Abstract
The complete chloroplast genome sequence of Xerophyta spekei Baker was reported in this study. The complete chloroplast genome showed a stereotypical quadripartite structure as observed in other angiosperms with a length of 155,235 bp and divided into four parts; a pair of IRs (27,109 bp) which is separated by a small single copy (SSC) region (17,388 bp) and a large single copy (LSC) region (83,629bp). The chloroplast genome had 132 genes, including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Seven protein-coding genes were identified to have RNA editing.
Keywords: Xerophyta spekei, Velloziaceae, chloroplast, genome sequence, phylogeny
Xerophyta spekei Baker is a much branched shrub that grows 2–5 m tall in rocky outcrops at elevations from 300–1900 m (Beentje 1994; Pócs and Luke 2007). It is distributed in Kenya, Tanzania, Zambia, Zimbabwe, and possibly also in Ethiopia (Beentje 1994; Behnke et al. 2013). Xerophyta spp. with the exception of Xerophyta elegans are known to be poikilochlorophyllous, i.e. they lose their chlorophyll during desiccation (Mello-Silva et al. 2011; Behnke et al. 2013; Farrant et al. 2015). They are better adapted to xeric environments with specific substrates for their growth and development hence enhancing their endemism (Behnke et al. 2013; Farrant et al. 2015). Xerophyta spekei is a useful traditional medicinal plant whereby the leaf is used in case of stiffness of neck or other body parts; a piece of cloth is put on the aching part, and the area is rubbed with the warmed leaf and also the ashes are used to treat burns and diabetes (Kareru, Kenji, and Gachanja 2007; Kisangau and Herrmann 2007). Additionally, the stem is pounded and made into very strong brooms and paintbrushes and unspecified plant parts are used for cleaning metal pans and utensils (Beentje 1994).
Leaf samples were collected from Kibwezi, Chyulu National Park (Chyulu Base II) (02″44′18.94S, 037″56′41.04E), Kenya. The sample includes fresh and young photosynthetic leaves of X. spekei (SAJIT-006336). The collected samples were kept in silica gel and stored at −80 °C in Wuhan Botanical Garden (HIB) until chloroplast DNA extraction. The total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle 1991) and sequenced using the Illumina platform at Novogene Company (Beijing, China). After filtering the low-quality data and adaptors, the obtained clean data were assembled by GetOrganelle-1.6.2 (Jin et al. 2018), and then manually corrected. Finally, we used the geneious to find the IR region and annotated using PGA. The complete chloroplast genome of Xerophyta spekei showed four-part annular structures similar to most land plants. The length of the Complete Cp genome of X. spekei was 155,235 bp with a quadripartite structure and contained a pair of IRs (27,109 bp) which is separated by a small single copy (SSC) region (17,388 bp) and a large single copy (LSC) region (83,629 bp). The X. spekei Cp genome had 132 genes, including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC content of Xerophyta spekei was 37.6%, with LSC, SSC and IR regions having 35.5, 31.8, and 42.5%, respectively.
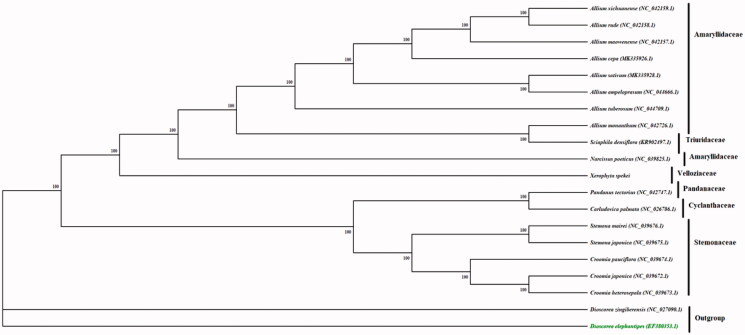
Phylogenetic analysis was performed using whole chloroplast genome for the families; Cyclanthaceae, Pandanaceae, Stemonaceae, Triuridaceae found in the order pandanales; with one species from family Dioscoreaceae used as an out-group based on a previous study (Mennes et al. 2013), and an additional species from a closely related family Amaryllidaceae. All the nine taxa were aligned by MAFFT and the phylogenetic relationships were reconstructed by means of maximum-likelihood (ML) performed by IQ-Tree that is integrated in Phylosuite (Zhang et al. 2018) a GUI-based software written in Python 3.6.7. The phylogenetic tree was divided into two groups with X. spekei showing a closer relationship to species in the families’ Triuridaceae and Amaryllidaceae (Figure 1).
Figure 1.
Phylogenetic tree based on maximum parsimony analysis of X. spekei with related species. The numbers above the branches are the bootstrap statistics values from 1000 replications.
Funding Statement
This work was supported by grants from the Backbone Talents Project of Wuhan Botanical Garden, CAS (Y655301M01),Sino-Africa Joint Research Center, CAS (SAJC201614) and the National Natural Science Foundation of China (31970211).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Beentje H. 1994. Kenya trees, shrubs, and Lianas. 1st ed. Nairobi, Kenya: National Museums of Kenya. [Google Scholar]
- Behnke HD, Hummel E, Hillmer S, Sauer-Gürth H, Gonzalez J, Wink M. 2013. A revision of African Velloziaceae based on leaf anatomy characters and RbcL nucleotide sequences. Bot J Linn Soc. 172(1):22–94. [Google Scholar]
- Doyle J. 1991. DNA protocals for plants. Molecular techniques in taxonomy. NATO ASI Series. 57: 283–293. [Google Scholar]
- Farrant JM, Cooper K, Hilgart A, Abdalla KO, Bentley J, Thomson JA, Dace HJW, Peton N, Mundree SG, Rafudeen MS. 2015. A molecular physiological review of vegetative desiccation tolerance in the resurrection plant Xerophyta viscosa (Baker). Planta. 242(2):407–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, Yu W-B, Yang J-B, Song Y, Yi T-S, Li D-Z. 2018. GetOrganelle: a simple and fast pipeline for de Novo Assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. :256479; doi: 10.1101/256479. [DOI] [Google Scholar]
- Kareru PG, Kenji GM, Gachanja A. 2007. Traditional medicines among the Embu and Mbeere peoples of Kenya[J]. African Journal of Traditional Complementary & Alternative Medicines, 4(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisangau DP, Herrmann TM. 2007. Utilization and conservation of medicinal plants used for primary health care in Makueni District, Kenya. IJBSM. 3(3):184–192. [Google Scholar]
- Mello-Silva R, Santos DYAC, Salatino MLF, Motta LB, Cattai MB, Sasaki D, Lovo J, Pita PB, Rocini C, Rodrigues CDN, et al. 2011. Five vicarious genera from Gondwana: the Velloziaceae as shown by molecules and morphology. Ann Bot. 108(1):87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes CB, Smets EF, Moses SN, Merckx VSFT. 2013. New insights in the long-debated evolutionary history of Triuridaceae (Pandanales). Mol Phylogenet Evol. 69(3):994–1004. [DOI] [PubMed] [Google Scholar]
- Pócs T, Luke Q. 2007. East African Bryophytes, Xxv: bryological records from the Chyulu Range, Kenya. J East Afr Nat Hist. 96(1):27–46. [Google Scholar]
- Zhang D, Gao F, Wen XL, Jakovlić I, Zou H, Zhang J, Gui TW. 2018. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. BioRxiv. :489088; doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]