Abstract
Ochetellus glaber (Mayr, 1867) is a dolichoderine ant found in the warm regions of Asia and Australia. We have determined the mitochondrial genome of O. glaber whose length is 16,259 bp including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large control region. The base composition was AT-biased (GC ratio is 17.8%). Gene order of O. glaber is identical to other species of the subfamily Dolichoderinae. Phylogenetic trees show that O. glaber is nested in other mitochondrial genomes of tribe Leptomyrmecini, implying the neotropical genera are ancestral to Australian genera such as Ochetellus.
Keywords: Ochetellus glaber, mitochondrial genome, Dolichoderinae, Formicidae, Korea
Ochetellus glaber, also known as the black household ant, is a minute ant found in the tropics and the subtropics of Asia and Australia (Janicki et al. 2016). It was first considered as an invasive species from Australia which spread out through human interactions. Further investigations on their morphologies, however, proved that O. glaber is a species complex indicating that the ants are, in fact, multiple species native to each region (Hoffmann et al. 2011). Still, boundaries between species are unclear and require additional in-depth investigations. As a first step in understanding the genetic background of this species, we successfully determined its complete mitochondrial genome.
Total DNA of O. glaber workers was extracted from the samples collected in Gageodo, Jeollanom-do, Republic of Korea (34°03′04.0″N; 125°07′48.0″E), using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeqX at Macrogen Inc., Korea, were filtered by Trimmomatic 0.33 (Bolger et al. 2014) and de novo assembled and confirmed by Velvet 1.2.10 (Zerbino and Birney 2008), SOAPGapCloser 1.12 (Zhao Q-Y et al. 2011), BWA 0.7.17 (Li et al. 2009), and SAMtools 1.9 (Li 2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on the other ant mitogenomes. ARWEN (Laslett and Canbäck 2008) was used to annotate tRNAs. DNA sample and specimen (95% ethanol) are deposited in the InfoBoss Cyber Herbarium (IN; J. Park, KFDS00124).
The mitochondrial genome of O. glaber (Genbank accession is MN044390) is 16,259 bp long and its GC ratio is 17.8%, which is the longest and most AT-biased Dolichoderine mitochondrial genome (Linepithema humile; 16,098 bp (Zhao E et al. 2017) and Dolichoderus sibiricus; 18.2% (Park et al. 2019)). It contains 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNAs, and a AT-rich control region. Gene order of O. glaber is identical to those of all other Dolichoderinae and closely related Pseudomyrmecinae (Vieira and Prosdocimi 2019). All PCGs start with typical ATN codon and end with a TAA or T– – except ND4L which stops with codon TAG.
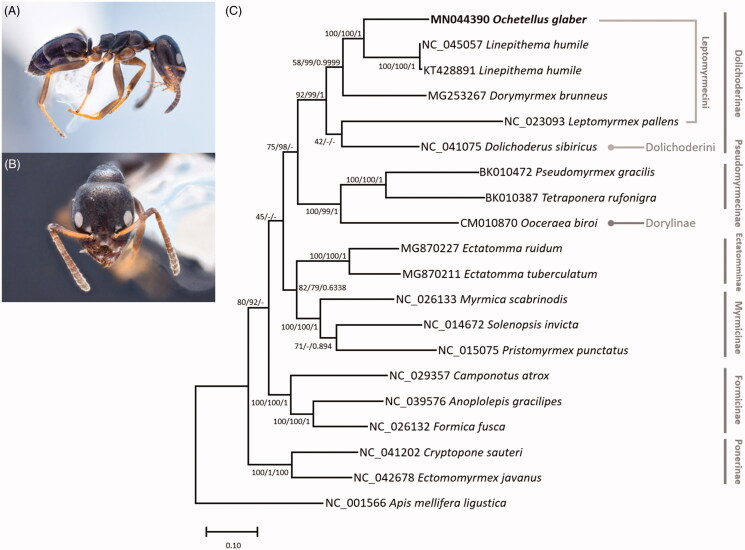
Thirteen PCGs and two rRNA genes from 19 ants mitogenomes including all available Dolichoderine species and an outgroup species, Apis mellifera ligustica, were aligned by MAFFT 7.450 (Katoh and Standley 2013) each. Bootstrapped maximum likelihood (bootstrap repeat is 1,000), neighbor joining (bootstrap repeat is 10,000), and bayesian inference (1,100,000 generations) trees were constructed using MEGA X (Kumar et al. 2018) and Mr. Bayes 3.2.6 (Huelsenbeck and Ronquist 2001). Phylogenetic trees show that O. glaber is located inside tribe Leptomyrmecini clade (Figure 1), congruent with former evolutionary study presenting old-world Leptomyrmecini genera such as Ochetellus arose from neotropical groups of this tribe, such as Dorymyrmex, Linepithema, and Leptomyrmex (Ward et al. 2010). However, maximum likelihood tree displays that Leptomyrmex pallens (NC_023093) is not clustered with the rest of the tribemates, but the other two trees show that it is placed at correct position (Figure 1). All subfamilies form monophyletic clade in the three trees; however, the inter-subfamily relation was inconsistent (Figure 1). These inconsistencies may be solved when more ant mitogenomes are available. The mitogenome of O. glaber will contribute in understanding the phylogenetics of both the species complex and family Formicidae.
Figure 1.
(A) Lateral view of an Ochetellus glaber worker (KFDS00124; J. Park; IN). (B) Head view of O. glaber worker (KFDS00124; J. Park; IN). (C) Maximum likelihood (bootstrap repeat is 1,000), neighbor joining (bootstrap repeat is 10,000), and bayesian inference (1,100,000 generations) phylogenetic trees of all available Dolichoderine ant mitochondrial genomes: Ochetellus glaber (MN044390 in this study), Linepithema humile (NC_045057 and KT428891), Dorymyrmex brunneus (MG253267), Leptomyrmex pallens (NC_023093), Dolichoderus sibiricus (NC_041075), as well as 13 ants form other subfamilies: Pseudomyrmex gracilis (BK010472), Tetraponera rufonigra (BK010387), Ooceraea biori (CM010870), Ectatomma ruidum (MG870227), Ectatomma tuberculatum (MG870211), Myrmica scabrinodis (NC_026133), Solenopsis invicta (NC_014672), Pristomyrmex punctatus (NC_015075), Camponotus atrox (NC_029357), Anoplolepis gracilipes (NC_039576), Formica fusca (NC_026132), Cryptopone sauteri (NC_041202), Ectomomyrmex javanus (NC_042678), and a honey bee, Apis mellifera ligustica (NC_001566) as an outgroup species. Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining trees, and posterior probability of bayesian inference tree, respectively.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by both InfoBoss Research Grant [IBG-0017] and Cooperative Research Program for Agriculture Science & Technology Development [Project No. PJ013389052019], Rural Development Administration, Republic of Korea.
Disclosure statement
The authors declare that they have no competing interests.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann BD, Andersen AN, Zhang X. 2011. Taxonomic confusion of two tramp ant species: Iridomyrmex anceps and Ochetellus glaber are really species complexes. Curr Zool. 57(5):662–667. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Janicki J, Narula N, Ziegler M, Guénard B, Economo EP. 2016. Visualizing and interacting with large-volume biodiversity data using client–server web-mapping applications: The design and implementation of antmaps. org. Ecological Informatics. 32:185–193. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint. 13033997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kwon W, Park J. 2019. The complete mitochondrial genome of Siberian odorous ant, Dolichoderus sibiricus Emery, 1889 (Hymenoptera: Formicidae). Mitochondrial DNA Part B. 4(1):525–526. [Google Scholar]
- Vieira GA, Prosdocimi F. 2019. Accessible molecular phylogenomics at no cost: obtaining 14 new mitogenomes for the ant subfamily Pseudomyrmecinae from public data. PeerJ. 7:e6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Brady SG, Fisher BL, Schultz TR. 2010. Phylogeny and biogeography of dolichoderine ants: effects of data partitioning and relict taxa on historical inference. Systemat Biol. 59(3):342–362. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Bi G, Yang J, Zhang Z, Liu G, Du Q, Shang E. 2017. Complete mitochondrial genome of the argentine ant, Linepithema humile (Hymenoptera: Formicidae). Mitochondrial DNA Part A. 28(2):210–211. [DOI] [PubMed] [Google Scholar]
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]