Abstract
The two complete mitochondrial genomes were sequenced from the freshwater monogonont rotifer Brachionus rubens. The genome sequences were 12,041 bp and 13,793 bp in size, and the gene order and contents were identical to those of the freshwater rotifer B. rubens China, but were different in three tRNA-Arg, tRNA-Ile, and tRNA-Leu between both B. rubens mitochondrial genomes, while B. calyciflorus had peculiar gene order in mitochondrial DNA I. Of 12 protein-coding genes (PCGs), one gene (ND5) had incomplete stop codons. Furthermore, the start codon of ND4 and CO2 gene was ATT, while the start codon of other PCGs was ATG. The base composition of 12 PCGs in B. rubens mitogenome showed 22.5% for A, 46.5% for T, 16.3% for C, and 14.7% for G, respectively.
Keywords: Monogonont rotifer, complete mitochondrial genome, Brachionus rubens
To date, only a few complete mitochondrial genomes have been published in the freshwater rotifer Brachionus sp.; Brachionus calyciflorus (Nie et al. 2016) and Brachionus rubens China (GenBank KJ489417 and KJ489418), while several marine rotifer Brachionus sp. mitogenomes were reported (Suga et al. 2008 for B. plicatilis; Hwang et al. 2014 for Brachionus koreanus, and Kim et al. 2017 for Brachionus rotundiformis). Brachionus rubens is one of the major cosmopolitan freshwater rotifers and was considered as a model for environmental toxicology in response to toxic cyanobacterium (Geng et al. 2006; Geng and Xie 2008; Pérez-Morales et al. 2014), temperature and metal (Azuara-García et al. 2006; Montúfar-Meléndez et al. 2007), suggesting that this species can be an important member of freshwater zooplankton community as a sentinel species for ecotoxicology. The analysis of B. rubens mitochondrial genome is important to identify field-sampled and laboratory stocks. In this study, we identified two complete mitochondrial genomes of the monogonont rotifer B. rubens Japan to better understand the phylogenetic placement of the freshwater rotifer Brachionus clade.
The adult B. rubens were collected from the freshwater catchment area at Urakami, Nagasaki in Japan (32°79′90.72″N, 129°86′55.32″E) in August 2004 and maintained at the Laboratory of Professor Atsushi Hagiwara, Nagasaki University in Japan. The type was deposited in the ichthyological collection of the Faculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Rot-0003. We sequenced 300 bp, 500 bp and 800 bp paired end library of B. rubens from whole body genomic DNA using the Illumina HiSeq 2500 platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted by SPAdes (version 3.13.0) (http://cab.spbu.ru/software/spades/) with K-mer auto. Of the assembled B. rubens 270,986 contigs with Newbler (version 2.9; identity 97) (http://www.454.com), seven mitochondrial contigs were obtained. After a manual curation of seven contigs with Consed (version 19.0) (http://www.phrap.org/consed/consed.html), two contigs were obtained to the mitochondrial DNA of B. rubens.
The complete mitochondrial genomes of B. rubens were 12,041 bp (mitochondrial DNA I; GenBank no. MN256531) and 13,793 bp (mitochondrial DNA II; GenBank no. MN256532) in size. The direction of 12 protein-coding genes (PGCs) of B. rubens Japan was identical to those of B. rubens China of the genus Brachionus, including the presence of nearly identical non-coding region (Identities: 5651/5731 = 98.25%) (Suga et al. 2008; Hwang et al. 2013). Of 12 PCGs, one gene (ND5) had incomplete stop codons. Furthermore, the start codon of ND4 and CO2 gene was ATT, while the start codon of other PCGs was ATG. The base composition of 12 PCGs in B. rubens Japan mitogenome showed 22.5% for A, 46.5% for T, 16.3% for C, and 14.7% for G, respectively. The mitochondrial genome A + T base composition (69.0%) of 12 PCGs was higher than G + C (31.0%), while the complete mitochondrial genome A + T base composition (67.5%) was higher than G + C (32.5%).
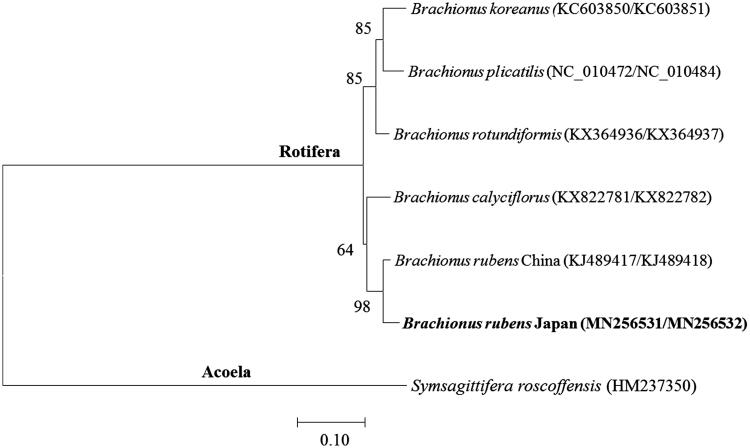
The placement of B. rubens Japan in the genus Brachionus with CO1 and Cytb was shown in Figure 1. B. angularis was clustered closely to B. rubens China. The gene order and contents of 12 PGCs were identical to those of the freshwater rotifer B. rubens China. However, interestingly, the order of tRNAs of three tRNA-Arg, tRNA-Ile, and tRNA-Leu was different between both B. rubens mitochondrial genomes, while the freshwater rotifer B. calyciflorus had peculiar gene order in mitochondrial DNA I. This indicates that the rearrangement of tRNAs is likely occurring in sporadic manner in the genus Brachionus (Hwang et al. 2014).
Figure 1.
Phylogenetic analysis of the rotifer Brachionus rubens Japan mitochondrial DNA. We conducted a comparison of two mitochondrial DNA genes (CO1 and Cytb) of Acoela and Rotifera. Two mitochondrial DNA genes CO1 and Cytb were aligned by ClustalW. Maximum likelihood (ML) analysis was performed by Raxml 8.2.8 (http://sco.h-its.org/exelixis/software.html) with GTR + γ+I nucleotide substitution model. The rapid bootstrap analysis was conducted with 1,000 replications with 48 threads running in parallel. The Acoela served as an outgroup. Ln=-5316.105. Modified from Choi et al. (2019).
Funding Statement
This work was supported by the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) [No. 20180430]. This work was also supported by a short-term invitation fellowship of the Japan Society of the Promotion of Science [JSPS ID: S19056] to Jae-Seong Lee.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Azuara-García R, Sarma SS, Nandini S. 2006. The combined effects of zinc and alga on the life table demography of Anuraeopsis fissa and Brachionus rubens (Rotifera). J Environ Sci Health A. 41(4):559–572. [DOI] [PubMed] [Google Scholar]
- Choi B-S, Lee YH, Hagiwara A, Lee J-S. 2019. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus calyciflorus (Rotifera, Brachionidae). Mitochondrial DNA B. 4(2):3593–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Xie P. 2008. Experimental studies on the effects of toxic Microcystis aeruginosa PCC7820 on the survival and reproduction of two freshwater rotifers Brachionus calyciflorus and Brachionus rubens. Ecotoxicology. 17(8):709–715. [DOI] [PubMed] [Google Scholar]
- Geng H, Xie P, Xu J. 2006. Effect of toxic Microcystis aeruginosa PCC7820 in combination with a green alga on the experimental population of Brachionus calyciflorus and B. rubens. Bull Environ Contam Toxicol. 76(6):963–969. [DOI] [PubMed] [Google Scholar]
- Hwang D-S, Dahms H-U, Park HG, Lee J-S. 2013. A new intertidal Brachionus and intrageneric phylogenetic relationship among Brachionus as revealed by allometry and CO1-ITS1 gene analysis. Zool Stud. 52(1):13. [Google Scholar]
- Hwang D-S, Suga K, Sakakura Y, Hagiwara A, Park HG, Rhee J-S, Lee J-S. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera: Brachionidae). Mitochondrial DNA. 25(1):29–30. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Hwang D-S, Kim H-J, Sakakura Y, Hagiwara A, Lee J-S. 2017. Complete mitochondrial genome of the monogonont rotifer Brachionus rotundiformis (Rotifera, Brachionidae). Mitochondrial DNA B. 2(1):39–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montúfar-Meléndez AI, Sánchez-Ortíz JR, Sarma SS, Nandini S. 2007. Combined effects of temperature and lead concentration on the competition between the rotifers Brachionus havanaensis and Brachionus rubens (Rotifera: Brachionidae). J Environ Sci Health A. 42(3):393–398. [DOI] [PubMed] [Google Scholar]
- Nie, Z-J, Gu, R-B, Du, F-K, Shao, N-L, Xu, P, Xu, G-C.. 2016. Monogonont rotifer, Brachionus calyciflorus, possesses exceptionally large, fragmented mitogenome. PLoS One 11(12):e0168263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Morales A, Sarma SS, Nandini S. 2014. Feeding and filtration rates of zooplankton (rotifers and cladocerans) fed toxic cyanobacterium (Microcystis aeruginosa). J Environ Biol. 35(6):1013–1020. [PubMed] [Google Scholar]
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25(6):1129–1137. [DOI] [PubMed] [Google Scholar]