SUMMARY
Obesity and type 2 diabetes (T2D) are metabolic disorders that are linked to microbiome alterations. However, their co-occurrence poses challenges in disentangling microbial features unique to each condition. We analyzed gut microbiomes of lean non-diabetic (n = 633), obese non-diabetic (n = 494), and obese individuals with T2D (n = 153) from German population and metabolic disease cohorts. Microbial taxonomic and functional profiles were analyzed along with medical histories, serum metabolomics, biometrics, and dietary data. Obesity was associated with alterations in microbiome composition, individual taxa, and functions with notable changes in Akkermansia, Faecalibacterium, Oscillibacter, and Alistipes, as well as in serum metabolites that correlated with gut microbial patterns. However, microbiome associations were modest for T2D, with nominal increases in Escherichia/Shigella. Medications, including antihypertensives and antidiabetics, along with dietary supplements including iron, were significantly associated with microbiome variation. These results differentiate microbial components of these interrelated metabolic diseases and identify dietary and medication exposures to consider in future studies.
Graphical Abstract

In Brief
Co-occurrence of obesity and type 2 diabetes poses challenges in assessing microbiota changes specific to each condition. By comparing gut microbiomes of lean, obese non-diabetic, and obese type 2 diabetic individuals, Thingholm et al. found that obesity, unlike type 2 diabetes, was associated with microbiome variation. Medications and dietary supplements were further linked to microbiota alterations.
INTRODUCTION
The incidences of both obesity and type 2 diabetes are increasing worldwide, and their comorbidities and medical requirements incur high and rising healthcare costs. Obesity is a risk factor for T2D, but while 86% of individuals with T2D are overweight or obese, not all obese individuals develop T2D (Daousi et al., 2006; Narayan et al., 2007). Multiple factors play a role in the development of these diseases, including genetics, lifestyle, and the gut microbiome, with an increasing body of evidence supporting the microbiome’s role in obesity and in T2D (Boulangé et al., 2016; Chobot et al., 2018; Peters et al., 2018; Trøseid et al., 2013; Turnbaugh et al., 2006; Vrieze et al., 2012). However, it remains difficult to disentangle the exact microbial features associated with obesity and T2D while also ruling out external confounders. Studies to identify distinguishing features of the microbiome that uniquely characterize each of the two conditions are needed in order to understand whether the progression from obesity to T2D is, in part, mediated by the gut microbiome.
Obesity and diabetes are both metabolic conditions associated with a range of physiological functions closely related to the gut and gut microbiota. Intestinal dysbiosis is a common observation in obesity, while the observation is less consistent in T2D (Aron-Wisnewsky et al., 2019; Le Chatelier et al., 2013; Forslund et al., 2015; Karlsson et al., 2013; Qin et al., 2012; Turnbaugh et al., 2009). Low-grade inflammation and altered levels of lipopolysaccharides (LPS) and short-chain fatty acid (SCFA) have also been associated with metabolic disease (Forslund et al., 2015; Karlsson et al., 2013; Qin et al., 2012). Together, these observations suggest that the development of obesity-associated T2D, characterized by dysregulated glucose metabolism and insulin resistance, could be related to progressive disruption of the gut microbiome after initiation by obesity.
While there is general agreement that ecological diversity and taxonomic composition of the gut microbiome are altered in obesity and T2D, associations with single microbes or microbial products have been inconsistent between studies. These deviations may potentially be due to small sample sizes, differing study designs, geography, and clinical factors (Falony et al., 2016; Finucane et al., 2014; Sze and Schloss, 2016; Yassour et al., 2016). The high inter-individual variability of the gut microbiome, and its sensitivity to environmental influences complicates population-scale microbial research in complex diseases generally, potentially explaining some of these inconsistencies (Falony et al., 2016; Zhernakova et al., 2016). Despite the complications of microbial studies, a number of consistent associations have been observed among them, including an altered abundance of butyrate producing bacteria (Le Chatelier et al., 2013; Forslund et al., 2015; Karlsson et al., 2013; Qin et al., 2012; Vrieze et al., 2012).
In the current study, we performed a detailed analysis of lean non-diabetic (“lean healthy”, LH), obese non-diabetic (ObH), and obese T2D (ObT2D) individuals to identify compositional and functional features of the gut microbiome that associate with obesity, as well as those which deviate between obese individuals with and without T2D. We included 16S rRNA gene (16S) sequencing data for 1,280 samples and shotgun metagenomic data for a subset of 201 samples, in addition to extensive covariates describing anthropometrics, nutritional behavior, and medications. All samples in the core analysis are from the northern German cohorts PopGen (Krawczak et al., 2006) and FoCus (Müller et al., 2015), while key findings were replicated in 880 additional individuals from the North-Eastern German SHIP (Study of Health in Pomerania) cohort (Völzke et al., 2011). We identified several associations between the gut microbiome, plasma metabolome, obesity and diabetes phenotypes, and environmental factors. These comprised associations with gut microbial taxa, including a decrease of Akkermansia, Oscillibacter, and Intestinimonas in obesity, and a nominal increase of Escherichia/Shigella specific to T2D, as well as circulating metabolite changes [including branched-chain amino acids (BCAA)]. Dietary supplements, including iron and magnesium, and medications also covaried with microbial composition and functional potential, such as the known strong association with metformin, and we identified further effects of antihypertensive and antiphlogistic medications on the gut microbiome. The effects of dietary iron on microbiome composition were confirmed in a mouse feeding experiment, in which the difference between high and sufficient iron intake recapitulated the diet-associated microbiome divergence observed in human populations.
RESULTS
Overview of the German Metabolic Disease Cohort
The current study included 1,280 individuals from the Northern German cohorts PopGen (Krawczak et al., 2006) and FoCus (Müller et al., 2015) (Figure 1; Table S1). The PopGen and Focus cohorts have recorded information on medication, diet, and dietary supplement usage, together with an extensive phenotypic characterization including values of age, gender, BMI, and fasting glucose levels (from the PopGen/P2N biobank; see STAR Methods, Table S1). Furthermore, untargeted serum metabolomic profiles were generated for 400 study participants using the Metabolon platform, comprising 390 identified metabolites (STAR Methods). Data and specimens from both cohorts were handled by the same biobank using the same study protocol. Compatibility of the 16S microbiome data between the two cohorts is good as described previously (Wang et al., 2016). All individuals were grouped into three phenotypes: (a) lean (BMI ≤ 25) without diabetes, inflammatory bowel disease (IBD), or irritable bowel syndrome (IBS), with fasting glucose level below 125 mg/dl (“lean healthy”, LH, n = 633); (b) obese (BMI > 30) with same criteria as LH except for BMI (“obese healthy,” ObH, n = 494); and (c) obese (BMI > 30) with diagnosed T2D or fasting glucose level above 125 mg/dl, and without IBD and IBS, respectively (ObT2D, n = 153; Table S1).
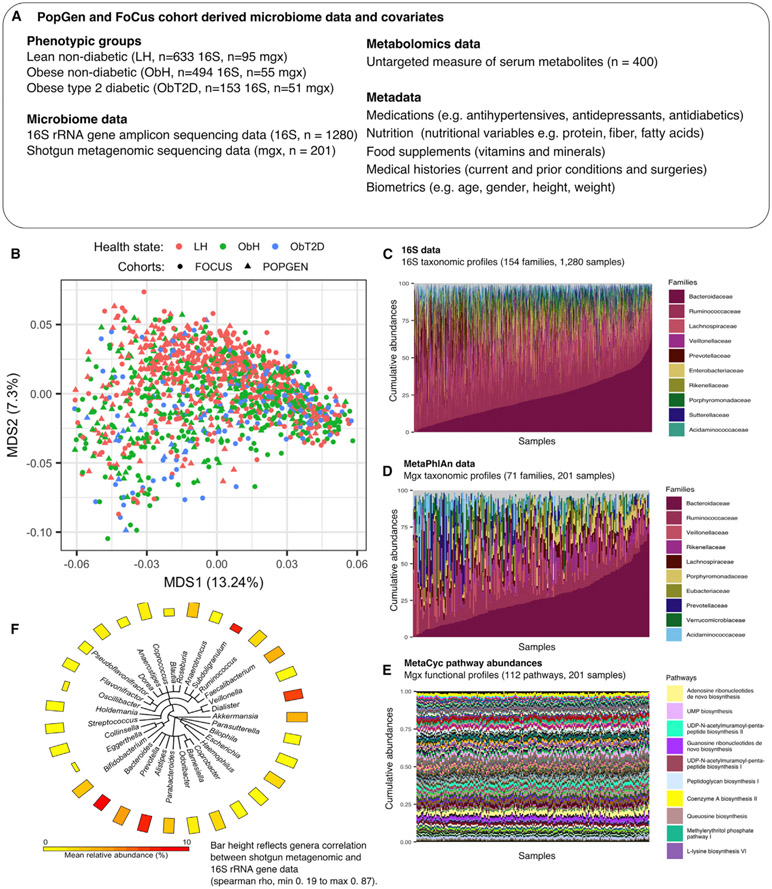
Figure 1. The Gut Microbiome in Obesity and Obesity-Associated type 2 Diabetes in Two Northern German Population Cohorts.
We investigated 1,280 individuals from two cohorts (popgen, n = 436, and focus, n = 844) to assess the role of the gut microbiome in T2D and obesity. In addition to extensive lifestyle, dietary, and environmental covariates recorded for these individuals, a stool sample from each participant was assayed using 16S rRNA gene sequencing, and a subset of these samples (n = 201) were metagenomically profiled.
(A) Overview of study data and metadata; detailed information in Table S1.
(B) Ordination of genus-level taxonomic profiles from 16S rRNA gene sequencing, using multidimensional scaling (MDS) of Bray-Curtis dissimilarities.
(C and D) Family-level taxonomic abundances for 16S (C) and metagenomic (D) data. The top 10 abundant families are annotated in the panel legend, while the remaining detected families are indicated in gray. Samples are ordered according to increasing relative abundance of bacteroidaceae.
(E) MetaCyc pathway abundances across 201 metagenomes (Caspi et al., 2014). The top 10 abundant pathways are colored and annotated in the panel legend, while the remaining core pathways (selected as top 50% mean abundant and top 50% variant) are indicated in gray.
(F) The phyloT-based (http://phylot.biobyte.de/) phylogeny of 31 genera well-detected in both the 16S and metagenomic profiles. The two measurement types generally agreed well (mean spearman ρ = 0.67). A total of 27 core genera from the shotgun data matched best with taxonomically identical taxa in the 16S data. The remaining four genera, eggerthella, blautia, oscillibacter, and subdoligranulum, showed highest correlation with a genus from the clostridiales order, predominantly in the unclassified clostridiales. See also Figure S1; Table S1.
A subset of 201 samples was selected for shotgun metagenomic sequencing. These were targeted to exclude several potential confounders during comparisons of the three populations (LH n = 95, ObH n = 55, and ObT2D n = 51), notably to achieve uniformity of cardiovascular measures (see STAR Methods, Table S1). The ObH group was selected to be generally healthy (despite being obese) as reflected by the uniformity of mean fasting glucose level and blood pressure between the lean and obese non-diabetic subjects. We identified both age and gender as significantly associated with microbiome composition (adonis PERMANOVA, p < 0.001, 16S data, STAR Methods and Supplemental Information), in agreement with previous findings (Falony et al., 2016; Oksanen et al., 2015; Wang et al., 2016; Zhernakova et al., 2016). Accordingly, we corrected for age and gender where possible in the following analyses. In initial comparisons of amplicon- and metagenome-based taxonomic profiles, abundances of genera within subject agreed well across taxa (mean Spearman ρ = 0.67, Figures 1 and S1). Exceptions to this included a small number of taxa (n = 5, of 31 total) with Spearman correlations below 0.5. Four of these five bacteria belonged to the clade Clostridiales, which has been subject to extensive reclassifications, potentially compromising consistent assignments between the two measurement types’ taxonomies (Yutin and Galperin, 2013).
In subsequent analyses, six microbiome feature types were studied (additional details in STAR Methods): taxonomic abundances from 16S rRNA gene sequencing [VSEARCH (Rognes et al., 2016)]; species-level taxonomic profiles from metagenomes [MetaPhlAn2 (Truong et al., 2015)]; functional profiles [HUMAnN2 (Franzosa et al., 2018)] summarized as MetaCyc (Caspi et al., 2014) and informative Gene Ontology (iGO) pathways (STAR Methods) and as KEGG Ontology (KO) (Kanehisa et al., 2014) and Enzyme Commission (EC) (Bairoch, 2000) gene families. For each of these six feature types, only the subset of features with the greatest overall mean abundance and standard deviation were analyzed (see STAR Methods), and the dimensionality of functional features (pathways and gene families) was further reduced prior to testing by hierarchical clustering. This resulted in 39 MetaCyc pathway groups, 35 iGO term groups, 38-EC groups, and 78 KO groups used for all subsequent tests (Table S1).
Obesity Is Associated with Specific Gut Microbial Taxa and Functional Capacity, while the T2D Status within Obese Individuals Is Associated with Lower Effect Size
To identify individual microbial features (taxa and functions) associated specifically with obesity and/or with T2D, and not with other covariates (e.g., diet, medications), we assessed each feature’s abundances comparing (a) LH with ObH and (b) ObH with ObT2D using generalized linear models in MaAsLin (Multivariate Association with Linear Models; Morgan et al., 2012), with automatic variable selection using boosting as a univariate prescreen (see STAR Methods). For the first analysis (LH versus ObH), covariates were selected from age, gender, fasting glucose levels, total iron intake (estimated from FFQ and dietary supplements), and medications summarized to the antihypertensive and analgesic classes. For the second analysis (ObH versus ObT2D), covariates were selected from age, gender and BMI, analgesics, antihypertensives, metformin and insulin medications, and the nutrients magnesium and iron intake. These analyses used transformed relative abundance data and implements corrections for sparse, compositional microbial feature data. Sparse (zero-inflated) models were used for taxonomic features with more than 30% zero elements.
One of the simplest summary statistics analyzed as a microbiome feature was alpha-diversity, which was significantly reduced specifically in obesity (p = 3.20×10−11 by robust regression, ObH versus LH) and not for ObT2D (p = 0.92, ObH versus ObT2D; Figure 2). This was also the case for composition (beta-diversity) of taxonomic and functional profiles (genera, GO, EC, KO, and MetaCyc pathways; adonis q < 0.1) in obesity, and for taxonomic evaluation of dispersion (genera, betadisper q < 0.1), although not that of functional features (GO, EC, KO, and MetaCyc pathways; betadisper q > 0.1; Figure 2; Table S2). In contrast, when comparing obese subjects with and without T2D, composition was not significantly different across microbial taxonomic profiles or functional features (genera, GO, EC, KO, and MetaCyc pathways; all adonis q > 0.1) after adjusting for diabetic medications. To avoid confounding from metformin and insulin usage, we next evaluated association of compositional variation with fasting glucose levels across all subjects not using metformin or insulin (subsampling from 561 samples). Neither taxonomic profiles nor functional capacity associated significantly with fasting glucose levels (GO, EC, KO, and pathways; adonis q > 0.1). Thus in our cohort, obesity (with normal fasting glucose levels) had a striking association both with taxonomic and functional features of the microbiome, while T2D (in contrast to non-diabetic obesity) had a weak association with microbiome features once diabetic medications were properly considered (Table S2).
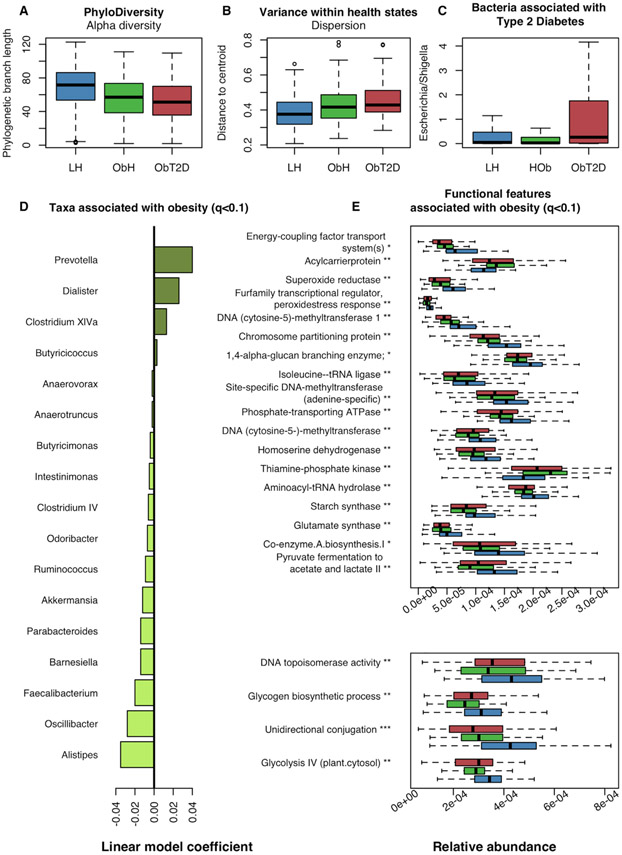
Figure 2. Functional and Microbial Deviations in Obesity and Type 2 Diabetes.
(A) Alpha-diversity (total unique phylogenetic branch length) was significantly reduced in obese subjects (compared to LH) (p = 3.20×10−11 by robust regression, 16S data) while not significantly different between obese with and without T2D (p = 0.92, ObH versus ObT2D).
(B) Microbial dispersion was significantly increased in obese subjects as compared to LH (q = 0.023), but not significant between ObH and ObT2D (q = 0.16; Table S2). The illustration was generated using all subjects with 16S data and the betadisper function in R package vegan.
(C) Analysis of individual taxa for association with ObT2D identified limited taxonomic variation in T2D (Table S3). After adjusting for insulin and metformin usage, no genera remained significantly associated after correcting for multiple testing. Escherichia possesses properties functionally relevant for T2D, and presented with a nominal increased abundance (p = 0.025) that however was not robust after multiple testing correction.
(D) Analysis of individual taxa for association with obesity identified 17 genera, including prevotella and alistipes (LH versus ObH, q < 0.1, Table S3). Associations were detected using 16S-based genera abundance profiles and MaAsLin generalized linear model (STAR Methods).
(E) Analysis of microbial processes (MetaCyc, iGO, EC, and KO) identified 22 clusters, comprising 97 features (see Table S3), associated with obesity (q < 0.1). Two plots are used for optimal visualization of all features (top and bottom). One feature from each associated cluster was selected to annotate the panel and may not be the cluster-representative feature. See Table S1 for a full overview of functional clusters. Associated processes were detected using MaAsLin-generalized linear model and metagenomic functional profiles (STAR Methods). Boxplots were made with R function boxplot with default settings (whiskers extend 1.5 times the interquartile range). ***: q < 0.01, **: 0.01 ≤ q < 0.05, *: 0.05 ≤ q < 0.1. Summary statistics and full lists of associated functional features and taxa are found in Table S3.
When analyzing the association of individual microbial genera (from 16S rRNA gene sequencing) with obesity or T2D, a total of 17 genera were significant with respect specifically to obesity (q < 0.1), including decreased Akkermansia, Faecalibacterium, Oscillibacter, and Alistipes (Table S3). The abundant anti-inflammatory species Faecalibacterium prausnitzii was decreased in obese individuals (q = 5.29×10−3) while unassociated with T2D (q > 0.1). Faecalibacterium prausnitzii, together with Bacteroides thetaiotaomicron (itself also obesity-associated), harbor most of the microbial functional features associated with obesity and may therefore be drivers of functional variation (Table S3). Of the 17 associations identified here, 15 were among the analyzed taxa in the independent SHIP cohort, and of these, 7 retained a significant association with obesity (q < 0.1) (Table S3).These obesity-associated taxa include replications of previous studies, thus lending further weight to a small but significant “core” of obesity-associated clades that are common among population contexts (in addition to more variable, population-specific associations). In contrast to obesity, no genera retained a significant robust association with T2D after incorporating the covariates above (i.e., remained differential between ObH and ObT2D individuals, q < 0.1, Figure S2; Table S3). Previous studies have found a positive association between Escherichia and both metformin usage and T2D, which we reproduced here in direction (i.e., enrichment) but without statistical significance (Pedersen et al., 2016; Qin et al., 2012; Table S3).
To understand how microbial functional capacities further relate to obesity and T2D, we first tested a group of pathways [from MetaCyc (Caspi et al., 2014) and the informative Gene Ontology, iGO (STAR Methods)] and gene families [as summarized by KO (Kanehisa et al., 2014) and ECs (Bairoch, 2000)] for association specifically with obesity (LH versus ObH). Sixteen gene family clusters and six pathway clusters, together comprising 97 functional features, were associated with obesity (q < 0.1, Table S3). This included a decreased capacity for unidirectional conjugation (Gene Ontology: GO:0009291, q = 3.79×10−4) and superoxide reductase (KEGG Orthology: K05919, q = 1.84×10−2). Conjugation is an important mechanism for bacteria, and the mechanism has been shown to play an important role for the gut microbiome (Sitaraman, 2018). Superoxide reductase catalyzes the conversion of the reactive oxygen species superoxide into the less toxic hydrogen peroxide. A reduction in this capacity indicates a microbiome-induced increase in reactive oxygen species in the intestine of obese subjects. Induction of oxygen stress by microbial dysbiosis has previously been suggested as one of the mechanisms by which the microbiome can cause weight gain and insulin resistance (Qin et al., 2012). Interestingly, while these functionally specific perturbations suggest microbial mechanisms contributing to weight gain, no pathways or gene families significantly differed in abundance between obese subjects with and without T2D after adjusting for multiple testing (q < 0.1, Table S3; Figure S2). This remained true with and without including metformin and insulin in the list of covariates. Thus, we detected little variation in either microbiome functional capacity or in the taxonomic profiles during T2D, after accounting for the separate effects of diabetic medications and of obesity itself.
Commonly Used Medications and Dietary Supplements Associate with Gut Microbiome to the same Extent as Nutritional Variables
Medication intake is increasingly being linked to microbiome composition, often to a degree as much or more than dietary components, highlighting the importance of considering these exposures in microbiome population studies (Falony et al., 2016; Forslund et al., 2015). We thus evaluated the associations of medication, dietary supplements, and dietary intake with microbiome structure in our cohort (STAR Methods, Figure S4; Table S1). Five main medication classes, comprising analgesics, antidepressants, antihypertensives, antiphlogistics, and antidiabetics, were evaluated and all five were found to be associated with both microbiome structure and functional capacity (summarized as MetaCyc pathways, using linear discriminant analysis (LDA) q < 0.1, see STAR Methods and Table 1). To ensure as equal sample sizes of users and non-users as possible in the analysis, antihypertensives were tested in ObH, while the remaining 4 medication classes were tested in ObT2D subjects (see percent users per group in Table S1). All associations with microbiome structure were supported by the additional independent cross-sectional cohort from North-Eastern Germany (SHIP; LDA q < 0.1). For the analysis of dietary supplements, usage (or not) of multivitamins; the minerals magnesium, iron, and calcium; and vitamin C was evaluated in all non-diabetic subjects. The LDA showed that overall gut microbiome taxonomic and functional profiles significantly associated (q < 0.1) with all tested dietary supplements (Table 1). Evaluation of specific taxa and microbial processes (MetaCyc, KO, EC and GO) identified very little specific variation associated with the evaluated dietary supplements and medications, potentially reflecting limited power for such stratified analyses (Table S4).
Table 1.
Association of Commonly Used Medications and Dietary Supplements with Gut Microbiome Composition
| A) Medication classes |
B) Dietary supplements |
||||
|---|---|---|---|---|---|
| Medication class |
p value |
q value |
Dietary supplement |
p value |
q value |
| Genera | Genera | ||||
| Antihypertensives | 3.85 × 10−4 | 9.13 × 10−4 (1.12 × 10−6) | Magnesium | 3.54 × 10−4 | 5.90 × 10−4 |
| Analgesics | 5.48 × 10−4 | 9.13 × 10−4 (3.12 × 10−5) | Multivitamins and mineral | 1.15 × 10−4 | 3.86 × 10−4 |
| Antidepressants | 6.88 × 10−3 | 8.60 × 10−3 (3.33 × 10−2) | Iron | 1.56 × 10−4 | 3.86 × 10−3 |
| Antiphlogistics | 1.25 × 10−2 | 1.25 × 10−2 (8.83 × 10−5) | Vitamin C | 7.68 × 10−3 | 7.68 × 10−3 |
| Antidiabetics | 6.73 × 10−5 | 3.37 × 10−4 (2.75 × 10−5) | Calcium | 1.02 × 10−3 | 1.28 × 10−3 |
| Pathways | Pathways | ||||
| Antihypertensives | 3.31 × 10−7 | 8.28 × 10−7 | Magnesium | 3.16 × 10−3 | 3.07 × 10−3 |
| Analgesics | 4.73 × 10−6 | 7.88 × 10−6 | Multivitamins and minerals | 1.01 × 10−3 | 2.15 × 10−3 |
| Antidepressants | 2.02 × 10−4 | 2.53 × 10−4 | Iron | 3.85 × 10−2 | 3.85 × 10−2 |
| Antiphlogistics | 2.55 × 10−4 | 2.55 × 10−4 | Vitamin C | 5.32 × 10−4 | 2.53 × 10−3 |
| Antidiabetics | 3.21 × 10−11 | 1.61 × 10−10 | Calcium | 2.69 × 10−2 | 3.36 × 10−2 |
Linear discriminant analysis (LDA) was performed to evaluate the ability of the gut microbiome to discriminate between users and non-users of (A) five commonly used medication classes and (B) five dietary supplements. P values and q values are given for analyses of samples in the Northern German cohorts (PopGen and FoCus) for medication classes and dietary supplements, for both bacterial abundance (genera) and functional capacity here represented by MetaCyc pathways. In brackets, q values are given for analyses of bacterial abundance in the supporting SHIP cohort (medication only). See STAR Methods for details on the LDA analyses. See also Tables S1 and S4.
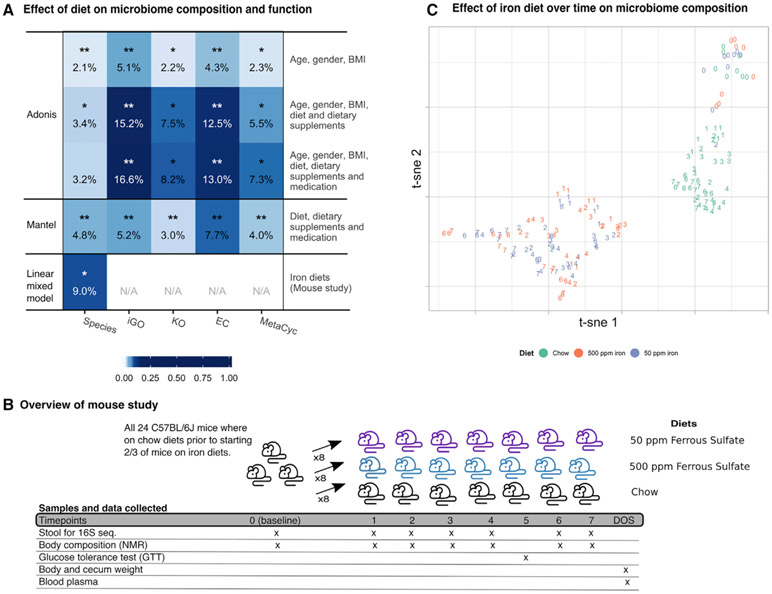
Evaluation of diet, comprising dietary supplements and nutritional variables obtained from food frequency questionaries (FFQ) data, showed a significant association between any measure of the gut microbiome, taxonomic or functional, and diet across the non-diabetic subjects (PERMANOVA, q < 0.05); however, only a small proportion of variance in the microbiome could be explained by long-term dietary factors, on the order of 1%–10% (Figure 3), in agreement with previous studies (Falony et al., 2016; Wang et al., 2016; Zhernakova et al., 2016). This is also in agreement with our own validation experiments in mice, in which iron supplementation corresponded with an 8.98% microbiome variation in effect size (see Figure 3; Table S6). Thus under this controlled setting in which mice were fed high or sufficient iron diets over seven weeks, the detectable effect of the change in a single dietary variable was substantially more pronounced. In previous work on the gut microbiome and inflammation, dietary iron intake has been shown to have a unique effect; this is often pro-inflammatory, and increased body iron storage is a risk factor for T2D (Bao et al., 2012; Jaeggi et al., 2015). However, neither total body mass nor glucose tolerance were associated with iron intake in the mice (linear mixed model q > 0.1, Table S6; Figure S3). In the more complex setting of our human population, nutrients, dietary supplements, and medication together corresponded to a similar extent with microbiome taxonomy and function, with limited variation explained by each individual variable (Figure S4). Importantly, these medication classes and dietary supplements are not unique to our cohort; nearly half of adults in similar populations receive at least one prescribed medication per month (CDC, 2017), making any effects on the microbiome of particular interest.
Figure 3. Overall Microbiome Composition and Functional Capacity Associate with Diet, Dietary Supplements, and Medication Intake.
(A) Evaluation of associations between external factors, comprising dietary nutrients, dietary supplements and medication classes, and the gut microbiome (metagenomic data), showed clear associations albeit with small effects. The percentage of variation explained and the significance of the associations between the gut microbiome (x axis) and external parameters, together with age, gender and bmi (y axis), was evaluated using adonis (top three rows). Furthermore, the correspondence between diet and medication profiles (four drug classes, dietary nutrients, and dietary supplements) and the microbiome, was evaluated using mantel (fourth row). Finally, the effect of high (500 ppm) and sufficient (50 ppm) iron diets on the microbiome composition was evaluated under controlled circumstances in a mouse study using a linear mixed model (fifth row). For adonis, variance explained was calculated using ω instead of r2 to limit overfitting, and significance was estimated using permutation of samples (see STAR Methods). Percentage variance explained was calculated for mantel analyses as mantel(r)-squared. Both mantel and adonis was performed across all non-diabetic subjects. ***q ≤ 0.001; **0.001 < q ≤ 0.01; *0.01 < q ≤ 0.05.
(B) Schematic overview of the mouse study and collected data. Three groups of eight mice were fed chow for one week (time-point (TP) 0, baseline), after which eight mice were started on an iron diet of 50 ppm ferrous sulfate and another eight mice were started on 500 ppm ferrous sulfate iron. Mice were kept on the respective diets for seven weeks during which stool was collected weekly for microbiome profiling (except week five, week of GTT). Furthermore, extensive phenotypic information was collected on a weekly basis.
(C) Non-linear dimensionality reduction of the mice stool microbiome(Rtsne function in r package Rtsne v0.15 based on OTU tables with Bray-Curtis dissimilarity and perplexity = 10) show clustering of mice on chow and the two iron diets [50 ppm and 500 ppm; (Krijthe, 2015)]. Points are colored by diet and numbered according to time-point (six time-points where mice were kept on different diets and stool collected). The microbiome of mice on different diets are clearly distinct, with the strongest separation between mice on chow versus the iron diets. For one mouse on chow, and one mouse on 50 ppm iron, the second timepoint cluster with the opposite diet group, suggesting that sample IDs for these two samples were swabbed during processing. ppm: parts per million; nmr: nuclear magnetic resonance; DOS: day of sacrifice; iGO: informative gene ontology; KO: kegg ontology; EC: enzyme commission. Figure relates to Figures S3 and S4, Tables 1 and S6.
Microbial Associations with Obesity Are Reflected in Serum Metabolite Profiles
The gut microbiome contribute to a range of metabolites that are detectable in serum, providing a direct link between variation in the gut microbiome and the profile of circulating compounds (Liu et al., 2017; Pedersen et al., 2016). Relatedly, a number of serum metabolites have been associated with the development and severity of metabolic diseases (Butte et al., 2015; Libert et al., 2018; Liu et al., 2017). We thus integrated serum metabolite profiles (comprising 390 identified metabolites from a subset of 638 curated peaks, STAR Methods) with microbiome profiles for a subset of the study participants (LH = 228, ObH = 145, and T2D = 27). Overall composition of serum metabolite profiles was significantly associated with that of the gut microbiome in non-diabetic subjects, albeit surprisingly with an extremely low effect size (Mantel test, r = 0.064, r2 = 0.41%, and p = 0.027). The limited gut microbial taxonomic variation explained by serum metabolites was less than the variation explained by environmental factors (i.e., diet and medication; r = 0.18, r2 = 3.43% in the 150 non-diabetic subjects with metagenomic data). These same environmental factors themselves also had a small direct effect on serum metabolites (r = 0.10, r2 = 1.0%, and p = 0.016), confirming significant but mainly indirect associations between serum metabolites and both the gut microbiome and dietary and medication factors.
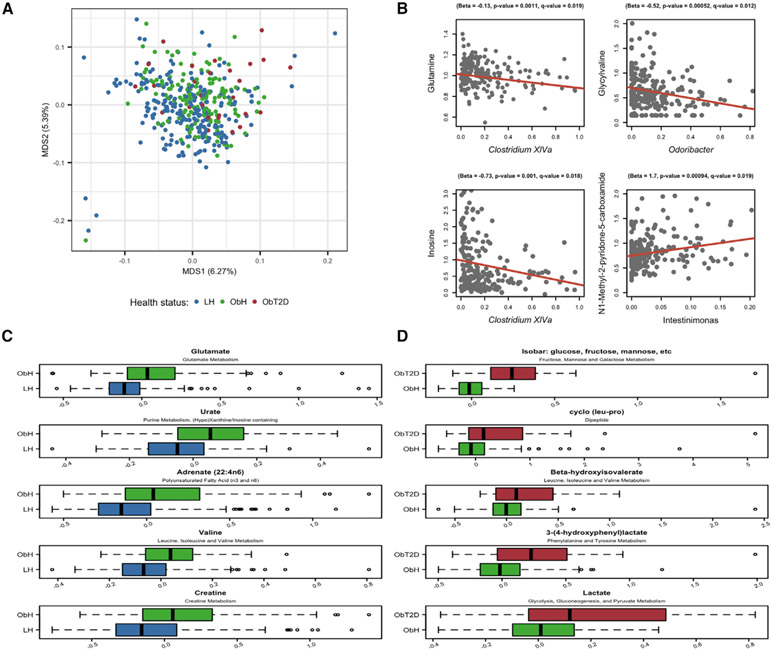
Overall, obesity and T2D were both significantly associated with overall serum metabolome composition (adonis, LH versus ObH p < 1.00 × 10−4, ObH versus ObT2D p = 1.5 ×10−3; Figure 4). Of the 390 identified metabolites, those with <50% prevalence across subjects were considered rare (n = 62) and the remaining abundant (n = 328). A total of 105 individual metabolites (of the 390 tested) were associated with obesity (100 abundant, five rare, q < 0.1; see STAR Methods, Figure 4; Table S5). Among rare metabolites (those with <50% prevalence), three specifically derived from hypertension treatments (oxypurinol, metoprolol, and hydrochlorothiazide) were significantly increased, likely due to comorbidity with obesity (Feig et al., 2008; Papademetriou et al., 2006). Among the abundant metabolites, glutamate was increased in obese individuals, of interest due to the association between free glutamate and appetite regulation ((Delgado, 2013; Ottosson et al., 2018), Table S5). The concentrations of 19 metabolites (18 abundant, one rare) were significantly different in ObT2D (in contrast to ObH subjects), where all but one were increased (Table S5). Of these metabolites, nine were specific to ObT2D. One metabolite, 3-hydroxyoctanoate, was significantly higher in ObT2D than in ObH, and significantly lower in ObH compared to LH (q < 0.1) (Table S5). 3-hydroxyoctanoate has been assigned anti-lipolytic activity, and found in fasting or diabetic ketoacidosis conditions to reach levels adequate to activate its receptor HCA3, indicating that 3-hydroxyoctanoate plays a role in regulating release of FFA from adipose tissue (Ahmed et al., 2009; Costa et al., 1998; Suzuki and Kaneko-Kawano, 2016). Further associations occurred within BCAA pathways, supporting previous observations associating them with insulin resistance ((Klein and Shearer, 2016; Newgard et al., 2009), Table S5). Intriguingly, the smaller number of specifically T2D-associated compounds corresponds with our findings for the gut microbiome, and may reflect a more restricted or specific molecular alteration overall between obese individuals with and without T2D. However, since our sample size for this comparison is limited to a group of 172 subjects, we acknowledge that power may also be more limited (see Discussion). Instead, we next focused on potential interactions between obesity-associated taxa and small molecules, as both BCAA and glutamate are recognized bacterial metabolites (Ottosson et al., 2018).
Figure 4. Serum Metabolite Profiles Associated with Obesity and Type 2 Diabetes.
(A) Ordination of 400 metabolite profiles across lean non-diabetic (LH), obese non-diabetic (ObH), and obese T2D (ObT2D) individuals, reflecting a shift in the profiles along the first ordination axis, using multidimensional scaling (MDS) based on gower’s index (capscale function in r package vegan).
(B) Correlation of four metabolites with single taxon abundance. All genera and metabolites tested for correlations were found in pre-analysis to associate with obesity. The results from MaAsLin analysis provide intercept and slope for the red line and association statistics are given over each plot. Samples are filtered as by maaslin (STAR Methods).
(C and D) A total of 105 metabolites were found to associate with obesity and 19 with T2D (linear model and chi-square test, q < 0.1, Table S5). (C) and (D) show the top five metabolites found by MaAsLin to associate with obesity (LH versus ObH) and T2D (ObH versus ObT2D), respectively. X axes show metabolite residuals after adjusting for age and gender. Boxplots were made with R function boxplot with default settings (whiskers extend 1.5 times the interquartile range). Summary statistics are in Table S5.
We first tested for direct associations of 19 medication-derived serum metabolites with gut microbial composition (as captured by Bray-Curtis dissimilarity). The levels of all three antihypertensive derivatives above were significantly associated with the microbiome among their respective users, as well as those of the analgesic metabolite salicylate (adonis q < 0.1, see STAR Methods, Table S5). Next, to assess whether individual microbes were detectably responsible for the associations during obesity (since few taxa were specifically associated with T2D), we directly performed correlation analyses for the 17 taxa and 100 abundant metabolites that were individually differential with respect to obesity (q < 0.1). We first did this, conservatively, only within the LH population using MaAsLin with metabolites as dependent variables (STAR Methods). Nineteen associations passed correction for multiple hypothesis testing (q < 0.1), including ten metabolites from the amino acid superpathway, glutamine, and four lipids. Nine of the metabolites associated with abundance of two taxa in the “anti-inflammatory” Clostridium clusters IV and XlVa (Table S5). Of interest, the association pattern within the ObH individuals differed from the pattern within lean: while the associations in the lean individuals were dominated by Clostridium, this taxon was not found among associations within the obese subjects. Instead, fifteen metabolite-microbe associations were significant within obese subjects, predominantly related to lipid metabolism (n = 8) and amino acids (n = 5) (Table S5). Together, these microbe-metabolite associations in obesity thus suggest a series of specific routes by which taxa and their associated functional roles in the gut might influence systemic signaling, nutrient uptake, and weight gain.
DISCUSSION
Ample evidence has shown the gut microbiome to be associated with obesity in humans, and studies in mice suggest that aspects of this association could be causal in its development and persistence. The gut microbiome has also been linked to insulin resistance and physiological parameters related to diabetes, but since these phenotypes are inevitably intertwined with each other and with concomitant medications in humans, the details have been difficult to establish. Here, we analyzed 1,280 gut microbiomes from a defined population in northern Germany to differentiate microbiome links to obesity, obesity-associated T2D, dietary supplements, medication usage, and serum metabolites. This design was uniquely able to determine components of the microbiome associated uniquely with T2D versus non-diabetic obesity. Comparing obese individuals with and without T2D showed only modest associations between the microbiome and T2D once medication and diet were also factored out, mostly characterized by a nominal increased abundance of Escherichia/Shigella. In contrast, comparing lean and obese individuals specifically without diabetes showed compositional differences, as well as differences in functional capacity. Finally, dietary factors, supplements, and medications were individually associated only weakly with components of the microbiome, but in aggregate accounted for a significant amount of variation; they thus have the potential to substantially affect a wide variety of additional microbiome population studies.
Human obesity has proven to be a particularly complex phenotype with respect to the microbiome (Finucane et al., 2014; Le Roy et al., 2018; Sze and Schloss, 2016). This may be due to the many factors that can influence BMI in human populations, including diet, medication, genetics, and systemic metabolism, which lead to heterogeneity of both clinical outcomes and comorbidities. This multifactorial character of obesity, combined with interpersonal variation in microbial composition and the small effect sizes of single-taxon associations, impede microbiome-obesity studies (Finucane et al., 2014; Sze and Schloss, 2016). A recent meta-analysis, for example, highlighted consistent obesity associations only at the level of whole-community alpha-diversity, and feature-specific classifiers trained on one cohort could not classify obesity in others (Sze and Schloss, 2016). In spite of this, some consistency is emerging, e.g., a decrease in members of Ruminococcaceae and Clostridia in obese subjects, across seven large cohorts including the current study (Peters et al., 2018; Le Roy et al., 2018). The same challenges hold true in associating T2D with the microbiome (Supplemental Information), and any such complex phenotype presents many of the same power, population structure, covariate, and statistical issues as do genetic association studies for complex traits (Falony et al., 2016; Zhernakova et al., 2016). This particularly affected our diabetic obese population, which numbered only 153, in contrast to a greater number of lean or obese non-diabetic subjects. The limited overlap in results among microbiome obesity population studies may both reflect factors such as insufficient power, phenotypic heterogeneity, microbial ecological variability, confounders such as medication, particular susceptibility to geographic or environmental factors (e.g., diet), as well as a broad instability of the disturbed microbiome of obese individuals.
A striking finding from this and several previous studies is the critical role of commonly used medications in modulating gut microbiome structure and function (Falony et al., 2016; Forslund et al., 2015; Imhann et al., 2016; Maier et al., 2018; Zhernakova et al., 2016). While the effects of any single non-antibiotic medication tend to be modest, the aggregate effect of several common medications per individual—and many such medications across a population—can be substantial. Similarly, while single dietary elements appears to be weakly associated with microbial variation (Falony et al., 2016; Zhernakova et al., 2016), in agreement with our study (adonis, 0.1%–0.3%, Figure S4), the combined association of overall diet with vitamin and mineral supplements with microbial variation becomes sizeable. In this space, the notable effect of dietary iron on the microbiome, and its known link with diabetes, prompted us to evaluate its effect in a controlled setting, specifically supplementary feeding in mice (Bao et al., 2012; Lee et al., 2017; Zhao et al., 2017). While direct comparison of affected taxa is not possible between specific-pathogen-free mice and humans, the results confirmed a causal effect of iron intake on the gut microbiome (effect size from linear mixed model, comparing 50 ppm to 500 ppm iron diets, 8.98%), providing support for the observed effect of long-term dietary iron in the human cohort. Thus, larger population studies enable the detection of significant, consistent associations, albeit with small effect sizes for individual compounds, as shown in the current study.
Obesity and T2D are alarming public health issues due to their rapid and continuing increase worldwide, and their etiologies are complex independently of the (also complex) gut microbiome. If host-microbiome interactions are to be targeted as an additional route of understanding these metabolic diseases, the scientific community must continue to study them in greater detail in larger, well-phenotyped cohorts and through additionally well-designed mechanistic studies. This will help to disambiguate the overlapping—but sometimes distinct—microbial effects of the two conditions, in addition to characterization of inter-individual variation in the microbiome as it relates to diet, medications, and other environmental exposures. Key microbial changes may ultimately prove to be context-specific, with distinct functional consequences depending on the host environment, genetics, and stage of temporal progression from health through obesity to T2D.
STAR★Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andre Franke (a.franke@mucosa.de). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study and Data Overview
The current study considered 1,280 individuals from the northern German cohorts PopGen (n=436; (Krawczak et al., 2006)) and FoCus (n=844; (Müller et al., 2015)). 16S rRNA amplicon sequencing data were generated for the total population, while 201 individuals were selected for shotgun metagenomic sequencing. The study further included data on medication, dietary intake and supplement usage, nutrition and a phenotypic characterization (Table S1). For analysis, samples were divided into three phenotypic groups: a) lean (BMI ≤ 25) without diabetes, (“lean healthy,” LH); b) obese (BMI >30) without diabetes (“obese healthy,” ObH); and c) obese (BMI >30) with T2D (ObT2D). To asses overall microbiome differences between the phenotypic groups, as well as association with fasting glucose levels and the role of antidiabetic medication, the PERMANOVA-like approach available through the adonis function and the betadisper function for evaluation of difference in dispersion between groups was used, both available in the R package vegan. To identify individual microbial features (taxa and processes) associated specifically with obesity and/or with obesity-associated T2D phenotypes, we assessed each feature’s abundance across (a) LH versus ObH and (b) ObH versus ObT2D using generalized linear models in MaAsLin (Multivariate Association with Linear Models) while controlling for clinical covariates (e.g. medication and age) (Morgan et al., 2012).
Further analyses were performed to assess the association of medication, nutrition and dietary supplements with features of the gut microbiome. This included evaluating if gut microbiome taxonomic and functional profiles could discriminate between users and non-users of commonly used medications and dietary supplements, including iron, magnesium, antihypertensives and antidiabetics. The evaluation was performed using LDA, and supported identification of covariates for the analysis of obesity and T2D. To further evaluate the associations between iron intake and the gut microbiome that we observed in the human cohorts, we performed a diet intervention study in 24 C57BL/6J mice. All 24 mice were fed a chow diet for 4 weeks and then separated into groups of 8 as they started on study diets; one group continued on the chow diet, another 8 mice were started on a 50 ppm ferrous sulfate diet, and the remaining 8 mice were started on a 500 ppm ferrous sulfate diet. The mice were kept on these diets for an additional seven weeks. Mouse body composition measurements and stool were collected just prior to starting the mice on the study diets and then weekly thereafter. The collected stool was subjected to 16S sequencing to further evaluate the associations between iron intake and the gut microbiome.
As the effect of a dysbiotic gut microbiome on the host may in part be reflected in serum metabolite profiles, and as many serum metabolites has previously been related to obesity and T2D, we further profiled the serum metabolome and related the profiles to health states and the associated microbial variations. Finally, 16S rRNA amplicon sequencing data for 880 individuals in a second independent cohort, SHIP, was included to evaluate the robustness of associations where data availability allowed. As combining different cohorts can be a source of confounding, all analysis of the SHIP cohort has been kept separate from the analysis of the main study population selected from the PopGen and Focus cohorts. In particular, no analyses rely on data that simultaneously spans PopGen/FoCus and SHIP; instead, primary analyses are carried out only in PopGen/FoCus, with validation tests run separately within SHIP. SHIP is an independent cohort recruited in the area of Greifswald (North-Eastern Germany), using the same protocol for sequencing however with deviations in preprocessing of samples as for PopGen/FoCus (Frost et al., 2019).
Kiel FoCus and PopGen Cohorts
Study samples were selected from the second examination cycle of the PopGen cohort (n=436) (Krawczak et al., 2006) and the cross-sectional and clinical subset of the FoCus cohort (n= 844) (Müller et al., 2015). Samples were collected as described in Wang et al. (Wang et al., 2016). Information on subjects age, sex, BMI etc. can be found in Table S1.
Samples were selected for 16S rRNA amplicon sequencing as follows. Samples with missing data for age, gender and BMI were excluded. From among the remaining samples, non-diabetic lean samples were selected by requiring individual BMI ≤ 25, no diagnosis of IBD or IBS, fasting glucose level ≤ 125 mg/dl and no self-reported physician diagnosis of diabetes. Non-diabetic obese individuals were filtered by the same filtering criteria except a BMI >30. Finally, obese individuals with T2D had no IBD or IBS diagnosis, BMI >30 and were diagnosed with T2D or has fasting glucose levels above >125 mg/dl. Medication data were curated, manually checked for spelling and consistency, and grouped into analgesics, antihypertensives, antidepressants, antibiotics, antiphlogistics, and antidiabetics (details provided in Supplemental Information).
A subset of samples from each group was selected for shotgun metagenomic sequencing as follows. To minimize potential confounding effects, individuals reporting chronic diarrhea, use of hormone replacement therapy, coronary heart disease, cardiac or venous insufficiency, or thrombotic embolism were excluded. Additional individuals were excluded if they reported a current illness, with the exception of a common cold lacking antibiotics treatment. For individuals where data on CRP was available, non-diabetic samples were selected for normal CRP levels (<5 mg/l). The resulting LH and ObH health groups had compatible fasting glucose levels, and all groups had compatible diastolic and systolic blood pressure levels with slightly lower systolic blood pressure in the LH group (Table S1).
Written, informed consent was obtained from all study participants, and all experiments were approved by the institutional ethical review committee in adherence with the Declaration of Helsinki Principles.
SHIP
Participants of SHIP (the Study of Health in Pomerania) (Völzke et al., 2011) were selected from among the 1,904 individuals with available fecal 16S rRNA amplicon sequencing data from both the SHIP-TREND and SHIP2 arm of the SHIP cohort. A subset of these samples that included medication data and the phenotypic variables age, gender, weight and height was selected for further study. Samples were grouped as obese or lean using the BMI-based definition introduced above. Samples were grouped as diabetic or not diabetic based on information from two variables in SHIP-TREND (T0) (‘nn_diab_01’ which informs whether diabetes has been diagnosed, and ‘diabetes_typ2_t0’ which informs whether T2D can be diagnosed based on the data) and one variable for SHIP (S2) (‘nn_diab_01’). We note that the variable ‘nn_diab_01’ does not distinguish between type 1 and T2D, however with the additional requirement of a BMI > 30 we anticipate the majority of individuals will have T2D and not type 1 diabetes. Three samples were removed due to antibiotic usage. These criteria resulted in the selection of 880 samples: 399 lean non-diabetic, 408 obese non-diabetic and 73 obese diabetic. All participants provided written informed consent and the study was approved by the ethics committee of the University Medicine Greifswald.
Mouse Iron-Feeding Study
All animal experiments were approved by the UCLA Animal Care and Use Committee, in accordance with PHS guidelines. The protocol number is ARC #1992–169 (Approval Period from 6/6/2016 through 11/25/2018). Twenty-four male C57BL/6J mice were obtained by the Lusis lab at UCLA from The Jackson Laboratory (Bar Harbor, ME) at 3 weeks of age. The mice were housed four per cage and fed a chow diet (LabDiet PicoLab Rodent Diet 20, cat #5R53*, St. Louis, MO) for one week. During this time, the bedding and feces were mixed together at the end of each day and redistributed among all the cages, in order to minimize variation in baseline intestinal microbial composition.
At 4 weeks of age, 2 cages of mice (8 mice in total) were continued on the chow diet, while 2 cages (8 mice total) were started on a 50 ppm ferrous sulfate diet, and 2 cages (8 mice total) were started on a 500 ppm ferrous sulfate diet. These latter two diets were AIN93M defined diets with microcrystalline cellulose as the fiber source (Dyets, cat #115838 and cat #115839, Bethlehem, PA). Mice were kept on these diets for an additional seven weeks. Mouse body composition measurements and stool were collected just prior to starting the mice on the study diets and then weekly thereafter. Body composition (total fat, muscle, and free fluid) was measured by MRI in a Bruker Minispec with software from Echo Medical Systems, Houston, TX. Total body weight (g), and percentage of fat, muscle and free fluid were calculated. For the stool collection, each mouse was placed in an individual cleaned cage with no bedding for ~5-10 minutes. Feces dropped in each cage were collected for each mouse and snap frozen. Stool and body composition measures were again collected the day before sacrifice (DBS).
After four weeks on the study diets, a glucose tolerance test (GTT) was performed. For the GTT, mice were fasted in clean cages with free access to water for six hours from 7am-1pm. The tip of a mouse’s tail was then nicked with a razor blade and the second blood drop was analyzed using an AlphaTrak2 blood glucose test strip with an AlphaTrak glucometer (Abbott laboratories). The mouse was then dosed IP with 1g glucose per kg mouse body weight with a sterile 10% solution of Beta-D(+) glucose in phosphate buffered saline (PBS). At 15, 30, 60, and 120 minutes after dosing, an additional blood sample from the nick was analyzed as described above. The GTT was performed on 3 mice per group on day 1, 3 mice per group on day 2, and 2 mice per group on day 3 after 4 weeks on the study diet). The week of the GTT, neither stool nor body composition measures were collected.
At the day of sacrifice (DOS), mice were anesthetized by inhaled isoflurane, and then body weight was measured. Blood was collected from the retroorbital plexus into lithium heparin tubes (BD Microtainer, catalog # 365971) to measure plasma insulin and glucose levels. Blood samples were centrifuged, and plasma was removed and snap frozen. The cecum was excised and weighed. Plasma insulin was measured using an ELISA insulin kit (Alpco catalog #80-INSMSU-E10). Plasma glucose was measured by an enzymatic assay (Stanbio, catalog # 1070-125).
METHOD DETAILS
Medication Variables for SHIP
Medication variables were defined based on ATC codes. The ATC codes used were: ’Analgesic’ (N02 and M01A); ’Antihypertensives’ (C02, C09 and C07); ’Antidepressant’ (N06A); ’Antibiotics’ (A07AA, G01AA, G01BA, J02AA, J04AB, L01DC, R02AB and S01AA); ’Antiphlogistics’ (A07E, S01B, V10A and L04A); ’Antidiabetics’ (A10).
Medication Variables for PopGen and FoCus
Information on medication usage was obtained for each study subject as free text. To convert this information into a data format that could be implemented in the statistical analyses, a classification tool was constructed and used to classify medication for samples with 16S rRNA gene sequencing data. The classification tool consists of scripts written in PERL and Visual Basic for Applications (VBA) in MS Excel.
Curation of free text and annotation of drug names:
In the first step, PERL scripts were used to extract and to annotate relevant data from the free text on medication usage. Stop words, abbreviations and other irrelevant words (e.g. names of pharmaceutical companies) were filtered out by several process steps. The remaining text was parsed through the German ABDA database to find drug names. Each hit was annotated by further information from ABDA and KEGG DRUGS, and resulting summaries saved in Excel sheets divided in three quality categories. The three categories were defined as a) “exact matches” given when the complete drug name was found in the free text, b) “matches” given when the main, relevant part of the drug name was found and c) “approximate matches” found by similarity search in order to correct words caused by typos in the origin text.
Assignment of drug classes:
In the next step, a VBA script was used to further classify the drug names according to a set of rules and the obtained annotations. The rules were generated in two steps. First, information from the manually classified medication data was used (medication data for samples with shotgun metagenomic data was manually classified), then the list of rules was extended by ‘training the tool’ - rules were added for unclassified drugs after each run. The rules were based on the annotations obtained from the searched databases and were designed as follows: An output field from a database was specified together with a search term e.g. “Look for the term ‘Analgesic’ in the field ‘Activity’ in the database ‘KEGG DRUGS’”. If a match was found, a result term was written to the field ‘High Level’ (HL). A similar rule, extended to include the content assigned to HL, assigned a term to the field ‘Low Level’ (LL). HL rules would assign a higher drug class such as ‘Antihypertensives’ or ‘Hormones’, while the LL rules would assign a subclass such as ‘Diuretics’ or ‘Thyroid hormones’.
Manual evaluation of classification tool:
Each drug was assigned a HL and LL term using each rule resulting in multiple result rows per drug. Identical rows were automatically concatenated into a single row, while drugs assigned deviating results was left for the subsequent manual evaluation. Output from the automated classification tool was manually evaluated by looking through rows for errors and unassigned drugs which were then assigned a HL and LL term by searching the databases manually. Based on this tool all drugs listed in the medication data which belonged to the drug classes of interest were identified, and each sample was assigned ‘1’ for ‘using’ if one or more of the drugs were reported for that sample, and ‘0’ if no match was found.
16S and Shotgun Metagenomic Sequencing
Fecal samples were subjected to 16S rRNA amplicon sequencing as described in Wang et al. (Wang et al., 2016) for FoCus and PopGen. In summary, bacterial genomic DNA was extracted using the QIAamp DNA Stool Mini Kit from QIAGEN and the V1-V2 region of 16S rRNA gene was sequenced on the MiSeq platform with MiSeq Reagent Kits V3. Stringent demultiplexing was carried out by allowing no mismatches in either index sequence. For a subset of 201 individuals, the same DNA extracts were subjected to shotgun metagenomic sequencing as described in Wang et al. (Wang et al., 2016). Briefly, these samples were prepared following the Illumina Nextera DNA Library Preparation Kit and sequenced on the HiSeq Platform as 2×125 bp paired-end reads. Preprocessing of SHIP samples was performed as described in (Frost et al., 2019) and with the same protocol for 16S rRNA amplicon sequencing as for the FoCus/PopGen samples.
16S rRNA Gene Data Processing
Raw 16S rRNA amplicon reads for the Focus and PopGen samples were end-trimmed using sickle in PE mode with a sliding window of 0.1 × read length to maintain average quality ≥20. Reads <100 nts after trimming were discarded. Paired reads were then stitched using VSEARCH (Rognes et al., 2016) with minimum length 280 and maximum length 350bp. VSEARCH was further used to filter reads with more than 1 expected error and FastX-Toolkit::fastq_quality_filter was used to filter reads with more than 5% of nucleotides with a quality score below 30. Remaining reads were converted to FASTA format and subjected to chimera filtering using VSEARCH and the gold.fa database. Remaining reads were classified using the UTAX algorithm with RDP database and reads classified as either chloroplast or not classified at domain level were removed.
OTU tables were generated using the UPARSE algorithm (Edgar, 2013) implemented in VSEARCH software and all reads across samples. For reads with multiple exact sequence copies (replicates) only one copy was kept, and read only occurring once across all samples (singletons) were removed, as they were considered likely to be sequencing artefacts. The remaining reads were then clustered at 97% similarity. OTU representative sequences were again checked for chimeras using VSEARCH in de-novo mode. All reads per sample were then mapped to representative sequences using VSEARCH to generate OTU abundance tables. Representative sequences were additionally taxonomically annotated using the RDP database and the RDP classifier (Wang et al., 2007) to the lowest possible level with minimum 80% bootstrap confidence. OTUs with identical taxonomic annotations were then grouped into taxonomic bins. Samples with fewer than 10,000 reads were removed. Overgrowth of oxygen-tolerant Proteobacteria was quality controlled by excluding samples which fell above the third quartile plus three times interquartile range (IQR) of phylum abundance (n=26).
Raw 16S rRNA amplicon reads for the SHIP cohort were processed as described above except that SINTAX (Edgar, 2016) was used for annotating sequences, and samples were rarefied to 10,000 reads per sample.
Shotgun Metagenomic Processing
Metagenomic sequencing data were quality controlled by requiring a minimum of 4 M paired-end reads per sample after Nextera library adaptor removal (Trimmomatic (Bolger et al., 2014)), trimming of low quality ends (Sickle (Joshi and Fass, 2011)), and removal of host-reads (DeconSeq (Schmieder and Edwards, 2011)). Quality control methods were run with default parameters. Metagenomic samples were taxonomically profiled using MetaPhlAn2 (Segata et al., 2012) and functionally profiled using HUMAnN2 v0.11.1 (Franzosa et al., 2018), both with default settings plus the addition of ‘–bt2_ps very-sensitive’ for the ‘—metaphlan-options’ provided to HUMAnN2. Species from taxonomic profiles were retained for further analysis if their mean relative abundance exceeded 0.005 (0.5%) across the dataset with a minimum abundance of 0.05 (5%) in at least one sample and non-zero abundance in at least 60% of samples.
Tables of pathway and gene family abundance obtained using HUMAnN2 were normalized to relative abundance using ’humann2_renorm_table –units relab’ including unmapped and unintegrated (i.e. not assigned to a pathway) read mass. Samples potentially affected by oxygen exposure were removed (identified as described below).
Pathway abundance files were joined into one abundance table and filtered to contain the pathways with top 50% mean abundance and top 50% variance (final n=112). To reduce redundant testing, pathways were clustered using Ward hierarchical clustering (R function hclust in package stats) with dissimilarity 1-Spearman correlation coefficient. Pathway clusters were defined at height 0.6 using the R function cutree in package stats and a representative pathway was selected for each cluster (defined as the pathway with the median mean abundance, Table S1).
Gene family files were regrouped using the humann2_regroup_table command with ’uniref90_level4ec’ (Enzyme Commission), ’uniref90_infogo1000’ (informative Gene Ontology, iGO) and ’uniref90_ko’ (KEGG Orthology) terms. iGO is computed specifically for HUMAnN2 as informative subsets of GO build on UniProt’s annotations and the structure of the GO hierarchy (Ashburner et al., 2000). Regrouping was performed in order to join gene families with similar function or properties, and thereby reduce both the number of low-abundance variables and the number of tests to be performed downstream. Regrouped, per-sample abundance profiles were joined into three abundance tables and filtered to contain features with top 25% mean abundance and top 25% variance. This filtering returned 453 KO terms, 110 iGO terms and 132 EC terms. For KO, the terms with ‘subunit.ribosomal.protein’ were removed before filtering in order to further reduce the multiple testing burden. Features from these tables were then clusters (and representative features selected) following the procedures outlined above for metabolic pathways, resulting in 38 EC, 35 iGO and 78 KO clusters (Table S1).
Nutrition Data
Dietary intake during the year preceding examination was assessed in participants of the FoCus and PopGen cohorts via a validated, self-administered food frequency questionnaire (FFQ) (Nöthlings et al., 2007). Nutrient intakes (e.g. dietary protein content) were obtained using the German Food Code and Nutrient Database (vII.3) and provided by the Department of Epidemiology of the German Institute of Human Nutrition Potsdam-Rehbrücke (Dehne et al., 1999). Vitamin and mineral intake from the dietary supplement data was incorporated in downstream analyses in one of two ways: Alone as categorical variable (e.g. in the LDA analyses), and as continuous variables of “total intake” obtained by combining dietary supplement data with the nutritional data derived from FFQ records of intake of food groups (e.g. as covariates in the MaAsLin analyses). As there exists some uncertainty as to the reproducibility of some nutrient groups when assessed with FFQs, and as many nutritional variables correlate, a subset of higher level variables were selected for inclusion in the downstream analyses (Martinez et al., 2013; Nagel et al., 2007). Selected variables with summary statistics are shown in Table S1.
The nutritional variables for mineral and vitamin intake analyzed for association with the microbiome using MaAsLin and adonis included intake originating from diet and supplements. Data on supplement usage was recorded as binary data, which we recoded to amount per day by estimating the typical amount of each compound in commonly used supplementary products in Germany. The supplements evaluated were multivitamin/mineral supplements, and individual supplements including folic acid, iron, zinc, magnesium, calcium, selenium, vitamin B6, vitamin B7 (biotin), vitamin B complex (containing all 8 B vitamins), vitamin E, carotene, vitamin A and vitamin C. For each type of supplement, we selected five products which are sold over the counter for adults and not aimed for treating any specific disease (except vitamin or mineral deficiency). We assumed users took the recommended amount, included all reported sources of each component (e.g. if a subject used both vitamin C tablets and multivitamin we included vitamin C from both sources) and added the median milligram to the amount obtained from nutrition. This resulted in the total (T) vitamin/mineral variables used through the study.
Serum Metabolites
Non-targeted metabolomics profiling analysis was performed for n=855 samples from the control subset of the PopGen. Metabolites in serum samples were extracted with 475 μL methanol and centrifugated. The supernatant was split into 4 aliquots of 100 μL, keeping two as reserve. The other two aliquots were used for LC-MS/MS analysis in positive and negative electrospray ionization mode. LC-MS/MS analysis was performed on a linear ion trap LTQ XL mass spectrometer (Thermo Fisher Scientific, Germany) coupled with a Waters Acquity UPLC system (Waters, Germany). Two different columns (2.1 x 100 mm Waters BEH C18, 1.7 μm particle-size) each optimized for the respective positive or negative electrospray ionization, were used. Metabolites were identified by comparing the recorded LC-MS/MS spectra with spectra found in Metabolon’s proprietary spectra library (Metabolon, USA). Further details can be found in Koch et al. (Koch et al., 2017). Of the 855 samples, 400 overlapped with the 1,280 study individuals (LH=228, ObH=145, T2D=27). For each metabolite (totaling 638 in the subset of 400 subjects), the levels measured were divided by the median value of the samples’ run day to account for technical day-to-day variations, and missing values were imputed with the minimum detected value.
The ComBat function in R package sva v3.30.0 was applied to correct remaining batch variation, with settings par.prior=F and mean.only = T (Leek et al., 2012). Prior to ComBat, metabolites with zero variance were removed. Among the 390 identified metabolites (based on included standards and manual curation), low-prevalence ("rare") metabolites were separated for handling by appropriate models, specifically those with >50% zero values in non-imputed data, of which a subset of 62 were identified. The remaining 328 identified were considered "abundant".
Microbiome Data from Mouse Iron-Feeding Study
Fecal pellets were transported from UCLA, USA, to Kiel, Germany, frozen on dry ice, processed and 16S sequenced as described for the human samples. The raw 16S data was quality controlled and processed as for the human data to obtain OUT and taxonomic abundance tables for statistical analyses. Following data processing and filtering, 167 samples remained (out of the initial 168).
QUANTIFICATION AND STATISTICAL ANALYSIS
Correcting for Multiple Hypothesis Testing
For all analyses described below and throughout the manuscript, nominal p-values are corrected for multiple hypothesis testing to false discovery rates (q-values) using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995).
Multivariate Linear Modeling Using MaAsLin
For most associations between microbiome and health outcomes we used a modified general linear model as implemented by MaAsLin (Multivariate microbial Association with Linear Models, see for details (Morgan et al., 2012)), which combines an arcsine-square root transformed analysis of relative abundances in a standard multivariable linear model with outlier removal, variable selection by boosting, and (for sparse features) zero inflation. Here, we required a minimum relative abundance of 10−10 for taxa and functional features (dMinAbd), and outliers removed >3 inter-quartile ranges past the 1st/3rd quartiles (dOutlierFence).
The full list of covariates to select from in models contrasting healthy and obese subjects (not T2D) were age, gender, fasting glucose levels, total iron intake, antihypertensive and analgesic medications. Only the covariate of health status was enforced. The full list of covariates to select from for models contrasting obese with and without T2D (not lean) were age; gender; BMI; the medications antihypertensives, analgesics, metformin and insulin; and the dietary variables total magnesium and total iron intake. Only the covariates of health status and BMI were enforced. However, to evaluate the effect of metformin/insulin on individual taxa, two models were applied to the taxonomic features that deviate from the above on the specification of metformin/insulin as follows: One without metformin/insulin covariates and one enforcing metformin while keeping insulin and metformin among the full list of covariates. Taxonomic features from 16S data were included as summed at the genus through phylum levels, and with features with >30% zero values were modeled as zero-inflated. The functional features assessed were pathway and gene family group representatives as defined above (using the default model). The specified models were further used to analyze the association between health states and the facultative-to-obligate anaerobe ratio, as well as targeted butyrate metabolizing pathways.
To evaluate the genera associated with obesity in the independent SHIP cohort, MaAsLin was run with same settings across all non-diabetic samples, with the covariates age, gender, antihypertensives and antidepressants (antihypertensives and antidepressants were the most common medications across samples at 49% and 30% users, respectively).
Further, MaAsLin models were used for the analysis of dietary supplement and medication intake. Non-diabetic samples were used except for analyses of antidiabetic medications (insulin and metformin), where only diabetic samples were used. Analyses of medication usage included the covariates age, gender and BMI. For the PopGen and FoCus samples (where dietary supplement data was available) the additional covariates total iron and total magnesium were included. Only the covariates medication and BMI were enforced. Analyses of supplement usage included the covariates age, gender, BMI, analgesic and antihypertensives (used by 22% and 30% of individuals, respectively), enforcing BMI and the evaluated supplement. Finally, MaAsLin was used to analyze associations of microbial taxa and functional processes with age and gender (enforcing those two variables in their respective analyses, as well as health state). Non-diabetic subjects were included and settings were as described above, with covariates selected as in the analysis of obesity.
Variance Explained by Diet and Medication
The function adonis in the R package vegan (Oksanen et al., 2015) was used to calculate the overall variation in microbiome composition and functional capacity that could be explained by both the combined variables, and the variation in microbiome composition that could be explained by any single nutritional variable. Analyses were performed for all non-diabetic subjects. The full list of nutritional variables (n=38), including daily intake from the dietary supplements, is listed in Table S1. The variables diet fibers, proteins, fats, carbohydrates and minerals, were excluded from the analysis of joined variance explained as they are summary measures of the lower-level nutritional variables included in the model. The adjusted coefficient of determination (ω2) was used as a measure of overall variance explained instead of the coefficient of determination (R2) to avoid overestimation (Kelly et al., 2015). The coefficient of determination (R2) was used when evaluating single nutritional variables. We used a permutated null distribution (n=999 samples) of overall variance explained (ω2) based on reshuffling of sample labels to estimate statistical significance. For calculation of the variance explained by each individual nutritional variable, each variable was analyzed in separate models while adjusting for BMI, age, gender and total energy intake.
Selection of Nutritional Covariates
The inclusion of iron intake in the analysis of both obesity and T2D, and of magnesium intake in analysis of T2D, was based on the difference in number of users and non-users between the health state groups (Table S1), these minerals’ overall effects as shown by LDA, and prior knowledge linking them to the respective health states (see main text and Supplemental information). The limited adjustment for nutrient and supplement covariates in the models described above were decided in part based on the analysis of variance: the overall variance explained by dietary intake, including supplements, was estimated at ~1–10% for taxonomic and functional abundance (Figure 3). While these results indicated a slightly greater potential for nutritional data to explain variation in functional capacity (as compared to bacterial abundance), we found that the limited explanatory power argued against inclusion of more dietary variables in the analysis of T2D. As diet is part of the obesity phenotype we also limited dietary covariates in this analysis.
Beta Diversity Association Analysis
To evaluate whether overall bacterial composition or functional capacity associated with obesity (LH versus ObH), BMI, fasting glucose levels or T2D (ObH versus ObT2D), the R function adonis in R package vegan was used with default settings except perm=999, and 16S data at genera level or shotgun metagenomic gene abundance data regrouped to iGO and KO terms, or MetaCyc and GO pathways. To ensure compatible power of the analysis of each trait, data were subsampled to compatible sample sizes (n=100 for metagenomic data, and n=300 for 16S data), and the analyses repeated 50 times to ensure robustness (using the sample function in R with replacement). Furthermore, to ensure compatibility of results, the number of considered covariates were kept at six for all adonis analyses except where diabetic medication was not included: Analysis of obesity and BMI included covariates age, gender antihypertensives, analgesics, total iron intake and fasting glucose levels. Analysis of fasting glucose levels included age, gender, antihypertensives, analgesics, total iron intake and BMI. Analysis of T2D included age, gender, BMI and total iron intake, with or without metformin and insulin as specified with results. Analysis of fasting glucose levels excluded subjects using T2D medication. For the analysis of age and gender, the LH subjects were included, and the analyses were adjusted for covariates as in the MaAsLin-based analysis of obesity. Finally, the function betadisper from the R package vegan was used with default settings to evaluate dispersion between groups of i) LH and ObH and ii) ObH and ObT2D subjects, subsampling and repetitions were used as described for adonis, and the anova function was used to estimate significance.
Alpha-Diversity Association Analysis
Phylogenetic diversity (total unique phylogenetic branch length) was used as the measure of alpha-diversity, and calculated using the phylogenetic tree built on the aligned OTU sequences using FastTree v2 (Price et al., 2010) in nucleotide mode (−nt) and applying a generally time-reversible model (−gtr). The resulting 16S OTU table and phylogenetic tree were used as input in mothur’s phylo.diversity function (Schloss et al., 2009). The significance level of the difference in alpha-diversity between groups was estimated using robust regression (function lmRob in R package robust v0.4 (Wang et al., 2017)) adjusting for the covariates listed for MaAsLin.
Medication Association with Microbiome
The importance of medication consumption for bacterial composition (16S data) was evaluated for analgesic, antidepressant, antiphlogistic and anti-diabetic medications in obese T2D samples and for antihypertensives in non-diabetic obese samples. This selection was based on percent medication users within groups and the group with closest to 50% medication users were selected. Individuals were grouped as users or non-users of the target medication classes, and linear discriminant analysis (LDA) was performed as follows; MDS ordination axis were calculated for the relative abundance of genera (16S, excluding genera with zero abundance in all selected samples) using capscale (R package vegan) with Bray-Curtis dissimilarity, and LDA was performed with the first 10 ordination axis, using the MASS package in R. The ordination axes were used instead of the microbial variables directly in order to avoid over-fitting. The obtained linear discriminant function was compared between medication users and non-users with Wilcoxon rank sum test. The same analysis approach was applied to pathway abundance data.
To support the findings in PopGen and FoCus that the gut microbiome could significantly discriminate between users and non-users of five commonly used medication classes, we repeated the LDA with the SHIP data. Analysis of antidiabetics was performed with ObT2D, of antidepressants with ObH and the remaining 3 medication classes (analgesics, antiphlogistics, antihypertensives) were evaluated with data from LH individuals.
Dietary Intake Association with Microbiome
We used LDA as described for medication usage above to evaluate the deviation in microbiome composition between supplement users and non-users of the selected supplements across non-diabetic subjects. We considered age to be the most likely confounding factor in this group of non-diabetic subjects as based on prior findings (see above) and prior publications of microbiome studies. We therefore first considered the distribution of supplement usage across ages and, by visual inspection, found no patterns to prompt the inclusion of age as a covariate.
Serum Metabolites’ Association Analysis
For analyses where covariates could not be specified (Mantel, Betadisper), metabolite residuals adjusted for age and gender were used. Residuals were calculated for each metabolite using a linear regression. For all linear models each metabolite was scaled by dividing by the metabolite standard deviation, and the covariates age and gender were included.
Differences in metabolic composition between health states were evaluated using adonis with default settings except method=”gower” and perm=104, with the covariates age, gender, fasting glucose levels, antihypertensives and analgesics for analysis of obesity, and the covariates age, gender, BMI, metformin, antihypertensives and analgesics for analysis of T2D. Similarly, dispersion between health states was evaluated using betadisper and significance calculated using permutest, both from R package vegan. Correspondence between the metabolite profile and gut microbiome composition (genera) or environment (nutrition, dietary supplements and medication) was evaluated using mantel.rtest in R package ade4 v1.7 (Chessel et al., 2004) across all non-diabetic subjects with metabolite data. Gower distances was used for metabolites and environmental variables to accommodate the mixed data.
The identified and abundant metabolites (n=328) were selected for analysis of single metabolite differences between health states using a linear model in MaAsLin with covariates as above (for adonis), enforcing only the health state and with no data transformation. The remaining 62 metabolites that were identified but with low abundance across the 400 subjects were evaluated for association with obesity or T2D using a chi-squared test (chisq.test function in R package stats) on the scaled, but non-imputed non-adjusted metabolites after conversion to presence/absence data. To obtain information on the direction of the association, e.g. increase or decrease in obese, we applied the Wilcoxon rank sum test in R package stats was used with conf.int=T and the estimate multiplied with −1.
The genera that associated with obesity (q<0.1) were selected and evaluated for association with the 100 abundant metabolites found to associate with obesity. For these, we first tested for an association across all LH samples and then across all obese non-diabetic samples (ObH). The analysis was performed using MaAsLin with covariates age, gender, antihypertensives and analgesics, enforcing only the taxa being evaluated.
Among the 28 metabolites assigned as medication metabolites, 19 metabolites were likely to originate from one of the five medication classes evaluated above. For these 19 metabolites, a generalized linear model with binomial distribution (glm function in R package stats) was applied across all subjects to detect metabolite-to-medication associations. Furthermore, adonis was applied to detect associations between the gut microbiome (genera) and medication metabolite levels (including medication users only and adjusting for health state).
Mouse Features Association with Iron Intake
To evaluate association between overall microbiome composition and iron intake, the microbiome of mice fed 50 ppm and 500 ppm ferrous sulfate were compared. The analysis included data from all time points were mice where on different diets, and was performed using a linear mixed model (function lmer in R package lme4 v1.1 (Bates et al., 2015), with the lmerTest v3.0.1 (Kuznetsova et al., 2017) package to estimate significance and r.squaredGLMM function from R package MuMln (Bartoń, 2018) to extract variance explained by fixed effect terms). Dissimilarities were calculated for the OTU table using function vegdist with method=“bray” in R package vegan. The model included diet (same or different) as fixed effect and intercepts for subjects and cage as random effects to consider dependence between time-points and cage effects, respectively. The same design, however including mice fed chow for the duration of the study, was used to evaluate the dissimilarity between mice on chow versus the respective iron diets.
Comparisons of body composition, glucose tolerance test results, and plasma glucose and insulin levels were performed using the linear mixed model as described above. The area under the glucose curve (AUC) for each mouse was calculated using the formula: AUC = 0.25 (fasting value) + 0.5 (30 minute value) + 0.75 (60 minute value) + 0.5 (120 minute value) per the Schonfeld1 project protocol (https://phenome.jax.org/projects/Schonfeld1/protocol).
R and Additional R-Packages Used
R environment v3.5, RStudio v1.1, grid v3.5 (R Core Team, 2018), gridExtra v2.3 (Baptiste and Antonov, 2017), gridBase v0.4 (Murrell, 2014), gridGraphics v0.3 (Murrell, 2015), reshape2 v1.4 (Wickham, 2007), metaphor v.2.0 (Viechtbauer, 2010), MASS 7.3 (Kafadar et al., 2002), data.table v1.11 (Dowle and Srinivasan, 2019), ggrepel v0.8 (Slowikowski, 2018), plyr v1.8 (Wickham, 2011), dplyr 0,7 (Wickham et al., 2019), stringr v1.3 (Wickham, 2015), caret v6.0, RColorBrewer v1.1 (Neuwirth, 2014), lattice v0.20 (Sarkar, 2008), graphics v3.5 (Murrell, 2018).
DATA AND CODE AVAILABILITY
All data related to the Kiel cohorts (PopGen and FoCus) is available upon application from the PopGen biobank (http://www.uksh.de/p2n/Information+for+Researchers.html). All data related to the Study of Health in Pomerania (SHIP) was obtained from the SHIP data management unit and can be applied for online (https://www.fvcm.med.uni-greifswald.de/dd_service/data_use_intro.php). Data related to the mouse study is available at the NCBI Sequence Read Archive (SRA). The accession number for the mouse data reported in this paper is BioProject: PRJNA550303.
Supplementary Material
KEY RESOURCES TABLE
Highlights.
Obesity, but not type 2 diabetes, associated with gut microbiome variation
Medications and dietary supplements associated with gut microbiome variation
High iron intake affected microbiome composition in mice
Microbiome variation was also reflected in serum metabolite profiles
ACKNOWLEDGMENTS
This study was supported by the German Ministry of Education and Research (BMBF) program e:Med sysINFLAME (http://www.gesundheitsforschung-bmbf.de/de/5111.php, no.: 01ZX1306A) and received infrastructure support from the Deutsche Forschungsgemeinschaft (DFG) Cluster of Excellence “Inflammation at Interfaces” (http://www.inflammation-at-interfaces.de, no: EXC306 and EXC306/2) and by National Institutes of Health NIDDK grant R24DK110499 to C.H. We thank the PopGen/P2N biobank in Kiel for infrastructure support. M.K. is a recipient of a Postdoctoral Research Fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft, KO 5187/1-1). The mouse study was funded by: BKF: Ruth L. Kirschstein National Research Service Award T32HL69766, AJL: NIH (HL28481) and Nih R01HL144651, and CDV: NIH (GM083198-01A1). We would also like to thank Ms. Ilona Urbach, Ms. Ines Wulf, and Mr. Tonio Hauptmann from the IKMB microbiome laboratory and Sarada Charugundla at UCLA, for excellent technical support. SHIP is part of the Community Medicine Research net of the University Medicine Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg-West Pomerania.
Footnotes
DECLARATION OF INTERESTS
L.B.T. is an employee and shareholder of BiomCare. C.H. is a scientific advisor for Seres Therapeutics, microbiome Insights, and ZOE.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2019.07.004.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Lazo de la Vega-Monroy et al., 2013; Singer and Geohas, 2006.
REFERENCES
- Ahmed K, Tunaru S, Langhans CD, Hanson J, Michalski CW, Kölker S, Jones PM, Okun JG, and Offermanns S (2009). Deorphanization of GPR109B as a receptor for the β-oxidation intermediate 3-OH-octanoic acid and its role in the regulation of lipolysis. J. Biol. Chem 284, 21928–21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, Verger EO, Hedjazi L, Bouillot JL, Chevallier JM, et al. (2019). Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut 68, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A (2000). The ENZYME database in 2000. Nucleic Acids Res. 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Rong Y, Rong S, and Liu L (2012). Dietary iron intake, body iron stores, and the risk of type 2 diabetes: a systematic review and meta-analysis. BMC Med. 10, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptiste A, and Antonov A (2017). gridExtra: miscellaneous functions for ‘“Grid”’graphics. R Packag. Version [Google Scholar]
- Bartoń K (2018). Package “MuMIn”: multi-model inference, Cran-R. [Google Scholar]
- Bates D, Mächler M, Bolker B, and Walker S (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw 67. [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc 57, 289–300. [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangé CL, Neves AL, Chilloux J, Nicholson JK, and Dumas ME (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte NF, Liu Y, Zakeri IF, Mohney RP, Mehta N, Voruganti VS, Göring H, Cole SA, and Comuzzie AG (2015). Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am. J. Clin. Nutr 102, 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42, D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2017). Therapeutic drug use. [Google Scholar]
- Chessel D, Dufour A-B, and Thioulouse J (2004). The ade4 package-I-one-table methods. Res. News [Google Scholar]
- Chobot A, Górowska-Kowolik K, Sokołowska M, and Jarosz-Chobot P (2018). Obesity and diabetes-not only a simple link between two epidemics. Diabetes Metab. Res. Rev 34, e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa CG, Dorland L, Holwerda U, De Almeida IT, Poll-The BT, Jakobs C, and Duran M (1998). Simultaneous analysis of plasma free fatty acids and their 3-hydroxy analogs in fatty acid β-oxidation disorders. Clin. Chem 44, 463–471. [PubMed] [Google Scholar]
- Daousi C, Casson IF, Gill GV, MacFarlane IA, Wilding JPHH, and Pinkney JH (2006). Prevalence of obesity in type 2 diabetes in secondary care: association with cardiovascular risk factors. Postgrad. Med. J 82, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehne LI, Klemm C, Henseler G, and Hermann-Kunz E (1999). The German food code and nutrient data base (BLS II.2). Eur. J. Epidemiol 15, 355–359. [DOI] [PubMed] [Google Scholar]
- Delgado TC (2013). Glutamate and GABA in appetite regulation. Front. Endocrinol. (Lausanne) 4, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowle M, and Srinivasan A (2019). Data.table: extension of ‘data.frame’. [Google Scholar]
- Edgar R (2016). SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv. [Google Scholar]
- Edgar RC (2013). Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. [DOI] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. (2016). Population-level analysis of gut microbiome variation. Science 352, 560–564. [DOI] [PubMed] [Google Scholar]
- Feig DI, Soletsky B, and Johnson RJ (2008). Effect of allopurinol on blood pressure of adolescents with newly diagnosed essential hypertension: A randomized trial. JAMA 300, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane MM, Sharpton TJ, Laurent TJ, and Pollard KS (2014). A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One 9, e84689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-silva S, Gudmundsdottir V, Pedersen HK, et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, et al. (2018). Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost F, Kacprowski T, Rühlemann M, Bülow R, Kühn JP, Franke A, Heinsen FA, Pietzner M, Nauck M, Völker U, et al. (2019). Impaired Exocrine Pancreatic Function Associates With changes in intestinal microbiota composition and diversity. Gastroenterology 156, 1010–1015. [DOI] [PubMed] [Google Scholar]
- Imhann F, Bonder MJ, Vich Vila AV, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJM, et al. (2016). Proton pump inhibitors affect the gut microbiome. Gut 65, 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeggi T, Kortman GAM, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Timmerman HM, Swinkels DW, Tjalsma H, et al. (2015). Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 64, 731–742. [DOI] [PubMed] [Google Scholar]
- Joshi NA, and Fass JN (2011). Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files, 1.33 version. [Google Scholar]
- Kafadar K, Koehler JR, Venables WN, and Ripley BD (2002). Modern applied statistics with S, Fourth edition. [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, and Bäckhed F (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103. [DOI] [PubMed] [Google Scholar]
- Kelly BJ, Gross R, Bittinger K, Sherrill-Mix S, Lewis JD, Collman RG, Bushman FD, and Li H (2015). Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 31, 2461–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MS, and Shearer J (2016). Metabolomics and Type 2 diabetes: translating basic research into clinical application. J. Diabetes Res 2016, 3898502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Freitag-Wolf S, Schlesinger S, Borggrefe J, Hov JR, Jensen MK, Pick J, Markus MRP, Höpfner T, Jacobs G, et al. (2017). Serum metabolomic profiling highlights pathways associated with liver fat content in a general population sample. Eur. J. Clin. Nutr 71, 995–1001. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Nikolaus S, Von Eberstein H, Croucher PJP, El Mokhtari NE, and Schreiber S (2006). PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- Krijthe JH (2015). T-distributed stochastic neighbor embedding using Barnes-hut [Google Scholar]
- Kuznetsova A, Brockhoff PB, and Christensen RHB (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Softw 82. [Google Scholar]
- Lazo de la Vega-Monroy ML, Larrieta E, German MS, Baez-Saldana A, and Fernandez-Mejia C (2013). Effects of biotin supplementation in the diet on insulin secretion, islet gene expression, glucose homeostasis and beta-cell proportion. J. Nutr. Biochem 24, 169–177. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. [DOI] [PubMed] [Google Scholar]
- Le Roy CI, Beaumont M, Jackson MA, Steves CJ, Spector TD, and Bell JT (2018). Heritable components of the human fecal microbiome are associated with visceral fat. Gut Microbes 9, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Clavel T, Smirnov K, Schmidt A, Lagkouvardos I, Walker A, Lucio M, Michalke B, Schmitt-Kopplin P, Fedorak R, et al. (2017). Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut 66, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, and Storey JD (2012). The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert DM, Nowacki AS, and Natowicz MR (2018). Metabolomic analysis of obesity, metabolic syndrome, and type 2 diabetes: amino acid and acylcarnitine levels change along a spectrum of metabolic wellness. PeerJ 6, e5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med 23, 859–868. [DOI] [PubMed] [Google Scholar]
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MF, Philippi ST, Estima C, and Leal G (2013). Validity and reproducibility of a food frequency questionnaire to assess food group intake in adolescents. Cad. Saude Publica 29, 1795–1804. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N, Schulte DM, Türk K, Freitag-Wolf S, Hampe J, Zeuner R, Schröder JO, Gouni-Berthold I, Berthold HK, Krone W, et al. (2015). IL-6 blockade by monoclonal antibodies inhibits apolipoprotein (a) expression and lipoprotein (a) synthesis in humans. J. Lipid Res 56, 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell P (2014). gridBase: integration of base and grid graphics. [Google Scholar]
- Murrell P (2015). gridGraphics: redraw base graphics using “grid” graphics. R packag, 0.1.5 version. [Google Scholar]
- Murrell P (2018). R graphics, Second edition. [Google Scholar]
- Nagel G, Zoller D, Ruf T, Rohrmann S, and Linseisen J (2007). Long-term reproducibility of a food-frequency questionnaire and dietary changes in the European prospective investigation into cancer and nutrition (EPIC)-Heidelberg cohort. Br. J. Nutr 98, 194–200. [DOI] [PubMed] [Google Scholar]
- Narayan KMV, Boyle JP, Thompson TJ, Gregg EW, and Williamson DF (2007). Effect of BMI on lifetime risk for diabetes in the U.S. Diabetes Care 30, 1562–1566. [DOI] [PubMed] [Google Scholar]
- Neuwirth E (2014). ColorBrewer palettes. Packag. RColorBrewer. [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthlings U, Hoffmann K, Bergmann MM, and Boeing H (2007). Fitting portion sizes in a self-administered food frequency questionnaire. J. Nutr 137, 2781–2786. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Guillaume, Blanchet F, Michael F, Roeland K, Pierre L, Dan M, Minchin PR, O’Hara RB, Simpson GL, et al. (2015). Vegan: community ecology package. [Google Scholar]
- Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, Melander O, and Orho-Melander M (2018). Connection between BMI related plasma metabolite profile and gut microbiota. J. Clin. Endocrinol. Metab 103, 1491–1501. [DOI] [PubMed] [Google Scholar]
- Papademetriou V, Hainer JW, Sugg J, and Munzer D; ATTACH Study Group (2006). Factorial antihypertensive study of an extended-release metoprolol and hydrochlorothiazide combination. Am. J. Hypertens 19, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, et al. (2016). Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381. [DOI] [PubMed] [Google Scholar]
- Peters BA, Shapiro JA, Church TR, Miller G, Trinh-Shevrin C, Yuen E, Friedlander C, Hayes RB, and Ahn J (2018). A taxonomic signature of obesity in a large study of American adults. Sci. Rep 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li SS, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. [DOI] [PubMed] [Google Scholar]
- R Core Team (2018). R: a language and environment for statistical computing (R Foundation for Statistical Computing). [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, and Mahé F (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D (2008). Lattice: multivariate data visualization with R (Springer), Use R! [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, and Edwards R (2011). Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6, e17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, and Huttenhower C (2012). Metagenomic microbial community profiling using unique clade- specific marker genes. Nat. Methods 9, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GM, and Geohas J (2006). The effect of chromium picolinate and biotin supplementation on glycemic control in poorly controlled patients with type 2 diabetes mellitus: a placebo-controlled, double-blinded, randomized trial. Diabetes Technol. Ther 8, 636–643. [DOI] [PubMed] [Google Scholar]
- Sitaraman R (2018). Prokaryotic horizontal gene transfer within the human holobiont: ecological-evolutionary inferences, implications and possibilities. Microbiome 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowikowski K (2018). Ggrepel: automatically position non-overlapping text labels with “ggplot2.”. [Google Scholar]
- Suzuki K, and Kaneko-Kawano T (2016). Biological roles and therapeutic potential of G protein-coupled receptors for free fatty acids and metabolic intermediates. J. Phys. Fit. Sports Med 5, 213–227. [Google Scholar]
- Sze MA, and Schloss PD (2016). Looking for a signal in the noise: revisiting obesity and the microbiome. MBio 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trøseid M, Nestvold TK, Rudi K, Thoresen H, Nielsen EW, and Lappegård KT (2013). Plasma lipopolysaccharide is closely associated with glycemic control and abdominal obesity: evidence from bariatric surgery. Diabetes Care 36, 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, and Segata N (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, and Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. [DOI] [PubMed] [Google Scholar]
- Viechtbauer W (2010). Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw 36. [Google Scholar]
- Völzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, Aumann N, Lau K, Piontek M, Born G, et al. (2011). Cohort profile: the study of health in Pomerania. Int. J. Epidemiol 40, 294–307. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7. [DOI] [PubMed] [Google Scholar]
- Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen FA, Rühlemann MC, Szymczak S, et al. (2016). Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet 48, 1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zamar R, Marazzi A, Yohai V, Salibian-Barrera M, Ricardo M, Zivot E, Rocke D, Martin D, Maechler M, et al. (2017). Robust: Port of the S+ “Robust Library.”. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, and Cole JR (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2007). Reshaping data with the reshape package. J. Stat. Softw 21. [Google Scholar]
- Wickham H (2011). The split-apply-combine strategy for data analysis. J. Stat. Softw 40. [Google Scholar]
- Wickham H (2015). Stringr: simple, consistent wrappers for common string operations. R Packag. Version [Google Scholar]
- Wickham H, Francois R, Henry L, and Müller K (2019). Package ‘dplyr’. A grammar of data manipulation. R Packag, 0.8.0.1 version [Google Scholar]
- Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, Lee K, Franzosa EA, Morgan XC, Gevers D, et al. (2016). Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, and Galperin MY (2013). A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol 15, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Lian J, Tian J, Shen Y, Ping Z, Fang X, Min J, and Wang F (2017). Dietary intake of heme iron and body iron status are associated with the risk of gestational diabetes mellitus: a systematic review and meta-analysis. Asia Pac. J. Clin. Nutr 26, 1092–1106. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. (2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data related to the Kiel cohorts (PopGen and FoCus) is available upon application from the PopGen biobank (http://www.uksh.de/p2n/Information+for+Researchers.html). All data related to the Study of Health in Pomerania (SHIP) was obtained from the SHIP data management unit and can be applied for online (https://www.fvcm.med.uni-greifswald.de/dd_service/data_use_intro.php). Data related to the mouse study is available at the NCBI Sequence Read Archive (SRA). The accession number for the mouse data reported in this paper is BioProject: PRJNA550303.