Abstract
Delphinium grandiflorum L. is a perennial herb, and has very high medicinal value. However, the evolutionary relationship analysis of D. grandiflorum is limited. Its cp genome was 157,339 bp in length, containing a pair of inverted repeated regions (52,304 bp), separated by a large single copy region of 88,098 bp, and a small single copy region of 16,937 bp. Moreover, a total of 117 functional genes were annotated, including 79 mRNA, 30 tRNA genes, and 8 rRNA genes. The phylogenetic relationships inferred that D. grandiflorum was closely related to Gymnaconitum gymnandrum. This study will provide a theoretical basis for species identification and biological research.
Keywords: Delphinium grandiflorum L., chloroplast genome, phylogenetic relationship
Delphinium grandiflorum L., belonging to the family Ranunculaceae, is a perennial herb mainly distributed in some regions of Siberia, the Northwest of China, and People’s Republic of Mongolia (Chen et al. 2017). Delphinium grandiflorum can be used as a folk medicine for the treatment of toothache and native pesticide (Zhang et al. 2012). However, the evolutionary relationship of D. grandiflorum is limited in study. Here, the complete cp genome of D. grandiflorum was sequenced on the Illumina NovaSeq platform (Benagen Tech Solution Co., Ltd, Wuhan, China), which will provide a theoretical basis for species identification and biological research.
The fresh leaves of D. grandiflorum were collected in Lanzhou Scientific Observation and Experiment Field Station of the Ministry of Agriculture for Ecological System in the Loess Plateau Area (36lecte, 103lected in Lanzho1700 m), Gansu, China, on 22 July 2019. The voucher specimen was kept in Herbarium of Lanzhou Institute of Husbandry and Pharmaceutical Science, Chinese Academy of Agricultural Sciences (CYSLS-DgDUAN20190722). The total genomic DNA of D. grandiflorum was extracted from the fresh leaves with a modified CTAB method (Li et al. 2018). One library (250 bp) was constructed using pure DNA according to the manufacturer’s instructions (NEBNext®UltraTM DNA Library Prep Kit for Illumina®). The library was performed with an Illumina NovaSeq platform (Benagen Tech Solution Co., Ltd, Wuhan, China) and 150 bp paired-end reads were generated. Approximately 6.4 GB of clean data were yielded. The assembled reads were joined into bidirectional iterative derivation using NOVOPlasty (version: 32, parameter: k-mer = 39) to obtain the whole-genome sequence (Dierckxsens et al. 2017). The assembled genome was annotated using GeSeq (Tillich et al. 2017). The circular gene map of D. grandiflorum was drawn using the OGDRAWv1.2 program (Loshe et al. 2007).
The complete cp genome of D. grandiflorum is 157,339 bp in length with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions (52,304 bp) that are separated by a large single copy (LSC) region of 88,098 bp, and a small single copy (SSC) region of 16,937 bp. The GC content of the whole cp genome was 38.08%. A total of 117 functional genes were annotated, including 79 protein-coding genes (mRNA), 30 tRNA genes, and 8 rRNA genes. The protein-coding genes, tRNA genes, and rRNA genes account for 67.52, 25.64, and 6.84% of all annotated genes, respectively.
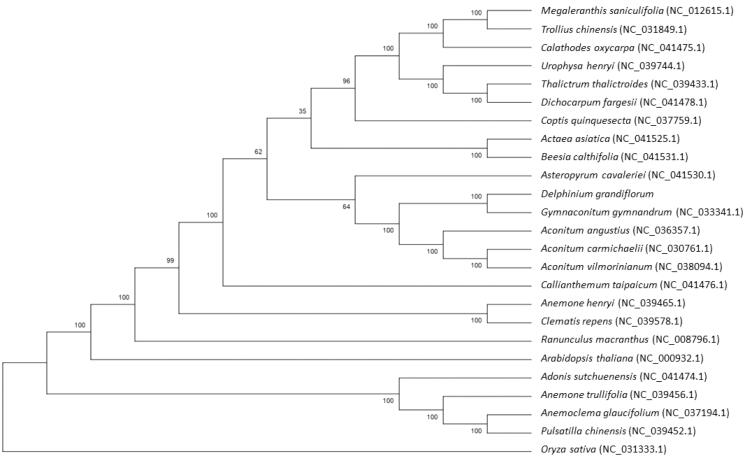
The phylogenetic tree was generated based on the complete cp genome of D. grandiflorum and other 24 species (Figure 1). The alignment was conducted using MAFFT (Katoh and Standley 2013). The phylogenetic tree was built using the neighbor-joining (NJ) method. The results showed that D. grandiflorum was closely related to Gymnaconitum gymnandrum.
Figure 1.
Phylogenetic relationships of 25 species based on complete chloroplast genome using the neighbor-joining methods. The bootstrap values were based on 1000 replicates and are shown next to the branches.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31700338], the Fundamental Research Funds for the Central Public-Interest Scientific Institution [1610322019012 and 1610322019013], and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences [CAASASTIP-2019-LIHPS-08].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen NH, Zhang YB, Li W, Li P, Chen LF, Li YL, Li GQ, Wang GC. 2017. Grandiflodines A and B, two novel diterpenoid alkaloids from Delphinium grandiflorum. RSC Adv. 7(39):24129–24132. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li YF, Zang MY, Li MZ, Fang YM. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Quercusacutissima. IJMS. 19(8):2443–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loshe M, Drechesel O, Bock R. 2007. OrganellarGenomeDraw(ORDRAW): a tool for easy generation of high quality customgraphical maps of plastid and mitochondrial genomes. Curr Genet. 52:15655. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YN, Li B, He M. 2012. Effects of saline and alkali stress on seed germination of Delphinium grandiflorum. Pratacult Sci. 29:1235–1239. Chinese. [Google Scholar]