Genetic analysis of the Arabidopsis VPS34 PI3K complex through the ATG14 or VPS38 subunits revealed a preferential role for ATG14 during autophagy and the widespread importance of PI3P in plants.
Abstract
Phosphatidylinositol 3-phosphate (PI3P) is an essential membrane signature for both autophagy and endosomal sorting that is synthesized in plants by the class III phosphatidylinositol 3-kinase (PI3K) complex, consisting of the VPS34 kinase, together with ATG6, VPS15, and either VPS38 or ATG14 as the fourth subunit. Although Arabidopsis (Arabidopsis thaliana) plants missing the three core subunits are infertile, vps38 mutants are viable but have aberrant leaf, root, and seed development, Suc sensing, and endosomal trafficking, suggesting that VPS38 and ATG14 are nonredundant. Here, we evaluated the role of ATG14 through a collection of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 and T-DNA insertion mutants disrupting the two Arabidopsis paralogs. atg14a atg14b double mutants were relatively normal phenotypically but displayed pronounced autophagy defects, including reduced accumulation of autophagic bodies and cargo delivery during nutrient stress. Unexpectedly, homozygous atg14a atg14b vps38 triple mutants were viable but showed severely compromised rosette development and reduced fecundity, pollen germination, and autophagy, consistent with a need for both ATG14 and VPS38 to fully actuate PI3P biology. However, the triple mutants still accumulated PI3P, but they were hypersensitive to the PI3K inhibitor wortmannin, indicating that the ATG14/VPS38 component is not essential for PI3P synthesis. Collectively, the ATG14/VPS38 mutant collection now permits the study of plants altered in specific aspects of PI3P biology.
INTRODUCTION
Polyphosphoinositides (PPIs) are inositol-containing phospholipids that provide key determinants in cellular trafficking and membrane/organelle identity through the phosphate moiety or moieties attached to their myo-inositol head group (Munnik and Nielsen, 2011; Balla, 2013; Noack and Jaillais, 2017). Especially prevalent are PPIs phosphorylated at the D-3, D-4, and/or D-5 positions, which are controlled by the combined actions of kinases and phosphatases and then recognized by specific PPI binding proteins. After docking to PPI-labeled membranes, these binding proteins direct vesicle formation, trafficking, and fusion, and they ultimately provide signatures for various intracellular compartments. Particularly important in plants is the use of specific PPIs to direct the transport of vesicles from the endoplasmic reticulum (ER) or plasma membrane to the trans-Golgi network for subsequent trafficking via early endosomes and multivesicular bodies to other compartments such as the vacuole (Munnik and Nielsen, 2011; Gerth et al., 2017).

Of interest here are the critical roles of PPIs bearing a phosphate at the D-3 position (phosphatidylinositol 3-phosphate [PI3P]) during endocytosis, endosomal sorting, and macroautophagy (referred to here as autophagy; Di Sansebastiano et al., 2017; Noack and Jaillais, 2017; Marshall and Vierstra, 2018). Despite representing a minor fraction of the PPI pool, PI3P is central to these transport processes through its recognition by a collection of proteins bearing either Fab1/YOTB/Vac1/EEA1 or Phox-Homology domains that recognize the PI3P moiety (Vermeer et al., 2006; Zhuang et al., 2013; Gao et al., 2015; Noack and Jaillais, 2017). PI3P is particularly important during autophagy, where it works in conjunction with the AUTOPHAGY-RELATED8 (ATG8) protein modified with the lipid phosphatidylethanolamine (PE) to uniquely decorate the engulfing autophagic membranes as they trap and transport appropriate cargo for final vacuolar breakdown (Marshall and Vierstra, 2018).
In animals, PI3P is synthesized by three types of phosphoinositide 3-kinases (PI3Ks) containing distinct subunit compositions, defined as class I, II, and III (Balla, 2013). In plants, yeast (Saccharomyces cerevisiae), and algae, only the class III PI3K/VACUOLAR PROTEIN SORTING34 (VPS34) type is evident by sequence comparisons. VPS34 specifically phosphorylates phosphatidylinositol at the D-3 position in complex with three accessory proteins: VPS15 and ATG6 (also known as Beclin 1 or VPS30) together with either ATG14 or VPS38 (the ortholog to mammalian UV RADIATION RESISTANCE-ASSOCIATED GENE [UVRAG]) as a fourth subunit, uniquely generating complex I or complex II isoforms, respectively (Gerth et al., 2017; Noack and Jaillais, 2017). Three-dimensional structures of the complex II isoform revealed two membrane contacts: one involving VPS34, which likely positions its kinase catalytic site near inositol head groups, and the other involving ATG6 and VPS38/UVRAG, which intertwine through their coiled-coil regions and bind lipids through related β-α repeated autophagy-specific (BARA) and BARA2 domains, respectively (Rostislavleva et al., 2015; Young et al., 2016). ATG14 is devoid of an obvious BARA-type domain but instead has a Cys-rich region N-terminal to the coiled-coil region that might promote ER association (Matsunaga et al., 2010). Whereas the yeast and mammalian versions of VPS38/UVRAG mainly control endosomal protein sorting, ATG14 appears more devoted to autophagy (Kihara et al., 2001; Itakura et al., 2008).
At present, our understanding of PI3K in plants, and the functions of its corresponding lipid product, are limited. Numerous genetic analyses with Arabidopsis (Arabidopsis thaliana) implied that PI3P is essential, based on findings that homozygous null mutants compromising either VPS34, ATG6, or VPS15 could not be generated due to failed reproduction (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Lee et al., 2008a, 2008b; Patel and Dinesh-Kumar, 2008; Xu et al., 2011; Wang et al., 2012). Whereas female gametogenesis proceeds normally in the mutants and male gametogenesis continues up to formation of the tube cell and the pair of sperm cells, a strong block in pollen germination arises that prevents transmission of the male gametes. Additional defects were also seen in Arabidopsis plants treated with the pharmacological PI3K inhibitors 3-methyladenine, LY294002, and wortmannin, which included compromised autophagy, cellulose biosynthesis, gravitropism, and the vacuolar sorting of seed storage proteins (Takatsuka et al., 2004; Joo et al., 2005; Takáč et al., 2013; Zhuang et al., 2013; Shin et al., 2014; Fujimoto et al., 2015; Huang et al., 2019). VPS34 antisense plants (Welters et al., 1994), and heterozygous atg6 plants or homozygous atg6 plants rescued with low expression levels from an ATG6-GREEN FLUORESCENT PROTEIN (GFP) reporter (Qin et al., 2007; Harrison-Lowe and Olsen, 2008), were also phenotypically aberrant, indicating that the functions of PI3P in plants extend beyond reproduction.
As an alternative to studying the core PI3K complex, we (Liu et al., 2018) and others (Lee et al., 2018; Yuan et al., 2018) have recently focused on the nonredundant VPS38 and ATG14 subunits as a strategy to define the roles of PI3P in plant development. Initial analyses of the predicted VPS38 subunit revealed that it is indeed part of complex II through its interactions with ATG6 and VPS34. Homozygous null vps38 mutants were viable, indicating that this isoform is not essential during reproduction. However, the mutant plants did exhibit a range of abnormal phenotypes associated with defects in vesicle trafficking and membrane identity, including dampened pollen germination despite apparently normal male gametogenesis, suppressed processing of seed storage proteins and development of storage vacuoles, dilated endosomes, and impaired gravitropism caused in part by aberrant endosomal cycling of the PIN-FORMED (PIN) family of auxin transporters (Lee et al., 2018; Liu et al., 2018; Yuan et al., 2018). Also obvious were defects in leaf morphology and vascular tissue development and increased seed abortion. Autophagy was also diminished modestly in the vps38 plants, as seen by the reduced accumulation of autophagic bodies inside vacuoles that in turn elicited a slight hypersensitivity to nitrogen and fixed-carbon starvation reminiscent of weak autophagy mutants (Liu et al., 2018).
To help define the unique functions of ATG14 and its associated VPS34 complex I kinase, we describe here the genetic, biochemical, and cell biological analysis of the two Arabidopsis paralogs encoding this component. Interaction studies demonstrated that ATG14 likely assembles into a PI3K complex through its interactions with ATG6 and displays expression patterns congruent with those for ATG6, VPS15, and VPS34. Through analysis of atg14a atg14b double null mutants resulting from T-DNA insertions or mutagenesis by clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-associated nuclease Cas9, we found both overlapping and distinct sets of phenotypes compared to null vps38 mutants, indicating that these subunits of the complex I and complex II isotypes have nonredundant roles in plants, like their yeast and mammalian counterparts (Kihara et al., 2001; Itakura et al., 2008). In particular, homozygous atg14a atg14b plants showed strongly impaired autophagy upon both nitrogen and fixed-carbon starvation, but relatively normal seed and leaf development, and transmission of the mutant alleles.
Furthermore, we successfully generated triple homozygous mutants missing ATG14a, ATG14b, and VPS38, strongly suggesting that the fourth subunit in the VPS34 PI3K complex is not essential. The resulting atg14a atg14b vps38 plants displayed a composite phenotype relative to atg14a atg14b and vps38 plants, including a strong suppression of male gamete transmission and autophagy. Surprisingly, the homozygous triple mutants still accumulated PI3P, albeit at reduced levels, but they were hypersensitive to wortmannin, strongly suggesting that the three core subunits of the VPS34 PI3K complex are sufficient to synthesize viable levels of PI3P. With this collection of atg14a, atg14b, and vps38 alleles, we now provide a venue to study PI3P function(s) throughout the Arabidopsis life cycle.
RESULTS
Identification of Arabidopsis ATG14
As a companion to previous studies on the Arabidopsis VPS34 complex II PI3K containing its signature subunit VPS38 (Lee et al., 2018; Liu et al., 2018; Yuan et al., 2018), we sought here to investigate the function(s) of complex I through the analysis of its distinguishing ATG14 subunit. The two possible paralogs in Arabidopsis (ATG14a and ATG14b) harbored similar domain architectures despite low amino acid sequence identities to each other (46%) and to their yeast counterpart (21 and 34%, respectively; Figure 1A; Lee et al., 2018; Liu et al., 2018). Subsequent searches detected ATG14 relatives in a variety of other plant species (35 to 100% identity) that were distinct from the VPS38 family and separated phylogenetically into distinct dicot, monocot, and seedless plant clades (Figure 1A; Supplemental Figure 1A). The ATG14a and ATG14b loci are located on chromosomes 1 and 4, respectively, outside of syntenic patches, suggesting that they originated from a relatively ancient gene duplication event within or before the Brassicaceae lineage.
Figure 1.
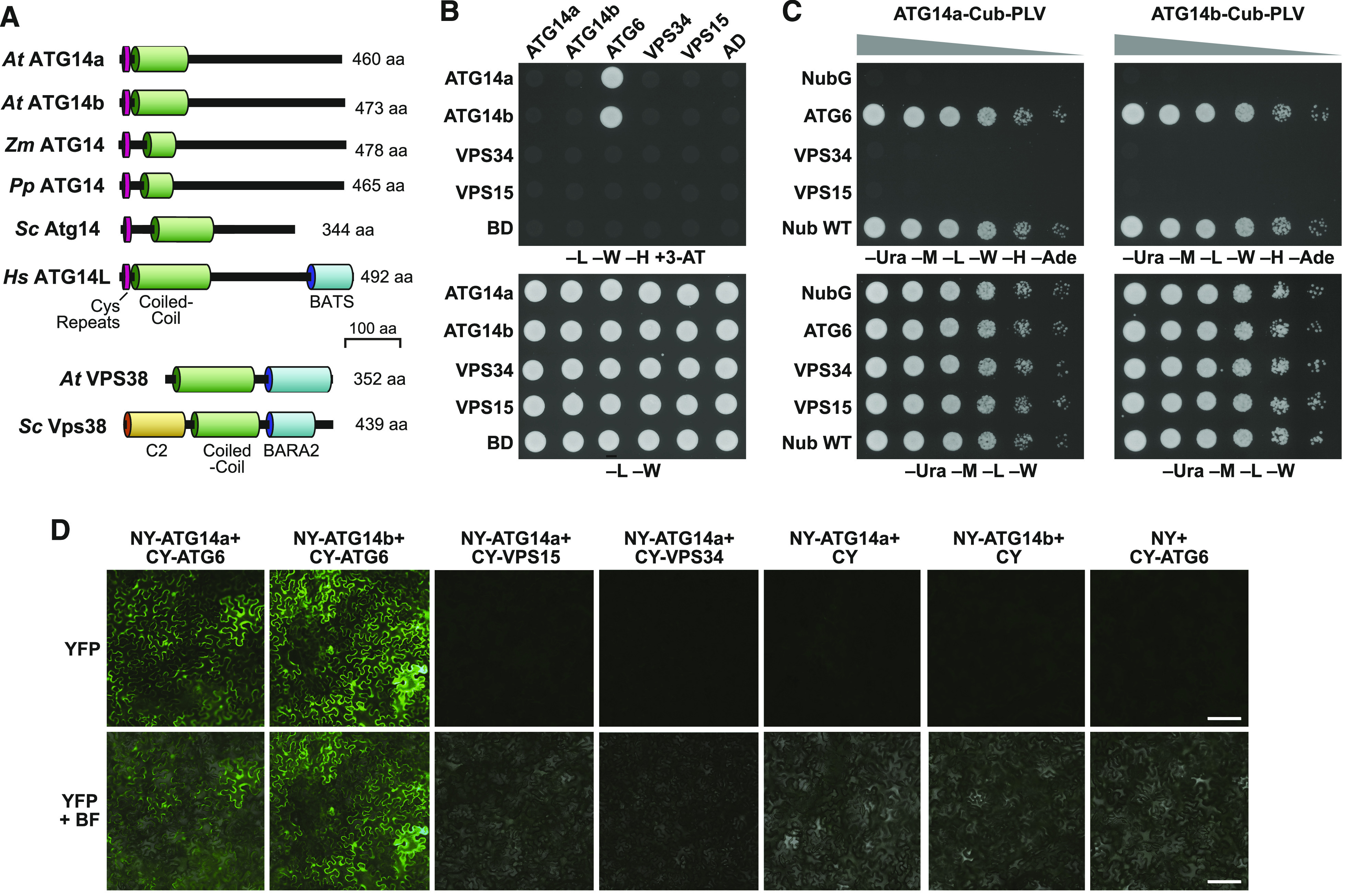
Description of the ATG14 Protein Family and Interaction Analysis of Arabidopsis ATG14 within the PI3K Complex.
(A) Architecture and domain organization of the ATG14 family. The Cys-repeat, coiled-coil, and BATS domains are highlighted. The amino acid (aa) length of each protein is indicated on the right. Arabidopsis and yeast VPS38, with their lipid binding C2, coiled-coil, and BARA2 domains, are included for reference. At, Arabidopsis thaliana; Hs, Homo sapiens; Pp, Physcomitrium patens; Sc, Saccharomyces cerevisiae; Zm, Zea mays.
(B) and (C) Pairwise Y2H assays between Arabidopsis ATG14 and other PI3K subunits. (B) Standard Y2H assays using the soluble Gal4-based system. Full-length ATG6, ATG14a, ATG14b, VPS15, and VPS34 fused to the C terminus of the activation domain (AD) or DNA binding domain (BD) of Gal4 were tested for binding by growth on synthetic dropout medium lacking Leu, Trp, and His, and containing 25 mM 3-AT (–L–W–H+3-AT). Empty AD or BD vectors served as negative controls. Viability of the cells was confirmed by growth on medium lacking Leu and Trp (–L–W). (C) Pairwise Y2H assays using the split-ubiquitin PLV system for membrane proteins. Full-length proteins were expressed as a fusion to either Cub-PLV as bait or Nub as prey and coexpressed in diploid yeast cells. Positive interactions were determined by growth of twofold serial dilutions on synthetic dropout medium lacking Ura, Met, Leu, Trp, His, and Ade (–Ura–M–L–W–H–Ade). Empty NubG and Nub-wild type (WT) vectors served as negative and positive controls, respectively (Stagljar et al., 1998). Viability of the cells was confirmed by growth on medium lacking Ura, Met, Leu, and Trp (–Ura–M–L–W).
(D) Pairwise in planta BiFC assays between Arabidopsis ATG14 and other PI3K subunits in N. benthamiana leaf epidermal cells. Each full-length protein was assembled as a fusion to either the NY or CY fragments of YFP. Reconstitution of the fluorescent signal was observed by confocal microscopic analysis 36 h after infiltration. YFP fluorescence and merged bright-field (BF) images are shown. Additional controls for the BiFC are presented in Supplemental Figure 4. Bar = 30 µm.
Unlike plant VPS38, which starts with an N-terminal coiled-coil region (Lee et al., 2018; Liu et al., 2018), the plant ATG14 sequences, like their yeast and mammalian brethren (Diao et al., 2015), contain a Cys-repeat region upstream of the coiled-coil region (Figure 1A). This 21–amino acid stretch is notable for the strict positional conservation of four cysteines (Supplemental Figure 2) that are required for ER localization and autophagy of their yeast and mammalian relatives (Matsunaga et al., 2010). Interestingly, the human Atg14L protein ends in a Barkor autophagosome-targeting sequence (BATS) domain that appears unique to mammals. Previous studies showed that this BATS region interacts with both PI3P and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], possibly through an amphipathic helix, to regulate assembly of the PI3K complex and mediate its association with autophagosomes (Fan et al., 2011; Tan et al., 2016). Whether the plant ATG14 orthologs have a cryptic BATS region with orthologous function(s) remains unclear, but a similar C-terminal helix bearing bulky hydrophobic residues is predicted (Supplemental Figure 2).
ATG14a and ATG14b Have Distinct Expression Patterns
Expression patterns gathered from the transcriptome variation analysis (TraVA) database (http://travadb.org/; Klepikova et al., 2016) revealed that ATG14a and ATG14b, like VPS38, are ubiquitously expressed in most Arabidopsis tissues (Supplemental Figure 1B). In general, transcripts encoding subunits of the VPS34 PI3K complex (except ATG14b) were consistently high in mature or senescing organs, as one might expect for roles of ATG14 and the rest of the complex during cell death and autophagy, as seen in other organisms (Cao and Klionsky, 2007; Lindmo et al., 2008; Jaber et al., 2012; Diao et al., 2015). Similar to mRNAs for the other subunits (ATG6, VPS15, VPS34, and VPS38), ATG14a mRNA was also abundant in anthers (Supplemental Figure 1B), which is consistent with a prominent role for this kinase during pollen development and/or germination (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Lee et al., 2008b; Xu et al., 2011; Wang et al., 2012), while ATG14b mRNA was abundant in immature and dry seeds, suggesting a specific role for this paralog during reproduction. Collectively, the expression data indicated that Arabidopsis ATG14a, ATG14b, and VPS38 have both overlapping and distinct functions.
We also examined connections among ATG14, VPS38, and other subunits of the PI3K complex and with other Arabidopsis proteins by coexpression analysis of the Arabidopsis transcriptome, using the ATTED-II version 10 platform (Obayashi et al., 2018). Whereas the VPS34, ATG6, and VPS15 loci displayed direct and indirect expression relationships with a number of other ATG loci, ATG14a displayed a direct coexpression relationship only with ATG18f, while ATG14b and VPS38 displayed direct and indirect coexpression relationships with only ATG13b, respectively (Supplemental Figure 1C), again suggesting nonredundant functions for ATG14 and VPS38. Interestingly, connections with a number of other Arabidopsis proteins, some of which were not yet functionally defined, were also detected that could represent novel autophagic components or regulators.
ATG14 Interacts with ATG6 from the VPS34 PI3K Complex
Given the low sequence homology of Arabidopsis ATG14 relative to its human/yeast counterparts, we attempted to confirm its orthology through complementation of a yeast Δatg14 deletion mutant. Using both the free GFP release assay based on GFP-Atg8 (Marshall et al., 2016), and activation of the Pho8Δ60 reporter to measure autophagic flux (Noda and Klionsky, 2008), neither full-length Arabidopsis ATG14a nor ATG14b tagged with hemagglutinin (HA) successfully rescued the Δatg14 strain, while the yeast ortholog was effective (Supplemental Figure 3). The Arabidopsis counterparts of Vps15, Vps30, and Vps34 (VPS15, ATG6, and VPS34, respectively) also failed to rescue the corresponding yeast deletion strains (Supplemental Figure 3), implying that the plant and fungal VPS34 PI3Ks have sufficiently diverged to prevent cross-kingdom complementation. However, we note that Welters et al. (1994) succeeded in rescuing a yeast Δvps34 strain with a chimera of Arabidopsis VPS34 and its yeast counterpart, suggesting that at least some portion(s) of the two proteins is functionally compatible.
As an alternative, we assessed whether ATG14 assembles with the VPS34 PI3K complex through binding studies. As a first attempt, we used standard Gal4-based yeast two-hybrid (Y2H) assays to test the interaction of ATG14a/b with their likely Arabidopsis partner ATG6, to which they are predicted to bind through their intertwined coiled-coil regions (Rostislavleva et al., 2015; Young et al., 2016). As shown in Figure 1B, clear interactions were detected based on colony growth on selection medium lacking Leu, Trp, and His and containing 3-amino-1,2,4-triazole (3-AT), for yeast harboring either ATG14a or ATG14b fused to the Gal4 DNA binding domain paired with ATG6 fused to the Gal4 activation domain. We note that ATG6 fused to the Gal4 DNA binding domain self-activated the assay (Lee et al., 2018; Liu et al., 2018). We failed to detect interactions among ATG6, VPS15, and VPS34 or between ATG14a/b and VPS15 or VPS34 using this Y2H system (Figure 1B).
Next, we tested binding using the split-ubiquitin Y2H system designed for membrane proteins, which exploits the reassociation of ubiquitin fragments to direct the proteolytic release of the membrane-anchored Protein A/LexA/VP16 transcription factor (PLV; Stagljar et al., 1998). Notably, this method successfully detected an interaction between ATG6 and VPS34, which was not seen using the Gal4-based system (Liu et al., 2018). Interactions between either ATG14a or ATG14b bearing the C-terminal fragment of ubiquitin fused to PLV (Cub-PLV), and ATG6 bearing the N-terminal fragment of ubiquitin (NubG) were robust on selection medium compared to the positive control Nub-wild type (Figure 1C). In contrast to VPS38, which bound both VPS34 and VPS15 by this assay (Liu et al., 2018), interactions between ATG14a/b and these two subunits were not evident, suggesting that ATG14 integrates differently into the VPS34 PI3K complex compared to VPS38.
Finally, we confirmed binding of ATG14a and ATG14b to ATG6 by bimolecular fluorescence complementation (BiFC) assays. We observed binding, as evidenced by strong cytoplasmic fluorescence of epidermal pavement cells from Nicotiana benthamiana leaves infiltrated with plasmids encoding the N-terminal half of yellow fluorescent protein (YFP) fused to ATG14a or ATG14b paired with a plasmid encoding the C-terminal half of YFP fused to ATG6. Control pairs of plasmids (nonrecombined pSITE vectors) or plasmids expressing YFP fragments fused to VPS15 or VPS34 (Figure 1D; Supplemental Figure 4) showed no fluorescence. Together, the binding studies confirmed the identity of ATG14a/b and were consistent with the architecture of the yeast and human class III PI3Ks, which intimately connects ATG14/VPS38 with ATG6 (Rostislavleva et al., 2015; Young et al., 2016).
Isolation of Mutants Compromising ATG14a and ATG14b
To study the function(s) of Arabidopsis ATG14 genetically, we searched the available T-DNA insertion collections for alleles disrupting either locus. Although no insertion lines were available for ATG14a, two lines were described for ATG14b in the Columbia-0 (Col-0) accession from the Arabidopsis Biological Resource Center (ABRC) collection (www.abrc.osu.edu). One harbored a T-DNA sequence within the 10th exon at nucleotide 2155 downstream of the start codon (SAIL_1207_H10) and was designated here as atg14b-1, while the second harbored a T-DNA sequence within the 1st exon at nucleotide 262 downstream of the start codon (SALK_145203) and was designated here as atg14b-2 (Figures 2A and 2B). We identified homozygous atg14b-1 and atg14b-2 individuals by PCR genotyping. RT-PCR analyses showed that these mutants fail to accumulate the full-length ATG14b transcript (Figure 2D), although partial transcripts were evident in atg14b-1, suggesting that shortened polypeptide(s) may be synthesized from this allele. As the hypothetical atg14b-1 protein might encompass most of the ATG14b protein (409 residues), including all of the Cys-repeat and the coiled-coil regions (Supplemental Figure 2), the atg14b-1 allele might be partially functional.
Figure 2.
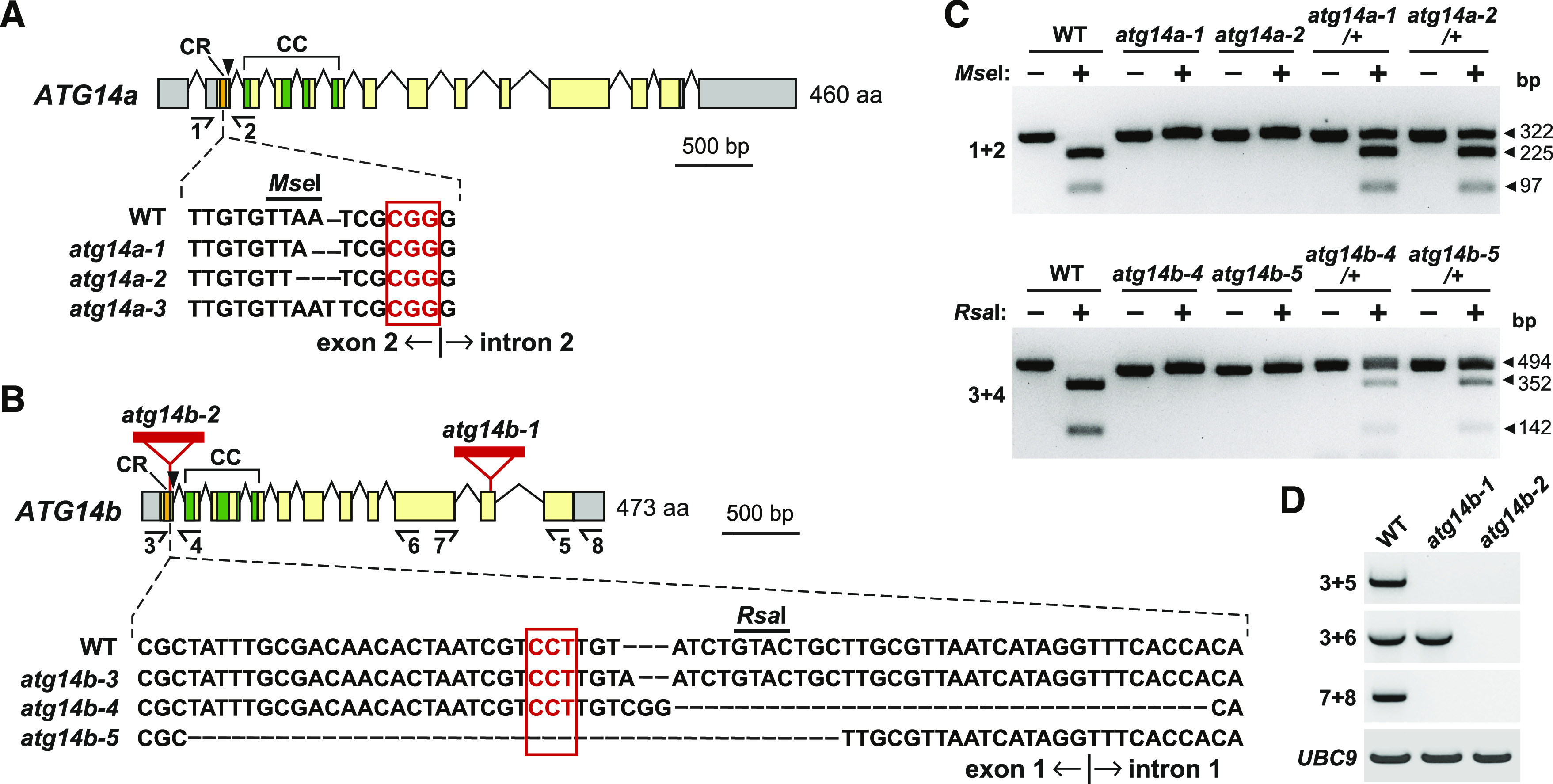
Construction of atg14a atg14b Mutant Combinations Using CRISPR/Cas9-Mediated Gene Editing.
(A) and (B) Diagram of the Arabidopsis ATG14a (A) and ATG14b (B) loci, indicating the positions and effects of the various atg14a and atg14b alleles. Colored boxes indicate coding regions and gray boxes indicate 5′ and 3′ untranslated regions. The sequences encoding the Cys-repeat (CR) and coiled-coil (CC) domains are in orange and green, respectively. Lines identify introns. Red triangles in (B) show the T-DNA insertion sites in the atg14b-1 and atg14b-2 mutants. Half arrows, primers used for genomic- or RT-PCR in (C) and (D), respectively. Positions of the NGG PAM targeting sites for CRISPR/Cas9 gene editing are indicated by the arrowheads. CRISPR/Cas9-induced deletions/insertions present in the atg14a or atg14b mutants and the position of the PAM triplet nucleotides (red box) are shown below the diagrams in comparisons to the wild-type (WT) sequences. Exon/intron boundaries are indicated. Exact sequence descriptions of the CRISPR alleles are presented in Supplemental Figure 5.
(C) Identification of the CRISPR/Cas9-derived atg14a and atg14b mutants by CAPS analysis. Genomic DNA extracted from mutagenized seedlings was amplified by PCR with the indicated primer pairs (see [A] and [B]), subjected to restriction endonuclease digestion with MseI (atg14a) or RsaI (atg14b) and then separated by gel electrophoresis. The arrowheads indicate DNA fragments before and after digestion, and their lengths (bp).
(D) RT-PCR analysis of the atg14b T-DNA insertion mutants. Total RNA isolated from the wild type (WT), atg14b-1, and atg14b-2 plants was subjected to RT-PCR using the primers indicated in (B). RT-PCR of UBC9 was included as an internal control.
To generate mutants disrupting ATG14a and additional alleles in ATG14b, we used CRISPR/Cas9-mediated mutagenesis (Xing et al., 2014; Wang et al., 2015), with guide RNAs that targeted DNA stretches in both ATG14a and ATG14b just upstream of a possible protospacer adjacent motif (PAM) NGG triplet needed to position Cas9 for cleavage (Figures 2A and 2B). This mutagenesis generated three independent alleles for ATG14a, which harbor either 1- or 2-bp deletions, or a 1-bp insertion just upstream of the PAM, and were designated here as atg14a-1, atg14a-2, and atg14a-3, respectively (Figure 2A; Supplemental Figure 5A). Importantly, all three mutations introduced a reading frameshift downstream of the Val-20 codon, which should substantially truncate the ATG14a protein if translated.
For ATG14b, we identified three CRISPR/Cas9 alleles that harbor either a 1-bp insertion, a 35-bp deletion combined with a 3-bp insertion, or a 39-bp deletion just upstream of the PAM, and were designated here as atg14b-3, atg14b-4, and atg14b-5, respectively (Figure 2B). The atg14b-3 allele introduced a frameshift downstream of the Cys-18 codon, which should substantially shorten the translated ATG14b protein (Figure 2B; Supplemental Figure 5B). The atg14b-4 allele eliminated the junction between the first exon and the first intron, resulting in the retention of the first intron. The atg14b-5 allele eliminated 14 codons after the Cys-8 codon, thus deleting the coding sequence for part of the Cys-repeat region. In all three cases, a substantially truncated, and likely functionally null, ATG14b protein should accumulate if translated (Supplemental Figure 5B).
Within the CRISPR/Cas9 population, we observed a variety of mutant combinations by DNA sequence analysis, including hemizygous individuals and individuals affected at both ATG14a and ATG14b loci. We separated the mutations by selfing and backcrossing to isolate single and double mutant combinations that disrupted both loci but lacked the Cas9 locus. To help track the atg14a-1, atg14a-2, atg14b-4, and atg14b-5 CRISPR/Cas9 alleles, we developed a cleaved amplified polymorphic sequence (CAPS) strategy (Konieczny and Ausubel, 1993) based on the loss of cut sites by the restriction enzymes MseI and RsaI in genomic PCR products derived from the induced alleles. As shown in Figure 2C, only the wild-type ATG14a and ATG14b PCR products were readily digested with MseI and RsaI, respectively. Ultimately, we used the strong, double-allele combinations atg14a-1 atg14b-4, atg14a-2 atg14b-2, and atg14a-3 atg14b-3 for subsequent studies.
Plants Missing ATG14 Senesce Early and Are Hypersensitive to Nutrient Deprivation
Like null mutants for VPS38, all our single and double homozygous mutants inactivating ATG14a and ATG14b were viable and capable of producing seeds (Supplemental Figure 6A), indicating that this subunit is not essential to Arabidopsis, in contrast to mutants eliminating the core PI3K complex components ATG6, VPS15, and VPS34 (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Lee et al., 2008b; Xu et al., 2011; Wang et al., 2012). Whereas the vps38 rosettes were severely stunted (Liu et al., 2018), the atg14a and atg14b mutant combinations developed phenotypically normal rosettes indistinguishable from those of wild type and other strong autophagy mutants (atg2-1, atg7-2, atg9-4, atg11-1, and atg13a-2 atg13b-2; Supplemental Figure 6A; see below).
However, like other atg mutants (Doelling et al., 2002; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010), the homozygous atg14a atg14b lines displayed a number of phenotypes consistent with a role for ATG14 in autophagy. Indeed, we observed early leaf senescence when grown in short days (8-h-light/16-h-dark cycle; Figure 3A), and a hypersensitivity to both nitrogen and fixed-carbon starvation, as evidenced by accelerated chlorosis of seedlings grown on liquid or solid medium in the light but without nitrogen (Figure 3B; Supplemental Figure 6B), or on Suc-free medium in extended darkness (Figure 3C), respectively. Quantification of seedling death caused by fixed-carbon starvation showed that while mutants missing core autophagy components (atg2-1, atg5-1, and atg7-2; Thompson et al., 2005; Chung et al., 2010; Shin et al., 2014) were strongly sensitive and died after 9 d in darkness, the atg14a atg14b double mutants had a weaker response even after 13 d of darkness (Figure 3D). This mild response was similar to that seen for autophagy mutations compromising the upstream ATG1 kinase complex (atg11-1, atg13a-2 atg13b-2, and the quadruple atg1a-2 atg1b-1 atg1c-1 atg1t-1 mutants; Suttangkakul et al., 2011; Li et al., 2014; Huang et al., 2019), or ATG9 needed for autophagic membrane assembly (atg9-4; Figure 3D; Shin et al., 2014). None of the single atg14a or atg14b mutants displayed abnormal phenotypes compared to the wild type either with or without nutrient stress, indicating the likely absence of secondary mutations and suggesting that the pair have strongly overlapping activities.
Figure 3.
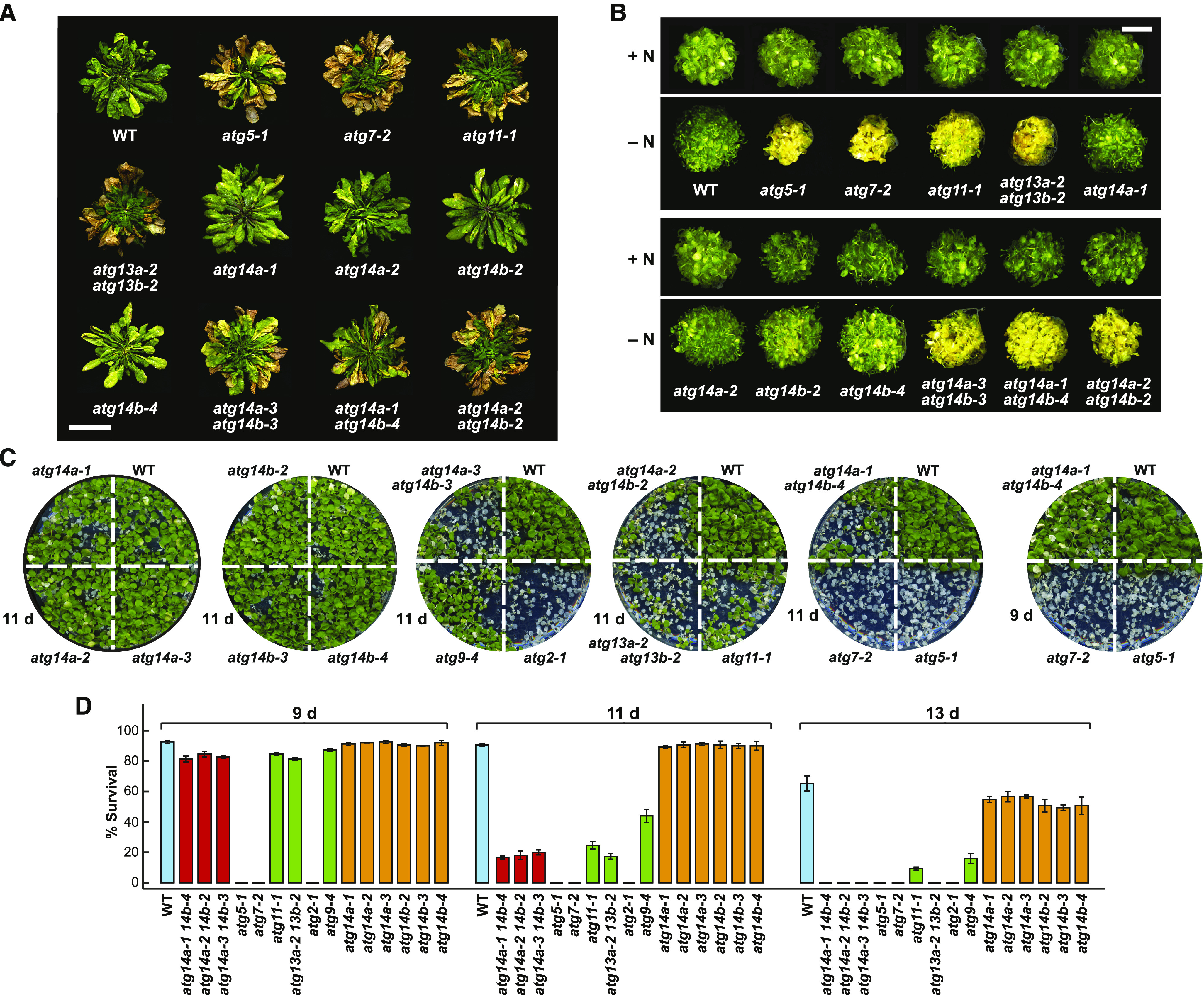
atg14a atg14b Mutants Have Accelerated Senescence and Increased Sensitivity to Nitrogen and Fixed-Carbon Starvation.
The homozygous atg14a and atg14b single or double mutants are described in Figure 2. Previously described homozygous autophagy mutants atg5-1, atg7-2, atg9-4, atg11-1, and atg13a-2 atg13b-2, and the wild-type Col-0 (WT) were included for comparisons.
(A) Accelerated senescence of plants grown under a short-day photoperiod (8-h-light/16-h dark). Plants were grown on soil at 23°C for 10 weeks. Bar = 1 cm.
(B) Enhanced sensitivity to nitrogen starvation. Plants were grown in constant white light in nitrogen-containing liquid Murashige and Skoog (MS) medium for 1 week and then transferred to either nitrogen-rich (+N) or nitrogen-deficient (–N) liquid medium for 2 weeks. Close-up images of the seedlings grown on solid medium are shown in Supplemental Figure 6B. Bar = 2 cm.
(C) Enhanced sensitivity to fixed-carbon starvation. Seedlings were grown on Suc-free (–Suc) solid MS medium under a long-day photoperiod (16-h-light/8-h-dark; LD) for 2 weeks, transferred to darkness for 9 or 11 d, and then allowed to recover for 12 d in LD.
(D) Quantification of the effects of fixed-carbon starvation based on the survival of seedlings after 9, 11, or 13 d in darkness followed by 12 d in LD. Each bar represents the average (±sd) survival percentage of 50 seedlings.
To ascertain where ATG14a/b influences the autophagy system, we compared levels of various ATG proteins, as well as the ATG12-ATG5 and ATG8-PE adducts. The addition of PE to ATG8 helps embed ATG8 into autophagic membranes, while the conjugation of ATG12 to ATG5 is essential to the ATG12-ATG5-ATG16 ligase complex that drives this lipidation (Marshall and Vierstra, 2018). Levels of ATG8, ATG5, and ATG1 were slightly elevated in the atg14a atg14b double mutants (Supplemental Figure 7A), consistent with prior studies with other atg mutants in which a block in autophagy upregulates expression of ATG genes and suppresses turnover of the encoded proteins (Contento et al., 2004; Sláviková et al., 2005; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010; Li et al., 2014; Huang et al., 2019).
To confirm the identity of the ATG8-PE species, we demonstrated its sensitivity to phospholipase D (Yoshimoto et al., 2004; Chung et al., 2010), which converted the faster migrating lipidated species, seen upon SDS-PAGE in the presence of 6 M urea, to the slower migrating free ATG8 species (Supplemental Figure 7B). Notably, assembly of the ATG12-ATG5 conjugate essential for ATG8 lipidation, and the accumulation of ATG8-PE itself, were not appreciably affected by the absence of ATG14 (Supplemental Figures 7A and 7B). We previously reported that vps38 mutants hyperaccumulate ubiquitin conjugates, possibly due to altered endomembrane dynamics (Liu et al., 2018); we observed no such increase for atg14a-1 atg14b-4 seedlings (Supplemental Figure 7C).
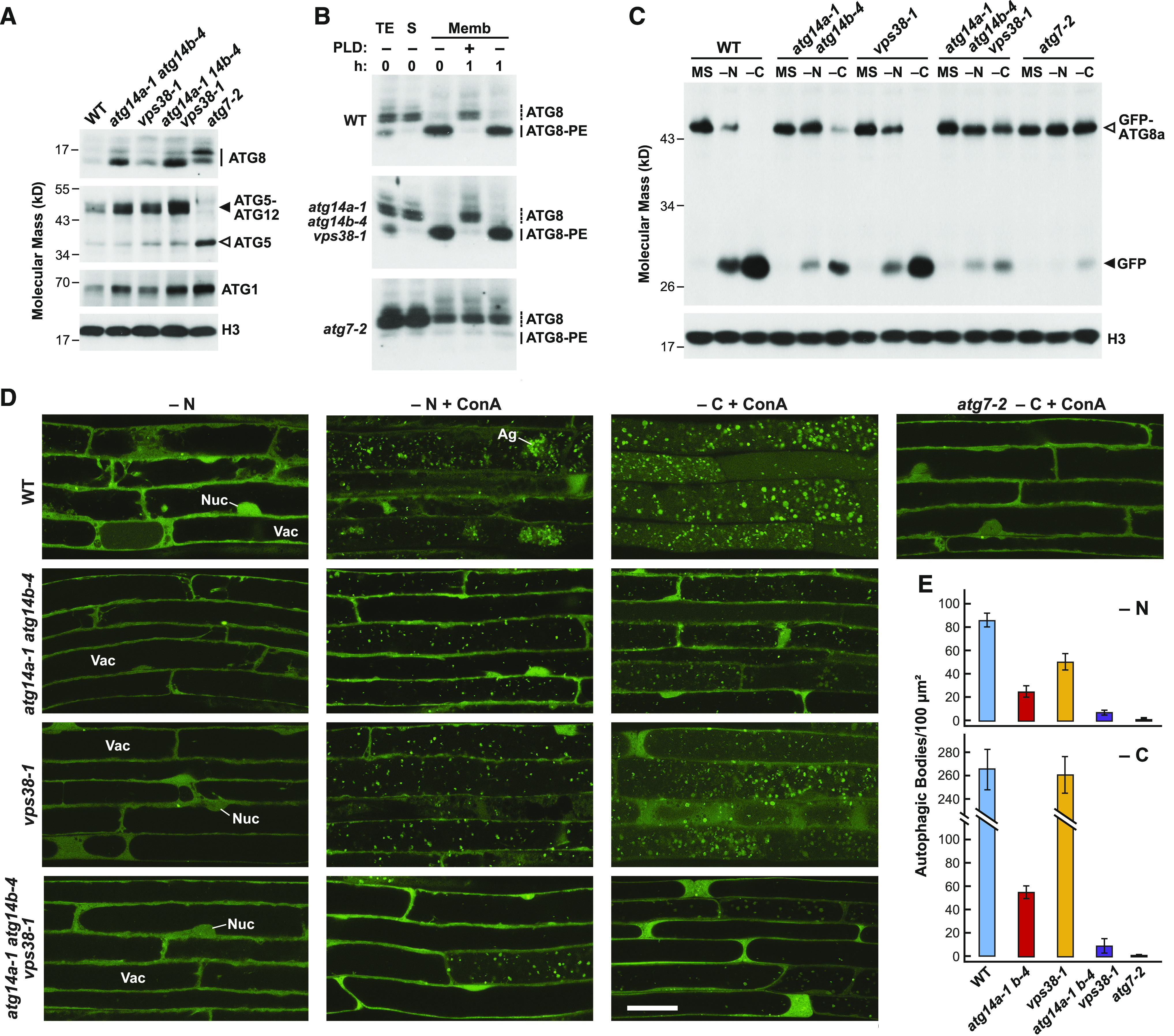
ATG14a/b Promote Autophagic Transport and Accumulation of Autophagic Bodies
Using a GFP-ATG8a reporter (Thompson et al., 2005), we compared the importance of ATG14 and VPS38 in autophagy by fluorescence confocal microscopy to assess autophagic body accumulation, and by the free GFP release assay that measures autophagy-dependent vacuolar transport (Yoshimoto et al., 2004; Chung et al., 2010; Suttangkakul et al., 2011; Marshall and Vierstra, 2018). Time-course studies of GFP-ATG8a roots exposed to nitrogen or fixed-carbon starvation and treated with concanamycin A (ConA) to suppress autophagic body degradation (Thompson et al., 2005) revealed that the atg14a atg14b double mutants effectively blocked vesicle accumulation relative to the wild type and the vps38-1 mutant, although not as effectively as the atg7-2 mutant (Figures 4A and 4B). In fact, under our treatment conditions, the density of autophagic bodies was sometimes so great in the wild-type root cells that vesicle clusters appeared (Figure 4C). Whereas the atg14a atg14b mutants strongly depressed autophagic body accumulation during both nitrogen and fixed-carbon starvation, the vps38-1 mutant was only mildly effective under nitrogen starvation and ineffective during fixed-carbon starvation, demonstrating that ATG14 is uniquely critical for autophagy (Figure 4B).
Figure 4.
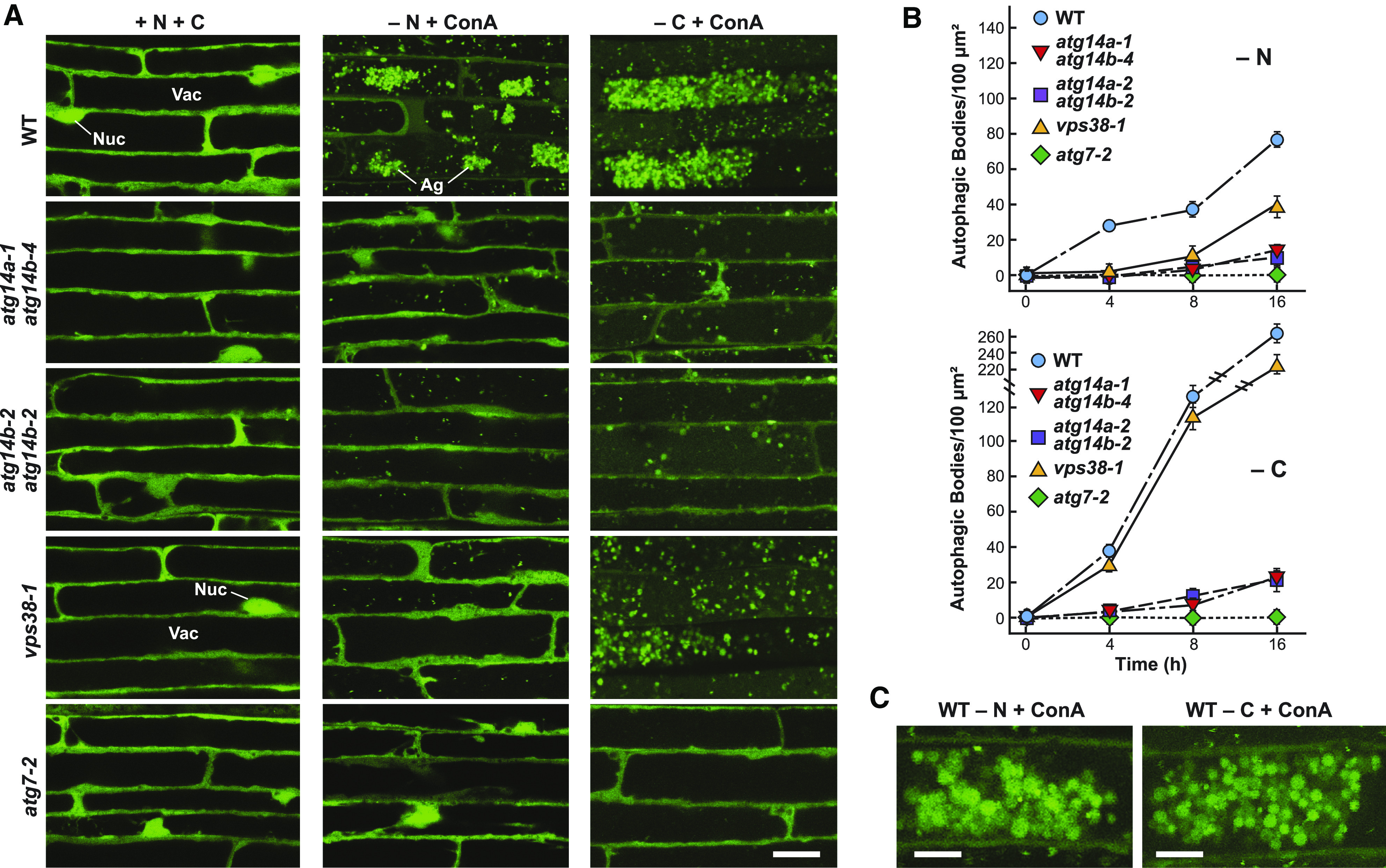
ATG14 Promotes the Vacuolar Accumulation of Autophagic Bodies.
(A) atg14a atg14b mutant plants accumulate fewer autophagic bodies upon nitrogen or fixed-carbon starvation. The 7-d-old wild-type (WT), atg14a atg14b double mutant, atg7-2, and vps38-1 seedlings accumulating the GFP-ATG8a reporter were grown in a LD photoperiod on nitrogen- and Suc-rich solid MS medium and transferred either to fresh MS liquid medium, to nitrogen-deficient liquid MS medium containing 1 µM ConA and incubated in the light for 16 h (−N), or to Suc-free liquid MS medium containing 1 µM ConA and incubated in darkness for 16 h (−C). Root cells were imaged for GFP-ATG8a by confocal fluorescence microscopy. Ag, autophagic body aggregates; Nuc, nucleus; Vac, vacuole. Bar = 20 µm.
(B) Time courses for the accumulation of autophagic bodies upon nitrogen or fixed-carbon starvation. Roots of the indicated genotypes were treated as described in (A), and the accumulation of autophagic bodies was quantified over time by confocal microscopy and is expressed as the mean (±sd) vacuolar density based on the analysis of 25 cells from multiple roots.
(C) Close-up images showing autophagic body aggregates that accumulate in the wild-type Col-0 (WT) upon nitrogen or fixed-carbon starvation. Bar = 10 µm.
Measurement of autophagic flux by the free GFP release assays validated the importance of ATG14a/b to autophagy. In contrast to the robust release of free GFP from the GFP-ATG8a reporter in the wild-type roots, free GFP was effectively absent in atg7-2 roots under both prolonged nitrogen and fixed-carbon starvation (Figure 5). The vps38-1 mutant only partially dampened free GFP release upon nitrogen starvation and was relatively ineffective upon fixed-carbon starvation. By contrast, we saw a strong inhibition of free GFP release in two sets of atg14a atg14b double mutants under both conditions (Figure 5). Collectively, the data implicate ATG14 and its class III complex I kinase as the main source of PI3P during autophagy.
Figure 5.
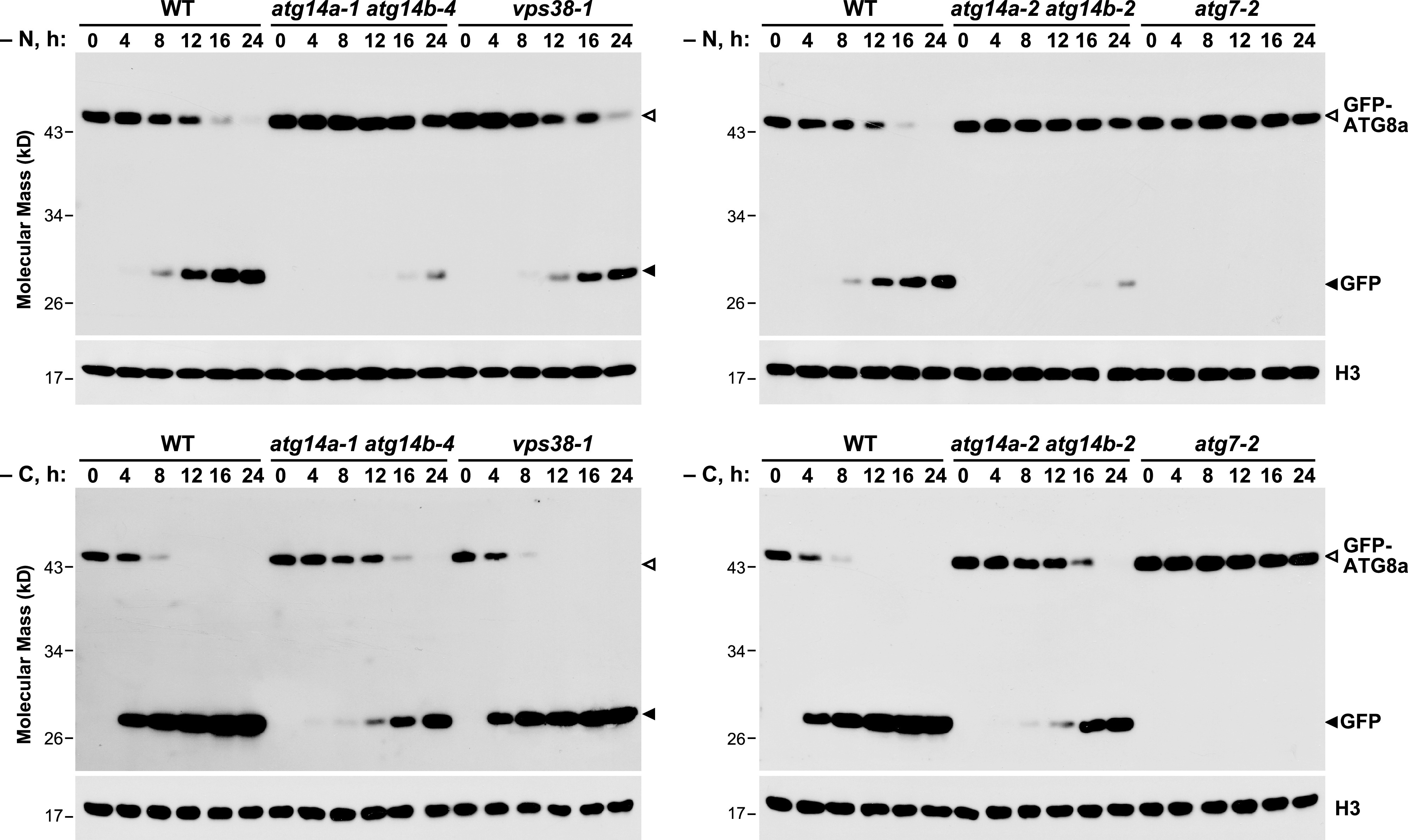
atg14a atg14b Mutants Have Defective Autophagic Transport as Judged by the Release of Free GFP from GFP-ATG8a.
The wild-type Col-0 (WT), atg14a-1 atg14b-4, atg14a-2 atg14b-2, vps38-1, and atg7-2 seedlings accumulating the GFP-ATG8a reporter were grown in constant light in nitrogen-rich MS liquid medium containing 1% (w/v) Suc, transferred to either nitrogen-deficient liquid MS medium under light (−N; top panels), or Suc-free liquid MS medium in darkness (−C; bottom panels), and then harvested at the indicated times. Total seedling extracts were immunoblotted with anti-GFP antibodies, using anti-histone H3 antibodies to confirm near equal protein loading. Open and filled arrowheads locate the GFP-ATG8a fusion and free GFP, respectively.
ATG14 Is Targeted for Autophagy through Its Association with Autophagic Bodies
To confirm the atg14a atg14b phenotypes through complementation and to examine whether ATG14, like other autophagy components, is degraded by autophagy upon binding autophagic membranes (Suttangkakul et al., 2011; Li et al., 2014), we created atg14a-1 atg14b-4 and atg7-2 lines constitutively expressing a translational fusion of ATG14b and GFP. Genomic PCR, RT-PCR, and CAPS analysis of the resulting homozygous plants detected the full-length GFP-ATG14b coding region with the correct sequence inserted into the respective genomes and transcribed into mRNA (Supplemental Figure 8). Importantly, this reporter successfully replaced ATG14 based on the increased survival of GFP-ATG14b atg14a atg14b seedlings upon fixed-carbon starvation (Figure 6B). However, we note that the complementation studies were not completely effective based on long-term fixed-carbon starvation assays (Supplemental Figure 9), suggesting either that insufficient levels of GFP-ATG14b accumulated or that the GFP moiety interfered with ATG14b function.
Figure 6.
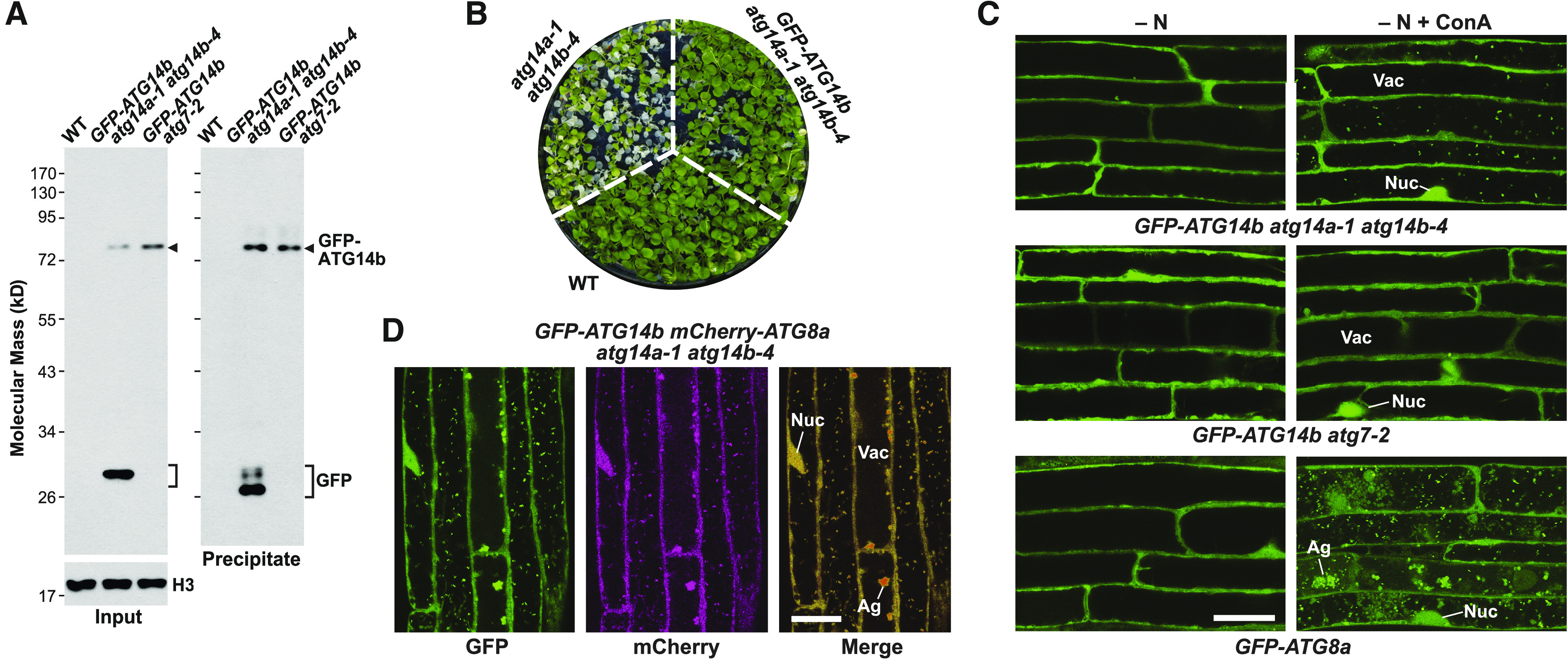
GFP-ATG14 is a Target of Autophagy and Associates with Autophagic Vesicles.
(A) Autophagic breakdown of the GFP-ATG14b reporter as determined by the free GFP release assay. Total extracts from Col-0 (WT), GFP-ATG14b atg14a-1 atg14b-4, and GFP-ATG14b atg7-2 seedlings grown under nutrient-rich conditions were concentrated by TCA precipitation and subjected directly to immunoblot analysis with anti-GFP antibodies, or immunoprecipitated with anti-GFP antibody beads after which the precipitates were subjected to immunoblot analysis. GFP-ATG14b and free GFP are indicated by the arrowhead and brackets, respectively. An immunoblot with anti-histone H3 antibodies was included to confirm near equal protein loading.
(B) The GFP-ATG14b reporter rescues the hypersensitivity of atg14a-1 atg14b-4 seedlings to fixed-carbon starvation. Col-0 (WT), atg14a-1 atg14b-4, and GFP-ATG14b atg14a-1 atg14b-4 seedlings were grown on Suc-free solid MS medium in a LD photoperiod for 10 d, placed in darkness for 10 d, and then returned to the LD photoperiod for 1 week before imaging. See Supplemental Figure 9 for additional survival data with the rescued line.
(C) GFP-ATG14b decorates vacuolar puncta during nitrogen starvation. Seven-day-old GFP-ATG14b atg14a-1 atg14b-4, GFP-ATG14b atg7-2, and GFP-ATG8a seedlings were grown in a LD photoperiod on nitrogen-rich solid MS medium, transferred to nitrogen-deficient liquid MS medium (−N) with or without 1 µM ConA, and incubated in the light for 16 h. Root cells were imaged by confocal fluorescence microscopy. Ag, autophagic body aggregates; Nuc, nucleus; Vac, vacuole. Bar = 20 µm.
(D) GFP-ATG14b colocalizes with mCherry-ATG8a in vacuolar autophagic bodies during nitrogen starvation. GFP-ATG14b mCherry-ATG8a atg14a-1 atg14b-4 roots were starved for nitrogen and treated with ConA as described in (C), and then imaged by confocal fluorescence microscopy. Shown are the GFP, mCherry, and merged channels. Bar = 20 µm.
Analysis of the GFP-ATG14b lines by the free GFP release assay revealed that the GFP-ATG14b reporter was targeted for autophagic turnover. While we detected both fused and free GFP forms in the atg14a-1 atg14b-4 background by immunoblot analysis of total protein extracts and immunoprecipitates generated with anti-GFP antibodies, we only observed the intact fusion in the atg7-2 background (Figure 6A), consistent with impaired ATG14b turnover. Subsequent confocal fluorescence microscopy of root cells detected autophagic bodies decorated with the GFP-ATG14b reporter. Whereas these fluorescent vacuolar puncta accumulated in nitrogen-starved GFP-ATG14b atg14a-1 atg14b-4 seedlings upon ConA treatment, they were absent in similarly treated GFP-ATG14b atg7-2 seedlings (Figure 6C). We then confirmed that these puncta were autophagic bodies by colocalization studies on nitrogen-starved atg14a-1 atg14b-4 roots expressing both GFP-ATG14b and mCherry-ATG8a; most vacuolar puncta were labeled with both reporters (Figure 6D).
We attempted to exploit the GFP-ATG14b fusion to confirm in planta assembly of a PI3K complex also containing VPS34, ATG6, and VPS15 by tandem mass spectrometric analysis of immunoprecipitates prepared with anti-GFP antibodies from total GFP-ATG14b atg14a-1 atg14b-4 seedling extracts. Unfortunately, we failed to detect any of these factors. Neither were ATG14a/b, VPS34, ATG6, VSP15, nor VSP38 detected by shot-gun mass spectrometry of total seedling extracts despite the identification of more than 3300 distinct Arabidopsis proteins, suggesting that one barrier to detection is that this complex is present at very low levels.
Arabidopsis Plants Missing Both ATG14 and VPS38 Are Viable
Given that homozygous Arabidopsis mutants missing the core subunits of the VPS34 PI3K complex could not be created due to defects in pollen development (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Xu et al., 2011; Wang et al., 2012), we predicted that triple mutants missing ATG14a, ATG14b, and VPS38 function would likewise be embryo or seedling lethal. As a first test, we allowed triple heterozygous atg14a-1 atg14b-4 vps38-1 plants to self and then cataloged the genotypes of the resulting progenies, using genomic PCR to identify the vps38-1 T-DNA insertion and our CAPS strategy to score the atg14a-1 and atg14b-4 mutations by MseI/RsaI sensitivity (Figure 2C). After genotyping 523 individuals, we failed to find plants that were homozygous for all three mutations (Figure 7A). Moreover, while most other allelic combinations segregated at the predicted frequencies, the exceptions were mutants homozygous for two of the loci and heterozygous for the third, strongly suggesting that haploid female and/or male gametes bearing the atg14a atg14b vps38 null combinations were compromised.
Figure 7.
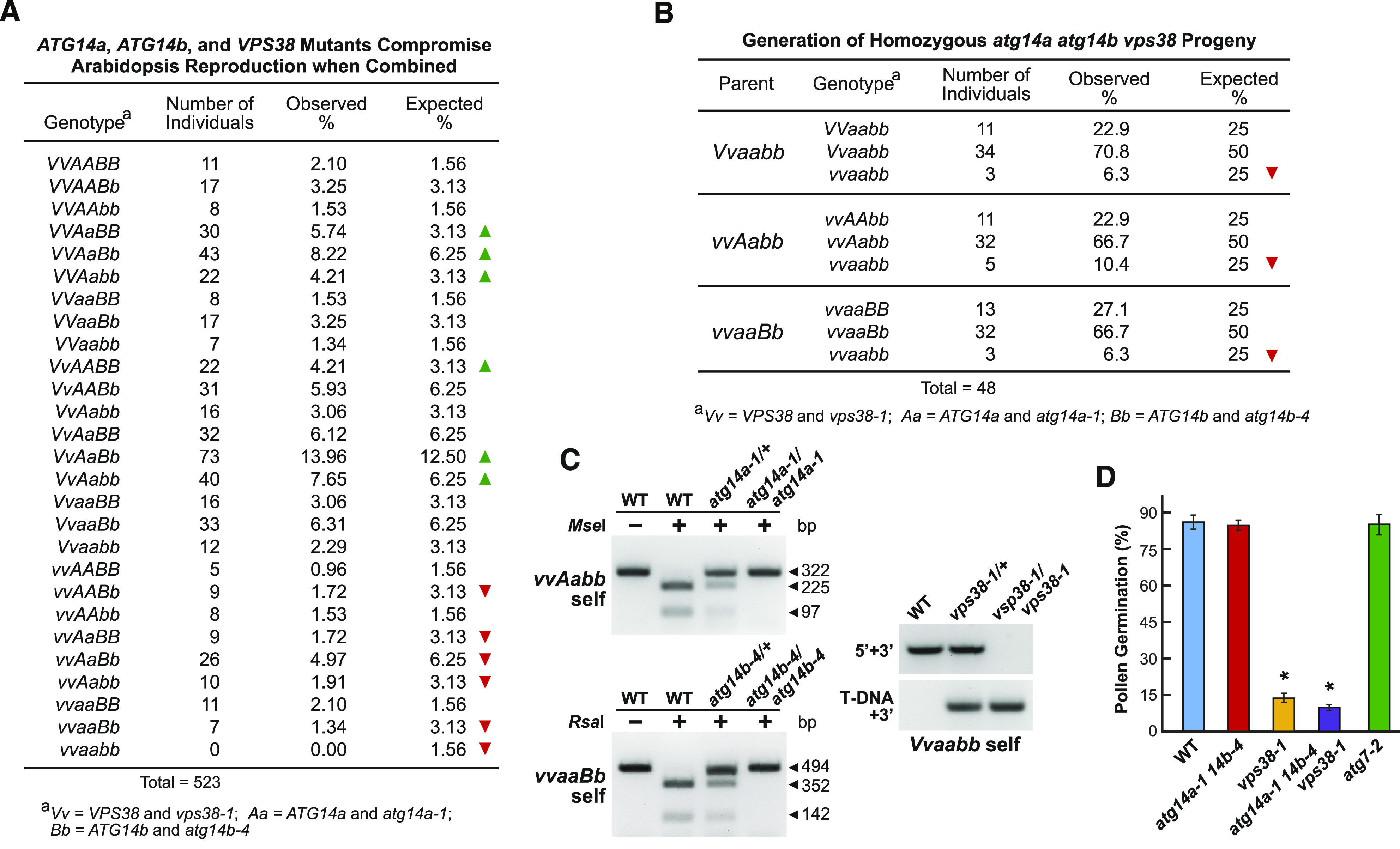
Combined Loss of ATG14a, ATG14b, and VPS38 Compromises Arabidopsis Reproduction.
(A) Homozygous atg14a-1 atg14b-4 vps38-1 triple mutant progenies appear at lower than expected frequencies in selfed heterozygous populations. Shown is the frequency of various genotypes in 523 progenies from a selfed triple heterozygous atg14a-1 (Aa) atg14b-4 (Bb) vps38-1 (Vv) parent. Genotypes were scored by genomic PCR as described in (C). Under- and over-represented genotypes, compared to the expected frequency based on a chi-squared test (P-values < 0.001), are indicated by the red and green arrowheads, respectively.
(B) Viable triple homozygous atg14a-1 atg14b-4 vps38-1 seedlings obtained by selfing of parents harboring various combinations of atg14a-1, atg14b-4, and vps38-1 in either the homozygous or heterozygous states. The Vvaabb, vvAabb, and vvaaBb parents were selected from the F2 generation described in (A) and allowed to self, and their progenies were genotyped as shown in (C). Lower than expected genotypes are indicated by the red arrowheads based on a chi-squared test (P-values < 0.001).
(C) CAPS and RT-PCR analyses confirm the homozygous vps38-1, atg14a-1, and atg14b-4 genotypes of progeny obtained from the three crosses shown in (B). The atg14a-1 and atg14b-4 alleles generated by CRISPR/Cas9 mutagenesis were identified by CAPS analysis, while the vps38-1 T-DNA insertion mutation was identified by genomic PCR (see Figures 2C and 2D, respectively). WT, wild type.
(D) atg14a-1 atg14b-4 vps38-1 triple mutant has depressed pollen germination beyond that seen for the vps38-1 mutant. Pollen grains collected at anthesis from homozygous plants were incubated on germination medium and scored for pollen tube emergence after 10 h. Each bar represents the average (±sd) of three independent analyses, each consisting of >300 pollen grains from 10 anthers. Asterisks indicate significant differences from the wild-type Col-0 (WT) by one-way ANOVA (P-value < 0.01). Images of pollen germination are presented in Supplemental Figure 10C.
As a second test, we screened 96 seedlings derived from a selfed atg14a atg14b vps38 plant that was homozygous for the vps38-1 allele based on the stunted rosette phenotype, and at least heterozygous for the atg14a-1 and atg14b-4 mutations as determined by genotyping. Remarkably, we identified one individual that was homozygous for all three mutations, strongly indicating that generating Arabidopsis plants null for ATG14a, ATG14b, and VPS38 was possible. As a third test to improve our odds, we allowed double homozygous/single heterozygous individuals affecting the three loci (Vvaabb, vvAabb, and vvaaBb, where V = VPS38, A = ATG14a, and B = ATG14b) to self and genotypically examined 48 progenies from each cross, with the calculation that one in four could be triple homozygous. Strikingly, we succeeded in identifying multiple triple homozygous atg14a atg14b vps38 individuals in each population (Figures 7B and 7C). Collectively, their repeated identification demonstrated that the fourth subunit of the VPS34 PI3K is not essential to Arabidopsis.
The actual frequencies for triple homozygous progeny from the third test were significantly lower than expected (6 to 10%; P-value = 0.0034; chi-squared test), suggesting that one or more aspects of reproduction were compromised, but not eliminated. Demonstrably affected was pollen germination (Figures 7D). While atg14a-1 atg14b-4 pollen germinated at high frequency, similar to that seen for the wild-type and atg7-2 plants, pollen from homozygous atg14a-1 atg14b-4 vps38-1 plants germinated poorly, with an even lower frequency than that seen for vps38-1 pollen (Figure 7D; Supplemental Figure 10C; Liu et al., 2018). However, triple mutant pollen had normal viability, as judged by Alexander’s stain, and underwent normal microsporogenesis, as judged by the presence of the tube nucleus and both sperm nuclei in pollen collected at anthesis (Supplemental Figures 10A and 10B).
Arabidopsis Missing Both ATG14 and VPS38 Are Phenotypically Compromised
When grown under nonstressed long-day (LD) conditions, triple homozygous atg14a atg14b vps38 plants displayed an exaggerated phenotype even compared to vps38 plants. The rosette leaves were more stunted and crinkled, and flowering was more strongly delayed (Figures 8A and 8B; Supplemental Figure 11A). The vps38-1 plants flowered ∼3 weeks later than the wild type, while the triple mutant plants flowered ∼4 weeks later. The triple mutants also senesced faster than vps38-1, were more sensitive to Suc deprivation, as judged by their slowed root elongation compared to the atg14a atg14b double and vps38 single mutants, and were more suppressed for hypocotyl elongation when kept in the dark (Supplemental Figure 12). The triple mutants also retained the mild gravitropism insensitivity reported previously for vps38-1 roots (Liu et al., 2018), as evidenced by their enhanced wavy root pattern when grown vertically on plates (Supplemental Figures 12B and 12D).
Figure 8.
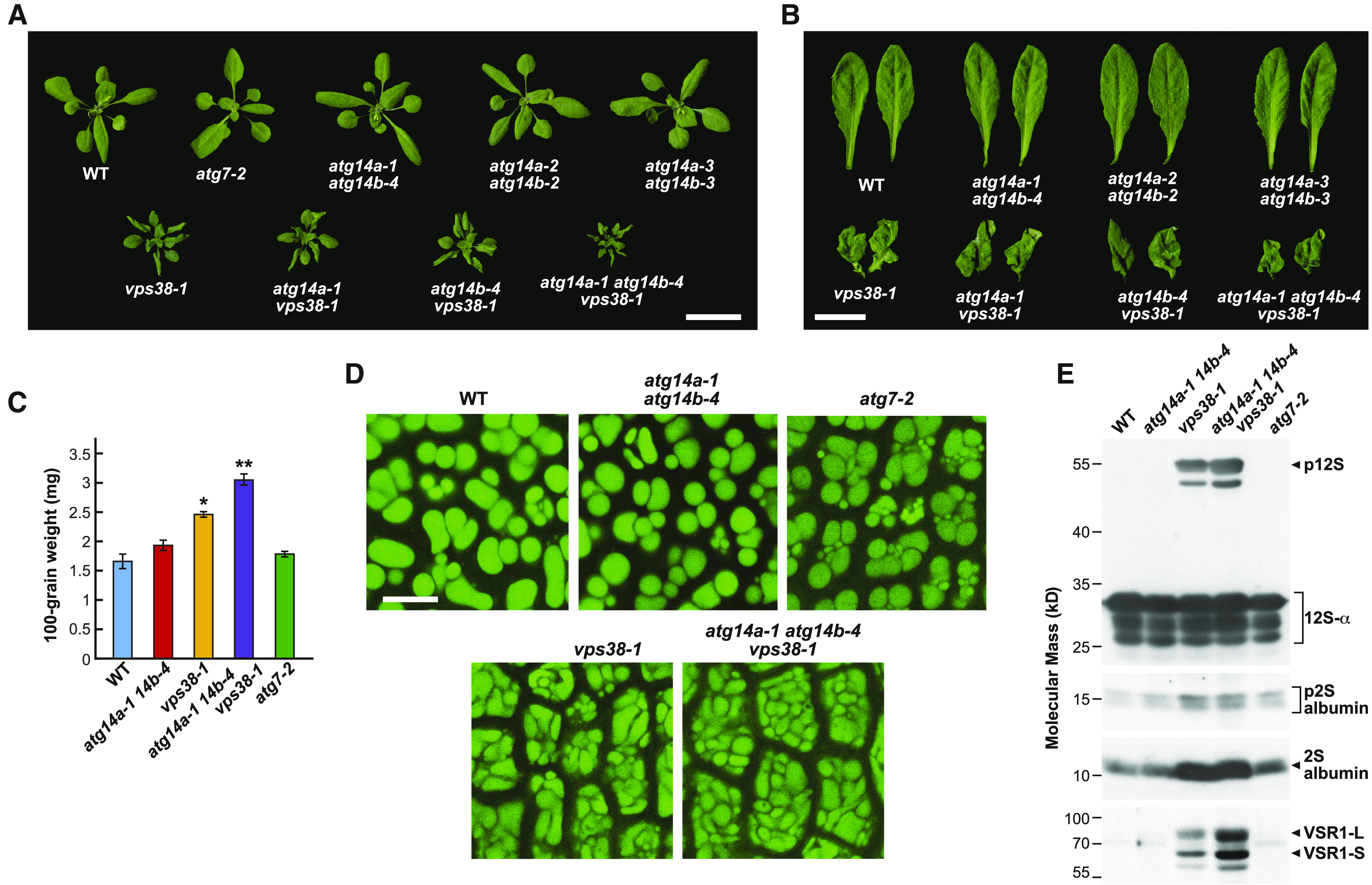
Triple Mutants Missing ATG14a, ATG14b, and VPS38 Are Compromised in Their Development.
(A) Images of 5-week-old plants grown on soil in a LD photoperiod showing the dwarfed rosettes and crinkled leaves of the vps38 single mutant, vps38 atg14a and vps38 atg14b double mutants, and vps38 atg14a atg14b triple mutant plants, compared to the wild-type Col-0 (WT). Bar = 2 cm.
(B) Close-up images of fully expanded leaves from the plants shown in (A). Bar = 1 cm.
(C) atg14a atg14b vps38 triple mutants produce larger seeds. Each bar represents the mean weight (±sd) of 100 seeds. Asterisks indicate significant differences from the wild-type Col-0 (WT) by one-way ANOVA; * and ** represent P-values of <0.05 and <0.01, respectively. Photographs of the seeds are available in Supplemental Figure 11B.
(D) atg14a atg14b vps38 cotyledons accumulate larger cells with irregularly shaped PSVs. Cotyledon PSVs were visualized by autofluorescence in dissected dry seeds using fluorescence confocal microscopy. Bar = 10 μm.
(E) atg14a atg14b vps38 triple mutant seeds hyperaccumulate the 12S globulin and 2S albumin precursors and VSR1. Total proteins from dry seeds of the selfed wild-type Col-0 (WT), atg14a-1 atg14b-4, vps38-1, atg14a-1 atg14b-4 vps38-1, and atg7-2 plants were subjected to immunoblot analysis with anti–12S globulin, anti–2S albumin, and anti-VSR1 antibodies. Brackets/arrowheads locate the p12S precursor of the 12S-α and 12S-β globulins (p12S), and the precursors to the 2S albumins (p2S), along with mature forms of these globulins and albumins. The 80-kD VSR1-L and 60-kD VSR1-S versions of VSR1 (Shimada et al., 2003) are indicated by the arrowheads. The SDS-PAGE profiles of total seed protein from the mutant collection after staining with Coomassie blue are presented in Supplemental Figure 11C.
Surprisingly, seeds from self-crossed atg14a-1 atg14b-4 vps38-1 plants, while not as prevalent, were larger than those from atg14a-1 atg14b-4 and vps38-1 plants and substantially larger than those from the wild type (Figure 8C; Supplemental Figure 11B). Imaging of embryonic cotyledons within dry seeds by autofluorescence revealed that the triple mutant contained cells more tightly packed with protein storage vacuoles (PSVs; Figure 8D). When seeds from the triple mutant combinations were subjected to SDS-PAGE and either stained for total protein or subjected to immunoblot analysis with antibodies against the 12S globulin and 2S albumin seed storage proteins (Figure 8E; Supplemental Figure 11C), we determined that unprocessed storage protein precursors accumulated, indicative of defects in membrane trafficking to PSVs, as seen previously for the vps38 mutants (Liu et al., 2018). Accordingly, the level of VACUOLAR SORTING RECEPTOR1 (VSR1) critical for depositing storage proteins into PSVs (Shimada et al., 2003; Di Sansebastiano et al., 2017) was also strongly elevated in atg14a-1 atg14b-4 vps38-1 seeds; moderately increased in vps38-1 seeds; but barely detected in the wild-type, atg14a-1 atg14b-4, or atg7-2 seeds (Figure 8E). Taken together, the phenotypes of the triple mutant appear additive to those missing ATG14 and VPS38 individually.
The atg14a atg14b vps38 Triple Mutant Has Strongly Dampened Autophagy
To assess how the lack of both ATG14 and VPS38 would impact autophagy, we compared various aspects of the triple mutant relative to the atg14a atg14b and vps38 mutant combinations. As predicted, atg14a-1 atg14b-4 vps38-1 seedlings accumulated more ATG8, ATG1, and the ATG12-ATG5 conjugate compared to the atg14a atg14b and vps38 seedlings (Figure 9A). By contrast, the ATG8-PE adduct formed and accumulated normally in the triple mutant (Figure 9B), indicating that lipidation was unaffected.
Figure 9.
ATG14 Preferentially Impacts Autophagy Compared To VPS38.
(A) atg14a atg14b double and the triple atg14a atg14b vps38 mutants hyperaccumulate ATG8, ATG1, and the ATG12-ATG5 conjugate. Total seedling extracts from the 14-d-old wild-type Col-0 (WT) or homozygous atg14a-1 atg14b-4, vps38-1, atg14a-1 atg14b-4 vps38-1, and atg7-2 seedlings were grown in a LD photoperiod in nitrogen-rich solid MS medium for 7 d and then subjected to immunoblot analysis with anti-ATG1, -ATG5, or -ATG8a antibodies. Free ATG5 and the ATG12-ATG5 adduct are located by the open and filled arrowheads, respectively. The family of ATG8 isoforms is indicated by a solid line. An immunoblot with anti-histone H3 antibodies was included to confirm near equal protein loading.
(B) atg14a atg14b vps38 triple mutant accumulates the ATG8-PE adduct normally. The wild-type Col-0 (WT), atg14a-1 atg14b-4 vps38-1, and atg7-2 seedlings were grown in constant light on nitrogen-rich liquid medium for 7 d and then exposed to nitrogen-deficient medium for 2 d before extraction. Total seedling extracts (TE) were separated into the soluble (S) and membrane (Memb) fractions by centrifugation, subjected to SDS-PAGE in the presence of 6 M urea, and assayed for ATG8-PE by immunoblot analysis. Dashed and solid lines locate free ATG8 and the ATG8-PE adduct, respectively.
(C) ATG14 preferentially contributes to the autophagic transport of GFP-ATG8a compared to VPS38, as seen by the free GFP release assay. Seedlings expressing GFP-ATG8a were grown in constant light in nitrogen-rich MS liquid medium, transferred to either fresh nitrogen-rich MS medium under light, nitrogen-deficient liquid MS medium under light (−N), or Suc-free liquid MS medium in darkness (−C), and then harvested after 16 h. Total seedling extracts were immunoblotted with anti-GFP antibodies, using the anti-histone H3 antibody to confirm near equal protein loading. Open and filled arrowheads locate the GFP-ATG8a fusion and free GFP, respectively.
(D) atg14a atg14b double and the triple atg14a atg14b vps38 mutants accumulate fewer autophagic bodies than the vps38 mutant upon nitrogen or fixed-carbon starvation. The 7-d-old wild-type Col-0 (WT), atg14a-1 atg14b-4 double mutant, vps38-1, and atg14a-1 atg14b-4 vps38-1 triple mutant seedlings expressing GFP-ATG8a were grown in a LD photoperiod on nitrogen-rich solid MS medium, transferred to either nitrogen-deficient liquid MS medium containing 1 µM ConA and incubated in the light for 16 h, or transferred to Suc-free liquid MS medium containing 1 µM ConA and incubated in darkness for 16 h. Root cells were imaged by confocal fluorescence microscopy. A confocal image of GFP-ATG8a atg7-2 roots starved for fixed carbon and treated with ConA is included for comparison. Ag, autophagic body aggregate; Nuc, nucleus; Vac, vacuole. Bar = 20 µm.
(E) Quantification of autophagic body accumulation in roots from the genotypes shown in (D) exposed to nitrogen or fixed-carbon starvation. Each bar represents the mean (±sd) vacuolar density based on the analysis of 25 cells from multiple roots.
When measuring autophagic flux with the GFP-ATG8a reporter, either by the free GFP release assay or by confocal fluorescence microscopy, the triple mutant appeared more compromised relative to the atg14a atg14b and vps38 backgrounds (Figures 9C to 9E). This suppression was seen by the dampened accumulation of free GFP upon either nitrogen or fixed-carbon starvation, and by the reduced accumulation of autophagic bodies relative to those seen in the wild type. Comparisons of the mutant panel indicated that, while ATG14 played a dominant role in autophagy, VPS38 also had an effect. However, the triple mutant did not compromise autophagy as much as in the atg7-2 background (Figure 9E), suggesting that autophagy can proceed slowly without the fourth subunit of the VPS34 PI3K complex.
Loss of ATG14 and VPS38 Suppresses PI3P Accumulation
We hypothesized that the phenotypes associated with the atg14a atg14b and/or vps38 mutants were generated by attenuated PPI synthesis. While prior studies with the vps38-1 mutant reported no effect on PPI levels generally, or PI3P levels specifically (Lee et al., 2018), it was unclear how atg14a atg14b double mutants or the triple mutant would respond. Here, we labeled the wild-type and mutant seedlings with [32P]PO43– and then analyzed their PPI content by thin layer chromatography (TLC). As shown in Figure 10A, we saw little to no change in the profiles of various phospholipids in atg14a atg14b vps38 seedlings versus the wild type, implying that the ATG14/VPS38 subunits of the VPS34 PI3K have little influence over phospholipid metabolism overall.
Figure 10.
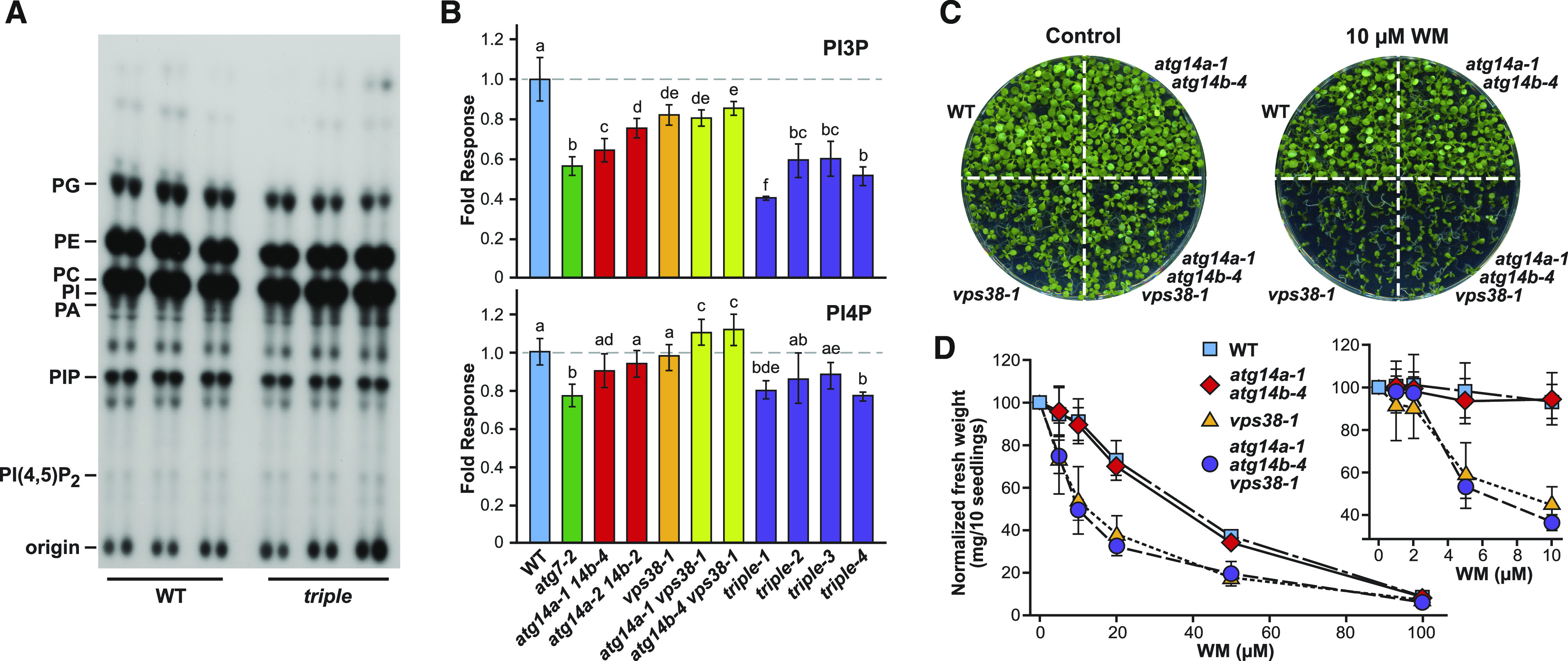
ATG14 and VPS38 Influence but Are Not Essential for PI3P Biosynthesis.
(A) and (B) Quantification of various Arabidopsis phospholipids by TLC. The 5-d-old green seedlings of the indicated genotypes were labeled with [32P]PO43– overnight, after which their lipids were extracted and analyzed by TLC or quantified for radioactivity by scintillation counting following HPLC separation. (A) Autoradiographic image after TLC. PA, phosphatidic acid; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PIP, phosphatidylinositol phosphate. (B) Levels of PI3P and PI4P normalized to those in the wild-type Col-0 (WT; dashed line). The triple mutant was analyzed using four independent seed batches from atg14a-1 atg14b-4 vps38-1 plants generated as in Figure 7B. Each bar represents the average (±sd) of six biological replicates with each replicate containing three seedlings. The letters represent significantly different means according to paired Student’s t test analysis (P value < 0.05). Levels of PI(4,5)P2 are available in Supplemental Figure 13.
(C) and (D) Sensitivity of the panel of atg14a atg14b, vps38, and triple atg14a atg14b vps38 mutants to the VPS34 PI3K inhibitor wortmannin (WM). Seedlings were grown on solid MS medium containing various WM concentrations or DMSO as the control for 10 d. (C) Photograph of the plates containing 10 µM WM or DMSO. (D) Concentration dependence of WM on growth inhibition. Each point represents the mean (±sd) fresh weight of three biological replicates each containing 10 seedlings, normalized to the seedling response without WM.
To specifically measure the levels of PI3P and its relatives phosphatidylinositol 4-phosphate (PI4P) and PI(4,5)P, we excised the PIP spots after TLC, removed their fatty acids by mono-methylamine deacylation, and separated the resulting glycerophosphoinositol phosphates by anion-exchange HPLC. We then quantified the levels of individual species by scintillation counting of the HPLC fractions for 32P (Munnik, 2013). The vps38-1 mutant only modestly reduced the levels of PI3P, with a slightly greater reduction evident for atg14a atg14b lines (Figure 10B). Surprisingly, when we examined the atg14a atg14b vps38 triple mutant, the accumulation of PI3P was still evident but was now dampened ∼50% compared to that seen in the wild type (Figure 10B). In fact, all four different triple mutant seed batches derived from the double homozygous single heterozygous parents described in Figure 7B (Vvaabb, vvAabb, and vvaaBb) accumulated significantly less PI3P than the atg14a atg14b double or the vps38 single mutants. We also observed similar but more modest reductions in PI4P levels in the triple mutant, while on the contrary, PI(4,5)P2 levels increased in all backgrounds that harbored the vps38-1 mutation (Figure 10B; Supplemental Figure 13), indicating that alterations in VPS34 PI3K activity perturbed the levels of related PPIs. Notably, parallel analysis of PPI levels in atg7-2 seedlings revealed that a general block in autophagy also modestly altered the pools of both PI3P, PI4P, and PI(4,5)P2 (Figure 10B). Taken together, the atg14a atg14b vps38 triple mutations reduced but did not eliminate PI3P.
vps38 Mutants Are Hypersensitive to Wortmannin
We speculated that the residual PI3P seen in the triple mutants was derived from the core VPS34 PI3K complex working even in the absence of ATG14 and VPS38. To indirectly test this hypothesis, we examined the sensitivity of the 10-d-old wild-type seedlings and various allelic combinations of atg14a-1, atg14b-4, and vps38-1 to wortmannin, a well-described inhibitor of VPS34 (Takatsuka et al., 2004; Huang et al., 2019). Whereas growth of the wild-type and atg14a-1 atg14b-4 seedlings was relatively immune to concentrations of wortmannin sufficient to suppress autophagy (Shin et al., 2014), growth of the vps38-1 and the triple atg14a-1 atg14b-4 vps38-1 seedlings was severely compromised at concentrations of 5 µM and above (Figures 10C and 10D). Collectively, these results suggested that the viability of vps38 mutants is derived from the VPS34/ATG6/VPS15 core complex working alone to synthesize PI3P, and indicated that PI3P associated with endosomal trafficking is more phenotypically critical to Arabidopsis growth and development than that associated with autophagy.
DISCUSSION
Defining the roles of PI3P in plant endosomal trafficking and autophagy has been challenging due to the lack of viable mutants missing the core components of the VPS34 PI3K complex responsible for synthesizing this phospholipid (Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Xu et al., 2011; Wang et al., 2012). We and others have overcome this hurdle in Arabidopsis by focusing on the fourth subunit encoded by either VPS38 (Lee et al., 2018; Liu et al., 2018; Yuan et al., 2018) or the ATG14a and ATG14b gene pair (this study). Presumably, the domains that distinguish ATG14 and VPS38 (Cys-repeat and BATS-like versus C2 and/or BARA2, respectively; Fan et al., 2011; Rostislavleva et al., 2015) allow them to selectively bind to their appropriate membrane surfaces and/or associated factors. In the absence of useful T-DNA insertion alleles for ATG14a, our genetic analyses were augmented by exploiting CRISPR/Cas9 mutagenesis to generate a suite of strong alleles affecting both isoforms. Particularly helpful was the development of CAPS markers to easily follow the CRISPR/Cas9-generated mutations.
Prior studies implicated PI3P in plant autophagy, based on the efficacy of the inhibitors 3-methyladenine, LY294002, and wortmannin (Takatsuka et al., 2004; Takáč et al., 2013; Zhuang et al., 2013; Shin et al., 2014; Huang et al., 2019), and more indirectly the involvement of PI3P binding Fab1/YOTB/Vac1/EEA1 proteins in this process (Zhuang et al., 2013; Gao et al., 2015). However, given the inherent secondary effects often associated with these inhibitors, the results presented here with ATG14 now directly connect this phosphoinositide to autophagy. Although sequence homology between Arabidopsis ATG14a/b (and other plant versions) and their animal and yeast counterparts is low (<20%), their identities were confirmed by Y2H and BiFC interaction assays with ATG6. Such low identities also exist among the three core subunits of the class III complex and were likely responsible for our failure to rescue any of the yeast mutants (Δatg14, Δvps15, Δvps30 (Δatg6), and Δvps34) with their Arabidopsis counterparts. We note that VPS34 and VPS15 failed to associate with each other or with ATG6 in our Gal4-based Y2H binding assays, but they were previously shown to associate by split-ubiquitin Y2H and/or BiFC assays (Liu et al., 2018), indicating that the interactions within the PI3K complex are highly sensitive to the binding assay chosen.
While prior analyses of Arabidopsis VPS38 revealed its prominent role during endosome trafficking (Lee et al., 2018; Liu et al., 2018; Yuan et al., 2018), our studies here showed that ATG14 has a prominent role during autophagy, consistent with these two subunit isoforms having largely nonredundant functions, like their yeast and mammalian orthologs (Kihara et al., 2001; Itakura et al., 2008). Such distinctions can be seen by comparing the phenotypes of homozygous atg14a atg14b and vps38 plants and by their strengths in perturbing autophagy, and were also implied by comparing the expression patterns of each isoform. Especially evident was the strongly compromised physiology of vps38 plants, which included deeply crinkled leaves, and defects in vascular tissue development, endomembrane morphology, gravitropism, pollen germination, and seed development as it relates to storage protein trafficking to PSVs (Lee et al., 2018; Liu et al., 2018; Yuan et al., 2018; this study). By contrast, atg14a atg14b plants were relatively normal phenotypically, except for strongly attenuated autophagy and the associated hypersensitivity to nitrogen and fixed-carbon starvation and premature rosette senescence. Whereas the collection of atg14a atg14b mutants effectively suppressed delivery of autophagic cargo to vacuoles (as seen by the free GFP release assay and confocal microscopy using the GFP-ATG8a reporter), in agreement with this isoform being the primary effector of such recycling, the process was also more subtly compromised in vps38 mutants, which was especially noticeable during nitrogen starvation. The reason behind this response to nitrogen starvation is unknown but may reflect additional roles for VPS38 and PI3P in plants under nitrogen stress.
From the analysis of the GFP-ATG14b reporter, it appears that ATG14 is also an autophagy substrate, similar to other regulatory components of autophagy such as the ATG1, ATG11, and ATG13 subunits of the ATG1 kinase complex (Suttangkakul et al., 2011; Li et al., 2014). Whereas the reporter was stable when expressed in the atg7-2 background (as judged by the free GFP release assay), its GFP moiety was released in the atg14a-1 atg14b-4 background, consistent with its delivery to vacuoles by the autophagic system. The fact that this release was seen in nonstressed plants indicates that GFP-ATG14b constitutively undergoes autophagy. This delivery was confirmed by confocal fluorescence microscopy, which showed the appearance of GFP fluorescence within vacuolar puncta decorated with mCherry-ATG8a by a process also requiring ATG7. It should be noted that expression of GFP alone in Arabidopsis, or of a truncated des-Gly version of GFP-ATG8 blocked in lipidation, does not lead to fluorescence visualization of autophagic bodies in vacuoles (Yoshimoto et al., 2004; Thompson et al., 2005; Marshall et al., 2015), thus discounting the possibility that the freed GFP became an autophagy substrate after cytoplasmic cleavage.
Presumably, the autophagic breakdown of the GFP-ATG14b reporter (and possibly the rest of the VPS34 PI3K complex), is driven by its association with developing autophagic membranes, which might trap the complex before enclosure. In fact, all three core subunits of the class III kinase complex in mammals have likely ATG8-interacting motifs (Birgisdottir et al., 2019) that could indirectly tether ATG14 to autophagic membranes before enclosure, while the Cys-repeat and BATS domains in ATG14 might directly promote membrane association (Fan et al., 2011).
To test whether simultaneous loss of both ATG14 and VPS38 would phenocopy loss of the essential core subunits of the class III PI3K complex (ATG6, VPS15, and VPS34; Fujiki et al., 2007; Qin et al., 2007; Harrison-Lowe and Olsen, 2008; Xu et al., 2011; Wang et al., 2012), we attempted to generate triple homozygous atg14a atg14b vps38 plants. While initial screens of progeny generated by selfing of triple heterozygous plants failed, despite the analysis of a population predicted to be sufficient (>500 progeny), we eventually succeeded by using various combinations of double homozygous/single heterozygous parents in self-crosses. Even with these populations, our success in finding triple homozygous progeny was well below expectations. We presume that this low success rate was at least partially explained by the severely attenuated germination of atg14a atg14b vps38 pollen, mainly caused by the absence of VPS38 impacting PI3P levels (Liu et al., 2018; this study). However, we note that phosphatidylinositol 3,5-bisphosphate, which is synthesized from PI3P through the FATTY ACID BIOSYNTHESIS1 (FAB1), FAB2, and VACUOLE MORPHOLOGY AND INHERITANCE MUTANT14 (VAC14) factors, has also been connected to proper pollen development (Whitley et al., 2009; Zhang et al., 2018). Although the consequences of fab1/2 and vac14 mutants on pollen appear distinct from those seen here for atg14a atg14b vps38 mutants, they do raise the possibility that the defects in pollen germination seen here actually reflect diminished phosphatidylinositol 3,5-bisphosphate levels.
Owing to the challenges in isolating null mutants in the core PI3K subunits, our analyses of homozygous atg14a atg14b vps38 Arabidopsis provide a compelling genetic demonstration of PI3P importance in plants. Notably, triple homozygous plants had a collection of abnormal phenotypes that appeared to combine the defects seen for atg14a atg14b and vps38 lines separately, again consistent with the conclusion that the two versions of the VPS34 PI3K complex mainly work nonredundantly. However, some overlap in functions was apparent by the increased effects of the triple mutant on phenotypes associated with either ATG14 or VPS38. These included accentuated defects in leaf morphology, pollen germination, seed size, and seed storage protein processing, which appear associated with VPS38, and accentuated defects in autophagy, which appear associated with ATG14. Interestingly, neither the atg14a atg14b mutants nor the triple mutant disturbed assembly of the ATG12-ATG5 and ATG8-PE adducts, suggesting that PI3P acts somewhere after decoration of the phagophore with ATG8-PE but before fusion of autophagosomes with the vacuole. Wherever PI3P acts, the suppressed autophagy seen for atg14a atg14b backgrounds confirms that autophagy vesicle dynamics requires both ATG8-PE and PI3P, presumably through autophagic adaptors that recognize both membrane surface signatures (e.g., the SH3 domain-containing protein2 [SH3P2]; Zhuang et al., 2013).
From quantitative analysis of various PPIs, we were surprised to see that the levels of total PPIs generally, and PI3P specifically, were not strongly compromised in the homozygous atg14a atg14b vps38 plants. At most, we saw only an ∼50% reduction in PI3P upon analysis of four different seed lines, along with a modest drop in PI4P and a slight increase in PI(4,5)P2 levels. The most parsimonious explanation is that ATG14 and VPS38 promote, but are not essential for, the activity of the VPS34 PI3K, possibly by helping tether the complex to appropriate membrane surfaces. The residual PI3P levels seen in atg14a atg14b vps38 plants may then reflect the ATG6/VPS15/VPS34 core acting alone but in a less directed fashion. The phenotypic hypersensitivity of the vps38 backgrounds to low concentrations of the VPS34-specific inhibitor wortmannin supports this notion, which we presume reflects suppression of residual VPS34 activity focused toward endosomal trafficking. That growth of the atg14a atg14b mutants was not appreciably compromised by wortmannin was consistent with the relatively normal phenotype of Arabidopsis autophagy-null mutants under nonstressed conditions (Supplemental Figure 6; Doelling et al., 2002: Thompson et al., 2005). However, we cannot discount the remote possibility that plants either use a novel PI3K type that is unrelated to complexes I and II typical of eukaryotic class III PI3Ks, or have additional regulators for the VPS34 PI3K complex beyond ATG14/VPS38 (e.g., mammalian Rubicon, for which a possible Arabidopsis homolog exists; Matsunaga et al., 2009; Heucken and Ivanov, 2018).
Why PI(4,5)P2 levels increased in the vps38 and atg14a atg14b vps38 backgrounds is currently unclear. In two simple scenarios, this increase might have been induced by a delay in Arabidopsis seeding development in the absence of PI3P, or it might represent an attempt by the mutants to compensate for reduced PI3P levels in an effort to maintain normal endomembrane dynamics and/or autophagy. Of interest to the latter scenario, Tan et al. (2016) recently connected PI(4,5)P2 to autophagy in mammalian cells through the ability of the ATG14 BATS domain to bind this PPI, which in turn promoted class III PI3K assembly. Alternatively, increased PI(4,5)P2 levels may reflect the accumulation of intermediates of membrane trafficking decorated with this lipid that are unable to reach their final destinations in the absence of PI3P. In any event, our assembly of a viable suite of Arabidopsis mutants missing various combinations of ATG14a, ATG14b, and VPS38 now permits the study of PI3P across the entire lifespan of Arabidopsis. When combined with various endomembrane markers and VPS34-specific PI3K inhibitors such as wortmannin, 3-methyladenine, and LY249002 (Takatsuka et al., 2004; Takáč et al., 2013; Zhuang et al., 2013; Shin et al., 2014; Huang et al., 2019), understanding the precise roles of this PPI in endomembrane dynamics and autophagy should now be possible.
METHODS
Sequence and Phylogenetic Analyses of ATG14
We identified possible ATG14 orthologs in other plant species by reciprocal Basic Local Alignment Search Tool for Protein (BLASTP) and Nucleotide (TBLASTN) searches performed on the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST/blast.cgi), using the coding regions of Arabidopsis (Arabidopsis thaliana) accession Col-0 ATG14a (At1g77890) and ATG14b (At4g08540) as queries (Lee et al., 2018; Liu et al., 2018). For phylogenetic analyses, the ATG14 protein sequences were aligned with EMBL-ESI Multiple Alignment using Fast Fourier Transform version 7, under default settings with yeast (Saccharomyces cerevisiae), fruit fly (Drosophila melanogaster), and various plant VPS38 sequences serving as the outgroup (Supplemental File 1). The alignment was used to generate an unweighted pair group method with arithmetic mean tree within MEGA7 (Kumar et al., 2016) with 1000 bootstrap replications, which we then visualized in TreeView (Supplemental File 2; Page, 1996). Identical and similar amino acids were displayed by BoxShade (http://www.embnet.vital-it.ch/software/BOX_form.html), and protein domains were predicted by PFAM (http://pfam.xfam.org).
Gene Expression Analysis
Transcript abundances were assessed in the TraVA database (http://travadb.org/; Klepikova et al., 2016) and visualized as a heatmap in R version 3.3.2 using hierarchical clustering. We normalized all read counts generated from RNA sequencing to values ranging from 0 to 1 by the median-of-ratio method in DESeq/DESeq2 (Love et al., 2014) and divided by maximum expression for each gene. Coexpression analyses used the NetworkDrawer tool in ATTED-II version 10 (https://atted.jp/top_draw/), using the ATG14a, ATG14b, VPS38, ATG6, VPS15, and VPS34 genes as queries (Obayashi et al., 2018). Parameters under the ath-u.c1-0 platform were as follows: Disp. type “Cytoscape,” Coex option “Add many genes,” and PPI option “Draw 2-path PPIs.” For RT-PCR analysis, total RNA was extracted using the plant RNeasy mini kit (QIAGEN) from 14-d-old seedlings grown on Murashige and Skoog (MS) medium. mRNAs were converted to cDNA via the SuperScript III first-strand synthesis system (Invitrogen) with oligo(dT)20 primers, before using them as templates for RT-PCR with gene-specific primers. Details of all oligonucleotide primers used are given in Supplemental Data Set 1.
Protein–Protein Interaction Assays
Standard Gal4-based Y2H assays relied on HIS3 expression, as described by Gingerich et al. (2007), with slight modifications. cDNAs encoding the full-length sequence of ATG14a, ATG14b, ATG6/VPS30, VPS15, and VPS34 were generated by PCR with the Fusion DNA polymerase (Thermo Fisher Scientific) using reverse-transcribed total seedling RNA isolated from the wild-type Col-0, before subcloning into the pDONR221 entry plasmid via Gateway BP clonase II reactions (Invitrogen). These fragments were recombined in frame into the pDEST22 plasmid harboring the Gal4 activation domain and the pDEST32 plasmid harboring the Gal4 DNA binding domain via Gateway LR clonase II reactions (Invitrogen). The plasmids were cotransformed pairwise into yeast strain MaV203, and the colonies were assayed for positive protein–protein interactions by 2-d growth at 30°C on synthetic dropout medium lacking His, Leu, and Trp and containing 25 mM 3-AT, using synthetic dropout medium lacking Leu and Trp as the growth control, and unrecombined pDEST22 and pDEST32 as negative interaction controls.
For Y2H assays using the split-ubiquitin Protein A/LexA/VP16 (PLV) system, positive interactions are based on association between the N- and C-terminal fragments of ubiquitin (NubG and Cub, respectively), which allows proteolytic release and nuclear import of the chimeric PLV transcription factor to then activate HIS3 and ADE2 expression (Stagljar et al., 1998; Liu et al., 2018). We introduced the relevant cDNAs into the pMETYCgate/pNXgate32/pXNgate22-based plasmid system by Gateway recombination as described above. Candidate baits were translationally fused to the N terminus of Cub-PLV and transformed into yeast strain THY.AP4 (mating type a), while candidate preys were fused to either the N terminus or C terminus of NubG and transformed into yeast strain THY.AP5 (mating type α). After mating, the diploid THY.AP4 THY.AP5 cells were plated on synthetic dropout medium lacking His, Leu, Trp, Ura, and Ade to test for protein–protein interactions by growth. The NubG and Nub-wild type empty vectors were used as negative and positive controls, respectively (Stagljar et al., 1998).
For BiFC, we recombined the entry plasmids described above into the pSITE-nEYFP-C1 vector (ABRC catalog no. CD3-1648) encoding the N-terminal fragment (residues 1 to 174) of YFP (NY) and the pSITE-cEYFP-C1 vector (ABRC catalog no. CD3-1649) encoding the C-terminal fragment (residues 175 to 239) of YFP (CY; Martin et al., 2009; Li et al., 2014). Pairwise plasmid combinations bearing these N-terminal NY/CY tags were transiently coexpressed in N. benthamiana leaves via Agrobacterium (Agrobacterium tumefaciens)–mediated infiltration as previously described by Grimsley et al. (1986) and Marshall et al. (2015). The unrecombined pSITE-nEYFP-C1 and pSITE-cEYFP-C1 vectors were used as negative controls. YFP fluorescence was assayed after 24 to 48 h by confocal fluorescence microscopy of the infiltrated leaf sectors.
Yeast Complementation by Free GFP Release and Pho8Δ60 Assays
We PCR amplified the coding sequences for the Arabidopsis or yeast ATG14, VPS15, ATG6/VPS30, and VPS34 genes from cDNAs generated from total RNA extracted from Col-0 seedlings or BY4741 yeast cells using the SuperScript III first-strand synthesis system as described above. Where necessary, sequences for an N-terminal HA tag were included in the relevant PCR amplification primer. The resulting PCR products were recombined into pDONR221 via Gateway BP clonase II reactions as described above, sequence confirmed, and then recombined into the pAG424-GPD-ccdB or pAG424-GPD-ccdB-HA vectors (Addgene product nos. 14152 and 14248, respectively) via Gateway LR clonase II reactions. The resulting plasmids were introduced into the yeast Δatg14, Δvps15, Δvps30, and Δvps34 strains (Dharmacon) expressing yeast GFP-Atg8 or Pho8Δ60, and cultured on synthetic dropout medium lacking Trp. Details of all yeast strains and oligonucleotide primers used are given in Supplemental Data Sets 1 and 2.
For nitrogen starvation experiments, we grew 15-mL liquid cultures in yeast-peptone-dextrose-adenine (YPDA) medium overnight at 30°C with vigorous shaking, diluted the cultures to an OD600 of 0.1 in 15 mL of YPDA medium, grew them for an additional 2 to 3 h until an OD600 of ∼0.5 was reached, and then switched to synthetic dropout medium lacking nitrogen (0.17% yeast nitrogen base without amino acids and ammonium sulfate and containing 2% Glc), followed by continued incubation at 30°C (Marshall et al., 2016). Cell aliquots were collected at the indicated times and immediately frozen in liquid nitrogen. Total protein extracts, obtained by homogenization in 500 μL of lysis buffer (0.2 N NaOH and 1% 2-mercaptoethanol), were precipitated with 50% (w/v) trichloroacetic acid (TCA) and resolubilized in 150 μL of hot 2× SDS-PAGE sample buffer (80 mM Tris-HCl, pH 6.8, 10% [v/v] glycerol, 4% [w/v] SDS, 4% [v/v] 2-mercaptoethanol, and 0.002% [w/v] bromophenol blue). Proteins were subjected to immunoblot analysis as described by Marshall et al. (2016), using anti-GFP, anti-HA, or anti-histone H3 antibodies (see Immunoblot for details) combined with goat anti-mouse or goat anti-rabbit horseradish peroxidase conjugates and the SuperSignal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific).
The Pho8Δ60 activity assays measuring autophagic flux were performed in yeast essentially as described by Noda and Klionsky (2008), with minor modifications by Marshall et al. (2016). We grew strains TN124 or RSM353-365 as 250-mL YPDA cultures, subjected them to nitrogen starvation for 8 h, and collected aliquots corresponding to 5.0 OD units at the indicated time points. Cell pellets were resuspended in 500 μL of lysis buffer (20 mM piperazine-N,Nʹ-bis-2-ethanesulfonic acid-KOH, pH 8.5, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 μM ZnSO4, 0.5% [v/v] Triton X‐100, and 1 mM phenylmethylsulphonyl fluoride), lysed by vigorous mixing in the presence of acid-washed glass beads, and clarified by centrifugation at 20,000g for 20 min at 4°C. Equal amounts of protein (20 μg) were then assayed spectrophotometrically for alkaline phosphatase activity using p-nitrophenyl phosphate as the substrate.
Plant Materials and Growth Conditions
The atg14b-1 (SAIL_1207_H10) and atg14b-2 (SALK_145203) T-DNA insertion mutants in the Arabidopsis Col-0 accession were obtained from the ABRC. We generated atg14a and additional atg14b mutants with an egg cell–specific promoter-controlled CRISPR/Cas9 genome editing system (Xing et al., 2014; Wang et al., 2015), using the wild type Col-0 as the recipient. Briefly, appropriate Cas9-targeting sequences in ATG14a and ATG14b (Supplemental Data Set 1), as located by the CRISPR-PLANT website (https://www.genome.arizona.edu/crispr/index.html; Xie et al., 2014) within the second exon of ATG14a and the first exon of ATG14b, were fused into the CRISPR cassette by PCR using pCBC-DT1T2 as the template. The PCR products were introduced into the pHEE401E plasmid by Golden Gate cloning with the restriction enzyme BsaI (New England Biolabs), resulting in pHEE401E-ATG14a/14b that was then introduced into the recipient plants by the Agrobacterium-mediated floral-dip method, with the resulting transformants selected by hygromycin resistance. We amplified DNA from resistant plants by genomic PCR using primers flanking each target site (Supplemental Data Set 1) for sequencing to define the nature of the mutations. Homozygous plants bearing the individual mutations in the progeny were selected from selfed T1 plants.
The genotypes of the T-DNA–bearing atg14b mutants were determined by genomic PCR using a left border T-DNA primer in combination with a primer 3′ to the T-DNA insertion site (Supplemental Data Set 1; Liu et al., 2018). Genotypes of the CRISPR/Cas9-derived atg14a and atg14b mutants were determined by genomic PCR of seedlings using primers that spanned the Cas9-targeting sequence (Supplemental Data Set 1). The presence of mutations around the targeting sequences was deduced by CAPS (Konieczny and Ausubel, 1993), using the loss of MseI and RsaI restriction sites in the atg14a and atg14b PCR products, respectively, as assayed by agarose gel electrophoresis of digested PCR fragments.
The vps38-1 mutant was previously described by Lee et al. (2018) and Liu et al. (2018). The various double and triple mutant combinations of atg14a, atg14b, and vps38-1 were identified by genomic PCR and/or the phenotypes of F2 progeny derived from crosses between homozygous atg14a atg14b and vps38-1 parents. Homozygous atg1abct (Huang et al., 2019), atg2-1 (Inoue et al., 2006), atg5-1 (Thompson et al., 2005), atg7-2 (Chung et al., 2010), atg9-4 (Shin et al., 2014), atg11-1 (Li et al., 2014), and atg13a-2 atg13b-2 (Suttangkakul et al., 2011) mutants, and plants harboring GFP-ATG8a expressed from the cauliflower mosaic virus 35S promoter (Thompson et al., 2005), all in the Col-0 background, were described previously. We introgressed the GFP-ATG8a transgene into the various mutant lines by genetic crosses (Liu et al., 2018; this study); homozygous offspring were then selected from the F3 backcross generations.
For atg14 complementation studies, we constructed a GFP-ATG14b transgene under the control of the 35S promoter from a cDNA encompassing the full-length coding region, which was introduced into the pMDC43 plasmid (Curtis and Grossniklaus, 2003) between the 35S promoter and the NOS 3′ untranslated region sequences using Gateway LR clonase II reactions. The resulting 35Spro:GFP-ATG14b construction was introduced into the homozygous atg14a-1 atg14b-4 and atg7-2 backgrounds by the floral-dip method, followed by selfing to obtain fully homozygous lines.
Plant Phenotypic Assays
We surface sterilized seeds with the vapor-phase method before sowing seeds in culture dishes containing full-strength MS medium supplemented with 2% (w/v) Suc, 1.4% (w/v) agar, and 2 mM MES-KOH, pH 5.7. Seeds were stratified for 3 to 4 d and then exposed to continuous white light (T5 54W 84IHO-FLOUR lamps, 150 µmol m−2 s−1) for 12 h at 4°C to induce uniform germination. For nitrogen starvation experiments, seedlings were grown for 7 d at 23°C in liquid MS medium under continuous white light irradiation (Suttangkakul et al., 2011). We then substituted the medium to MS medium containing or lacking nitrogen for an additional 14 d. For fixed-carbon starvation experiments, seedlings were grown for 14 d under white light in a LD (16-h-light/8-h-dark) photoperiod on solid MS medium lacking Suc and containing 1% agar before transfer to continuous darkness for the indicated number of days and then allowed to recover under LD lighting for 12 d (Chung et al., 2010). Percent survival rate was determined by the extent of regreening of chlorotic seedlings after the indicated times. For senescence assays, we grew plants in soil under a short-day photoperiod (8-h-light/16-h-dark) at 23°C for 10 weeks.
To assay wortmannin sensitivity, we stratified seeds at 4°C for 5 d before germination on solid GM medium (3.2 g/L Gamborg’s B5 basal medium with minimal organics, 1% [w/v] Suc, 0.05% [w/v] MES-KOH, pH 5.7, and 0.7% [w/v] agar) supplemented with the indicated concentration of wortmannin (catalog no. W1628; Sigma-Aldrich) or equivalent volumes of DMSO as the control. After 10 d of growth at 21 to 23°C under a LD photoperiod, we measured the fresh weight of 10 seedlings.
Immunoblot and Immunoprecipitation Assays
We detected ATG1, ATG8, ATG5, the ATG12-ATG5 conjugate, GFP-ATG8a, and ubiquitin conjugates by immunoblot assays using total seedling extracts prepared from 100 mg of seedlings homogenized in 200 μL of 2× SDS-PAGE sample buffer (100 mM Tris-HCl, pH 6.8, 4% [w/v] SDS, 0.2% [w/v] bromophenol blue, and 20% [v/v] glycerol), followed by heating the samples to 95°C for 2 min and then clarifying the extracts by brief centrifugation at 14,000g for 10 min at room temperature. Antibodies against plant ubiquitin (van Nocker and Vierstra, 1993) and Arabidopsis ATG1a (Suttangkakul et al., 2011), ATG5, and ATG8a (Thompson et al., 2005) were as described. Anti-histone H3 antibodies were obtained from Abcam (product no. AB1791; 1:10,000 dilution), and monoclonal antibodies against HA and GFP were obtained from Sigma-Aldrich (product nos. H6908 [1:3000 dilution] and 11814460001 [1:5000 dilution], respectively). Goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase or alkaline phosphatase and goat anti-mouse secondary antibodies conjugated to horseradish peroxidase were purchased from SeraCare (product nos. 5220-0336, 5220-0353, and 5220-0341, respectively). Blots were developed using either the SuperSignal West Pico Chemiluminescent Substrate or the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific), according to the manufacturer’s instructions.
ATG8a lipidation was monitored according to Chung et al. (2010). Membrane fractions were collected from total seedling extracts by centrifugation at 100,000g for 10 min at 4°C. The pellets were solubilized in 0.5% (v/v) Triton X-100 in TNPI buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 10 mM iodoacetamide) and clarified, and the resulting supernatants were incubated for 1 h at 37°C with or without the addition of Streptomyces chromofuscus phospholipase D (product no. BML-SE301; Enzo Life Sciences). Samples were subjected to SDS-PAGE in the presence of 6 M urea and immunoblotted with anti-ATG8a antibodies. For the GFP-ATG8a cleavage assays (Chung et al., 2010; Suttangkakul et al., 2011; Huang et al., 2019), we grew seedlings expressing GFP-ATG8a in continuous white light in nitrogen-containing liquid MS medium before transfer to MS medium lacking nitrogen, or to MS medium lacking Suc and placed in darkness. Immunoblots were performed on total seedling extracts with anti-GFP antibodies as described above.
For the immunoprecipitation of GFP-ATG14b, we grew seedlings of the indicated genotype (Col-0 [WT], GFP-ATG14b atg14a-1 atg14b-4, or GFP-ATG14b atg7-2) for 7 d in 50 mL of liquid GM medium at 21 to 23°C in constant white light with gentle shaking, with ∼100 mg of dry seeds used per culture, resulting in ∼5 g of fresh weight tissue. Frozen tissue was ground to a fine powder in liquid nitrogen, and proteins were then extracted on ice for 20 min with 1.5 volumes of extraction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% [v/v] glycerol, 5 mM DTT, 2 mM phenylmethylsulfonyl fluoride, 0.1% [v/v] Triton X-100, and 1× plant protease inhibitor cocktail). Extracts were filtered through two layers of Miracloth and clarified at 30,000g for 20 min at 4°C. The resulting supernatants were immediately applied three times at 4°C over a 12-mL PolyPrep chromatography column containing 100 μL (equal to a 50-μL bead volume) of GFP-Trap_A beads (product no. gta-20; ChromoTek) pre-equilibrated in extraction buffer. The column was washed five times with wash buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% [v/v] glycerol, and 2 mM DTT), and the remaining bound proteins were eluted with 100 μL of 200 mM Gly-HCl, pH 2.5, and immediately neutralized with 20 μL of 1 M Tris-HCl, pH 8.0. The input and elution samples were analyzed by SDS-PAGE followed by immunoblotting with anti-GFP or anti-histone H3 antibodies. The input sample was concentrated 200-fold by precipitation with 50% (w/v) TCA prior to analysis. The GFP-ATG14b immunoprecipitates and total cell lysates were trypsinized prior to HPLC-coupled tandem mass spectrometricanalysis as described by Marshall et al. (2019).
Seed protein extraction and immunoblot assays were performed as previously described by Shimada et al. (2003) and Reguera et al. (2015). Briefly, we homogenized dry seeds (60 each) in 150 μL of 2× SDS-PAGE sample buffer and clarified them by centrifugation at 14,000g for 10 min at 4°C. The supernatants were subjected to SDS-PAGE, and either stained for protein with Coomassie Brilliant Blue R-250 or immunoblotted with antibodies against either Arabidopsis 2S albumin, 12S globulin α-subunit, or VSR1 (Reguera et al., 2015).
Confocal Fluorescence Microscopy and Image Analysis
Fluorescent cell images were collected with a Nikon A1Si laser scanning confocal microscope, using 358-nm light combined with a 461-nm filter for 4′,6-diamidino-2-phenylindole, 488-nm light combined with 500 to 550-nm filters for GFP, YFP, and seed autofluorescence, and 543-nm light combined with a 565 to 615-nm filter for mCherry. The images were processed using the NIS Elements software (Nikon) and converted to TIFF files for use in the figures. We quantified the accumulation of autophagic bodies using ImageJ (www.imagej.nih.gov/ij) by counting their numbers in 100-µm2 areas of the central vacuole of multiple cells from multiple roots (25 cells total) after 4, 8, or 16 h of nutrient starvation.
Pollen Viability Assays
For pollen viability tests, we incubated anthers dissected from freshly opened flowers for several hours with Alexander’s stain (1 mL of 1% [w/v] malachite green in 95% [v/v] ethanol, 5 mL of 1% [w/v] acid fuchsin in water, 0.5 mL of 1% [w/v] orange G in water, 10 mL of ethanol, 25 mL of glycerol, 5 g of phenol, 5 g of chloral hydrate, and 4 mL of acetic acid, all dissolved in 50 mL of water; Alexander, 1969 ). Pollen germination was examined after a 10-h incubation of fresh pollen in medium containing 0.01% (v/v) boric acid, 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 18% (w/v) Suc, and 0.5% (v/v) agar at pH 7.0 (Lee et al., 2008b). We identified pollen nuclei by fluorescence microscopic examination of pollen grains under UV light after staining with 4′,6-diamidino-2-phenylindole.
Phospholipid Analysis
Five-day-old, light-grown seedlings of the indicated genotypes were labeled overnight using [32P]PO43– (32Pi; PerkinElmer) as detailed by Munnik (2013), using 5 to 10 μCi per sample (PerkinElmer). The next day, lipids were extracted and separated by TLC using an alkaline solvent, and the TLC plates were exposed to x-ray film for visualization and quantification by phosphorimaging (Typhoon FLA 7000 laser scanner; GE Healthcare).
To distinguish PI3P from PI4P, we scraped the phosphatidylinositol monophosphate (PIP) spots from the TLC plates and subjected them to deacetylation by mono-methylamine (product no. 426466; Sigma-Aldrich) as described by Munnik (2013). The resulting glycerophosphoinositol phosphates were desalted through cation exchange (AG 50W-X8, 200 to 400 mesh; Bio-Rad) and separated by HPLC using a Partisil 10 strong anion-exchange column (4.5 × 250 mm; Sigma-Aldrich) and a gradient of NaH2PO4, pH 3.7. Fractions (collected every 20 s) were quantified for radioactivity by liquid scintillation counting. The final results for different PIP isomers were obtained by quantifying the PIP spots from the TLC by phosphorimaging and then normalizing the results with respect to the ratios of PI3P to PI4P obtained by HPLC analysis.
Statistical Analysis
Data reported in this study are means (±sd) of at least three independent experiments, each consisting of three technical replicates, unless otherwise noted. The P-values were determined by one-way analysis of variance (ANOVA) or two-tailed Student’s t tests (Supplemental Data Set 3). The outcomes of the genetic crosses were evaluated by the chi-squared test (P < 0.05).
Accession Numbers
DNA and derived protein sequence data from this article are available in the Arabidopsis Information Resource database under the following accession numbers: ATG14a (At1g77890); ATG14b (At4g08540); ATG6 (At3g61710); VPS15 (At4g29380); VPS34 (At1g60490); VPS38 (At2g32760).
Supplemental Data
Supplemental Figure 1. Phylogeny, expression patterns, and co-expression analysis of Arabidopsis ATG14 and VPS38.
Supplemental Figure 2. Amino acid sequence alignment of ATG14-related proteins in various plant, animal, and fungal species.
Supplemental Figure 3. Arabidopsis components of the VPS34 PI3K complex-I failed to rescue their corresponding yeast mutants.
Supplemental Figure 4. BiFC assays with ATG14a, ATG14b, and ATG6 testing various control combinations.
Supplemental Figure 5. DNA sequence analysis of the atg14a and atg14b mutants generated by CRISPR/Cas9 mutagenesis.
Supplemental Figure 6. Growth of atg14a atg14b seedlings on nutrient-rich and nitrogen-deficient media.
Supplemental Figure 7. atg14a atg14b double mutants accumulate the ATG12-ATG5 and ATG8-PE adducts normally but have elevated levels of ATG8 and ATG1.
Supplemental Figure 8. Genotypic analysis of Arabidopsis expressing the GFP-ATG14b reporter.
Supplemental Figure 9. The GFP-ATG14b reporter only partially rescues the fixed-carbon starvation hypersensitivity of atg14a-1 atg14b-4 seedlings.
Supplemental Figure 10. Triple atg14a atg14b vps38 pollen mature normally but have dampened germination.
Supplemental Figure 11. Triple mutants missing ATG14a, ATG14b, and VPS38 have compromised seed and plant development.
Supplemental Figure 12. Triple atg14a atg14b vps38 plants show accelerated senescence and hypersensitivity to nitrogen and fixed-carbon starvation.
Supplemental Figure 13. VPS38 influences the levels of PI(4,5)P2.
Supplemental Data Set 1. Oligonucleotide primers used in this study.
Supplemental Data Set 2. Saccharomyces cerevisiae strains used in this study.
Supplemental Data Set 3. Details of statistical analyses.
Supplemental File 1. Fasta file of ATG14 protein alignment used for the phylogeneticanalysis.
Supplemental File 2. Newick format of the ATG14 phylogenetic tree.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Qijun Chen for the CRISPR/Cas9 reagents. This work was supported by the National Science Foundation, Plant Genome Research Program (grants IOS-1339325 and IOS-1840687) and the National Institutes of Health, National Institute of General Medical Sciences (grant R01-GM124452 to R.D.V.); the National Natural Science Foundation of China (grant 31770356 to F. Li); the China Scholars Fellowship Program (predoctoral fellowship to F. Liu.); and the Netherlands Organization for Scientific Research (grant NWO 867.15.020 to T.M.).
AUTHOR CONTRIBUTIONS
F. Liu, F. Li, and R.D.V. designed the research. F. Liu, W.H., F. Li, R.S.M., and X.Z. performed the research. F. Liu, W.H., R.S.M., X.Z., T.M., and R.D.V. analyzed the data. F. Liu and R.D.V. wrote the article with input from all authors.
Footnotes
Articles can be viewed without a subscription.
References
- Alexander M.P.(1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Balla T.(2013). Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 93: 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Mouilleron S., Bhujabal Z., Wirth M., Sjøttem E., Evjen G., Zhang W., Lee R., O’Reilly N., Tooze S.A., Lamark T., Johansen T.(2019). Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 15: 1333–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Klionsky D.J.(2007). Physiological functions of Atg6/Beclin 1: A unique autophagy-related protein. Cell Res. 17: 839–849. [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D.(2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Contento A.L., Kim S.J., Bassham D.C.(2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135: 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U.(2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano G.P., Barozzi F., Piro G., Denecke J., de Marcos Lousa C.(2017). Trafficking routes to the plant vacuole: Connecting alternative and classical pathways. J. Exp. Bot. 69: 79–90. [DOI] [PubMed] [Google Scholar]
- Diao J., et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520: 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D.(2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Fan W., Nassiri A., Zhong Q.(2011). Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 108: 7769–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y.(2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143: 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Suda Y., Vernhettes S., Nakano A., Ueda T.(2015). Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol. 56: 287–298. [DOI] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Cui Y., Fu X., He Y., Zhao Q., Zeng Y., Shen J., Luo M., Jiang L.(2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 112: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth K., Lin F., Menzel W., Krishnamoorthy P., Stenzel I., Heilmann M., Heilmann I.(2017). Guilt by association: A phenotype-based view of the plant phosphoinositide network. Annu. Rev. Plant Biol. 68: 349–374. [DOI] [PubMed] [Google Scholar]
- Gingerich D.J., Hanada K., Shiu S.-H., Vierstra R.D.(2007). Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19: 2329–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley N., Hohn B., Hohn T., Walden R.(1986). “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. USA 83: 3282–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe N.J., Olsen L.J.(2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4: 339–348. [DOI] [PubMed] [Google Scholar]
- Heucken N., Ivanov R.(2018). The retromer, sorting nexins and the plant endomembrane protein trafficking. J. Cell Sci. 131: 203695. [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2019). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and -independent autophagic routes during fixed-carbon starvation. Plant Cell 31: 2973–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y.(2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652. [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N.(2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19: 5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Dou Z., Chen J.-S., Catanzaro J., Jiang Y.-P., Ballou L.M., Selinger E., Ouyang X., Lin R.Z., Zhang J., Zong W.-X.(2012). Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 109: 2003–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J.H., Yoo H.J., Hwang I., Lee J.S., Nam K.H., Bae Y.S.(2005). Auxin-induced reactive oxygen species production requires the activation of phosphatidylinositol 3-kinase. FEBS Lett. 579: 1243–1248. [DOI] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y.(2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A.(2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M.(1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K.(2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.N., Zarza X., Kim J.H., Yoon M.J., Kim S.-H., Lee J.-H., Paris N., Munnik T., Otegui M.S., Chung T.(2018). Vacuolar trafficking protein VPS38 is dispensable for autophagy. Plant Physiol. 176: 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Bak G., Choi Y., Chuang W.-I., Cho H.-T., Lee Y.(2008a). Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol. 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim E.-S., Choi Y., Hwang I., Staiger C.J., Chung Y.-Y., Lee Y.(2008b). The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 147: 1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D.(2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K., Brech A., Finley K.D., Gaumer S., Contamine D., Rusten T.E., Stenmark H.(2008). The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy 4: 500–506. [DOI] [PubMed] [Google Scholar]
- Liu F., Hu W., Vierstra R.D.(2018). The Vacuolar Protein Sorting-38 subunit of the Arabidopsis phosphatidylinositol 3-kinase complex plays critical roles in autophagy, endosome sorting, and gravitropism. Front. Plant Sci. 9: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., Hua Z., Mali S., McLoughlin F., Vierstra R.D.(2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177: 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D.(2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R.S., McLoughlin F., Vierstra R.D.(2016). Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 16: 1717–1732. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Vierstra R.D.(2018). Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69: 173–208. [DOI] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M.(2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N.T., Izumi T., Noda T., Yoshimori T.(2010). Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190: 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11: 385–396. [DOI] [PubMed] [Google Scholar]
- Munnik T.(2013). Analysis of D3-,4-,5-phosphorylated phosphoinositides using HPLC In Plant Lipid Signaling Protocols, Munnik T., and Heilmann I., eds (Totowa, NJ: Humana Press; ), pp. 17–24. [DOI] [PubMed] [Google Scholar]
- Munnik T., Nielsen E.(2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14: 489–497. [DOI] [PubMed] [Google Scholar]
- Noack L.C., Jaillais Y.(2017). Precision targeting by phosphoinositides: How PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40: 22–33. [DOI] [PubMed] [Google Scholar]
- Noda T., Klionsky D.J.(2008). The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 451: 33–42. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Aoki Y., Tadaka S., Kagaya Y., Kinoshita K.(2018). A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 59: 400. [Google Scholar]
- Page R.D.M.(1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S.P.(2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27. [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D.(2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.-J., Gu H.(2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17: 249–263. [DOI] [PubMed] [Google Scholar]
- Reguera M., Bassil E., Tajima H., Wimmer M., Chanoca A., Otegui M.S., Paris N., Blumwald E.(2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27: 1200–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J.E., Masson G.R., Johnson C., Steyaert J., Ktistakis N.T., Williams R.L.(2015). Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350: aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I.(2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K.D., Lee H.N., Chung T.(2014). A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol. Cells 37: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G.(2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849. [DOI] [PubMed] [Google Scholar]
- Stagljar I., Korostensky C., Johnsson N., te Heesen S.(1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95: 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D.(2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáč T., Pechan T., Samajová O., Šamaj J.(2013). Vesicular trafficking and stress response coupled to PI3K inhibition by LY294002 as revealed by proteomic and cell biological analysis. J. Proteome Res. 12: 4435–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka C., Inoue Y., Matsuoka K., Moriyasu Y.(2004). 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 45: 265–274. [DOI] [PubMed] [Google Scholar]
- Tan X., Thapa N., Liao Y., Choi S., Anderson R.A.(2016). PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc. Natl. Acad. Sci. USA 113: 10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D.(2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Vierstra R.D.(1993). Multiubiquitin chains linked through lysine 48 are abundant in vivo and are competent intermediates in the ubiquitin proteolytic pathway. J. Biol. Chem. 268: 24766–24773. [PubMed] [Google Scholar]
- Vermeer J.E., van Leeuwen W., Tobeña-Santamaria R., Laxalt A.M., Jones D.R., Divecha N., Gadella T.W. Jr., Munnik T.(2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47: 687–700. [DOI] [PubMed] [Google Scholar]
- Wang W.-Y., Zhang L., Xing S., Ma Z., Liu J., Gu H., Qin G., Qu L.-J.(2012). Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J. Genet. Genomics 39: 81–92. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J.(2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters P., Takegawa K., Emr S.D., Chrispeels M.J.(1994). AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc. Natl. Acad. Sci. USA 91: 11398–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P., Hinz S., Doughty J.(2009). Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol. 151: 1812–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Zhang J., Yang Y.(2014). Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7: 923–926. [DOI] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J.(2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Gao X.-Q., Zhao X.Y., Zhu D.Z., Zhou L.Z., Zhang X.S.(2011). Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 77: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y.(2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.N., Cho K., Lawrence R., Zoncu R., Hurley J.H.(2016). Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. Proc. Natl. Acad. Sci. USA 113: 8224–8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Lan J., Fang Y., Yu H., Zhang J., Huang J., Qin G.(2018). The Arabidopsis USL1 controls multiple aspects of development by affecting late endosome morphology. New Phytol. 219: 1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.T., Li E., Guo Y.K., Yu S.X., Wan Z.Y., Ma T., Li S., Hirano T., Sato M.H., Zhang Y.(2018). Arabidopsis VAC14 is critical for pollen development through mediating vacuolar organization. Plant Physiol. 177: 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Wang H., Lam S.K., Gao C., Wang X., Cai Y., Jiang L.(2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]


