Abstract
Neurotensin (Nts) is a neuropeptide implicated in the regulation of many facets of physiology, including cardiovascular tone, pain processing, ingestive behaviors, locomotor drive, sleep, addiction and social behaviors. Yet, there is incomplete understanding about how the various populations of Nts neurons distributed throughout the brain mediate such physiology. This knowledge gap largely stemmed from the inability to simultaneously identify Nts cell bodies and manipulate them in vivo. One means of overcoming this obstacle is to study NtsCre mice crossed onto a Cre-inducible green fluorescent reporter line (NtsCre;GFP mice), as these mice permit both visualization and in vivo modulation of specific populations of Nts neurons (using Cre-inducible viral and genetic tools) to reveal their function. Here we provide a comprehensive characterization of the distribution and relative densities of the Nts-GFP populations observed throughout the male NtsCre;GFP mouse brain, which will pave the way for future work to define their physiologic roles. We also compared the distribution of Nts-GFP neurons with Nts-In situ Hybridization (Nts-ISH) data from the adult mouse brain. By comparing these data sets we can distinguish Nts-GFP populations that may only transiently express Nts during development but not in the mature brain, and hence which populations may not be amenable to Cre-mediated manipulation in adult NtsCre;GFP mice. This atlas of Nts-GFP neurons will facilitate future studies using the NtsCre;GFP line to describe the physiological functions of individual Nts populations and how modulating them may be useful to treat disease.
Keywords: lateral hypothalamus, parabrachial nucleus, periaqueductal gray, central amygdala, thalamus, nucleus accumbens, preoptic area, olfactory tubercle, galanin
Introduction
The tridecapeptide Neurotensin (Nts) was first identified from the bovine hypothalamus (Carraway and Leeman, 1973), suggesting its potential function as a neuropeptide. Yet, Nts is also produced peripherally by intestinal enteroendocrine N-cells and the adrenal gland, and these sources account for the large pool of circulating Nts (Grunddal et al., 2016; Mustain et al., 2011; Rokaeus et al., 1984). Since its discovery, Nts has been implicated in regulating a host of physiologic responses, including feeding, locomotor activity, social behavior, analgesia, sleep, and response to addictive drugs (Benmoussa et al., 1996; Boules et al., 2011; Brown et al., 2017; Cape et al., 2000; Demeule et al., 2014; Ferraro et al., 2016; Fitzpatrick et al., 2012; Gammie et al., 2009; Hawkins et al., 1989; Lambert et al., 1995; Levine et al., 1983; Merullo et al., 2015; Nemeroff et al., 1977; Smith et al., 2012). How Nts mediates these effects remains unclear, and this is particularly true when considering whether and to what extent these effects are attributable to the Nts produced within the brain versus the periphery. Moreover, central and peripheral Nts may exert opposing control over some processes, such as those impacting body weight. Peripheral Nts mediates intestinal fat absorption and smooth muscle tone important for moving nutrients through the intestine (Carraway and Leeman, 1973; Li et al., 2016); thus, the high circulating pro-Nts levels observed in obese individuals could be predicted to facilitate the fat absorption that underlies weight gain. Contradictorily, gastric bypass surgery further elevates circulating Nts and the number of Nts-producing intestinal cells, suggesting that Nts signaling may contribute to the pronounced weight loss evoked by these procedures (Mumphrey et al., 2012; Ratner et al., 2016). A potential mechanism that reconciles these data is that local increases in intestinal Nts might be sufficient to access the central nervous system (CNS) via vagal afferents, which may support pro-weight loss behaviors. Indeed, some circulating Nts can access blood brain barrier-adjacent regions implicated in suppressing homeostatic feeding (Gevaert et al., 2016; Ratner et al., 2016); however, this Nts does not reach deeper Nts receptor-containing brain regions that suppress motivated feeding, such as the ventral tegmental area (VTA) (Ratner et al., 2016; Woodworth et al., 2017b, 2017a). Intriguingly, Nts administration to the VTA or central activation of specific Nts neurons that project there suppresses feeding and promotes physical activity behaviors that support weight loss (Cador et al., 1986; Hawkins, 1986; Kalivas and Duffy, 1990; Patterson et al., 2015; Woodworth et al., 2017b). Given the rapid turnover of circulating Nts (Aronin et al., 1982; Ratner et al., 2016) and its limited penetrance into deep brain structures, it is likely that some Nts-mediated physiology is solely regulated via Nts neurons within the brain and that there are distinct mechanisms by which peripheral and central Nts modify body weight and other physiology. Thus, defining the central sources of Nts is an important first step to understand how Nts mediates a diverse repertoire of physiology, including regulation of feeding and body weight.
Nts also exerts site-specific effects within the brain, hinting that distinct Nts populations coordinate specific behaviors. For instance, infusion of Nts into the periaqueductal gray (PAG), the rostral ventromedial medulla, central amygdala (CEA), posterior hypothalamic nucleus (PH), nucleus accumbens (Acb), or medial preoptic nucleus (MPO) results in decreased pain sensation with no effects on feeding (Al-Rodhan et al., 1991; Benmoussa et al., 1996; Kalivas et al., 1982; Neubert et al., 2004). Activation of Nts neurons in the MPO also modulates social interaction (McHenry et al., 2017). By contrast, Nts administered into in the VTA suppresses feeding and promotes locomotor activity that can support weight loss (Cador et al., 1986; Elliott and Nemeroff, 1986; Kalivas and Taylor, 1985). Thus, it is imperative to identify and systematically test how each Nts-expressing population in the brain contributes to physiology and behavior, as this information could inform the development of precision-treatments for chronic pain, social anxiety, obesity, or eating disorders.
The technical challenge of identifying Nts neurons, however, has hindered discovery of how they coordinate normal physiology. In situ hybridization (ISH) is suitable to identify Nts-expressing neurons but can’t be used to modulate them in vivo, as necessary to reveal their physiologic roles. Antibody-mediated Nts immunoreactivity (Nts-IR) only identifies fibers in the CNS, indicating axons of passage or terminals via which Nts is released, but fails to identify cell bodies (Uhl et al., 1977; Woodworth et al., 2018). Pre-treating animals with colchicine disrupts the microtubule network required for anterograde transport of peptides and effectively concentrates Nts within soma to permit their detection via Nts-IR; this method has been used to reveal Nts perikarya within the nucleus of the solitary tract (NTS), parabrachial nucleus (PB), dorsal raphe nucleus (DR), PAG, VTA, paraventricular hypothalamic nucleus (PVH), rostral arcuate nucleus (Arc), lateral hypothalamic area (LHA), CEA, MPO, and bed nucleus of the stria terminalis (BNST) (Ibata et al., 1984; Jennes et al., 1982; Kahn et al., 1982, 1980; Triepel et al., 1984; Uhl et al., 1979). Problematically, colchicine causes neuronal dysfunction that may alter gene expression and it is lethal, prohibiting further studies to define how these Nts populations contribute to normal physiology or disease states.
To overcome the limitations of conventional Nts detection methods, investigators have begun to use NtsCre;GFP mice that permit visualization and manipulation of all Nts-expressing neurons using Cre-LoxP technology (McHenry et al., 2017; Naganuma et al., 2019; Woodworth et al., 2017b). The fidelity of the NtsCre;GFP line has been confirmed using ISH and colchicine-mediated Nts-IR, verifying that the line reliably identifies Nts neurons in known Nts-expressing brain regions including the LHA, MPO, and Acb (Brown et al., 2019; McHenry et al., 2017; Woodworth et al., 2018). We subsequently used the NtsCre;GFP line to determine which Nts neurons provide afferents to the VTA, highlighting the Nts neurons anticipated to exert the anorectic or social behaviors mediated via Nts in this region (McHenry et al., 2017; Woodworth et al., 2018, 2017b). During our analysis, we also noted substantial populations of Nts neurons throughout the brain that did not engage the VTA and, hence, were beyond the scope of that study. Yet, we reasoned that any substantial population of Nts neurons is a likely contributor to Nts-mediated physiology and that identifying these populations will open the door for future studies to reveal their functions. As a first step toward this goal, we have conducted a brain-wide assessment of the distribution of Nts-GFP neurons throughout the brains of NtsCre;GFP mice. Additionally, we compared the distribution of Nts-GFP neurons with Nts-ISH (Lein et al., 2007). This comparison is important to identify any Nts-GFP populations that, despite expressing GFP, do not actively express Nts in adulthood. This would occur in cells that transiently expressed Nts during development, resulting in recombination and permanent GFP expression, even if these cells do not continue to express Nts (or Cre) during maturity. Our work thus provides a comprehensive “Nts-GFP atlas” that will be useful to identify Nts-containing populations in developing and adult mice. This resource will enable investigators to identify Nts populations of interest so that they may be systematically studied in the future using NtsCre;GFP mice to finally reveal how various Nts populations mediate diverse physiology.
Materials and Methods
Animals
C57/BL6 backcrossed NtsCre mice (The Jackson Laboratory, stock # 017525) were crossed with Rosa26EGFP-L10a mice (Krashes et al., 2014) to visualize all Nts neurons via their Cre-mediated induction of green fluorescent protein (GFP). Within the confines of this text an “Nts neuron” refers to any neuron that expresses Nts (as signified by GFP or Nts-ISH labeling) and is simply a shorthand to identify these cells. Designation as an “Nts neuron” does not mean that a cell exclusively expresses Nts or solely signals via it, as neurons may contain multiple neuropeptides and/or neurotransmitters. NtsCre mice have an IRES-Cre sequence located within the 3’-UTR of the Nts gene, which permits robust Cre recombinase expression sufficient to induce Cre-mediated reporter labeling in any Nts-expressing cell. The fidelity of the NtsCre mice for identifying Nts-expressing neurons has been validated previously (Brown et al., 2019; Woodworth et al., 2018). We also verified that the Rosa26EGFP-L10a line is not “leaky” and does not exhibit GFP expression without Cre-mediated recombination (Woodworth et al., 2017a) and Supplementary Figure 1. Thus, male progeny heterozygous for NtsCre and GFP were utilized throughout this study to identify Nts neurons and are referred to as NtsCre;GFP mice. Mice were bred and housed in a 12hr light/12 hr dark cycle. All procedures were approved by the Institutional Animal Care and Use Committee at Michigan State University in accordance with Association for Assessment and Accreditation of Laboratory Animal Care and National Institutes of Health guidelines. Tail biopsies were taken between 2–3 wk of age and genotyped using standard polymerase chain reaction (PCR). The following primers were used to identify NtsCre mice: common forward, 5’ ATA GGC TGC TGA ACC AGG AA; cre reverse, 5’ CCA AAA GAC GGC AAT ATG GT; and WT reverse, 5’ CAA TCA CAA TCA CAG GTC AAG AA. Primers used to detect the presence of Rosa26EGFP-L10a include: mutant forward, 5’ TCT ACA AAT GTG GTA GAT CCA GGC; WT forward, 5’ GAG GGG AGT GTT GCA ATA CC, and common reverse, 5’ CAG ATG ACT ACC TAT CCT CCC.
Immunohistochemistry and Immunofluorescence
NtsCre;GFP mice were anesthetized at 23 weeks of age with intraperitoneal pentobarbital and transcardially perfused, first with 1X phosphate-buffered saline (PBS) and then with 10% formalin (Fisher Scientific, Pittsburgh, PA). Brains were removed, stored in 10% formalin overnight, and then dehydrated with a 30% sucrose solution. Brains were cut coronally into four series of 30 μm sections using a freezing microtome (Leica, Buffalo Grove, IL). To enhance visualization of Nts-GFP neurons, sections from one series were incubated in primary antibody for GFP (Abcam, chicken, 1:1000; RRID: AB_300798). To examine the CEA additional primary antibodies were used to detect Protein Kinase C-Delta (PKC-δ, BD Biosciences, mouse, 1:1000; RRID: AB_397781). After overnight incubation at room temperature in primary antibodies, brain sections were washed 6 times in PBS. Next, species-specific Alexa-488 conjugated (Jackson ImmunoResearch, 1:200; RRID: AB_2340375) or Alexa-568 conjugated antibodies (LifeTech, 1:200; RRIDs: AB_2534013 and AB_2534017) were applied for 2 hours at room temperature. Sections were finally washed with PBS to remove any non-specific binding and were then mounted onto slides and coverslipped with ProLong Antifade mounting media. Immunolabeled brain sections were analyzed using an Olympus BX53 fluorescence microscope outfitted with FITC and Texas Red filters, and images were collected using Cell Sens software and a Qi-Click 12 Bit cooled camera. Images were subsequently analyzed using Photoshop software (Adobe) to identify anatomical landmarks, then were referenced to the Franklin and Paxinos mouse brain atlas (Franklin and Paxinos, 2013) to determine the bregma coordinate that best matched the image. Supplemental Figure 1 shows antibody controls in ++;GFP mice and NtsCre;GFP mice, verifying that GFP expression is indeed Cre-dependent and that the immunolabeling protocol specifically detects GFP in NtsCre;GFP mice.
Three separate male NtsCre;GFP mice were analyzed to map the location of Nts soma throughout the entire brain and qualitatively assess the density of Nts-GFP populations within each brain region. Due to the design of the reporter strain (a GFP-L10a ribosomal protein fusion) Cre-induced GFP was robustly localized within ribosome-rich structures such as the soma. Neuronal processes were not readily visible since these structures have few ribosomes and hence accrue little GFP-L10a. We therefore restricted our study to assessing the location of GFP-labeled soma. Nts-GFP images were analyzed “by eye” to rate the relative density of Nts neurons as follows, from most to least dense: ++++ = Very dense distribution of soma that nearly overlap; +++/++++ = dense distribution of soma but no overlap; +++ = Numerous distributed soma; ++/+++ = Intermediate; ++ = Moderate density of soma; +/++ = Some scattered soma; + = Sparse soma; 0/+ = Very few if any soma noted. Density ratings with two figures (e.g. ++/+++) indicate that the Nts density is somewhat between the higher and lower rating. For example, a ++/+++ means that there are moderate-to-numerous neurons in this area, but not quite as many as a +++ region. Data were compared between the three brains, and an average rating of Nts-GFP cell density was assigned for each brain structure. While there was some variability in Nts cell density between the three brains analyzed (one brain each with low, intermediate and higher relative number of Nts soma), they all had comparable, consistent distributions of Nts neurons. For example, the range of Nts neurons observed from the LHA and BNST of the three brains is shown in Supplementary Figure 2 and confirms the minor variability between brains. The average density rating was taken from the brain with the intermediate density of Nts-GFP neurons, as it represents the conservative “middle ground” of Nts cell density: Supplemental Table 1 lists these the average relative densities of Nts-GFP neurons throughout the brain.
Additionally, we searched the database of coronally-sectioned, adult mouse brain Nts-ISH images courtesy of the Allen Brain Atlas (Lein et al., 2007) to identify a section with similar landmarks/bregma level to the Nts-GFP image. The entire Nts-ISH dataset can be referenced via the Allen Brain Atlas website at http://mouse.brain-map.org/experiment/show/73788032 and images obtained from it are identified in our figures as “Image Credit: Allen Institute”. Nts ISH data from the Allen Brain atlas was performed using a 56 week-old male C57BL/6J mouse. All Nts-ISH images were assigned a bregma coordinate to permit comparison of similar brain areas from Nts-GFP images. Due to differences in sectioning or fixation the assigned bregma coordinates may not be matched with exact precision. Yet, bregma coordinates are provided as a helpful starting point for investigators who may want to target specific Nts populations in the future via stereotaxic manipulations.
Fluorescence In Situ Hybridization (ISH)
C57Bl/6J wild type mice (Jackson Labs, n=3 per experiment) were perfused with fixative and brains were extracted, post-fixed and sectioned as above. For each mouse, three free floating sections spanning a brain region of interest were selected for analysis using Advanced Cell Diagnostics RNAScope single-plex or multiplex fluorescent kits (catalog #322360 and 323100), per the manufacturer’s protocol. Briefly, sections were dried onto slides in a 40 °C oven for 1 hour, incubated in hydrogen peroxide solution for 10–60 min at room temperature, washed in distilled water and dried. After incubation in 100°C 1X Target Retrieval Reagent (7–10 minutes), sections were rinsed, dried and dipped in 100% alcohol. Dry sections were incubated in Protease III solution for 15 minutes at 40 °C and washed in distilled water. Samples were then incubated with probes for 2 hours in a humidified oven at 40°C. Samples singly labeled with Nts probe (catalog no. 420441) were visualized with Fast-Red-A and -B (60:1). Samples simultaneous hybridized with Nts and Galanin (Gal; Mm-Gal-C2, cat. no. 400961-C2) probes were visualized with TSA plus fluorescein or Cy3, respectively. Sections were finally rinsed, dipped in xylene for 2 min and coverslipped with antifade mounting media.
Results
General Observations
We observed many Nts-GFP cells scattered throughout the brain, which are described in Supplemental Table 1 by their location across the caudal-rostral brain axis and relative density. In many cases Nts-GFP cells were evenly distributed throughout a brain region, but we also observed sites in which Nts-GFP neurons were visibly grouped together in clusters; we refer to the latter as Nts-GFP populations. Figures include representative images from NtsCre;GFP mice of the brain areas with the largest density of Nts-GFP cells (those with qualitative density ratings of +++/++++ or ++++, see Supplemental Table 1 and Table 1) across the entire caudal-rostral axis of the brain. Each Nts-GFP image was assigned a Bregma level according to the mouse brain atlas (Franklin and Paxinos, 2013) to permit identification of Nts-GFP containing brain regions using stereotaxic coordinates, and thus how to target specific Nts-GFP populations for future manipulations. Corresponding images of Nts-ISH from an adult mouse (courtesy of the Allen Brain Atlas (Lein et al., 2007)) are presented alongside each Nts-GFP image to distinguish whether mature neurons in these regions actively express Nts. This is important because Cre-mediated recombination will occur during whichever stage Nts is expressed in the NtsCre;GFP mice, inducing permanent GFP expression. Thus, any developmentally-expressing Nts cells will undergo recombination and remain GFP-labeled throughout the lifespan, and this is independent of whether or not such cells actively express Nts in the mature brain. Comparing Nts-GFP and Nts-ISH data from adult mice suggests which Nts-GFP cells expressed Nts developmentally versus during adulthood. Table 1 summarizes regions in which we observed a higher density of Nts-GFP neurons than Nts-ISH (average density ratings differing by more than one “+”), and which may exhibit developmentally-restricted expression of Nts.
Table 1:
Brain Regions with Differing Density of Nts-GFP Neurons and Nts-ISH. Caudal to rostral list of brain regions in which the average density rating of Nts-GFP neurons and Nts-ISH neurons differed by one or more ‘+’ rating units and the Bregma coordinates at which they were found. The relative density of Nts-ISH was assessed in each of these regions from the publicly accessible Allen Brain dataset of coronal Nts-ISH images, and their corresponding Bregma coordinates are given (Lein et al., 2007) (© 2004 Allen Institute for Brain Science. Allen Mouse Brain Atlas. Available from: mouse.brain-map.org).
| Abbreviation | Structure | Relative Density of Nts-GFP cells | Relative Density of Nts-ISH Cells (Allen Brain) |
|---|---|---|---|
| MdV | Medullary reticular nucleus, ventral part | ++ | 0/+ |
| IOA/IOB | Inferior olive, subnucleus A and B of the medial nucleus | +++ | 0/+ |
| Sp5I | Spinal trigeminal nucleus, interpolar part | +++ | + |
| IOC | Inferior olive, subnucleus C of medial nucleus | ++/+++ | 0/+ |
| IOD | Inferior olive, dorsal nucleus | ++/+++ | 0 |
| IODM | Inferior olive, dorsomedial cell group | +/++ | 0 |
| 7VM, 7DM, 7DI, 7DL, 7L, 7VI | Facial nucleus subnuclei | +++ | +/++ |
| P7 | Perifacial zone | +/++ | 0 |
| VCA | Ventral cochlear nucleus, anterior part | +++ | 0 |
| LPBD | Lateral parabrachial nucleus, dorsal part | +++/++++ | 0/+ |
| 5ADi | Motor trigeminal nucleus, anterior digastric part | ++ | 0/+ |
| CnF | Cuneiform nucleus | +++/++++ | ++/+++ |
| Su5 | Supratrigeminal nucleus | ++/+++ | + |
| PR5VL | Principal sensory trigeminal nucleus, ventrolateral part | +++/++++ | 0 |
| Sag | Sagulum nucleus | +++/++++ | + |
| DCIC | Dorsal cortex of the inferior colliculus | ++/+++ | 0/+ |
| PR5 | Principal sensory trigeminal nucleus | ++++ | 0 |
| LPAG | Lateral periaqueductal gray | +++/++++ | +/++ |
| DMPAG | Dorsomedial periaqueductal gray | +++/++++ | 0 |
| DLPAG | Dorsolateral periaqueductal gray | +++/++++ | 0 |
| DpGi | Deep gray layer of the superior colliculus | +++/++++ | + |
| DpWh | Deep white layer of the superior colliculus | +++/++++ | 0/+ |
| InG | Intermediate gray layer of the superior colliculus | ++/+++ | + |
| InWh | Intermediate white layer of the superior colliculus | ++/+++ | 0/+ |
| PBG | Parabigeminal nucleus | +++/++++ | + |
| 3N | Oculomotor nucleus | ++/+++ | 0/+ |
| 3PC | Oculomotor nucleus, parvicellular part | ++/+++ | + |
| PRh | Perirhinal Cortex | +/++ | 0 |
| rs | Rubrospinal tract | ++ | 0/+ |
| Op | Optic nerve layer of the superior colliculus | ++ | 0/+ |
| RSG/RSD | Retrosplenial Granular/Dysgranular Cortex | ++++ | 0 |
| DpGi | Deep Gray Layer of the Superior Colliculus | ++/+++ | 0 |
| DpWh | Deep White Layer of the Superior Colliculus | ++ | 0 |
| MGV | Medial Geniculate nucleus, Ventral part | +++ | 0 |
| InC/InCSh | Interstitial nucleus of Cajal w/ shell region | ++ | 0 |
| APir | Amygdalopiriform transition area | ++/+++ | 0 |
| Dk | Nucleus of Darkschewitsch | +++ | + |
| PoT | Posterior Thalamic nucleus, Triangular | +++/++++ | + |
| LT | Lateral terminal nucleus acc optic tract | ++ | 0 |
| APT | Anterior pretectal nucleus | +/++ | 0 |
| ZIC | Zona incerta, caudal | ++/+++ | 0/+ |
| p1Rt | p1 reticular formation | +/++ | 0 |
| MCPC | Magnocellular nucleus post comm | +/++ | 0 |
| ML | Medial mammillary nucleus, lateral | +++ | 0 |
| PBP | Parabrachial pigmented nucleus of the Ventral Tegmental Area | +/++ | 0 |
| PLi | Posterior Limitans Thalamic nucleus | ++ | 0 |
| LM | Lateral mammillary nucleus | +++/++++ | 0 |
| rmx | Retromammillary decussation | +/++ | 0 |
| PAG | Periaqueductal Gray | +++/++++ | + |
| RML | Retromammillary nucleus, Lateral | ++/+++ | 0 |
| SNC/SNR | Substantia Nigra Compacta/Reticular | +++ | 0/+ (SNC), + (SNR) |
| LPMC | LP Thalamic nucleus, Mediocaudal | ++/+++ | 0 |
| Py | Pyramidal Cell Hippocampus | ++++ | 0/+ |
| LPLR | LP Thalamic nucleus, Laterorostral | +/++ | 0 |
| LPMR | LP Thalamic nucleus, Mediorostral | +/++ | 0 |
| PR | Prerubral Field | ++/+++ | 0 |
| FF | Fields of Forel | ++/+++ | 0 |
| pv | Paraventricular fiber system | +++ | +/++ |
| ArcLP/ArcMP | Caudal Arcuate Hypothalamic nucleus | ++ | 0 |
| PH | Posterior Hypothalamic nucleus | +++ | +/++ |
| MD, MDL, MDC, MDM | Mediodorsal Thalamic nucleus | ++ | 0 |
| PF | Parafascicular Thalamic nucleus | ++ | 0 |
| CM | Central Medial Thalamic nucleus | ++/+++ | 0 |
| PVP | Paraventricular Thalamic nucleus, Posterior | +++ | + |
| PV | Paraventricular Thalamic nucleus | +++ | 0 |
| Po | Posterior Thalamic nuclear group | ++ | 0 |
| SPF | Subparafascicular Thalamic nucleus | +++ | 0 |
| PoMn | Posteromedian Thalamic nucleus | ++ | 0 |
| VMH | Ventromedial Hypothalamic nucleus | ++ | 0 |
| MePV | Medial Amygdalar nucleus, posteroventral | ++ | 0 |
| DMV | Dorsomedial Hypothalamic nucleus, Ventral | ++ | 0 |
| CL | Centrolateral Thalamic nucleus | ++ | 0 |
| PC | Paracentral Thalamic nucleus | ++ | 0 |
| ArcD/ArcL | Arcuate hypothalamic nucleus, Dorsal/Lateral | ++/+++ | 0 |
| Xi | Xiphoid Thalamic nucleus | ++ | 0/+ |
| IMD | Intermediodorsal Thalamic nucleus | ++/+++ | 0 |
| Arc | Rostral Arcuate Hypothalamic nucleus | ++++ | +/++ |
| BMA | Basomedial Amygdalar nucleus, anterior | ++/+++ | 0 |
| AV | Anteroventral thalamic nucleus | +/++ | 0 |
| RchL | Retrochiasmatic, Lateral | +/++ | 0 |
| EP/MGP | Entopeduncular nucleus | ++/+++ | 0 |
| MeAD | Medial Amygdalar nucleus, Anterodorsal | ++/+++ | 0 |
| AHP | Anterior Hypothalamic Area, Posterior | ++/+++ | 0 |
| RCh | Retrochiasmatic Area | ++ | 0 |
| IAD | Interanterodorsal thalamic nucleus | +/++ | 0 |
| IAM | Interanteromedial thalamic nucleus | +/++ | 0 |
| MeAV | Medial Amygdalar nucleus, Anteroventral | +++ | +/++ |
| IM | Intercalated amygdalar nucleus, main | ++/+++ | 0 |
| BAOT | Bed nucleus Access of the Olfactory tract | +/++ | 0 |
| AA | Anterior amygdalar area | ++ | 0 |
| AHC | Anterior Hypothalamic Area, Central | ++ | 0/+ |
| VLH | Ventrolateral hypothalamic nucleus | +/++ | 0 |
| Cg | Cingulate Cortex | ++++ | 0 |
| EAC/EAM/EA | Sublenticular extended amygdala | ++/+++ | 0 |
| A14 | A14 Dopamine cells | ++ | 0 |
| AVPV | Anteroventral Periventricular nucleus | ++++ | ++/+++ |
| VLPO | Ventrolateral Preoptic nucleus | ++/+++ | + |
| StA | Strial part of the Preoptic Area | ++ | 0 |
| AcbC | Nucleus Accumbens, Core | *between + and +++ | 0/+ |
| SIB | Substantia innominata | ++/+++ | 0 |
| AcbSh | Nucleus Accumbens, Shell | ++++ | ++/+++ |
| ICjM | Island of Cajella, Major Island | +++ | 0 |
| DTT | Dorsal Tenia Tecta | ++ | 0 |
| PrL | Prelimbic Cortex | ++ | 0 |
| IL | Infralimbic Cortex | ++ | 0 |
| SHi | Septohippocampal nucleus | +++ | 0 |
| Nv | Navicular Postolfactory nucleus | ++++ | + |
| VTT | Ventral Tenia Tecta | ++/+++ | 0 |
| AOM | Anterior Olfactory Area, Medial Part | ++/+++ | 0 |
| MO | Medial Orbital Cortex | +/++ | 0 |
Relative density ratings:
Very dense distribution of soma that nearly overlap;
dense distribution of soma but no overlap
Numerous distributed soma
Intermediate
Moderate density of soma
Some scattered soma
Sparse soma
Very few if any soma noted.
Hindbrain
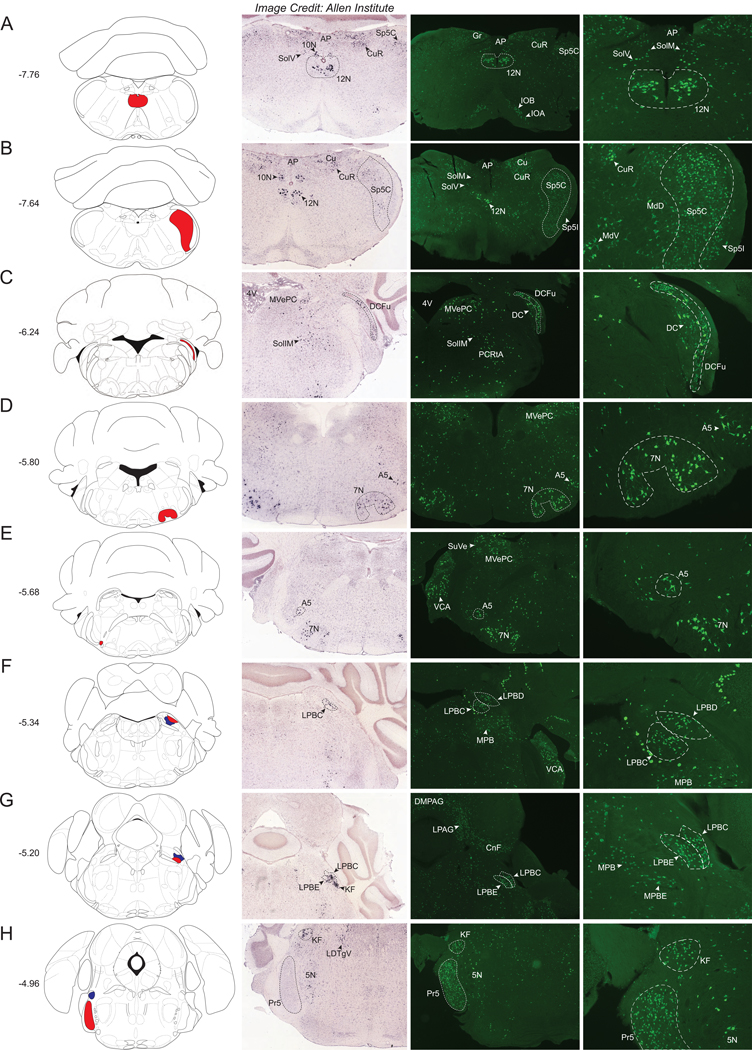
Starting caudally, the most notable structure containing a dense population of Nts-GFP neurons is the hypoglossal nucleus (12N), which also contains robust Nts-ISH (Fig 1A). Other hindbrain regions containing sizable Nts-GFP populations and dense Nts-ISH include the caudal portion of the spinal trigeminal nucleus (Sp5C) (Fig 1B), the fusiform region of the dorsal cochlear nucleus (DCFu) (Fig 1C), the facial nucleus (7N) (Fig 1D), the A5 group of noradrenaline cells (Fig 1D, E), and the dorsal, external, and central parts of the lateral parabrachial nucleus (LPBD, LPBE, LPBC) (Fig 1F–G). The Koelliker-fuse nucleus (KF) and the principal sensory trigeminal nucleus (Pr5) contain many Nts-GFP neurons, and while a comparable level of Nts-ISH exists in the KF, there is an absence of detectable Nts-ISH signal in the Pr5 (Fig 1H), suggesting that Nts is transiently expressed by Pr5 cells at some stage of development, but not in adult Nts-GFP neurons.
Figure 1.
Nts-GFP and Nts-ISH in the Hindbrain. From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Red and blue shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma −7.76, Red shading corresponds to the Hypoglossal nucleus (12N), B) Bregma −7.64, Red shading corresponds to the Spinal trigeminal tract, caudal part (Sp5C), C) Bregma −6.24, Red shading corresponds to the Dorsal cochlear nucleus, fusiform layer (DCFu), D) Bregma −5.80, Red shading corresponds to the Facial nucleus (7N), E) Bregma −5.68, Red shading corresponds to the A5 noradrenaline cell group (A5), F) Bregma −5.34, Red shading corresponds to the Lateral parabrachial nucleus, dorsal part (LPBD) whereas blue shading corresponds to the Lateral parabrachial nucleus, central part (LPBC), G) Bregma −5.20, Red shading corresponds to the Lateral parabrachial nucleus, external part (LPBE) while blue shading corresponds to the LPBC, H) Bregma −4.96, Red shading corresponds to the Principal sensory trigeminal nucleus (Pr5) whereas blue shading corresponds to the Koelliker-Fuse nucleus (KF). AP= Area postrema, 10N = dorsal motor nucleus of the vagus, Gr= Gracile nucleus, CuR= Cuneate nucleus, Cu= Cuneate nucleus and fasciculus, Sp5I= Spinal trigeminal tract, interpolar part, IOB= Inferior olive subnucleus B of the medial nucleus, IOA= Inferior olive subnucleus A of the medial nucleus, SolM= Solitary nucleus, medial part, SolV= Solitary nucleus, ventral part, MdV= Medullary reticular nucleus, ventral part, MdD= Medullary reticular nucleus, dorsal part, 4V= 4th ventricle, MVePC= Medial vestibular nucleus, parvicellular part, DC= Dorsal cochlear nucleus, SollM= Solitary nucleus, intermediate part, PCRtA= Parvicellular reticular nucleus, SuVe= Superior vestibular nucleus, VCA= Ventral cochlear nucleus, anterior part, MPB= Medial parabrachial nucleus, DMPAG= Dorsomedial periaqueductal gray, LPAG= Lateral periaqueductal gray, CnF= Cuneiform nucleus, 5N= Motor trigeminal nucleus.
Other hindbrain structures contained more diffuse, but readily identifiable populations of Nts-GFP neurons with qualitative density ratings of ++/+++ or +++, as per Supplemental Table 1. Some of these moderately dense populations of Nts-GFP neurons are shown in Figure 1 and include the gracile (Gr) and cuneate nuclei (Cu and CuR), the inferior olivary complex (IOA and IOB), the caudal aspect of the interpolar spinal trigeminal nucleus (SP5I), the area postrema (AP), the parvicellular part of the medial vestibular nucleus (MVePC), the anterior aspect of the ventral cochlear nucleus (VCA), and the medial parabrachial nucleus (MPB and MPBE). We observed other moderately sized Nts-GFP populations (not pictured but described in Supplemental Table 1) within the retractor bulbi part of the adbucens nucleus (6RB), the supratrigeminal nucleus (Su5), and the laterodorsal tegmental nucleus (LDTg). Nts-ISH was detected in a similar distribution and density as the Nts-GFP cells within the Gr, Cu, AP, MVePC, 6RB, MPB and LDTg.
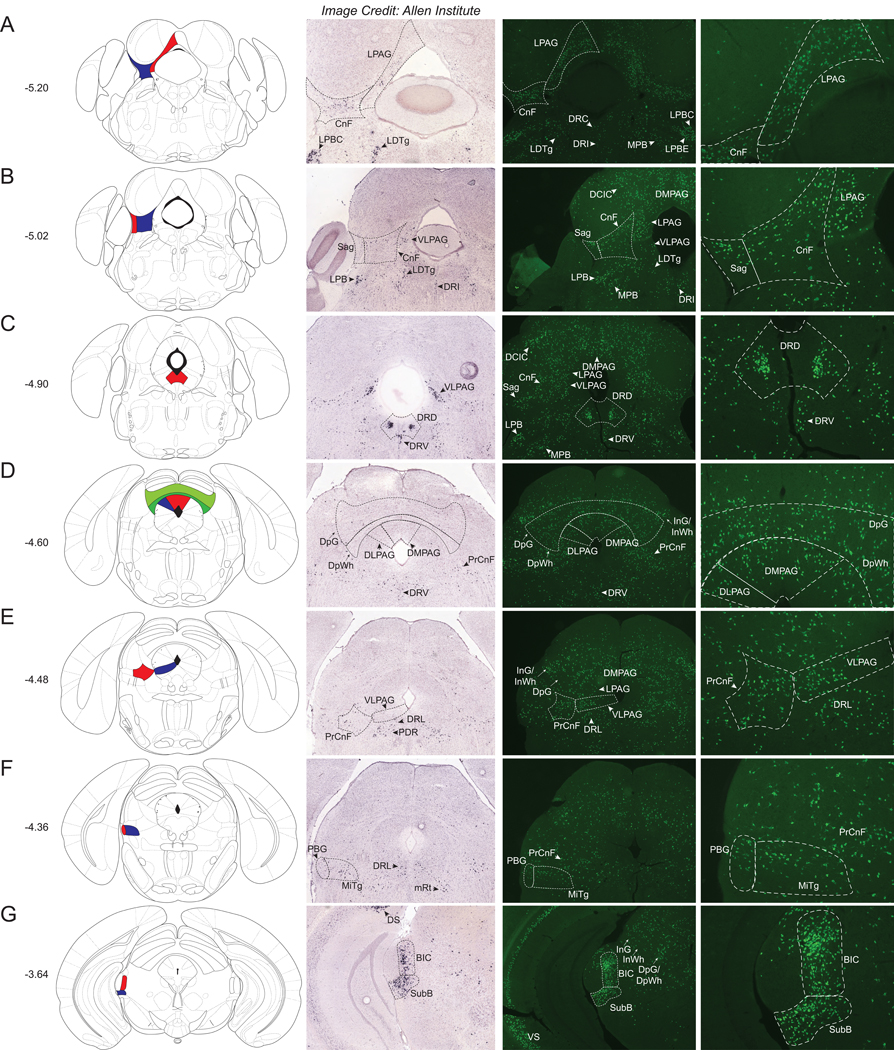
Midbrain
We observed many Nts-GFP neurons evenly scattered throughout the periaqueductal gray (PAG), including within the lateral (LPAG- Fig 2A), dorso-lateral and dorso–medial (DLPAG/DMPAG - Fig 2D), and the ventrolateral (VLPAG- Fig 2E) sub-regions. Interestingly, only the caudal VLPAG exhibited significant Nts-ISH (Fig 2B and C), whereas Nts-ISH was absent from the DLPAG and DMPAG (Fig 2D). This discrepancy between the distributions of Nts-GFP and Nts-ISH may signify that Nts is transiently expressed throughout most of the PAG during development, but is only in adult neurons of the VLPAG. In contrast, the adjacent cuneiform nucleus (CnF) contains a dense population of Nts-GFP neurons as well as Nts-ISH (Fig 2A and 2B). A large, dense cluster of Nts-GFP neurons and Nts-ISH was found within the dorsal aspect of the dorsal raphe nucleus (DRD) that lies ventral to the cerebral aqueduct (Fig 2C). Nts-GFP neurons and corresponding Nts-ISH were also found, but more evenly distributed, within the lateral and ventral aspects of the dorsal raphe (DRL and DRV) (Fig 2C–E). Two regions with particularly dense distributions of Nts-GFP neurons and Nts-ISH included the subbrachial nucleus (SubB) and the nucleus of the brachium of the inferior colliculus (BIC) (Fig 2G). Other midbrain regions with sizable, yet evenly dispersed Nts-GFP neurons and Nts-ISH, include the Sagulum (Sag) (Fig 2B), the deep gray and white layers of the superior colliculus (DpG) (Fig 2D), the precuneiform area (PrCnF) (Fig 2E), the parabigeminal nucleus (PBG) (Fig 2F), and the microcellular tegmental nucleus (MiTg) (Fig 2F). Midbrain structures with more moderate densities of Nts-GFP neurons (with qualitative density ratings of ++/+++ or +++) include the dorsal cortex of the inferior colliculus (DCIC), lateral lemniscus (ll), subpeduncular tegmental nucleus (SPTg), located just beneath the decussation of the superior cerebellar peduncle (scp), the intermediate gray and white layers of the superior colliculus (InG / InWh), the oculomotor nucleus (3N) and associated structures, the mesencephalic reticular formation (mRT) and the nucleus of Darkschewitsch (Dk) (Supplemental Table 1). At the transition between the midbrain and caudal hypothalamus we also observed scattered Nts-GFP neurons and Nts-ISH within the Substantia Nigra Compacta (SNC); this is best observed in (Fig 4A).
Figure 2.
Nts-GFP and Nts-ISH in the Midbrain. From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Red, blue, and green shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma −5.20, Red shading corresponds to the Lateral periaqueductal gray (LPAG) while blue shading corresponds to the Cuneiform nucleus (CnF), B) Bregma −5.02, Red shading corresponds to the Sagulum nucleus (Sag) whereas blue shading corresponds to the CnF, C) Bregma −4.90, Red shading corresponds to the Dorsal raphe nucleus, dorsal part (DRD), D) Bregma −4.60, Red shading corresponds to the Dorsomedial periaqueductal gray (DMPAG), blue shading corresponds to the Dorsolateral periaqueductal gray (DLPAG), dark green shading corresponds to the Deep white layer of the superior colliculus (DpWh), and light green shading corresponds to the Deep gray layer of the superior colliculus (DpG), E) Bregma −4.48, Red shading corresponds to the Precuneiform area (PrCnF) whereas blue shading corresponds to the ventrolateral periaqueductal gray (VLPAG), F) Bregma −4.36, Red shading corresponds to the Parabigeminal nucleus (PBG) while blue shading corresponds to the Microcellular Tegmental nucleus (MiTg), G) Bregma −3.64, Red shading corresponds to the Brachium of the Inferior Colliculus (BIC) whereas blue shading corresponds to the Subbrachial nucleus (SubB). LDTg= Laterodorsal tegmental nucleus, LPBC= Lateral parabrachial nucleus, central part, LPBE=Lateral parabrachial nucleus, external part, MPB= Medial parabrachial nucleus, DRC= Dorsal raphe nucleus, caudal part, DRI=Dorsal raphe nucleus, interfascicular part, MPB= Medial parabrachial nucleus, DCIC= Dorsal cortex of the inferior colliculus, DRV= Dorsal raphe nucleus, ventral part, DRL= Dorsal raphe nucleus, lateral part, InG/InWh= Intermediate gray or white layer of the superior colliculus, PDR= Posterodorsal raphe nucleus, mRt=Mesencephalic reticular formation, DS= Dorsal Subiculum, VS=Ventral Subiculum.
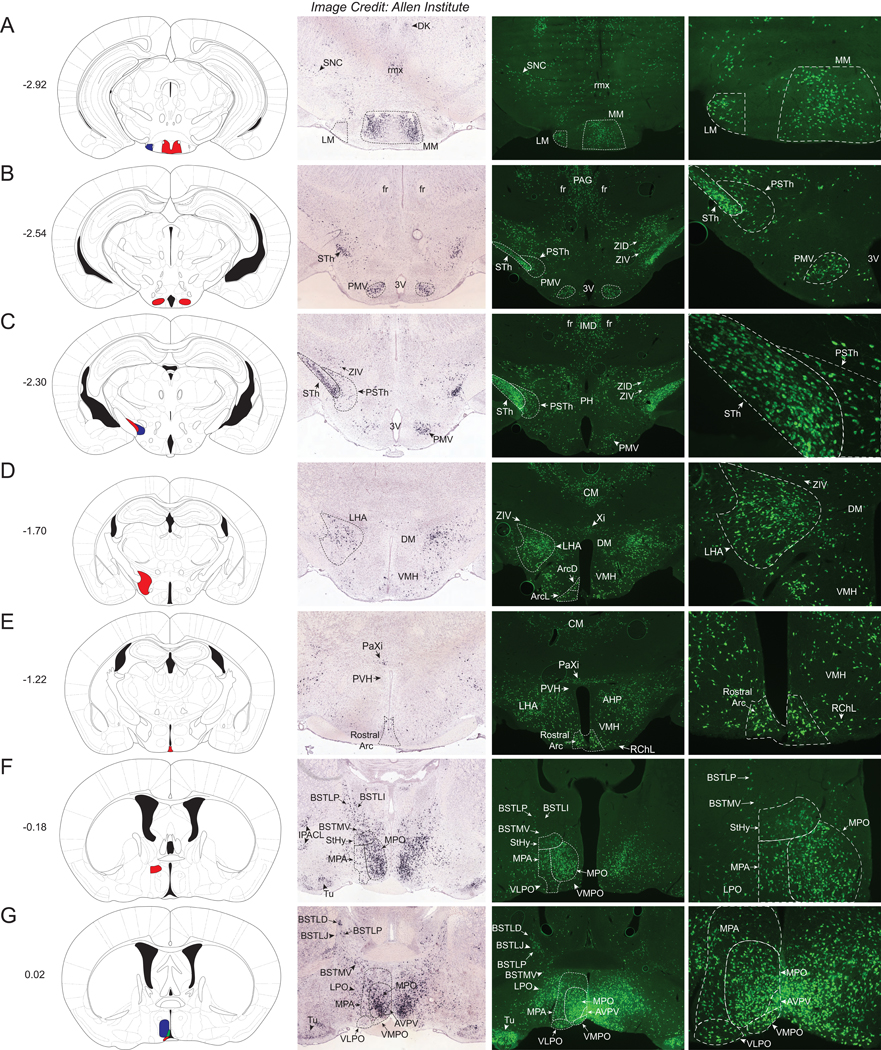
Figure 4. Nts-GFP and Nts-ISH in the Hypothalamus.
From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma −2.92, Red shading corresponds to the medial mammillary nucleus, medial part (MM) whereas blue shading corresponds to the lateral mammillary nucleus (LM), B) Bregma −2.54, Red shading corresponds to the Premammillary nucleus, ventral (PMV), C) Bregma −2.30, Red shading corresponds to the Subthalamic nucleus (STh) while blue shading corresponds to the Parasubthalamic nucleus (PSTh), D) Bregma −1.70, Red shading corresponds to the Lateral Hypothalamus (LHA), E) Bregma −1.22, Red shading corresponds to the Rostral Arcuate nucleus (Arc), F) Bregma −0.18, Red shading corresponds to the Striohypothalamic nucleus (StHy), G) Bregma +0.02, Red shading corresponds to the Ventromedial Preoptic nucleus (VMPO), blue shading corresponds to the Medial Preoptic nucleus (MPO), and green shading corresponds to the Anteroventral Periventricular nucleus (AVPV). DK= Nucleus of Darkschewitsch, rmx= Retromammillary decussation, SNC= Substantia Nigra Compacta, fr= Fasciculus retroflexus, PSTh= Parasubthalamic nucleus, STh= Subthalamic nucleus, 3V= 3rd ventricle, PAG= Periaqueductal Gray, ZID= Zona Incerta, Dorsal, ZIV= Zona Incerta, Ventral, IMD= Intermediodorsal Thalamic nucleus, PH= Posterior Hypothalamic nucleus, DM= Dorsomedial Hypothalamic nucleus, VMH=Ventromedial Hypothalamic nucleus, CM= Central Medial Thalamic nucleus, Xi= Xiphoid Thalamic nucleus, PaXi= Paraxiphoid nucleus of Thalamus, ArcD= Arcuate hypothalamic nucleus, Dorsal, ArcL= Arcuate hypothalamic nucleus, Lateral, PVH= Paraventricular nucleus of the Hypothalamus, AHP= Anterior Hypothalamic Area, Posterior, RChL= Retrochiasmatic, Lateral, BSTLI= Bed nucleus of the stria terminalis, lateral division, intermediate part, BSTLP= Bed nucleus of the stria terminalis, lateral division, posterior part, BSTMV= Bed nucleus of the stria terminalis, medial division, ventral part, BSTLD= Bed nucleus of the stria terminalis, lateral division, dorsal part, BSTLJ= Bed nucleus of the stria terminalis, lateral division, juxtacapsular part, IPACL= Interstitial nucleus of the posterior limb of the anterior commissure, MPA= Medial Preoptic Area, Tu= Olfactory tubercle, VLPO= Ventrolateral Preoptic nucleus, LPO= Lateral Preoptic nucleus.
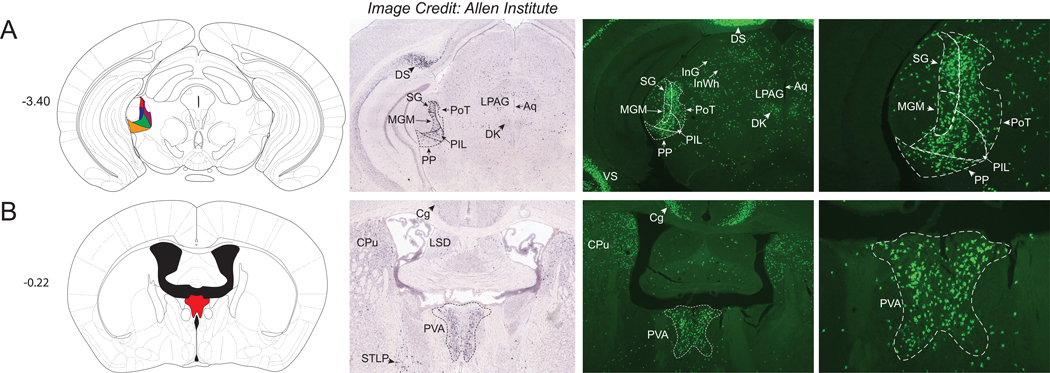
Thalamus
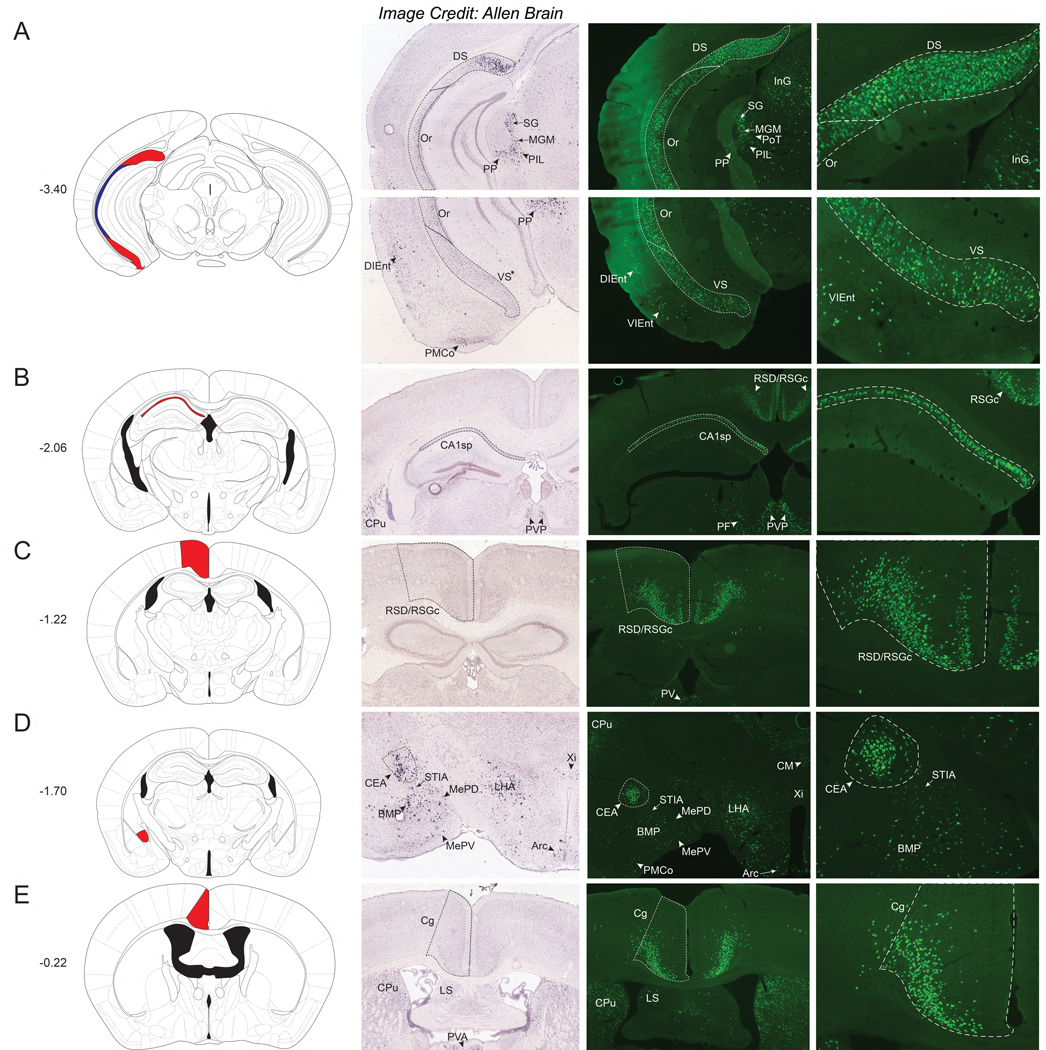
There were few significant Nts-GFP clusters observed in the thalamus of NtsCre;GFP mice compared to other brain areas. Dense populations of Nts-GFP neurons were observed within the medial aspect of the medial geniculate nucleus (MGM), the triangular posterior thalamic nucleus (PoT), the posterior intralaminar thalamic nucleus (PIL), the peripeduncular nucleus (PP), and the suprageniculate thalamic nucleus (SG) (Fig 3A). Apart from the PoT, these thalamic structures contained ample Nts-ISH and, hence, actively express Nts in the adult brain. Another concentrated population of Nts-GFP neurons and a similar distribution of Nts-ISH was observed within the anterior paraventricular thalamic nucleus (PVA) (Fig 3B). The density of Nts-GFP neurons increased over the caudal to rostral extent of the PVA, such that the Nts-GFP neurons were most abundant in the rostral aspect. Other thalamic regions contained more modest populations of Nts-GFP neurons, and these areas included the ventral part of the medial geniculate nucleus (MGV), the mediocaudal LP thalamic nucleus (LPMC), the central medial thalamic nucleus (CM), the subparafascicular thalamic nucleus (SPF), and the intermediodorsal thalamic nucleus (IMD) (refer to Supplemental Table 1). While the intermediodorsal (IMD) and central medial (CM) thalamic nuclei contain many Nts-GFP neurons, these regions lack comparable Nts-ISH (best observed in Fig 4C–E). Despite reports indicating the presence of Nts-IR fibers within these medial thalamic structures (Jennes et al., 1982; Ray and Price, 1990), the failure to detect Nts-IR soma or Nts ISH in these sites together with our data suggests that Nts is only transiently expressed during the development of these thalamic neurons. One notable exception are the visible clusters of Nts-GFP and Nts-ISH cells observed in the Xiphoid and Paraxiphoid nuclei of Thalamus (Xi and PaXi); however, these structures are more closely associated with the hypothalamus (Fig 4D–E). Hence, Nts may play an important role in the development of the thalamus but signaling may only be maintained during adulthood in select thalamic cells.
Figure 3.
Nts-GFP and Nts-ISH in the Thalamus. From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma −3.40, Red shading corresponds to the Suprageniculate Thalamic nucleus (SG), blue shading corresponds to the Medial Geniculate nucleus, medial part (MGM), yellow shading corresponds to the Peripeduncular nucleus (PP), green shading corresponds to the Posterior Intralaminar Thalamic nucleus (PIL), and maroon shading corresponds to the Posterior Thalamic nucleus, Triangular (PoT), B) Bregma −0.22, Red shading corresponds to the Paraventricular Thalamic nucleus, Anterior (PVA). DS= Dorsal Subiculum, LPAG=Lateral periaqueductal gray, Aq=Aqueduct, DK= Nucleus of Darkschewitsch, InG/InWh= Intermediate gray or white layer of the superior colliculus, CPu= Caudate Putamen, Cg= Cingulate Cortex, LSD= Lateral Septal Nucleus, dorsal part, BSTLP= Bed nucleus of the stria terminalis, lateral division, posterior part.
Hypothalamus and Adjacent Regions
Starting at the caudal extent of the hypothalamus, we observed many Nts-GFP neurons in the medial and lateral regions of the mammillary nucleus (MM and LM), yet, comparable Nts-ISH was only observed in the MM and was absent from the LM (Fig 4A). The ventral premammillary nucleus (PMV) contained a distinct cluster of Nts-GFP neurons consistent with Nts-ISH data (Fig 4B). In addition, a densely packed population of Nts-GFP neurons and comparable Nts-ISH were apparent within the subthalamic nucleus (STh) (Fig 4C). The adjacent parasubthalamic nucleus (PSTh) contained more sparsely distributed Nts-GFP and Nts-ISH-identified neurons (Fig 4C). Just above these regions lie the ventral and dorsal portions of the Zona Incerta (ZIV, ZID), which contained sparse Nts-GFP neurons and similar distributions of Nts-ISH (Fig 4C). Moving rostrally, the next large population of Nts-GFP neurons and Nts-ISH was found within the lateral hypothalamic area (LHA) (Fig 4D). Nts-GFP neurons were also noted within the rostral arcuate nucleus (Arc), a region essential for regulating energy balance; however, sparse Nts-ISH was observed in this structure (Fig 4E). Other mediobasal areas that modulate energy balance, such as the ventromedial and dorsomedial hypothalamic nuclei (VMH and DM), contained scattered Nts-GFP neurons but little observable Nts-ISH (Fig 4D). Notably, the paraventricular nucleus of the hypothalamus (PVH) was virtually devoid of Nts-GFP cells and Nts ISH (Fig 4E), which is interesting given the known cellular heterogeneity of this brain area and its importance in energy balance.
The rostral-medial hypothalamus harbored abundant Nts-GFP neurons, notably within the striohypothalamic nucleus (StHy), the medial preoptic nucleus (MPO), the ventromedial preoptic nucleus (VMPO), and the anteroventral periventricular nucleus (AVPV) (Fig 4F and 4G). Indeed, the sheer density of tightly-packed Nts-GFP cells in the MPO and AVPV made it difficult to resolve individual neurons. The Nts-ISH distribution matches that of the Nts-GFP cells within the MPO, but is less pronounced in the StHy, VMPO, and AVPV (Fig 4F and 4G). More modestly-sized populations of Nts-GFP neurons were found within the posterior aspect of the anterior hypothalamic area (AHP), posterior hypothalamus (PH), lateral preoptic nucleus (LPO), ventrolateral preoptic nucleus (VLPO), septohypothalamic nucleus (SHy), and parastrial nucleus (PS) and comparably less Nts-ISH was present in these regions relative to Nts-GFP cells (Supplemental Table 1).
The bed nucleus of the stria terminalis (BNST) complex lies dorsal to the preoptic area (POA) and contained a moderate number of Nts-GFP and Nts-ISH labeled cells. These cells were mostly scattered throughout the lateral division, including the intermediate (BSTLI), posterior (BSTLP), dorsal (BSTLD) and juxtacapsular (BSTLJ) parts (Fig 4F and 4G). The medial division of the ventral aspect of the BNST (BSTMV) also contained considerable dispersed Nts-GFP and Nts-ISH-labeled cells. Intriguingly, the BNST was the rare brain region in which Nts-ISH labeled cells were slightly more abundant than the corresponding Nts-GFP labeled cells; this was particularly true for the BSTLD and BSTLI (Fig 4F).
Cerebral Cortex and Amygdala
Compared to the broad distribution of Nts-GFP neurons throughout the bulk of the hypothalamus, the hippocampus contained more restricted populations of Nts-GFP neurons. Notably, the prosubiculum, dorsal subiculum (DS) and ventral subiculum (VS) harbor numerous Nts-GFP neurons (Fig 5A). Nts-ISH is also prominent within the prosubiculum and DS, but not the VS (Fig 5A). The hippocampal CA1 pyramidal cell layer encompasses many Nts-GFP neurons, and these neurons are localized primarily within the caudal aspects of the structure up through the level of the DM (Fig 5B). Since much less Nts-ISH is apparent in the CA1 region from the Allen Brain Atlas, there may be a reduction in the number of mature neurons that continue to express Nts in this region (Fig 5B). The markedly higher density and distribution of Nts-GFP cells in the VS and CA1 compared to Nts-ISH suggests that Nts is expressed developmentally throughout the hippocampus, but expression is not sustained in the VS or CA1 of adult mice.
Figure 5. Nts-GFP and Nts-ISH in the Cortex.
From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma −3.40, Red shading corresponds to the Dorsal and Ventral Subiculum (DS and VS) while blue shading corresponds to the Oriens layer of the hippocampus (Or), B) Bregma −2.06, Red shading corresponds to Field CA1 of the hippocampus (CA1sp) C) Bregma −1.22, Red shading corresponds to the Retrosplenial Dysgranular and Granular Cortex (RSD/RSGc), D) Bregma −1.70, Red shading corresponds to the Central Amygdalar nucleus (CEA), E) Bregma −0.22, Red shading corresponds to the Cingulate Cortex (Cg). SG= Suprageniculate Thalamic nucleus, MGM=Medial Geniculate nucleus, PIL= Posterior Intralaminar Thalamic nucleus, PP= Peripeduncular nucleus, PoT= Posterior Thalamic nucleus, InG= Intermediate gray layer of the superior colliculus, DIEnt= Dorsointermedial Entorhinal Cortex, VIEnt= Ventral Intermediate Entorhinal Cortex, PMCo= Posteromedial cortical amygdalar nucleus, CPu= Caudate Putamen, PVP= Paraventricular Thalamic nucleus, Posterior, PF= Parafascicular Thalamic nucleus, PV= Paraventricular Thalamic nucleus, STIA= ST, intraamygdalar division, BMP= Basomedial Amygdalar nucleus, posterior, MePD= Medial Amygdalar nucleus, posterodorsal, MePV= Medial Amygdalar nucleus, posteroventral, LHA= Lateral Hypothalamus, Arc= Arcuate hypothalamic nucleus, Xi= Xiphoid Thalamic nucleus, CM= Central Medial Thalamic nucleus, LS=Lateral Septal Nucleus.
The distribution of cortical Nts-GFP was also fairly circumscribed, as it was limited to the retrosplenial (RSD and RSGc) and cingulate (Cg) regions (Fig 5B, C and E). Sizable populations of Nts-GFP neurons were observed in these regions, but Nts-ISH was undetectable (Fig 5B, C and E and Fig 3B). A striking, large population of Nts-GFP neurons was confined within the Cg, but Nts ISH was very low and virtually undetectable in this region (Fig 5E and 5B). As with the hippocampus, these data hint that Nts provides a primarily developmental role in the cortex and that it is not an active neuropeptide signal within the adult cortical regions.
Within the amygdala, only the CEA possessed a significant cluster of Nts-GFP neurons (Fig 5D). In agreement, intense Nts-ISH labeling was observed within the caudal CEA (Fig 5D). A number of amygdala-associated structures contained more moderate, but still considerable amounts, of Nts neurons (++/+++ to +++). These structures included the amygdalopiriform transition area (APir), ventral aspect of the basolateral amygdalar nucleus (BLV), anterior aspect of the basomedial amygdalar nucleus (BMA), cortical amygdalar area (COA), intraamygdalar division of the stria terminalis (STIA), cortex-amygdala transition area (CxA), anterodorsal and anteroventral medial amygdalar nucleus (MeAD/MeAV), main intercalated amygdalar nucleus (IM), and sublenticular extended amygdala (EA) (Supplemental Table 1). Of these structures, comparable Nts-ISH labeling within the Allen Brain Atlas was observed for the BLV, BMA, and STIA, while slightly lower levels of Nts ISH (+/++) was detected within the COA, CxA, and MeAV (Supplemental Table 1).
Striatum, Pallidum, and Forebrain
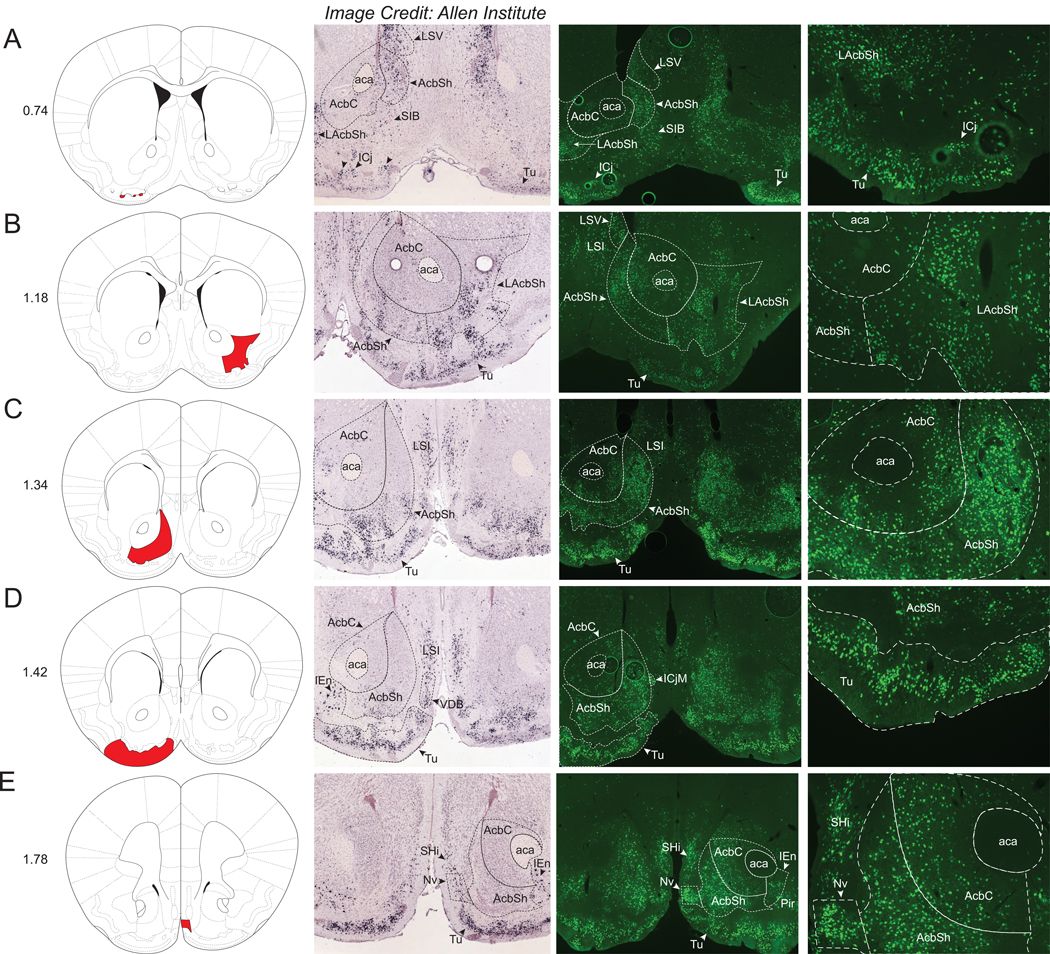
We observed extensive Nts-GFP and Nts-ISH in the mouse ventral striatum, which broadly consists of the olfactory tubercle (Tu) and the nucleus accumbens (Acb). Abundant Nts-GFP neurons were found throughout the rostrocaudal extent of the olfactory tubercle (Tu), including within clusters of neurons known as the islands of cajella (ICj) (Fig 6A). Nts-ISH was very intense and mostly similar in distribution to the Nts-GFP neurons throughout the Tu and ICj. The Acb also contained numerous Nts-GFP neurons and Nts-ISH that was predominantly located within the medial and lateral shell (AcbSh and LAcbSh) with a more minor population residing in the nucleus accumbens core (AcbC) (Fig 6A–E). The density of Nts-GFP neurons was greatest at the very medial aspect of the AcbC (+++), whereas this density is much lower within the lateral core (+). Other structures within the striatum and pallidum contained smaller but still considerable densities of Nts-GFP neurons (++/+++ or +++). Notably, these structures include the caudate putamen (CPu), globus pallidus (internal)/entopeduncular nucleus (EP), anterior and ventral aspects of medial portion of the bed nucleus of the stria terminalis (BSTMA/BSTMV), interstitial nucleus of the posterior limb of the anterior commissure (IPAC), lateral septal nucleus (LS), the substantia innominata (SIB), and septohippocampal nucleus (SHi) (Supplemental Table 1). Within the CPu, the highest density of Nts-GFP neurons (+++) was found in the caudal aspect of the region spanning between the levels of the rostral Arc and the caudal MPO (Fig 3B and Fig 5D, E). The CPu, STMA and STMV, IPAC, and LS sub-regions had Nts-ISH densities comparable to the observed distribution of Nts-GFP neurons (Supplemental Table 1). The navicular postolfactory nucleus (Nv) was the rostral-most structure with a large Nts-GFP population (Fig 6E).
Figure 6. Nts-GFP and Nts-ISH in the Forebrain.
From left to right, each row contains a Bregma-numbered atlas image (Franklin and Paxinos, 2013), an image of Nts-ISH at the same Bregma level, courtesy of the Allen Brain Atlas (Lein et al., 2007), a 4x image of Nts-GFP neurons, and a 10x image of Nts-GFP neurons from the same area. Shaded areas in the atlas image are outlined in the Nts-GFP images. A) Bregma +0.74, Red shading corresponds to the Islands of Cajella (ICj), B) Bregma +1.18, Red shading corresponds to the Nucleus Accumbens, lateral shell (LAcbSh), C) Bregma +1.34, Red shading corresponds to the Nucleus Accumbens, Shell (AcbSh), D) Bregma +1.42, Red shading corresponds to the Olfactory tubercle (Tu), E) Bregma +1.78, Red shading corresponds to the Navicular Postolfactory nucleus (Nv). LSV=Lateral Septal Nucleus, ventral part, LSI=Lateral Septal Nucleus, intermediate part, aca=anterior commissure, AcbC=Nucleus Accumbens, Core, ICjM=Island of Cajella, Major Island, SIB=Substantia innominata, VDB=Nucleus of the Vertical Limb of the Diagonal Band, SHi=Septohippocampal nucleus, Pir=Piriform Cortex.
The only notable discrepancy between forebrain Nts-GFP and Nts-ISH distributions was in the ventral tenia tecta (VTT) and the medial portion of the anterior olfactory area (AOM), which are olfactory structures contained within the rostral-most aspect of the brain (Supplemental Table 1). While the VTT and AOM contained moderate densities of Nts-GFP neurons, no detectable Nts-ISH was present in either structure.
Secondary Confirmation of Nts-ISH Within Brain Regions
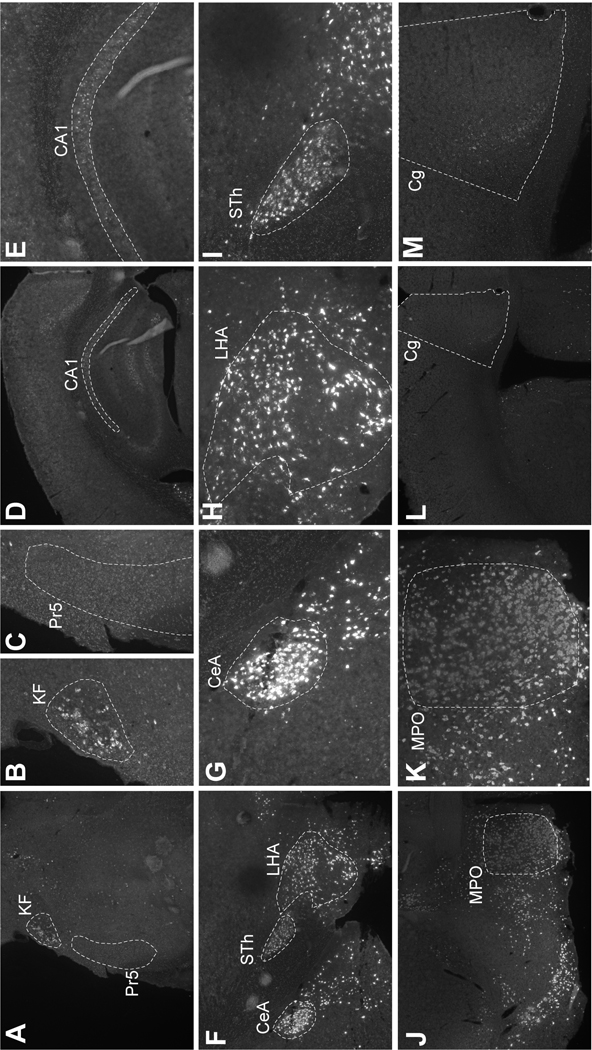
Brain regions with differential distributions of Nts-GFP and Nts-ISH from the Allen Brain Atlas may indicate sites that express Nts transiently, but not in adulthood. Yet, the Nts-ISH from the Allen Brain Atlas only provides a snapshot of Nts-ISH from a single male mouse and may not reflect biological variability in Nts expression. An apparent lack of expression could also result from an experimental artifact, for example if the Nts probe was not fully distributed throughout a section. To overcome these possible confounds, we performed RNA-Scope Nts-ISH in specific brain regions in which we’d found concordance or discordance between the Nts-GFP and Allen Brain Nts-ISH signals. For example, the KF and Pr5 in the hindbrain both had ample Nts-GFP neurons, but Allen Brain Nts-ISH was only noted in the KF (Fig 1H). Similarly, RNA-Scope analysis identified Nts-ISH in the KF but not the Pr5 (Fig 7A–C). We also confirmed that there is minimal Nts-ISH in the CA1 and Cg, which is similar to the Allen Brain atlas and contrasts to the numerous Nts-GFP neurons in these regions (CA1: Fig 7D–E vs. Fig 5B; Cg: Fig 7L–M vs. Fig 5E). These data lend further support to the idea that Nts is transiently expressed at high levels in the Pr5, CA1 and Cg, but that this expression is absent in the adult brain. We also performed RNA-Scope Nts-ISH in brain regions that had abundant Nts-GFP signal that was concordant with the Allen Brain Atlas Nts-ISH. Indeed, RNA-Scope analysis confirmed high Nts-ISH in the CeA, STh, LHA and MPO, similar to the Allen Brain and Nts-GFP distributions. (CeA: Fig 7F–G vs. Fig 5D; LHA: Fig 7F,H vs. Fig 4D; STH: Fig 7F,I vs. Fig 4C; MPO: Fig 7J,K vs. Fig 4F). These findings suggest that the Allen Brain Nts-ISH is a reliable indicator of Nts expression in the adult mouse brain and can be utilized for comparison to the Nts-GFP distributions.
Figure 7. Verification of Nts-ISH in Specific Brain Regions.
RNA Scope ISH for Nts (white) in wild type male mice (n=3). Panels show representative images of findings. A) 4x image of section containing the Pr5 and KF. B) KF (20x) and C) Pr5 (20x). CA1 region of the hippocampus at D) 4x and E) 20x. F) 4x image of section containing the CeA, LHA and STh. 20x images of from this section of the G) CeA, H) LHA and I) STh. Abundant Nts is found in the J) 4x image of the MPO and K) can be seen in numerous MPO cell bodies at 20x. By contrast, note the dearth of Nts in the L) 4x image and K) 20x image of the Cg. Pr5=Principal sensory trigeminal nucleus, KF=Koelliker-Fuse nucleus (KF), CA1=Field CA1 of the hippocampus, CeA=central amygdala, LHA= lateral hypothalamic area, STh= subthalamic nucleus, MPO=medial preoptic area, Cg=cingulate cortex.
Heterogeneity of Nts Neurons Within Brain Regions
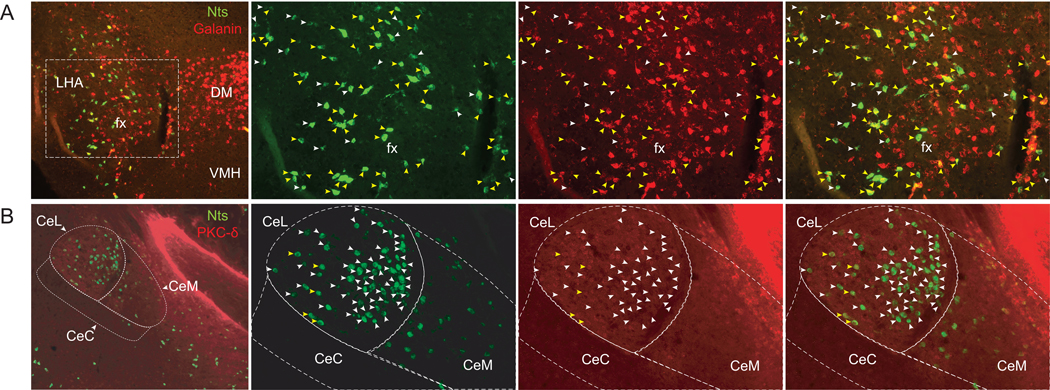
We were struck by the observation of very dense populations of Nts-GFP neurons within the LHA and CEA, regions known to contain multiple molecularly-distinct neuronal populations that exert unique modulation of feeding. We therefore reasoned that Nts-expressing neurons in the LHA and CEA may not be homogeneous and might differ in their expression of other neuropeptides or molecular markers that would provide clues as to their function. We first tested this hypothesis in the LHA by examining the co-distribution of the neuropeptides Nts and Gal. LHA neurons expressing anorectic Nts are alleged to overlap with the same neuronal population that expresses orexigenic Gal, which was determined by analyzing IR for Nts and Gal in colchicine-treated mice (Laque et al., 2013). Yet, subsequent studies showed that LHA Nts and LHA Gal neurons differ in projection targets and physiologic regulation of feeding and behavior, suggesting they may not be a fully overlapping population (Qualls-Creekmore et al., 2017; Woodworth et al., 2017b). To examine this possibility, we performed dual ISH for Nts and Gal (Fig 8A), thus bypassing the requirement for colchicine treatment, and potentially interrupted anterograde transport that might jeopardize cell health and alter gene expression. Using dual ISH, we observed robust Gal throughout the DM, but no Nts was found in this structure (Fig 8A). These findings are consistent with prior ISH (Lein et al., 2007) and the dearth of Nts-GFP cells in the DM (Fig 4). By contrast, we noted ample distributions of Gal-positive and Nts-positive cells within the LHA. While many LHA neurons contained high levels of both Gal and Nts (Fig 8A, yellow arrows), many Gal neurons did not overlap with Nts neurons. Moreover, we also identified Nts-labeled neurons that completely lacked or had negligible Gal signal (white arrowheads) (Fig 8A, white arrows). These data suggest that LHA Nts neurons are heterogeneous and that there are at least two subpopulations of Nts neurons in this structure, one of which robustly co-expresses Gal and the other of which does not. These anatomical findings also concur with recent single-cell molecular analysis of LHA populations, which found populations of Nts neurons containing Gal as well as Nts neurons that did not (Mickelsen et al., 2019).
Figure 8. Heterogeneity of Nts Neurons Within the LHA and CEA.
From left to right, each row contains a 10x image of merged red and green channels, followed by 20X images of green, red, and merged channels. A) RNA Scope dual-fluorescent ISH for Nts (green) and Galanin (Gal, red) in the LHA. Yellow arrows identify neurons expressing Nts and robust levels of Gal. White arrows identify neurons expressing Nts and negligible Gal. B) Section of the CEA from an NtsCre;GFP mouse immunostained for GFP (Nts-GFP, green) and PKC-δ (red). Yellow arrows identify the few neurons co-labeled with Nts-GFP and PKC-δ, which lie primarily within the lateral aspect of the CEA (CeL). White arrows identify CEA neurons that contain Nts but no detectable PKC-δ. LHA= Lateral Hypothalamus, fx=fornix, DM=Dorsomedial Hypothalamic nucleus, VMH=Ventromedial Hypothalamic nucleus, CeL=Central Amygdalar nucleus, lateral, CeC=Central Amygdalar nucleus, central, CeM=Central Amygdalar nucleus, medial.
We next examined the CEA, where Nts and protein kinase c-δ (PKC-δ) are localized, and both are implicated in anorexic behavior (Cai et al., 2014; Cooke et al., 2009; Levine et al., 1983; Woodworth et al., 2017b). To investigate whether these purported anorectic proteins overlap spatially, we examined PKC-δ immunoreactivity (IR) within the CEA of NtsCre;GFP mice. This analysis revealed a few CEA Nts-GFP cells that also contained PKC-δ-IR (Fig 8B, yellow arrows), but the majority of CEA Nts-GFP and PKC-δ neurons were largely separate and did not overlap (Fig 8B, white arrows). Interestingly, most Nts-GFP neurons were found within the medial aspect of the CeL subregion of the CEA, and these did not colocalize with PKC-δ-IR. A small number of Nts-GFP neurons found within the lateral aspect of the CeL, however, did co-express PKC-δ (Fig 8). These data corroborate recent literature showing that very little Nts-ISH overlaps with PKC-δ within the CeL (Mccullough et al., 2018) and further demonstrates that NtsCre;GFP mice can be useful to both identify Nts neurons and to define their molecular phenotype.
Discussion
Importance of Assessing the Distribution of Nts Neurons in the Mouse Brain
A major goal of neuroscience is to understand how molecularly- and regionally-specified neuronal populations coordinate behavior and physiology. Because central Nts mediates a diverse array of physiologic responses depending on where it is administered in the brain (analgesia, regulation of body temperature, suppression of feeding, locomotor activity, vasodepressor response), it is likely that regionally-defined populations of Nts neurons coordinate specific functions. Characterizing the roles of these distributed Nts populations requires the ability to identify and then manipulate them in vivo to reveal how they mediate behavior and biology. While the use of ISH and colchicine-mediated Nts-IR has been valuable to identify Nts neurons, primarily in rats, these methods don’t permit subsequent manipulation of neurons of interest. By contrast, the recombinase-mediated labeling of Nts neurons that occurs in NtsCre;GFP mice facilitates Nts neuron detection and permits their manipulation using widely available Cre-Lox tools. Indeed, this approach has already been successful in establishing that Nts neurons in the POA vs. the LHA modulate social and feeding behaviors, respectively (McHenry et al., 2017; Woodworth et al., 2017b). Much, however, remains to be learned about Nts-mediated physiology. Given the differences in Nts expression and brain architecture between rodents (Schroeder and Leinninger, 2018; Smits et al., 2004), prior descriptions of Nts-expressing neurons in rats may not translate to NtsCre;GFP mice to guide function-directed studies. Our work here thus fills a critical gap by providing an “Nts-GFP atlas” that investigators can use to identify Nts populations and then systematically test their function in NtsCre;GFP mice.
Important Considerations in Using NtsCre;GFP mice to Study Nts Neurons
NtsCre mice are engineered so that Cre expression is an excellent proxy of which cells are actively expressing Nts. However, as with any knock-in recombinase mouse model, once NtsCre mice are bred onto a Cre-inducible reporter line, Cre expressed at any point during development causes recombination and permanent reporter labeling. Thus, while Cre-inducible expression of reporters like GFP are ideal to permit cell detection, immediate recombination upon Cre expression prevents discrimination of which cells transiently expressed Nts/Cre during development vs. those that actively express them in adult cells. Additionally, the GFP reporter line does not indicate Nts expression at the level of individual neurons, as constitutive GFP expression will occur in cells that have undergone recombination, regardless of the degree to which Nts is actively expressed. These confounds must be considered when examining NtsCre;GFP mice. Since Nts expression differs within the neonatal, postnatal, and adult brain of rats (Bissette et al., 2006; Wolfson et al., 1985; Yamano et al., 1984), it is likely that there is also some Nts-dependent ontogeny in the mouse brain. Moreover, because Nts receptors are broadly expressed in the developing rat brain but their expression becomes more circumscribed in maturity (Lépée-Lorgeoux et al., 1999; Palacios et al., 1988), it is possible that Nts exerts different functions during the formation of neural circuits as compared to Nts signaling in the mature brain. Hence, prior to performing any manipulations of Nts-GFP neurons in adult NtsCre;GFP mice, it is important to verify whether the cells in question are actively expressing Cre/Nts, or whether they were labeled during development. Only cells actively expressing Cre can be modulated using Cre-Lox methodologies. For this reason, we compared the distribution of Nts-GFP neurons with adult Nts-ISH provided by the Allen Brain Institute (Lein et al., 2007), reasoning that any sites of Nts-GFP neurons that lack Nts-ISH represent Nts-GFP populations that transiently expressed Nts and underwent recombination during development but do not actively express Nts in adulthood. We noted several brain areas with discrepant Nts-GFP and Nts ISH profiles (see Table 1) and have pointed them out in the text. We acknowledge, that the Allen Brain Nts-ISH may not perfectly represent Nts expression, as it is derived from a single sample and inherent biological variability or technical artifacts could result in under-detection of Nts-expressing neurons. However, our assessment of specific brain areas across three male mice via RNA-Scope revealed comparable Nts-ISH to that of the Allen Brain Atlas, lending support to its utility to predict RNA expression. Yet, since we could not evaluate all areas with discordant expression, the absence of Nts-ISH in areas with Nts-GFP neurons should not be taken as absolute confirmation of their “developmental” profile or that that they do not express Cre/Nts during maturity. In addition, only three NtsCre;GFP brains were evaluated, and while they were generally consistent, there was some expected biological variability in the distribution and relative densities of Nts populations between them (Supplemental Figure 2). Hence, given the small sample sizes of these studies, investigators should cautiously and conservatively interpret the data as a “snap-shot” of regions that contain Nts, and should confirm levels of Cre/Nts expression in their brain region, age and physiological context of interest. This can easily be done by injecting Cre-inducible reagents into adult NtsCre;GFP mice at sites of interest, and only neurons actively expressing Nts will express Cre and undergo recombination.
The fidelity of the NtsCre;GFP mice has been previously characterized. Dual ISH for Nts and Cre performed in NtsCre mice demonstrated that Cre recombinase is solely co-expressed by Nts neurons (McHenry et al., 2017). In addition, we have previously verified that Nts-immunoreactive neurons faithfully colocalize with GFP cells in NtsCre;GFP mice (Brown et al., 2019; Woodworth et al., 2018). While nearly all GFP-expressing neurons overlapped with Nts-immunoreactive cells in regions with known dense populations of Nts neurons, such as the LHA, POA, and Acb, there was a minor portion of GFP cells that did not exhibit Nts expression (Woodworth et al., 2018). This indicates that, even in regions in which densities of Nts-ISH correspond well with densities of GFP neurons, there may still be a few GFP neurons within the NtsCre;GFP mouse that are not actively expressing Nts. Thus, those interested in using these mice should take care to fully characterize the Nts vs. GFP expression in their region of interest, to understand to what extent the GFP identifies active Nts-expressing cells. However, Cre expression reflects whether or not a neuron is actively expressing Nts. Thus, injectable viral Cre-dependent technologies, such AAV-delivered activating or inhibitory DREADDs, will result in activation and inhibition only in Cre/Nts-expressing neurons, and has been used for this purpose in the LHA (Naganuma et al., 2019; Woodworth et al., 2017b). This technique can be used to verify the extent of neurons in a specific area that are actively expressing Nts/Cre, as any modulations are only of “real” actively expressing Nts neurons.
One additional consideration is that we characterized the distribution of Nts-GFP neurons from the brains of adult male NtsCre;GFP mice, but it is possible that the distribution and/or relative density of Nts-GFP populations may differ in females. A number of brain regions have already been reported to display enhanced Nts expression in females relative to males, likely due to estradiol-induced effects, including the arcuate nucleus, medial preoptic nucleus, anteroventral periventricular nucleus (Alexander, 1993; Alexander et al., 1991; Vastagh and Liposits, 2017). Going forward, investigators should validate the distributions of Nts neurons in areas of interest in both sexes, particularly if they are studying the role of Nts in physiology with known sex differences. For example, loss of function Nts variants have been discovered in individuals with anorexia nervosa, a type of eating disorder that is more prevalent in females than males. It is possible that differences in Nts expression or function might contribute to the development and sex difference of eating disorders, though this has yet to be mechanistically examined (Lutter et al., 2017). Additionally, males and females also exhibit differences in pain processing; hence, there may be differences in Nts signaling that underline sexual dimorphism in pain sensing and analgesia (Loyd and Murphy, 2009). In light of the established sex differences in Nts expression within select brain regions, (Alexander, 1999, 1993; Alexander et al., 1991; Axelson et al., 1992; Herbison and Theodosis, 1992; Vastagh and Liposits, 2017), investigators should characterize Nts expression in female NtsCre;GFP mice before using the line to assess the role of Nts in reproduction or sex differences. This is particularly crucial when considering that our study, Allen Brain ISH, and most other Nts studies were carried out in males.
Lastly, the method used to identify Nts neurons robustly labeled soma but prohibited detection of Nts-containing fibers. This was an inherent caveat of the Cre-mediated GFP:L10a reporter line used for the study: GFP is fused to the ribosomal protein unit L10a (Rpl10a) so any induced GFP is restricted to ribosomes within the cell body. Since axons have few ribosomes little GFP-L10a will be found in these structures. Going forward, however, investigators can easily reveal the projection targets of any Nts population of interest using NtsCre;GFP mice and Cre-mediated tract tracers.
Summary
The goal of this work was to provide a descriptive map of Nts neurons throughout the NtsCre;GFP mouse brain. We documented numerous brain regions that contain Nts-GFP neurons, some of which were not thought to possess Nts neurons prior to this report, and the Nts neurons at many of these sites are plausible participants in the regulation of various aspects of physiology, spanning from analgesia, locomotor activity, cardiovascular response, social behavior, addiction, learning, memory, and feeding. These findings emphasize the wealth of information yet to be mined about how Nts neurons contribute to biology and behavior. It is our hope that this work will facilitate subsequent studies designed to both understand the roles of these Nts populations in normal physiology and to determine whether these populations may be tractable targets to improve maladaptive behaviors or disease states.
Supplementary Material
Acknowledgements
We thank David P. Olson (University of Michigan) for graciously sharing the Cre-inducible RosaeGFP-L10a reporter line used in these studies. Cristina Rivera Quilles, Angela Garcia and Crystal Colon-Ortiz contributed to this project as undergraduates thanks to support from the NIH-NINDS Bridge to the PhD in Neuroscience (BPNP)- ENDURE Program (R25-NS090989). This research was supported by a grant from the NIH to GML (R01-DK103808).
Footnotes
Disclosure Summary: The authors have no conflicts of interest to declare.
References
- Al-Rodhan NRF, Richelson E, Gilbert JA, McCormick DJ, Kanba KS, Pfenning MA, Nelson A, Larson EW, Yaksh TL, 1991. Structure-antinociceptive activity of neurotensin and some novel analogues in the periaqueductal gray region of the brainstem. Brain Res. 557, 227–235. 10.1016/0006-8993(91)90139-M [DOI] [PubMed] [Google Scholar]
- Alexander MJ, 1999. Colocalization of Neurotensin Messenger Ribonucleic Acid (mRNA) and Progesterone Receptor mRNA in Rat Arcuate Neurons under Estrogen-Stimulated Conditions1. Endocrinology 140, 4995–5003. [DOI] [PubMed] [Google Scholar]
- Alexander MJ, 1993. Estrogen-regulated synthesis of neurotensin in neurosecretory cells of the hypothalamic arcuate nucleus in the female rat. Endocrinology 133, 1809–1816. [DOI] [PubMed] [Google Scholar]
- Alexander MJ, Kiraly ZJ, Leeman SE, 1991. Sexually dimorphic distribution of neurotensin/neuromedin N mRNA in the rat preoptic area. J. Comp. Neurol. 311, 84–96. 10.1002/cne.903110107 [DOI] [PubMed] [Google Scholar]
- Aronin N, Carraway RE, Ferris CF, Hammer RA, Leeman SE, 1982. The stability and metabolism of intravenously administered neurotensin in the rat. Peptides 3, 637–642. 10.1016/0196-9781(82)90164-4 [DOI] [PubMed] [Google Scholar]
- Axelson JF, Shannon W, Van Leeuwen FW, 1992. Immunocytochemical localization of estrogen receptors within neurotensin cells in the rostral preoptic area of the rat hypothalamus. Neurosci. Lett. 136, 5–9. 10.1016/0304-3940(92)90634-J [DOI] [PubMed] [Google Scholar]
- Benmoussa M, Chait A, Loric G, De Beaurepaire R, 1996. Low doses of neurotensin in the preoptic area produce hyperthermia. Comparison with other brain sites and with neurotensin-induced analgesia. Brain Res. Bull. 39, 275–279. 10.1016/0361-9230(95)02138-8 [DOI] [PubMed] [Google Scholar]
- Bissette G, Richardson C, Kizer JS, Nemeroff CB, 2006. Ontogeny of Brain Neurotensin in the Rat: A Radioimmunoassay Study. J. Neurochem. 43, 283–287. 10.1111/j.1471-4159.1984.tb06711.x [DOI] [PubMed] [Google Scholar]
- Boules M, Oliveros A, Liang Y, Williams K, Shaw A, Robinson J, Fredrickson P, Richelson E, 2011. A neurotensin analog, NT69L, attenuates intravenous nicotine self-administration in rats. Neuropeptides 45, 9–16. 10.1016/j.npep.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Brown JA, Bugescu R, Mayer TA, Gata-Garcia A, Kurt G, Woodworth HL, Leinninger GM, 2017. Loss of Action via Neurotensin-Leptin Receptor Neurons Disrupts Leptin and Ghrelin-Mediated Control of Energy Balance. Endocrinology 158, 1271–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Wright A, Bugescu R, Christensen L, Olson DP, Leinninger GM, 2019. Distinct Subsets of Lateral Hypothalamic Neurotensin Neurons are Activated by Leptin or Dehydration. Sci. Rep. 9, 1–16. 10.1038/s41598-018-38143-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cador M, Kelley AE, Le Moal M, Stinus L, 1986. Ventral tegmental area infusion of substance P, neurotensin and enkephalin: Differential effects on feeding behavior. Neuroscience 18, 659–669. 10.1016/0306-4522(86)90061-8 [DOI] [PubMed] [Google Scholar]
- Cai H, Haubensak W, Anthony TE, Anderson DJ, 2014. Central amygdala PKC-[delta]+ neurons mediate the influence of multiple anorexigenic signals. Nat Neurosci 17, 1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cape EG, Manns ID, Alonso A, Beaudet A, Jones BE, 2000. Neurotensin-Induced Bursting of Cholinergic Basal Forebrain Neurons Promotes γ and θ Cortical Activity Together with Waking and Paradoxical Sleep. J. Neurosci. 20, 8452 LP-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway R, Leeman SE, 1973. The Isolation of a New Hypotensive Peptide, Neurotensin, from Bovine Hypothalami. J. Biol. Chem. 248, 6854–6861. [PubMed] [Google Scholar]
- Cooke JH, Patterson M, Patel SR, Smith KL, Ghatei MA, Bloom SR, Murphy KG, 2009. Peripheral and Central Administration of Xenin and Neurotensin Suppress Food Intake in Rodents. Obesity 17, 1135–1143. 10.1038/oby.2008.652 [DOI] [PubMed] [Google Scholar]
- Demeule M, Beaudet N, Régina A, Besserer-Offroy É, Murza A, Tétreault P, Belleville K, Ché C, Larocque A, Thiot C, Béliveau R, Longpré J-M, Marsault É, Leduc R, Lachowicz JE, Gonias SL, Castaigne J-P, Sarret P, 2014. Conjugation of a brain-penetrant peptide with neurotensin provides antinociceptive properties. J. Clin. Invest. 124, 1199–1213. 10.1172/JCI70647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott PJ, Nemeroff CB, 1986. Repeated neurotensin administration in the ventral tegmental area: Effects on baseline and d-amphetamine-induced locomotor activity. Neurosci. Lett. 68, 239–244. 10.1016/0304-3940(86)90149-7 [DOI] [PubMed] [Google Scholar]
- Ferraro L, Tiozzo Fasiolo L, Beggiato S, Borelli AC, Pomierny-Chamiolo L, Frankowska M, Antonelli T, Tomasini MC, Fuxe K, Filip M, 2016. Neurotensin: A role in substance use disorder? J. Psychopharmacol. 30, 112–127. 10.1177/0269881115622240 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick K, Winrow CJ, Gotter AL, Millstein J, Arbuzova J, Brunner J, Kasarskis A, Vitaterna MH, Renger JJ, Turek FW, 2012. Altered Sleep and Affect in the Neurotensin Receptor 1 Knockout Mouse. Sleep 35, 949–956. 10.5665/sleep.1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G, 2013. Paxinos and Franklin’s The mouse brain in stereotaxic coordinates. [Google Scholar]
- Gammie SC, D’Anna KL, Gerstein H, Stevenson SA, 2009. Neurotensin inversely modulates maternal aggression. Neuroscience 158, 1215–1223. 10.1016/j.neuroscience.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert B, Wynendaele E, Stalmans S, Bracke N, D’Hondt M, Smolders I, van Eeckhaut A, De Spiegeleer B, 2016. Blood-brain barrier transport kinetics of the neuromedin peptides NMU, NMN, NMB and NT. Neuropharmacology 107, 460–470. 10.1016/j.neuropharm.2016.03.051 [DOI] [PubMed] [Google Scholar]
- Grunddal KV, Ratner CF, Svendsen B, Sommer F, Engelstoft MS, Madsen AN, Pedersen J, Nøhr MK, Egerod KL, Nawrocki AR, Kowalski T, Howard AD, Poulsen SS, Offermanns S, Bäckhed F, Holst JJ, Holst B, Schwartz TW, 2016. Neurotensin Is Coexpressed, Coreleased, and Acts Together With GLP-1 and PYY in Enteroendocrine Control of Metabolism. Endocrinology 157, 176–194. [DOI] [PubMed] [Google Scholar]
- Hawkins MF, 1986. Aphagia in the rat following microinjection of neurotensin into the ventral tegmental area. Life Sci. 38, 2383–2388. 10.1016/0024-3205(86)90606-5 [DOI] [PubMed] [Google Scholar]
- Hawkins MF, Baker JD, Baumeister AA, 1989. Neurotensin-induced polydipsia: a structure-activity study. Brain Res. 487, 188–191. 10.1016/0006-8993(89)90957-8 [DOI] [PubMed] [Google Scholar]
- Herbison AE, Theodosis DT, 1992. Localization of oestrogen receptors in preoptic neurons containing neurotensin but not tyrosine hydroxylase, cholecystokinin or luteinizing hormone-releasing hormone in the male and female rat. Neuroscience 50, 283–298. 10.1016/0306-4522(92)90423-Y [DOI] [PubMed] [Google Scholar]
- Ibata Y, Kawakami F, Fukui K, Okamura H, Obata-Tsuto HL, Tsuto T, Terubayashi H, 1984. Morphological survey of neurotensin-like immunoreactive neurons in the hypothalamus. Peptides 5, Supplem, 109–120. 10.1016/0196-9781(84)90270-5 [DOI] [PubMed] [Google Scholar]
- Jennes L, Stumpf WE, Kalivas PW, 1982. Neurotensin: Topographical distribution in rat brain by immunohistochemistry. J. Comp. Neurol. 210, 211–224. 10.1002/cne.902100302 [DOI] [PubMed] [Google Scholar]
- Kahn D, Abrams GM, Zimmerman EA, Carraway R, Leeman SE, 1980. Neurotensin neurons in the rat hypothalamus: an immunocytochemical study. Endocrinology 107, 47–54. 10.1210/endo-107-1-47 [DOI] [PubMed] [Google Scholar]
- Kahn D, Hou-Yu A, Zimmerman EA, 1982. LOCALIZATION OF NEUROTENSIN IN THE HYPOTHALAMUS*. Ann. N. Y. Acad. Sci. 400, 117–131. 10.1111/j.1749-6632.1982.tb31564.x [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Duffy P, 1990. Effect of acute and daily neurotensin and enkephalin treatments on extracellular dopamine in the nucleus accumbens. J. Neurosci. 10, 2940 LP-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Gau BA, Nemeroff CB, Prange AJ, 1982. Antinociception after microinjection of neurotensin into the central amygdaloid nucleus of the rat. Brain Res. 243, 279–286. 10.1016/0006-8993(82)90251-7 [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Taylor S, 1985. Behavioral and neurochemical effect of daily injection with neurotensin into the ventral tegmental area. Brain Res. 358, 70–76. 10.1016/0006-8993(85)90949-7 [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, Liberles SD, Lowell BB, 2014. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert PD, Gross R, Nemeroff CB, Kilts CD, 1995. Anatomy and Mechanisms of Neurotensin-Dopamine Interactions in the Central Nervous System. Ann. N. Y. Acad. Sci. 757, 377–389. 10.1111/j.1749-6632.1995.tb17496.x [DOI] [PubMed] [Google Scholar]
- Laque A, Zhang Y, Gettys S, Nguyen T-A, Bui K, Morrison CD, Münzberg H, 2013. Leptin receptor neurons in the mouse hypothalamus are colocalized with the neuropeptide galanin and mediate anorexigenic leptin action. Am. J. Physiol. - Endocrinol. Metab. 304, E999–E1011. 10.1152/ajpendo.00643.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chi Chin M, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong H-W, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf K-R, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Feng Yuan X, Zhang B, Zwingman TA, Jones AR, 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Lépée-Lorgeoux I, Betancur C, Rostène W, Pélaprat D, 1999. Differential ontogenetic patterns of levocabastine-sensitive neurotensin NT2 receptors and of NT1 receptors in the rat brain revealed by in situ hybridization. Dev. Brain Res. 113, 115–131. 10.1016/S0165-3806(99)00009-7 [DOI] [PubMed] [Google Scholar]
- Levine AS, Kneip J, Grace M, Morley JE, 1983. Effect of centrally administered neurotensin on multiple feeding paradigms. Pharmacol. Biochem. Behav. 18, 19–23. 10.1016/0091-3057(83)90244-7 [DOI] [PubMed] [Google Scholar]
- Li J, Song J, Zaytseva YY, Liu Y, Rychahou P, Jiang K, Starr ME, Kim JT, Harris JW, Yiannikouris FB, Katz WS, Nilsson PM, Orho-Melander M, Chen J, Zhu H, Fahrenholz T, Higashi RM, Gao T, Morris AJ, Cassis LA, Fan TW-M, Weiss HL, Dobner PR, Melander O, Jia J, Evers BM, 2016. An obligatory role for neurotensin in high-fat-diet-induced obesity. Nature 533, 411–415. 10.1038/nature17662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyd DR, Murphy AZ, 2009. The Role of the Periaqueductal Gray in the Modulation of Pain in Males and Females: Are the Anatomy and Physiology Really that Different? Neural Plast. 2009, 462879. 10.1155/2009/462879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter M, Bahl E, Hannah C, Hofammann D, Acevedo S, Cui H, McAdams CJ, Michaelson JJ, 2017. Novel and ultra-rare damaging variants in neuropeptide signaling are associated with disordered eating behaviors. PLoS One 12, e0181556. 10.1371/journal.pone.0181556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccullough KM, Morrison FG, Ressler KJ, Hartmann J, Carlezon WA, 2018. Quantified Coexpression Analysis of Central Amygdala Subpopulations 01, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry JA, Otis JM, Rossi MA, Robinson JE, Kosyk O, Miller NW, McElligott ZA, Budygin EA, Rubinow DR, Stuber GD, 2017. Hormonal gain control of a medial preoptic area social reward circuit. Nat. Neurosci. 20, 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merullo DP, Cordes MA, Susan DeVries M, Stevenson SA, Riters LV, 2015. Neurotensin neural mRNA expression correlates with vocal communication and other highly-motivated social behaviors in male European starlings. Physiol. Behav. 151, 155–161. 10.1016/j.physbeh.2015.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen LE, Bolisetty M, Chimileski BR, Fujita A, Beltrami EJ, Costanzo JT, Naparstek JR, Robson P, Jackson AC, 2019. Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat. Neurosci. 22, 642–656. 10.1038/s41593-019-0349-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumphrey MB, Patterson LM, Zheng H, Berthoud H-R, 2012. Roux-en-Y gastric bypass surgery increases number but not density of CCK-, GLP-1-, 5-HT-, and neurotensin-expressing enteroendocrine cells in rats. Neurogastroenterol. Motil. 25, e70–e79. 10.1111/nmo.12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustain WC, Rychahou PG, Evers BM, 2011. The role of neurotensin in physiologic and pathologic processes. 10.1097/MED.0b013e3283419052 [DOI] [PubMed]
- Naganuma F, Kroeger D, Bandaru SS, Absi G, Madara JC, Vetrivelan R, 2019. Lateral hypothalamic neurotensin neurons promote arousal and hyperthermia. PLOS Biol. 17, e3000172. 10.1371/journal.pbio.3000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff CB, Bissette G, Prange AJ Jr., Loosen PT, Steven Barlow T, Lipton MA, 1977. Neurotensin: Central nervous system effects of a hypothalamic peptide. Brain Res. 128, 485–496. 10.1016/0006-8993(77)90173-1 [DOI] [PubMed] [Google Scholar]
- Neubert MJ, Kincaid W, Heinricher MM, 2004. Nociceptive facilitating neurons in the rostral ventromedial medulla. Pain 110, 158–165. 10.1016/j.pain.2004.03.017 [DOI] [PubMed] [Google Scholar]
- Palacios JM, Pazos A, Dietl MM, Schlumpf M, Lichtensteiger W, 1988. The ontogeny of brain neurotensin receptors studied by autoradiography. Neuroscience 25, 307–317. 10.1016/0306-4522(88)90028-0 [DOI] [PubMed] [Google Scholar]
- Patterson CM, Wong JMT, Leinninger GM, Allison MB, Mabrouk OS, Kasper CL, Gonzalez IE, Mackenzie A, Jones JC, Kennedy RT, Myers MG, 2015. Ventral tegmental area neurotensin signaling links the lateral hypothalamus to locomotor activity and striatal dopamine efflux in male mice. Endocrinology 156, 1692–1700. 10.1210/en.2014-1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualls-Creekmore E, Yu S, Francois M, Hoang J, Huesing C, Bruce-Keller A, Burk D, Berthoud H-R, Morrison CD, Münzberg H, 2017. Galanin-Expressing GABA Neurons in the Lateral Hypothalamus Modulate Food Reward and Noncompulsive Locomotion. J. Neurosci. 37, 6053 LP-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner C, Skov LJ, Raida Z, Bächler T, Bellmann-Sickert K, Le Foll C, Sivertsen B, Dalbøge LS, Hartmann B, Beck-Sickinger AG, Madsen AN, Jelsing J, Holst JJ, Lutz TA, Andrews ZB, Holst B, 2016. Effects of Peripheral Neurotensin on Appetite Regulation and Its Role in Gastric Bypass Surgery. Endocrinology 157, 3482–3492. [DOI] [PubMed] [Google Scholar]
- Ray JP, Price JL, 1990. Postnatal changes in the density and distribution of neurotensin-like immunoreactive fibers in the mediodorsal nucleus of the thalamus in the rat. J. Comp. Neurol. 292, 269–282. 10.1002/cne.902920209 [DOI] [PubMed] [Google Scholar]
- Rokaeus ÅKE, Fried G, Lundberg JANM, 1984. Occurrence, storage and release of neurotensin-like immunoreactivity from the adrenal gland. Acta Physiol. Scand. 120, 373–380. 10.1111/j.1748-1716.1984.tb07397.x [DOI] [PubMed] [Google Scholar]
- Schroeder LE, Leinninger GM, 2018. Role of central neurotensin in regulating feeding: Implications for the development and treatment of body weight disorders. Biochim. Biophys. Acta - Mol. Basis Dis. 1864, 900–916. 10.1016/j.bbadis.2017.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KE, Boules M, Williams K, Richelson E, 2012. NTS1 and NTS2 mediate analgesia following neurotensin analog treatment in a mouse model for visceral pain. Behav. Brain Res. 232, 93–97. 10.1016/j.bbr.2012.03.044 [DOI] [PubMed] [Google Scholar]
- Smits SM, Terwisscha van Scheltinga AF, van der Linden AJA, Burbach JPH, Smidt MP, 2004. Species differences in brain pre-pro-neurotensin/neuromedin N mRNA distribution: the expression pattern in mice resembles more closely that of primates than rats. Mol. Brain Res. 125, 22–28. 10.1016/j.molbrainres.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Triepel J, Mader J, Weindl A, Heinrich D, Forssmann WG, Metz J, 1984. Distribution of NT-IR perikarya in the brain of the guinea pig with special reference to cardiovascular centers in the medulla oblongata. Histochemistry 81, 509–516. 10.1007/BF00489528 [DOI] [PubMed] [Google Scholar]
- Uhl GR, Goodman RR, Snyder SH, 1979. Neurotensin-containing cell bodies, fibers and nerve terminals in the brain stem of the rat: Immunohistochemical mapping. Brain Res. 167, 77–91. 10.1016/0006-8993(79)90264-6 [DOI] [PubMed] [Google Scholar]
- Uhl GR, Kuhar MJ, Snyder SH, 1977. Neurotensin: immunohistochemical localization in rat central nervous system. Proc. Natl. Acad. Sci. U. S. A. 74, 4059–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastagh C, Liposits Z, 2017. Impact of Proestrus on Gene Expression in the Medial Preoptic Area of Mice. Front. Cell. Neurosci. 11, 183 10.3389/fncel.2017.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson B, Manning RW, Davis LG, Baldino F, 1985. A quantitative, immunochemical demonstration of the postnatal development of neurotensin in the medial preoptic area of the rat. Dev. Brain Res. 18, 241–250. 10.1016/0165-3806(85)90268-8 [DOI] [PubMed] [Google Scholar]
- Woodworth HL, Batchelor HM, Beekly BG, Bugescu R, Brown JA, Kurt G, Fuller PM, Leinninger GM, 2017a. Neurotensin Receptor-1 Identifies a Subset of Ventral Tegmental Dopamine Neurons that Coordinates Energy Balance. Cell Rep. 20, 1881–1892. 10.1016/j.celrep.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth HL, Beekly BG, Batchelor HM, Bugescu R, Perez-Bonilla P, Schroeder LE, Leinninger GM, 2017b. Lateral Hypothalamic Neurotensin Neurons Orchestrate Dual Weight Loss Behaviors via Distinct Mechanisms. Cell Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth HL, Brown JA, Batchelor HM, Bugescu R, Leinninger GM, 2018. Determination of neurotensin projections to the ventral tegmental area in mice. Neuropeptides 68, 57–74. 10.1016/j.npep.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano M, Inagaki S, Tateishi N, Hamaoka T, Tohyama M, 1984. Ontogeny of neuropeptides in the nucleus ventromedialis hypothalami of the rat: An immunohistochemical analysis. Dev. Brain Res. 16, 253–262. 10.1016/0165-3806(84)90030-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.