Abstract
Despite advances in cancer therapeutics and the integration of personalized medicine, the development of chemoresistance in many patients remains a significant contributing factor to cancer mortality. Upon treatment with chemotherapeutics, the disruption of homeostasis in cancer cells triggers the adaptive response which has emerged as a key resistance mechanism. In this review, we summarize the mechanistic studies investigating the three major components of the adaptive response, autophagy, endoplasmic reticulum (ER) stress signaling, and senescence, in response to cancer chemotherapy. We will discuss the development of potential cancer therapeutic strategies in the context of these adaptive resistance mechanisms, with the goal of stimulating research that may facilitate the development of effective cancer therapy.
Keywords: Cancer, adaptive response, chemoresistance, chemotherapy, autophagy, ER stress signaling, senescence
Introduction
The high prevalence and mortality rate of cancer is a major burden to human health worldwide1. Unfortunately, despite extensive efforts and advances in cancer research, only a slight decrease in the cancer death rate has been observed2. The limited efficacy of chemotherapy, which is one of the principal modes of cancer treatment, is considered to be a major hindrance to our ability to effectively treat and manage the disease. In order to improve current cancer therapies and to develop novel treatment strategies, a better understanding of the mechanisms underlying the limitations of chemotherapy is urgently needed.
The factors contributing to the limited success of chemotherapy are complicated and multifactorial, and our inability to accurately predict how cancer patients will respond to drug treatment is significant. Recent technological advances have facilitated the molecular understanding of cancers and the identification of targets for therapeutic interventions3 via computational analysis. However, these methods are limited by intra-tumor heterogeneity, as characteristics of the major tumor cell type may not necessarily predict the features of mixed populations4. Furthermore, rare mutations in tumors are often undetected due to the limitations of sequencing technology. For example, sequencing at the initiation of treatment may fail to detect cancer cells harboring mutations that confer resistance to chemotherapeutics, such as mutations in KRAS. Over the course of treatment, selective pressure results in the expansion and proliferation of drug-resistant cells5. In addition, therapeutics may have differing levels of efficacy and toxicity in individuals with varied genetic backgrounds. For instance, mutations in TP53 have been shown to contribute to the risk of treatment failure in patients with relapsed childhood acute lymphoblastic leukemia6. These complications exemplify the need for individualized and tailored cancer treatment in order to maximize efficacy and minimize unwanted side effects. However, the field of personalized medicine is still under development, and a myriad of obstacles must be overcome before it can be applied in clinics7.
The development of drug resistance in cancer cells is arguably one of the most challenging factors limiting the success of chemotherapy. Chemoresistance can be broadly categorized into two types: (1) intrinsic resistance and (2) acquired resistance. The two groups differ in the origin of resistance: intrinsic resistance pre-exists within the cancer (cancer cells are resistant to initial treatments by chemotherapeutic agents) while acquired resistance emerges in response to treatment (resistance develops in cancer cells after initiation of treatment with chemotherapeutic agents). However, they share common mechanisms of resistance including reduced drug transport, altered drug targets, metabolic adaptations, dysregulation of DNA damage repair pathways, defective apoptotic signaling, activation of pro-survival signaling, pro-tumorigenic microenvironments, and cellular adaptive responses8.
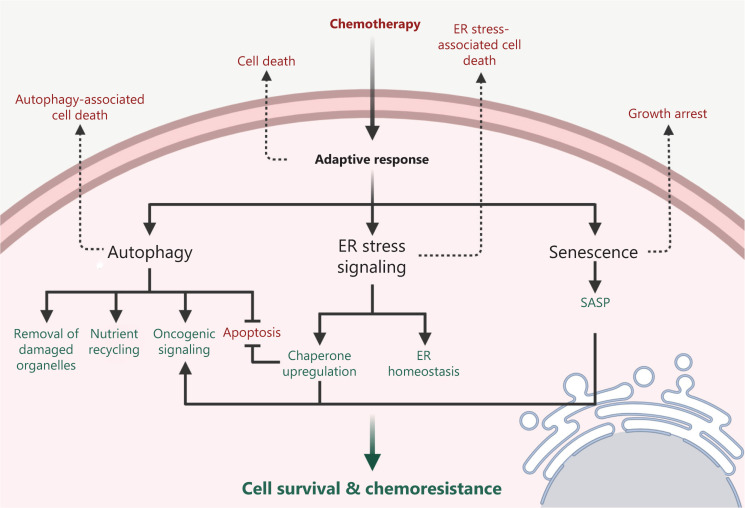
The cellular response to stress can lead to either the activation of cell death pathways or the adaptive response that maintains the survival of the cells. The adaptive response is the ability of a cell, tissue, or organism to better resist stress damage by prior exposure to a sublethal stress, including changes in temperature, oxygen tension, redox potential, extracellular signals, and chemical insults such as chemotherapeutic drugs9,10. During the adaptive process, cells undergo dramatic metabolic and physiological adaptations to prevent cellular damage and to maintain homeostasis. This is accomplished through the concerted action of diverse molecular signaling including autophagy, ER stress signaling, and senescence10. Accumulating evidence has revealed that these adaptive responses are crucial for tumorigenesis, tumor survival, and tumor progression11–13. This review will focus on the mechanisms by which autophagy, ER stress signaling, and senescence promote cell survival and contribute to the resistance in cancer cells exposed to targeted therapies (Table 1) and chemotherapeutic drugs (Figure 1).
Table 1.
List of clinical targeted therapeutic agents inducing an adaptive response in cancer cells
| Adaptive response | Drugs (generic name) | Trade name | Drug type | Target protein | Cancer | Reference |
|---|---|---|---|---|---|---|
| Autophagy | Afatinib | Gilotrif | Tyrosine kinase inhibitor | EGFR | NSCLC | 204 |
| Bevacizumab | Avastin | Monoclonal antibody | VEGF-A | CRC | 30 | |
| GBM | 205 | |||||
| Bortezomib | Velcade | Proteasome inhibitor | 26S proteasome | Breast cancer | 97,206 | |
| Cetuximab | Erbitux | Fv (variable, antigen-binding) regions of monoclonal antibody | EGFR | Lung cancer, CRC | 59 | |
| Dasatinib | Sprycel | Tyrosine kinase inhibitor | BCR/ABL, Src family | NSCLC | 207 | |
| Gefitinib | Iressa | Tyrosine kinase inhibitor | EGFR | Breast cancer | 65 | |
| Idelalisib | Zydelig | PI3K inhibitor | P110 delta | CML | 98 | |
| Lapatinib | Tykerb | Tyrosine kinase inhibitor | EGFR, HER2 | Breast cancer | 208 | |
| HCC | 209 | |||||
| Osimertinib | Tagrisso | Tyrosine kinase inhibitor | EGFR | NSCLC | 210 | |
| Sorafenib | Nexavar | Tyrosine kinase inhibitor | Raf, PDGF, VEGFR2/3, Kit | RCC | 211 | |
| Sunitinib | Sutent | Tyrosine kinase inhibitor | PDGFR, VEGFR, KIT | HCC | 212 | |
| mRCC | 213 | |||||
| PanNET | 214 | |||||
| Trametinib | Mekinist | MEK kinase inhibitor | MEK1/2 | Melanoma | 215 | |
| Leukemia | 216 | |||||
| Trastuzumab | Herceptin | Monoclonal antibody | HER2 | Breast cancer | 217–219 | |
| Vemurafenib | Zelboraf | Competitive kinase inhibitor | BRAF (V600E) | Melanoma | 220 | |
| Vismodegib | Erivedge | Cyclopamine-competitive antagonist | SMO | CML | 221 | |
| ER stress | Bortezomib | Velcade | Proteasome inhibitor | 26S proteasome | Breast cancer | 97 |
| Trastuzumab | Herceptin | Monoclonal antibody | HER2 | Breast cancer | 222 | |
| SASP | Sunitinib | Sutent | Tyrosine kinase inhibitor | PDGFR, VEGFR, KIT | Breast cancer | 193 |
EGFR, epidermal growth factor receptor; VEGF-A, vascular endothelial growth actor A; HER2, human epidermal growth factor receptor 2; MEK1/2, mitogen-activated protein kinase kinase 1/2; VEGFR, vascular endothelial growth factor receptor; SMO, smoothened; NSCLC, non-small cell lung cancer; CRC, colorectal cancer; GBM, glioblastoma; CML, chronic myeloid leukemia; HCC, hepatocellular carcinoma; mRCC, metastatic renal cell carcinoma; PanNET, pancreatic neuroendocrine tumors.
Figure 1.
Mechanisms of adaptive response induced by chemotherapy in the cancer cells. The administration of chemotherapy may lead to the disruption of cellular homeostasis followed by the activation of multiple signaling pathways including autophagy, endoplasmic reticulum (ER) stress signaling, and senescence. Cellular homeostasis may be restored by autophagy through the removal of damaged organelles and the recycling of nutrients, or by the induction of ER stress signaling through the recovery of ER homeostasis and the upregulation of chaperones. Some oncogenic signaling pathways can also be activated through the autophagy, ER stress signaling, or in the senescent cells with the senescence-associated secretory phenotype (SASP), thereby promoting cell survival and chemoresistance. On the contrary, depending on the type, intensity, and duration of the therapy-associated stress, adaptive response signaling may fail to be activated and eventually lead to cell death or growth arrest.
Autophagy
Macroautophagy (hereafter denoted as autophagy) is a dynamic process in which double-membrane vesicles, or autophagosomes, are formed to sequester cytoplasm or organelles. Autophagosomes are then targeted to lysosomes where the autophagosomal cargo is degraded and recycled for the needs of the cell. Autophagy is an important mechanism for maintaining intracellular homeostasis. Unfolded proteins or dysfunctional mitochondria can be eliminated through selective autophagy, thereby preventing the excessive production of reactive oxygen species (ROS), which cause genome instability and elicit tumorigenesis. In addition, autophagy is considered a distinct type of programmed cell death (type II programmed cell death).
Due to its involvement in cell death, autophagy may be a tumor-suppressive mechanism. Indeed, the monoallelic deletion of BECN1 (encoding Beclin1), a gene essential for autophagy, is observed in 40%–75% of human ovarian, breast, and prostate cancer tissues14. The heterozygous disruption of BECN1 has also been shown to promote tumorigenesis in a mouse model14. However, there is evidence demonstrating that autophagy actually supports tumorigenesis in some settings and may promote tumor growth and cancer cell survival in established tumors15,16. The tumor-promoting activity of autophagy may partly come from its ability to restore nutritional and oxidative homeostasis under stress conditions including hypoxia, tumor acidosis, extracellular matrix detachment, and oncogene-induced transformation. Importantly, several studies have demonstrated that inhibition of autophagy may be a therapeutic strategy for cancer patients of certain stages16,17.
The paradoxical involvement of autophagy in both tumor suppression and progression is also in line with its complex role in the cellular response to chemotherapy. Upregulation of autophagy has been found in drug-resistant cells and has been shown to be a protective mechanism against therapeutic stress18 (Table 2). Alternatively, enhancing autophagy could potentially lead to autophagy-associated cell death, synergizing with chemotherapy to suppress tumor growth. As a context-dependent mediator of chemotherapeutic responses, the role of autophagy is influenced by different factors such as the tumor stroma and oncogenic signaling in cancer cells, as detailed below.
Table 2.
List of clinical chemotherapeutic drugs inducing autophagy in cancer cells
| Drug (generic name) | Trade name | Drug type | Cancer | Reference |
|---|---|---|---|---|
| 5-Fluorouracil | Adrucil | Antimetabolite | CRC | 83,223–225 |
| HCC | 82 | |||
| Asparaginase | Elspar/Kidrolase | Enzyme | ALL | 226 |
| Cisplatin | Platinol | Alkylating agent | Ovarian cancer | 227 |
| Esophageal cancer | 228 | |||
| Desmoid tumors | 229 | |||
| NSCLC | 230 | |||
| HGSOC | 231 | |||
| Dexamethasone | Decadron | Glucocorticosteroid | Lymphoid malignancy | 232 |
| Docetaxel | Taxotere | Plant alkaloid/taxane/antimicrotubule agent | Prostate cancer | 233 |
| Doxorubicin | Adriamycin/Rubex | Anthracycline antibiotic | Leukemia | 234 |
| Multiple myeloma | 235 | |||
| Melanoma | 236 | |||
| CRPC | 237 | |||
| Enzalutamide | Xtandi | Anti-androgen | Prostate cancer | 238 |
| Epirubicin | Ellence | Anthracycline antibiotic | TNBC | 239 |
| Breast cancer | 240 | |||
| HCC | 241 | |||
| Etoposide | Etopophos | Anthracycline antibiotic | SCLC | 242 |
| Gemcitabine | Gemzar | Antimetabolite | Pancreatic cancer | 243 |
| Irinotecan (CPT-11) | Camptosar | Plant alkaloid/topoisomerase I inhibitor | CRC | 244 |
| Oxaliplatin | Eloxatin | Alkylating agent | CRC | 30,245 |
| HCC | 82 | |||
| Paclitaxel | Taxol/Onxal | Plant alkaloid/taxane/antimicrotubule agent | Cervical cancer | 246 |
| Ovarian cancer | 247 | |||
| Breast cancer | 248 | |||
| Pemetrexed | Alimta | Antimetabolite | HCC | 249 |
| Vincristine | Oncovin | Plant alkaloid | Leukemia | 234 |
CRC, colorectal cancer; HCC, hepatocellular carcinoma; ALL, acute lymphocytic leukemia; NSCLC, non-small cell lung cancer; CRPC, castration-resistant prostate cancer; TNBC, triple negative breast cancer; SCLC, small cell lung cancer.
Autophagy under the influence of the tumor stroma
Endothelial cells
The tumor stroma includes (i) the extracellular matrix, (ii) mesenchymal cells such as fibroblast and myofibroblast/cancer-associated fibroblasts (CAFs), (iii) blood and lymphoid vessels, and (iv) nerve and inflammatory cells. It is a complex, three-dimensional compartment that surrounds the parenchyma and influences tumor growth, metastasis, and therapeutic responses19,20. Anti-angiogenic cancer therapies directly target the tumor stroma by destroying the tumor vasculature, thereby depriving the tumor of oxygen and nutrients21. Although preclinical and clinical trials afford demonstrable efficacy of anti-angiogenic therapy, the benefits are at best transitory and are followed by a restoration of tumor growth and progression22. Multiple resistance mechanisms to anti-angiogenic therapy have been proposed and recent studies have indicated that the activation of autophagy is one of them23,24. Hypoxia, resulting from the anti-angiogenic therapy-mediated devascularization of tumors, was shown to activate autophagy via pathways controlled by hypoxia-inducible factor 1α (HIF-1α) and adenosine monophosphate (AMP)-activated protein kinase (AMPK)24. HIF-1α activation led to the expression of genes encoding the Bcl-2 homology 3 (BH3)-only proteins BCL2/adenovirus E1B 19-kDa-interacting protein 3 (BNIP3) and BNIP3-like (BNIP3L). The atypical BH3 domains of these proteins can induce autophagy through disruption of the Bcl-2–Beclin1 complex, thereby releasing Beclin1, a major autophagy activator25,26. AMPK has been shown to activate Unc-51-like kinase 1 (ULK1), a key initiator of autophagy, through phosphorylation and inhibition of mammalian target of rapamycin complex 1 (mTORC1) activity, leading to the induction of autophagy in cancer cells27.
Autophagy promotes tumor cell survival under anti-angiogenic treatment by clearing damaged organelles, reducing oxidative metabolism, and providing nutrients when blood perfusion is limited25,26. Preclinical studies have shown that autophagy-related genes were upregulated upon treatment with the vascular endothelial growth factor (VEGF)-neutralizing antibody bevacizumab in glioblastoma, hepatocarcinoma, and colon cancer28–30. In addition, in the glioblastoma xenograft model, in vivo targeting of the essential autophagy gene ATG7 resulted in tumor suppression when combined with bevacizumab28. Importantly, tumors from glioblastoma patients resistant to bevacizumab were shown to have increased regions of hypoxia and elevated levels of BNIP3 expression compared with the pretreatment specimens from the same patients28. Furthermore, the combination of the autophagy inhibitor chloroquine with bevacizumab significantly increased apoptosis of cancer cells under hypoxia, suggesting that the combinatorial treatment may be effective in curbing resistance to anti-angiogenic therapy29,30.
Interestingly, recent studies have reported that the induction of autophagy in both tumor and endothelial cells may negatively regulate angiogenesis31–34. While the exact mechanisms remain to be elucidated, some studies indicate that it may be in part due to the degradation of angiogenic factors through autophagy. For example, gastrin-releasing peptide (GRP) is a gut neuropeptide that promotes endothelial cell proliferation and stimulates angiogenesis in various cancers35. Following the induction of autophagy, enhanced degradation of GRP and subsequent inhibition of endothelial cell proliferation and tubule formation were observed32. However, it remains to be seen whether the inhibition of angiogenesis by treatment-induced autophagy will exacerbate hypoxic conditions of the tumor microenvironment, thereby establishing a vicious cycle that further increases chemoresistance.
Fibroblast
Fibroblasts that acquire activated phenotypes in response to the pro-fibrotic factors secreted by cancer cells, such as transforming growth factor-β (TGF-β), platelet-derived growth factor (PDGF), and fibroblast growth factor 2 (FGF2), are termed myoblasts or CAFs19. CAFs have been implicated in tumorigenesis, tumor progression, and chemoresistance in several cancers36,37. Their role and mechanisms in cancers have been recently reviewed38,39. The loss of caveolin-1 (Cav-1) in CAFs is associated with poor prognosis and tamoxifen resistance in human breast cancer patients40,41. Martinez-Outschoorn et al.42,43 demonstrated that breast cancer cells induced high oxidative stress and activated autophagy in CAFs, leading to their autophagic degradation of Cav-1 via HIF-1α and nuclear factor kappa B (NFκB) activation. Proteomic analysis revealed that the loss of Cav-1 in the stroma further elevated oxidative stress in CAFs and promoted their transformation44. CAFs have also recently been shown to increase the cell viability of tongue squamous cell carcinoma treated with cisplatin45. In addition, autophagic CAFs have been shown to contribute to chemoresistance by upregulating the expression of TP53-induced glycolysis regulatory phosphatase (TIGAR) in adjacent cancer cells46. TIGAR is a novel p53-inducible protein that has been shown to decrease intracellular ROS levels and reduce the sensitivity of the cell to p53 and other ROS-associated apoptotic signals induced by drugs43,46.
Autophagy under the influence of oncogenic signaling
Aberrant activation of signaling pathways contributes to tumorigenesis and tumor progression and mediates the cellular response to chemotherapy in many human malignancies. Several studies have demonstrated that cancer cells utilize autophagy to cope with oncogenic stress47. The links between autophagy and oncogenic signaling pathways, and how this interplay regulates chemoresistance in cancer cells, are gradually emerging. Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that is over-activated in most epithelial cancers due to EGFR mutations and/or overexpression48. Small-molecule receptor tyrosine kinase inhibitors (TKI), such as erlotinib and gefitinib, have been shown to be effective in blocking EGFR activity, especially in non-small cell lung cancers (NSCLCs) with an in-frame deletion in exon 19 or the single base substitution resulting in an L858R mutation49,50. However, approximately 30% of NSCLC patients demonstrated intrinsic resistance to TKI therapy, partly explained by the clonal MET amplification51,52. Those responding to therapy inevitably developed drug resistance via various mechanisms such as the T790M secondary mutation50. Recently, the non-genetic resistance through the activation of Aurora A and Aurora B has been reported to be associated with acquired resistance to EGFR TKIs53,54, and the treatment with Aurora kinase inhibitors could enhance and prolong the EGFR inhibitor response in preclinical models53,54. Studies have also indicated that autophagy is implicated in TKI resistance. In lung cancer cell lines with either wild-type or mutant EGFR, treatment with erlotinib and gefitinib induced autophagy, and the degree of induction was greater in resistant cells, suggesting that autophagy is involved in both innate and acquired resistance to EGFR-target therapy55–58. In addition, higher basal autophagy levels have been demonstrated in geftinib- resistant cell lines compared to parental cells57. Similarly, the EGFR-blocking antibody cetuximab has been shown to induce autophagy in lung cancer cell lines, promoting cell survival59,60. Treatment with TKIs and cetuximab was shown to induce autophagy through the inhibition of the class I phosphatidylinositol 3-kinase (PtdIns3K)/protein kinase B (Akt)/mTOR pathway as well as activating the class III PtdIns3K (hVps34)/Beclin1 autophagic pathway55,60. Of note, glioblastoma patients expressing EGFRvIII, an EGFR mutation variant associated with therapy resistance, were recently found to be particularly responsive to treatment with the autophagy inhibitor chloroquine61, suggesting the involvement of autophagy in the EGFRvIII+ tumors.
The mechanisms by which TKI inhibitors induce resistance by regulating autophagy are emerging. As mentioned already, TKI treatment can induce autophagy activation through its inhibition of the phosphatidylinositol-3-kinase (PI3K)/Akt/mTOR pathway. Some studies have also demonstrated that EGFR itself directly interacts with the core autophagy machinery, and the treatment of TKI may disrupt this interaction, leading to the activation of autophagy and cell survival. For example, Wei et al.62 demonstrated that active EGFR mediates Beclin1 phosphorylation, leading to its inhibition and decreased autophagy. TKI therapy, however, disrupts Beclin1 phosphorylation, restoring autophagy. Furthermore, K721A kinase-dead (KD) EGFR, which mimics the blocking of EGFR by TKIs, was able to cooperate with lysosomal-associated transmembrane protein 4B (LAPTM4B) and Sec5 at endosomes to cause the disassociation of the autophagic inhibitor Rubicon from Beclin1, thereby initiating autophagy63. These data suggest that EGFR inhibition may directly promote autophagy, and that co-targeting EGFR and autophagy may be a promising approach for overcoming chemoresistance under TKI treatment. Indeed, some preclinical studies have reported synergism between the autophagy inhibitors chloroquine or hydroxychloroquine and EGFR TKIs in NSCLC, breast cancer, and glioblastoma55,56,64,65.
Another common mutation that can be targeted therapeutically is the V600E mutation in the v-RAF murine sarcoma viral oncogene homolog B1 (BRAF), which constitutively activates the BRAF kinase, resulting in sustained extracellular signal-related kinase (ERK) signaling and increased tumor growth66. In melanoma cells with BRAFV600E, hyperactivation of ERK induced higher levels of autophagy67. In BRAFV600E-driven lung tumors in mice, autophagy was essential for mitochondria metabolism and tumor growth, and deletion of an essential autophagy gene prolonged survival compared to the wild-type control mice68. The defect in mitochondrial respiration in autophagy-deficient BRAFV600E cancer cells was shown to be rescued by the addition of exogenous glutamine, which suggests that autophagy promotes cancer survival in part through sustaining mitochondrial metabolism and by providing essential amino acids. This phenomenon is termed “autophagy addiction”69. Furthermore, the activation of autophagy may also promote drug resistance in cancer cells treated with the BRAF inhibitor (BRAFi) vemurafenib, as the biopsies from BRAFi-resistant melanoma patients exhibited increased levels of autophagy compared with baseline70. Importantly, both melanoma and brain tumor cells with BRAFV600E (but not wild type) displayed synergy when chloroquine was combined with vemurafenib, and tumor regression was observed in BRAFi-resistant xenografts and cancer patients, suggesting that combination therapies may delay the acquisition of resistance and improve patient outcomes70,71. Li et al.72 demonstrated that treatment with BRAFi induced autophagy via a transcriptional program coordinating lysosomal biogenesis and function mediated by transcription factor EB (TFEB), providing a clue to the mechanism of BRAFi-induced autophagy.
Kirsten rat sarcoma viral oncogene homolog (K-Ras) is an upstream regulator of BRAF and the mutant KRAS is associated with aggressive cancer phenotypes and poor patient prognosis73. The expression of KRASV12 has been shown to upregulate basal autophagy, which was required to maintain the pool of functional mitochondria needed to deal with the high levels of oxidative phosphorylation and ROS in K-Ras-driven tumors15,74. Ablation of autophagy was also shown to decrease K-Ras-mediated adhesion-independent transformation, proliferation, and cell survival in several KRASG12D-driven cancers, such as pancreatic, breast, and lung cancer cells15,74–77.
Autophagy and non-coding RNA
The advancements in genomic technology have altered our perception of non-coding RNAs (ncRNAs) from non-functional (sometimes referred to as junk) to regulatory molecules that modulate cellular processes and play important roles in diseases like cancer78,79. ncRNA species include microRNAs (miRNAs), circular RNAs (circRNAs), and long ncRNAs (lncRNAs)79. Recent studies have demonstrated that lncRNAs may influence the sensitivity of cancer cells to chemotherapy via autophagy80,81. For example, both lncRNAs HULC (highly upregulated in liver cancer) and H19 were found to be associated with 5-fluorouracil resistance via their modulation of autophagy regulator sirtuin 1 (SIRT1)82,83. Their expression levels were found to increase in liver and colorectal cancers, respectively, and were found to be associated with cancer recurrence83. Similarly, gallbladder cancer drug resistance-associated lncRNA1 (GBCDRlnc1) was found to be upregulated in both gallbladder cancer tissues and doxorubicin-resistant gallbladder cancer cells. Knockdown of GBCDRlnc1 may increase the sensitivity of Dox-resistant cancer cells via inhibiting autophagy84. Importantly, lncRNAs have also been found to increase chemoresistance by modulating the expression of miRNAs and thus the downstream autophagic signaling. For instance, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a ncRNA known to be a regulator of metastasis in NSCLC85, has recently been shown to promote autophagy and increase chemoresistance by miR-124 downregulation and miR-23b-3p sequestration86,87. On the other hand, lncRNAs have also been observed to reduce chemoresistance in cancer cells by suppressing autophagy. For instance, lncRNA growth arrest-specific 5 (GAS5) was found to increase the sensitivity to cisplatin in glioma cells by activating mTOR signaling, thereby inhibiting autophagy88.
Taken together, these studies suggest an emerging role of lncRNA in autophagy regulation and chemoresistance in cancer cells. The lncRNA profiles of cancer patients should be taken into consideration when designing therapeutic strategies, especially via autophagy inhibition.
ER stress signaling
The ER is a perinuclear, cytosolic organelle which is required for cell survival and normal cellular function. The functions of ER include intracellular calcium homeostasis, lipid biosynthesis and protein folding, modification, and secretion. Disturbances, such as nutrient deprivation, hypoxia, ER Ca2+ depletion, and oxidative stress can impair glycosylation or protein disulfide bond formation, leading to the accumulation of unfolded proteins in the ER, triggering an evolutionarily conserved protein quality control mechanism termed unfolded protein response (UPR) or ER stress signaling pathways89. Canonical UPR signaling includes three pathways, each mediated by a different ER stress sensor: protein kinase RNA-like ER kinase (PERK), inositol-requiring kinase 1α (IRE1α), and the activating transcription factor 6 (ATF6). Glucose-regulated protein, 78 kDa (GRP78), a major ER chaperone, acts as a master regulator of the UPR through direct binding to all three sensors, keeping them in an inactive form. Upon ER stress, GRP78 preferentially binds the accumulating misfolded proteins, leading to the activation of the three sensors and the transduction of UPR signals across the ER membrane to the cytosol and ultimately the nucleus90. Triggering UPR can lead to cell survival or cell death, depending on the cellular context, the intensity of stress, and the length of exposure. The function of UPR is to restore ER homeostasis, mainly by suppressing global mRNA translation via PERK-mediated phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), leading to reduced influx of new proteins into the ER. As the stress continues, adaptive mechanisms become activated. The adaptive response primarily involves the activation of autophagy and transcriptional programs that induce expression of genes for enhancing ER protein folding and ER-assisted degradation (ERAD). ERAD facilitates removal of the unfolded proteins in the ER and export to the cytosol for degradation91. However, when adaptive mechanisms fail, cell death pathways will eventually be induced, eliminating cells beyond repair89,90,92.
Rapidly proliferating cancer cells are often subject to ER stress. This is because (1) fast-growing cells place higher demand on ER activity for protein production93; (2) inadequate vascularization of tumors leads to hypoxia and nutrition deprivation, which results in inadequate protein glycosylation and ATP production required for proper protein folding94,95; and (3) the hypoxic tumor microenvironment leads to disturbances in cellular redox regulation in which oxidizing or reducing agents lead to improper protein folding and the activation of UPR pathways. Indeed, the expression of ER stress markers has been shown to increase in many cancers and has been associated with aggressive phenotypes and therapeutic resistance96–99. Furthermore, ER stress was found to increase after treatment with chemotherapeutic drugs in various cancers100,101 (Table 3).
Table 3.
List of clinical chemotherapeutic drugs inducing ER stress in cancer cells
| Drugs (generic name) | Trade name | Drugs type | Cancer | Reference |
|---|---|---|---|---|
| Cisplatin | Platinol | Alkylating agent | CRC | 100 |
| HCC | 250 | |||
| Doxorubicin | Ariamycin PFS/Adriamycin RDF/Rubex | Anthracycline antibiotic | Breast cancer | 251 |
| Epirubicin | Ellence | Anthracycline antibiotic | Breast cancer | 251 |
| Gemcitabine | Gemzar | Antimetabolite | PDAC | 252 |
| Ixabepilone | Ixempra | Plant alkaloid/eopthilones/antimicrotubule agent | RCC | 253 |
| Paclitaxel | Taxol | Plant alkaloid/taxane/antimicrotubule agent | Breast cancer | 254–256 |
| Melanoma | 257 | |||
| Vinblastine | Velban | Plant alkaloid | Breast cancer | 254 |
CRC, colorectal cancer; HCC, hepatocellular carcinoma; PDAC, pancreatic ductal carcinoma; RCC, renal cell carcinoma.
UPR and chaperones in chemoresistance
Several mechanisms have been proposed to account for UPR-mediated therapeutic resistance. For example, some pro-survival proteins were found to be induced by UPR signaling. Wroblewski et al.102 demonstrated that treatment with the BH3 mimetics Obatoclax and ABT-737 induced anti-apoptotic Bcl-2 family proteins Mcl-2 via UPR in melanoma. The drug resistance-related protein prion was also reported to be regulated through UPR pathways in breast cancer103.
A major resistance mechanism generated by UPR is through the upregulation of catalysts that accelerate protein folding and molecular chaperones that inhibit protein aggregation under ER stress. These proteins facilitate protein folding, thereby attenuating ER stress and preventing the induction of apoptosis104. Downregulation of the ER stress chaperones by small interfering RNA (siRNA) has been shown to increase apoptosis induced by chemotherapeutic drugs105. In addition to their function in restoring ER homeostasis, chaperone proteins have also been implicated in several mechanisms of chemoresistance104,106,107. For example, protein disulfide isomerase (PDI) is an enzyme that catalyzes disulfide bond formation and isomerization, and is a chaperone facilitating proper folding of many secretory proteins108. Inhibition of PDI has been shown to increase apoptosis in response to ER stress109. In addition, the overexpression of PDIA4 and PDIA6 was found in cisplatin-resistant NSCLC cell lines as well as biopsies from lung cancer patients110. The inhibition of PDI4 resulted in the reactivation of the classical mitochondrial pathway, while downregulation of PDIA6 led to a non-canonical cell death pathway with some necroptotic features110.
Another major family of chaperones involved in chemoresistance is heat shock proteins (HSPs). HSPs are induced by cell stress and are known to facilitate tumor progression via controlling the stability and function of their target proteins104,111,112. HSP90 has an indispensable role in regulating mitogenesis and cell cycle progression helping to stabilize fragile structures of receptors, protein kinases, and transcription factors necessary for cell growth104,113. For example, HSP90 has been shown to protect the androgen receptor in castration-resistant prostate cancer (CRPC) cells from degradation, and to stabilize multiple components involved in the development and/or maintenance of CRPC including Akt, receptor tyrosine-protein kinase erbB-2 (ERBB2), and cyclin-dependent kinases (CDKs). As a result, blocking HSP90 suppressed the growth and survival signaling of resistant cells, leading to apoptosis and cell cycle arrest114. Moreover, the combination of HSP90 inhibitors with other chemotherapeutic drugs has been shown to generate synergistic effects in overcoming chemoresistance both in vitro and in vivo115,116. In a study on mantle cell lymphoma, the combination of the HSP90 inhibitor IPI-504 and bortezomib, an inhibitor of the 26S proteasome, overcame bortezomib resistance by inhibiting UPR and promoting the Noxa-mediated apoptotic pathway115. Importantly, in an NSCLC patient with crizotinib resistance, the HSP90 inhibitor ganetespib was able to induce marked tumor shrinkage after one cycle of monotherapy116.
HSP27, HSP47, and HSP70 are also involved in cancer drug resistance primarily through preventing apoptosis under cellular stress conditions111,117,118. These HSPs have been shown to inhibit proteolytic maturation of caspases, cleavage of their substrate, and apoptosome formation104,119–121. Recent studies have demonstrated that OGX-427, a second-generation antisense oligonucleotide, was able to inhibit HSP27 expression both in vitro and in xenograft mice in vivo122. OGX-427 has been shown to suppress tumor progression and enhance the efficacy of gemcitabine, a nucleoside analog chemotherapy, in pancreatic cancer in vitro and in vivo122. Interestingly, many HSPs play overlapping roles in sustaining tumor survival and inhibiting cell death pathways. Simultaneous inhibition of HSP90 and HSP27 has been shown to synergistically increase tumor suppression effects through enhanced apoptosis and ER stress123,124. The results of phase II clinical trials using OGX-427 alone for the treatment of CRPC, metastatic NSCLC, pancreatic cancer, and bladder cancer do not look promising, further suggesting that a combinatorial targeting of HSPs may be necessary for effective cancer treatment125–128.
GRP78 is a member of the HSP70 superfamily that plays a critical role in cell proliferation, survival, and angiogenesis of various cancers93. The induction of GRP78 under ER stress has been regarded as a substantial contributor to chemoresistance in cancer cells129. GRP78 has been shown to promote cell survival under therapeutic stress through various mechanisms including through the cytoprotective branches of UPR induction. The expression level of the gene encoding GRP78 is greatly induced under ER stress and like other molecular chaperones, GRP78 prevents the aggregation of misfolded proteins, which can cause toxicity and trigger apoptosis130. In addition, GRP78 upregulation has been shown to lead to the stress-dependent activation of p38 and PERK signaling, which promote survival and drug resistance in dormant carcinoma cells131. The activation of UPR signaling can also lead to ER stress-associated cell death and as a result, GRP78 induction of UPR is tightly controlled. For example, SPARC (secreted protein acidic and rich in cysteine) was identified as a GRP78-binding partner and was shown to interfere with the association between GRP78 and PERK and the activation of the downstream ER stress signaling in response to chemotherapy132. This suggests that a complicated protein network is involved and that further mechanistic studies are required to understand the ER stress-associated chemoresistance.
A subpopulation of GRP78 was found to bind to and inactivate pro-apoptotic proteins to prevent the induction of apoptosis. For example, GRP78 was shown to form a complex with caspase-7 or -12 at the ER surface, preventing their activation and release133,134. As capase-12 mediates ER stress-induced apoptosis, inactivation by GRP78 led to resistance of cancer cells to proteasome inhibitors and DNA-damaging agents135–137. Moreover, under stress conditions, elevated levels of GRP78 have been shown to suppress cell death signaling by sequestering B-cell lymphoma 2 (BCL-2) from binding with BCL-2-interacting killer (BIK), which is a pro-apoptotic BH3-only protein138.
Importantly, the induction of GRP78 by ER stress not only led to an increase in GRP78 in the ER compartment, but also promoted GRP78 relocalization to other cellular locations, including the cell surface, cytosol, mitochondria, nucleus, and the exterior of the cell through secretion139. Cell surface GRP78 (sGRP78) was found to act as a co-receptor mediating tumor cell signal transduction, mainly through Akt signaling, to promote cell survival and drug resistance140–143. For example, sGRP78 was demonstrated to initiate mitogen-activated protein kinase (MAPK) and AKT-dependent signaling and to downregulate the apoptotic pathway through the binding of active α2-macroglobulin (α2-M*), a serum proteinase inhibitor144,145. In prostate cancer, a serum protein complex composed of native α2-M and prostate-specific antigen was shown to bind GRP78, resulting in the activation of mitogen-activated protein kinase kinase 1/2 (MEK1/2), ERK1/2, S6K, and Akt pro-survival pathways and the increase of DNA and protein synthesis146,147. Additionally, sGRP78 was also shown to modulate T-cadherin signaling via Akt, promoting pro-survival effects in endothelial cells148. In addition, sGRP78 can form a complex with oncoprotein cripto to activate MAPK/PI3K signaling and promote tumor cell proliferation149. sGRP78 was demonstrated to couple with PI3K, facilitating PIP3 formation and the activation of PI3K/AKT signaling in breast and prostate cancer cells resistant to hormonal therapy150.
GRP78 translocation to other cellular location under stress may also promote drug resistance. For instance, mitochondrial GRP78 has been shown to stabilize Raf-1 on the outer membrane of mitochondria, thereby maintaining mitochondrial permeability and preventing the activation of apoptosis under stress151. Moreover, a variety of bortezomib-resistant solid tumor cell lines, but not the sensitive myeloma cell lines, were shown to secrete high amounts of GRP78. Secreted GRP78 induced pro-survival signaling by phosphorylation of ERK and suppressed p53-mediated expression of pro-apoptotic Bok and Noxa proteins, leading to bortezomib resistance in endothelial cells152. Notably, GRP78 was also induced in non-stressed cells by the treatment with a histone deacetylase (HDAC) inhibitor, which removes the transcription repression exerted by HDAC1 on the ER stress response elements of GRP78, leading to chemoresistance153,154.
UPR and chemoresistance in cancer stem cells (CSCs)
The significance of the stress response and molecular chaperones in stem cell oncogenesis is gradually emerging155,156. Several studies indicate that ER stress signaling is involved in the maintenance of stemness properties of CSCs, which is thought to be a mechanism of chemoresistance and tumor recurrence157–161. CSCs are a subpopulation of neoplastic cells within a tumor that have an increased ability to seed new tumors upon experimental implantation in appropriate animal hosts162. While most chemotherapeutic agents kill the bulk of tumors, CSCs have been shown to survive and proliferate after chemotherapy. For example, DNMT3A-mutant hematopoietic stem cells (HSCs) were reported to exist and expand in the remission samples of patients with acute myeloid leukemia163. Similarly, the pool of glioma stem cells has also been shown to expand over time under the exposure to therapeutic doses of temozolomide in both patient-derived and established glioma cell lines164.
Although CSCs have been connected with drug inefficiency for years, the exact molecular mechanisms of resistance caused by CSCs are not completely understood. The slow cycling characteristic of CSCs and the overexpression of drug transporters, anti-apoptotic proteins, and DNA damage enzymes only partially explain the entire resistance spectrum162,165. There is accumulating evidence showing that the stress response is crucial for sustaining stem-like properties in both normal and neoplastic stem cells, and that the inhibition of ER stress sensors sensitizes sphere-forming cells to apoptosis166. van Galen et al.167 proposed a model for how stress signaling is integrated within tissue hierarchies and how it coordinates with stemness in HSCs, revealing an indispensable role of UPR in sustaining HSC’s clonal integrity. In addition, the UPR modulator GRP78 has been shown to be necessary for the survival of embryonic stem cell precursors and was also highly expressed in HSCs168. Furthermore, the inducible knockout of GRP78 in the hematopoietic system resulted in a significant reduction of HSCs, common lymphoid and myeloid progenitors, and lymphoid cell populations in the mutant mice169. Consistent with these findings, UPR has been shown to play an important role in normal stem cells. For example, GRP78 anchored at the plasma membrane has been proposed as a surface marker for CSCs from colon cancer and head and neck squamous cell carcinoma (HNSCC)170,171. Knockdown of GRP78 markedly decreased the self-renewal ability and expression of stemness genes, but inversely promoted cell differentiation and apoptosis in CSCs from HNSCC170. Moreover, it has been reported that in breast CSCs, GRP78 mediates chemoresistance through β-catenin/ATP-binding cassette super-family G member 2 (ABCG2) signaling172. However, as most of the study focused on the association between GRP78 and CSCs, it would be of great importance to know how the individual UPR pathways participate in the regulation of CSC stemness and how it is coordinated with existing stem cell pathways to enable therapeutic targeting of CSCs and chemoresistance.
UPR and the epithelial-to-mesenchymal transition (EMT) in chemoresistance
Activation of the EMT program in carcinoma cells has been shown to give rise to cells with stem-like properties173,174. In human mammary epithelial cells, the induction of EMT resulted in the acquisition of mesenchymal traits and the expression of stem cell markers. Cells undergoing EMT were also shown to form mammospheres, soft agar colonies, and tumors more efficiently173. Indeed, EMT activation has been linked to increased anti-apoptotic ability and chemoresistance in various cancers175–179. Chemotherapeutic drugs such as cisplatin and doxorubicin have been shown to activate ER stress and subsequent EMT signaling pathways180. Interestingly, emerging evidence indicates that there is cross-talk between UPR and EMT activation. UPR induction was shown to potentiate EMT and vice versa181–183. Feng et al.184 demonstrated that cancer cells undergoing EMT had particularly high sensitivity to ER stress-induced death, suggesting that UPR is constitutively activated in EMT cells. Moreover, EMT gene expression strongly correlated with the PERK-eIF2α axis of UPR and the blocking of PERK signaling pathway interfered with the ability of EMT cells to invade and metastasize. These data suggest that UPR and EMT signaling are closely related and that interfering with the interplay between these two pathways is a potential strategy to suppress CSCs and tumor progression.
Senescence
Cellular senescence is a status where cells stably exit the cell cycle at the end of the cellular lifespan or in response to different stresses. Senescence is a heterogeneous phenotype. Depending on the type of stimulus, organism of origin, and cellular context, senescent cells may display various senescence markers which contribute to the phenotype to different extents185. For example, in addition to a lack of proliferation, senescent cells may show large and flat morphologies, higher senescence-associated β-galactosidase activity (SA-β-Gal), and the appearance of senescence-associated heterochromatin foci (SAHF), a facultative heterochromatin domain that contributes to the silencing of proliferation-promoting genes186,187. Although senescent cells remain arrested even when treated with growth factors, these cells are in fact metabolically active and were shown to exert non-cell-autonomous activities by secreting soluble signaling factors. The profound changes in the secretome of senescent cells are termed the senescence-associated secretory phenotype (SASP)185,188.
Although it has long been accepted that senescence is a tumor-suppressive mechanism that permanently arrests cells at risk for malignant transformation, accumulating evidence suggests that senescent cells can actually drive tumor progression. They do this through modulation of the tumor microenvironment by altering the secretion of interleukins, inflammatory cytokines, protease, and growth factors in SASP188. It has been shown that SASP enhances cell proliferation and motility, regulates tumor immunology, and promotes the emergence of the cancer-stem-like cells189. It has also been shown that after cyclic stimulus of senescence-inducing androgen deprivation, senescence-resistant, androgen-refractory cells were generated. These cells were characterized with notable chemoresistance and enhanced pro-survival mechanisms190. Several studies also demonstrated that the SASP is associated with therapeutic resistance in the current cancer treatment (Table 4). For example, SASP induced by cisplatin administration in melanoma cells may promote the proliferation of non-senescent cells through the activation of ERK1/2–ribosomal S6 kinase 1 (RSK1) pathway191. Hepatocellular carcinoma cells resistant to sorafenib were found to be associated with the SASP-related p16/IL6 axis192. Interestingly, Mastri et al.193 found a temporary cellular change similar to SASP after the withdrawal of anti-angiogenic therapy (SASP-mimicking anti-angiogenic therapy-induced secretome, ATIS) in cancer cells. The senescence hallmarks observed in these cells ultimately reversed after long drug withdrawal periods. This incomplete induction of the SASP phenomenon may explain the highly diverse treatment efficacy observed in patients receiving anti-angiogenic therapy193.
Table 4.
List of clinical chemotherapeutic drugs inducing SASP in cancer cells
| Drug (generic name) | Trade name | Drugs type | Cancer | Reference |
|---|---|---|---|---|
| Cisplatin | Platinol | Alkylating agent | Melanoma | 191 |
| Doxorubicin | Ariamycin PFS/Adriamycin RDF/Rubex | Anthracycline antibiotic | CRC | 258 |
| Gemcitabine | Gemzar | Antimetabolite | PDAC | 259 |
| Temozolomide | Temodar/Temodal/Temcad | Alkylating agent | Melanoma | 260 |
CRC, colorectal cancer; PDAC, pancreatic ductal carcinoma.
Importantly, resistance induced by the SASP was also observed in clinics. Malignant pleural mesothelioma (MPM) patients with upregulated senescence marker were shown to have a worse prognosis after receiving platinum-based therapy194. To explore the potential mechanism by which senescence induces chemoresistance, Canino et al.195 used MPM as a model and found that conditioned media from pemetrexed-treated senescent MPM cells induced the emergence of EMT-like, clonogenic, and chemoresistant cell subpopulations with high levels of aldehyde dehydrogenase (ALDH) activity. It was shown that these SASP-cytokine-induced chemoresistant cells could be targeted by STAT3 knockdown or HSP90 inhibition, resulting in a reduction of the population of high ADLH-expressing cells and EMT genes expression both in vitro and in vivo195,196. This suggests that SASP signaling-mediated pathways may be a potential target in anti-cancer therapy.
Conclusion
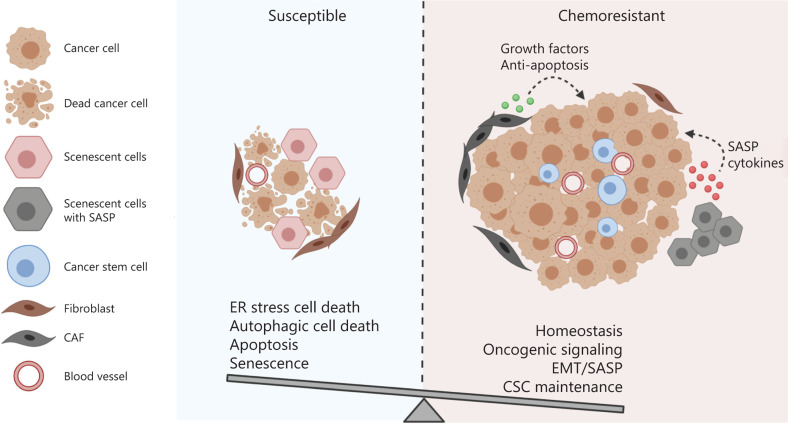
The adaptive response is essential for cell and organism survival of sublethal cellular damage and disruption of homeostasis. The transient and reversible adjustments in response to stress can be mediated by biochemical or post-translational mechanisms, or rely on alterations in gene expression197. The activation of autophagy and ER stress signaling have been shown to play important roles in the restoration of cellular homeostasis, while senescence may contribute to growth arrest, thereby extending the life span of an organism under stress. Nevertheless, as discussed already, these adaptive responses may in turn promote resistance to chemotherapy (Figure 2). One of the common characteristics of autophagy and ER stress signaling is that their activation can either lead to cell survival or death, depending on the levels and types of stress. However, the specific factors that activate the death program still remain to be elucidated in both autophagy and ER stress signaling. Several mechanisms have been proposed to be involved, and cell signaling pathways may interact in different ways to have specific outcomes in different cell types91,198. This may partly explain the variable efficacy of bortezomib and hydroxychloroquine treatment in cancer patients199,200. Therefore, understanding how cancer cells integrate information from different signaling pathways, and the establishment of a system quantitatively monitoring the stress intensity and duration and its subsequent effects on cell fate following chemotherapeutic drugs administration, are particularly needed to overcome chemoresistance. Tumor heterogeneity and the influence of the tumor microenvironment may pose a challenge for establishing a standard treatment protocol. However, due to the recent advances in computational biology and single-cell multi-omics approaches, this may be attainable201–203. Collaboration between oncologists, cancer biologists, and bioinformaticians is necessary to overcome the immense challenge of chemotherapy resistance.
Figure 2.
The balance between the integrated cell survival and death signaling determines the cell fate of cancer cells receiving chemotherapy. Multiple cellular adaptive response may be induced in cancer cells following chemotherapy. The cell fate is determined by the balance between the integrated pro-survival and pro-death adaptive response. If the pro-survival signaling overrides the death-triggering signaling, the cancer cells are resistant to the treatment and may continue to proliferate and advance in the patients.
Acknowledgments
This study was supported by the Canadian Institutes of Health Research (CIHR) (Grant #MOP-82881); CIHR New Investigator salary award to Isabella T. Tai (MSH-95344). Figures were created using biorender.com.
Footnotes
Conflict of interest statement No potential conflicts of interest are disclosed.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: are we ready. Nat Rev Genet. 2012;13:818–24. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer. Nat Rev Cancer. 2012;12:323–34. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hof J, Krentz S, van Schewick C, Körner G, Shalapour S, Rhein P, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol. 2011;29:3185–93. doi: 10.1200/JCO.2011.34.8144. [DOI] [PubMed] [Google Scholar]
- 7.Hamburg M, Collins F. The path to personalized medicine. N Engl J Med. 2010;363:301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 8.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 9.Crawford DR, Davies KJ. Adaptive response and oxidative stress. Environ Health Perspect. 1994;102(Suppl):25–8. doi: 10.1289/ehp.94102s1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–27. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 12.Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 17.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Y, Xu Z, Dai S, Qian L, Sun L, Gong Z. Targeting autophagy to sensitive glioma to temozolomide treatment. J Exp Clin Cancer Res. 2016;35:23. doi: 10.1186/s13046-016-0303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 20.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21:488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 22.Jain RK, Duda DG, Willett CG, Sahani DV, Zhu AX, Loeffler JS, et al. Biomarkers of response and resistance to antiangiogenic therapy. Nat Rev Clin Oncol. 2009;6:327–38. doi: 10.1038/nrclinonc.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singleton DC, Harris AL. Microenvironmental induced essentiality of autophagy. Clin Cancer Res. 2013;19:2791–3. doi: 10.1158/1078-0432.CCR-13-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu YL, Jahangiri A, DeLay M, Aghi MK. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012;72:4294–9. doi: 10.1158/0008-5472.CAN-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72:1773–83. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo XL, Li D, Sun K, Wang J, Liu Y, Song JR, et al. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med. 2013;91:473–83. doi: 10.1007/s00109-012-0966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvakumaran M, Amaravadi RK, Vasilevskaya IA, O’Dwyer PJ. Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res. 2013;19:2995–3007. doi: 10.1158/1078-0432.CCR-12-1542. [DOI] [PubMed] [Google Scholar]
- 31.Kim KW, Paul P, Qiao J, Chung DH. Autophagy mediates paracrine regulation of vascular endothelial cells. Lab Invest. 2013;93:639–45. doi: 10.1038/labinvest.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KW, Paul P, Qiao J, Lee S, Chung DH. Enhanced autophagy blocks angiogenesis via degradation of gastrin-releasing peptide in neuroblastoma cells. Autophagy. 2013;9:1579–90. doi: 10.4161/auto.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SY, Kim NH, Cho YS, Lee H, Kwon HJ. Convallatoxin, a dual inducer of autophagy and apoptosis, inhibits angiogenesis in vitro and in vivo. PLoS One. 2014;9:e91094. doi: 10.1371/journal.pone.0091094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poluzzi C, Casulli J, Goyal A, Mercer TJ, Neill T, Iozzo RV. Endorepellin evokes autophagy in endothelial cells. J Biol Chem. 2014;289:16114–28. doi: 10.1074/jbc.M114.556530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel O, Shulkes A, Baldwin GS. Gastrin-releasing peptide and cancer. Biochim Biophys Acta. 2006;1766:23–41. doi: 10.1016/j.bbcan.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Loeffler M, Krüger JA, Niethammer AG, Reisfeld RA. Targeting tumor-associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J Clin Invest. 2006;116:1955–62. doi: 10.1172/JCI26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 38.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol. 2017;10:76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y, Chen X, Wang X, Zhao Z, Hu W, Zeng S, et al. The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. J Exp Clin Cancer Res. 2019;38:171. doi: 10.1186/s13046-019-1172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witkiewicz AK, Dasgupta A, Sotgia F, Mercier I, Pestell RG, Sabel M, et al. An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am J Pathol. 2009;174:2023–34. doi: 10.2353/ajpath.2009.080873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witkiewicz AK, Casimiro MC, Dasgupta A, Mercier I, Wang C, Bonuccelli G, et al. Towards a new “stromal-based” classification system for human breast cancer prognosis and therapy. Cell Cycle. 2009;8:1654–8. doi: 10.4161/cc.8.11.8544. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: a new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–76. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: role of hypoxia, HIF1 induction and NFκB activation in the tumor stromal microenvironment. Cell Cycle. 2010;9:3515–33. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 45.Liao J-K, Zhou B, Zhuang X-M, Zhuang PL, Zhang DM, Chen WL. Cancer-associated broblasts confer cisplatin resistance of tongue cancer via autophagy activation. Biomed Pharmacother. 2018;97:1341–8. doi: 10.1016/j.biopha.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 46.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 47.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–9. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 49.Paez J, Jänne P, Lee J, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 50.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:225–35. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Jänne PA, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–60. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai GGY, Lim TH, Lim J, Liew PJR, Kwang XL, Nahar R, et al. Clonal MET amplification as a determinant of tyrosine kinase inhibitor resistance in epidermal growth factor receptor-mutant non-small-cell lung cancer. J Clin Oncol. 2019;37:876–84. doi: 10.1200/JCO.18.00177. [DOI] [PubMed] [Google Scholar]
- 53.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–8. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Giménez-Capitán A, Cantero F, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10:1812. doi: 10.1038/s41467-019-09734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou Y, Ling Y-H, Sironi J, Schwartz EL, Perez-Soler R, Piperdi B. The autophagy inhibitor chloroquine overcomes the innate resistance of wild-type EGFR non-small-cell lung cancer cells to erlotinib. J Thorac Oncol. 2013;8:693–702. doi: 10.1097/JTO.0b013e31828c7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakuma Y, Matsukuma S, Nakamura Y, Yoshihara M, Koizume S, Sekiguchi H, et al. Enhanced autophagy is required for survival in EGFR-independent EGFR-mutant lung adenocarcinoma cells. Lab Invest. 2013;93:1137–46. doi: 10.1038/labinvest.2013.102. [DOI] [PubMed] [Google Scholar]
- 58.Jutten B, Rouschop KMA. EGFR signaling and autophagy dependence for growth, survival, and therapy resistance. Cell Cycle. 2014;13:42–51. doi: 10.4161/cc.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1a and Bcl-2 and activating the Beclin 1/hVps34 complex. Cancer Res. 2010;70:5942–52. doi: 10.1158/0008-5472.CAN-10-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Lu Y, Pan T, Fan Z. Roles of autophagy in cetuximab-mediated cancer therapy against EGFR. Autophagy. 2010;6:1066–77. doi: 10.4161/auto.6.8.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jutten B, Keulers TG, Peeters HJM, Schaaf MBE, Savelkouls KGM, Compter I, et al. EGFRvIII expression triggers a metabolic dependency and therapeutic vulnerability sensitive to autophagy inhibition. Autophagy. 2018;14:283–95. doi: 10.1080/15548627.2017.1409926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–84. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell. 2015;160:145–60. doi: 10.1016/j.cell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eimer S, Belaud-Rotureau MA, Airiau K, Jeanneteau M, Laharanne E, Véron N, et al. Autophagy inhibition cooperates with erlotinib to induce glioblastoma cell death. Cancer Biol Ther. 2011;11:1017–27. doi: 10.4161/cbt.11.12.15693. [DOI] [PubMed] [Google Scholar]
- 65.Dragowska WH, Weppler SA, Wang JC, Wong LY, Kapanen AI, Rawji JS, et al. Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS One. 2013;8:1–20. doi: 10.1371/journal.pone.0076503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 67.Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V, et al. Induction of autophagy and inhibition of melanoma growth in vitro and in vivo by hyperactivation of oncogenic BRAF. J Invest Dermatol. 2010;130:1657–67. doi: 10.1038/jid.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strohecker AM, White E. Targeting mitochondrial metabolism by inhibiting autophagy in Braf-driven cancers. Cancer Discov. 2014;4:766–72. doi: 10.1158/2159-8290.CD-14-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–17. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levy JMM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, et al. Autophagy inhibition improves chemosensitivity in BRAFV600E brain tumors. Cancer Discov. 2014;4:773–80. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S, Song Y, Quach C, Guo H, Jang GB, Maazi H, et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun. 2019;10:1693. doi: 10.1038/s41467-019-09634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murugan AK, Grieco M, Tsuchida N. RAS mutations in human cancers: roles in precision medicine. Semin Cancer Biol. 2019;59:23–35. doi: 10.1016/j.semcancer.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 74.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–32. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–15. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 79.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Z, Yan Y, Qian L, Gong Z. Long non-coding RNAs act as regulators of cell autophagy in diseases (Review) Oncol Rep. 2017;37:1359–66. doi: 10.3892/or.2017.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, Avendaño-Félix M, Silva-Benítez E, López-Camarillo C, et al. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front Oncol. 2019;9:1008. doi: 10.3389/fonc.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–40. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 83.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, et al. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18:82. doi: 10.1186/s12943-019-1016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119:3853–63. doi: 10.1002/jcb.26464. [DOI] [PubMed] [Google Scholar]
- 87.YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huo J-F, Chen X-B. Long noncoding RNA growth arrest-specific 5 facilitates glioma cell sensitivity to cisplatin by suppressing excessive autophagy in an mTOR-dependent manner. J Cell Biochem. 2019;120:6127–36. doi: 10.1002/jcb.27900. [DOI] [PubMed] [Google Scholar]
- 89.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–30. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 90.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–18. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69:169–81. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 92.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 93.Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 94.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Görlach A, Klappa P, Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal. 2006;8:1391–418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- 96.McConkey DJ, Zhu K. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist Updat. 2008;11:164–79. doi: 10.1016/j.drup.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 97.Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–23. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 98.Mahoney E, Lucas DM, Gupta SV, Wagner AJ, Herman SE, Smith LL, et al. ER stress and autophagy: new players in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor flavopiridol. Blood. 2012;120:1262–73. doi: 10.1182/blood-2011-12-400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yadav RK, Chae S-W, Kim H-R, Chae HJ. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19:75–88. doi: 10.15430/JCP.2014.19.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mandic A, Hansson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem. 2003;278:9100–6. doi: 10.1074/jbc.M210284200. [DOI] [PubMed] [Google Scholar]
- 101.Fribley A, Zeng Q, Wang C. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24:9695–704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wroblewski D, Jiang CC, Croft A, Farrelly ML, Zhang XD, Hersey P. OBATOCLAX and ABT-737 induce ER stress responses in human melanoma cells that limit induction of apoptosis. PLoS One. 2013;8:1–13. doi: 10.1371/journal.pone.0084073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Déry M-A, Jodoin J, Ursini-Siegel J, Aleynikova O, Ferrario C, Hassan S, et al. Endoplasmic reticulum stress induces PRNP prion protein gene expression in breast cancer. Breast Cancer Res. 2013;15:R22. doi: 10.1186/bcr3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–72. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 105.Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, et al. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. Br J Cancer. 2007;96:1062–71. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 107.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 109.Lovat PE, Corazzari M, Armstrong JL, Martin S, Pagliarini V, Hill D, et al. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 2008;68:5363–9. doi: 10.1158/0008-5472.CAN-08-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tufo G, Jones AWE, Wang Z, Hamelin J, Tajeddine N, Esposti DD, et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014;21:685–95. doi: 10.1038/cdd.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol Cell Biol. 1997;17:5317–27. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garrido C, Ottavi P, Fromentin A, Hammann A, Arrigo AP, Chauffert B, et al. HSP27 as a mediator of confluence-dependent resistance to cell death induced by anticancer drugs. Cancer Res. 1997;57:2661–7. [PubMed] [Google Scholar]
- 113.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–24. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 114.Azad A, Zoubeidi A, Gleave M, Chi KN. Targeting heat shock proteins in metastatic castration-resistant prostate cancer. Nat Rev Urol. 2015;12:26–36. doi: 10.1038/nrurol.2014.320. [DOI] [PubMed] [Google Scholar]
- 115.Roue G, Perez-Galan P, Mozos A, López-Guerra M, Xargay-Torrent S, Rosich L, et al. The Hsp90 inhibitor IPI-504 overcomes bortezomib resistance in mantle cell lymphoma in vitro and in vivo by down-regulation of the prosurvival ER chaperone BiP/Grp78. Blood. 2011;117:1270–9. doi: 10.1182/blood-2010-04-278853. [DOI] [PubMed] [Google Scholar]
- 116.Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov. 2013;3:430–43. doi: 10.1158/2159-8290.CD-12-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huot J, Houle F, Spitz DR, Landry J. HSP27 phosphorylation-mediated resistance against actin fragmentation and cell death induced by oxidative stress. Cancer Res. 1996;56:273–9. [PubMed] [Google Scholar]
- 118.Chern Y, Zhang P, Ju H, Tai IT. Heat shock protein 47 promotes tumor survival and therapy resistance by modulating AKT signaling via PHLPP1 in colorectal cancer. Cancer Biol Med. 2020;17:343–56. doi: 10.20892/j.issn.2095-3941.2019.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jego G, Hazoumé A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2013;332:275–85. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 120.Calderwood SK, Gong J. Molecular chaperones in mammary cancer growth and breast tumor therapy. J Cell Biochem. 2012;113:1096–103. doi: 10.1002/jcb.23461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett. 2012;325:117–24. doi: 10.1016/j.canlet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 122.Baylot V, Andrieu C, Katsogiannou M, Taieb D, Garcia S, Giusiano S, et al. OGX-427 inhibits tumor progression and enhances gemcitabine chemotherapy in pancreatic cancer. Cell Death Dis. 2011;2:e221. doi: 10.1038/cddis.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lamoureux F, Thomas C, Yin MJ, Zoubeidi A, Gleave ME. Suppression of heat shock protein 27 using OGX-427 induces endoplasmic reticulum stress and potentiates heat shock protein 90 inhibitors to delay castrate-resistant prostate cancer. Eur Urol. 2014;66:145–55. doi: 10.1016/j.eururo.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lelj-Garolla B, Kumano M, Beraldi E, Nappi L, Rocchi P, Ionescu DN, et al. Hsp27 inhibition with OGX-427 sensitizes non-small cell lung cancer cells to erlotinib and chemotherapy. Mol Cancer Ther. 2015;14:1107–16. doi: 10.1158/1535-7163.MCT-14-0866. [DOI] [PubMed] [Google Scholar]
- 125.Yu EY, Ellard SL, Hotte SJ, Gingerich JR, Joshua AM, Gleave ME, et al. A randomized phase 2 study of a HSP27 targeting antisense, apatorsen with prednisone versus prednisone alone, in patients with metastatic castration resistant prostate cancer. Invest New Drugs. 2018;36:278–87. doi: 10.1007/s10637-017-0553-x. [DOI] [PubMed] [Google Scholar]
- 126.Spigel DR, Shipley DL, Waterhouse DM, Jones SF, Ward PJ, Shih KC, et al. A randomized, double-blinded, phase II trial of carboplatin and pemetrexed with or without apatorsen (OGX-427) in patients with previously untreated stage IV non-squamous-non-small-cell lung cancer: the SPRUCE trial. Oncologist. 2019;24:e2409–16. doi: 10.1634/theoncologist.2018-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ko AH, Murphy PB, Peyton JD, Shipley DL, Al-Hazzouri A, Rodriguez FA, et al. A randomized, double-blinded, phase II trial of gemcitabine and nab-paclitaxel plus apatorsen or placebo in patients with metastatic pancreatic cancer: the RAINIER trial. Oncologist. 2017;22:1427–e129. doi: 10.1634/theoncologist.2017-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rosenberg JE, Hahn NM, Regan MM, Werner L, Alva A, George S, et al. Apatorsen plus docetaxel versus docetaxel alone in platinum-resistant metastatic urothelial carcinoma (Borealis-2) Br J Cancer. 2018;118:1434–41. doi: 10.1038/s41416-018-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gifford JB, Huang W, Zeleniak AE, Hindoyan A, Wu H, Donahue TR, et al. Expression of GRP78, master regulator of the unfolded protein response, increases chemoresistance in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2016;15:1043–52. doi: 10.1158/1535-7163.MCT-15-0774. [DOI] [PubMed] [Google Scholar]
- 130.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 131.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–11. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chern Y-J, Wong JCT, Cheng GSW, Yu A, Yin Y, Schaeffer DF, et al. The interaction between SPARC and GRP78 interferes with ER stress signaling and potentiates apoptosis via PERK/eIF2alpha and IRE1alpha/XBP-1 in colorectal cancer. Cell Death Dis. 2019;10:504. doi: 10.1038/s41419-019-1687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rao RV, Peel A, Logvinova A, del Rio G, Hermel E, Yokota T, et al. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514:122–8. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–24. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 135.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 136.Pyrko P, Schönthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–16. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 137.Mozos A, Roué G, López-Guillermo A, Jares P, Campo E, Colomer D, et al. The expression of the endoplasmic reticulum stress sensor BiP/GRP78 predicts response to chemotherapy and determines the efficacy of proteasome inhibitors in diffuse large B-cell lymphoma. Am J Pathol. 2011;179:2601–10. doi: 10.1016/j.ajpath.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou H, Zhang Y, Fu Y, Chan L, Lee AS. Novel mechanism of anti-apoptotic function of 78-kDa glucose-regulated protein (GRP78): endocrine resistance factor in breast cancer, through release of B-cell lymphoma 2 (BCL-2) from BCL-2-interacting killer (BIK) J Biol Chem. 2011;286:25687–96. doi: 10.1074/jbc.M110.212944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181–8. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin Y, Wang Z, Liu L, Chen L. Akt is the downstream target of GRP78 in mediating cisplatin resistance in ER stress-tolerant human lung cancer cells. Lung Cancer. 2011;71:291–7. doi: 10.1016/j.lungcan.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 141.Zhang LH, Yang XL, Zhang X, Cheng JX, Zhang W. Association of elevated GRP78 expression with increased astrocytoma malignancy via Akt and ERK pathways. Brain Res. 2011;1371:23–31. doi: 10.1016/j.brainres.2010.11.063. [DOI] [PubMed] [Google Scholar]
- 142.Uckun FM, Qazi S, Ozer Z, Garner AL, Pitt J, Ma H, et al. Inducing apoptosis in chemotherapy-resistant B-lineage acute lymphoblastic leukaemia cells by targeting HSPA5, a master regulator of the anti-apoptotic unfolded protein response signalling network. Br J Haematol. 2011;153:741–52. doi: 10.1111/j.1365-2141.2011.08671.x. [DOI] [PubMed] [Google Scholar]
- 143.Gray MJ, Mhawech-Fauceglia P, Yoo E, Yang W, Wu E, Lee AS, et al. AKT inhibition mitigates GRP78 (glucose-regulated protein) expression and contribution to chemoresistance in endometrial cancers. Int J Cancer. 2013;133:21–30. doi: 10.1002/ijc.27994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Misra UK, Gonzalez-Gronow M, Gawdi G, Wang F, Pizzo SV. A novel receptor function for the heat shock protein Grp78: silencing of Grp78 gene expression attenuates a2M*-induced signalling. Cell Signal. 2004;16:929–38. doi: 10.1016/j.cellsig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 145.Misra UK, Deedwania R, Pizzo SV. Activation and cross-talk between Akt, NF-kappaB, and unfolded protein response signaling in 1-LN prostate cancer cells consequent to ligation of cell surface-associated GRP78. J Biol Chem. 2006;281:13694–707. doi: 10.1074/jbc.M511694200. [DOI] [PubMed] [Google Scholar]
- 146.Christensson A, Laurell CB, Lilja H. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine proteinase inhibitors. Eur J Biochem. 1990;194:755–63. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]
- 147.Misra UK, Payne S, Pizzo SV. Ligation of prostate cancer cell surface GRP78 activates a proproliferative and antiapoptotic feedback loop: a role for secreted prostate-specific antigen. J Biol Chem. 2011;286:1248–59. doi: 10.1074/jbc.M110.129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Philippova M, Ivanov D, Joshi MB, Kyriakakis E, Rupp K, Afonyushkin T, et al. Identification of proteins associating with glycosylphosphatidylinositol- anchored T-cadherin on the surface of vascular endothelial cells: role for Grp78/BiP in T-cadherin-dependent cell survival. Mol Cell Biol. 2008;28:4004–17. doi: 10.1128/MCB.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kelber JA, Panopoulos AD, Shani G, Booker EC, Belmonte JC, Vale WW, et al. Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene. 2009;28:2324–36. doi: 10.1038/onc.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang Y, Tseng C-C, Tsai Y-L, Fu X, Schiff R, Lee AS. Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PLoS One. 2013;8:e80071. doi: 10.1371/journal.pone.0080071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shu C, Sun F, Cho J, Lin CC, Liu PF, Chen PY, et al. GRP78 and Raf-1 cooperatively confer resistance to endoplasmic reticulum stress-induced apoptosis. J Cell Physiol. 2008;215:627–35. doi: 10.1002/jcp.21340. [DOI] [PubMed] [Google Scholar]
- 152.Kern J, Untergasser G, Zenzmaier C, Sarg B, Gastl G, Gunsilius E, et al. GRP-78 secreted by tumor cells blocks the antiangiogenic activity of bortezomib. Blood. 2009;114:3960–7. doi: 10.1182/blood-2009-03-209668. [DOI] [PubMed] [Google Scholar]
- 153.Baumeister P, Dong D, Fu Y, Lee AS. Transcriptional induction of GRP78/BiP by histone deacetylase inhibitors and resistance to histone deacetylase inhibitor-induced apoptosis. Mol Cancer Ther. 2009;8:1086–94. doi: 10.1158/1535-7163.MCT-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–76. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mousa SA, Sudha T, Dyskin E, Dier U, Gallati C, Hanko C, et al. Stress resistant human embryonic stem cells as a potential source for the identification of novel cancer stem cell markers. Cancer Lett. 2010;289:208–16. doi: 10.1016/j.canlet.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 156.Kang J, Shakya A, Tantin D. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 2009;34:491–9. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 157.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 158.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 159.Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, et al. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–50. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102:1276–83. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cammareri P, Scopelliti A, Todaro M, Eterno V, Francescangeli F, Moyer MP, et al. Aurora-A is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer Res. 2010;70:4655–65. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- 162.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose. Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119–31. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13:109–18. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fujimoto A, Kawana K, Taguchi A, Adachi K, Sato M, Nakamura H, et al. Inhibition of endoplasmic reticulum (ER) stress sensors sensitizes cancer stem-like cells to ER stress-mediated apoptosis. Oncotarget. 2016;7:51854–64. doi: 10.18632/oncotarget.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–72. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 168.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–97. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wey S, Luo B, Lee AS. Acute inducible ablation of GRP78 reveals its role in hematopoietic stem cell survival, lymphogenesis and regulation of stress signaling. PLoS One. 2012;7:e39047. doi: 10.1371/journal.pone.0039047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu M-J, Jan C-I, Tsay Y-G, Yu YH, Huang CY, Lin SC, et al. Elimination of head and neck cancer initiating cells through targeting glucose regulated protein78 signaling. Mol Cancer. 2010;9:283. doi: 10.1186/1476-4598-9-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chiu C, Lee L, Li Y, Chen YJ, Lu YC, Li YL, et al. Grp78 as a therapeutic target for refractory head-neck cancer with CD24(-)CD44(+) stemness phenotype. Cancer Gene Ther. 2013;20:606–15. doi: 10.1038/cgt.2013.64. [DOI] [PubMed] [Google Scholar]
- 172.Wang N, Wang Z, Peng C, You J, Shen J, Han S, et al. Dietary compound isoliquiritigenin targets GRP78 to chemosensitize breast cancer stem cells via β-catenin/ABCG2 signaling. Carcinogenesis. 2014;35:2544–54. doi: 10.1093/carcin/bgu187. [DOI] [PubMed] [Google Scholar]
- 173.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Morel A-P, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lehmann K, Janda E, Pierreux CE, Rytömaa M, Schulze A, McMahon M, et al. Raf induces TGF/xDF production while blocking its apoptotic but not invasive responses: a mechanism leading to increased malignancy in epithelial cells. Genes Dev. 2000;14:2610–22. doi: 10.1101/gad.181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Angela Nieto M. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–53. doi: 10.1158/1078-0432.CCR-06-0038. [DOI] [PubMed] [Google Scholar]
- 178.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 179.Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, et al. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int J Oncol. 2007;31:277–83. [PubMed] [Google Scholar]
- 180.Shah PP, Dupre TV, Siskind LJ, Beverly LJ. Common cytotoxic chemotherapeutics induce epithelial-mesenchymal transition (EMT) downstream of ER stress. Oncotarget. 2017;8:22625–39. doi: 10.18632/oncotarget.15150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tanjore H, Cheng D-S, Degryse AL, Zoz DF, Abdolrasulnia R, Lawson WE, et al. Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J Biol Chem. 2011;286:30972–80. doi: 10.1074/jbc.M110.181164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zeindl-Eberhart E, Brandl L, Liebmann S, Ormanns S, Scheel SK, Brabletz T, et al. Epithelial-mesenchymal transition induces endoplasmic-reticulum-stress response in human colorectal tumor cells. PLoS One. 2014;9:e87386. doi: 10.1371/journal.pone.0087386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shen X, Xue Y, Si Y, Wang Q, Wang Z, Yuan J, et al. The unfolded protein response potentiates epithelial-to-mesenchymal transition (EMT) of gastric cancer cells under severe hypoxic conditions. Med Oncol. 2014;32:447. doi: 10.1007/s12032-014-0447-0. [DOI] [PubMed] [Google Scholar]
- 184.Feng Y, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JHL, Proia TA, et al. Epithelial-to-mesenchymal transition activates PERK-eIF2a and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014;4:702–15. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 185.Acosta JC, Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol. 2012;22:211–9. doi: 10.1016/j.tcb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 186.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cahu J, Bustany S, Sola B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 2012;3:e446. doi: 10.1038/cddis.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Burton DGA, Giribaldi MG, Munoz A, Halvorsen K, Patel A, Jorda M, et al. Androgen deprivation-induced senescence promotes outgrowth of androgen-refractory prostate cancer cells. PLoS One. 2013;8:e68003. doi: 10.1371/journal.pone.0068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun X, Shi B, Zheng H, Min L, Yang J, Li X, et al. Senescence-associated secretory factors induced by cisplatin in melanoma cells promote non-senescent melanoma cell growth through activation of the ERK1/2-RSK1 pathway. Cell Death Dis. 2018;9:260. doi: 10.1038/s41419-018-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Niu L-L, Cheng C, Li M-Y, Yang SL, Hu BG, Chong CCN, et al. ID1-induced p16/IL6 axis activation contributes to the resistant of hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018;9:852. doi: 10.1038/s41419-018-0926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mastri M, Tracz A, Lee CR, Dolan M, Attwood K, Christensen JG, et al. A transient pseudosenescent secretome promotes tumor growth after antiangiogenic therapy withdrawal. Cell Rep. 2018;25:3706–20.e8. doi: 10.1016/j.celrep.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 194.Sidi R, Pasello G, Opitz I, Soltermann A, Tutic M, Rehrauer H, et al. Induction of senescence markers after neo-adjuvant chemotherapy of malignant pleural mesothelioma and association with clinical outcome: an exploratory analysis. Eur J Cancer. 2011;47:326–32. doi: 10.1016/j.ejca.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 195.Canino C, Mori F, Cambria A, Diamantini A, Germoni S, Alessandrini G, et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 2012;31:3148–63. doi: 10.1038/onc.2011.485. [DOI] [PubMed] [Google Scholar]
- 196.Martino S di, Amoreo CA, Nuvoli B, Galati R, Strano S, Facciolo F, et al. HSP90 inhibition alters the chemotherapy-driven rearrangement of the oncogenic secretome. Oncogene. 2018;37:1369–85. doi: 10.1038/s41388-017-0044-8. [DOI] [PubMed] [Google Scholar]
- 197.Davies KJA. Adaptive homeostasis. Mol Aspects Med. 2016;49:1–7. doi: 10.1016/j.mam.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fitzwalter BE, Thorburn A. Recent insights into cell death and autophagy. FEBS J. 2015;282:4279–88. doi: 10.1111/febs.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–8. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sonneveld P, Schmidt-Wolf IGH, van der Holt B, El Jarari L, Bertsch U, Salwender H, et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: results of the randomized phase III HOVON-65/GMMG-HD4 trial. J Clin Oncol. 2012;30:2946–55. doi: 10.1200/JCO.2011.39.6820. [DOI] [PubMed] [Google Scholar]
- 201.Huntsman DG, Ladanyi M. The molecular pathology of cancer: from pan-genomics to post-genomics. J Pathol. 2018;244:509–11. doi: 10.1002/path.5057. [DOI] [PubMed] [Google Scholar]
- 202.Gorgoulis VG, Pefani D-E, Pateras IS, Trougakos IP. Integrating the DNA damage and protein stress responses during cancer development and treatment. J Pathol. 2018;246:12–40. doi: 10.1002/path.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–3. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 204.Hu X, Shi S, Wang H, Yu X, Wang Q, Jiang S, et al. Blocking autophagy improves the anti-tumor activity of afatinib in lung adenocarcinoma with activating EGFR mutations in vitro and in vivo. Sci Rep. 2017;7:4559. doi: 10.1038/s41598-017-04258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Abdul Rahim SA, Dirkse A, Oudin A, Schuster A, Bohler J, Barthelemy V, et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A . Br J Cancer. 2017;117:813–25. doi: 10.1038/bjc.2017.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Muñoz-Galván S, Gutierrez G, Perez M, Carnero A. MAP17 (PDZKIP1) expression determines sensitivity to the proteasomal inhibitor bortezomib by preventing cytoprotective autophagy and NF. B activation in breast cancer. Mol Cancer Ther. 2015;14:1454–65. doi: 10.1158/1535-7163.MCT-14-1053. [DOI] [PubMed] [Google Scholar]
- 207.Rupniewska E, Roy R, Mauri FA, Liu X, Kaliszczak M, Bellezza G, et al. Targeting autophagy sensitises lung cancer cells to Src family kinase inhibitors. Oncotarget. 2018;9:27346–62. doi: 10.18632/oncotarget.25213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yeh ES, Abt MA, Hill EG. Regulation of cell survival by HUNK mediates breast cancer resistance to HER2 inhibitors. Breast Cancer Res Treat. 2015;149:91–8. doi: 10.1007/s10549-014-3227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pan H, Wang Z, Jiang L, Sui X, You L, Shou J, et al. Autophagy inhibition sensitizes hepatocellular carcinoma to the multikinase inhibitor linifanib. Sci Rep. 2014;4:6683. doi: 10.1038/srep06683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li L, Wang Y, Jiao L, Lin C, Lu C, Zhang K, et al. Protective autophagy decreases osimertinib cytotoxicity through regulation of stem cell-like properties in lung cancer. Cancer Lett. 2019;452:191–202. doi: 10.1016/j.canlet.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 211.Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459:234–9. doi: 10.1016/j.bbrc.2015.02.084. [DOI] [PubMed] [Google Scholar]
- 212.Lu S, Yao Y, Xu G, Zhou C, Zhang Y, Sun J, et al. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death Dis. 2018;9:646. doi: 10.1038/s41419-018-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Luo P, Xu Z, Li G, Yan H, Zhu Y, Zhu H, et al. HMGB1 represses the anti-cancer activity of sunitinib by governing TP53 autophagic degradation via its nucleus-to-cytoplasm transport. Autophagy. 2018;14:2155–70. doi: 10.1080/15548627.2018.1501134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wiedmer T, Blank A, Pantasis S, Normand L, Bill R, Krebs P, et al. Autophagy inhibition improves sunitinib efficacy in pancreatic neuroendocrine tumors via A lysosome-dependent mechanism. Mol Cancer Ther. 2017;16:2502–15. doi: 10.1158/1535-7163.MCT-17-0136. [DOI] [PubMed] [Google Scholar]
- 215.Ojha R, Leli NM, Onorati A, Piao S, Verginadis II, Tameire F, et al. ER translocation of the MAPK pathway drives therapy resistance in BRAF-mutant melanoma. Cancer Discov. 2019;9:396–415. doi: 10.1158/2159-8290.CD-18-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Altman JK, Szilard A, Goussetis DJ, Sassano A, Colamonici M, Gounaris E, et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin Cancer Res. 2014;20:2400–9. doi: 10.1158/1078-0432.CCR-13-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Cuyàs E, López-Bonet E, et al. The anti-malarial chloroquine overcomes primary resistance and restores sensitivity to trastuzumab in HER2-positive breast cancer. Sci Rep. 2013;3:2469. doi: 10.1038/srep02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Urruticoechea A, Martin-Castillo B, et al. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget. 2012;3:1600–14. doi: 10.18632/oncotarget.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Martin S, Dudek-Perić AM, Maes H, Garg AD, Gabrysiak M, Demirsoy S, et al. Concurrent MEK and autophagy inhibition is required to restore cell death associated danger-signalling in Vemurafenib-resistant melanoma cells. Biochem Pharmacol. 2015;93:290–304. doi: 10.1016/j.bcp.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 221.Zeng X, Zhao H, Li Y, Fan J, Sun Y, Wang S, et al. Targeting Hedgehog signaling pathway and autophagy overcomes drug resistance of BCR-ABL-positive chronic myeloid leukemia. Autophagy. 2015;11:355–72. doi: 10.4161/15548627.2014.994368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kumandan S, Mahadevan NR, Chiu K, DeLaney A, Zanetti M. Activation of the unfolded protein response bypasses trastuzumab-mediated inhibition of the PI-3K pathway. Cancer Lett. 2013;329:236–42. doi: 10.1016/j.canlet.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sui X, Kong N, Wang X, Fang Y, Hu X, Xu Y, et al. JNK confers 5-fluorouracil resistance in p53-deficient and mutant p53-expressing colon cancer cells by inducing survival autophagy. Sci Rep. 2014;4:4694. doi: 10.1038/srep04694. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 224.Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900–9. doi: 10.1016/j.ejca.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 225.Ou J, Peng Y, Yang W, Zhang Y, Hao J, Li F, et al. ABHD5 blunts the sensitivity of colorectal cancer to fluorouracil via promoting autophagic uracil yield. Nat Commun. 2019;10:1078. doi: 10.1038/s41467-019-08902-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Takahashi H, Inoue J, Sakaguchi K, Takagi M, Mizutani S, Inazawa J, et al. Autophagy is required for cell survival under L-asparaginase-induced metabolic stress in acute lymphoblastic leukemia cells. Oncogene. 2017;36:4267–76. doi: 10.1038/onc.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–73. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yu L, Gu C, Zhong D, Shi L, Kong Y, Zhou Z, et al. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355:34–45. doi: 10.1016/j.canlet.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 229.Braggio D, Koller D, Jin F, Siva N, Zewdu A, Lopez G, et al. Autophagy inhibition overcomes sorafenib resistance in S45F-mutated desmoid tumors. Cancer. 2019;125:2693–703. doi: 10.1002/cncr.32120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pan X, Chen Y, Shen Y, Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10:429. doi: 10.1038/s41419-019-1660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ma H, Li Y, Wang X, Wu H, Qi G, Li R, et al. PBK, targeted by EVI1, promotes metastasis and confers cisplatin resistance through inducing autophagy in high-grade serous ovarian carcinoma. Cell Death Dis. 2019;10:166. doi: 10.1038/s41419-019-1415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jiang L, Xu L, Xie J, Li S, Guan Y, Zhang Y, et al. Inhibition of autophagy overcomes glucocorticoid resistance in lymphoid malignant cells. Cancer Biol Ther. 2015;16:466–76. doi: 10.1080/15384047.2015.1016658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tan Q, Joshua AM, Saggar JK, Yu M, Wang M, Kanga N, et al. Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy. Br J Cancer. 2015;112:832–40. doi: 10.1038/bjc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 234.Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, et al. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9:e97242. doi: 10.1371/journal.pone.0097242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 235.Jaganathan S, Malek E, Vallabhapurapu S, Vallabhapurapu S, Driscoll JJ. Bortezomib induces AMPK-dependent autophagosome formation uncoupled from apoptosis in drug resistant cells. Oncotarget. 2014;5:12358–70. doi: 10.18632/oncotarget.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Armstrong JL, Corazzari M, Martin S, Pagliarini V, Falasca L, Hill DS, et al. Oncogenic B-RAF signaling in melanoma impairs the therapeutic advantage of autophagy inhibition. Clin Cancer Res. 2011;17:2216–26. doi: 10.1158/1078-0432.CCR-10-3003. [DOI] [PubMed] [Google Scholar]
- 237.Hu F, Zhao Y, Yu Y, Fang JM, Cui R, Liu ZQ, et al. Docetaxel-mediated autophagy promotes chemoresistance in castration-resistant prostate cancer cells by inhibiting STAT3. Cancer Lett. 2018;416:24–30. doi: 10.1016/j.canlet.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 238.Nguyen HG, Yang JC, Kung H-J, Shi XB, Tilki D, Lara PN, Jr, et al. Targeting autophagy overcomes Enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33:4521–30. doi: 10.1038/onc.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin Cancer Res. 2014:3159–73. doi: 10.1158/1078-0432.CCR-13-2060. [DOI] [PubMed] [Google Scholar]
- 240.Sun W-L, Chen J, Wang Y-P, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035–44. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 241.Song B, Bian Q, Shao CH, Li G, Liu AA, Jing W, et al. Ulinastatin reduces the resistance of liver cancer cells to epirubicin by inhibiting autophagy. PLoS One. 2015;10:e0120694. doi: 10.1371/journal.pone.0120694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pan B, Chen Y, Song H, Xu Y, Wang R, Chen L. Mir-24-3p downregulation contributes to VP16-DDP resistance in small-cell lung cancer by targeting ATG4A. Oncotarget. 2015;6:317–31. doi: 10.18632/oncotarget.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ma T, Chen W, Zhi X, Liu H, Zhou Y, Chen BW, et al. USP9X inhibition improves gemcitabine sensitivity in pancreatic cancer by inhibiting autophagy. Cancer Lett. 2018;436:129–38. doi: 10.1016/j.canlet.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 244.Zhang J, Zhang S, Song J, Sun K, Zong C, Zhao Q-D, et al. Autophagy inhibition switches low-dose camptothecin-induced premature senescence to apoptosis in human colorectal cancer cells. Biochem Pharmacol. 2014;90:265–75. doi: 10.1016/j.bcp.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 245.Liu W, Zhang Z, Zhang Y, Chen X, Guo S, Lei Y, et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol Ther. 2015;16:511–7. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, et al. Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-a-mediated signaling. Cell Death Dis. 2014;5:e1367. doi: 10.1038/cddis.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Taylor P. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:37–41. doi: 10.1080/15548627.2014.998931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wen J, Yeo S, Wang C, Chen S, Sun S, Hass MA, et al. Autophagy inhibition re-sensitizes pulse stimulation-selected paclitaxel-resistant triple negative breast cancer cells to chemotherapy-induced apoptosis. Breast Cancer Res Treat. 2015;149:619–29. doi: 10.1007/s10549-015-3283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tong Y, Huang H, Pan H. Inhibition of MEK/ERK activation attenuates autophagy and potentiates pemetrexed-induced activity against HepG2 hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2015;456:86–91. doi: 10.1016/j.bbrc.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 250.Chen R, Dai RY, Duan CY, Liu YP, Chen SK, Yan DM, et al. Unfolded protein response suppresses cisplatin-induced apoptosis via autophagy regulation in human hepatocellular carcinoma cells. Folia Biol (Praha) 2011;57:87–95. doi: 10.14712/fb2011057030087. [DOI] [PubMed] [Google Scholar]
- 251.Sisinni L, Maddalena F, Lettini G, Condelli V, Swann D, Esposito F, et al. TRAP1 role in endoplasmic reticulum stress protection favors resistance to anthracyclins in breast carcinoma cells. Int J Oncol. 2014;44:573–82. doi: 10.3892/ijo.2013.2199. [DOI] [PubMed] [Google Scholar]
- 252.Wang L, Zhang Y, Wang W, Zhu Y, Chen Y, Tian B. Gemcitabine treatment induces endoplasmic reticular (ER) stress and subsequently upregulates urokinase plasminogen activator (uPA) to block mitochondrial-dependent apoptosis in Panc-1 cancer stem-like cells (CSCs) PLoS One. 2017;12:e0184110. doi: 10.1371/journal.pone.0184110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Roemeling CA, von, Marlow LA, Kennedy WP, Kennedy GT, Copland JA, Menefee ME. Preclinical evaluation of the mTOR inhibitor, temsirolimus, in combination with the epothilone B analog, ixabepilone in renal cell carcinoma. Am J Cancer Res. 2013;3:390–401. [PMC free article] [PubMed] [Google Scholar]
- 254.Wang J, Yin Y, Hua H, Li M, Luo T, Xu L, et al. Blockade of GRP78 sensitizes breast cancer cells to microtubules-interfering agents that induce the unfolded protein response. J Cell Mol Med. 2009;13:3888–97. doi: 10.1111/j.1582-4934.2009.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Maddalena F, Sisinni L, Lettini G, Condelli V, Matassa DS, Piscazzi A, et al. Resistance to paclitxel in breast carcinoma cells requires a quality control of mitochondrial antiapoptotic proteins by TRAP1. Mol Oncol. 2013;7:895–906. doi: 10.1016/j.molonc.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Notte A, Rebucci M, Fransolet M, Roegiers E, Genin M, Tellier C, et al. Taxol-induced unfolded protein response activation in breast cancer cells exposed to hypoxia: ATF4 activation regulates autophagy and inhibits apoptosis. Int J Biochem Cell Biol. 2015;62:1–14. doi: 10.1016/j.biocel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 257.Mhaidat NM, Thorne R, Zhang XD, Hersey P. Involvement of endoplasmic reticulum stress in Docetaxel-induced JNK-dependent apoptosis of human melanoma. Apoptosis. 2008;13:1505–12. doi: 10.1007/s10495-008-0276-8. [DOI] [PubMed] [Google Scholar]
- 258.Strzeszewska A, Alster O, Mosieniak G, Ciolko A, Sikora E. Insight into the role of PIKK family members and NF-кB in DNAdamage-induced senescence and senescence-associated secretory phenotype of colon cancer cells. Cell Death Dis. 2018;9:44. doi: 10.1038/s41419-017-0069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Toste PA, Nguyen AH, Kadera BE, Duong M, Wu N, Gawlas I, et al. Chemotherapy-induced inflammatory gene signature and protumorigenic phenotype in pancreatic CAFs via stress-associated MAPK. Mol Cancer Res. 2016;14:437–47. doi: 10.1158/1541-7786.MCR-15-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Martínez J, Tarallo D, Martínez-Palma L, Victoria S, Bresque M, Rodríguez-Bottero S, et al. Mitofusins modulate the increase in mitochondrial length, bioenergetics and secretory phenotype in therapy-induced senescent melanoma cells. Biochem J. 2019;476:2463–86. doi: 10.1042/BCJ20190405. [DOI] [PMC free article] [PubMed] [Google Scholar]