Abstract
Here we investigated the consequences of PCR amplification errors in the identification of intraindividual mtDNA variation. The bumblebee Bombus morio was chosen as model for the COI gene amplification tests with two DNA polymerases (Taq and Q5) presenting different error rates. The amplifications using Taq resulted in a significant increase of singleton haplotypes per individual in comparison to Q5. The sequence characteristics indicated that Taq resulted haplotypes are mostly due to amplification errors. Studies focusing on intraindividual variability should address special attention to the DNA polymerase fidelity to avoid overestimation of heteroplasmic haplotypes.
Keywords: Taq DNA polymerase, Q5 DNA polymerase, heteroplasmy, NUMTs
Introduction
Taq DNA polymerase (Taq-pol) is the most common polymerase used in polymerase chain reaction (PCR) (Yamagami et al. 2014). This enzyme was isolated first from the thermophilic bacterium Thermus aquaticus (Taq) (Saiki et al. 1988) and is thermostable. This characteristic was decisive for the automation of PCR (Reiss et al. 1990), besides the increase in specificity and efficiency of the reaction (Saiki et al. 1988). Over time, other thermostable DNA polymerases have been isolated from other microorganisms (e.g. Pfu and Vent®) or developed from protein engineering (e.g. Phusion® and Q5®). Some of these enzymes have 3′-5′ proofreading exonuclease activity, absent in Taq-pol, which promotes the checking and removing mismatched nucleotides during the polymerization (Kunkel 1992; Kunkel and Bebenek 2000) and, therefore, are considered as high-fidelity DNA polymerases.
The fidelity of a DNA polymerase is usually expressed by the mean error rate per base per duplication (Keohavong and Thilly 1989). Taq-pol has an error rate between 2 × 10−4 (Saiki et al. 1988; Keohavong and Thilly 1989) and 2 × 10−5 (Eckert and Kunkel 1990; McInerney et al. 2014). Proofreading activity of the high-fidelity polymerases drops error rates to 10−6 or even lower (Cline 1996; Li et al. 2006; McInerney et al. 2014). Despite the higher error rate of Taq-pol compared to high-fidelity polymerases, the frequency of each error in the population of amplified molecules is still considered low (Lin et al. 2002). However, these errors may hinder the analysis of coamplification of different sequences (Pascual et al. 1994; Bracho et al. 1998), as in studies of heteroplasmy.
In a previous study, heteroplasmy was unveiled for the bumblebee Bombus morio (Hymenoptera, Apidae, Bombini) (Françoso et al. 2016). Thus B. morio was our model species to evaluate the degree of DNA polymerases error effects in the identification of heteroplasmic haplotypes. Here, we tested two DNA polymerases (Taq-pol and Q5), each presenting distinct error rates, for the amplification of a mitochondrial fragment encompassing the cytochrome C oxidase subunit I (COI) gene.
Material and methods
Samples and DNA extraction
Six individuals of B. morio (1BM, 2BM, 3BM, 4BM, 5BM, and 6BM), from four Brazilian locations, were used for DNA extraction following the protocol described by Françoso et al. (2015). Samples were obtained from the cryogenic collection (−80 °C) of the Laboratório de Genética e Evolução de Abelhas from Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil (see Supplementary file 1).
Amplification
A 676 bp fragment of COI gene was amplified with a Taq-pol and a high-fidelity polymerase for each individual in a Mastercycler pro (Eppendorf, Germany), using the primers BarbeeF (Françoso and Arias 2013) and mtD9 (Simon et al. 1994).
Amplifications with Taq DNA polymerase
The amplifications were set up with 4.0 μl of DNA template in 20 μl final volume containing 1X PCR buffer, 0.4 μM each primer, 0.2 mM each dNTP, 1.5 mM MgCl2, and 1.5 U of Platinum® Taq DNA Polymerase (Thermo Fisher Scientific, Waltham, MA). Reactions consisted of an initial denaturation at 94 °C for 2 min followed by 35 cycles at 94 °C for 45 s, 48 °C for 45 s, and 64 °C for 50 s, and a final extension at 64 °C for 5 min. The error rate of Platinum Taq is 2.28 × 10−5 according to the manufacturer.
Amplifications with high-fidelity DNA polymerase
Amplifications were run in a 25 µl volume containing 12.5 µl of Q5® High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA), 0.8 µM of each primer and 4.0 µl of DNA template. PCR reactions were conducted according to Q5 manufacturer’s recommendations, and included an initial denaturation at 98 °C for 30 s followed by 35 cycles at 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 2 min. The error rate of Q5 polymerase is 5.3 × 10−7 (Potapov and Ong 2017).
Cloning and sequencing
PCR products from each polymerase amplification were cloned in the pGEM plasmid vector (Promega, Madison, WI) and used to transform competent Escherichia coli DH5-α cells. For each B. morio sample and DNA polymerase tested, around 48 colonies were picked, added to a 20 μl of TE buffer (0.5X) and boiled at 99 °C for 10 min. The cloned inserts were amplified using the pUC/M13 primers Forward (17mer) (−40) (5′-GTTTTCCCAGTCACGAC-3′) and Reverse (17mer) (5′-CAGGAAACAGCTATGAC-3′), following the conditions previously described for Taq-pol. PCR products were purified with the ExoProStar (GE Healthcare Life Sciences, Little Chalfont, UK) and sequenced at the Macrogen sequencing service (Macrogen Inc., Seoul, South Korea).
Quantification of intraindividual haplotypes
Sequences were aligned using the algorithm MUSCLE (Edgar 2004) in GENEIOUS 9.1.6 software (Kearse et al. 2012). The number of haplotypes per individual was quantified using PEGAS 0.9 package (Paradis 2010) in the R 3.3.1 (R Core Team 2016). Haplotype networks were constructed in the POPART 1.7 program (Leigh and Bryant 2015), using the median-joining algorithm (Bandelt et al. 1999).
Frequency of errors and characterization of base substitutions
The frequency of PCR products with an amplification error was estimated using the following equation adapted from Smith and Modrich (1997):
| (1) |
where f is the expected frequency of molecules exhibiting an error, l the size of the amplified fragment (in bp), n the number of cycles used in the amplification, and a the polymerase error rate. These estimates were used to calculate the number of expected sequences with an amplification error, as follows:
| (2) |
were es is the number of expected sequences with an amplification error, f the expected frequency of molecules exhibiting an error, and N the number of sequenced clones. These estimates were compared with the number of intraindividual haplotypes to test whether the observed results can be explained by amplification errors. These comparisons were performed using Student’s t-test implemented in the R 3.1.1.
The number of synonymous and non-synonymous substitutions between intraindividual haplotypes was calculated by DNAsp 5.10.01 (Librado and Rozas 2009). Transitions and transversions were also quantified to verify if the amount of substitutions is related to the mutational spectrum of the polymerases.
Results and discussion
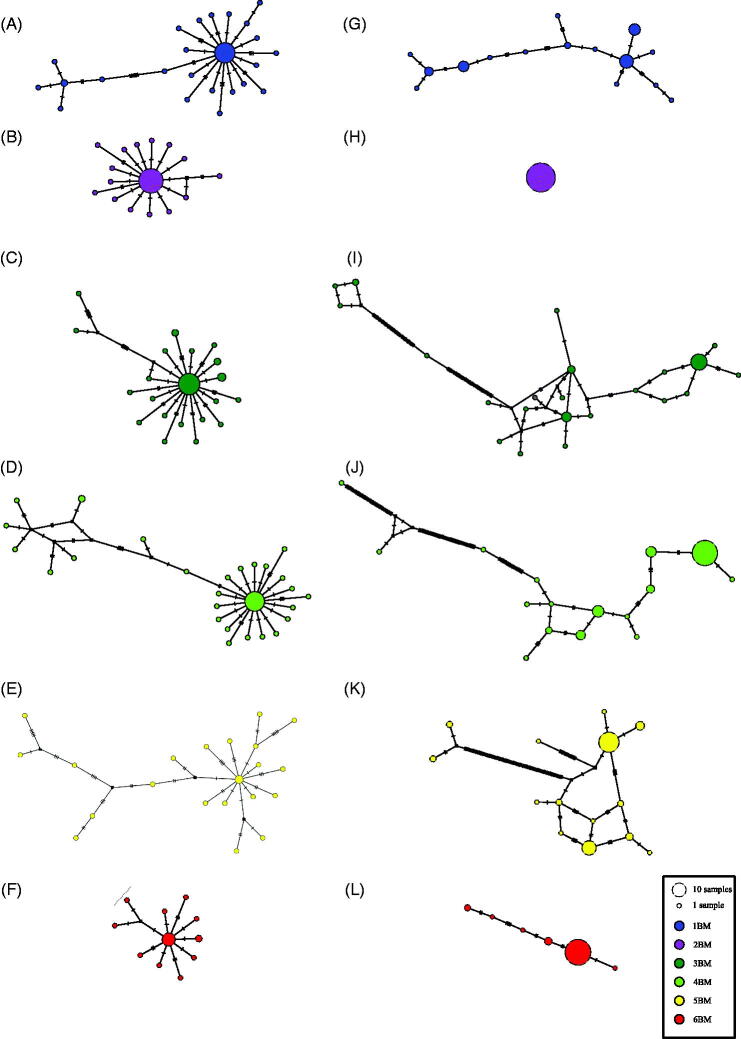
The two polymerases tested recovered several COI haplotypes per individual (GenBank accession: MK994547-MK994748). However, the results indicate that most of the singletons (haplotypes represented by a single intraindividual sequence) obtained after Taq-pol probably are due to amplification errors. First, 90% of intraindividual haplotypes are singletons (Table 1), and most present only a single base substitution in relation to the most frequent intraindividual haplotype, leading to a star-like topology network (Figure 1(A–F)). Second, no significant difference was verified between the number of intraindividual haplotypes and the expected number of sequences with amplification errors (Table 1). This statistical significance remained when only singletons were considered. And third, most of the observed substitutions among singleton were A→G/T→C transitions (61.4%) (Supplementary file 2). Previous studies have reported that about 57 to 66% of the errors generated by Taq-pol are A→G/T→C transitions (Dunning et al. 1988; Ennis et al. 1990; Bracho et al. 1998; Potapov and Ong 2017), values similar to those observed in this work. Besides that, the number of non-synonymous substitutions was also higher among PCR products amplified with Taq-pol (mean: 18 Taq, 3 Q5). In addition, indels were observed in several sequences (Table 1). Usually the substitutions verified in heteroplasmic sequences are synonymous and occur in the third base of the codon. Amplification errors, in turn, are randomly distributed in the sequence, regardless the codon position (Bracho et al. 1998). Indels may also indicate amplification errors, especially in coding regions due to reading frameshift, leading to nonfunctional gene products, normally.
Table 1.
Individual identification and respective number of sequenced clones (N seq), haplotypes (h), singletons (s), type of substitutions (synonymous [syn] and non-synonymous [nsyn]) and indels recovered after DNA amplification using Taq and Q5. Singletons frequencies in relation to the total haplotype number (%), and the number of expected sequences considering the amplification error (es) are presented.
|
Taq DNA polymerase |
High-fidelity polymerase (Q5) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | N seq | h | s | es | syn | nsyn | indels | N seq | h | s | es | syn | nsyn | indels |
| 1BM | 43 | 24 | 22 (92%) | 23 | 19 | 17 | 39 | 15 | 10 (67%) | <1 | 12 | 5 | ||
| 2BM | 42 | 19 | 18 (95%) | 23 | 9 | 10 | 5 | 44 | 1 | 0 (0%) | <1 | |||
| 3BM | 45 | 21 | 17 (81%) | 24 | 17 | 26 | 2 | 42 | 23 | 19 (83%) | <1 | 12a | 6a | 1a |
| 4BM | 45 | 30 | 28 (93%) | 24 | 17 | 24 | 4 | 59 | 17 | 11 (65%) | <1 | 8a | 4a | 1a |
| 5BM | 23 | 21 | 20 (95%) | 12 | 21 | 20 | 2 | 54 | 14 | 6 (43%) | <1 | 9a | 2a | 0a |
| 6BM | 19 | 11 | 9 (82%) | 10 | 5 | 9 | 1 | 43 | 6 | 3 (62%) | <1 | 9 | 0 | |
| Total/Mean | 217/36 | 126/21 | 114/19 | 117/19 | 88/15 | 106/18 | 14/2 | 281/46 | 76/12 | 49/8 | 4/ | 50/8 | 17/3 | 2/0 |
aSequences treated as NUMTs were excluded to avoid bias, since they presented high number of base substitutions.
Figure 1.
The six intraindividual (see aside legend) haplotype networks obtained from COI sequences cloned of Bombus morio (GenBank accession: MK994547-MK994748). COI amplifications were performed using two different polymerases: Platinum Taq DNA Polymerase (A–F) and Q5 DNA polymerase (G–L). Circles represent intraindividual haplotypes, and the size is proportional to their frequency. Crossbars indicate number of nucleotide substitution. Black squares represent a missing intermediate haplotype.
Conversely, the clones sequencing data from high-fidelity polymerase Q5 presented fewer singletons in comparison to Taq-pol. The difference between the singletons frequency obtained with Taq-pol and with Q5 (90 and 53%, respectively) was significant (p-value = 0.00384). Also, the star-like pattern was not observed in the Q5 haplotype networks (Figure 1(G–L)). Strikingly, the individual 2BM showed only a single haplotype (Figure 1(H)). The singletons obtained with the Q5 usually presented more than one substitution relative to the most frequent intraindividual haplotype, but the genetic distances typically did not exceed 2%. Considering the Q5 error rate, the frequency of singletons for most samples was higher than expected (Table 1). Moreover, most of the singletons presented several substitutions in relation to the most frequent haplotypes. Thus, it is unlikely that the singletons are due to amplification errors solely. They may represent real mitochondrial haplotypes. It is noteworthy that some singletons from individual 3BM, 4BM and 5BM presented a high number of base substitutions (Figure 1(I–K)), and the intraindividual genetic distances ranging from 2.6 to 11.8%. In addition, these singletons showed many non-synonymous substitutions relative to the other sequences. It is quite likely that these singletons represent NUMTs, copies of mitochondrial regions present in the nuclear genome (Richly and Leister 2004), since the differences in relation to the other sequences cannot be explained solely by amplification errors.
Finally, the number of singletons presented a narrow and inverse relation to the fidelity of the polymerase, especially the ones linked to the most frequent haplotype by just one nucleotide substitution. Therefore, these singletons probably result from amplification errors and may lead to misinterpretation concerning mtDNA variation (false positives). Singletons with many substitutions probably represent intraindividual haplotypes, since the accumulation of many errors in the single sequence is unlikely, even using low fidelity polymerases. Thus, in order to reduce false positives and the chances of removing real intraindividual haplotypes we suggest (1) the use of a high-fidelity enzyme, which can greatly reduce the number of singletons, and (2) the removal of singletons, but only those linked by a single substitution to the frequent haplotypes.
Supplementary Material
Acknowledgments
The authors acknowledge Susy Coelho for lab maintenance, Alexandre R. Zuntini and Natália S. Araujo for discussion.
Funding Statement
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES - Finance Code 001); Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Proc. 2013/12530-4 and 2016/24669-5); and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, research sponsorship to MCA, Proc. 306932/2016-4).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Bracho MA, Moya A, Barrio E. 1998. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J Gen Virol. 79(12):2921–2928. [DOI] [PubMed] [Google Scholar]
- Cline J. 1996. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24(18):3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Talmud P, Humphries SE. 1988. Errors in the polymerase chain reaction. Nucl Acids Res. 16(21):10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert KA, Kunkel TA. 1990. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucl Acids Res. 18(13):3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis HL, Zemmour J, Salter RD, Parham P. 1990. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. Proc Natl Acad Sci. 87(7):2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Françoso E, Arias MC. 2013. Cytochrome c oxidase I primers for corbiculate bees: DNA barcode and mini-barcode. Mol Ecol Resour. 13(5):844–850. [DOI] [PubMed] [Google Scholar]
- Françoso E, Gomes F, Arias MC. 2015. A protocol for isolating insect mitochondrial genomes: a case study of NUMT in Melipona flavolineata (Hymenoptera: Apidae). Mitochondrial DNA. 27:1–4. [DOI] [PubMed] [Google Scholar]
- Françoso E, Zuntini AR, Carnaval AC, Arias MC. 2016. Comparative phylogeography in the Atlantic forest and Brazilian savannas: Pleistocene fluctuations and dispersal shape spatial patterns in two bumblebees. BMC Evol Biol. 16(1):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P, Thilly WG. 1989. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci. 86(23):9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. 1992. DNA replication fidelity. J Biol Chem. 267(26):18251–18254. [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. 2000. DNA replication fidelity. Annu Rev Biochem. 69(1):497–529. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. 2015. Popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116. [Google Scholar]
- Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. 2006. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 3(2):95–97. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. 2002. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 11(2):133–145. [DOI] [PubMed] [Google Scholar]
- McInerney P, Adams P, Hadi MZ. 2014. Error rate comparison during Polymerase Chain Reaction by DNA polymerase. Mol Biol Int. 2014:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 26(3):419–420. [DOI] [PubMed] [Google Scholar]
- Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. 1994. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 180(1):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V, Ong JL. 2017. Examining sources of error in PCR by single-molecule sequencing. PLoS One. 12(1):e0169774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Reiss J, Krawczak M, Schloesser M, Wagner M, Cooper DN. 1990. The effect of replication errors on the mismatch analysis of PCR-amplified DNA. Nucl Acids Res. 18(4):973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. 2004. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 21(6):1081–1084. [DOI] [PubMed] [Google Scholar]
- Saiki R, Gelfand D, Stoffel S, Scharf S, Higuchi R, Horn G, Mullis K, Erlich H. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 239(4839):487–491. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 87(6):651–701. [Google Scholar]
- Smith J, Modrich P. 1997. Removal of polymerase-produced mutant sequences from PCR products. Proc Natl Acad Sci. 94(13):6847–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami T, Ishino S, Kawarabayasi Y, Ishino Y. 2014. Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering. Front Microbiol. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.