Abstract
Insolubility of recombinant proteins in Escherichia coli is a major impediment to their production for structural and functional studies. One way around this problem is to fuse an aggregation-prone protein to a highly soluble partner. E. coli maltose-binding protein (MBP) is widely recognized as a premier solubilizing agent. In this chapter, we describe how to construct dual His6-MBP-tagged fusion proteins by Gateway® recombinational cloning and how to predict their yield and solubility. We also describe a simple and rapid procedure to test the ability of a His6-MBP fusion protein to bind to Ni-NTA resin and to be digested by tobacco etch virus (TEV) protease, along with a method to assess the solubility of the target protein after it has been separated from His6-MBP.
Keywords: Maltose-binding protein, MBP, Fusion protein, Solubility enhancer, TEV protease, Tobacco etch virus protease, Hexahistidine tag, His-tag, His6-MBP, Gateway® cloning, Recombinational cloning
1. Introduction
When successful, producing recombinant proteins in Escherichia coli is relatively inexpensive and straightforward for most molecular biology laboratories. However, many potentially interesting proteins aggregate into an insoluble and unusable form when produced via simple expression schemes [1]. Before abandoning bacterial expression in favor of more complicated and costly eukaryotic systems, we suggest employing a simple strategy that combines the solubility-enhancing benefit conferred by E. coli maltose-binding protein (MBP) [2, 3] with the powerful advantage of immobilized metal affinity chromatography (IMAC) [4], made possible by the use of a polyhistidine tag in a tandem configuration with MBP (His6-MBP) [5]. In this chapter, we describe how to construct His6-MBP fusion proteins and conduct a few simple pilot experiments that are reliable predictors of protein production success prior to extensive resource investment.
2. Materials
2.1. Construction of Expression Vectors by Recombinational Cloning
The Gateway® destination vector pDEST-HisMBP (see Fig. 1).
PCR Reagents, including thermostable DNA polymerase (see Note 1).
Synthetic oligodeoxyribonucleotide primers for PCR amplification (see Fig. 2).
TE buffer: 10 mM Tris–HCl, pH 8.0, 1 mM EDTA.
Agarose, buffer, and an apparatus for submarine gel electrophoresis of DNA (see Note 2).
MinElute Gel Extraction Kit (Qiagen, Valencia, CA) for the extraction of DNA from agarose gels.
Chemically competent ccdB Survival™ 2 T1R cells (Life Technologies, Grand Island, NY) for propagating pDEST-HisMBP, pDONR201, or any vector with a Gateway® cloning cassette.
Competent gyrA+ cells (e.g., DH5α, MC1061, HB101) (see Note 3).
Gateway® PCR Cloning System (Life Technologies).
LB medium and LB agar plates containing ampicillin (100 μg/ml). LB medium: Add 10 g Bacto Tryptone, 5 g Bacto Yeast Extract, and 5 g NaCl to 1 l of H2O and sterilize by autoclaving. For LB agar, also add 12 g of Bacto Agar before autoclaving. To prepare plates, allow medium to cool until flask or bottle can be held in hands without burning, then add 1 ml ampicillin stock solution (100 mg/ml in H2O, filter sterilized), mix by gentle swirling, and pour or pipet ca. 30 ml into each sterile petri dish (100 mm dia.).
Reagents for small-scale plasmid DNA isolation (see Note 4).
An incubator set at 37 °C.
Fig. 1.
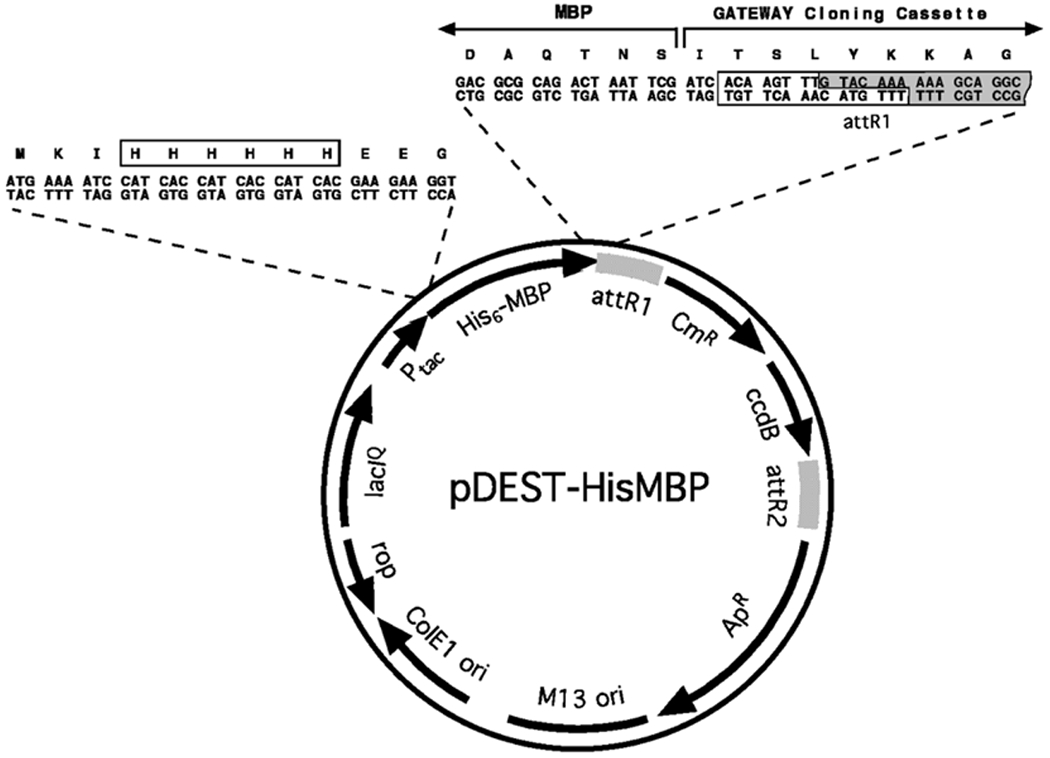
Schematic representation of the Gateway® destination vector pDEST-HisMBP. This vector can be recombined with an entry vector that contains an ORF of interest, via the LR reaction, to generate a His6-MBP fusion protein expression vector
Fig. 2.
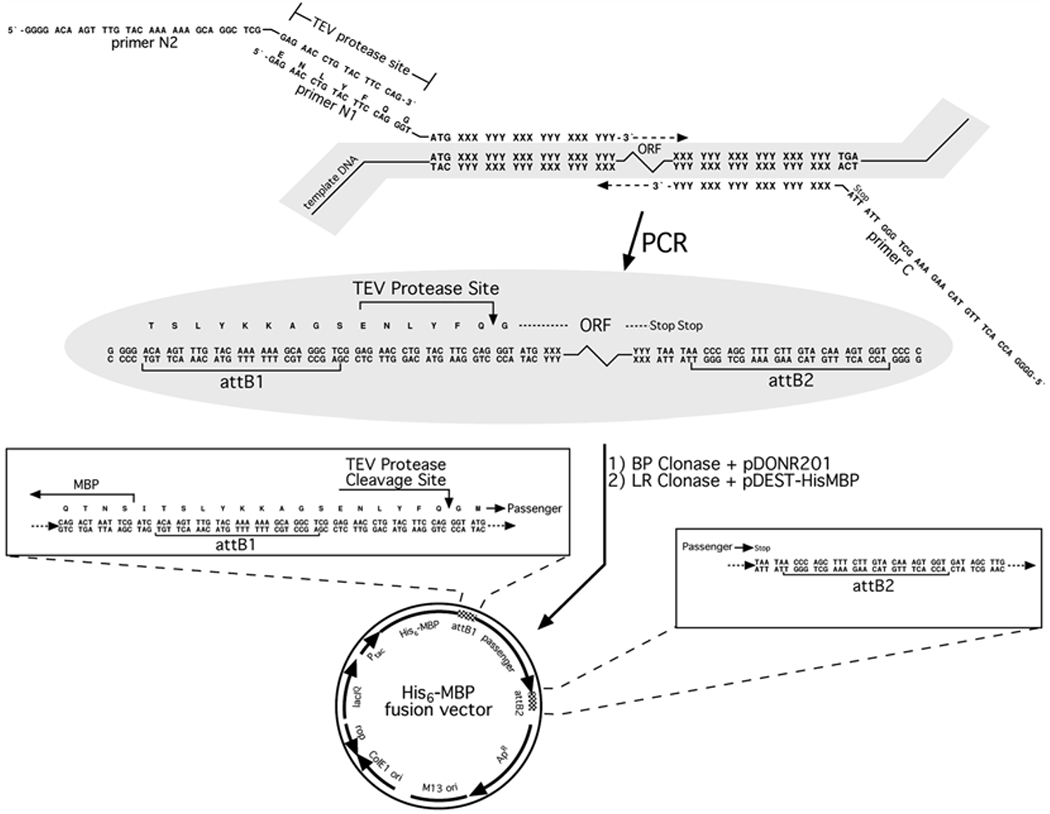
Construction of a His6-MBP fusion vector using PCR and Gateway® cloning technology. The ORF of interest is amplified from the template DNA by PCR, using primers N1, N2, and C. Primers N1 and C are designed to base-pair to the 5′ and 3′ ends of the coding region, respectively, and contain unpaired 5′ extensions as shown. Primer N2 base pairs with the sequence that is complementary to the unpaired extension of primer N1. The final PCR product is recombined with the pDONR201 or pDONR221 vector to generate an entry clone, via the BP reaction. This entry clone is subsequently recombined with pDEST-HisMBP using LR Clonase to yield the final His6-MBP fusion vector
2.2. Pilot Expression, Protease Cleavage, and Solubility Testing
Competent Rosetta™ 2(DE3) (EMD Millipore, Billerica, MA) (see Notes 5 and 6).
A derivative of pDEST-HisMBP that produces a His6-MBP fusion protein with a TEV protease recognition site in the linker between MBP and the passenger protein (see Subheading 3.1).
LB agar plates and broth containing both ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml). Prepare a stock solution of 30 mg/ml chloramphenicol in ethanol. Store at −20 °C for up to 6 months. (See Subheading 2.1, item 10 for LB broth, LB agar, and ampicillin stock solution recipes.) Dilute antibiotics 1,000-fold into LB medium or molten LB agar.
Isopropyl-thio-β-d-galactopyranoside (IPTG), Dioxane-free, Ultra Pure. Prepare a stock solution of 200 mM in H2O and filter-sterilize. Store at −20 °C.
Shaker/incubator.
Sterile baffled-bottom flasks.
B-PER with Enzymes Bacterial Protein Extraction Kit (Pierce Protein Biology Products, Thermo Fisher Scientific, Rockford, IL).
Ni-NTA Agarose.
AcTEV protease (Life Technologies), or TEV protease produced and purified as described [6].
Two IMAC-compatible buffers that contain 25 mM (for Buffer A) and 500 mM (for Buffer B) imidazole. For example, Buffer A: 25 mM Tris-HCl, 200 mM NaCl, 25 mM imidazole, pH 7.2; Buffer B: 25 mM Tris, 200 mM NaCl, 500 mM imidazole, pH 7.2.
4× SDS-PAGE sample buffer (Life Technologies) and 2-mercaptoethanol.
SDS-PAGE gel, electrophoresis apparatus, and running buffer (see Note 7).
Gel stain (e.g., Gelcode® Blue from Pierce Protein Biology Products, Thermo Fisher Scientific, or PhastGel™ Blue R from GE Healthcare Life Sciences, Piscataway, NJ).
Spectrophotometer.
1.5 ml microcentrifuge tubes.
3. Methods
3.1. His6-MBP Fusion Vector Recombinational Cloning
The Gateway® recombinational cloning system is based on the site-specific recombination reactions that mediate the integration and excision of bacteriophage lambda into and from the E. coli chromosome, respectively. For detailed information about this system, the investigator is encouraged to consult the technical literature supplied by Invitrogen, Inc. (http://www.invitrogen.com).
3.1.1. pDEST-HisMBP
To utilize the Gateway® system for the production of His6-MBP fusion proteins, one must first construct or obtain a suitable “destination vector.” An example of a destination vector that can be used to produce His6-MBP fusion proteins (pDEST-HisMBP), which is available from the authors or the Addgene plasmid repository (www.addgene.org), is shown in Fig. 1. pDEST-HisMBP was constructed by inserting an in-frame hexahistidine-coding sequence between codons 3 and 4 of MBP in pKM596 [4].
The Gateway® cloning cassette in pDEST-HisMBP carries a gene encoding the DNA gyrase poison CcdB, which provides a negative selection against the destination vector and the donor vector so that only the desired recombinant is obtained when the end products of the recombinational cloning reaction are transformed into E. coli and grown in the presence of ampicillin. pDEST-HisMBP and other vectors that carry the ccdB gene must be propagated in a host strain with a gyrA mutation (e.g., E. coli DB3.1) that renders the cells immune to the action of CcdB or, alternatively, in a strain that produces the CcdB antidote CcdA (e.g., ccdB Survival™ 2 T1R cells).
3.1.2. Gateway® Cloning Protocol
To construct a His6-MBP fusion expression vector, we amplify the target open reading frame (ORF) by PCR, incorporating into the primers elements necessary for Gateway® cloning and downstream protein production, then perform successive BP and LR reactions. The 3′ ends of the primers include a sufficient number of nucleotides that are complementary to the template sequence to result in a 69 °C melting temperature (by modified Breslauer’s method, see http://www.thermoscientificbio.com/webtools/tmc/). This enables 2-step PCR cycling using 72 °C as both the annealing and extension temperature. Proximal to the ORF-specific part of the forward primer, we add a sequence that encodes a TEV protease cleavage site preceded by an attB1 site to enable recombination. Because shorter primers are less expensive and because the TEV- and attB1-containing sequences are common to many of our experimental designs, we often use two overlapping forward primers, only one of which is ORF-specific (Fig. 2). An attB2 recombination site is added to the 5′ end of the ORF-specific portion of the reverse PCR primer. During early rounds of cycling, the inner, ORF-specific forward primer acts with the reverse primer to create a template amplified by N2 and the same reverse primer in later rounds. To favor full-length product accumulation, the concentration of N1 is 20-fold lower than that of N2 and C (see Note 8).
The PCR reaction mix is prepared as follows (see Note 9): 1 pg–10 ng template DNA, 10 μl 2× Phusion Flash PCR Master Mix (contains all necessary reaction components except primers and template, 0.025 μM primer N1, 0.5 μM primer N2, 0.5 μM primer C, H2O (to 20 μl total volume).
The reaction is placed in a thin-walled tube in a thermal cycler with an appropriate program, such as the following: initial denaturation for 3 min at 98 °C; 30 cycles of 98 °C for 10 s and 72 °C for 15 s, and final extension at 72 °C for 60 s (see Note 10); hold at 4 °C.
Purification of the PCR amplicon by agarose gel electrophoresis (see Note 2) is recommended.
- To create the His6-MBP fusion vector, the PCR product is recombined first into a donor vector, such as pDONR221 or pDONR201, to yield an entry clone intermediate (BP reaction), and then into pDEST-HisMBP (LR reaction; see Note 11).
- Add to a microcentrifuge tube: 100 ng of the PCR product in 1–5 μl TE or H2O, 1.3 μl of 150 ng/μl pDONR vector DNA, and enough TE to bring the total volume to 12 μl Mix well.
- Thaw BP Clonase II enzyme mix on ice (2 min) and then vortex briefly (2 s) twice (see Note 12).
- Add 3 μl of BP Clonase II enzyme mix to the components in (a) and vortex briefly; incubate the reaction at room temperature for at least 4 h (see Note 13).
- Add to 10 μl of BP reaction: 2 μl of 150 ng/μl destination vector (pDEST-HisMBP) and 3 μl of LR Clonase II enzyme mix (see Note 12). Mix by vortexing briefly.
- Incubate the reaction at room temperature for 2 h.
- Add 2 μl of the proteinase K stop solution and incubate for 10 min at 37 °C.
- Transform 1 μl of the reaction into 50 μl of appropriate competent E. coli, such as electrocompetent DH5α cells (see Note 3).
Plasmid DNA is isolated from saturated cultures started from individual ampicillin-resistant colonies and screened by sequencing putative clones to ensure that there are no PCR-induced mutations.
3.2. Pilot Fusion Protein Expression, Small-Batch Purification, TEV Protease Cleavage, and Solubility Testing
Before investing time and resources on the large-scale expression and purification of a protein, we perform a series of pilot experiments to assess protein production, fusion protein-IMAC resin binding, TEV protease cleavage, and target protein solubility. First, we transform the sequence-verified expression plasmid into an appropriate expression strain and induce production of the fusion protein. Following chemical disruption of the cells, we confirm that the His6-MBP fusion protein is present in the soluble fraction of the crude cell lysate. After passing this checkpoint, we model the purification steps in a microcentrifuge tube to establish successful binding of fusion protein to Ni-NTA resin. Following elution in this small-batch format, we test for successful cleavage from His6-MBP and sustained solubility of the protein of interest following its liberation. A problem at any of these steps can be addressed before scale-up.
3.2.1. Protein Expression
Transform competent Rosetta™ 2(DE3) (see Notes 5 and 6) with the His6-MBP fusion protein expression vector and spread them on an LB agar plate containing ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml). Incubate the plate overnight at 37 °C.
Inoculate 5 ml of LB medium containing ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml) in a culture tube with a single colony from the plate. Grow to saturation overnight at 37 °C with shaking.
The next morning, inoculate 50 ml of the same medium in a 250 ml baffled-bottom flask with 0.5 ml of the saturated overnight culture.
Grow the cells at 37 °C with shaking to mid-log phase (OD600nm ~ 0.5).
Adjust the temperature to 30 °C (see Note 15) and add IPTG (1 mM final concentration).
After 4 h, measure the OD600nm of the cultures (dilute cells 1:10 in LB to obtain an accurate reading). An OD600nm of about 3–3.5 is normal, although lower or higher densities are possible. If the density of the culture is much lower than this, it may be necessary to adjust the volume of the samples that are analyzed by SDS-PAGE.
Transfer 10 ml to a 15 ml conical centrifuge tube and pellet the cells by centrifugation (4,000 × g) at 4 °C.
Store the cell pellet at −80 °C. Alternatively, the cells can be disrupted immediately and the procedure continued without interruption, as described below.
3.2.2. B-PER Chemical Solubilization and Sample Preparation
Thaw the cell pellet on ice.
Chemically disrupt the cells with B-BER Bacterial Protein Extraction Reagent according to the manufacturer’s instructions. Briefly, resuspend the pellet in 500 μl B-PER plus 1 μl lysozyme and 1 μl DNaseI. Incubate the suspension at room temperature for 15 min.
Prepare a sample of the total intracellular protein (“T”) for SDS-PAGE by mixing 50 μl of the B-PER suspension with 50 μl of 2× SDS-PAGE sample buffer containing 10 % (v/v) 2-mercaptoethanol.
Pellet the insoluble cell debris and proteins by centrifuging the B-PER suspension at 15,000 × g in a microcentrifuge for 5 min.
Prepare a sample of the soluble intracellular protein (“S”) for SDS-PAGE by mixing 50 μl of the supernatant from step 4 with 50 μl of 2× SDS-PAGE sample buffer containing 10 % (v/v) 2-mercaptoethanol.
3.2.3. Small-Batch Purification and TEV Protease Cleavage
Centrifuge 250 μl of Ni-NTA Agarose slurry at 4,000 × g for 1 min in a microcentrifuge tube; remove and discard the supernatant.
Wash the resin by adding 400 μl of Buffer A; mix by pipetting up and down. Centrifuge at 4,000 × g for 1 min; remove and discard the supernatant. Repeat wash.
Bind protein by adding B-PER lysate to Ni-NTA; mix by pipetting up and down. Incubate for 1 h at 4 °C with rocking.
Centrifuge at 4,000 × g for 1 min; remove the supernatant and retain it as the flow-through (“FT”) fraction (by analogy with column chromatography; represents unbound proteins).
Wash the resin 3 times with 400 μl Buffer A; centrifuge at 4,000 × g for 1 min each time and retain the supernatants from each wash separately for electrophoretic analysis (samples “W1,” “W2,” and “W3”).
Elute bound protein from the Ni-NTA resin with 3 separate 50-μl volumes of Buffer B. Centrifuge at 4,000 × g for 1 min each time and retain the supernatants from each separately as samples “E1,” “E2,” and “E3.”
After removing aliquots for electrophoretic analysis, combine fractions E1-E3 for pilot TEV protease cleavage. Add approximately 0.1 mg of TEV protease per 15 mg of fusion protein. Mix and remove an aliquot for overnight incubation at room temperature; incubate remaining reaction at 4 °C overnight.
3.2.4. SDS-PAGE
We typically use precast NuPAGE gradient gels for SDS-PAGE to assess the yield and solubility of MBP fusion proteins (see Note 7). The investigator may choose any appropriate SDS-PAGE formulation appropriate for the protein size and laboratory preference.
Prepare samples from each step of the small-batch purification by mixing an aliquot of each with an equal volume of 2× SDS-PAGE sample buffer containing 10 % (v/v) 2-mercaptoethanol.
Heat the T, S, and purification samples at 90 °C for about 5 min and then spin them at maximum speed in a microcentrifuge for 5 min.
Assemble the gel in the electrophoresis apparatus, fill it with SDS-PAGE running buffer, load the samples (4–20 μl) and carry out the electrophoretic separation according to standard lab practices. T and S samples are loaded in adjacent lanes to allow easy assessment of solubility. Molecular weight standards may also be loaded on the gel, if desired.
Stain the proteins in the gel with GelCode® Blue reagent, PhastGel™ Blue R, or a suitable alternative.
3.2.5. Interpreting the Results
The overexpressed MBP fusion should be apparent as the predominant protein present on the gel. Examining the heaviest band relative to a molecular weight standard should confirm that the fusion is about the size of the protein of interest plus 42 kDa, the approximate size of MBP. Placing the total and soluble fractions next to each other on the gel allows for easy comparison and determination of the fusion’s solubility.
Figure 3 illustrates the benefit of using MBP as a solubility enhancer. Lane 1 indicates that upon induction, the Rosetta 2(DE3) expression strain was able to produce Yersinia pestis dihydrofolate reductase (DHFR) from a plasmid encoding the His6-tagged protein. However, lane 2 reveals that most of the His6-tagged protein is not found in the soluble fraction. In contrast, lanes 3 (total protein) and 4 (soluble protein) clearly demonstrate that virtually all of the His6-MBP-DHFR fusion protein is soluble when produced in the same strain.
Fig. 3.
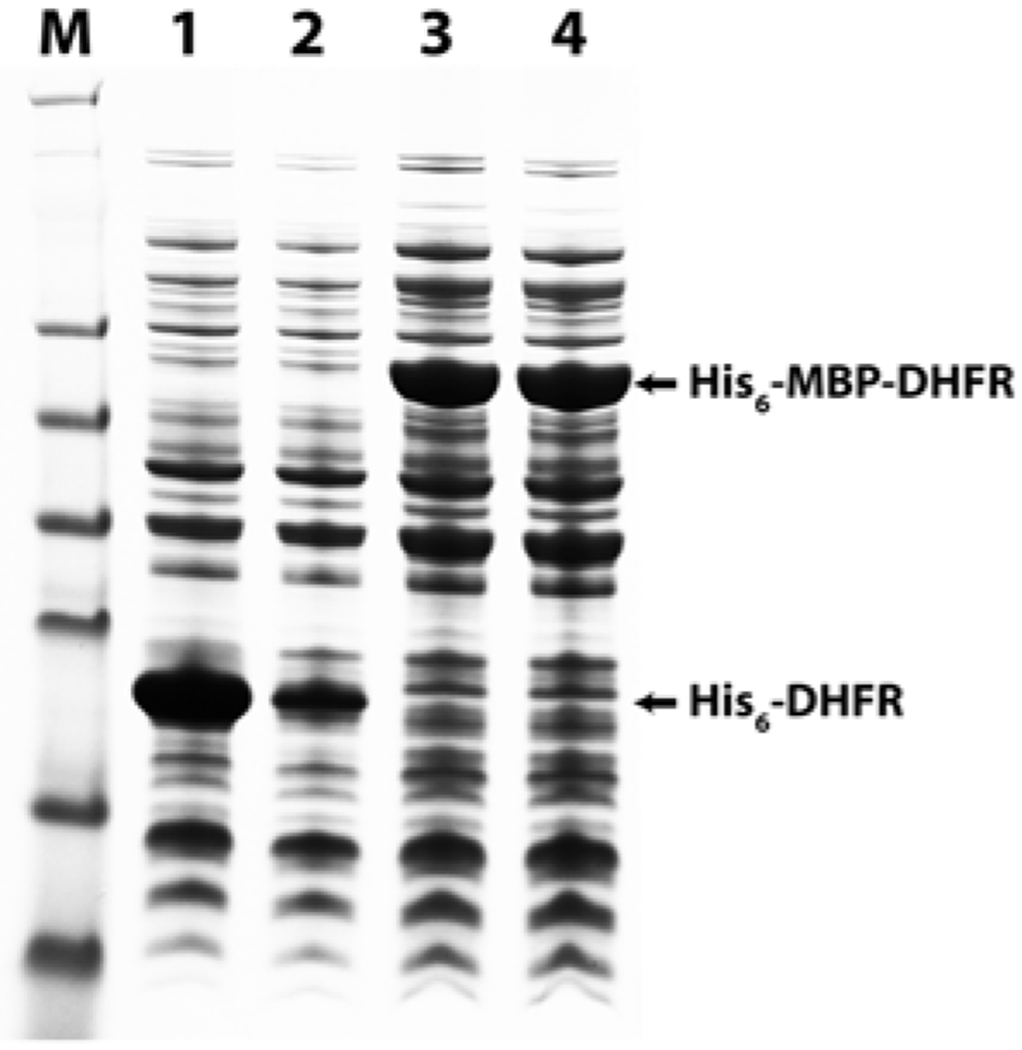
Comparison of the solubility of His6-tagged and His6-MBP-tagged Yersinia pestis dihydrofolate reductase (DHFR). Lanes 1–4 represent protein extracted from Rosetta 2(DE3) cells expressing either His6-tagged DHFR or His6-MBPDHFR from the appropriate plasmid. Lane M: SeeBlue Plus2 pre-stained marker standards. Lane 1: His6-DHFR total protein. Lane 2: His6-DHFR soluble protein. Lane 3: His6-MBP-DHFR total protein. Lane 4: His6-MBP-DHFR soluble protein
Performing a small-batch purification using Ni-NTA agarose in a microcentrifuge tube can establish whether or not the soluble His6-MBP fusion protein will bind to a similar column during scale-up, thereby enabling the investigator to identify at an early stage those rare cases in which the His6-tag is inaccessible and therefore unavailable for binding. Figure 4 shows the results of such a pilot purification, again using Y. pestis DHFR. The total and soluble intracellular proteins from cells overproducing the His6-MBP-DHFR fusion protein are shown in lanes 1 and 2, respectively. After incubation of the soluble fraction with Ni-NTA agarose, nearly all of the fusion protein was bound to the resin and removed by centrifugation at this stage (lane 3; “flow-through”). Had the His6-MBP-DHFR fusion protein failed to bind to the resin, a heavy band migrating at the size of the fusion would have appeared in the “flow-through” lane, predicting potential trouble with nickel column binding during scale-up. Most other intracellular proteins were separated from the resin during the three wash steps (lanes 4–6). The predominant band in the three elution fractions (lanes 7–9) is the His6-MBP fusion. Note that the protein is not absolutely pure at this stage, but it is significantly enriched relative to the starting material.
Fig. 4.
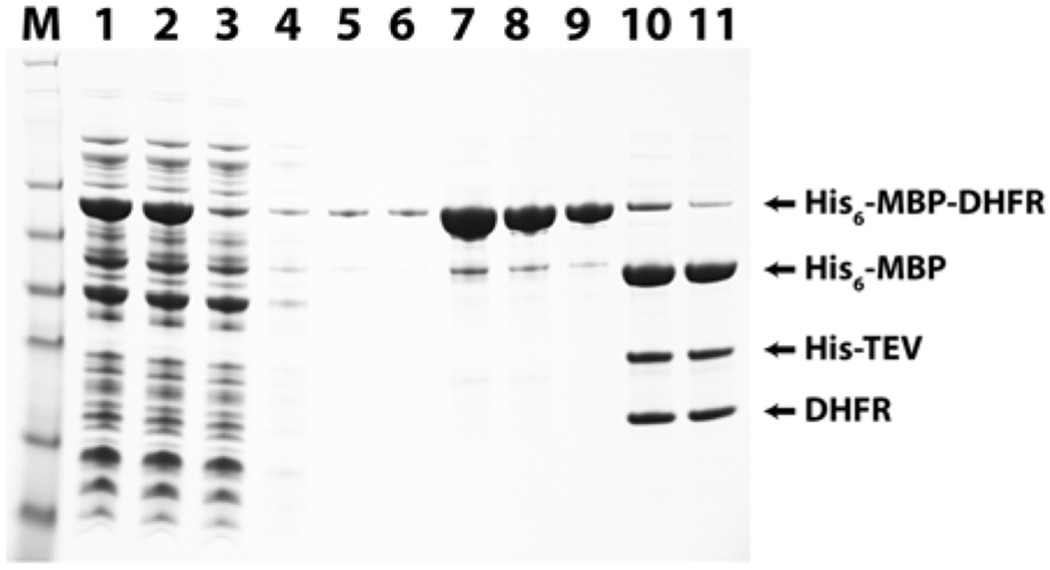
Small-scale pilot expression, purification, and digestion of fusion protein with TEV protease. Yersinia pestis DHFR was expressed from a derivative of pDEST-HisMBP in Rosetta 2(DE3) cells as described (see Subheading 3.2 ). Lane M: SeeBlue Plus2 pre-stained marker standards. Lane 1: His6-MBP-DHFR total protein. Lane 2: His6-MBP-DHFR soluble protein. Lane 3: Unbound/“flow-through” fraction. Lanes 4–6: Buffer A washes 1–3. Lanes 7–9: Elution fractions 1–3. Lane 10: TEV protease digest, total protein. Lane 11: TEV protease digest, soluble protein (see Subheadings 3.2 , items 4 and 5)
The product of the small-batch purification trial is sufficiently pure to test its ability to be cleaved by TEV protease in vitro. In lanes 10 and 11 of Fig. 4, which correspond to total and soluble products of the cleavage reaction, the band representing the fusion protein has largely disappeared, and three significant bands have materialized: a 42-kDa band for His6-MBP, a 28-kDa band for His-TEV protease, and an 18-kDa band migrating at the expected size of DHFR. The nearly identical intensities of the DHFR bands in the total and soluble samples indicate that DHFR remains soluble after cleavage from MBP, suggesting that it is probably properly folded. Had the TEV protease failed to cleave the fusion or had the target protein become insoluble after cleavage, troubleshooting would have been necessary. Otherwise, having successfully passed these diagnostic tests, the production and purification of the protein may now be scaled up as described [4].
3.2.6. Troubleshooting
Not every MBP fusion protein will be highly soluble. However, solubility usually can be increased by reducing the temperature of the culture from 30 to 18 °C during the time that the fusion protein is accumulating in the cells (i.e., after the addition of IPTG). In some cases, the improvement can be quite dramatic. It may also be helpful to reduce the IPTG concentration to a level that will result in partial induction of the fusion protein. Under these conditions, longer induction times (18–24 h) are required to obtain a reasonable yield of fusion protein.
If the fusion protein is a poor substrate for TEV protease in the small batch experiment, then the same is likely to be true during scale-up. However, in most cases it is still possible to obtain a sufficient quantity of the pure passenger protein by scaling up production (e.g., 4–6 l of cells or more). In especially problematic cases, the efficiency of the protease digest can be improved by inserting additional amino acid residues between the TEV protease recognition site and the N-terminus of the passenger protein. We have used both polyglycine and a FLAG-tag epitope in this position with good results [7].
Occasionally, a passenger protein may accumulate in a soluble but biologically inactive form after intracellular processing of an MBP fusion protein. Exactly how and why this occurs is unclear, but we suspect that fusion to MBP somehow enables certain proteins to evolve into kinetically trapped folding intermediates that are no longer susceptible to aggregation. Therefore, although solubility after intracellular processing is generally a useful indicator of a passenger protein’s folding state, it is not absolutely trustworthy. For this reason, we strongly recommend that a biological assay be employed (if available) at an early stage to confirm that the passenger protein is in its native conformation.
Acknowledgements
We thank Di Zhang for constructing the His6-MBP-DHFR expression vector. This research was funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
We recommend a processive, high-fidelity polymerase such as Phusion (Thermo Fisher, Waltham, MA or New England Biolabs, Ipswich, MA, USA) to reduce cycling times and minimize the occurrence of mutations during PCR.
We typically purify fragments by horizontal electrophoresis in 1–2 % Certified Molecular Biology Agarose (Bio-Rad, Hercules, CA) gels run in sodium boric acid [8] using standard submarine equipment. DNA fragments are extracted from slices of the ethidium bromide-stained gel using a MinElute Gel Extraction Kit (Qiagen) in accordance with the instructions supplied with the product.
Any gyrA+ strain of E. coli can be used. We prefer ElectroMAX™ DH5α-E™ Competent Cells (Life Technologies) because they are easy to use and very efficient.
We prefer the QIAprep™ Spin miniprep kit (Qiagen), but similar kits can be obtained from a wide variety of vendors.
Chemically competent cells are transformed according to the manufacturer’s instructions. Electrocompetent cells can be purchased or prepared: briefly, the cells are grown in 1 l of LB medium (with antibiotics, if appropriate) to mid-log phase (OD600 ~ 0.5) and then chilled on ice. The cells are pelleted at 4 °C, resuspended in 1 l of ice-cold H2O and then pelleted again. After several such washes with H2O, the cells are resuspended in 3–4 ml of 10 % glycerol, divided into 50 μl aliquots, and then immediately frozen in a dry ice-ethanol bath. Competent cells are stored at −80 °C. Electrotransformation procedures can be obtained from the electroporator manufacturers (e.g., Bio-Rad, BTX, Eppendorf). Immediately prior to electrotransformation, the cells are thawed on ice and mixed with 10–100 ng of DNA (e.g., a plasmid vector or a Gateway® reaction). The mixture is placed into an ice-cold electroporation cuvette and electroporated according to the manufacturer’s recommendations (e.g., a 1.5 kV pulse in a cuvette with a 1 mm gap). 1 ml of SOC medium [9] is immediately added to the cells and they are allowed to grow at 37 °C with shaking (ca. 250 rpm) for 1 h. 5–200 μl of the cells is then spread on an LB agar plate containing the appropriate antibiotic(s).
If the open reading frame encoding the passenger protein contains codons that are rarely used in E. coli (http://www.doe-mbi.ucla.edu/cgi/cam/racc.html), this can adversely affect the yield of an MBP fusion protein. In such cases, it is advisable that the expression strain carry an additional plasmid that codes for the cognate tRNA genes for rare codons. The pRIL plasmid (Stratagene, La Jolla, CA) is a derivative of the p15A replicon that carries the E. coli argU, ileY, and leuW genes, which encode the cognate tRNAs for AGG/AGA, AUA, and CUA codons, respectively. pRIL is selected for by resistance to chloramphenicol. In addition to the tRNA genes for AGG/AGA, AUA, and CUA codons, the pRARE accessory plasmid in the Rosetta™ host strain (Novagen, Madison, WI) also includes tRNAs for the rarely used CCC and GGA codons. Like pRIL, the pRARE plasmid is a chloramphenicol-resistant derivative of the p15A replicon. Both of these tRNA accessory plasmids are compatible with derivatives of pDEST-HisMBP.
We find it convenient to use precast SDS-PAGE gels, running buffer, molecular weight standards and electrophoresis supplies from Life Technologies.
Alternatively, the PCR reaction can be performed in two separate steps, using primers N1 and C in the first step and primers N2 and C in the second step. The PCR amplicon from the first step is used as the template for the second PCR. All primers are used at the typical concentrations for PCR in the two-step protocol.
The PCR reaction can be modified in numerous ways to optimize results, depending on the nature of the template and primers. See [9] (Vol. 2, Chapter 8) for more information.
PCR cycle conditions can also be varied based on reagents and consumables chosen, template complexity, and length of gene. For example, when using Phusion Flash High-Fidelity PCR Master Mix, extend for 15 s/kb of DNA. Consult the directions provided by the manufacturer of your thermostable polymerase.
This “one-tube” Gateway® protocol bypasses the isolation of an “entry clone” intermediate. However, the entry clone may be useful if the investigator intends to experiment with additional Gateway® destination vectors, in which case the BP and LR reactions can be performed sequentially in separate steps; detailed instructions are included with the Gateway® PCR kit. Alternatively, entry clones can easily be regenerated from expression clones via the BP reaction, as described in the instruction manual.
Clonase enzyme mixes should be thawed according to the manufacturer’s directions.
At this point, we remove a 5 μl aliquot from the reaction and add it to 0.5 μl of proteinase K stop solution. After 10 min at 37 °C, we transform 2 μl into 50 μl of competent DH5α cells (see Note 3) and spread 100–200 μl on an LB agar plate containing kanamycin (25 μg/ml), the selective marker for pDONR201. From the number of colonies obtained, it is possible to gauge the success of the BP reaction. Additionally, entry clones can be recovered from these colonies in the event that no transformants are obtained after the subsequent LR reaction.
If very few or no ampicillin-resistant transformants are obtained after the LR reaction, the efficiency of the process can be improved by incubating the BP reaction overnight.
We have found that decreasing the induction temperature to 30 °C increases the quality and solubility of the fusion protein without significantly decreasing the yield, especially in the presence of glucose.
References
- 1.Waugh DS (2005) Making the most of affinity tags. Trends Biotechnol 23(6):316–320. doi: 10.1016/j.tibtech.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Kapust RB, Waugh DS (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8(8):1668–1674. doi: 10.1110/ps.8.8.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox JD, Routzahn KM, Bucher MH, Waugh DS (2003) Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett 537 (1–3):53–57 [DOI] [PubMed] [Google Scholar]
- 4.Tropea JE, Cherry S, Nallamsetty S, Bignon C, Waugh DS (2007) A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol Biol 363:1–19. doi: 10.1007/978-1-59745-209-0_1 [DOI] [PubMed] [Google Scholar]
- 5.Routzahn KM, Waugh DS (2002) Differential effects of supplementary affinity tags on the solubility of MBP fusion proteins. J Struct Funct Genomics 2(2):83–92 [DOI] [PubMed] [Google Scholar]
- 6.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14(12):993–1000 [DOI] [PubMed] [Google Scholar]
- 7.Fox JD, Waugh DS (2003) Maltose-binding protein as a solubility enhancer. Methods Mol Biol 205:99–117 [DOI] [PubMed] [Google Scholar]
- 8.Brody JR, Kern SE (2004) Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques 36(2):214–216 [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]


