Abstract
The complete mitochondrial genome of the leafhopper Limassolla lingchuanensis was determined. It is 15,716 bp in length and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 rRNA genes, and a putative control region. ATN and TTG were initiation codons, and TAA, TAG, and T were termination codons. The phylogenetic relationships based on the neighbor-joining method were revealed using 13 PCGs with 10 leafhopper species of family Cicadellidae, which agree with the conventional taxonomy.
Keywords: Limassolla lingchuanensis, leafhopper, mitochondrial genome
The leafhopper species Limassolla lingchuanensis (Chou and Zhang 1985) belongs to the subfamily Typhlocybinae (Hemiptera: Cicadellidae), the genus Limassolla includes 44 known species distributed across the world (Md et al. 2019). Many leafhopper species in this genus are all herbivorous insects with a broad diet and can feed on a variety of plants (Zhang et al. 2011). In this study, all examined samples were collected from Luodian in Guizhou Province of China (N25°56′, E106°86′). The whole body specimen was preserved in ethanol and stored in the insect specimen room of Guizhou Normal University with an accession number GZNU-ELS-2019001.
The overall base composition is 43.18% A, 35.64% T, 11.62% C, and 9.56% G, with A + T the complete mitogenome of L. lingchuanensis (GenBank accession number MN605256) is a typical closed-circular molecule of 15,716 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 rRNA genes (rrnL and rrnS), and a putative control region. The orientation and gene order of L. lingchuanensis are identical with other leafhoppers’ mitogenomes (Xing and Wang 2019; Yang et al. 2019). Thirteen genes were coded on the minor strand (J-strand), whereas the others were oriented on the major strand (N-strand). Gene overlaps in the L. lingchuanensis mitogenome were found at 16 gene junctions and involved a total of 45 bp with length varying from 1 to 8 bp. The longest overlap was situated between trnC and trnW. There are 11 intergenic spacer sequences in a total of 47 bp and the largest overlap is 26 bp long intergenic spacer, which is located between nad5 and trnH.
The overall nucleotide composition was A (43.18%), T (35.64%), C (11.62%), and G (9.56%), The AT-skew and GC-skew of this genome were 0.0957 and −0.0972, respectively. The A + T content of the 13 PCGs ranged from 70.37% (cox1) to 83.01% (atp8). The length of 22 tRNAs ranged from 53 bp (trnH) to 71 bp (trnK), and A + T content varied from 73.02% (trnS1) to 89.06% (trnG). The rrnL and rrnS were spaced by trnV, with a length of 1168 and 743 bp, respectively. And all of them were encoded on the J-strand. The control region was 1416 bp length and 91.38% A + T content.
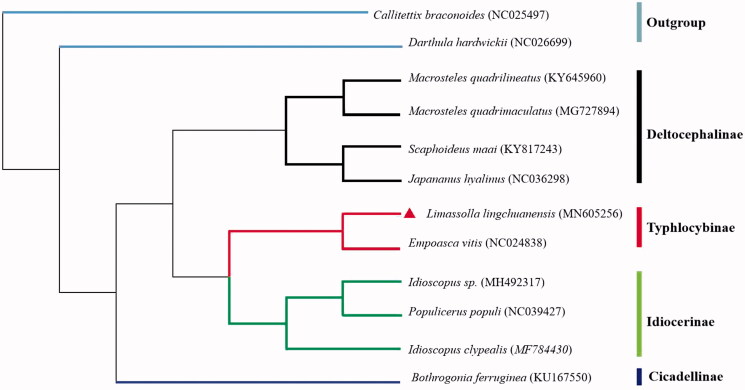
In the mitogenome of L. lingchuanensis, the total length of 13 PCGs is 10,787 bp, which accounts for 75.26% of the total genome. The nad4 initiated with A as the start codon, atp8 began with TTG, nad3, nad5, and nad6 started with ATA, cox2, nad1, and nad4l started with ATT, and remaining six PCGs started with ATG. Nine PCGs including atp6, atp8, cox3, nad1, nad3, nad4l, nad5, nad6, and cob are terminated with TAA as stop codon, cox1 and nad2 end with TAG, cox2 and nad4 end with a single T residue. We analyzed the amino acid sequences of 13 PCGs with neighbor-joining method to reveal the phylogenetic relationship of L. lingchuanensis with other leafhoppers in family Cicadellidae. The result shows that L. lingchuanensis forms a clade with Empoasca vitis (Figure 1), both of them belongs to the subfamily Typhlocybinae. It is consistent with the traditional classification.
Figure 1.
The neighbor-joining phylogenetic tree of L. lingchuanensis and 9 other leafhoppers in inner group. Callitrttix braconoides and Darthula hardwickii were used as an outgroup. GenBank accession numbers of each species are listed in the tree.
Funding Statement
This study was supported in part by the Project of Innovation Program for Postgraduate Education of Guizhou Province: Xiong Kangning’s Studio of Postgraduate Supervisors for the Karst Environment of Guizhou Province [201604], the Project for National Top Discipline Construction of Guizhou Province: Geography in Guizhou Normal University [201785-01], the Guizhou Provincial Science and Technology Foundation [(2018)1411], the Training Program for High-level Innovative Talents of Guizhou Province [(2016)4020] and the Program for First-class Discipline Construction in Guizhou Province [(2017)85], and the Guizhou Science and Technology Support Project ([2019]2855).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Chou Y, Zhang YL. 1985. On the tribe Zyginellini from China (Hemiptrea: Cicadellidae: Typhlocybinae). Entomotaxonomia. 4:287–296. [Google Scholar]
- Md SH, Jin HK, Sang JS, Yong JK. 2019. Taxonomic revision of the microleafhopper genus Limassolla Dlabola from Korea (Hemiptrea: Cicadellidae: Typhlocybinae). Zootaxa. 3:1175–5326. [DOI] [PubMed] [Google Scholar]
- Xing JC, Wang JJ. 2019. Complete mitochondrial genome of Paralaevicephalus gracilipenis (Hemiptera: Cicadellidae: Deltocephalinae) from China. Mitochondrial DNA Part B. 4(1):1372–1373. [Google Scholar]
- Yang WJ, Gao YR, Li C, Song YH. 2019. The complete mitochondrial genome of Chlorotettix nigromaculatus (Hemiptera: Cicadellidae: Deltocephalinae) with phylogenetic consideration. Mitochondrial DNA Part B. 4(1):624–625. [Google Scholar]
- Zhang YL, Gao X, Huang M. 2011. Two new species of Yangisunda Zhang (Hemiptera: Cicadellidae: Typhlocybinae: Zyginellini) from China, with a key to species. Zootaxa. 3097(1):45–52. [Google Scholar]