Abstract
Achyranthes bidentata (Amarathaceae) has been commonly used as a traditional Chinese medicine in the treatment of osteoporosis and bone nonunion. Here, the complete chloroplast genome of A. bidentata was assembled and characterized. The cp genome is 151,451 bp in length, composed of a pair of 25,150 bp inverted repeat (IR) regions separated by a large single-copy (LSC) region of 83,899 bp and a small single-copy (SSC) region of 17,252 bp. The whole cp genome of A. bidentata contains 130 genes(85 protein-coding genes, 37 tRNAs and eight rRNAs) and the overall GC content is 36.5%. Phylogenetic analysis based on the cp genome data showed that A. bidentata was close to Cyathula capitata.
Keywords: Achyranthes bidentata, chloroplast genome, medicinal plant, phylogenetic analysis
Achyranthes bidentata Blume (Amarathaceae), has been used for thousands of years as a blood-activating and stasis-resolving medicine for the treatment of osteoporosis in China and India (He et al. 2017). Moreover, its major component, A. bidentata alcohol, has anti-asthmatic, anti-inflammatory, anti-pyretic, anti-rheumatic, and diuretic activities (Hua and Zhang 2019), and A. bidentata polypeptides (ABPP) possess neuroprotective activity (Shen et al. 2008; Peng et al. 2018). In this study, we aim to establish and characterize the complete chloroplast (cp) genome of A. bidentata, and assess its phylogenetic position within Amaranthaceae.
Fresh and clean leaves of A. bidentata were sampled from Longping town of Jianshi county, Hubei, China (N30°48′24″, E110°1′47″, 1,750 m). The voucher specimen (HSN12316) was deposited in the herbarium of South-Central University for Nationalities (HSN). The total genomic DNA was extracted and used for sequencing on Illumina HiSeq 4000 Platform at the Beijing Novogene Bioinformatics Technology Co., Ltd. (Nanjing, China). About 2 GB raw data were used to de novo assemble the complete cp genome using SPAdes (Bankevich et al. 2012). The complete genome sequence was annotated using PGA (Qu et al. 2019) with manual adjustments. The sequence of cp genome was deposited in GenBank (accession numbers MN652923).
The circular cp genome of A. bidentata is 151,451 bp in size, and exhibits a typical quadripartite structure found in most land plants which is made up of a large single-copy region (LSC) of 83,899 bp, a small single-copy region (SSC) of 17,252 bp, isolated by a pair of identical inverted repeat (IR) regions of 25,150 bp. The total GC content of the whole sequence is 36.5%. The complete cp genome encodes 130 genes, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Most of the genes occurred in a single copy, while four rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA), seven tRNA genes (i.e. trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and six protein-coding genes (i.e. ndhB, rpl2, rpl23, rps7, rps12, and ycf2) occurred in double. Among the 113 unique genes, 14 had one intron, and three had two introns (clpP, rps12, and ycf3).
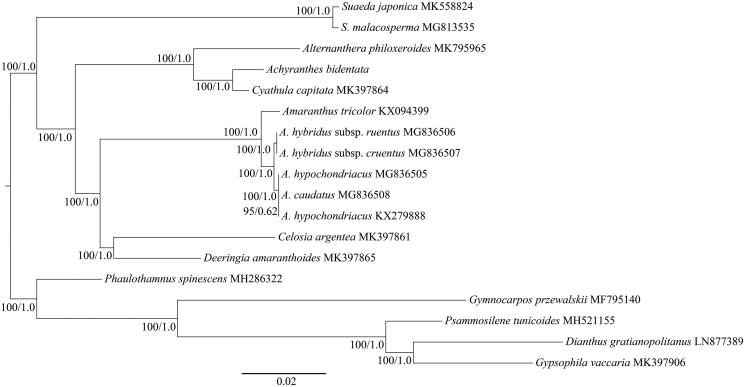
The phylogenetic position of A. bidentata was analyzed based on the complete cp genomes of this species and other seventeen species belonging to Achatocarpaceae, Amaranthaceae and Caryophyllaceae. The sequences were aligned with MAFFT (Katoh and Standley 2013). The maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were reconstructed using RAxML (Stamatakis 2014) and MrBayes (Ronquist et al. 2012). The ML and BI analyses generated the same tree topology (Figure 1). As shown in the phylogenetic tree (Figure 1), A. bidentata was closely related to Cyathula capitata with 100% bootstrap and 1.0 posterior probability support, respectively. Our findings will provide a foundation for further investigation of cp genome evolution and phylogenetic studies of Achyranthes.
Figure 1.
The maximum-likelihood (ML) tree of Amaranthaceae inferred from the complete chloroplast genome sequences. Numbers at nodes correspond to ML bootstrap percentages (1000 replicates) and Bayesian inference (BI) posterior probabilities.
Funding Statement
This study was supported by the National Natural Science Foundation of China [31870513] and the Project on the Integration of Industry, Education and Research of Liupanshui [52020-2018-04-12].
Disclosure statement
No potential conflict of interest was reported by the authors
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XR, Wang XX, Fang JC, Chang Y, Ning N, Guo H, Huang LH, Huang XQ. 2017. The genus Achyranthes: a review on traditional uses, phytochemistry, and pharmacological activities. J Ethnopharmacol. 203:260–278. [DOI] [PubMed] [Google Scholar]
- Hua S, Zhang XX. 2019. Effects of Achyranthes bidentata alcohol on proliferation capacity of osteoblasts and miRNA in Runx2. Exp Ther Med. 18:1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Wang CP, Ma JY, Jiang KT, Jiang YH, Gu XS, Sun C. 2018. Achyranthes bidentata polypeptide protects dopaminergic neurons from apoptosis in Parkinson’s disease models both in vitro and in vivo. Brit J Pharmacol. 175(4):631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Yuan Y, Ding F, Liu J, Gu X. 2008. The protective effects of Achyranthes bidentata polypeptides against NMDA-induced cell apoptosis in cultured hippocampal neurons through differential modulation of NR2A- and NR2B-containing NMDA receptors. Brain Res Bull. 77(5):274–281. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]