Abstract
W922, a novel PI3K/Akt/mTOR pathway inhibitor, exhibits efficient anti-tumor effects on HCT116, MCF-7 and A549 human cancer cells compared with other synthesized compounds. The present study aimed to investigate its anti-tumor effects on colorectal cancer cells. A total, of seven different colorectal cell lines were selected to test the anti-proliferation profile of W922, and HCT116 was found to be the most sensitive cell line to the drug treatment. W922 inhibited HCT116 cell viability and cell proliferation in vitro in concentration- and time-dependent manners. Furthermore, W922 suppressed the tumor growth in a xenograft mouse model and exhibited low toxicity. The proteomic alterations in W922-treated HCT116 cells were found to be associated with cell cycle arrest, negative regulation of signal transduction and lysosome-related processes. W922 caused cell cycle arrest of HCT116 cells in G0-G1 phase, but only triggered slight apoptosis. In addition, the PI3K/Akt/mTOR signaling proteins were dephosphorylated upon W922 treatment. It has been reported that inhibition of mTOR is relevant to autophagy, and the present results also indicated that W922 was involved in autophagy induction. An autophagy inhibitor, chloroquine, was used to co-treat HCT116 cells with W922, and it was identified that the cell cycle arrest was impaired. Moreover, co-treatment of W922 and chloroquine led to a significant population of apoptotic cells, thus providing a promising therapeutic strategy for colorectal cancer.
Keywords: W922, PI3K/Akt/mTOR inhibitor, colorectal cancer, cell cycle arrest, autophagy
Introduction
Colorectal cancer is the third most commonly diagnosed cancer in men and the second in women worldwide, with ~1.4 million cases and 693,000 mortalities in 2012. The incidence rates are higher in Europe, Australia and Northern American (1). The incidence of CRC is likely to increase as the world becomes richer, as more individuals shift to unhealthy diets and life styles (2). Currently, chemotherapy and surgery are the main treatment strategies for malignant tumors that form in the colon tissues (3). Chemotherapy can be used either before surgery to shrink the tumor before removal, or after surgery for patients with advanced CRC (4). While the overall survival duration of patients with advanced CRC has improved in the last decade due to the various chemotherapy regimens, drug resistance still occurs in nearly all patients with CRC and ultimately leads to chemotherapy failure (5). Thus, investigation of the molecular mechanisms of CRC progression and the development of novel targeted drugs are essential.
Widespread-usage of high-throughput sequencing allows for the identification of mutations present in CRC; however, progress in targeted therapies for CRC has been slow (6). There has been significant progress in the understanding of the molecular pathways regarding tumorigenesis. For instance, several pathways are dysregulated by common mutations that have been identified via CRC sequencing, including the WNT, MAPK, PI3K and p53 pathways (7). In total, ~60% of sequenced CRC tissues have potentially targetable genetic aberrations in the PIK3CA, BRAF and PTEN signaling pathway, and early studies have revealed promising effects for CRC by focusing on these markers (8,9).
PI3K/AKT/mTOR (PAM) signaling acts as a pivotal transduction pathway in tumor progression, which is involved in controlling cancer cell proliferation, differentiation, motility and survival (10). A mutation of PI3Kα is present in >18% of metastatic CRC cases, and thus, small-molecular compounds that can inhibit the PAM pathway may have great potential in the treatment of CRC (11). VS-5584, a highly selective PI3K/mTOR kinase inhibitor, demonstrates robust anti-tumor effects both in vitro and in vivo (12). Moreover, VS-5584 has 10-fold selectivity for cancer stem cells (13). These findings suggest that medicinal chemists should identify and develop novel and potent PI3K inhibitors for cancer treatment.
It has been reported that there is a close connection between mTOR and autophagy (14). It was first discovered in yeast that genetic or pharmacological inhibition of mTOR complex 1 could induce autophagy (15). Autophagy is the major cellular digestion process that is essential for cellular development and homeostasis. In addition, it is critical to provide energy in response to nutrient and environmental stress by recycling macromolecules (16). Autophagy induction could have pro-survival or pro-death properties depending on tumor types or treatment strategies (17). For instance, autophagy may facilitate tumor development when nutrients are limited (18). Thus, inhibition of autophagy may sensitize cancer cells to stress, leading to cell death (19). On the other hand, autophagy may induce autophagy-mediated cell death, which is also known as type II programmed cell death (20). Given the potential dual functions of autophagy in tumor progression, elucidating the precise function of autophagy in individual cancer types induced by PAM pathway inhibitors is essential for cancer therapy.
Our previous study synthesized a compound W922 (refers to compound A7) based on the structure of PI3K/mTOR dual inhibitor VS-5584 by replacing the imidazole ring with a triazine skeleton (21). It has been reported that triazine skeleton has potential anti-tumor bioantivity (22). In the present study aimed to evaluate the anti-proliferative effects of W922 both in vitro and in vivo, and identify the underlying mechanism of W922's anti-tumor effect. Data in this study will provide a pharmacological basis for W922's potential application, and will also provide information for further development of PI3K/AKT pathway inhibitors and triazine skeletal derivatives.
Materials and methods
Chemicals
W922 was synthesized in Department of Pharmaceutical Biochemistry, Xi'an Jiaotong University, and characterized using 1H NMR (nuclear magnetic resonance hydrogen spectrum), 13C NMR (nuclear magnetic resonance carbon spectrum) as previously described (21). VS-5584 (cat. no. A3927) and Choloroquine (CQ) diphosphate (cat. no. A8628) were purchased from APExBIO Technology LLC.
For in vitro assays, chemicals were prepared by dissolving into sterilized DMSO to a final concentration of 0.01 M and stored at −20°C before use. For the xenograft experiment, W922 and VS-5584 were dissolved in a mixture containing DMSO, PEG400 and ddH2O (ratio as 1:7:2).
Cell lines and cell culture conditions
CRC cell lines HCT116, HT29, RKO, Colo205, SW620, DLD1 and LOVO were purchased from American Type Culture Collection. All cell lines were maintained in DMEM (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin antibiotics (Beijing Solarbio Science & Technology Co., Ltd.), and cultured at 37°C in a humidified incubator with 5% CO2.
Reagents and antibodies
MTT, crystal violet solution, PI, RNAse A and DAPI were purchased from Beijing Solarbio Science & Technology Co., Ltd. Western blotting reagents were obtained from Beyotime Institute of Biotechnology. An Annexin V/PI apoptosis detection kit was purchased from 7 Sea Biotech (http://www.7seapharmtech.com/).
Primary antibodies against the following proteins were purchased from Cell Signaling Technology, Inc. (CST): AKT (cat. no. 9272), phosphorylated (p)-AKT (Ser473; cat. no. 4060), mTOR (cat. no. 2983), p-mTOR (Ser2448; cat. no. 5536), p70S6K (cat. no. 9202), p-p70S6 kinase (p70S6K; Ser371; cat. no. 9208), eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1; cat. no. 9644), p-4E-BP1 (Thr37/46; cat. no. 2855), Beclin-1 (cat. no. 3495), p-Beclin-1 (Ser93; cat. no. 14717), Cyclin D1 (cat. no. 2978), Cyclin E1 (cat. no. 4129), cleaved caspase-3 (cat. no. 9664), autophagy related 5 (ATG5; cat. no. 9980) and GAPDH (cat. no. 5174). LC3-I/II (cat. no. ABC929) was purchased from Sigma-Aldrich (Merck KGaA) and ki67 (cat. no. ab15580) was obtained from Abcam. Secondary horseradish peroxidase-linked antibodies against rabbit (cat. no. 7074) and mouse (cat. no. 7076) were purchased from CST. Small interfering RNA (si)ATG5 was purchased from Shanghai GenePharma Co., Ltd., and Lipofectamine® 2000 was obtained from Thermo Fisher Scientific, Inc.
Cell mutation search
The mutation data were obtained from the Catalogue Of Somatic Mutations In Cancer website (https://cancer.sanger.ac.uk/cosmic).
Cell viability assay
The MTT assay was used to measure cell viability. The CRC cells (HCT116, RKO, COLO205, SW620, DLD1, HT29 and LOVO) were seeded in a 96-well plate (5,000 cells/well) and incubated at 37°C overnight. The following day, the cells were treated with W922 (10 µM) and cultured for 24 h at 37°C. Then, MTT solution (5 mg/ml; 20 µl) was added into each well, and the plate was incubated for another 4 h at 37°C. The supernatant was removed and the purple formazan crystals were dissolved in 150 µl DMSO. The cell viability was calculated according to the absorbance values at 570 nm, measured via a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
The long-term effect of W922 on cell proliferation was evaluated using the colony formation assay in HCT116 cells. Cells were plated in a 6-well plate (1,000 cells/well). After overnight incubation, the cells were treated with W922 (0.1, 0.3, 1 and 3 µM) or DMSO and cultured at 37°C for ~3 weeks until colonies were visible. The colonies in each well of the 6-well plate were fixed with pure methanol (2 ml) for 20 min and stained with crystal violet solution (2 ml) for 20 min at room temperature. Then, the plates were washed with PBS three times, and the colonies were counted.
Xenograft antitumor study
A total of 30 BALB/c nude mice (age, 4 weeks; weight, 18 g; female) were purchased from Beijing Vital River Laboratory Animal Technology Co, Ltd., and were fed in the enviroument of 25°C, 60% humidity, 12 h light/dark cycle, and free access to food and water in Xi'an Jiatong University Health Science Center. HCT116 cells (1×106) resuspended in 100 µl PBS were subcutaneously injected into the right flanks of 4-week-old BALB/c nude mice. Once a palpable tumor was presented, the mice were randomly grouped and treated daily intraperitoneally with either vehicle solvent, W922 (10, 30 and 100 mg/kg) or VS-5584 (10 mg/kg) for 18 days. Tumor volumes were measured every 3 days using a caliper and calculated as 0.5 × (length × width2). The mice were sacrificed 18 days after transplanting, and the tumors were isolated and weighed.
Flow cytometric analysis
Cell cycle analysis was performed via PI staining. The cells were seeded in 12-well plates (5×104/well) and treated with W922 (0.1, 1 and 10 µM) for 24 h at 37°C. After harvesting, the cells were fixed in 70% ethanol at 4°C overnight and stained with PI solution (50 µl PI and 50 µl RNAse A in 10 ml PBS) for 30 min at room temperature before measurement. The data were obtained using a flow cytometer (Beckman Coulter, Inc.) and analyzed using the ModFitLT software (Version 5.0; www.mybiosoftware.com).
Cell apoptosis was measured using a Annexin V/PI double staining assay. The cells were seeded in 12-well plates (5×104/well) overnight and treated with W922 (0.1, 1 and 10 µM) for 24 and 48 h at 37°C. Then, the cells were harvested and subjected to the assay according to the manufacturer's instruction. The data were collected from a flow cytometer (Beckman Coulter, Inc.) and analyzed using FlowJo software (Version 10; FlowJo LLC).
Western blotting
HCT116 cells were seeded in 6-well plates (105 cells/well) and allowed to attach. The following day, cells were treated with W922 (0.1, 0.3, 1, 3 and 10 µM) at 37°C for 24 h. After treatment, the cells were harvested, and the proteins were collected with RIPA buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitor cocktail for 20 min on ice. The protein concentrations were determined using the BCA method. The same amount of proteins (20 µg) were separated via a 12% SDS-PAGE gel and transferred to a PVDF membrane. Subsequently, the membrane was blocked in 5% skim milk in room temperature for 2 h, and then incubated with primary antibodies (antibody: 5% skim milk =1:1,000) at 4°C overnight. The membranes were incubated with the secondary antibodies for 1 h. The protein expression levels were visualized using an enhanced chemiluminescence system (EMD Millipore), and the densitometry was detected using Image J software (Version 1.48; imagej.net).
Immunofluorescence microscopy
The cells were grown on 13-mm coverslips (10,000/well), placed in 24-well culture plate and treated with W922 (0.1, 1 and 10 µM) at 37°C for 24 h. for desired time periods. After treatment, the cells were rinsed in PBS, fixed with pure methanol for 20 min on ice and washed twice with PBS. Subsequently, the cells were blocked in PBS with 5% FBS at room temperature for 1 h, and then incubated with anti-LC3-I/II antibody, anti-Ki67 antibody and anti-Cleaved caspase-3 antibody overnight at 4°C. Cells were washed with PBS and incubated with Alexa Fluor 488-labeled and Alexa Fluor 555-labeled secondary antibodies in room temperature for 2 h (Invitrogen; Thermo Fisher Scientific, Inc.; cat. nos. A32790, A21208 and A32794; 1:1,000). The nuclei were stained with Hoechst 33342 (1 µg/ml) at room temperature for 30 min. After washing with PBS, the coverslips with cells were mounted onto slides with aqueous mounting medium. The cells were imaged using ZEISS LSM700 (Zeiss GmbH) confocal microscope system (magnification, ×20).
CQ pretreatment
CQ was used to inhibit autophagy. CQ (10 and 20 µM) was added to cells at 37°C for 2 h, and then W922 (1 µM) was used to treat HCT116 cells at 37°C for 24 h to distinguish whether W922 was an autophagy inhibitor or inducer. For proteomics assay, 50 µM choloroquine was used to pre-stimulate HCT116 cells for 2 h at 37°C before W922 (10 µM) treatment (24) to examine the protective or suppressive role served by autophgy.
Transfection experiment
siRNA targeting the ATG5 cDNA sequencing (5′-GACGUUGGUAACUGACAAATT-3′) was purchased from Shanghai GenePharma Co., Ltd. And siRNA NC was also supplied by GenePharma company and was taken as the negative control. HCT116 cells were plated in 6-well plates at a density of 2×105/well and cultured overnight. ATG5 siRNA at a concentration of 80 nM (dissolved in FBS-free medium) was transfected into the cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 6 h after transfection, the FBS-free medium was removed and replaced with complete medium, and cells were cultured for another 24 h. Successfully transfected HCT116 cells were used in the corresponding experiments.
Proteomics sample preparation
HCT116 cells were plated in 6-well plates (3×105) and treated by W922 or treated with the combination of choloroquine and W922 for 24 h at 37°C. After treatment, the cells were collected and lysed in RIPA buffer supplemented with PMSF and protease inhibitor on ice. The cell lysate was then centrifugated at 15,500 × g for 15 min at 4°C and the supernatant was collected into a new Eppendorf tube. After protein reduction with 8 mM 1,4-dithiothreitol (Sigma-Aldrich; Merck KGaA) at 55°C for 45 min and alkylation using 25 mM iodoacetamide (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min in the dark, the proteins were precipitated using pure acetone at -20°C overnight. Subsequently, the proteins were digested with Lys C (FUJIFILM Wako Pure Chemical Corporation; Lys C:protein amount=1:75) at 30°C for 8 h and trypsin (Promega Corporation) at 37°C overnight. Then, desalting of the peptides was conducted using C18 Sep-pak (WAT054960; Waters Corportation) at room temperature, and the peptides in each sample were analyzed via nano liquid chromatography (LC)-mass spectrometry (MS)/MS, as customary in shotgun proteomics (23).
LC-MS/MS analysis
LC-MS/MS analysis was performed on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Inc.). The instrument was equipped with an EASY ElectroSpray Source and connected to an UltiMate 3000 RSLCnano UPLC system (Thermo Fisher Scientific, Inc.). The injected sample fractions were preconcentrated and desalted online using a PepMap C18 nanotrap column (Thermo Fisher Scientific, Inc.) with a flow rate of 3 µl/min for 5 min. Peptide separation was performed on an EASY-Spray C18 reversed-phase nano-LC column (Thermo Fisher Scientific, Inc.) at 55°C and a flow rate of 300 nl/min. Peptides were separated by a binary solvent system, and were eluted with a gradient of 4-26% B in 120 min and 26-95% B in 10 min. The instrument was operated in the positive ion mode for data-dependent acquisition of MS/MS spectra with a dynamic exclusion time of previously selected precursor ions of 30 sec. Mass spectra were acquired in a mass-to-charge (m/z) range of 375-1,500 with a resolution of 120,000 at m/z 200, and at a resolution of 60,000 with a target value of 2×105 ions and a maximum injection time of 120 msec. The fixed first m/z was 100, and the isolation window was 1.2 m/z units.
The extracted data were searched via the Andromeda search engine in MaxQuant 1.5.6.5 software (www.maxquant.org). Gene names corresponding to upregulated and down-regulated proteins were submitted into STRING website (http://string-db.org) to analyze the potential network among proteins. The data of Gene Ontology (GO) analysis was also collected from STRING website.
Statistical analysis
Each sample has three biological replicates. Data are presented as the mean ± SEM. Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference. Data were analyzed using an unpaired Student's t-test, and one-way or two-way ANOVA followed by Dunnett's test.
Results
W922 inhibits tumor growth in vivo via the regulation of viability
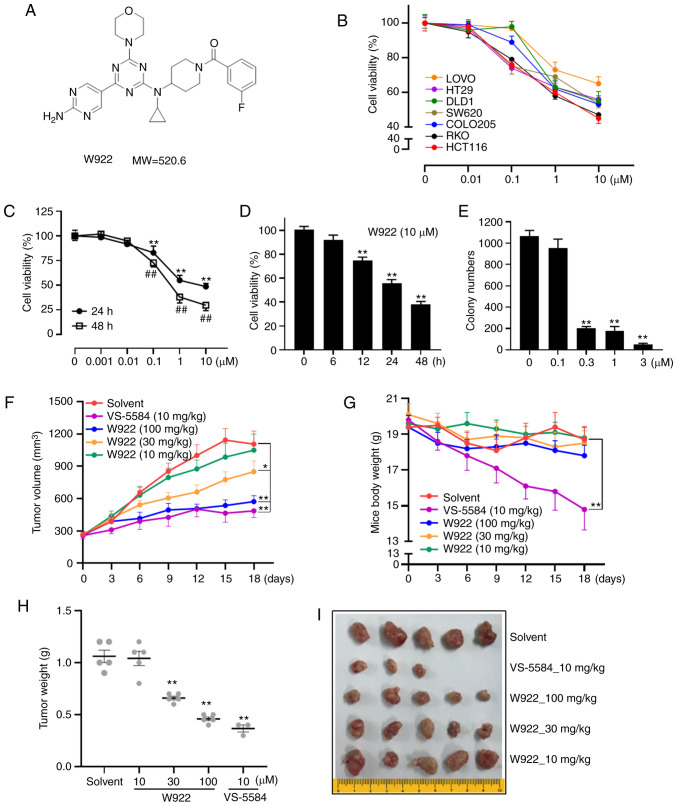
The chemical structure of W922 is presented in Fig. 1A. The effects of W922 on cell viability were first examined using seven different CRC cell lines. The mutant sites in the tested seven cell lines are summarized in Table I. HCT116 had mutation sites on PIK3CA and KRAS. Among these CRC cell lines, HCT116 was the most sensitive cell line to the drug treatment (Fig. 1B). Thus, all subsequent experiments were performed with this cell line.
Figure 1.
W922 inhibits tumor growth in vivo via the regulation of proliferation. (A) Chemical structure of W922. (B) Anti-proliferation effect of W922 on seven types of colorectal cancer cells after 24 h treatment (n=3) (C) W922 inhibited HCT116 cells in a concentration-dependent manner (n=3; one-way ANOVA followed by Dunnett's test). **P<0.01 vs. 0 µM-24 h treatment; ##P<0.01 vs. 0 µM-48 h treatment. (D) W922 time dependently inhibited HCT116 cells (n=3; one-way ANOVA followed by Dunnett's test. **P<0.01 vs. 0 h treatment. (E) Long-term cell viability was measured via clonogenic formation assay. HCT116 cells were seeded and treated with indicated concentrations of W922 for 2 weeks, and the number of the colonies were counted and quantified (n=3; one-way ANOVA followed by Dunnett's test). **P<0.01 vs. 0 µM treatment. (F) W922 inhibited the growth of HCT116 solid tumors in vivo. The nude mice were transplanted with HCT116 cells and randomly grouped when tumor volume reached ~200 mm3. Then, the mice were treated with solvent, W922 or positive control (VS-5584) for 18 days, and the tumor volumes were measured every 3 days using a caliper. (G) Body weight of mice during treatment. (H) On the final day of treatment, the mice were sacrificed and the solid tumors were excised and weighed [n=3-5; two-way ANOVA (F and G) and one-way ANOVA (H) followed by Dunnett's test]. *P<0.05, **P<0.01 vs. solvent group. (I) Image of the excised tumors. MW, molecular weight.
Table I.
Mutant sites in tested colorectal cancer cell lines.
| Cell line | ATCC no. | Mutant gene |
|---|---|---|
| LOVO | CCL-229 | PIK3CB; RAF1; KRAS |
| HT29 | HTB-38 | PIK3CA; PIK3R1; BRAF; TP53 |
| DLD1 | CCL-221 | KRAS |
| SW620 | CCL-227 | KRAS |
| COLO205 | CCL-222 | BRAF; TP53 |
| RKO | CRL-2577 | PIK3CA; BRAF |
| HCT116 | CCL-247 | PIK3CA; KRAS |
ATCC, American Type Culture Collection.
W922 treatment markedly decreased the viability of HCT116 cells in concentration- and time-dependent manners (Fig. 1C and D). Consistently, colony formation assay results indicated that the cells treated with W922 formed significantly smaller and fewer colonies compared with the untreated cells (Fig. 1E), suggesting the long-term anti-tumor effect of W922.
Next, a HCT116 cell xenograft nude mouse model was used to investigate the effect of W922 on CRC in vivo model. Significant suppression of tumor growth was observed when the dose of W922 was ≥30 mg/kg (Fig. 1F, H and I). While VS-5584 exhibited robust anti-tumor potency, it had significant toxicity, as the weight of the mice decreased significantly and two mice died in the VS-5584 group (Fig. 1G and I).
W922 negatively regulates the cell cycle processes
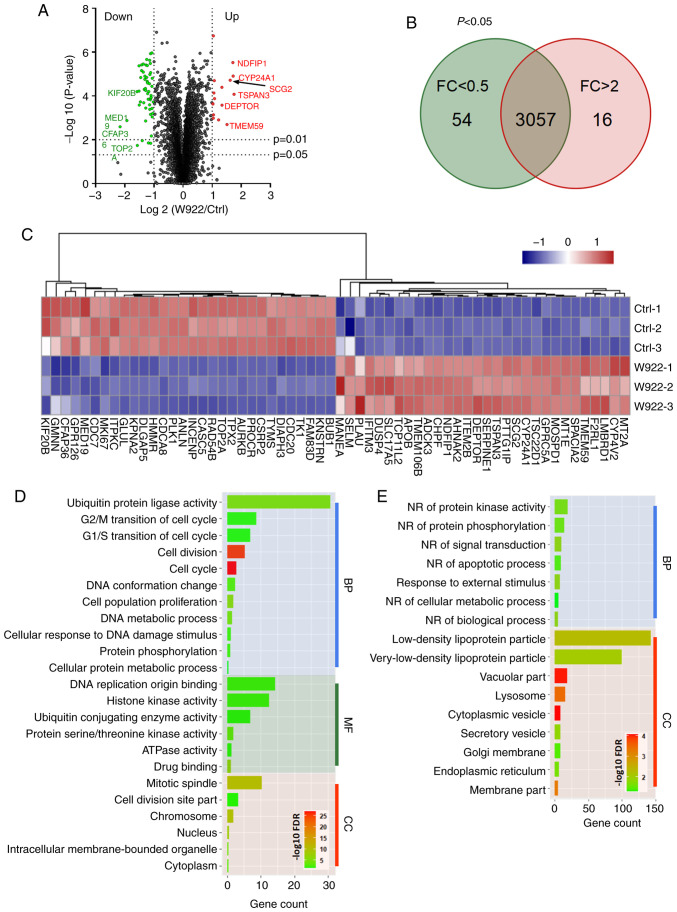
In order to investigate the mechanism via which W922 regulates tumor formation, MS analysis was performed. HCT116 cells were treated with W922 at 10 µM for 24 h, and >7,000 proteins were identified and quantified in all the replicates of treated and untreated cells (Fig. 2A). Among the common proteins in W922-treated and the control cells, 54 proteins were significantly downregulated (fold change <0.5) and 16 proteins were significantly upregulated (fold change >2) after W922 treatment (Fig. 2B). The top 30 up- and downregulated proteins are exhibited in Fig. 2C. Subsequently, all the significantly down- or upregulated proteins were classified using STRING online tool. W922 exhibited evident inhibitory effect on cell cycle regulation. As presented in Fig. 2C, >20 downregulated proteins, such as CDC20, CDC7, CDCA8, polo like kinase 1 and TPX2, caused by W922 treatment were associated with cell cycle regulation. Besides, the cell cycle markers in G1/S and G2/M transitions were found to be downregulated (Fig. 2D). W922 negatively regulated the signal transduction and cell biological process. Moreover, upregulation of lysosome-related proteins were identified after W922 treatment (Fig. 2E). These results verified the function of W922 as a signal transduction inhibitor, and it was suggested that the inhibitory effects on tumor cells may be connected with cell cycle arrest.
Figure 2.
W922 negatively regulates the cell cycle processes. (A) Volcano plot for protein abundances in HCT116 cells after W922 treatment. Red and green color points indicate significant up (FC >2)- and downregulated (FC <0.5) proteins, respectively. The lines represent P-values of 0.01 and 0.05 in two-tailed unpaired t-test. (B) Venn plot for proteins (P<0.05) in HCT116 cells after W922 treatment. (C) Heatmap of top 30 up (right)- and downregulated (left) proteins in W922 treatment. (D) Gene Ontology term enrichment of all significant downregulated proteins (FC <0.5; P<0.05) in W922 treated HCT116 cells. (E) Gene Ontology term enrichment of all significant upregulated proteins (FC >2; P<0.05) in W922 treated HCT116 cells. (n=3-5, unpaired Student's t-test). FC, fold change; Ctrl, control; BP, Biological process; MF, molecular function; CC, Cellular component; NR, negative regulation.
W922 inhibits HCT116 cell proliferation mainly via cell cycle arrest
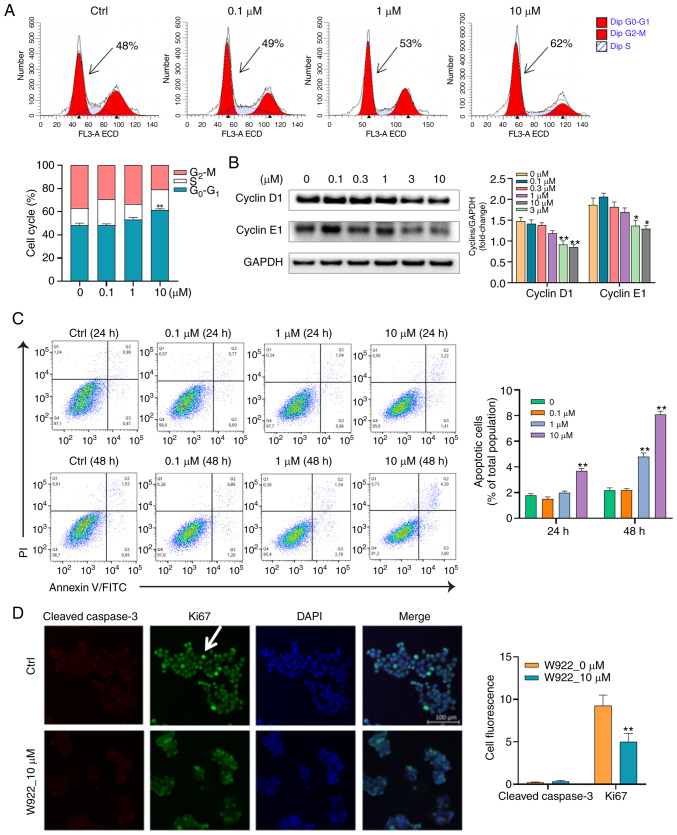
To further validate the proteomic results showing that W922 regulated proteins are cell cycle-related, flow cytometry was conducted to analyze the cell cycle distribution in W922-treated HCT116 cells. W922 led to G0-G1 phase accumulation in a concentration-dependent manner (Fig. 3A). To evaluate the molecular mechanism of the cell cycle arrest, immunoblot analysis was used to investigate the expression of cell cycle regulatory proteins. Cyclins are among the most important core cell cycle regulators. For instance, downregulation of cyclin D1 and E1 expression levels could block the cell cycle at G0-G1 stage and stop cell progression turning into S phase (31). It was identified that the protein expression levels of cyclin D1 and E1 were significantly concentration- dependently decreased in W922-treated cells compared with those in untreated cells (Fig. 3B).
Figure 3.
W922 inhibits HCT116 cell proliferation mainly by cell cycle arrest. (A) HCT116 cells were treated with W922 for 24 h, then the DNA contents were stained using PI and were analyzed via flow cytometry. The percentage of cells at G0-G1, S and G2-M phases are presented. (n=3; G0-G1 phase was analyzed via one-way ANOVA followed by Dunnett's test). **P<0.01 vs. 0 µM treatment. (B) HCT116 cells were treated with W922 for 24 h and the cell lysates were immunoblotted with antibodies against cyclin D1 and cyclin E1. (n=3; one-way ANOVA followed by Dunnett's test). *P<0.05, **P<0.01 vs. corresponding 0 µM. (C) Cells were treated with W922 for 24 or 48 h, and then were double stained with Annexin V-FITC and PI. Data were collected via flow cytometry (n=3; one-way ANOVA followed by Dunnett's test). **P<0.01 vs. 0 µM of each time point. (D) HCT116 cells were treated with either DMSO (Ctrl) or 10 µM W922 for 48 h. The expression levels of cleaved caspase-3 (red) and ki67 (green) were visualized via cell immunofluorescence (magnification, ×20) and were quantified using ImageJ (n=3; unpaired Student's t-test). **P<0.01 vs. 0 µM treatment. Ctrl, control.
Considering that apoptosis is the major form of cell death induced by chemotherapeutic agents, it was examined whether the anti-CRC effect of W922 was apoptosis dependent. Therefore, apoptosis of W922-induced HCT116 cells was measured using Annexin V/PI double staining method. Following the treatment of W922 (10 µM), the proportions of apoptotic HCT116 cells significantly increased by up to 2 and 6% in 24 and 48 h-treatment, respectively (Fig. 3C). Subsequently, immunofluorescence was performed in order to detect the different expression levels of ki67 and cleaved caspase-3 between W922-treated and untreated cells. The ki67 protein is a marker of proliferation and the expression of cleaved caspase-3 reflects the apoptotic level (24). It was found that the fluorescence intensity of ki67 in treated cells was significantly lower compared with that in untreated cells; however, no cleaved caspase-3 was identified in treated HCT116 cells (Fig. 3D). These results indicated that W922 inhibited HCT116 cell proliferation mainly by inducing cell cycle arrest rather than apoptosis.
W922 suppresses the PAM signaling pathway
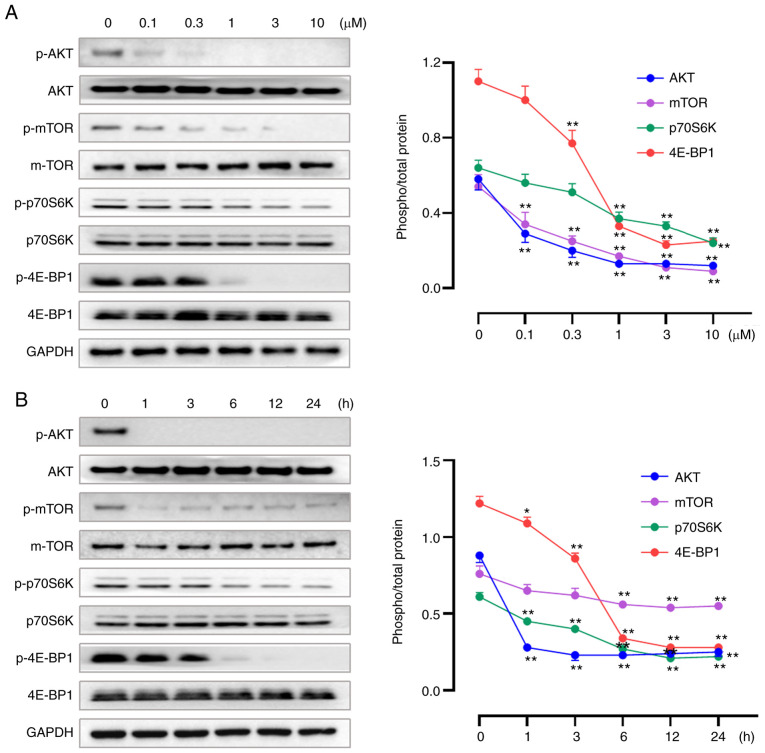
Our previous study reported W922 as an inhibitor against PI3K and mTOR enzymes, demonstrating the selective inhibition of W922 against PI3Kα extracellularly, with an IC50 of 15.1 nM. Moreover, potent activities of W922 against PI3Kβ, PI3Kγ, PI3Kδ and mTOR were revealed (21). To verify the inhibition of W922 on PAM pathway, western blotting assays were performed to determine the expression levels of PAM-related proteins. W922 suppressed the expression levels of p-AKT, p-mTOR, p-p70S6K and p-4E-BP1 in time- and concentration-dependent manners, while no alteration was found in the corresponding total proteins (Fig. 4). The proteomics results also indicated that W922 has a negative effect on the regulation of protein phosphorylation (Fig. 2E).
Figure 4.
W922 suppresses the PI3K/Akt/mTOR signaling pathway. HCT116 cells were treated with (A) gradient concentrations of W922 for 24 h, or with (B) W922 at 10 µM for gradient time periods. Then, the cells were collected and lysed. Cell lysates were immunoblotted with antibodies against PI3K/Akt/mTOR-related total and p- proteins. (n=3; one-way ANOVA followed by Dunnett's test). *P<0.05, **P<0.01 vs. 0 µM or 0 h treatment. P-, phosphory-lated; p70S6K, p70S6 kinase; 4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1.
W922 induces autophagy in HCT116 cells
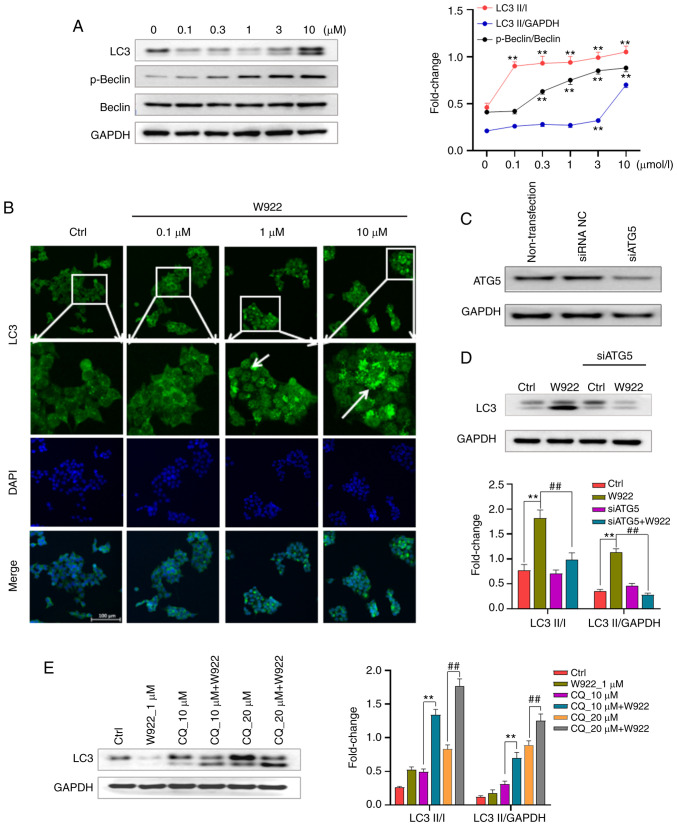
The PAM pathway is a well-established pathway that acts as a key negative regulator of autophagy (14). Since W922 downregulates the phosphorylation of mTOR, it was investigated whether W922 could induce autophagy in HCT116 cells. The results demonstrated that W922 treatment caused the induction of autophagy, as evidenced by concentration-dependent increase of LC3-I to LC3-II conversion (Fig. 5A). The phosphorylation of Beclin, an autophagy marker, was examined after W922 treatment, and it was identified that Beclin was significantly phosphorylated (Fig. 5A). Next, HCT116 cells were stained with LC3 antibody and visualized via immunofluorescence. Representative images indicated a weak staining in the untreated cells, while a moderate to strong intensity of LC3 fluorescence was observed in W922 treated groups (Fig. 5B).
Figure 5.
W922 induces autophagy in HCT116 cells. (A) HCT116 cells were treated with desired concentrations of W922 for 24 h. Collected cells were lysed and subsequently the lysates were immunoblotted with anti-LC3, anti-p-Beclin or anti-Beclin antibodies (n=3; one-way ANOVA followed by Dunnett's test). **P<0.01 vs. 0 µM treatment. (B) HCT116 cells were treated with W922 for 24 h, fixed and stained with anti-LC3 antibody (green) and with DAPI (blue), magnification, ×20. Quantification on the right showed LC3 fluorescence determined by ImageJ (n=3). **P<0.01 vs. Ctrl. (C) Transfection efficacy detection. HCT116 cells were transfected with siATG5 or siRNA NC, and the expression of ATG5 was detected. (D) HCT116 cells were successfully transfected with siATG5 and the expression of LC3 after W922 treatment was measured via western blotting (n=3; one-way ANOVA followed by Dunnett's test). **P<0.01 vs. Ctrl; ##P<0.01 vs. W922 group. (E) Autophagic flux was examined via co-treatment of CQ, an autophagosome-lysosome fusion inhibitor. HCT116 cells were treated with W922 and/or CQ for 24 h and the expression of LC3 was determined via western blotting (n=3; unpaired Student's t-test). **P<0.01 vs. 10 µM CQ treatment; ##P<0.01 vs. 20 µM CQ treatment. CQ, chloroquine; ATG5, autophagy related 5; Ctrl, control; siRNA, small interfering RNA.
To further investigate the autophagy induction by W922, the autophagy signaling was blocked via silencing ATG5. The results suggested that the conversion of LC3-I to II induced by W922 was inhibited in ATG5-silenced cells (Fig. 5C).
It remains difficult to distinguish autophagy inducers and inhibitors as both could increase the expression of LC3-II level (34). Therefore, in order to further investigate the role of W922 on autophagy, the impact on autophagic flux was evaluated via co-treatment with the autophagosome-lysosome fusion inhibitor, CQ. An enhanced LC3II/I ratio was observed in cells treated with W922 combined with CQ compared with W922 treatment alone (Fig. 5D and E). These results suggested that W922 functioned as an autophagy inducer in HCT116 cells.
Autophagy contributes to W922-induced cell cycle arrest
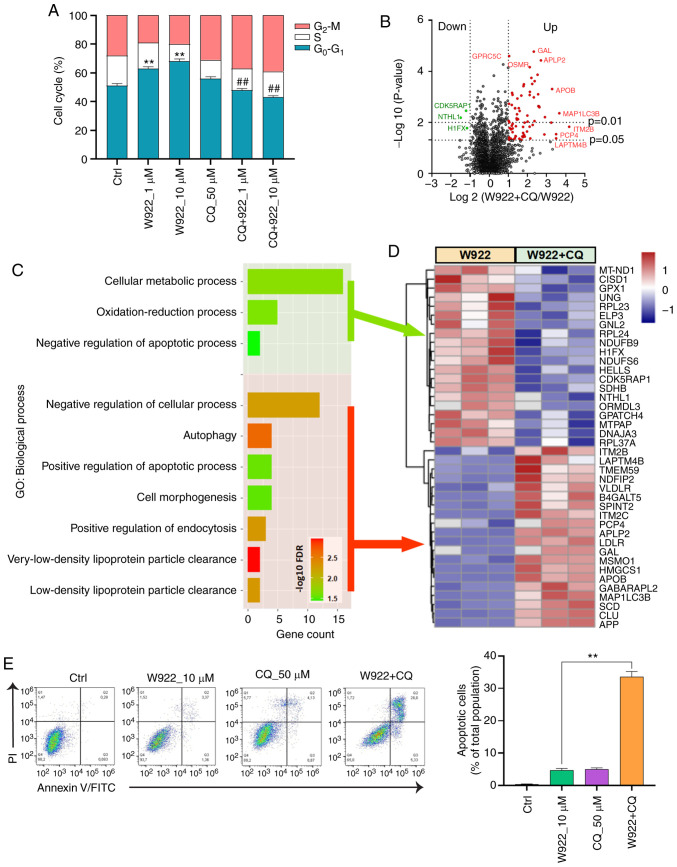
An autophagy inhibitor, CQ, was used to investigate whether W922-induced autophagy was involved in cell cycle arrest. As aforementioned, W922 treatment resulted in cell cycle arrest in the G0-G1 phase; however, such phenomenon was impaired by CQ co-treatment. CQ treatment alone did not demonstrate a significant impact on the cell cycle distribution of HCT116 cells, but CQ co-treatment significantly decreased >20% of G0-G1 phase proportion in W922-treatment cells (Fig. 6A).
Figure 6.
Autophagy contributes to W922-induced cell cycle arrest. (A) HCT116 cells were pretreated with 50 µM CQ before 24 h-W922 treatment, and were stained with PI and analyzed using flow cytometry (n=3; G0-G1 phase was analyzed by two-way ANOVA followed by Dunnett's test). **P<0.01 vs. Ctrl; ##P<0.01 vs. CQ single treatment. (B) Volcano plot for protein abundances in HCT116 cells pre-treated with 20 µM CQ before W922 treatment. (C) Gene Ontology term enrichment of top 20 up- and downregulated proteins in W922 and CQ co-treated HCT116 cells. (D) Heatmap of top20 up- (bottom) and downregulated (top) proteins in W922 and CQ co-treated HCT116 cells. (n=3-5, unpaired Student's t-test). (E) HCT116 cells were co-treated with 10 µM W922 and 50 µM CQ for 24 h, the apoptotic cells was detected via an Annexin V-FITC/PI double stain assay. (n=3; unpaired Student's t-test). **P<0.01 vs. 10 µM W922 treatment. CQ, chloroquine; Ctrl, control.
Next, the alterations of proteomics induced by CQ were detected via MS (Fig. 6B). Top 20 up- and downregulated proteins were sorted and classified using STRING analysis tool (Fig. 6D). CQ co-treatment led to a suppression in cellular metabolic process and oxidation-reduction process (Fig. 6C). Moreover, CQ enhanced the apoptosis of W922-treated cells. The data collected from flow cytometry assay also confirmed this result. Co-treatment of W922 with CQ led to ~30% apoptotic cells out of a total cellular population (Fig. 6E). These findings provide potential evidence for the combined treatment of W922 and CQ in CRC therapy.
Discussion
The present study demonstrated that the novel PAM pathway inhibitor, W922, inhibited tumor formation by causing cell cycle arrest. As aforementioned, W922 also induced autophagy in HCT116 cells. It was found that W922-induced autophagy contributed to W922-induced cell cycle arrest, since inhibition of autophagy by CQ impaired the cell cycle arrest. PAM is one of the pivotal transduction pathways regulating cellular basic activities (25,26). Thus, targeting this pathway may contribute to the tumor growth suppression. Our previous study developed a series of inhibitors against this cascade, and among which the most effective compound named W922 was selected. According to the previous enzymatic results, W922 was found to exhibit the strongest enzymatic inhibition against the PI3Kα enzyme compared with other synthesized compounds (21). In the current study, it was verified that W922 has robust ability to dephosphorylate PAM signaling proteins.
Inhibition of PI3K/Akt signaling has been suggested as potential therapeutic strategy for CRC. For example, previous studies have reported the association of the PI3K/Akt pathway with colon tumorigenesis (27,28). Moreover, the PAM pathway is well known as an important regulatory pathway of autophagy. PI3K/Akt regulates autophagy mainly via modulation of mTOR activity, and thus decreased mTOR activity by inhibitors could trigger autophagy (14). Autophagy is an evolutionarily highly conserved catabolic pathway, and the capacity of autophagy to maintain cellular metabolism improves the survival of cells (17). The autophagy pathway has a broad implication in numerous physiological and pathological processes, including carcinogenesis (29). Although upregulation and downregulation of autophagy have both been revealed to serve important roles in carcinogenesis, in general, autophagy is identified as a pro-survival mechanism in cancer cells by removing damaged organelles and recycling nutrients in response to chemotherapy stress (30). Therefore, autophagy is considered to be associated with drug resistance as it may protect cells from the cytotoxicity of chemotherapy drugs.
The present results suggested that W922 caused cell cycle arrest at the G0-G1 phase as a single agent; however, only slight apoptosis was observed after W922 treatment. The cell cycle represents a series of tightly integrated events that allow cells to proliferate (31). Therefore, cell cycle arrest contributes to the anti-tumor effect of W922. In response to stressful microenvironment, cells are able to arrest the cell cycle, which helps regulate cell proliferation and prevents the expansion of potential harmful cell populations (31,32). Cancer frequently represents a dysregulation of the cell cycle, and the cell cycle can be arrested under stress to protect the cells from DNA damage (33). Moreover, autophagy is induced in response to a variety of stress conditions, where it serves an essential role in protecting cellular viability (17). While the correlation between autophagy and cell cycle arrest has been widely studied, several signaling pathways exhibit opposite effects on the regulation of autophagy and cell cycle, and increasing evidence suggests that the opposite regulation may be rational and coordinated, since there is an interaction between the two processes (34,38,39).
The present study observed that W922 treatment led to cell cycle arrest and the upregulation of LC3-II. However, both autophagy inducers and inhibitors can cause an increase of LC3-II expression level (34). According to the guidelines for the use and interpretation of assays for monitoring autophagy, it is necessary to block lysosomal degradation of the protein to truly measure in vivo autophagic flux using LC3-II as a biomarker (40). Thus, co-treatment of CQ is necessary to examine whether W922 is an autophagy inducer or inhibitor. CQ is an autophagy inhibitor that can inhibit autophagosome degradation by blocking autophagic flux (34). If the LC3-II expression in the combination treatment is higher compared with that in W922 treatment alone, then W922 is an autophagy inducer. An enhanced LC3II/I ratio observed in combination treatment demonstrated the autophagy induction characteristic of W922. The cell cycle arrest induced by W922 was impaired when cells were co-treated with the autophagy inhibitor CQ, suggesting that autophagy may facilitate W922-induced cell cycle arrest. Therefore, the cell cycle arrest induced by W922 may be a protective action caused by the induction of autophagy.
Increasing preclinical evidence suggests that targeting autophagy can improve the efficacy of numerous cancer treatments (35). While various compounds have been developed to block the different stages of autophagy, the only clinically approved autophagy inhibitor is hydroxychloroquine (HCQ) (36). Preclinical trials have reported that HCQ can enhance tumor shrinkage either by single or combination treatment (36,37). Thus, there are multiple ongoing clinical trials combining HCQ with other chemo-drugs in cancer therapy. HCQ enhances the effect of temsirolimus on cytotoxicity and promotes apoptosis in renal cell carcinoma lines (37). The combination of HCQ and tamoxifen exhibits improved therapeutic efficacy compared with monotherapy in estrogen receptor-positive breast cancer cell lines (36). These findings suggest that combination therapy of a PAM signaling inhibitor with an autophagy inhibitor could be a novel method for cancer treatment. Therefore, the present study detected the combination therapy of W922 with autophagy blockers in vitro. The results demonstrated that CQ enhanced the inhibitory effects of W922 on cells, indicating that W922 and autophagy blockers promote mutual sensitization. These results suggested that W922 co-treatment with autophagy blockers may be a novel therapeutic strategy for CRC.
In conclusion, the present study first examine the function of a novel PAM inhibitor, W922, in the regulation of the proliferation in HCT116 CRC cells. W922 exerted its anti-tumor effects as a single agent mainly via the induction of cell cycle arrest. W922 also induced autophagy in HCT116 cells, and W922- induced cell cycle arrest was impaired when the cells were pre-treated with CQ, suggesting that autophagy may contribute to W922-induced cell cycle arrest. However, the aforementioned results were only obtained from in vitro experiments, and the same measurements should be collected in vivo to further verify the mechanism of the anti-cancer effects of W922. The current study demonstrated that simultaneously treating the cells with CQ led to an increase in the number of apoptotic cells. To verify the enhanced effect of W922 and autophagy inhibitor co-treatment, the same experiment should be performed in vivo in future studies. Collectively, the present results support the therapeutic potential of W922 in the treatment of CRC.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81670001) and Natural Science Basic Research Project of Shaanxi Province (grant no. 2018JQ8042).
Availability of data and materials
Data in this study are availabe from the corresponding author upon reasonable request.
Authors' contributions
JW performed most of the experiments, analyzed the data and was responsible for writing the manuscript. DL helped with the in vivo experiments and cell immunoflorescence assay. XPZ conducted the mass spectrometry technique. CFH participated in western blotting assay. LC participated in data analysis and manuscript revision. SQZ synthesized W922. XX analyzed data. YXC and XX provided the funding. SJL and YXC designed the experiment, revised the manuscript and gave final approval of the version to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were performed in accordance with the National Guidelines for Experimental Animal Welfare and with approval of the Ethics Committee of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Brody H. Colorectal cancer. Nature. 2015;521:S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 3.Stein A, Atanackovic D, Bokemeyer C. Current standards and new trends in the primary treatment of colorectal cancer. Eur J Cancer. 2011;47(Suppl 3):S312–S314. doi: 10.1016/S0959-8049(11)70183-6. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, II, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hara MH, Hamilton SR, O'Dwyer PJ. Molecular triage trials in colorectal cancer. Cancer J. 2016;22:218–222. doi: 10.1097/PPO.0000000000000199. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendell JC, Varghese AM, Hyman DM, Bauer TM, Pant S, Callies S, Lin J, Martinez R, Wickremsinhe E, Fink A, et al. A First-in-human phase 1 study of LY3023414, an oral PI3K/mTOR dual inhibitor, in patients with advanced cancer. Clin Cancer Res. 2018;24:3253–3262. doi: 10.1158/1078-0432.CCR-17-3421. [DOI] [PubMed] [Google Scholar]
- 9.Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745–1755. doi: 10.3748/wjg.v22.i5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Dienstmann R, Rodon J, Serra V, Tabernero J. Picking the point of inhibition: A comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 2014;13:1021–1031. doi: 10.1158/1535-7163.MCT-13-0639. [DOI] [PubMed] [Google Scholar]
- 12.Hart S, Novotny-Diermayr V, Goh KC, Williams M, Tan YC, Ong LC, Cheong A, Ng BK, Amalini C, Madan B, et al. VS-5584, a novel and highly selective PI3K/mTOR kinase inhibitor for the treatment of cancer. Mol Cancer Ther. 2013;12:151–161. doi: 10.1158/1535-7163.MCT-12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolev VN, Wright QG, Vidal CM, Ring JE, Shapiro IM, Ricono J, Weaver DT, Padval MV, Pachter JA, Xu Q. PI3K/mTOR dual inhibitor VS-5584 preferentially targets cancer stem cells. Cancer Res. 2015;75:446–455. doi: 10.1158/0008-5472.CAN-14-1223. [DOI] [PubMed] [Google Scholar]
- 14.Kim YC, Guan KL. mTOR: A pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 16.Parzych KR, Klionsky DJ. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 18.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33:610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsujimoto Y, Shimizu S. Another way to die: Autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):S1528–S1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 21.Wang HY, Shen Y, Zhang H, Hei YY, Zhao HY, Xin M, Lu SM, Zhang SQ. Discovery of 2-(aminopyrimidin-5-yl)- 4-(morpholin-4-yl)-6-substituted triazine as PI3K and BRAF dual inhibitor. Future Med Chem. 2018;10:2445–2455. doi: 10.4155/fmc-2018-0145. [DOI] [PubMed] [Google Scholar]
- 22.Singla P, Luxami V, Paul K. Triazine as a promising scaffold for its versatile biological behavior. Eur J Med Chem. 2015;102:39–57. doi: 10.1016/j.ejmech.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 23.McDonald WH, Yates JR., III Shotgun proteomics: Integrating technologies to answer biological questions. Curr Opin Mol Ther. 2003;5:302–309. [PubMed] [Google Scholar]
- 24.Silva MN, Leite JS, Mello MF, Silva KV, Corgozinho KB, de Souza HJ, Cunha SC, Ferreira AM. Histologic evaluation of Ki-67 and cleaved caspase-3 expression in feline mammary carcinoma. J Feline Med Surg. 2017;19:440–445. doi: 10.1177/1098612X16634150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandurangan AK. Potential targets for prevention of colorectal cancer: A focus on PI3K/Akt/mTOR and Wnt pathways. Asian Pac J Cancer Prev. 2013;14:2201–2205. doi: 10.7314/APJCP.2013.14.4.2201. [DOI] [PubMed] [Google Scholar]
- 26.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 27.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 28.Luo Q, Chen D, Fan X, Fu X, Ma T, Chen D. KRAS and PIK3CA bi-mutations predict a poor prognosis in colorectal cancer patients: A single-site report. Transl Oncol. 2020;13:100874. doi: 10.1016/j.tranon.2020.100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Zhang Y, Qu J, Xu L, Hou K, Zhang J, Qu X, Liu Y. β-Elemene-induced autophagy protects human gastric cancer cells from undergoing apoptosis. BMC Cancer. 2011;11:183. doi: 10.1186/1471-2407-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grana X, Reddy EP. Cell cycle control in mammalian cells: Role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 32.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 33.Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther. 2013;138:255–271. doi: 10.1016/j.pharmthera.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Zheng K, He Z, Kitazato K, Wang Y. Selective autophagy regulates cell cycle in cancer therapy. Theranostics. 2019;9:104–125. doi: 10.7150/thno.30308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook KL, Warri A, Soto-Pantoja DR, Clarke PA, Cruz MI, Zwart A, Clarke R. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin Cancer Res. 2014;20:3222–3232. doi: 10.1158/1078-0432.CCR-13-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HO, Mustafa A, Hudes GR, Kruger WD. Hydroxychloroquine destabilizes phospho-S6 in human renal carcinoma cells. PLoS One. 2015;10:e131464. doi: 10.1371/journal.pone.0131464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neufeld TP. Autophagy and cell growth-the yin and yang of nutrient responses. J Cell Sci. 2012;125:2359–2368. doi: 10.1242/jcs.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathiassen SG, De Zio D, Cecconi F. Autophagy and the cell cycle: A complex landscape. Front Oncol. 2017;7:51. doi: 10.3389/fonc.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data in this study are availabe from the corresponding author upon reasonable request.