations of exposure.
Several examples and case studies have considered the possibility of using high-throughput data in various regulatory decision contexts, from prioritization, to test replacement, to use in chemical-specific assessment (Browne et al., 2015; Cote et al., 2016; Judson et al., 2014; Judson et al., 2011; Kleinstreuer et al., 2017; Paul Friedman et al., 2016; Pradeep et al., 2017; Thomas et al., 2013). Progress has been made in the acceptance of NAMs for prioritization of chemicals subject to the US EPA Endocrine Disruptor Screening Program (EDSP) (Browne, et al., 2015; Kleinstreuer, et al., 2017) and as alternatives for existing in vivo endocrine disruptor-related test guidelines (EFSA, 2018; USEPA, 2015). Further regulatory acceptance of NAMs is demonstrated by the development of defined approaches for assessment of skin sensitization, with the goal of developing an internationally-recognized test guideline using an integrated set of NAMs to predict human skin sensitization hazard potential (Casati et al., 2018). The use of NAMs for determination of either the dose that may alter specific biological pathway activities of interest (e.g. nuclear receptor signaling) or general in vitro bioactivity has also demonstrated promise for prioritizing substances (Judson, et al., 2014; Judson, et al., 2011; Wetmore, et al., 2013). However, as with some in vivo toxicity studies, for many substances it may not be possible to identify a specific human health outcome, predominant mode-of-action (MoA), or adverse outcome pathway (AOP) based on the in vitro bioactivity data. Hence, screening-level assessment may require identification of a threshold dose at which no bioactivity would be observed in assays covering a broad biological space (Thomas, et al., 2013). Prioritization based on the integration of bioactivity data and predicted exposures has been suggested as a path forward for addressing the problem of thousands of chemicals with limited information for assessment by many groups, including Health Canada in their approach to the Chemical Management Plan (ECCC/HC, 2016a), academics, government scientists, and chemical industry scientists (Becker et al., 2015; Embry et al., 2014; Perkins et al., 2017; Sipes, et al., 2017b). The retrospective case study presented herein advances the integrated use of NAMs for in vitro bioactivity and exposure by addressing the following questions: can the proposed workflow to derive a PODNAM be shown to be broadly protective for potential application to screening-level chemical assessments independent of the biological events or adverse outcome pathways involved? Further, does in vitro bioactivity combined with exposure estimates provide a useful risk-informed prioritization metric?
Importantly, this work asks these questions as viewed through a multi-agency, international lens. The Accelerating the Pace of Chemical Risk Assessment (APCRA) initiative is an international cooperative collaboration of government agencies convened to address barriers and opportunities for the use of NAMs in chemical risk assessment (Kavlock, 2016; Kavlock et al., 2018). This initiative includes participants from across offices of the EPA, ECHA, EFSA, the U.S. National Toxicology Program (NTP), CalEPA, Health Canada, the European Commission’s Joint Research Center (JRC),the Organisation for Economic Cooperation and Development (OECD), France’s INERIS, the Australian National Industrial Chemicals Notification and Assessment Scheme (NICNAS), the National Institute for Public Health and the Environment (RIVM) of the Netherlands, the Japanese Ministry of Health, Welfare, and Labour, Korea’s Ministry of Environment, Singapore’s Agency for Science and Technology Research (A*STAR), and the Taiwanese Safety and Health Technology Center (SAHTECH). An initial goal of this group was to directly identify and address obstacles to adoption of NAMs in regulatory decision-making, considering the geographic differences in regulatory perspectives and requirements while understanding that generation and analysis of the data for substances of concern can be shared. This first APCRA case study is the result of a collaborative discourse within APCRA that aims to evaluate how the PODNAM compares to the PODtraditional across 448 chemicals with high-throughput hazard and toxicokinetic information in the context of screening-level assessments. We evaluate whether the PODNAM can serve as a “lower bound” estimate of the PODtraditional. In addition, this case study incorporates high-throughput exposure information to examine the BER as a potential metric for prioritization. This work intends to increase confidence in NAM-based workflows that could be used in regulatory decision making by presenting a case study of how this might be applied.
2. Methods
2.1. Overview of the approach
This section gives a brief overview of the approach, as illustrated in Figure 1 and with additional details provided in subsequent sections of the Methods and the Supplemental Appendix.
Figure 1. Overall workflow of the case study.
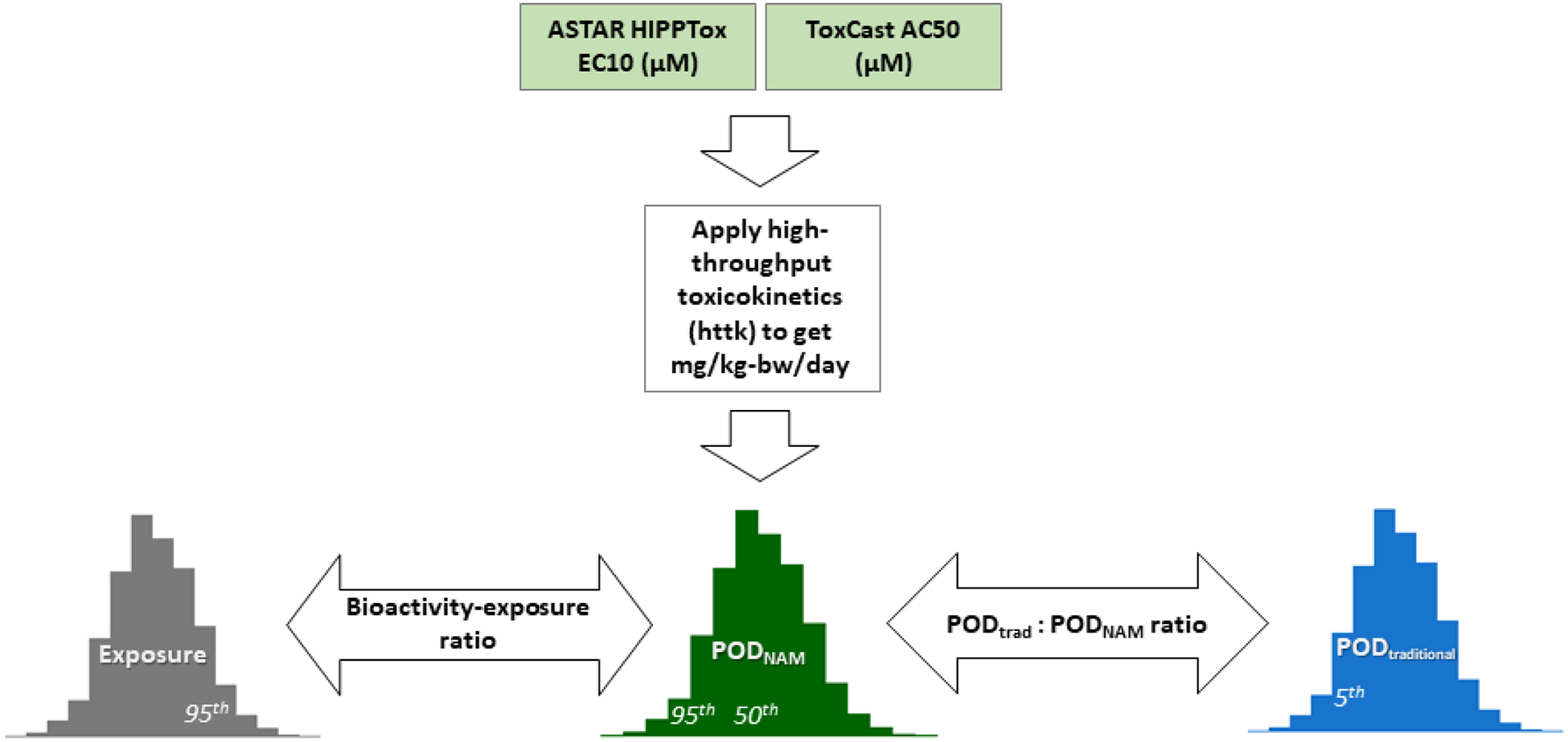
This case study includes 448 substances with exposure predictions, in vitro assay data, HTTK information, and in vivo hazard information. The 50th and 95th percentile from the Monte Carlo simulation of inter-individual toxicokinetic variability were used to estimate AEDs, and the minimum of either the ToxCast or HIPPTox-based AEDs were selected as the PODNAM, 50 or PODNAM, 95. The PODNAM estimates were compared to the 5th percentile from the distribution of the PODtraditional values obtained from multiple sources to obtain the log10POD ratio. The log10BER was obtained by comparing the PODNAM estimates to exposure predictions. All values used for computation were in log10-mg/kg-bw/day units
First, in vitro bioactivity data were aggregated to develop PODNAM estimates. Data were available from ToxCast for all 448 substances, and high-throughput phenotypic profiling toxicity (HIPPTox) data from A*STAR was available for 57 substances in this case study. For each ToxCast substance, a 5th percentile was calculated based on the distribution of 50% maximal activity concentration (AC50) values. For the HIPPTox data, a POD was defined differently (and referred to as the HIPPTox-POD). The HIPPTox-POD attempts to identify the lowest concentrations for any change in the measured cellular phenotypes of three cell models and uses EC10 to represent a threshold for this activity. For the HIPPTox data for kidney, liver, and/or lung toxicity, the minimum 10% effect concentration (min EC10) was calculated. The intent in selecting the minimum of either the 5th percentile of the ToxCast AC50 values or the minimum HIPPTox value was to provide a “lower bound” estimate on a bioactive concentration in vitro. The lower value of either the ToxCast 5th percentile or the HIPPTox min EC10 was assumed to represent the steady state plasma concentration that was then used to calculate the administered equivalent dose (AED) values using high throughput toxicokinetic (HTTK) information from the httk R package (Pearce et al., 2017). The HTTK model (built into the R package) used Monte Carlo simulation to incorporate population variability. The PODNAM values used in this work correspond to the 50th and 95th percentile in the population distribution of steady state AED values and are referred to as the PODNAM,50 and the PODNAM,95, respectively.
Second, after derivation of PODNAM values based on in vitro data, a series of comparisons to other values were made. The intersection of CASRN between ToxCast and HTTK information was used to obtain PODtraditional from ToxValDB and sources from the case study partners, including ECHA, EFSA, and Health Canada. The PODNAM was compared to the PODtraditional to derive a POD ratio (log10 PODtraditional:PODNAM) for both PODNAM,50 and PODNAM,95. Exposure predictions for the total U.S. population from the ExpoCast Systematic Empirical Evaluation of Models version 2 (SEEM2) framework (Wambaugh, et al., 2014) were used to derive a bioactivity:exposure ratio (BER). To understand how NAMs in this work compared to the TTC (HealthCanada, 2016; Kroes et al., 2004; Patlewicz et al., 2008), the PODNAM,95 was also compared to a TTC to derive a PODNAM,95:TTC ratio. The PODtraditional:PODNAM ratios, the BER, and the PODNAM,95:TTC ratio, expressed as logarithms in base 10, are the main metrics employed to evaluate the hypotheses in this study.
All of the data sources used in this case study, including chemical use type, high-throughput bioactivity data, HTTK, in vivo data, and exposure information, are summarized in Table 1, including the version and citations if applicable. Supplemental File 1 contains all of the in vivo POD information used, and Supplemental File 2 contains all of the values derived in this work (including BER and POD ratio). The software (written using R version 3.5.1) and all required source files are available via the US EPA GitHub repository (https://github.com/USEPA/-Examining-the-Utility-of-In-Vitro-Bioactivity) and FTP (ftp://newftp.epa.gov/COMPTOX/NCCT_Publication_Data/FriedmanPaul_K/APCRA_retrospective).
Table 1.
Description of data sources used.
| Data stream | Source | Version | Notes |
|---|---|---|---|
| Functional Use Categories | EPA’s Aggregated Computational Toxicology Online Resource (ACToR) | 2014 | Broad use categories (Dionisio et al., 2015; Wambaugh et al., 2014) used in ExpoCast SEEM2 were also used to describe the functional diversity of the 448 substances in this case study. |
| High-throughput bioactivity data | ToxCast | Invitrodb_v3 | This is the public release of invitrodb dated September 2018 (EPA, 2018). These data were fit using the ToxCast Data Pipeline approach (tcpl R package v2). The data used in this case study are available as Supplemental File X. |
| In vitro phenotypic profiles of lung, kidney, and liver cell models (HIPPTox) | Performed by A*STAR for this case study | The cell models and phenotypic readouts were described previously (Lee et al., 2018; Su et al., 2016). All phenotypic readouts (not limited to those predictive of tissue-specific adversary effects) were used in computation of the HIPPTox-POD. | |
| Toxicokinetics | High-throughput toxicokinetic (httk) data | Httk R package v1.8 | Httk R package v1.8 is available from CRAN (https://cran.r-project.org/web/packages/httk/index.html) |
| In vivo PODsa | ToxValDB in vivo toxicity information | Development v5 (May 2018) | This database includes summary point-of-departure information from multiple databases (as described in text) and study types, and is public in the CompTox Chemicals Dashboard. |
| ECHA | Repeated dose study results via the oral route in REACH registration dossiers | These data are publicly available at https://echa.europa.eu/ | |
| EFSA | Published human health risk assessments in support of EU food law 158/2002 | These data include PODs from multiple study types, mostly from acute, subchronic, chronic, and reproduction toxicity studies. | |
| Health Canada | Published risk assessments conducted for existing substances under the Canadian Environmental Protection Act, 1999 | Information was retrieved based on the availability of a published risk assessment conducted under various phases of Canada’s Chemicals Management Plan and earlier initiatives as well as corresponding availability of ToxCast and HTTK data. Point of departure information was extracted from oral repeat-dose studies (of various durations) as well as from developmental and reproductive toxicity studies cited within the assessments. Where possible, both the NO(A)EL and LO(A)EL for each study were collected and the basis for the effect level is described (ECCC/HC, 2016). | |
| Exposure | ExpoCast predictions | Systematic Empirical Evaluation of Models version 2 (SEEM2) | The median and 95th percentile on the credible interval for the total US population exposure estimates were used (Wambaugh, et al., 2014). |
| Health Canada | Published risk assessments conducted for existing substances under the Canadian Environmental Protection Act, 1999 | Exposure estimates were extracted from the same assessments as their respective in vivo POD values. This included the estimated daily intakes from environmental media as well as intakes from use of certain sentinel consumer products (ECCC/HC, 2016). |
All in vivo POD data from source databases were concatenated and are available in Supplemental File 1.
References for Table 1.
Dionisio, K. L., Frame, A. M., Goldsmith, M. R., Wambaugh, J. F., Liddell, A., Cathey, T., Smith, D., Vail, J., Ernstoff, A. S., Fantke, P., et al. (2015). Exploring consumer exposure pathways and patterns of use for chemicals in the environment. Toxicol Rep 2, 228–237.
ECCC/HC (2016). Chemicals Management Plan. In (H. C. Environment and Climate Change, Ed.), Vol. https://www.canada.ca/en/health-canada/services/chemical-substances/chemicals-management-plan.html. Government of Canada, Ottawa (ON).
EPA, U. (2018). ToxCast & Tox21 Data from invitrodb_v3. In (Retrieved from http://www2.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data.
Lee, J. J., Miller, J. A., Basu, S., Kee, T. V., and Loo, L. H. (2018). Building predictive in vitro pulmonary toxicity assays using high-throughput imaging and artificial intelligence. Arch Toxicol 92(6), 2055–2075.
Su, R., Xiong, S., Zink, D., and Loo, L. H. (2016). High-throughput imaging-based nephrotoxicity prediction for xenobiotics with diverse chemical structures. Arch Toxicol 90(11), 2793–2808.
Wambaugh, J. F., Wang, A., Dionisio, K. L., Frame, A., Egeghy, P., Judson, R., and Setzer, R. W. (2014). High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ Sci Technol 48(21), 12760–7.
2.2. Comparison of high-throughput bioactivity, in vivo point-of-departure, and exposure information
2.2.1. Substance identification
Compilation of the data for this case study resulted in a total of 448 chemicals with the requisite in vitro bioactivity, high-throughput toxicokinetic, exposure prediction, and traditional animal in vivo toxicity values. Each CASRN from the intersection of data sources was mapped to a registered substance identifier (DTXSID) in EPA’s DSSTox database through the Batch Search feature of the EPA CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/dsstoxdb/batch_search) (Williams, et al., 2017). Mapping to DTXSID enabled mapping from substance to identifiers that indicate specific structure(s) needed for use in evaluation of enrichment of structural features and generation of TTC values. Linking data records to DTXSID promotes data interoperability and clarity on the specific chemical structures used as databases including ToxCast, ToxValDB, and HTTK databases, among others, rapidly evolve.
The substance use categories utilized in ExpoCast SEEM2 modeling (Dionisio, et al., 2015; Wambaugh, et al., 2014) and available via the Aggregated Computational Toxicology Online Resource (ACToR) were retrieved to evaluate the functional diversity of the 448 substances examined in this case study. In some cases, a substance may be associated with multiple functional uses.
2.2.2. In vitro bioactivity data
In vitro bioactivity data from two sources were used: ToxCast data from the US EPA ToxCast program and HIPPTox data from the A*STAR program. The details of data extraction and selection of an in vitro bioactivity concentration to use for in vivo-to-in vitro extrapolation of AEDs is described in detail below. Briefly, the minimum of either the 5th percentile of the filtered ToxCast AC50 values or the HIPPTox-POD, if available, was used as the in vitro bioactive concentration for each substance in the case study.
2.2.2.1. ToxCast data
The ToxCast high-throughput bioactivity data were obtained from the MySQL database, invitrodb (version 3) (EPA, 2018b), for all 448 chemicals. Only multi-concentration screening data were used, as single concentration screening data were not considered quantitatively informative of a PODNAM. The structure of information in invitrodb and the R package used to maintain the database and perform curve-fitting are described in detail elsewhere (Filer et al., 2017; NCCT, 2018; Watt et al., 2018). The data retrieved included the AC50 and hit-call determination (from level 5 of invitrodb), caution flags on the curve-fitting for each AC50 (from level 6), and quantitative uncertainty associated with the curve-fitting (from level 7). The caution flag and uncertainty information from levels 6 and 7 were used to filter the ToxCast dataset, with the intent of removing AC50 values from the dataset that originate from curve-fits that may be less informative on a quantitative basis. As the ToxCast data pipeline is a semi-automated, first-tier analysis tool for heterogeneous data, using these data on a single substance-basis presents a challenge as a subset of the potency values may be from curve fits that may be artefacts of the curve-fitting workflow. In this work, we implemented a filtering of the curves available for each substance prior to estimation of the 5th percentile on the distribution of ToxCast AC50 values by substance.
Level 6 caution flag information denotes curve behavior that may indicate a less quantitatively-informative AC50 value, such as curves based on a single active concentration, AC50 value lower than lowest concentration screened, borderline activity, efficacy less than 50%, and general indicators of excessive noise and overfitting. There are currently 10 possible caution flags, and the curves for the substances in this case study had zero to six flags associated with them prior to filtering (Supplemental Appendix). Level 7 uncertainty information was generated (Brown et al., in prep) using bootstrap resampling to define the reproducibility of the curve fits (Watt and Judson, 2018; toxboot R package v0.2.0). Briefly, toxboot uses smooth, nonparametric bootstrap resampling to add random normally distributed noise to give a resampled set of concentration-response values. The resampled data is fit to the three ToxCast models (constant, Hill, gain-loss), repeated 1000 times, and the variables relating to model fitting parameters are stored in a Mongo database. The resulting data were used to generate point estimates, winning model, and hitcall for each of the 1000 resamples. Summary statistics (hit percent, median AC50, and AC50 95% confidence interval) were generated based on the toxboot resampling. Hit percent is the probability of a positive hitcall given the collection of resampled data. Filtering criteria using level 5, 6, and 7 information were as follows: curves were required to have less than 3 flags, and AC50 value greater than the lowest concentration screened, and a hit percent of greater than or equal to 50%.
Prior to curve filtering, for the 448 chemicals in this case study, the number of ToxCast concentration-response assay endpoints in which each substance was screened varied from 211 to 4557, with a median of 883 assay endpoints; the differences in numbers of assays screened may affect the observed positive hit rate. The filtering criteria described above reduced the total number of curves used from 54,048 to 46,735 (approximately 14% removed). The remaining curves are each associated with zero to 2 caution flags, and a median hit percent of 100, indicating that post-filtering most curves were highly reproducible (ranging 13 to 100). Following filtering, the number of positive hitcalls per substance ranged from 0 to 1351, with a median of 56 positive hitcalls per substance. Most substances (297 out of 448) had hitcall sums of less than 100, and only 36 substances had 5 or fewer positive hitcalls. One substance, phenobarbital (CASRN 50-06-6), was active in 4 out of 290 assay endpoints screened, but all of these were dropped during the filtering process. Thus, for phenobarbital, a single AC50 of 100 μM was used as a representative AC50 at the maximum concentration screened in ToxCast to derive a threshold AED. For phenobarbital, a concentration of 100 μM is actually fairly consistent with in vitro bioactivity from other reports that typically use phenobarbital to induce CYP2B6 in vitro at concentrations ranging from the 100 μM to 2 mM (Faucette et al., 2004; Hariparsad et al., 2017). No cytotoxicity filtering of the ToxCast data was performed. All of the positive data in ToxCast for a given chemical, after the curve filtering described here, were included in the AC50 distribution. The 5th percentile of that distribution was used to identify a minimum bioactive concentration for each chemical in ToxCast, regardless of the specific biological pathways involved. The effects of filtering the ToxCast AC50 values (Supplemental Figures 1–2) and of using the 5th percentile versus the minimum AC50 (Supplemental Figure 3) are further described in the Supplemental Appendix.
2.2.2.2. HIPPTox data
In vitro bioactivity data from the high-content-imaging-based HIPPTox platform were also included for a subset of 57 chemicals examined in this case study. Three human cell lines were tested with the chemicals. They include a bronchial epithelial cell line, BEAS-2B (Lee, et al., 2018), a proximal tubule cell line, HK-2 (Su, et al., 2016), and a hepatocarcinoma cell line, HepG2. Up to 165 phenotypic readouts (Lee, et al., 2018) were measured from the images of the cell models using the cellXpress software v1.4.2 (Laksameethanasan et al., 2013). For each cell model, a series of multivariate classifiers were trained to distinguish the cells treated with a chemical at seven concentrations (0.87 to 500 μM) from the cells treated with DMSO. The classifiers used multivariate phenotypic profiles constructed from all the readouts, and produced a series of classification accuracy values at all the tested concentrations (Loo et al., 2007). Then, the values were fitted using a standard log-logistic model and a flat constant model. The best fitted curve was determined using the Akaike information criterion (AIC). An EC10 was derived from the best fitted curve for each cell model. For curves based on the flat constant model, an arbitrary large number (namely, 105 μM) was used. Finally, the minimum EC10 across the three cell models was supplied for use in this case study as the HIPPTox-POD. Calculation of AEDs and calculation of PODNAM
The minimum of the ToxCast 5th percentile of the AC50 distribution or the HIPPTox-POD was converted to administered equivalent doses (AEDs) using the concept of reverse dosimetry and HTTK information, largely from in vitro experiments. The approach taken using the httk R package (v1.8) was similar to the approach used by Wetmore et al. (2012, 2014), as represented by the following Equation 1:
| Eq 1. |
Where the Css is the steady state plasma concentration estimated based on a 3-compartment steady state model assuming 100% bioavailability. Monte Carlo simulation was used to vary the pharmacokinetic parameters to represent inter-individual variability in a population. Population variability was incorporated into the first-order hepatic metabolic clearance, plasma protein binding, liver blood flow, and the rate of clearance via the kidney (Pearce, et al., 2017; Wetmore, et al., 2012). Dosing assumes oral infusion at a constant rate (Pearce, et al., 2017). More specifically, the AEDs were calculated programmatically using the “calc_mc_oral_equivalent” function in the httk R package (v1.8), with the following options: the 95th quantile (which.quantile = c(0.95)); restrictive clearance (restrictive.clearance=T); selection of species (species=‘Human’); direct resampling of the population data (method=‘dr’); a correction for the amount of unbound chemical in whole blood versus plasma (well.stirred.correction=T); the default 3 compartment model (model=‘3compartmentss’); the output unit as mg/kg-bw/day (specific in httk as output.units=‘mg’). Although many AEDs could be calculated, the PODNAM,50 and PODNAM,95 were derived from AEDs that resulted from the 50th and 95th percentile, respectively, of the Monte Carlo simulation of Css. For clarity, the PODNAM,95 is a lower AED than PODNAM,50. Additionally, the maximum AED (max AED) achievable was calculated using the 95th percentile Css prediction and the typical maximum in vitro concentration screened in ToxCast of 100 μM.
2.2.3. Selection of the PODtraditional
The largest source of summarized in vivo point-of-departure (POD) information that was publicly available for this case study was the US EPA Toxicity Value Database (ToxValDB) (Table 1). ToxValDB includes summary study information and POD information from 40 sub-sources, including sources such as: COSMOS, ToxRefDB, HPVIS, HESS, and PPRTVs. This database is currently publicly viewable on a single-substance basis using the CompTox Chemicals Dashboard (Williams, et al., 2017), under the Hazard tab and the Point-of-departure sub-tab. Additionally, as part of the efforts of APCRA, POD information for a subset of the chemicals in this case study was contributed by collaborators from ECHA (61 substances), EFSA (46 substances), and Health Canada (29 substances), which generally increased the amount of hazard data for chemicals already in ToxValDB, but also expanded the chemical space overall by 6 chemicals.
Following this data aggregation step, several filters were applied. First, only oral exposures in units of mg/kg-bw or mg/kg-bw/day, or units that could be converted to mg/kg-bw/day values such as parts per million or parts per billion in the diet or mg/kg in the diet, were used, thereby including systemic exposures and excluding inhalation and dermal routes. The factors used to convert parts per million in diet to mg/kg-bw/day units were as follows: 0.05 (rat); 0.15 (mouse); 0.025 (dog); and, 0.03 (rabbit). Study type was not constrained to allow for inclusion of the highest number of substances in the case study; acute, chronic, developmental/reproductive, neurotoxicity and developmental neurotoxicity, and other repeat dose study designs were all included (though there were only 66 records associated with an acute exposure design, which is less than 0.3% of the 22627 total study records included). Only the following POD types were included: no observable or no observable adverse effect levels (NOEL, NOAEL) or lowest observable or lowest observable adverse effect levels (LOEL, LOAEL). The PODtraditional was then calculated as the 5th percentile of the distribution of PODs from all sources for a given substance, in an effort to approximate a reasonable low POD value. Given that the number and distribution of POD records vary by substance, and that the majority of substances are associated with fewer than 100 POD observations, the 5th percentile was calculated using a discontinuous function with averaging between discontinuities (see type 2 for the quantile() function in the R stats package).
2.2.5. Calculation of the POD ratio
The log10POD ratio indicates whether the PODNAM is less than the PODtraditional. A log10POD ratio of less than zero indicates that the PODNAM is greater than the PODtraditional, whereas a log10POD ratio of greater than zero indicates that the PODNAM was less than the PODtraditional. Using POD values in log10-(mg/kg-day) units, the log10POD ratio is given by the difference between the log10PODtraditional and the log10PODNAM as in Equation 2:
| Eq. 2 |
Where the log10PODNAM employed may be the log10PODNAM,50 or log10PODNAM,95, resulting in log10POD ratio50 or log10POD ratio95, respectively. In this work, the log10POD ratio50 was computed for comparison with log10POD ratio95, but log10POD ratio95 was used as the primary value for further analyses study type enrichment and chemotype enrichment. As the ratios calculated in this work are on a log10 scale, it is important to note that the log of a ratio (log10(x/y)) is the difference of the logs (log10(x) – log10(y)).
2.2.6. Allometric scaling of the PODtraditional
To at least partially address cross-species differences in the PODtraditional values, a second iteration of the case study was performed using allometrically-scaled human equivalent doses for POD information from mouse, rat, guinea pig, rabbit, dog, and hamster studies. Allometric scaling was performed based on data adapted and modified from the U.S. Food and Drug Administration (FDA) guidelines (Nair et al., 2016) using the following scaling factors for each species to convert mg/kg/day values to human equivalent doses: mouse (0.081), rat (0.162), guinea pig (0.216), rabbit (0.324), dog (0.541), and hamster (0.135). The POD ratio was then recalculated using these allometrically scaled PODs, per equation 3 below:
| Eq. 3 |
Where F is the species-specific scaling factor as indicated above.
2.2.7. Exposure data
To enable exposure comparison for the largest number of substances possible, exposure predictions from the US EPA ExpoCast program Systematic Empirical Evaluation of Models version 2 (SEEM2) model (Wambaugh, et al., 2014) were used for all 448 substances in the case study (the SEEM2 model was run de novo for a single substance, raloxifene hydrochloride, that did not appear in the 2014 publication). The ExpoCast SEEM2 model was calibrated to existing human exposure predictions inferred from human biomonitoring data, and further relies on production volume and four binary use categories from the ACToR use database that indicate if a substance had industrial and consumer product use, consumer produce use alone, industrial use without consumer product use, and/or use as a pesticide active or inactive ingredient. The model can be used to generate predictions for a large number of substances, but these predictions are associated with large credible intervals. From the SEEM2 model, the “US Total Exposure” median and 95th percentile on the credible interval for the median prediction were used in calculation of the BER, as described in section 2.2.7.
Additionally, Health Canada provided exposure values from published screening level risk assessments conducted for existing substances under the Canadian Environmental Protection Act (1999) for consumer product and environmental exposures to the Canadian population; there were 18 chemicals in this case study with these values. These data were used only as a comparison to ExpoCast SEEM2 exposure values, and not in computation of the BER. Such a comparison is challenging due to differences in pathways, populations and metrics underlying the Health Canada traditional estimates and SEEM2 predictions. The Health Canada estimates used in screening level assessments for environmental media consider exposure for an individual from all sources, whereas screening level assessments for consumer products consider exposures for the users of such products on a product by product basis. Both environmental media and consumer product exposure estimates often make use of conservative assumptions. Further, the SEEM2 model prediction in the case study is based on the U.S population median and the credible interval around this median value. With these differences in mind, a comparison between the two was conducted as follows. For the Health Canada environmental exposure data, the total, or aggregate exposure from multiple media, for the 20–59 years age group were considered. For the consumer exposure data, daily exposure estimates for adults from the use of certain sentinel consumer products, where use was considered to be chronic, were examined where available (e.g. personal care products, cleaning products, textile, foam, plastics). The consumer product resulting in the highest exposure estimate was carried forward for analysis and no aggregation of exposure estimates across consumer products was performed. The highest exposure estimate from the combined data set of the selected consumer product and environmental media intakes were used for comparison to the 95th percentile on the credible interval for the median general population exposure estimate from ExpoCast SEEM2.
2.2.8. Calculation of the BER
Using the PODNAM and 95th percentile on the prediction of the median exposure from ExpoCast, both in log10(mg/kg-day) units, the log10BER95, is given by the difference between the log10PODNAM,95 and the log10ExpoCast95 prediction (Equation 4):
| Eq. 4 |
2.2.9. Enrichment calculations
2.2.9.1. Chemotype enrichment
Enrichment of chemical structural features for substances for which the log10POD ratio95 is less than zero makes it possible to investigate possible limitations in the NAM-based approach that might lead to PODNAM values greater than PODtraditional. For this purpose, a recently developed chemotype-enrichment workflow (CTEW) was utilized, based on the ToxPrint structure feature set developed by Altamira (Altamira, Columbus, OH USA) and Molecular Networks (Molecular Networks, Erlangen, GmbH) under contract from the U.S. Food and Drug Administration (Yang et al., 2015). Chemotype enrichment calculations were carried out by defining chemicals with a log10POD ratio95 of less than zero as the “positive” enriched space of interest, relative to the remaining case study set, i.e., the “negative” space. The general approach has been previously described (Strickland et al., 2018; Wang et al., 2019). The set of DTXSIDs corresponding to the 448 CASRN in this case study provide input to the CTEW and were used to retrieve DSSTox structures and compute a ToxPrint feature fingerprint for each structure using a Linux implementation of the CORINA software (Molecular Networks, GmbH). Of the 448 substances, 445 were mapped to a single DSSTox structure and further processed. The mixtures dipropylene glycol monomethyl ether (CASRN 34590-94-8) and abamectin (CASRN 71751-41-2) could not be mapped to a single structure, nor could the isomeric mixture 3-[(dimethoxyphosphinyl)oxy]-2-butenoic acid, methyl ester (CASRN 7786-34-7). ToxPrint chemotype (CT) enrichment statistics were evaluated for presence in the “positive” space. Enrichment was based on a computed odds ratio (OR) for each CT according to the following logic: a true positive indicates a chemical in the log10POD ratio95 < 0 space contained the CT; a true negative indicates a chemical in log10POD ratio95 that did not contain the CT; false positive indicates a chemical in the log10POD ratio95 > 0 space that contained the CT; and, false negative indicates a chemical in the log10POD ratio95 < 0 space that did not contain the CT. Quantitative metrics were used to evaluate the resultant confusion matrix and the significance of any enrichment. The Fischer’s exact test (as implemented in Python, scipy.stats, alternative=greater) was used to compute the significance of the enrichments as indicated by p-value, which tends to yield greater weight to enrichments of CTs that are associated with a higher number of chemicals. To identify the most interesting associations, OR values ≥3 and p-value ≤0.05 thresholds were used to filter the CT results for significance and further examination. This statistical test does not account for activating or deactivating effects when multiple CTs are present and is only indicative of the chemical features that may lead to an underestimation of potential hazard by the log10PODNAM,95.
2.2.9.2. Study type enrichment
To understand the possibility that certain in vivo endpoints, as represented by study types, might drive PODtraditional values for which the corresponding PODNAM values were not lower, an analysis of whether certain study types included as described in Section 2.2.4, might disproportionately define the log10POD ratio95 < 0 space was undertaken. The study types were programmatically reduced to: acute toxicity studies; repeat dose toxicity studies, defined by any study from 7 to 90 days in duration, including subacute and subchronic studies; chronic/carcinogenesis, defined by any repeat dose study in adult animals for greater than or equal to one year; reproductive/developmental, defined by any study including more than one generation, including developmental, reproductive, multigeneration reproductive studies, or similar designs; and, neurotoxicity studies. Following this programmatic simplification and standardization of study type, a Fischer’s exact test (R stats package) was used to indicate the significance, or p-value, of any enrichment of study type underlying the minimum PODtraditional value for the log10POD ratio95 < 0 space. Separate tests were run to understand potential enrichment of (1) reproductive/developmental studies and (2) chronic/carcinogenesis studies for the log10POD ratio95 < 0 space. The confusion matrices were defined per the following logic: true positive indicated that the minimum PODtraditional value for a given substance with log10POD ratio95 < 0 was derived from a reproductive/developmental study or chronic/carcinogenesis study; true negative indicated that the minimum PODtraditional value for a given substance with log10POD ratio95 > 0 was not derived from a reproductive/developmental study or chronic/carcinogenesis study; false positive indicated that the minimum PODtraditional value for a given substance with log10POD ratio95 > 0 was derived from a reproductive/developmental or chronic/carcinogenesis study; and, a false negative indicated that the minimum PODtraditional value for a given substance with log10POD ratio95 < 0 was not derived from a reproductive/developmental or chronic/carcinogenesis study. Like the CTEW described above, significance thresholds of a p ≤0.05 and OR ≥ 3 were used to determine significance of any association.
2.2.10. TTC values
The PODNAM,95 was compared to the TTC approach that is often proposed for rapidly screening chemicals for priority (EFSA, 2012; HealthCanada, 2016; WHO, 2016). TTC values for the substances that could be associated with distinct structures were assigned using the software ToxTree [v2.6.6] (Patlewicz, et al., 2008) which implements the TTC decision-tree as described in Kroes et al., 2004. The DSSTox chemical structure-data (SD) file generated within the CompTox Chemicals Dashboard was converted from V3000 to V2000 format using ACD/Spectrus DB 2017.2 and where necessary organic substances with counter ions (e.g. sodium salts) were converted to their neutral form with KNIME (v 3.2.1) and the RDKit salt stripper node. The structure file was imported into ToxTree, where the Kroes TTC decision tree was run in batch mode. The daily intake was set at > 90 μg/day for each chemical to run through the entire decision tree. A separate approach was required for organophosphates (OPs) since ToxTree does not correctly interpret the Kroes decision tree for these chemicals. First, each OP was screened using the carcinogenicity and mutagenicity rule-base by ISS within ToxTree to screen for genotoxicity alert (GA). If an OP triggered a GA then it was assigned a TTC value of 0.0025 μg/kg bw/day; otherwise, the OP was assigned the default Kroes TTC value for this class of chemicals, which is 0.3 μg/kg bw/day. Moreover, custom structural profilers built in OASIS LMC Pipeline Profiler [v1.0.53] were used to exclude benzidines, steroids and organo-silicon compounds from TTC value assignment. More recent scientific opinions related to TTC have recommended expansion of the original Kroes et al. 2004 exclusion criteria to maintain the conservative nature of the approach and/or that these compounds are not well represented in the dataset from which the TTC values were derived. Likewise, based on these opinions, carbamate substances were assigned a TTC value of 0.3 μg/kg bw/day (EFSA, 2012; HealthCanada, 2016; WHO, 2016).
3. Results
Al of the inputs and calculated metrics are summarized in Table 2.
Table 2.
Inputs and metrics.
| Input or Metric | Description | Rationale |
|---|---|---|
| In vitro concentration used | Minimum of 5th percentile of ToxCast AC50 values OR the HIPPTox-POD | The goal was to use a value that represents a “lower bound” for in vitro bioactivity while accounting for experimental error/variability in these in vitro models. |
| PODNAM,50 | This metric uses the 50th percentile (median) from the distribution of AED values based on the in vitro concentration used. | In the HTTK modeling, Monte Carlo simulation was used to vary pharmacokinetic parameters to represent inter-individual variability in a population for calculation of steady state concentration (Css). Use of PODNAM,50 results in a NAM-based POD value that is 1.7 to 19-fold higher than PODNAM,95. |
| PODNAM,95 | See PODNAM,50; this metric uses the 95th percentile (median) from the distribution. | PODNAM,95 accounts for inter-individual variability and indicates that a lower oral dose than the PODNAM,50 would be needed to achieve the Css indicated by the bioactive concentrations in vitro. |
| ExpoCast SEEM2 50th percentile | Using “US Total Exposure” median on the credible interval prediction. | This is a lower exposure value in mg/kg-bw/day that accounts for less uncertainty in the prediction. |
| ExpoCast SEEM2 95th percentile | Using “US Total Exposure” 95th percentile on the credible interval for a median exposure prediction. | This is a higher exposure value in mg/kg-bw/day, accounting for more uncertainty in the prediction. This value can be ~ 2 orders of magnitude greater than the ExpoCast SEEM2 50th percentile. |
| PODtraditional | 5th percentile of available in vivo PODs, including oral NOAEL, NOEL, LOAEL, LOEL values from mammalian toxicity studies in mg/kg-bw/day. | Use of the 5th percentile is intended to represent a lower bound for in vivo adverse effects in the available database. |
| Log10POD ratio95 | Log10POD ratio using the PODtraditional and the PODNAM,95 | This logic results in the PODNAM appearing protective for the PODtraditional 89% of the time in this case study. The median log10POD ratio95 was 2. |
| Log10POD ratio50 | Log10POD ratio using the PODtraditional and the PODNAM,50. | This logic results in the PODNAM appearing protective for the PODtraditional 80% of the time in this case study. The median log10POD ratio50 was 1.2. |
| BER95, 95th %ile | Bioactivity:exposure ratio; bioactivity = PODNAM,95; exposure = 95th percentile from the credible interval to predict median total US population exposure from ExpoCast SEEM2 | This BER is the most protective. It includes the highest amount of uncertainty from inter-individual variability in pharmacokinetic parameters in the IVIVE and the highest amount of uncertainty in the ExpoCast SEEM2 exposure prediction. Using this BER will make more substances appear to be of greater priority for review. |
| BER95, 50th %ile | Bioactivity:exposure ratio; bioactivity = PODNAM,50; exposure = 95th percentile from the credible interval to predict median total US population exposure from ExpoCast SEEM2 | This BER tends to be, approximately, 10 times greater than the BER95, 95th %ile. |
| BER50, 95th %ile | Bioactivity:exposure ratio; bioactivity = PODNAM,95; exposure = 50th percentile from the credible interval to predict median total US population exposure from ExpoCast SEEM2 | This BER tends to be, approximately, 10 times greater than the BER95, 50th %ile and 100 times greater than the BER95, 95th %ile. |
| BER50, 50th %ile | Bioactivity:exposure ratio; bioactivity = PODNAM,50; exposure = 50th percentile from the credible interval to predict median total US population exposure from ExpoCast SEEM2 | This BER tends to be, approximately, 10 times greater than the BER50, 95th %ile and 1000 times greater than the BER95, 95th %ile. |
Inputs and the resultant metrics used in this case study are consolidated and described, along with notes on the impact of selection of the input or metric in this analysis.
3.1. Substance diversity
The extent of substance diversity in this case study was demonstrated using the same functional use categories that inform the ExpoCast SEEM2 exposure model. Multiple general functional use categories (Dionisio, et al., 2015; Wambaugh, et al., 2014) may be associated with a given substance. The possible functional use categories included: industrial process with no consumer use; pesticide active with no consumer use; pesticide inert; consumer and industrial process; personal care product; flame retardant, consumer and no industrial process; pesticide active with consumer use; herbicide; colorant; fertilizer; petrochemical; food additive; and fragrance. Examination of these use categories demonstrated that substances with at least one use as a pesticide active (categories denoted as: pesticide active no consumer, pesticide active and consumer, herbicide, and/or antimicrobial) comprised nearly 70% (314/448) of the case study substances (Figure 2). This result is expected because the ToxCast Phase I chemical library was originally selected (Richard et al., 2016) in part to maximize the overlap with the ToxRefDB (Martin, et al., 2009a; Martin, et al., 2009b). Hence, pesticide active ingredients represent a significant percentage of the union of the ToxRefDB and ToxCast phase 1 libraries and supply much of the POD information available from summaries of registrant-submitted toxicity studies, known as data evaluation records (DERs), from the U.S. EPA’s Office of Pesticide Programs (OPP). Further, HTTK information are available largely for the ToxCast phase 1 and phase 2 chemical libraries (Pearce, et al., 2017).
Figure 2. Substance diversity.
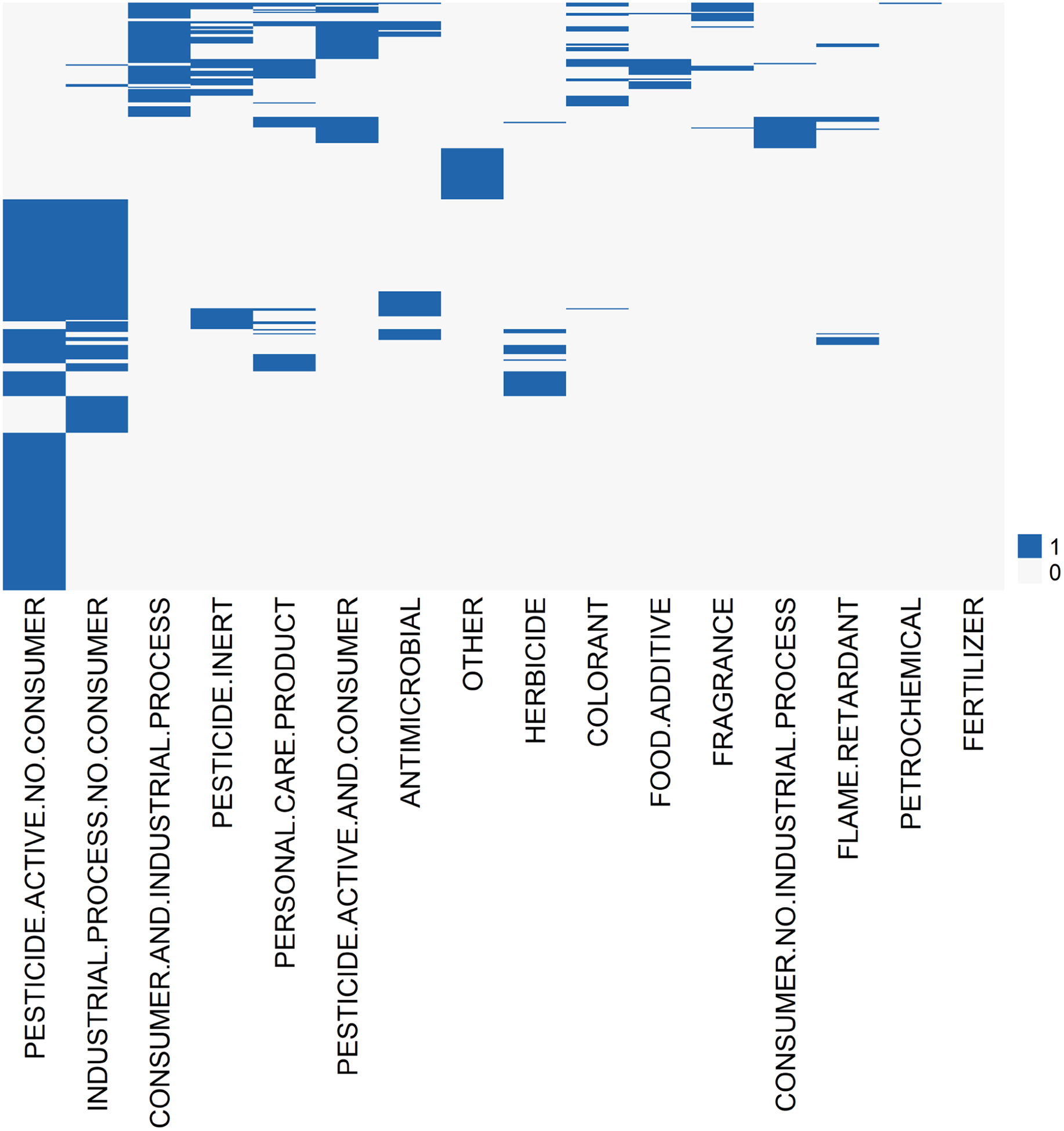
Generic functional use categories from ACToR for the 448 case study substances are illustrated. One substance, represented as a row in the heatmap, may be associated with multiple use categories
3.2. BER for the 448 chemicals
The exposure predictions from ExpoCast, the PODNAM estimates based on ToxCast and HIPPTox data, and the PODtraditional information are compared and visualized in Figure 3, all on a log10-mg/kg-bw/day basis. In this comparison, two estimates of the PODNAM have been included: the PODNAM,50 and the PODNAM,95, with the PODNAM,95 representing a lower dose and therefore more conservative estimate. For the majority of substances, the upper 95th percentile on the credible interval for the median total US exposure from the ExpoCast SEEM2 model corresponded to a daily log10-mg/kg-bw/day dose well below that anticipated to have bioactivity as well as the log10-mg/kg-bw/day dose at which effects were observed in traditional animal studies. Even using the PODNAM,95 estimate 95th percentile estimate from ExpoCast (Figure 4A black line), only 11 substances had a log10-BER95 of less than zero, indicating the potential for exposure to occur within the dose range that was bioactive in vitro (Figure 4B). Further examination of Figure 4 suggests that using the 95th percentile from the credible interval for the median total US exposure, rather than the predicted median or 50th percentile, significantly decreased the log10BER (shifting the BER95 values and BER50 values approximately 2 log10 orders of magnitude to the left in Figure 4A). Of course, given that the BER juxtaposes exposure and bioactivity predictions, uncertainty in the IVIVE methods applied to bioactivity can also result in a “shifting” of the BER estimate; using the PODNAM,50 results in a log10BER95 that is “right-shifted” in comparison to the log10-BER values from the PODNAM,95, as expected since for the substances in this case study the PODNAM,50 was 1.7 to 19-fold higher than the PODNAM,95 (see Supplemental Appendix Figure 5 for more details). Although the BER values provide an indication of risk-based priority, the BER values may be smaller given the nature of the ExpoCast predictions, i.e. predictions that have large uncertainty may result in the prediction of high exposures at the 95th percentile, and in vitro bioactivity data, i.e. very low AC50 values that were the result of permissive approaches in curve-fitting. The nature of the BER values were further explored in Figure 5 via 3 comparisons: ExpoCast versus PODNAM,95, followed by a side-by-side comparison of ExpoCast and ToxCast in vitro bioactivity data for the 11 substances with log10BER95 < 0 to the distribution of these data for the entire case study set of 448 substances. In Figure 5A, the 11 substances identified with low BER values are labeled and appear to demonstrate exposures that are generally greater than the median ExpoCast (95th percentile) estimate for the 448 substances, and all the PODNAM,95 are less than the median PODNAM,95 value for the case study substances. This is interrogated further in panels 5B and 5C. A distribution of the 95th percentile ExpoCast prediction for all 448 chemicals is used to understand if the 11 substances with log10BER95 <0 had high exposure predictions. A similar demonstration of the AC50 used to calculate the PODNAM is provided, where a distribution of all AC50 values for the 448 chemicals in the top panel is compared to AC50 values for the chemicals with log10BER <0. Several characteristics become apparent for the 11 substances with log10BER < 0: one, that many of these substances demonstrated relatively potent in vitro activity; two, that the most potent PODNAM,95 values, based on the combination of in vitro bioactivity and IVIVE, tended to drive lower BER values; and, three, that ExpoCast SEEM2 95th percentile estimates higher than the median in the case study seemed to contribute to lower BER values.
Figure 3. Comparison of the Exposure, PODNAM, and PODtraditional.
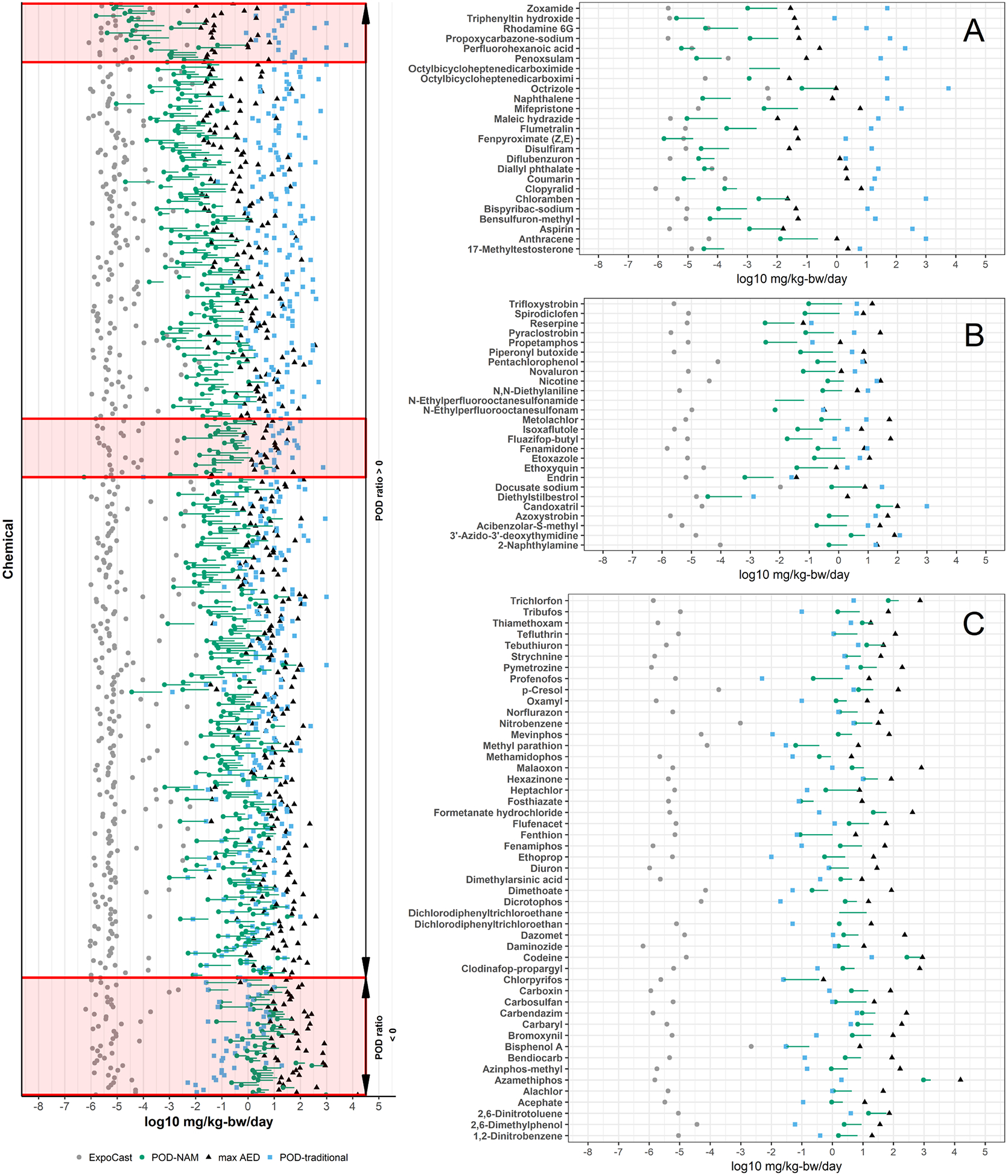
Comparison of ExpoCast (gray circles), PODNAM (green circles), maximum AED (black triangles), and PODtraditional values (blue boxes) for 448 substances. The green line segment indicates the PODNAM,95 to PODNAM,50. Inset images A, B, and C correspond to the red boxes overlaid on the main plot. Image 3A provides a magnification on the substances with the largest log10POD ratio values. Image 3B displays a sample of substances that approach the median log10POD ratio. Image 3C includes all 48 substances for which the PODNAM, 95 > PODtraditional.
Figure 4: Illustration of the log10-bioactivity-exposure-ratio (BER).
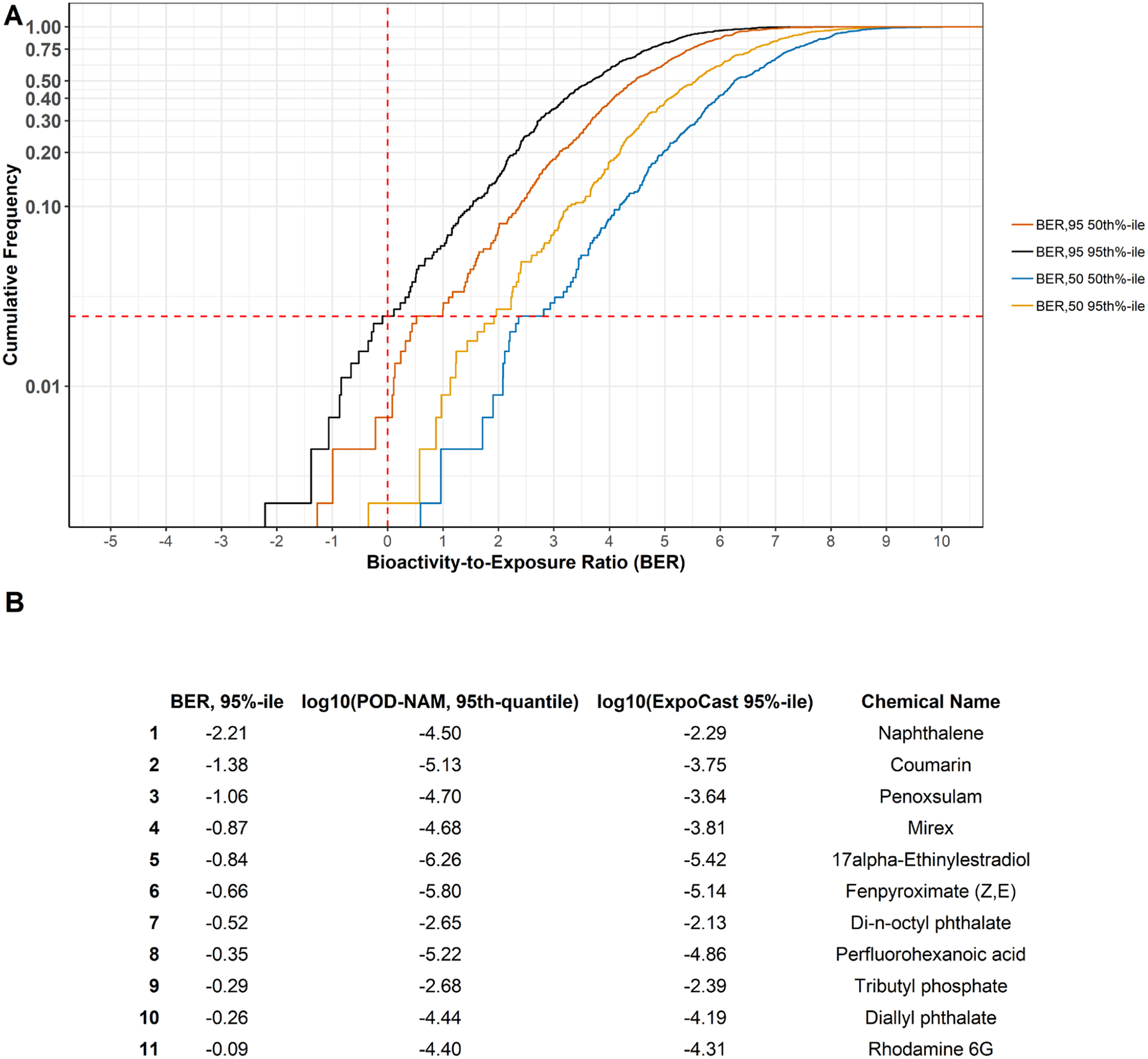
A) The cumulative frequency distributions for BER estimates are plotted. The BER95 values used the 95th percentile from the credible interval to predict the median total US population exposure from ExpoCast, whereas the BER50 values used the median exposure estimate. BER95 and BER50 values were calculated as the “95th%-ile” and “50th%-ile,” using the PODNAM,95 and PODNAM,50, respectively. Orange line = BER95 using PODNAM,50; black line = BER95 using PODNAM,95; blue line = BER50 using PODNAM,50; gold line = BER50 using PODNAM,95. B) Eleven chemicals had a BER95, 95th%-ile < 0, indicating overlap between the PODNAM,95 and the 95th percentile exposure prediction. Dashed red lines indicate where BER95, 95th%-ile = 0.
Figure 5: Exposure and in vitro bioactivity that defined chemicals with log10BER < 0.
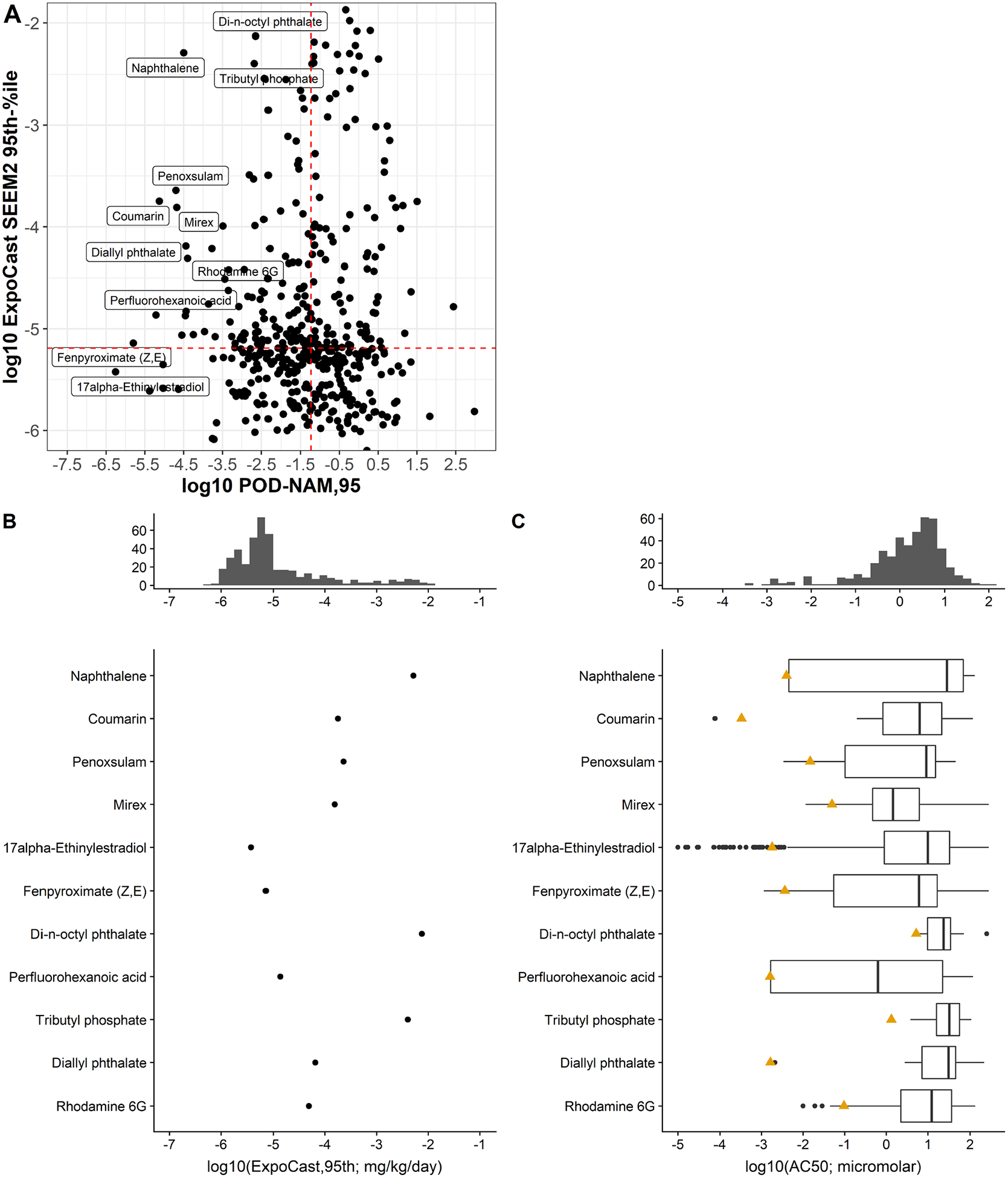
In (A), a scatterplot of log10 ExpoCast SEEM2 95th percentile value versus the PODNAM,95, with dotted red lines for the respective median values. The names of the 11 substances with log10BER95 < 0 are labeled. In (B) and (C), distributions of the exposure and the ToxCast AC50 data for all 448 substances are shown in the middle panels (gray histograms). Below these histograms in (B) and (C), side by side boxplots (showing the 1st quartile, median, and 3rd quartile) of the log10 ExpoCast SEEM2 95th percentile values and the ToxCast AC50 values are illustrated for the 11 substances with log10BER95 < 0. In (C), gold triangles indicate the 5th percentile of the AC50 distribution.
The performance of the ExpoCast SEEM2 model has been previously evaluated and described (Wambaugh, et al., 2014). In this case study, ExpoCast predictions for a relatively small subset of 18 chemicals were compared to manually curated values from Health Canada human health risk evaluations. The curated exposure values for these 18 chemicals had consumer product or environmental media exposure values from Health Canada assessments that could be compared to the median and 95th percentile on the credible interval for prediction of the median total US exposure from ExpoCast. The results illustrate for a limited chemical space that, as expected, the 95th percentile-ExpoCast values were within a range of the higher of either the consumer product or environmental Health Canada exposure values (Figure 6). The majority of the residuals (from the first to third quartile) for this comparison fall within ± 0.75 log10, indicating that the 95th percentile ExpoCast SEEM2 values may be a reasonable estimate of exposure in the absence of more refined models. One substance in particular, catechol (CASRN 120-80-9), stands out for larger differences between ExpoCast SEEM2 and Health Canada estimates. The ExpoCast SEEM2 model relies heavily on the ACToR use database, and catechol is suggested as a food additive and as having industrial (with no consumer) use. For the Health Canada catechol exposure estimate, dietary intake represents the majority of environmental media exposure with the predominant source being the natural occurrence of catechol in foods, and conservative estimates were derived using literature on maximum concentrations found in various food groups (EC/HC, 2008). Accounting for natural occurrence in food is not included as a use type in the ExpoCast SEEM2 model which may explain the discrepancy. Two other substances, di(2-ethylhexyl) adipate [DEHA] (CASRN 103-23-1) and chlorohexidine diacetate (CASRN 56-95-1), respectively, are consumer product chemicals that demonstrated slightly higher residuals. For DEHA, the Health Canada consumer product exposure estimate used to compare to ExpoCast SEEM2 represents the highest concentration reported in body lotion although a considerable range across products was reported (0.1 to 6%) which may partly explain the higher residual when compared to ExpoCast SEEM2 which represents the median of the U.S. population. Moreover, the Health Canada exposure estimate used in the comparison is the applied dose and it is known that DEHA exhibits low dermal absorption (the screening assessment adjusted the applied dose to estimate an internal dose using a dermal absorption value of 10% for risk characterization) (EC/HC, 2011). Likewise, the Health Canada estimate for chlorohexidine acetate is the applied dose, and it also exhibits low dermal absorption (EC/HC, 2017). The ExpoCast SEEM2 exposure estimates may be lower for poorly absorbed chemicals in near-field exposures such as topical application in part because the ExpoCast SEEM2 model was calibrated using human biomonitoring data.
Figure 6: Comparison of Exposure Predictions from ExpoCast and Health Canada Evaluations.
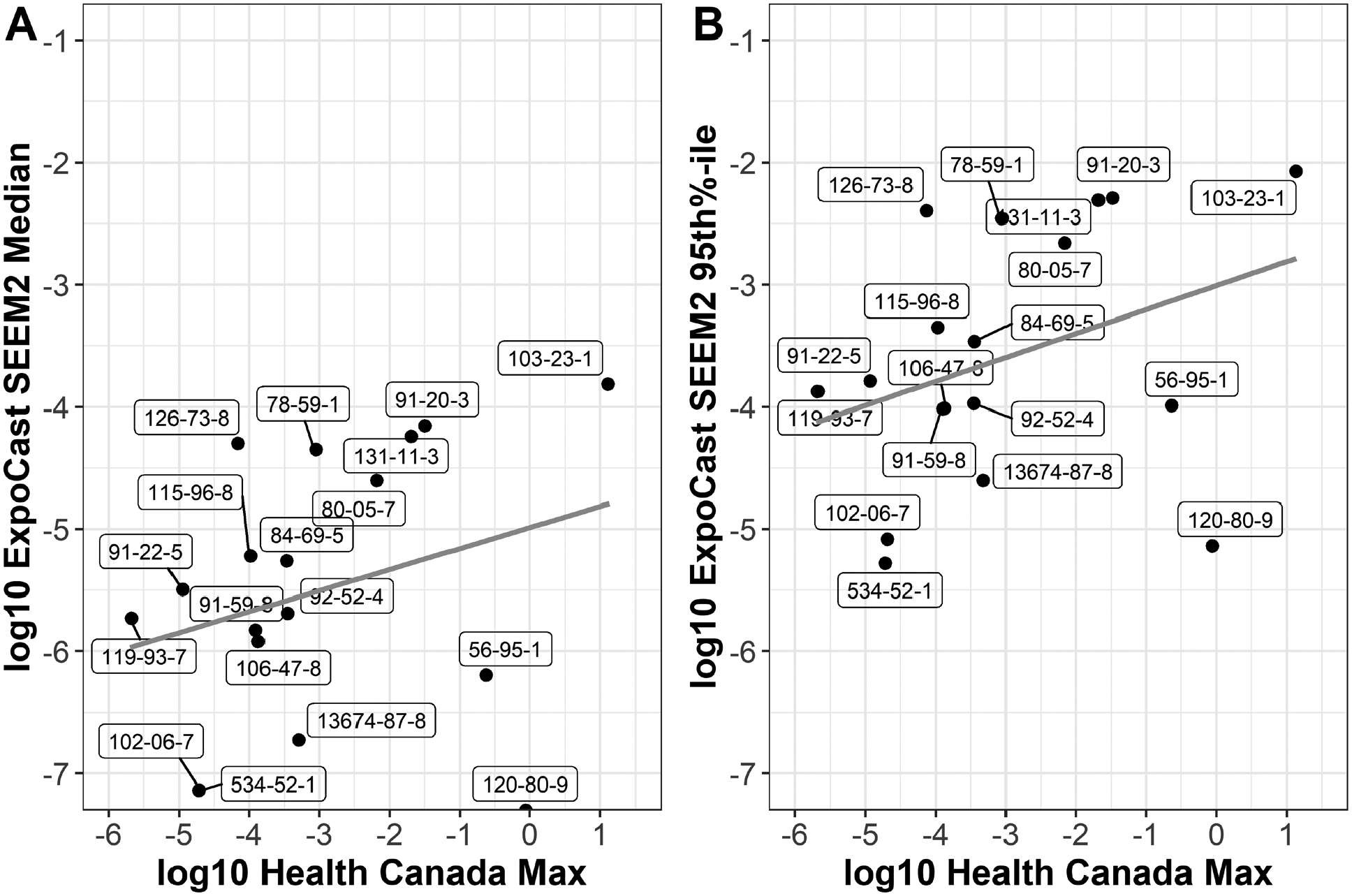
The total maximum values (in log10-mg/kg/day units) curated from Health Canada exposure assessments for 18 substances in this case study were compared to the ExpoCast (A) median and (B) 95th percentile predictions (in log10-mg/kg/day units), respectively. CASRN for these substances are labeled. The gray line shows a linear relationship. All CASRN and substance identifiers, including substance name, can be found in Supplemental File 2.
3.3. POD ratio for the 448 chemicals
The POD ratio depends on the PODNAM and the PODtraditional. In accounting for uncertainty from inter-individual variability, the PODNAM,50 and PODNAM,95 were both computed for comparison. The PODNAM,50 for substances in this case study are 1.7 to 19 times higher than the PODNAM,95, dependent on the substance, with the differences based on estimation of population differences in metabolic and renal clearance and/or plasma protein binding (see Supplemental Appendix, Figure 5). The log10POD ratio indicates whether a PODNAM is lower than the estimate of a dose associated with in vivo effects (PODtraditional). A log10POD ratio < 0 means that the log10PODNAM is greater than the log10PODtraditional. The log10POD ratio95 was < 0 for 48 of the 448 substances, or approximately 11% of the total (Figure 3C). Conversely, for 400 of 448 chemicals (89%), the PODNAM,95 is less than the PODtraditional (Figure 3). As the log10PODNAM,50 is greater than the log10PODNAM,95, the log10POD ratio50 is < 0 for a higher percentage of substances in this case study (20%, or 92 of 448 substances). Further examination of the distribution of the log10POD ratio95 demonstrates a range of −2.7 to 7.5, and a median of 2 (Figure 7A), indicating the median distance between the PODtraditional and PODNAM,95 on an arithmetic scale would be approximately 100-fold. Only three substances, all of which are organophosphate insecticides, dicrotophos (CASRN 141-66-2, DTXSID9023914), azamethiphos (CASRN 35575-96-3, DTXSID9034818), and mevinphos (CASRN 7786-34-7, DTXSID2032683), had a log10 POD ratio95 of less than −2., with a log10POD ratio95 values of −2.1, −2.7, and −2.2, respectively (Table 4). For the log10POD ratio50, the median was 1.2 and the range was −2.9 to 7.0.
Figure 7. Further understanding of the POD ratio distribution.
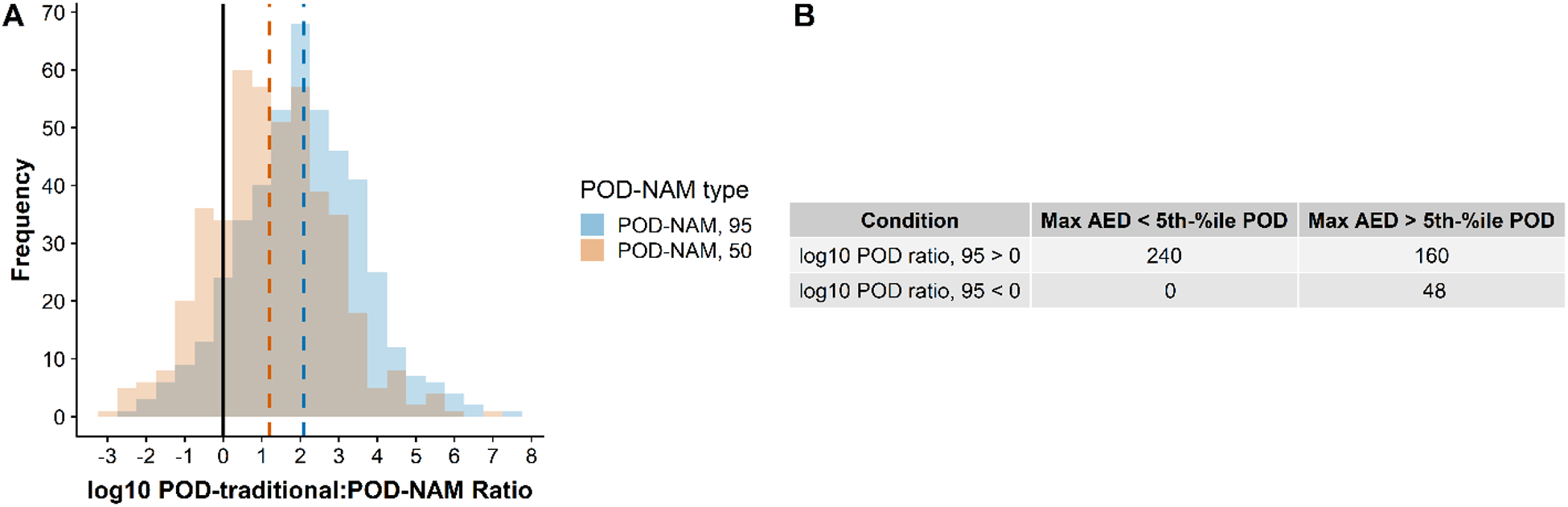
(A) The log10POD ratio is illustrated for the PODNAM,95 and the PODNAM, 50. The solid black line indicates where the log10-POD ratio95 is 0. Using the more conservative (i.e., lower) PODNAM,95, 48 of the 448 substances (10.7%) demonstrated a log10POD ratio < 0 (to the left of the dashed vertical line), whereas 92 of the 448 substances (20.5%) demonstrated a log10-POD ratio < 0 using the PODNAM,50. The medians of the log10-POD ratio distributions are indicated by dashed lines for PODNAM, 95 and PODNAM, 50 as 2 and 1.2, respectively. (B) Maximum AED (max AED) was less than the PODtraditional (5th-%ile POD) in 60% of the cases where the log10POD ratio95 > 0 (using PODNAM, 95). For the 48 chemicals with log10POD ratio95 < 0, the max AED was within the range of PODtraditional.
Table 4.
Details on the 48 substances with log10POD ratio95 < 0.
| # | DTXSID | CASRN | Name | Log10 PODNAM,95 | Log10 PODNAM,50 | Log10 POD ratio95 | Log10 POD ratio50 | BER95, 95th %ile | BER50, 95th %ile | BER95, 50th %ile | BER50, 50th %ile |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DTXSID9034818 | 35575-96-3 | Azamethiphos | 2.98 | 3.22 | −2.68 | −2.92 | 8.79 | 10.75 | 9.04 | 10.99 |
| 2 | DTXSID2032683 | 7786-34-7 | Mevinphos | 0.19 | 0.65 | −2.15 | −2.61 | 4.49 | 6.19 | 4.94 | 6.65 |
| 3 | DTXSID9023914 | 141-66-2 | Dicrotophos | 0.42 | 0.80 | −2.12 | −2.50 | 4.72 | 6.41 | 5.10 | 6.79 |
| 4 | DTXSID4032405 | 23422-53-9 | Formetanate hydrochloride | 1.34 | 1.78 | −1.77 | −2.21 | 6.67 | 8.47 | 7.10 | 8.90 |
| 5 | DTXSID4032611 | 13194-48-4 | Ethoprop | −0.25 | 0.42 | −1.75 | −2.42 | 4.98 | 6.90 | 5.65 | 7.57 |
| 6 | DTXSID3032464 | 41198-08-7 | Profenofos | −0.63 | 0.34 | −1.68 | −2.64 | 4.51 | 6.23 | 5.48 | 7.20 |
| 7 | DTXSID9024063 | 576-26-1 | 2,6-Dimethylphenol | 0.39 | 0.96 | −1.61 | −2.18 | 4.82 | 6.73 | 5.39 | 7.30 |
| 8 | DTXSID4020375 | 50-29-3 | Dichlorodiphenyltrichloroethane | 0.23 | 1.12 | −1.53 | −2.42 | 5.33 | 7.17 | 6.22 | 8.07 |
| 9 | DTXSID9032327 | 22781-23-3 | Bendiocarb | 0.42 | 0.94 | −1.32 | −1.84 | 5.75 | 7.50 | 6.27 | 8.02 |
| 10 | DTXSID3024102 | 22224-92-6 | Fenamiphos | 0.26 | 0.98 | −1.27 | −1.99 | 6.13 | 8.01 | 6.85 | 8.72 |
| 11 | DTXSID1024174 | 78-48-8 | Tribufos | 0.18 | 0.90 | −1.18 | −1.90 | 5.15 | 6.90 | 5.87 | 7.62 |
| 12 | DTXSID3022162 | 1689-84-5 | Bromoxynil | 0.65 | 1.27 | −1.18 | −1.79 | 5.90 | 7.72 | 6.51 | 8.33 |
| 13 | DTXSID2020341 | 76-57-3 | Codeine | 2.44 | 2.88 | −1.15 | −1.59 | 7.22 | 9.19 | 7.66 | 9.63 |
| 14 | DTXSID0021389 | 52-68-6 | Trichlorfon | 1.83 | 2.18 | −1.13 | −1.48 | 7.69 | 9.66 | 8.04 | 10.00 |
| 15 | DTXSID6021086 | 23135-22-0 | Oxamyl | 0.12 | 0.47 | −1.12 | −1.47 | 5.88 | 8.00 | 6.23 | 8.34 |
| 16 | DTXSID8023846 | 30560-19-1 | Acephate | −0.02 | 0.34 | −0.94 | −1.30 | 5.46 | 7.23 | 5.83 | 7.59 |
| 17 | DTXSID6024177 | 10265-92-6 | Methamidophos | −0.43 | −0.05 | −0.87 | −1.25 | 5.22 | 6.89 | 5.60 | 7.27 |
| 18 | DTXSID6032354 | 105512-06-9 | Clodinafop-propargyl | 0.34 | 0.73 | −0.83 | −1.22 | 5.54 | 7.24 | 5.92 | 7.62 |
| 19 | DTXSID3020122 | 86-50-0 | Azinphos-methyl | −0.03 | 0.51 | −0.79 | −1.34 | 5.71 | 7.64 | 6.25 | 8.18 |
| 20 | DTXSID0023951 | 5234-68-4 | Carboxin | 0.62 | 1.18 | −0.72 | −1.28 | 6.57 | 8.41 | 7.12 | 8.97 |
| 21 | DTXSID7020508 | 75-60-5 | Dimethylarsinic acid | 0.28 | 0.65 | −0.68 | −1.05 | 5.91 | 7.58 | 6.28 | 7.95 |
| 22 | DTXSID9020790 | 1634-78-2 | Malaoxon | 0.65 | 1.03 | −0.65 | −1.03 | 5.87 | 7.60 | 6.26 | 7.98 |
| 23 | DTXSID7020479 | 60-51-5 | Dimethoate | −0.67 | −0.14 | −0.64 | −1.16 | 3.48 | 5.38 | 4.01 | 5.90 |
| 24 | DTXSID3020679 | 76-44-8 | Heptachlor | −0.22 | 0.85 | −0.61 | −1.68 | 4.95 | 6.66 | 6.01 | 7.73 |
| 25 | DTXSID4024066 | 528-29-0 | 1,2-Dinitrobenzene | 0.20 | 0.82 | −0.60 | −1.22 | 5.23 | 7.13 | 5.86 | 7.75 |
| 26 | DTXSID5020528 | 606-20-2 | 2,6-Dinitrotoluene | 1.18 | 1.78 | −0.58 | −1.18 | 6.23 | 8.05 | 6.82 | 8.64 |
| 27 | DTXSID2032552 | 142459-58-3 | Flufenacet | 0.55 | 1.20 | −0.47 | −1.12 | 5.67 | 7.48 | 6.32 | 8.13 |
| 28 | DTXSID2032637 | 123312-89-0 | Pymetrozine | 0.93 | 1.46 | −0.44 | −0.96 | 6.85 | 8.80 | 7.38 | 9.33 |
| 29 | DTXSID2034962 | 153719-23-4 | Thiamethoxam | 0.98 | 1.33 | −0.37 | −0.72 | 6.69 | 8.73 | 7.04 | 9.08 |
| 30 | DTXSID7024902 | 533-74-4 | Dazomet | 0.37 | 0.85 | −0.35 | −0.83 | 5.21 | 7.22 | 5.69 | 7.70 |
| 31 | DTXSID1020855 | 298-00-0 | Methyl parathion | −1.20 | −0.43 | −0.32 | −1.10 | 2.90 | 4.73 | 3.67 | 5.51 |
| 32 | DTXSID3024316 | 34014-18-1 | Tebuthiuron | 1.12 | 1.70 | −0.28 | −0.86 | 6.56 | 8.22 | 7.14 | 8.80 |
| 33 | DTXSID9020247 | 63-25-2 | Carbaryl | 0.83 | 1.35 | −0.23 | −0.74 | 6.25 | 7.98 | 6.77 | 8.49 |
| 34 | DTXSID4024729 | 10605-21-7 | Carbendazim | 0.97 | 1.40 | −0.17 | −0.60 | 6.85 | 8.99 | 7.28 | 9.42 |
| 35 | DTXSID7021869 | 106-44-5 | p-Cresol | 0.86 | 1.34 | −0.16 | −0.64 | 4.58 | 6.33 | 5.06 | 6.81 |
| 36 | DTXSID9020370 | 1596-84-5 | Daminozide | 0.21 | 0.56 | −0.12 | −0.48 | 6.40 | 8.21 | 6.76 | 8.56 |
| 37 | DTXSID8020620 | 55-38-9 | Fenthion | −1.05 | 0.01 | −0.10 | −1.17 | 4.10 | 5.95 | 5.16 | 7.02 |
| 38 | DTXSID5023950 | 55285-14-8 | Carbosulfan | 0.10 | 1.12 | −0.10 | −1.12 | 5.31 | 7.04 | 6.33 | 8.06 |
| 39 | DTXSID0034930 | 98886-44-3 | Fosthiazate | −1.03 | −0.62 | −0.06 | −0.48 | 4.33 | 6.08 | 4.74 | 6.49 |
| 40 | DTXSID8024234 | 27314-13-2 | Norflurazon | 0.24 | 0.83 | −0.04 | −0.63 | 5.46 | 7.31 | 6.06 | 7.90 |
| 41 | DTXSID1022265 | 15972-60-8 | Alachlor | 0.03 | 0.64 | −0.03 | −0.64 | 5.42 | 7.17 | 6.02 | 7.78 |
| 42 | DTXSID6023600 | 57-24-9 | Strychnine | 0.43 | 0.93 | −0.03 | −0.53 | 6.25 | 8.14 | 6.74 | 8.63 |
| 43 | DTXSID3020964 | 98-95-3 | Nitrobenzene | 0.73 | 1.31 | −0.03 | −0.61 | 3.74 | 5.67 | 4.32 | 6.25 |
| 44 | DTXSID7020182 | 80-05-7 | Bisphenol A | −1.50 | −0.75 | −0.02 | −0.77 | 1.16 | 3.10 | 1.91 | 3.85 |
| 45 | DTXSID4024145 | 51235-04-2 | Hexazinone | 1.02 | 1.50 | −0.02 | −0.50 | 6.38 | 8.07 | 6.86 | 8.54 |
| 46 | DTXSID5032577 | 79538-32-2 | Tefluthrin | 0.05 | 0.82 | −0.01 | −0.78 | 5.09 | 6.91 | 5.85 | 7.68 |
| 47 | DTXSID4020458 | 2921-88-2 | Chlorpyrifos | −1.59 | −0.43 | −0.01 | −1.17 | 4.02 | 6.19 | 5.19 | 7.35 |
| 48 | DTXSID0020446 | 330-54-1 | Diuron | −0.11 | 0.55 | −0.01 | −0.66 | 5.88 | 7.80 | 6.53 | 8.45 |
Substances in this table are ordered based on the log10POD ratio, from smallest to largest for substances with log10POD ratio95 < 0 (column in gray). Note that for 33 of the 48 substances, the log10POD ratio95 is within one log10. The full table for all substances is available as Supplemental File 2.
As the concentration range evaluated in the ToxCast assays may limit the upper bound of the PODNAM, a comparison between the PODtraditional and the maximum AED possible from high-throughput screening was also calculated. The maximum AED, using the 95th percentile prediction for the Css and using 100 μM as the input concentration, was calculated. This maximum AED (Figure 3, Figure 7B) was based on the general assumption that no ToxCast library substances would be screened at nominal concentrations that exceeded 100 μM. For the 48 substances with a log10POD ratio95 < 0, the maximum AED exceeded the minimum PODtraditional in all cases. In contrast, for the remaining 400 substances with log10POD ratio95 > 0, 60% had a maximum AED that was less than the minimum PODtraditional (Figure 7B).
Similar to evaluation of the chemicals with log10BER95 < 0, hypotheses regarding why substances demonstrated a log10POD ratio95 < 0 were considered. First, the chemical domain was considered via calculation of statistical enrichment of ToxPrint chemical structure features, or chemotypes (CTs) (Strickland, et al., 2018; Yang, et al., 2015) (Table 3). Through this analysis, six CTs were identified as enriched, with OR ≥ 3 and p-value ≤ 0.05. The local balanced accuracy (BA) values (within a CT subspace) ranged from 0.57 to 0.62, with the bond:P=0_phosphate_thio CT completely contained within the log10POD ratio95 < 0 space. Of the 48 substances with log10POD ratio95 < 0, half (24 substances) contained one or more enriched CTs, corresponding to structural features indicative of organophosphate or carbamate related chemistries. Twenty-one of these 24 substances have clear indication of being a carbamate or organophosphate pesticide.
Table 3.
Chemical features enriched in the log10POD ratio95 < 0 set.
| ChemoType Information | Appearance of the ToxPrint | Metrics | ChemoType Information | Appearance of theToxPrint | Metrics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | ToxPrint | Total | POD ratio SO | POD ratio ≤0 | BA | OR | p-value | Label | ToxPrint | Total | POD ratio ≤ 0 | POD ratio > 0 | BA | OR | p-value |
| bond:P=O_phosphorus_oxo |  |
18 | 12 | 6 | 0.62 | 22 | 7.4E-09 | bond:P~N_generic |  |
5 | 4 | 1 | 0.54 | 36 | 0.00055 |
| bond:P=O_phosphate_thio | 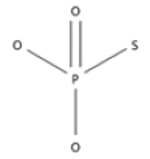 |
3 | 3 | 0 | 0.53 | NA | 0.0012 | bond:C(=O)N_carbamate | 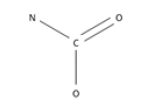 |
20 | 6 | 14 | 0.54 | 3.9 | 0.014 |
| bond:P~S_generic | 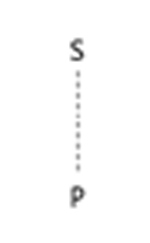 |
27 | 13 | 14 | 0.62 | 10 | 3.5E-7 | bond:CS_sulfide |  |
53 | 15 | 38 | 0.61 | 4.3 | 0.00011 |
The enriched chemical structural features, as represented by ToxPrints, for the log10POD ratio95 < 0 set. BA = balanced accuracy; OR = odds ratio; POD ratio = log10POD ratio95.
As ToxCast assay endpoints do not completely cover biological space, and generally include only short-term assays, the hypothesis that ToxCast data failed to identify chemicals that demonstrated critical effects in developmental/reproductive or chronic studies was tested via enrichment analysis of the study type that underpinned the minimum PODtraditional for each substance. Using a Fisher’s exact test, we failed to observe any significant enrichment of developmental/reproductive studies (grouped for this analysis, p < 0.87) or chronic studies (p-value < 0.69) in the log10POD ratio95 < 0 space. The frequency of developmental/reproductive and chronic studies defining the minimum PODtraditional is illustrated in the matrices in Figure 8. Though there were no significant enrichments of study type for the minimum PODtraditional for chemicals with log10POD ratio95 < 0, it is interesting to note that chronic toxicity data appeared to generate the minimum PODtraditional more often, in general (for 272 out of 448 chemicals). However, this may be due to the fact that not all chemicals in the case study had all study types, introducing a major caveat to this analysis, such that broader inferences about the importance of study type are limited.
Figure 8. Study types enriched in the log10POD ratio95 < 0 set.
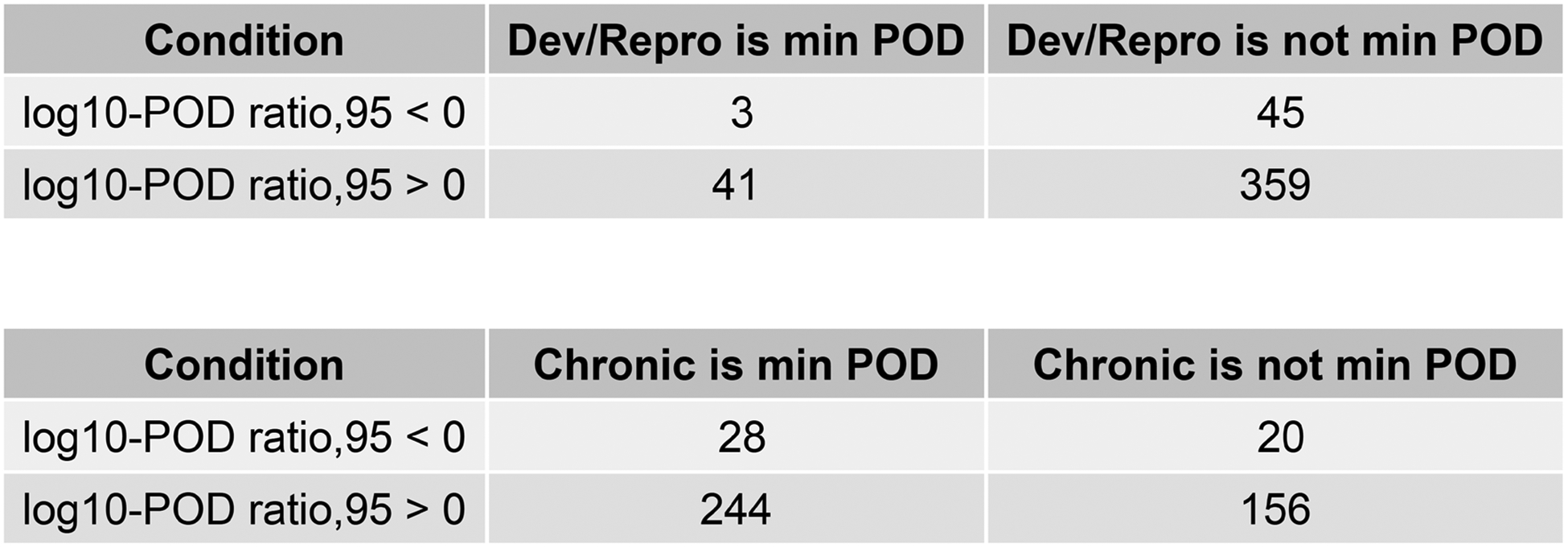
The matrices used to evaluate study type enrichment are shown. Neither developmental/reproductive (grouped together) (p = 0.88) nor chronic (p = 0.45) study types appeared to be enriched in the log10POD ratio95 < 0 subset.
3.4. Comparison of PODNAM,95 to a TTC approach
Considering that, in the absence of HTS data, one approach to screening level risk assessment might be to employ a TTC to help in prioritization (HealthCanada, 2016), the PODNAM was further compared to a TTC developed using ToxTree (Patlewicz, et al., 2008; ToxTree, 2015). For the 448 substances in this case study, the following TTC values were defined: 141 substances were designated as 0.0025 μg/kg-bw/day (potential genotoxic chemical threshold), 36 substances were designated as 0.3 μg/kg-bw /day (OP or carbamate), 212 substances were designated as 1.5 μg/kg-bw /day (Cramer Class III), 29 substances were designated as 30 μg/kg-bw /day (Cramer Class I), and 5 substances were designated as 9 μg/kg/day (Cramer Class II). Twelve substances demonstrated exclusion criteria (6 steroids, 2 metals, 2 benzidines, 1 organosilicon, 1 N-nitroso), and finally, 3 substances lacked a defined structure as previously described.
The primary observation from this comparison is that the PODNAM generally appeared to be greater than the TTC value, with the PODNAM,95 greater than the TTC for 87% of the substances (389/448) and the PODNAM,50 greater than the TTC for 92% of the substances (413/448). The median log10PODNAM,95:TTC ratio was 2.25, suggesting that on average there is approximately a 100-fold difference between these two predictions (Figure 9A). This finding may be partly explained by the methods used to develop the TTC values for each chemical class which are analogous to the approaches used in quantitative cancer risk assessment or in the development of a reference dose. For potential genotoxic chemicals the TTC value was developed through the use of linear lose dose extrapolation to 1 in 106 lifetime risk-based on reference chemicals in a carcinogenicity database (Kroes, et al., 2004). For the non-cancer portion of the decision tree (Cramer classifications, OPs and carbamates) the TTC values were developed including the application of an uncertainty factor (UF) (e.g. UF of 100 applied to the 5th percentile from a distribution of NOELs from Cramer classified substances making up a repeat dose reference database (Kroes, et al., 2004). No UFs are applied to the PODNAM (or the PODtraditional) in this analysis. In addition, we failed to observe a linear relationship between the log10TTC value and the log10PODNAM,95 value, with considerable variability in the log10PODNAM,95 values reported within each TTC value category (Figure 9B). As the BER has been suggested as a prioritization metric, and BER is the quotient of the PODNAM and the exposure prediction, we also examined how replacing log10PODNAM,95 with TTC would have affected the log10BER95 for the 11 substances with log10BER95 < 0. Interestingly, for the 11 substances with log10BER95 < 0, the log10TTC was greater than the log10PODNAM,95 for 8 substances, and only one substance (napthalene, DTXSID8020913) still had a log10BER < 0 when using the log10TTC instead of the log10PODNAM,95.
Figure 9. PODNAM,95 compared to the TTC.
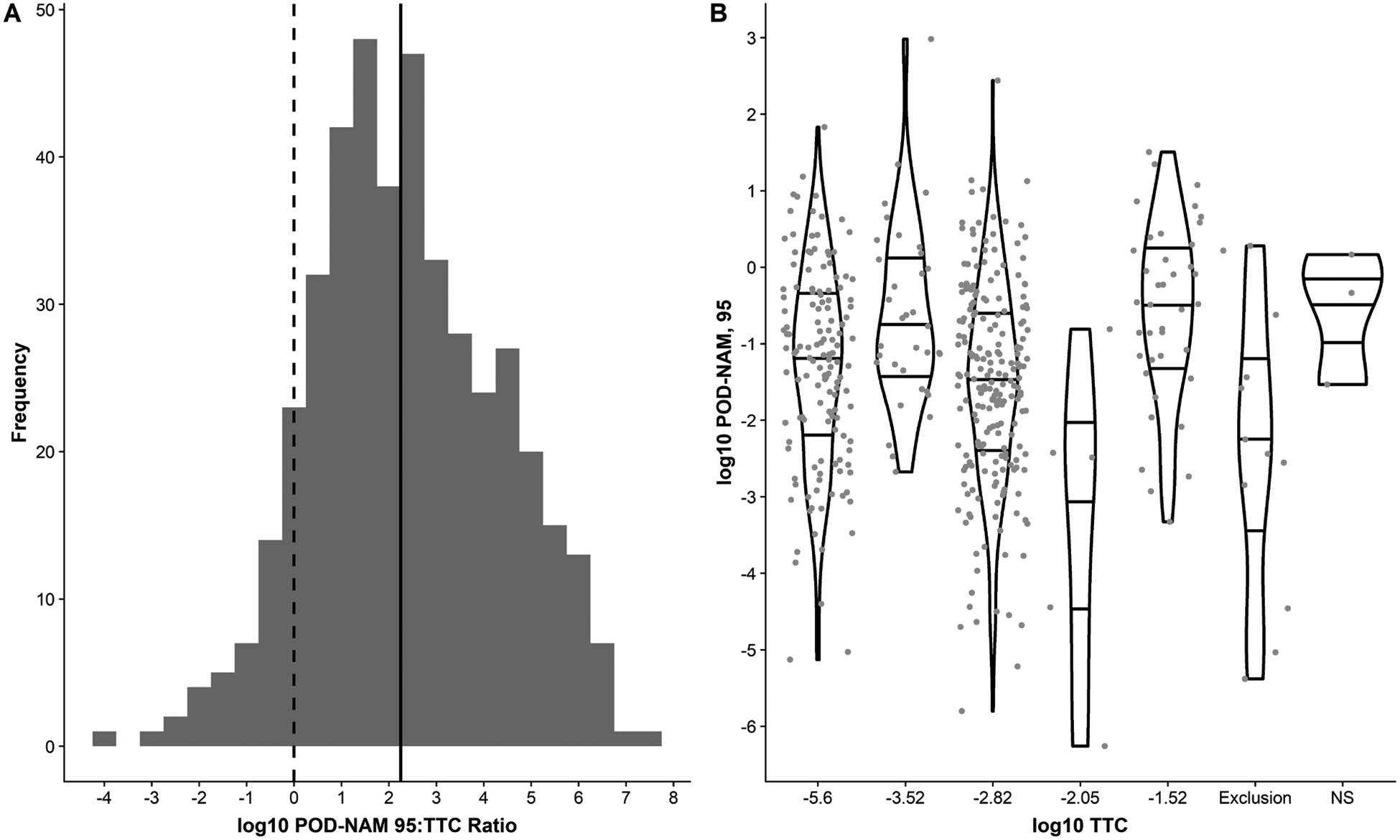
The log10TTC: PODNAM,95 ratio is illustrated for the 448 case study chemicals in (A). In (B), the log10 TTC value bin is compared to the log10PODNAM,95, in units of log10-mg/kg/day; dots represent all points and violin plots capture the shape of the distribution.
3.5. Comparison of PODNAM to PODtraditional from allometrically scaled data
For the main case study, PODtraditional data were collected from any in vivo toxicology study, regardless of species or strain, and then grouped to derive a 5th percentile from the distribution by chemical. In contrast, the PODNAM was derived from an AED that was calculated using human HTTK parameters and in vitro bioactivity (mostly from human cell lines and proteins). To address the issue of combining multiple species in this analysis, we compared the log10PODNAM,95, derived from an AED calculated using human HTTK parameters, to a human equivalent dose, i.e. human PODtraditional, based on allometric scaling of the PODtraditional data. Limiting to studies in mouse, rat, guinea pig, rabbit, dog, and hamster, 447 of the 448 chemicals could be included in this comparison. The human PODtraditional is derived from PODtraditional via multiplication by a factor less than one (derived based on body surface area by species) (Nair, et al., 2016). This led to 82 substances (18%) having log10PODNAM,95 higher than the human PODtraditional (compared to 48 when using the animal-based PODtraditional), and 158 substances for which the log10PODNAM,50 was higher than the human PODtraditional (compared to 92 when using the animal-based PODtraditional). The median human log10POD ratio95 was 1.33, with a range of −3.3 to 6.7 (Supplemental Appendix, Figure 7). Given that the mechanism(s) of toxicity and metabolic processes may differ considerably across species, a consideration of differences in dosing based on allometric scaling (i.e., surface area) provides limited information regarding uncertainty based on interspecies differences.
4. Discussion
Herein we present a retrospective analysis to address two key questions: (1) would using in vitro bioactivity data from HTS programs such as ToxCast provide a “lower bound” estimate of a POD when compared to traditional toxicology approaches? And, (2) is the BER, using in vitro bioactivity across a broad range of assays, a useful tool for prioritization of substances? This analysis is the largest of its kind presented to date, with information for 448 substances included. A major premise of this work is that the minimal concentration corresponding to in vitro bioactivity is likely to be a threshold for any specific effects or toxicities that might be observed in vivo. The primary conclusion of our work is that for 89% of the chemicals in this case study, the HTS approach to derivation of a PODNAM,95 for screening and prioritization purposes produced a value less than or equal to the PODtraditional from in vivo toxicology studies. Further, we found that BER may be a useful data-driven metric for prioritization that can be customized to the resources available for follow-up, i.e. different choices in calculation of the BER can be made depending on how much uncertainty is acceptable and how many substances can be further evaluated given resource constraints. The customizable decisions in the HTS approach employed herein are demonstrated in adjustments in the amount of uncertainty in (1) the IVIVE that is included in development of the PODNAM and (2) the exposure predictions, highlighting that for different screening applications differing amounts of uncertainty can be included in this workflow. As demonstrated, metrics that account for more of these uncertainties (e.g., use of a 95th percentile rather than a median on exposure predictions, or use of a PODNAM,95 instead of a PODNAM,50) can be used in a screening and prioritization application (see Table 2). The context for use of the PODNAM was further examined via comparison to a TTC approach, ultimately demonstrating that there may be some advantages to combining these approaches for preliminary screening of substances for safety. The collaborative, international consideration of these issues in screening level assessments demonstrates the current state-of-the-science and presents a transparent and adaptable basis for utilization of HTS information.
A potential concern regarding the approach used in the case study might be whether it is overly conservative, i.e. whether the PODNAM values are too low or the exposure predictions are too high. To begin to address such a question, it is necessary to consider the impacts of the selections and assumptions made in the current approach. First, we consider the uncertainty that we consider in the use of NAM-based exposure predictions; the ExpoCast SEEM2 model is akin to a low-tier exposure assessment tool, and grappling with uncertainty in exposure prediction is more familiar to traditional safety evaluation (EPA, ExpoBox). The exposure predictions (Wambaugh, et al., 2014) from the ExpoCast SEEM2 analysis have wide credible intervals, as demonstrated by the shifting of the log10BER95 by approximately 2 log10 units based on selection of either the median or 95th percentile exposure predictions. In selecting the 95th percentile exposure prediction for most of the comparative analyses in this work, we are using a value that includes more of the uncertainty, and the median could be used instead depending on the number of substances that should be prioritized and the degree of certainty that is desired for a given application of these data. The fact that only 2.5% of the 448-chemical case study would have a log10BER95 less than zero, and only 15% with a log10BER95 less than 2, indicates that though the approach the log10BER95 attempts to account for more uncertainty, it does not necessarily indicate a high priority for all substances in the case study. There are caveats to this conclusion in that the exposure model was informed by functional use categories, and pesticide actives, which comprised 70% of the substances with sufficient data for inclusion in this case study, were generally predicted to have lower exposure than substances associated with other use categories (Wambaugh, et al., 2014). It is possible that using a substance list that included more substances associated with uses as pesticide inerts, personal care products, or other functional uses (Dionisio, et al., 2015) that more substances would be prioritized using the BER. In comparing exposure estimates from the ExpoCast SEEM2 model and curated exposure assessments from Health Canada, we found that the ExpoCast 95th percentile was a reasonable surrogate for 18 substances. Of these substances, the few that demonstrated larger differences (i.e., greater than 1 log10mg/kg-bw/day different) were likely related to reliance on different use and exposure scenarios between the ExpoCast SEEM2 model and the exposure assessments, supporting refinements in understanding of use and exposure scenario as essential for improved confidence in utilization of high-throughput exposure estimates (Biryol et al., 2017; Brandon et al., 2018; Dionisio et al., 2018; Ring et al., 2018). Moving forward with safety evaluation of substances across geographies, international collaboration and data-sharing regarding substance use categories and exposure scenarios or pathways will support progress in exposure prediction for thousands of substances.
As used in the aggregate in this case study, the in vitro bioactivity data was used to define a concentration range for any activity and does not necessarily support hypotheses regarding specific toxicological effects. Disruption of molecular targets as described by the PODNAM is necessary, but not sufficient, for producing adverse effects. In contrast, the PODtraditional is intended to represent a threshold for adversity; as such, we might anticipate that the PODNAM would in many cases be lower than an estimate of PODtraditional. Caveats in the use of ToxCast data include the possibility that, despite an attempt to filter these data for more reliable curve-fits (see Methods and Supplemental Appendix), we have not completely eliminated results from noisy curve-fits or assay interference, thereby resulting in the possible inclusion of some AC50 values that are not reproducible and/or biologically meaningful. This may bias the distribution of AC50 values available for substances, and thus detailed examination of the in vitro bioactivity data may be warranted when deriving screening level assessment values or following up on specific substances identified as a priority based on the BER. The use of a point estimate for the potency value (i.e., AC50) without use of confidence bounds (Watt, et al., 2018) may produce a higher AED value than using a threshold concentration based on a confidence interval. The need for applied research on quality control processes for improved filtering of any HTS data to include more reproducible and/or more reliable curves clearly emerges from this case study. Use of the 5th percentile for the ToxCast AC50 values per substance ameliorates some concerns with the appearance of low concentration outliers in the AC50 distribution (likely from less reproducible curve fits) (see Supplemental Appendix, Supplemental Figure 3), but also provides a “lower bound” estimate of a threshold concentration for bioactivity in vitro. The number of detailed, and thereby customizable, decisions made in determining an in vitro concentration for use in IVIVE is apparent.
Consideration of how much uncertainty the PODNAM should include in the IVIVE approach used is needed. We assumed steady state conditions and complete bioavailability when converting the in vitro potency values to AED values (Wetmore, et al., 2014; Wetmore, et al., 2015; Wetmore, et al., 2013; Wetmore, et al., 2012). The assumption of steady state conditions may be fairly accurate for pharmaceuticals and may also work for some but not all of diverse, environmentally-relevant chemicals (Sipes et al., 2017a; Wambaugh, et al., 2018; Wambaugh et al., 2015; Wang, 2010; Wetmore, et al., 2015; Yoon et al., 2014). However, beyond the assumption of steady state conditions, other assumptions in the HTTK and IVIVE approach may err on the side of lower AED predictions due to the lack of extrahepatic metabolic clearance; use of suspended hepatocyte model for capturing hepatic metabolism; and, lack of information about bioavailability (Wetmore, et al., 2015). Available high-throughput IVIVE methods are being continuously improved and more data is being added to reduce assumptions in the modeling (e.g., estimates of bioavailability, extrahepatic clearance). For this analysis, we demonstrated how inter-individual variability can be accounted for by using the 95th percentile estimate of the Css from a Monte Carlo simulation, which may result in a relatively low estimate of the PODNAM,95. We compared this to the PODNAM,50 because, dependent on the screening level application, it may be desirable to exclude inter-individual variability from initial computation of the PODNAM. The difference between the PODNAM,50 and PODNAM,95 is variable by substance, i.e. the range of potential Css values observed across a theoretical population is dependent on metabolic and renal clearance, plasma protein binding, and other features that are substance-dependent (for additional consideration, see Supplemental Appendix, Supplemental Figure 5). Ultimately, despite a number of approximations and uncertainties, the current workflow demonstrates the potential utility of the approach for calculation of the PODNAM using data and tools that are currently available.
For the substances in this case study, the PODNAM,50 and PODNAM,95 were lower or approximately equal for the PODtraditional 80% and 89% of the substances, respectively. The PODNAM,95 and resultant log10POD ratio95 and log10BER95 were selected for further detailed analyses in this work because this represented an approach that included consideration of more uncertainty and narrowed the substances with log10POD ratio and log10BER values < 0 to clearly identify possible limitations in the NAM-based approach. One limitation evident from the CT enrichment work is that the current NAM battery in ToxCast and HIPPTox, combined with IVIVE, failed to quantitatively capture the potency of effects expected for substances that contain structural features of carbamate and organophosphate insecticides. ToxCast does contain assays responsive to acetylcholinesterase inhibitors (Padilla et al., 2012; Sipes et al., 2013); however, it has been previously suggested that these assays lack the ability to accurately reflect acetylcholinesterase inhibition potency (Aylward et al., 2011). Additionally, often OP metabolites are more potent acetylcholinesterase inhibitors, and this would not be well-captured in assays with limited to no metabolism. For substances with the structural features of carbamate and/or organophosphate insecticides, a TTC approach like the one employed herein (which includes a separate workflow for these chemistries) may provide a more useful PODNAM value that would be less than or equal to the PODtraditional.
Another potential limitation of the NAM-based approach included in this work is that the ToxCast and HIPPTox assays are short-term in duration. One hypothesis as to why a substance would demonstrate a log10POD ratio95 < 0 was that the PODtraditional may have been effects observed in vivo following exposures of longer duration or involving specific susceptible lifestages. Interestingly, the minimum PODtraditional was not associated with a particular study type, as a surrogate for phenotype (i.e. developmental/reproductive or chronic studies), more frequently for substance with log10POD ratio95 < 0. Conclusions from this analysis of study type are necessarily limited because not all substances in the case study included all study types. To allow for the largest data set possible, each substance included in the case study may have had traditional toxicity information available from multiple sources and study types, and in some instances, it is possible that multiple records may correspond to the same study.
Although the in vitro bioactivity approach to definition of a PODNAM has some associated uncertainties, in the absence of other information, the PODNAM could be used as a “lower bound” or protective estimate. To reduce potential uncertainties, the data used to derive the PODNAM could always be further reviewed and refined in a number of ways, at the expense of performing a more automated analysis for many substances. First, manual or semi-automated curation of ToxCast/Tox21 data to select the in vitro bioactivity concentration based only on assays and curve-fits with high reproducibility could be performed for substances identified as priority from automated analyses. Second, the use of metabolically-competent in vitro models to predict the bioactivity of parent and metabolite(s) may account for potential bioactivated toxicants (DeGroot et al., 2018; Ramaiahgari et al., 2017). Additional bioactivity screening using metabolically-competent in vitro models would be needed to understand the potential impacts of metabolism on a workflow like the one employed herein. Third, the specific data or assay data-based predictions to indicate specific potential adversities, modeling neurotoxicity, hepatotoxicity, reproductive, or developmental toxicity, in an expansion of the HIPPTox and ToxCast approaches included here, could be added. This also relates to a limitation in that not all substances in this case study were tested in all available assays in ToxCast or HIPPTox. Fourth, the addition of high data content in vitro assays, e.g. high-throughput transcriptomics and/or cellular phenotypic profiling data like HIPPTox, may help comprehensively cover the biological pathways possibly disrupted by the test substances. Finally, further refinement of the IVIVE approach may also improve the utility of the PODNAM. Though currently the nominal media concentrations from assay endpoints are used in calculation of AED values that form the basis of the PODNAM, refinement of the IVIVE modeling procedure to account for differential in vitro partitioning could reduce uncertainty (Fischer et al., 2017).
Comparison of the in vitro bioactivity approach to a TTC-based approach for definition of a PODNAM provides some practical insight for screening-level evaluation. When available, the PODNAM, which uses data generated for a particular target substance, generally provides a more refined estimate of a POD than the TTC. Further, the PODNAM values are likely to improve and change over time as more sophisticated and comprehensive HTS tools are developed. Though the TTC value is generally lower than the PODNAM, there may be advantages to using PODNAM and TTC in concert. In some cases, a TTC cannot be easily generated due to exclusion criteria or the presence of multiple structures in an undefined mixture, whereas a PODNAM may be generated. In the fraction of cases where the log10TTC value was greater than the log10PODNAM,95, the log10TTC could be used as a check on low PODNAM values. Indeed, Health Canada recently applied the TTC approach to a group of substances amongst the remaining priorities under the CMP. A preliminary qualitative characterization of uses and exposure potential indicated that 237 of the CMP substances were good candidates for the TTC approach as exposure to the general population was expected to be limited. However, after developing quantitative exposure estimates only 89 of the 237 substances (38%) had exposures below their respective TTC values (HealthCanada, 2016). Thus, having an approach such as the use of PODNAM to develop a BER may have proven useful as an additional screening level tool, and a suite of in silico and in vitro NAMs, used collectively, provide an informed, multidimensional screening level approach.
There are several considerations when using the PODtraditional. For the PODtraditional data themselves, toxicologists generally accept that animal studies conducted at different times, by different laboratories, with a different cohort of perhaps the same strain of animal, may yield results that are qualitatively and/or quantitatively distinct (Gottmann et al., 2001; Kleinstreuer, et al., 2017; Ward et al., 2017; Wolf et al., 2017). Though we do not quantify this variability here, the resolution of the difference between the PODNAM and PODtraditional may be limited by the variability in the animal study results (Casati, et al., 2018) in addition to the variability in the in vitro bioactivity methods. A continuing challenge and necessity for comparing the results of NAM will be establishing the variability in reference set data. An additional limitation in interpreting this work is comparison of PODNAM, largely from human in vitro data, to PODtraditional, which is based on several different mammalian species. Though allometric scaling based on differences in animal surface area was performed for comparison in this work, the value of this exercise is limited as species-specific absorption, distribution, metabolism, and excretion processes might be anticipated on a substance by substance basis. Since humans are the species of interest for this case study, PODNAM based on mostly human bioactivity data were used. In the risk assessment process, an uncertainty factor of 10 might be used when considering interspecies differences.
Related issues (made apparent in this case study) in using traditional toxicology information as a reference for the NAM-based approach are the challenges of data curation and interoperability. For instance, clarification of records that summarize the same original study, or identification of specific effects observed, is limited in current publicly available databases of toxicity information (e.g., ToxValDB, ToxRefDB, eChemPortal, etc.) because of a lack of controlled semantic ontology for study features and biological effects. For international collaborations such as this one, it is not straightforward to identify the unique toxicological studies nor specific effects labeled using different terminology. Moreover, the basis for establishing the PODs from the original studies, regardless of their origin, may not be fully structured or reported, and differences in POD selection from different reviewers may arise. Consistent identification of the substance tested, and controlled vocabularies describing study designs and the reported effects, are needed to share curated data across databases, and across the world, in order to leverage the largest dataset possible for improved understanding of traditional toxicity information for regulatory toxicology. Efforts to create such a large, consistent, and reliable dataset are an ongoing interest within the APCRA initiative, among others. Improved curation and digitization of traditional toxicity information for comparison with NAMs across a broad range of endpoints and study types will require a significant investment of resources.
We have presented herein a retrospective analysis to demonstrate the utility of the BER in identifying potential priority chemicals for further evaluation, as well as the conservativism of a PODNAM when compared to PODtraditional. The result bolsters confidence that decisions based on a PODNAM can be health protective in screening level assessments, lending support for using approaches like this one to rapidly evaluate substances and potential needs for further screening information. Using relatively conservative assumptions of predicted exposure and bioactivity, only 15% of the substances in this case study demonstrated a log10BER95 < 2, suggesting that this approach may provide a data-informed and reasonable approach to identifying chemicals of interest for further testing and assessment. A primary goal of the APCRA collaboration supporting this work is to identify and resolve impediments to adoption of NAMs in safety assessment. In demonstrating the state-of-the-science for generation of BER and POD ratio values, we have shown in practice for 448 chemicals a way to accelerate screening and assessment using NAMs for hazard and exposure. Further, we have identified a number of areas for refinement in this workflow to be considered in subsequent application of this NAM-based approach to regulatory toxicology questions. Ongoing work will be needed to demonstrate that a workflow like the one demonstrated herein is generalizable to substances with little to no available traditional POD information. As continuing improvements in HTS approaches and availability and interoperability of database resources are made, confidence in the utilization of NAMs for screening level assessments will be bolstered by scientific quality, relevance, and transparency.
Supplementary Material
6. Acknowledgments
The authors would like to thank John Cowden, John Wambaugh, and Barbara Wetmore of the US EPA for their insightful comments on a previous version of this manuscript.
Footnotes
Supplemental Files
Supplemental Appendix: A text file containing Supplemental Figures 1–7 and supporting text.
Supplemental File 1: A spreadsheet containing all of the in vivo POD values used to derive the PODtraditional.
Supplemental File 2: A spreadsheet containing all of the summary information (including BER and POD ratio values)
Publisher's Disclaimer: Disclaimer: The United States Environmental Protection Agency (U.S. EPA) through its Office of Research and Development has subjected this article to Agency administrative review and approved it for publication. Mention of trade names or commercial products does not constitute endorsement for use. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of A*STAR, US EPA, EFSA, ECHA, Health Canada, or the JRC.
7 References
- Aylward LL, and Hays SM (2011). Consideration of dosimetry in evaluation of ToxCast data. J Appl Toxicol 31(8), 741–51. [DOI] [PubMed] [Google Scholar]
- Becker RA, Friedman KP, Simon TW, Marty MS, Patlewicz G, and Rowlands JC (2015). An exposure:activity profiling method for interpreting high-throughput screening data for estrogenic activity--proof of concept. Regul Toxicol Pharmacol 71(3), 398–408. [DOI] [PubMed] [Google Scholar]
- Bell SM, Chang X, Wambaugh JF, Allen DG, Bartels M, Brouwer KLR, Casey WM, Choksi N, Ferguson SS, Fraczkiewicz G, et al. (2017). In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicol In Vitro 47, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryol D, Nicolas CI, Wambaugh J, Phillips K, and Isaacs K (2017). High-throughput dietary exposure predictions for chemical migrants from food contact substances for use in chemical prioritization. Environ Int 108, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon N, Dionisio KL, Isaacs K, Tornero-Velez R, Kapraun D, Setzer RW, and Price PS (2018). Simulating exposure-related behaviors using agent-based models embedded with needs-based artificial intelligence. J Expo Sci Environ Epidemiol doi: 10.1038/s41370-018-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, and Thomas RS (2015). Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ Sci Technol 49(14), 8804–14. [DOI] [PubMed] [Google Scholar]
- Casati S, Aschberger K, Barroso J, Casey W, Delgado I, Kim TS, Kleinstreuer N, Kojima H, Lee JK, Lowit A, et al. (2018). Standardisation of defined approaches for skin sensitisation testing to support regulatory use and international adoption: position of the International Cooperation on Alternative Test Methods. Arch Toxicol 92(2), 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commission E (2007). Registration, Evaluation, Authorisation and Restriction of Chemicals. In Regulation (EC) No 1907/2006 of the European Parliament and of the Council ( [Google Scholar]
- Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, Bois FY, Burgoon LD, Chiu WA, Crawford-Brown D, et al. (2016). The Next Generation of Risk Assessment Multi-Year Study-Highlights of Findings, Applications to Risk Assessment, and Future Directions. Environ Health Perspect 124(11), 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGroot DE, Swank A, Thomas RS, Strynar M, Lee MY, Carmichael PL, and Simmons SO (2018). mRNA transfection retrofits cell-based assays with xenobiotic metabolism. J Pharmacol Toxicol Methods 92, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio KL, Frame AM, Goldsmith MR, Wambaugh JF, Liddell A, Cathey T, Smith D, Vail J, Ernstoff AS, Fantke P, et al. (2015). Exploring consumer exposure pathways and patterns of use for chemicals in the environment. Toxicol Rep 2, 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio KL, Phillips K, Price PS, Grulke CM, Williams A, Biryol D, Hong T, and Isaacs KK (2018). The Chemical and Products Database, a resource for exposure-relevant data on chemicals in consumer products. Sci Data 5, 180125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EC/HC (2008). Screening assessment for the Challenge: 1,2-benzenediol (catechol), Chemical Abstracts Service Registry Number 120-80-9. In (Vol. http://www.ec.gc.ca/eseees/default.asp?lang=En&n=04FDC10E-1#a5. Government of Canada, Ottawa (ON). [Google Scholar]
- EC/HC (2011). Screening assessment for the Challenge: Hexanedioic acid, bis(2-ethylhexyl) ester (DEHA) Chemical Abstracts Service Registry Number 103-23-1. In (Vol. http://www.ec.gc.ca/ese-ees/default.asp?lang=En&n=39958D25-1#a9 Government of Canada, Ottawa (ON). [Google Scholar]
- EC/HC (2017). Updated draft screening assessment: Chlorhexidine and its Salts. In (Vol. http://www.ec.gc.ca/ese-ees/default.asp?lang=En&n=59BDF713-1#toc-071 Government of Canada, Ottawa (ON). [Google Scholar]
- ECCC/HC (2016a). Chemicals Management Plan (CMP) Science Committee Objectives Paper Meeting no. 5 - Integrating New Approach Methodologies within the CMP: Identifying Priorities for Risk Assessment, Existing Substances Risk Assessment Program. In (Vol. Retrieved from http://www.ec.gc.ca/eseees/default.asp?lang=En&n=172614CE-1. Government of Canada, Ottawa (ON). [Google Scholar]
- ECCC/HC (2016b). Chemicals Management Plan In (H. C. Environment and Climate Change, Ed.), Vol. https://www.canada.ca/en/health-canada/services/chemical-substances/chemicals-management-plan.html. Government of Canada, Ottawa (ON). [Google Scholar]
- ECHA New Approach Methodologies in Regulatory Science, Proceedings of a scientific workshop, April 19–20, 2016 2016, Helsinki, Finland: Available at: https://echa.europa.eu/documents/10162/22816069/scientific_ws_proceedings_en.pdf/a2087434-0407-4705-9057-95d9c2c2cc57. Accessed September 3, 2018. [Google Scholar]
- ECHA (2017). ECHA Strategic Plan 2019–2023. In ( [Google Scholar]
- EFSA (2012). Scientific Opinion on Exploring Options for Providing Advice about Possible Human Health Risks Based on the Concept of Threshold of Toxicological Concern (TTC). In (Vol. 10, EFSA Journal. [Google Scholar]
- EFSA (2018). Guidance for the identification of endocrine disruptors in the context of Regulations (EU) No 528/2012 and (EC) No 1107/2009. In (doi: 10.2903/j.efsa.2018.5311, https://www.efsa.europa.eu/it/efsajournal/pub/5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeghy PP, Judson R, Gangwal S, Mosher S, Smith D, Vail J, and Cohen Hubal EA (2012). The exposure data landscape for manufactured chemicals. Sci Total Environ 414, 159–66. [DOI] [PubMed] [Google Scholar]
- Embry MR, Bachman AN, Bell DR, Boobis AR, Cohen SM, Dellarco M, Dewhurst IC, Doerrer NG, Hines RN, Moretto A, et al. (2014). Risk assessment in the 21st century: roadmap and matrix. Crit Rev Toxicol 44 Suppl 3, 6–16. [DOI] [PubMed] [Google Scholar]
- EPA, U. (2018a). Final Strategic Plan to Promote Development and Implementation of Alternative Test Methods Supporting Toxic Substances Control Act. In (Vol. 83, pp. 30167–30168. Federal Register, Washington, D.C. [Google Scholar]
- EPA, U. (2018b). ToxCast & Tox21 Data from invitrodb_v3. In (Retrieved from http://www2.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data. [Google Scholar]
- EPA, U. (ExpoBox). EPA ExpoBox: Exposure Assessment Tools by Tiers and Types - Screening-Level and Refined. Available at: https://www.epa.gov/expobox/exposure-assessment-tools-tiers-and-types-screening-level-and-refined. Accessed Febuary 13, 2019. [Google Scholar]
- Faucette SR, Wang H, Hamilton GA, Jolley SL, Gilbert D, Lindley C, Yan B, Negishi M, and LeCluyse EL (2004). Regulation of CYP2B6 in primary human hepatocytes by prototypical inducers. Drug Metab Dispos 32(3), 348–58. [DOI] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, and Martin MT (2017). tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics 33(4), 618–620. [DOI] [PubMed] [Google Scholar]
- Fischer FC, Henneberger L, Konig M, Bittermann K, Linden L, Goss KU, and Escher BI (2017). Modeling Exposure in the Tox21 in Vitro Bioassays. Chem Res Toxicol 30(5), 1197–1208. [DOI] [PubMed] [Google Scholar]
- Gottmann E, Kramer S, Pfahringer B, and Helma C (2001). Data quality in predictive toxicology: reproducibility of rodent carcinogenicity experiments. Environ Health Perspect 109(5), 509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariparsad N, Ramsden D, Palamanda J, Dekeyser JG, Fahmi OA, Kenny JR, Einolf H, Mohutsky M, Pardon M, Siu YA, et al. (2017). Considerations from the IQ Induction Working Group in Response to Drug-Drug Interaction Guidance from Regulatory Agencies: Focus on Downregulation, CYP2C Induction, and CYP2B6 Positive Control. Drug Metab Dispos 45(10), 1049–1059. [DOI] [PubMed] [Google Scholar]
- HealthCanada (2016). Science Approach Document: Threshold of Toxicological Concern (TTC)-based Approach for Certain Substances In (E. S. R. A. Bureau, Ed.). Government of Canada, Ottawa, Ontario. [Google Scholar]
- Jamei M, Dickinson GL, and Rostami-Hodjegan A (2009). A framework for assessing inter-individual variability in pharmacokinetics using virtual human populations and integrating general knowledge of physical chemistry, biology, anatomy, physiology and genetics: A tale of ‘bottom-up’ vs ‘top-down’ recognition of covariates. Drug Metab Pharmacokinet 24(1), 53–75. [DOI] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Knudsen T, Thomas RS, Sipes N, Shah I, Wambaugh J, and Crofton K (2014). In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme. Basic Clin Pharmacol Toxicol 115(1), 69–76. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, et al. (2009). The toxicity data landscape for environmental chemicals. Environ Health Perspect 117(5), 685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, and Dix DJ (2011). Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem Res Toxicol 24(4), 451–62. [DOI] [PubMed] [Google Scholar]
- Kavlock R (2016). Practitioner Insights: Bringing New Methods for Chemical Safety into the Regulatory Toolbox; It is Time to Get Serious. Bloomberg BNA Daily Environment REport(223 B-1). [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, et al. (2012). Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25(7), 1287–302. [DOI] [PubMed] [Google Scholar]
- Kavlock RJ, Bahadori T, Barton-Maclaren TS, Gwinn MR, Rasenberg M, and Thomas RS (2018). Accelerating the Pace of Chemical Risk Assessment. Chem Res Toxicol 31(5), 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D, et al. (2017). Development and Validation of a Computational Model for Androgen Receptor Activity. Chem Res Toxicol 30(4), 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes R, Renwick AG, Cheeseman M, Kleiner J, Mangelsdorf I, Piersma A, Schilter B, Schlatter J, van Schothorst F, Vos JG, et al. (2004). Structure-based thresholds of toxicological concern (TTC): guidance for application to substances present at low levels in the diet. Food Chem Toxicol 42(1), 65–83. [DOI] [PubMed] [Google Scholar]
- Laksameethanasan D, Tan R, Toh G, and Loo LH (2013). cellXpress: a fast and user-friendly software platform for profiling cellular phenotypes. BMC Bioinformatics 14 Suppl 16, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberg FR (2016). Frank R. Lautenberg Chemical Safety for the 21st Century Act. In (Congress t. U., Ed.), pp. 114–182. Public Law. [Google Scholar]
- Lee JJ, Miller JA, Basu S, Kee TV, and Loo LH (2018). Building predictive in vitro pulmonary toxicity assays using high-throughput imaging and artificial intelligence. Arch Toxicol 92(6), 2055–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LH, Wu LF, and Altschuler SJ (2007). Image-based multivariate profiling of drug responses from single cells. Nat Methods 4(5), 445–53. [DOI] [PubMed] [Google Scholar]
- Martin MT, Judson RS, Reif DM, Kavlock RJ, and Dix DJ (2009a). Profiling chemicals based on chronic toxicity results from the U.S. EPA ToxRef Database. Environ Health Perspect 117(3), 392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MT, Mendez E, Corum DG, Judson RS, Kavlock RJ, Rotroff DM, and Dix DJ (2009b). Profiling the reproductive toxicity of chemicals from multigeneration studies in the toxicity reference database. Toxicol Sci 110(1), 181–90. [DOI] [PubMed] [Google Scholar]
- Nair AB, and Jacob S (2016). A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 7(2), 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCT (2018). tcpl version 2.0. CRAN. [Google Scholar]
- Padilla S, Corum D, Padnos B, Hunter DL, Beam A, Houck KA, Sipes N, Kleinstreuer N, Knudsen T, Dix DJ, et al. (2012). Zebrafish developmental screening of the ToxCast Phase I chemical library. Reprod Toxicol 33(2), 174–87. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Jeliazkova N, Safford RJ, Worth AP, and Aleksiev B (2008). An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. Sar and Qsar in Environmental Research 19(5–6), 495–524. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Papineni S, Marty MS, Yi KD, Goetz AK, Rasoulpour RJ, Kwiatkowski P, Wolf DC, Blacker AM, and Peffer RC (2016). A predictive data-driven framework for endocrine prioritization: a triazole fungicide case study. Crit Rev Toxicol 46(9), 785–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RG, Setzer RW, Strope CL, Sipes NS, and Wambaugh JF (2017). httk: R Package for High-Throughput Toxicokinetics. Journal of Statistical Software 79(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins EJ, Habib T, Escalon BL, Cavallin JE, Thomas L, Weberg M, Hughes MN, Jensen KM, Kahl MD, Villeneuve DL, et al. (2017). Prioritization of Contaminants of Emerging Concern in Wastewater Treatment Plant Discharges Using Chemical:Gene Interactions in Caged Fish. Environ Sci Technol 51(15), 8701–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep P, Mansouri K, Patlewicz G, and Judson R (2017). A systematic evaluation of analogs and automated read-across prediction of estrogenicity: A case study using hindered phenols. Comput Toxicol 4, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, Waidyanatha S, Dixon D, DeVito MJ, Paules RS, and Ferguson SS (2017). Three-Dimensional (3D) HepaRG Spheroid Model With Physiologically Relevant Xenobiotic Metabolism Competence and Hepatocyte Functionality for Liver Toxicity Screening. Toxicol Sci 160(1), 189–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, et al. (2016). ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol 29(8), 1225–51. [DOI] [PubMed] [Google Scholar]
- Ring CL, Arnot J, Bennett DH, Egeghy P, Fantke P, Huang L, Isaacs KK, Jolliet O, Phillips K, Price PS, et al. (2018). Consensus Modeling of Median Chemical Intake for the U.S. Population Based on Predictions of Exposure Pathways. Environ Sci Technol doi: 10.1021/acs.est.8b04056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, and Knudsen TB (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol 26(6), 878–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Wambaugh JF, Pearce R, Auerbach SS, Wetmore BA, Hsieh J-H, Shapiro AJ, Svoboda D, DeVito MJ, and Ferguson SS (2017a). An Intuitive Approach for Predicting Potential Human Health Risk with the Tox21 10k Library. Accessed. [DOI] [PMC free article] [PubMed]
- Sipes NS, Wambaugh JF, Pearce R, Auerbach SS, Wetmore BA, Hsieh JH, Shapiro AJ, Svoboda D, DeVito MJ, and Ferguson SS (2017b). An Intuitive Approach for Predicting Potential Human Health Risk with the Tox21 10k Library. Environ Sci Technol 51(18), 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland JD, Martin MT, Richard AM, Houck KA, and Shafer TJ (2018). Screening the ToxCast phase II libraries for alterations in network function using cortical neurons grown on multi-well microelectrode array (mwMEA) plates. Arch Toxicol 92(1), 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R, Xiong S, Zink D, and Loo LH (2016). High-throughput imaging-based nephrotoxicity prediction for xenobiotics with diverse chemical structures. Arch Toxicol 90(11), 2793–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Paules RS, Simeonov A, Fitzpatrick SC, Crofton KM, Casey WM, and Mendrick DL (2018). The US Federal Tox21 Program: A strategic and operational plan for continued leadership. ALTEX 35(2), 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Philbert MA, Auerbach SS, Wetmore BA, Devito MJ, Cote I, Rowlands JC, Whelan MP, Hays SM, Andersen ME, et al. (2013). Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol Sci 136(1), 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, and Bucher JR (2013). Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121(7), 756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ToxTree (2015). ToxTree Prediction Module, Version 2.6.13. [Google Scholar]
- USEPA (2015). Use of High-Throughput Assays and Computational Tools; Endocrine Disruptor Screening Program; Notice of Availability and Opportunity for Comment. In (Federal Register. [Google Scholar]
- Wambaugh JF, Hughes MF, Ring CL, MacMillan DK, Ford J, Fennell TR, Black SR, Snyder RW, Sipes NS, and Wetmore BA (2018). Evaluating In Vitro-In Vivo Extrapolation of Toxicokinetics. Toxicological sciences : an official journal of the Society of Toxicology 163(1), 152–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh JF, Wang A, Dionisio KL, Frame A, Egeghy P, Judson R, and Setzer RW (2014). High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ Sci Technol 48(21), 12760–7. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Wetmore BA, Pearce R, Strope C, Goldsmith R, Sluka JP, Sedykh A, Tropsha A, Bosgra S, Shah I, et al. (2015). Toxicokinetic Triage for Environmental Chemicals. Toxicological Sciences 147(1), 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hallinger D, Murr A, Buckalew A, Lougee R, Richard AM, S. L, and T. S (2019). High-Throughput Screening and Chemotype-Enrichment Analysis of ToxCast Phase II Chemicals Evaluated for Human Sodium-Iodide Symporter (NIS) Inhibition. Environ Int. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H (2010). Confidence Assessment of the Simcyp Time-Based Approach and a Static Mathematical Model in Predicting Clinical Drug-Drug Interactions for Mechanism-Based CYP3A Inhibitors. Drug Metabolism and Disposition 38(7), 1094–1104. [DOI] [PubMed] [Google Scholar]
- Ward JM, Schofield PN, and Sundberg JP (2017). Reproducibility of histopathological findings in experimental pathology of the mouse: a sorry tail. Lab Anim (NY) 46(4), 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt ED, and Judson RS (2018). Uncertainty quantification in ToxCast high throughput screening. PLoS One 13(7), e0196963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore BA, Allen B, Clewell HJ 3rd, Parker T, Wambaugh JF, Almond LM, Sochaski MA, and Thomas RS (2014). Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicol Sci 142(1), 210–24. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Allen B, Ferguson SS, Sochaski MA, Setzer RW, Houck KA, Strope CL, Cantwell K, Judson RS, et al. (2015). Incorporating High-Throughput Exposure Predictions With Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicol Sci 148(1), 121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Li L, Clewell HJ 3rd, Judson RS, Freeman K, Bao W, Sochaski MA, Chu TM, et al. (2013). Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicol Sci 132(2), 327–46. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, Clewell HJ 3rd, Dix DJ, Andersen ME, Houck KA, et al. (2012). Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicological sciences : an official journal of the Society of Toxicology 125(1), 157–74. [DOI] [PubMed] [Google Scholar]
- WHO, E. a. (2016). Review of the Threshold of Toxicological Concern (TTC) approach and development of new TTC decision tree. In (Vol. EN=1006, pp. 50 EFSA Event Report. [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, et al. (2017). The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 9(1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JC, and Maack G (2017). Evaluating the credibility of histopathology data in environmental endocrine toxicity studies. Environ Toxicol Chem 36(3), 601–611. [DOI] [PubMed] [Google Scholar]
- Yang C, Tarkhov A, Marusczyk J, Bienfait B, Gasteiger J, Kleinoeder T, Magdziarz T, Sacher O, Schwab CH, Schwoebel J, et al. (2015). New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model 55(3), 510–28. [DOI] [PubMed] [Google Scholar]
- Yoon M, Efremenko A, Blaauboer BJ, and Clewell HJ (2014). Evaluation of simple in vitro to in vivo extrapolation approaches for environmental compounds. Toxicology in Vitro 28(2), 164–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


