Chloroplast m-type thioredoxins interact with PGRL1 and regulate the PGR5/PGRL1-dependent PSI cyclic pathway.
Abstract
In addition to linear electron transport, photosystem I cyclic electron transport (PSI-CET) contributes to photosynthesis and photoprotection. In Arabidopsis (Arabidopsis thaliana), PSI-CET consists of two partially redundant pathways, one of which is the PROTON GRADIENT REGULATION5 (PGR5)/PGR5-LIKE PHOTOSYNTHETIC PHENOTYPE1 (PGRL1)–dependent pathway. Although the physiological significance of PSI-CET is widely recognized, the regulatory mechanism behind these pathways remains largely unknown. Here, we report on the regulation of the PGR5/PGRL1-dependent pathway by the m-type thioredoxins (Trx m). Genetic and phenotypic characterizations of multiple mutants indicated the physiological interaction between Trx m and the PGR5/PGRL1-dependent pathway in vivo. Using purified Trx proteins and ruptured chloroplasts, in vitro, we showed that the reduced form of Trx m specifically decreased the PGR5/PGRL1-dependent plastoquinone reduction. In planta, Trx m4 directly interacted with PGRL1 via disulfide complex formation. Analysis of the transgenic plants expressing PGRL1 Cys variants demonstrated that Cys-123 of PGRL1 is required for Trx m4-PGRL1 complex formation. Furthermore, the Trx m4-PGRL1 complex was transiently dissociated during the induction of photosynthesis. We propose that Trx m directly regulates the PGR5/PGRL1-dependent pathway by complex formation with PGRL1.
INTRODUCTION
In the light reactions of photosynthesis, light energy is utilized by photosystem II (PSII) to oxidize water, generating protons, oxygen, and electrons. These electrons are transferred to photosystem I (PSI) via the cytochrome b6f complex and ultimately used for the reduction of NADP+ to NADPH in the stroma. Electron transport through the cytochrome b6f complex is coupled to proton transfer from the stroma into the lumen, and the resulting ΔpH is used for ATP synthesis. Linear electron transport from water to NADP+ requires both photosystems and produces both NADPH and ATP. By contrast, cyclic electron transport around PSI (PSI-CET) is driven solely by PSI. In PSI-CET, electrons are recycled from ferredoxin (Fd) to plastoquinone (PQ) and pass through the cytochrome b6f complex, generating ΔpH without net production of NADPH.
PSI-CET was discovered by Arnon and co-workers more than 60 years ago, before the concept of linear electron transport was even established (Arnon et al., 1954). In angiosperms, PSI-CET consists of two pathways: the first pathway, known as the PROTON GRADIENT REGULATION5 (PGR5) and the PGR5-LIKE PHOTOSYNTHETIC PHENOTYPE1 (PGRL1)–dependent pathway, is sensitive to antimycin A (Tagawa et al., 1963; Munekage et al., 2002; DalCorso et al., 2008), whereas the second pathway, the NADH dehydrogenase like (NDH) complex–dependent pathway, is antimycin A insensitive (Joët et al., 2001; Yamamoto et al., 2011). In Arabidopsis (Arabidopsis thaliana), the PGR5/PGRL1-dependent pathway is the main route of PSI-CET and contributes more significantly to ΔpH generation than the NDH-dependent pathway (Munekage et al., 2002; DalCorso et al., 2008). The Arabidopsis pgr5 mutant, for example, cannot induce the energization-dependent quenching (qE) component of nonphotochemical quenching (NPQ) and the downregulation of the cytochrome b6f complex termed photosynthetic control, due to impaired acidification of the thylakoid lumen (Munekage et al., 2002; Joliot and Johnson, 2011; Yamamoto and Shikanai, 2019). The qE reflects the size of thermal dissipation of the excess light energy from PSII (Krause and Weis, 1991). Since pgr5 cannot fully activate these photoprotective mechanisms, it is sensitive to high light and cannot survive under fluctuating light conditions (Munekage et al., 2002; Suorsa et al., 2012; Yamamoto and Shikanai, 2019). By contrast, Arabidopsis mutants defective in the chloroplast NDH complex such as chlororespiratory reduction 2-2 (crr2-2) do not exhibit any growth phenotypes (Hashimoto et al., 2003). The NDH-dependent pathway appears to play a compensatory function in the pgr5 mutant background. The crr2-2 pgr5 double mutant shows severely impaired photosynthesis and growth even under constant low light conditions (Munekage et al., 2004). Despite the widespread recognition of the physiological significance of PSI-CET, its regulatory mechanism remains unknown.
Several factors have been suggested to regulate the activity of PSI-CET. ATP concentration in the stroma is one of the possible factors. During the induction of photosynthesis, PSI-CET operates efficiently to supply ATP to the Calvin–Benson cycle (Joliot and Joliot, 2006). The stromal redox state has also been proposed as a regulator of PSI-CET. A higher stromal reduction state enhances PSI-CET (Breyton et al., 2006; Okegawa et al., 2008).
More recently, several papers reported the chloroplastic thioredoxin (Trx) systems to be the most promising candidates for the regulation of PSI-CET (Courteille et al., 2013; Hertle et al., 2013; Nikkanen et al., 2018). Chloroplasts have two Trx systems (Buchanan, 2016; Yoshida and Hisabori, 2016; Cejudo et al., 2019). In the classical Trx system, Trxs are reduced by photo-reduced Fd, via Fd-Trx reductase (FTR), and regulate many stromal enzymes, including the Calvin–Benson cycle enzymes (Geigenberger and Fernie, 2014; Buchanan, 2016). Another system, the NADPH-Trx reductase C (NTRC) pathway, was newly found to be a unique system in the chloroplast (Serrato et al., 2004; Pérez-Ruiz et al., 2006). Since NTRC utilizes NADPH as an electron donor, it can function even in the dark. In Arabidopsis, typical Trxs in the stroma are classified into five types: two f-type (Trxs f1 and f2), four m-type (Trxs m1, m2, m3, and m4), one x-type (Trx x), two y-type (Trxs y1 and y2), and one z-type (Trx z; Buchanan, 2016). Trx m was the most abundant type, accounting for 69.1% of the total stromal Trx content, where Trxs m1 (24.8%), m2 (21.8%), m3 (0.9%), and m4 (21.6%) made up the balance (Okegawa and Motohashi, 2015). On the basis of the mutant phenotype, Courteille et al. (2013) reported that Trx m4 regulated both PSI-CET pathways, although targets of Trx m4 have not yet been identified. In our previous report, we also suggested PSI-CET to have been enhanced in a mutant defective in three Trx m isoforms (Trxs m1, m2, and m4; Okegawa and Motohashi, 2015). Based on in vitro experiments, Hertle et al. (2013) proposed a schematic model for the PGR5/PGRL1-dependent pathway in Arabidopsis. The dimeric, inactive form of PGRL1 was demonstrated to be reduced to a monomer and activated by Trx m (Hertle et al., 2013). However, a regulatory mechanism in vivo remains unknown. NTRC has also been proposed to be involved in the regulation of PSI-CET, since its overexpression stimulated the NDH-dependent pathway (Nikkanen et al., 2018). Thus, two Trx systems are suggested to regulate PSI-CET, but their targets and the physiological significance of their regulation in vivo are largely unclear.
Knowledge of the regulation mechanism of PSI-CET is essential for understanding photosynthesis and photoprotection. In this study, using both genetic and biochemical approaches, we show that Trx m4 directly downregulates the PGR5/PGRL1-dependent pathway via disulfide complex formation with PGRL1 in Arabidopsis. Herein, we discuss the physiological significance of Trx m4–dependent regulation of PSI-CET.
RESULTS
Growth Defects of trx m124-2 Were Alleviated by Defects in PSI-CET Pathways
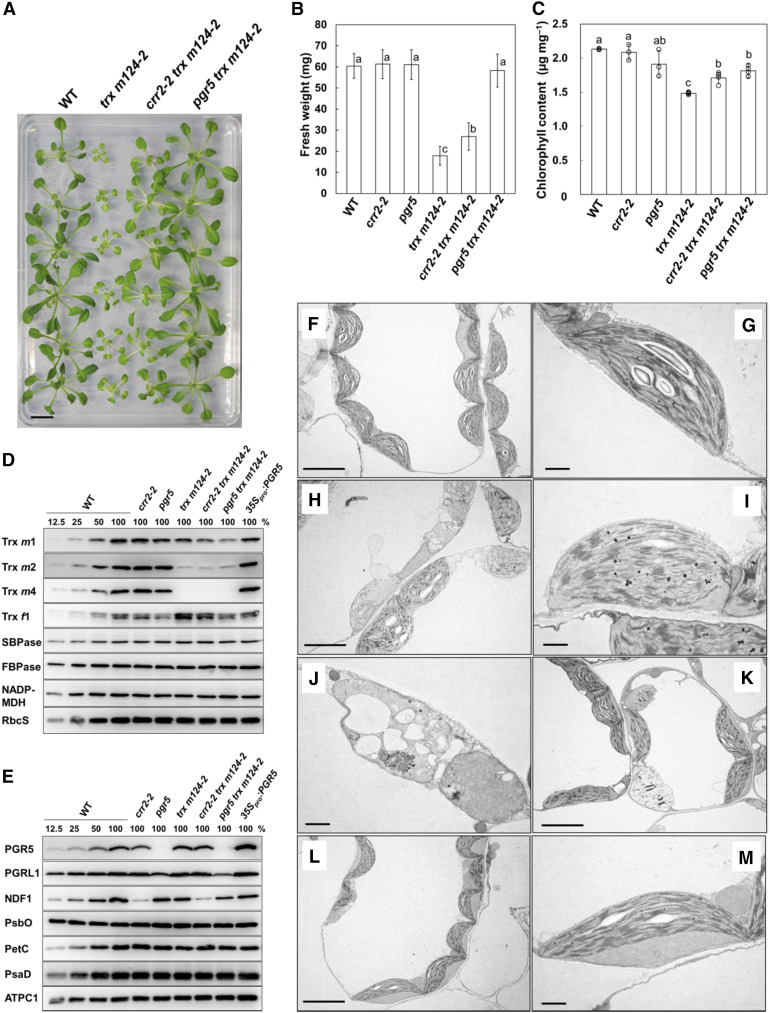
We previously reported that the Arabidopsis trx m124-2 mutant exhibited growth defects (Okegawa and Motohashi, 2015), characterized by decreased fresh weight and chlorophyll content (Figures 1A to 1C). The trx m124-2 mutant is a triple mutant, which was obtained by crossing the T-DNA knockdown mutant lines trx m1 and trx m2-2 and the T-DNA knockout mutant line trx m4. In trx m124-2, Trx m4 did not accumulate at all, but the protein levels of Trxs m1 and m2 were 52 and 13% of the wild-type level, respectively (Okegawa and Motohashi, 2015). Therefore, the amount of Trx m isoform protein in trx m124-2 was 23% of that in the wild type (Figure 1D; Okegawa and Motohashi, 2015). To investigate the physiological link between Trx m and PSI-CET, we tested the genetic interaction between the mutants, by generating the quadruple mutants crr2-2 trx m124-2 and pgr5 trx m124-2 (Figures 1A, 1D, and 1E). Seedlings were grown under continuous light conditions at a light intensity of 50 µmol photons m–2 s–1. In crr2-2 trx m124-2, the leaves were still pale green, and the petioles were white as in trx m124-2, but the growth defects were partially alleviated in terms of the seedling size and fresh weight (Figures 1A and 1B). In the pgr5 mutant background, the growth phenotype of trx m124-2 was suppressed more significantly. The pgr5 trx m124-2 mutant grew similarly to the wild type (Figures 1A and 1B), and its chlorophyll content also recovered to similar levels as in pgr5 (Figure 1C), although the pgr5 single mutant had a slightly lower chlorophyll content than the wild type (Figure 1C; Munekage et al., 2002). These results indicated that a lack of PSI-CET pathway recovered the growth defect of trx m124-2 and the pgr5 defect suppressed their phenotype more effectively than the crr2-2 defect. The recovery of growth defects in trx m124-2 was also observed under short-day and higher light (200 µmol photons m–2 s–1) conditions (Supplemental Table 1). Furthermore, the growth of the trx m124-1 mutant, which is a weak allele mutant of trx m124-2 (Okegawa and Motohashi, 2015), was also recovered by defects in PSI-CET (Supplemental Table 2). To further interrogate the genetic interaction between Trx m and PSI-CET, the growth phenotype of trx m124-2 was analyzed also in the pgrl1ab mutant background, which is another mutant lacking the PGR5/PGRL1-dependent pathway (DalCorso et al., 2008). Similar to pgr5 trx m124-2, the pgrl1ab trx m124-2 quintuple mutant grew like the wild type (Supplemental Figure 1), suggesting that the growth defects of trx m124-2 are recovered in the absence of the PGR5/PGRL1-dependent pathway. In trx m124-2, PSI-CET pathways may have not been regulated, leading to the growth defects.
Figure 1.
Phenotypes of the Wild Type, trx m124-2, crr2-2 trx m124-2, and pgr5 trx m124-2 Mutants.
Seedlings were grown on MS medium in growth chambers (50 µmol photons m–2 s–1; continuous light; 23°C) for 3 to 4 weeks.
(A) Photographs of the plants. WT, wild type. Bar = 10 mm.
(B) Fresh weight of seedlings. Each value is shown as the mean ± sd (n = 10 to 15 independent plants). Columns with the same letters are not significantly different between genotypes according to one-way ANOVA with Tukey–Kramer multiple comparison test, P < 0.05 (Supplemental Table 5). WT, wild type.
(C) Chlorophyll content of seedlings, per unit fresh weight. Each value is the mean ± sd (n = 4 to 6 independent plants). Columns with the same letters are not significantly different between genotypes according to one-way ANOVA with Tukey–Kramer multiple comparison test, P < 0.05 (Supplemental Table 6). WT, wild type.
(D) and (E) Immunoblot analysis in the wild type (WT), crr2-2, pgr5, trx m124-2, crr2-2 trx m124-2, pgr5 trx m124-2, and 35Spro:PGR5. Chloroplasts were fractionated into the stromal fractions and thylakoid membranes. Trxs m1, m2, and m4, Trx f1, SBPase, FBPase, NADP-MDH, and RbcS were immunodetected using specific antibodies in the stromal fractions (D). PGR5, PGRL1, NDF1, PsbO, PetC, PsaD, and ATPC1 were immunodetected using specific antibodies in the thylakoid membranes (E). Protein samples corresponding to 1.0 µg chlorophyll were loaded in each lane.
(F) to (M) Chloroplasts in the wild type (WT), trx m124-2, and pgr5 trx m124-2. Chloroplast ultrastructure of WT (F) and (G), trx m124-2 (see [H] to [K]), and pgr5 trx m124-2 (see [L] and [M]). Seedlings were observed using TEM. Bar in (F), (H), (K), and (L) = 5 µm; bar in (G), (I), (J), and (M) = 1 µm.
The ultrastructure of the chloroplast was characterized using transmission electron microscopy (TEM). Wild-type chloroplasts contained well-developed thylakoid membranes with normally stacked granal thylakoids and starch granules (Figures 1F and 1G). In some parts of trx m124-2 leaves, chloroplasts swelled to different extents (Figure 1H). Their thylakoid membranes were severely distorted and detected as fragments, although granal stacks were partially observed (Figures 1H and 1I). Some plastids of trx m124-2 lacked developed thylakoid membranes and granal stacks and contained vacuolated membrane structures (Figures 1H and 1J). Conversely, a number of well-developed chloroplasts were observed in the green parts of trx m124-2 leaves, although aberrant plastids were still present (Figure 1K). By contrast, chloroplasts of pgr5 trx m124-2 contained normally developed thylakoid membrane structure (Figures 1L and 1M) and starch granules, just like the wild type. This observation may explain the recovery of plant growth in pgr5 trx m124-2 (Figures 1A and 1B).
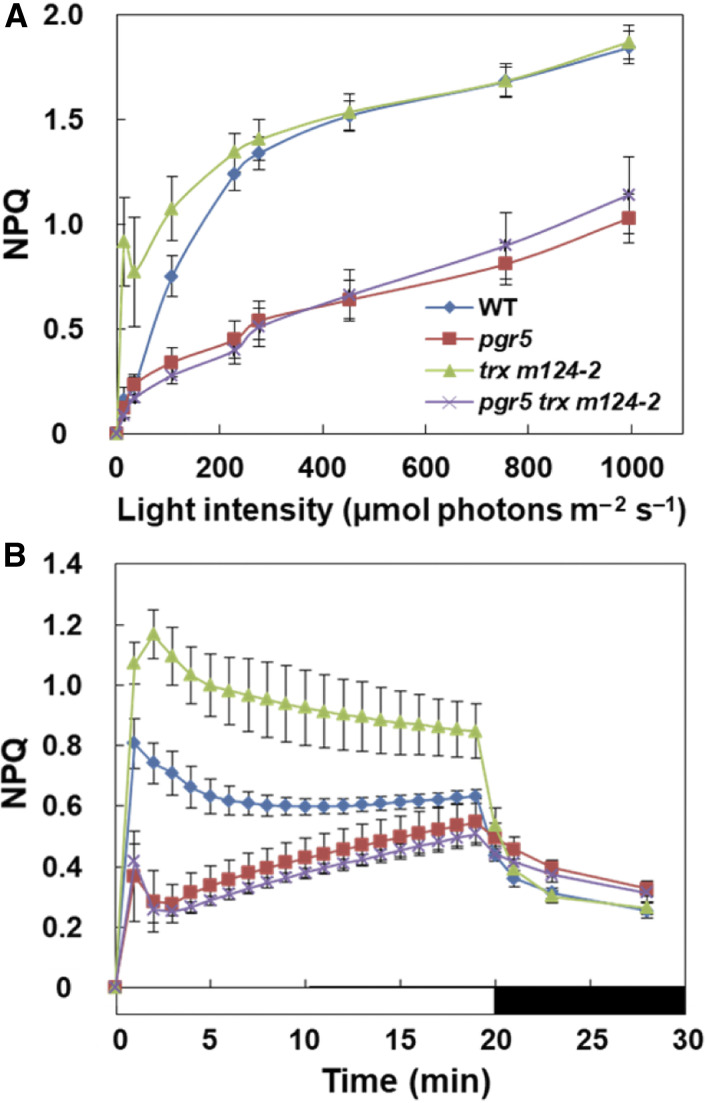
The trx m124-2 mutant induced higher NPQ than the wild type at light intensities of less than 200 µmol photons m–2 s–1 (Figure 2A; Okegawa and Motohashi, 2015). The qE component of NPQ is induced by the acidification of the thylakoid lumen (Krause and Weis, 1991). The PGR5/PGRL1-dependent PSI-CET contributes to luminal acidification (Munekage et al., 2002; DalCorso et al., 2008). The pgr5 mutant showed decreased NPQ compared to the wild type (Figures 2A and 2B). To investigate whether the high NPQ phenotype observed in trx m124-2 was dependent on the PGR5/PGRL1-dependent pathway, the light-intensity dependence of NPQ was analyzed (Figure 2A; Supplemental Figure 2). In the wild type, NPQ was induced at light intensities of more than 50 µmol photons m–2 s–1 (Figure 2A). Similar to trx m124-2, crr2 trx m124-2 induced higher NPQ than the wild type at light intensities of less than 200 µmol photons m–2 s–1 (Supplemental Figure 2). By contrast, NPQ was not induced in pgr5 trx m124-2 as in pgr5 (Figure 2A).
Figure 2.
In Vivo Analysis of Chlorophyll Fluorescence.
(A) Dependence of the NPQ on the light intensity in the wild type (WT), pgr5, trx m124-2, and pgr5 trx m124-2. NPQ values were recorded 2 min after a shift to each light intensity.
(B) Time courses of the NPQ induction and relaxation. NPQ values were measured during the actinic illumination with 62 µmol photons m–2 s–1 for 20 min, followed by relaxation in the dark for 8 min. Each data point represents the mean ± sd (n = 5 independent plants).
The qE component is characterized by its relatively fast induction and relaxation kinetics on a physiological time scale of seconds to several minutes (Horton et al., 1996). For a detailed characterization of the qE component, the time course of NPQ induction and relaxation was analyzed at a light intensity of 62 µmol photons m–2 s–1 and subsequently in the dark (Figure 2B). During the induction of photosynthesis, a relatively high NPQ was induced even in the wild type, which relaxed to a steady state level within several minutes (Figure 2B). In trx m124-2, however, high NPQ was retained for a longer time, without fully relaxing—to the wild-type level—even during the steady state photosynthesis (Figure 2B). By contrast, in pgr5 trx m124-2, the NPQ level was almost identical to that in pgr5 (Figure 2B). These results indicate that the high NPQ induced in trx m124-2 was dependent on the PGR5/PGRL1-dependent pathway.
Impaired Light-Dependent Reduction of Thiol Enzymes in trx m124-2 Was Partially Recovered by the pgr5 Mutation
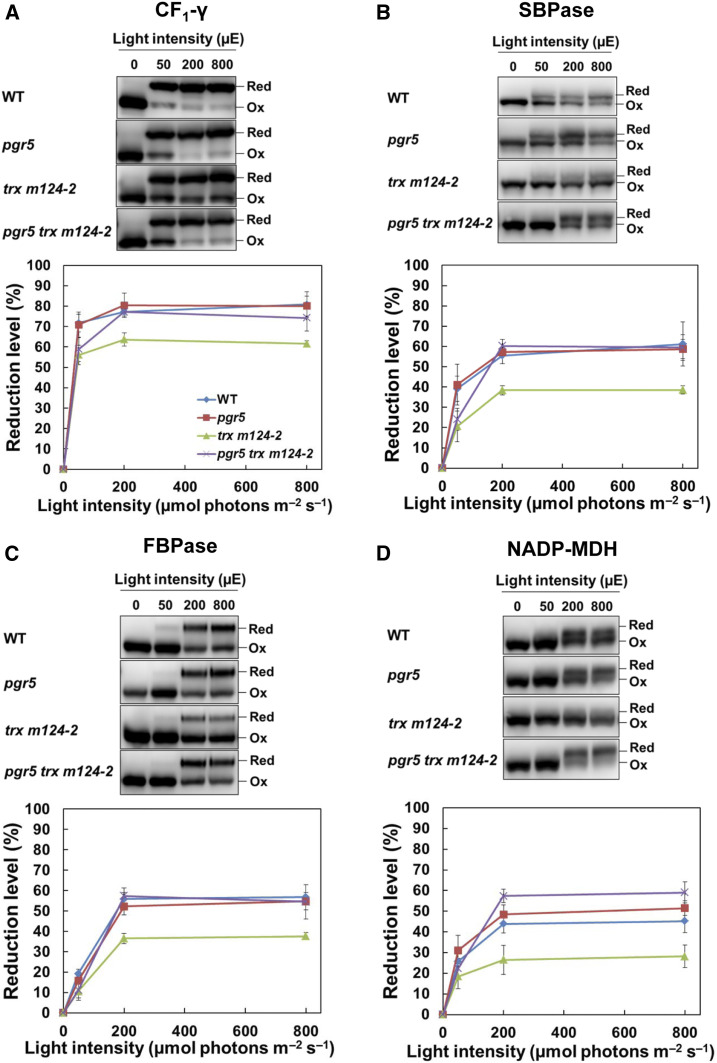
Most thiol enzymes, including the Calvin–Benson cycle enzymes, are activated by the reduction of the disulfide bonds by Trxs (Geigenberger and Fernie, 2014). The growth retardation in trx m124-2 was suggested to be caused by the insufficient activation of the Calvin–Benson cycle and the resulting decrease in carbon assimilation (Okegawa and Motohashi, 2015). In pgr5 trx m124-2, the growth defects of trx m124-2 were significantly recovered (Figures 1A and 1B). Therefore, we examined the reduction level of several thiol enzymes in pgr5 trxm124-2. The protein levels of thiol enzymes in pgr5 trx m124-2 were indistinguishable from those in the other genotypes, including the wild type (Figures 1D and 1E). The redox state of thiol enzymes was determined by labeling the free thiols with the thiol-reactive 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) reagent. Samples were extracted from plants adapted to dark, or to different light intensities for 1 h. In the dark, all enzymes were oxidized in all the genotypes (Figure 3); however, at a light intensity of 50 µmol photons m–2 s–1 ∼70% of the ATP synthase CF1-γ (ATPC1) subunit was reduced in the wild type, compared to a slightly increased level under higher light conditions (>200 µmol photons m–2 s–1; Figure 3A). Under all light intensities, trx m124-2 had a lower reduction level of CF1-γ than the wild type. In pgr5 trx m124-2, the reduction rate in the CF1-γ was still lower than in the wild type at 50 µmol photons m–2 s–1, but it was recovered to the wild type level under higher light conditions (Figure 3A). This tendency was also observed when the redox state of the Calvin–Benson cycle enzymes was examined. The reduction level of sedoheptulose-1,7-bisphosphatase (SBPase) and fructose-1,6-bisphosphatase (FBPase) in trx m124-2 was lower than in the wild type under all light intensities (Figures 3B and 3C). Under higher light conditions, their reduction levels in pgr5 trx m124-2 were restored to the level of the wild type (Figures 3B and 3C). The photo-reduction of NADP-malate dehydrogenase (NADP-MDH), which is the key enzyme of the malate valve, was also investigated (Figure 3D). Compared to trx m124-2, NADP-MDH was more reduced in pgr5 trx m124-2 (Figure 3D). These results indicate that the insufficient activation of thiol enzymes in trx m124-2 was partially recovered by the pgr5 mutation.
Figure 3.
Photo-Reduction of the Thiol Enzymes in the Wild Type and the pgr5, trx m124-2, and pgr5 trx m124-2 Mutants under Different Light Intensities.
(A) to (D) After 8 h in dark, seedlings were subjected to illuminations of 50, 200, and 800 µmol photons m–2 s–1 (µE), for 1 h each, in a stepwise manner. Samples were collected at the indicated light intensities and modified with AMS, after which 35 µg of proteins (per sample) were subjected to nonreducing SDS-PAGE. Redox states of the ATPC1 (A), SBPase (B), FBPase (C), and NADP-MDH (D) were detected by immunoblot analysis (top). The reduction levels of thiol enzymes were indicated as the percentage of the reduced versus the total protein (bottom). Each value represents the mean ± sd (n = 5 independent plants). Ox, oxidized; Red, reduced; WT, wild type.
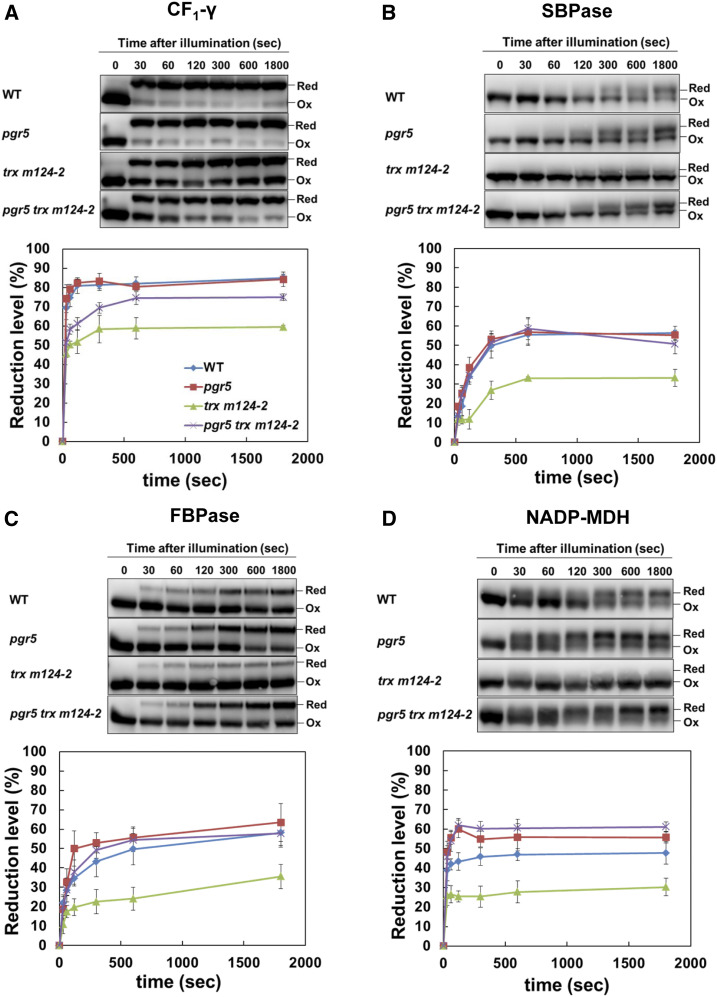
Next, the redox state of the thiol enzymes was kinetically analyzed during the induction of photosynthesis (Figure 4). Dark-adapted plants were exposed to a light intensity of 200 µmol photons m–2 s–1 for 1800 s. In the wild type, all enzymes were rapidly reduced upon illumination and achieved different steady state reduction levels after 600 s of illumination (Figure 4). Consistent with the results of light intensity dependence (Figure 3), the reduction level of all enzymes was lower in trx m124-2 than in the wild type (Figure 4). In pgr5 trx m124-2, the reduction levels for these enzymes were higher than in trx m124-2 (Figure 4), although the reduction level of the CF1-γ was significantly lower than in the wild type, for at least a few minutes after the exposure to light (Figure 4A). Thus, the impaired photo-reduction of the thiol enzymes in trx m124-2 was partially recovered by the pgr5 mutation. These results suggest that the suppression of growth defects in pgr5 trx m124-2 can be caused by the recovery of normal chloroplast development (Figure 1), notably by the activation of the Calvin–Benson cycle enzymes.
Figure 4.
Activation of Thiol Enzymes during the Induction of Photosynthesis.
(A) to (D) Seedlings were dark adapted for 8 h and exposed to light (200 µmol photons m–2 s–1). Samples were collected at the indicated time points. Redox states of the ATPC1 (A), SBPase (B), FBPase (C), and NADP-MDH (D) were detected by immunoblot analysis (top). The reduction levels of thiol enzymes were indicated as the percentage of the reduced versus the total protein (bottom). Each value shows the mean ± sd (n = 5 independent plants). Ox, oxidized; Red, reduced; WT, wild type.
Reduced Trx m Suppressed the Fd-Dependent PQ Reduction Activity in Ruptured Chloroplasts
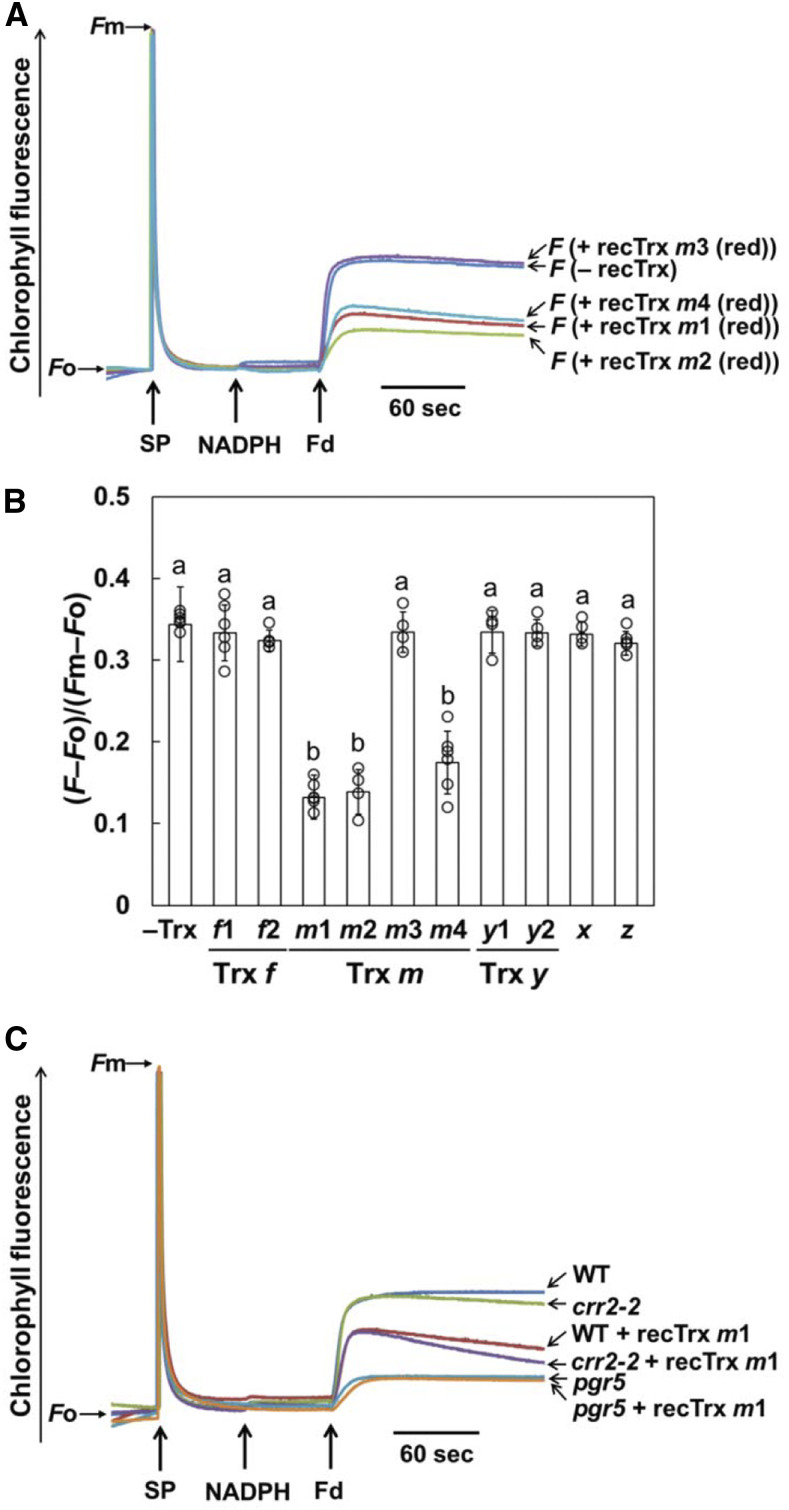
We next evaluated the effect of Trx m on Fd-dependent PQ reduction in ruptured chloroplasts using purified Trx proteins. Disulfide reduction activity of the purified Trx proteins was confirmed using the insulin reduction assay (Supplemental Figure 3). Fd-dependent PQ reduction was monitored as an increase in the chlorophyll fluorescence. By the addition of NADPH and Fd, PQ was rapidly reduced due to the electron input activity to PQ via both PSI-CET pathways, without the addition of Trxs (Figure 5A, – recTrx). Since the concentration of Trx proteins was ∼5.8 µM in the chloroplast stroma (Okegawa and Motohashi, 2015), 5.0 µM of reduced Trx m proteins was added to ruptured chloroplasts before the measurement. In the presence of reduced Trxs m1, m2, and m4, the rate and final level of PQ reduction were lower than in the absence of Trxs, while the addition of reduced Trx m3 did not affect the chlorophyll fluorescence level (Figure 5A). Activity of Fd-dependent PQ reduction was also estimated using the equation (F − Fo)/(Fm − Fo) (Figure 5B; Taira et al., 2013). In addition, the influence of seven other Trx isoforms localized in the stroma was examined in a similar way, but they did not affect the Fd-dependent PQ reduction activity (Figure 5B; Supplemental Figures 4A and 4B). We also measured the Fd-dependent PQ reduction activity using the oxidized Trx m proteins, but the oxidized Trxs m1, m2, and m4 had no suppression effect on this activity (Supplemental Figure 4C). These results indicated that reduced Trx m (except for Trx m3) specifically impaired Fd-dependent PQ reduction in ruptured chloroplasts.
Figure 5.
Fd-Dependent PQ Reduction Assay in Ruptured Chloroplasts.
(A) Increases in the chlorophyll fluorescence, by the addition of NADPH (250 µM) and Fd (5 µM) under a weak light (1.0 µmol photons m–2 s–1), were monitored in ruptured chloroplasts (20 µg chlorophyll mL–1) from the wild type in the absence (– recTrx) or presence (+ recTrx) of reduced Trx m proteins. Ruptured chloroplasts were incubated with 5.0 µM of the prereduced Trx m before the measurement. The fluorescence levels were standardized by the Fm levels.
(B) Trx isoform-specific suppression effect on the Fd-dependent PQ reduction in ruptured chloroplasts from the wild type. Activity of Fd-dependent PQ reduction was evaluated using the equation (F – Fo)/(Fm – Fo) (Figure 5A; Supplemental Figures 4A and 4B). Each value represents the mean ± sd of four to six independent chloroplast preparations. Columns with the same letters are not significantly different between genotypes (Tukey–Kramer test, P < 0.05).
(C) Effect of the reduced Trx m on the Fd-dependent PQ reduction in ruptured chloroplasts from the crr2-2 and pgr5 mutants. Prereduced Trx m1 (5.0 µM) was added to ruptured chloroplasts before the measurement. WT, wild type.
To examine which PSI-CET pathway was affected by Trx m, the crr2-2 and pgr5 mutants were used for this assay. Similar to the wild type, the addition of the reduced Trx m1 to the crr2-2 chloroplasts decreased the level of chlorophyll fluorescence (Figure 5C). In pgr5, however, the Fd-dependent PQ reduction activity was not affected further by the addition of the reduced Trx m1 (Figure 5C). Addition of the reduced Trx m2 or m4 produced the same results as Trx m1 (Supplemental Figures 4D and 4E). These results suggest that Trx m specifically downregulates the PGR5/PGRL1-dependent pathway in a redox-dependent manner.
Trx m4 Interacted with PGRL1 in Vivo
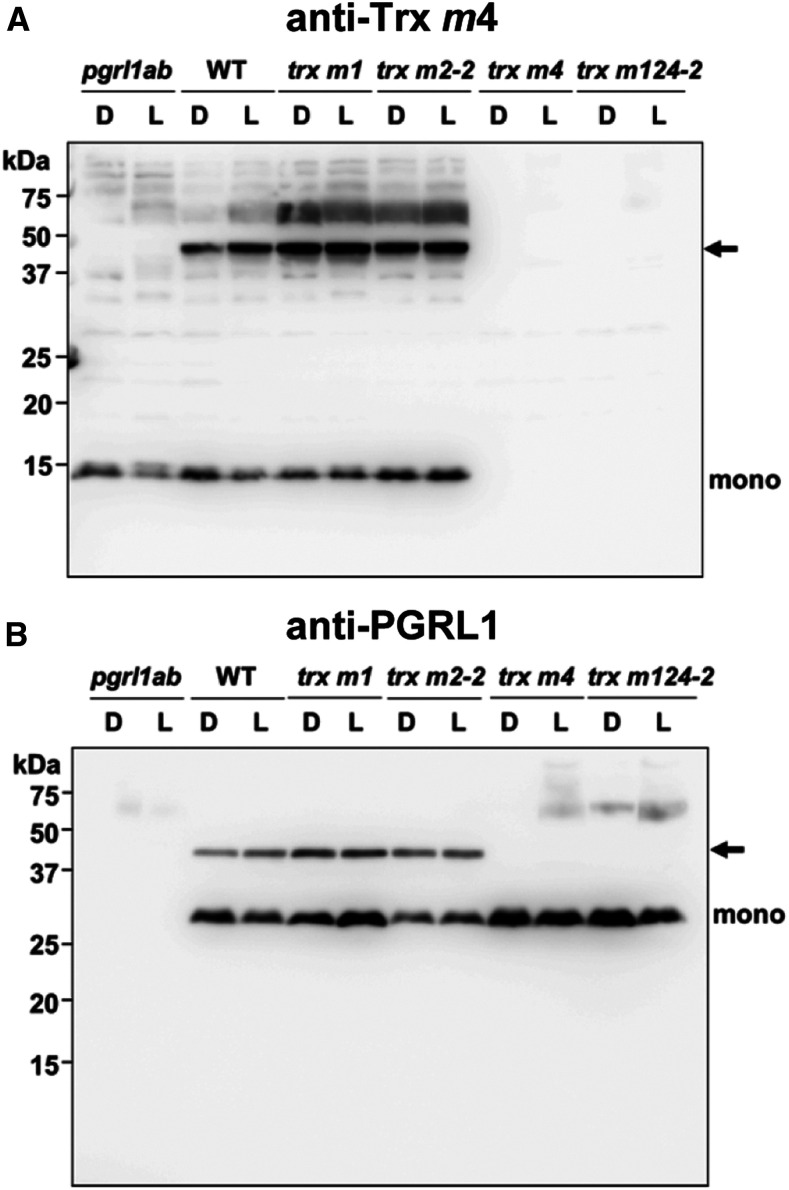
The PGR5/PGRL1-dependent pathway was suggested to be redox regulated by Trx m; however, the molecular mechanism by which this is done remains unknown. To identify the target proteins in this regulation, we tried to isolate proteins interacting with Trx m in vivo. Total proteins extracted from leaves adapted to dark or growth light were subjected to nonreducing SDS-PAGE, and each Trx m isoform was detected by immunoblotting. When Trxs m1 and m2 antibodies were used, only a monomer of each Trx was detected (Supplemental Figure 5A), but when Trx m4 antibody was used, an additional band—besides Trx m4 monomer (13 kD)—was observed at ∼40 kD (Figure 6A, arrow). Interestingly, this additional band was not present in pgrl1ab (Figure 6A). This result suggests that this additional band is in some way related to the PGR5/PGRL1-dependent pathway. This was also supported by the fact that this band was enhanced in the PGR5 overexpression lines (35Spro:PGR5), in which PSI-CET is elevated (Okegawa et al., 2007; Supplemental Figure 5B). Under reducing conditions, this additional band disappeared (Supplemental Figure 5C), indicating that Trx m4 forms a heterodimeric complex with other proteins via a disulfide bond in vivo.
Figure 6.
Intermolecular Disulfide Complex Formation of Trx m4 and Its Target Proteins.
(A) Detection of the complex containing Trx m4. Protein samples were extracted from the wild type (WT), pgrl1ab, trx m1, trx m2-2, trx m4, and trx m124-2 mutants that were either dark adapted (D) or subjected to growth light (L; 50 µmol photons m–2 s–1). Fifty micrograms of proteins was separated by SDS-PAGE under nonreducing conditions. In addition to Trx m4 monomer (mono), Trx m4 complex was immunodetected using Trx m4 antibody (arrow).
(B) Detection of the complex containing PGRL1. The same protein samples as in (A) were immunodetected using PGRL1 antibody. Arrow indicates the Trx m4-PGRL1 complex. WT, wild type.
On the basis of the molecular mass of this heterodimeric complex, a likely candidate for Trx m4-interacting protein is PGRL1 (29 kD). To test this possibility, immunoblot analysis was performed using PGRL1 antibody. In the wild type, an additional band—besides PGRL1 monomer—was observed at the same position as detected by Trx m4 antibody (Figure 6B, arrow). Moreover, this band was missing in trx m4 and trx m124-2 (Figure 6B), which is consistent with the result obtained using Trx m4 antibody (Figure 6A). Consequently, these findings strongly suggest that PGRL1 is the likely target of Trx m4 and that Trx m4 directly regulates PGRL1 via disulfide bond formation in vivo.
Cys-123 of PGRL1 Was Involved in the Interaction with Trx m4 and the Suppression of the PGR5/PGRL1-Dependnet Pathway by Trx m4
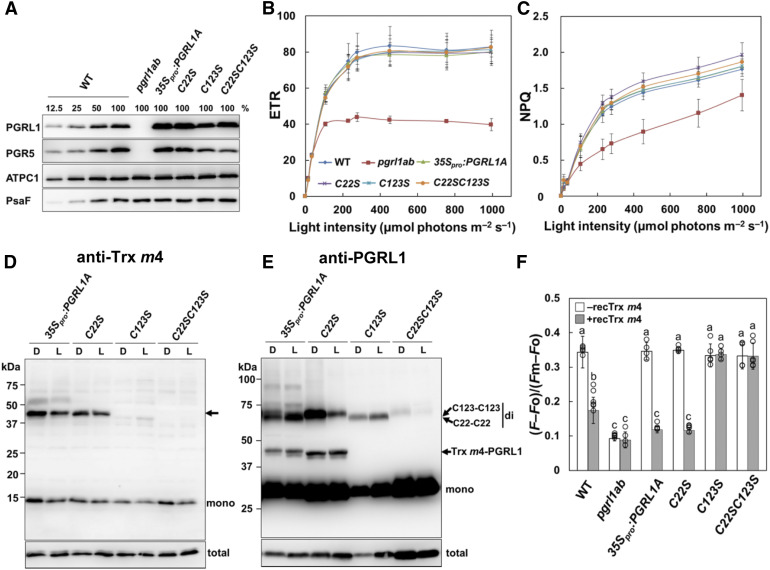
Mature Arabidopsis PGRL1 (PGRL1A and PGRL1B) has six well-conserved Cys residues (Hertle et al., 2013). To determine which Cys residue is responsible for the interaction with Trx m4, transgenic plants were generated, expressing wild-type PGRL1A (35Spro:PGRL1A) and mutant forms of PGRL1A (C22S, C123S, C22SC123S, C212SC215S, and C240SC243S), where one or two Cys residues in PGRL1A were replaced by Ser. Amino acids were numbered excluding the transit peptide sequence, as reported previously (Hertle et al., 2013). All Cys variants were expressed in the pgrl1ab mutant background, under the control of the cauliflower mosaic virus 35S promoter. Accumulation of PGRL1 protein was tested by immunoblot analysis. The 35Spro:PGRL1A, C22S, C123S, and C22SC123S lines had stable accumulation of the expressed PGRL1 proteins in their thylakoid membranes (Figure 7A). Despite repeated attempts, we could not obtain transgenic plants accumulating PGRL1 variants of C212SC215S and C240SC243S. Hertle et al. (2013) reported that Cys-212/215 is essential for Fe binding and that Cys-212/215 and Cys-240/243 interact with PGR5 in the in vitro experiment using the recombinant PGRL1 Cys variants. Hence, the substitution of Cys-212/215 and Cys-240/243 with Ser may destabilize the protein structure of PGRL1A.
Figure 7.
Characterization of PGRL1 Cys Variants.
(A) Immunoblot analysis of PGRL1 in the 35Spro:PGRL1A Cys variant lines. PGRL1, PGR5, ATPC1, and PsaF were immunodetected using specific antibodies. Lanes were loaded with thylakoid membranes corresponding to 1.0 µg of chlorophyll (100%) from the wild type (WT), pgrl1ab, and the 35Spro:PGRL1A, C22S, C123, and C22SC123S Cys variant lines, along with a dilution series of WT sample.
(B) In vivo analysis of electron transport activity. Light intensity dependence of ETR. Each data point represents the mean ± sd (n = 5 independent plants). WT, wild type.
(C) Light intensity dependence of NPQ of chlorophyll fluorescence. Each data point represents the mean ± sd (n = 5 independent plants).
(D) and (E) Detection of the Trx m4-PGRL1 complex on nonreducing SDS-PAGE. Protein samples were extracted from dark-adapted (D) or growth light–incubated (L; 50 µmol photons m–2 s–1) leaves and immunodetected using Trx m4 (D) and PGRL1 (E) antibodies. Arrow in (D) indicates the Trx m4-PGRL1 complex. “mono” and “di” indicate monomer and dimer, respectively. C22-C22 and C123-C123 indicate dimers formed via intermolecular disulfide bonds between Cys-22 and Cys-22 or Cys-123 and Cys-123 of two PGRL1 monomers, respectively. The total amount of Trx m4 or PGRL1 protein (Total) was analyzed on reducing SDS-PAGE to cleave disulfide bonds of complexes, in the same volume used in the detection of the Trx m4-PGRL1 complex. To detect the Trx m4-PGRL1 complex, the chemiluminescence signal of PGRL1 monomer was oversaturated.
(F) Effect of the reduced Trx m4 on the Fd-dependent PQ reduction. Activity of Fd-dependent PQ reduction (Supplemental Figures 6B to 6D) was evaluated using the equation (F – Fo)/(Fm – Fo). White bars indicate chloroplast preparations without Trx m4; gray bars indicate chloroplast preparations with 5 µM of reduced Trx m4. Each value represents the mean ± sd of four to six independent ruptured chloroplast preparations. Columns with the same letters are not significantly different between genotypes (Tukey–Kramer test, P < 0.05). WT, wild type.
To assess the impact of mutations in each Cys residue on photosynthesis, chlorophyll fluorescence parameters were analyzed in the transgenic plants. As reported previously by DalCorso et al. (2008), pgrl1ab showed lower electron transport rate (ETR) and NPQ than the wild type (Figures 7B and 7C). In all transgenic lines, the ETR and NPQ were restored to the same levels as in the wild type (Figures 7B and 7C), indicating that the substitution of Cys-22 and/or Cys-123 with Ser did not affect the fundamental function of PGRL1A under the studied conditions.
To investigate how these substitutions of PGRL1 affect the formation of the Trx m4-PGRL1 complex in vivo, the total proteins were directly extracted from transgenic plants. Interestingly, unlike the wild type (Figure 6B), in the 35Spro:PGRL1A line, dimers of PGRL1 were detected in addition to monomers of PGRL1 and the Trx m4-PGRL1 complex (Figure 7E). This is probably because the protein level of PGRL1 was higher in the 35Spro:PGRL1A line than in the wild type (Figure 7A). Under reducing conditions, these dimers disappeared (Supplemental Figure 6A), indicating that PGRL1 forms a homodimer via a disulfide bond in vivo, similar to the Trx m4-PGRL1 complex. Dimers of PGRL1 were detected also in the C22S and C123S lines, but not in the C22SC123S line. These results are consistent with the previous finding that Cys-22/123 is involved in the dimerization of PGRL1 in vitro (Hertle et al., 2013). As in the 35Spro:PGRL1A line, the Trx m4-PGRL1 complex was detected in the C22S line, using both Trx m4 and PGRL1 antibodies (Figures 7D and 7E); however, in the C123S and C22SC123S lines, the band for the Trx m4-PGRL1 complex was lacking (Figures 7D and 7E). These results revealed that Cys-123 in PGRL1A is required for formation of a heterodimeric complex with Trx m4.
Next, we evaluated the effect of the reduced Trx m4 on the Fd-dependent PQ reduction in ruptured chloroplasts in the 35Spro:PGRL1A Cys variant lines. In the absence of the reduced Trx m4, the activity of the Fd-dependent PQ reduction in all transgenic lines was almost identical to that in the wild type (Figure 7F, white bar). Adding reduced Trx m4 to the 35Spro:PGRL1A and C22S chloroplasts decreased the value of (F – Fo)/(Fm – Fo) (Figure 7F, gray bar; Supplemental Figure 6B). By contrast, the C123S and C22SC123S lines were insensitive to the reduced Trx m4 (Figure 7F; Supplemental Figures 6C and 6D). These results showed that Cys-123 of PGRL1A was a target residue of Trx m4–dependent suppression of the Fd-dependent PQ reduction. Cys-123 of PGRL1A was suggested to be required for both a complex formation with Trx m4 and the downregulation of the PGR5/PGRL1-dependent pathway by Trx m4.
To elucidate whether the PGR5/PGRL1-dependent pathway is in fact downregulated via complex formation of the Trx m4-PGRL1, ruptured chloroplasts used for the in vitro Fd-dependent PQ reduction assay were subjected to immunoblot analysis. Consistent with the in vivo experiment (Figure 7E), dimers of PGRL1 were observed (except in the C22SC123S lines) and these dimers were decreased in the presence of reduced Trx m4 (Supplemental Figure 6E). Without the reduced recombinant Trx m4, the endogenous Trx m4-PGRL1 complex was not detected in any transgenic lines (Supplemental Figure 6E). The endogenous Trx m4-PGRL1 complex observed in vivo (Figure 6) appeared to be dissociated during the isolation of chloroplasts for the in vitro Fd-dependent PQ reduction assay. An addition of the reduced Trx m4 caused formation of the Trx m4-PGRL1 complex in the 35Spro:PGRL1A and C22S lines, but not in the C123S and C22SC123S lines (Supplemental Figure 6E). This was consistent with the results of the in vivo (Figures 7D and 7E) and in vitro (Figure 7F; Supplemental Figures 6B to 6D) experiments. These results suggest that the PGR5/PGRL1-dependent pathway was downregulated by the formation of a complex via the disulfide bond between Trx m4 and Cys-123 of PGRL1.
In ruptured chloroplasts, not only Trx m4, but also Trxs m1 and m2, suppressed the Fd-dependent PQ reduction (Figure 5A), although Trxs m1 and m2 did not form a complex with PGRL1 in vivo (Supplemental Figure 5A). To evaluate the complex formation of Trxs m1 and m2 with PGRL1 in the in vitro experiment (Figure 5A), immunoblot analysis was performed using ruptured chloroplasts isolated from the wild type (Supplemental Figure 7). Unlike the in vivo experiment (Figure 6B), dimers of PGRL1 were observed in ruptured chloroplasts (Supplemental Figure 7, –Trx). Furthermore, oxidized and reduced forms of PGRL1 monomer were detected. As well as Trx m4, the addition of reduced recombinant Trxs m1 and m2 to ruptured chloroplasts caused the formation of a complex with PGRL1. By contrast, Trx f1, which had no effect on Fd-dependent PQ reduction (Figure 5B), did not interact with PGRL1 (Supplemental Figure 7). These results indicate that Trx m downregulates the PGR5/PGRL1-dependent pathway by forming a complex with PGRL1, suggesting that Trxs m1 and m2 act in the same way as Trx m4 in vitro.
Trx m4-PGRL1 Complex Was Transiently Dissociated during the Induction of Photosynthesis
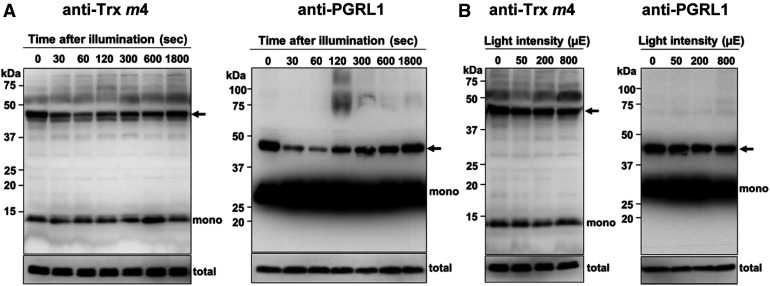
The PGR5/PGRL1-dependent pathway was suggested to be downregulated via formation of the Trx m4-PGRL1 complex, but the question arises on how Trx m4 would regulate the PGR5/PGRL1-dependent pathway in vivo. To address this, the formation of the Trx m4-PGRL1 complex was investigated during the induction of photosynthesis. The dark-adapted wild-type plants were exposed to a light intensity of 200 µmol photons m–2 s–1 for 1800 s. In the dark (time 0), the Trx m4-PGRL1 complex was detected (Figure 8A). During the early induction of photosynthesis (30 to 120 s), the level of the Trx m4-PGRL1 complex transiently decreased (Figure 8A, arrows); however, this complex was gradually reformed after 300 s in the light, reaching the same level as that in the dark after 1800 s. The total amount of Trx m4 and PGRL1 did not change significantly during the induction of photosynthesis (Figure 8A, total). The dynamics of Trx m4-PGRL1 complex formation during the induction of photosynthesis were also observed at a lower light intensity (80 µmol photons m–2 s–1; Supplemental Figure 8A). Dissociation and association of Trx m4-PGRL1 complex was coincident with the time course of induction and relaxation of transiently induced NPQ (Supplemental Figure 8B). These results suggest that the downregulation of the PGR5/PGRL1-dependent pathway by Trx m4 was transiently relaxed during the induction of photosynthesis.
Figure 8.
Dynamics of Complex Formation between Trx m4 and PGRL1 during Photosynthesis In Vivo.
(A) Transient dissociation of the Trx m4-PGRL1 complex during the induction of photosynthesis. Seedlings of the wild type were dark adapted for 8 h and exposed to 200 µmol photons m–2 s–1 for the indicated time periods, before harvesting the plant materials. Next, 50 µg of protein samples was subjected to nonreducing SDS-PAGE. The Trx m4-PGRL1 complex was immunodetected using Trx m4 and PGRL1 antibodies. Arrows indicate the Trx m4-PGRL1 complex. The total amount of Trx m4 or PGRL1 protein (total) was analyzed on reducing SDS-PAGE to cleave disulfide bonds of complexes, in the same volume used in the detection of Trx m4-PGRL1 complex. At least three independent experiments using different plants were performed, and the representative results are shown.
(B) Trx m4-PGRL1 complex formation, under different light intensities. Seedlings were first dark adapted (8 h) and then illuminated at 50, 200, and 800 µmol photons m–2 s–1, for 1 h each, in a stepwise manner. Samples were collected at the indicated light intensities. Other details are as described in (A). To detect the Trx m4-PGRL1 complex, the chemiluminescence signal of PGRL1 monomer was oversaturated.
Formation of the Trx m4-PGRL1 complex was assessed at increasing light intensities also. The Trx m4-PGRL1 complex could be detected at similar levels under all the examined light intensities (Figure 8B, arrows). These results indicate that Trx m4 downregulates the PGR5/PGRL1-dependent pathway during the steady state of photosynthesis, as well as in the dark.
DISCUSSION
Trx m Downregulates PGR5/PGRL1-Dependent PQ Reduction during Chloroplast Development
The assembly of the PSI and PSII protein complexes generally occurs during de-etiolation, but Trx m proteins, together with components of PSI-CET pathways, accumulated already in the etioplasts (Supplemental Figure 9; Kanervo et al., 2008; Plöscher et al., 2011). The trx m124-2 mutants exhibited low chlorophyll content and retarded growth because of disturbed chloroplast development (Figure 1). As observed in the variegated Arabidopsis immutans mutant defective in plastid terminal oxidase (PTOX; Carol et al., 1999; Wu et al., 1999), the oxidized PQ pool is necessary for carotenoid biosynthesis and normal chloroplast development. PTOX oxidizes PQ, and its collaboration with the two PSI-CET pathways is required to optimize the redox state of the PQ pool for normal chloroplast development (Okegawa et al., 2010; Kambakam et al., 2016). Because PSI is not fully assembled during early chloroplast development (Supplemental Figure 9; Kanervo et al., 2008; Plöscher et al., 2011), the mode of electron transport is not cyclic but linear to O2 (chlororespiration). Notably, the 35Spro:PGR5 lines exhibited leaf variegation (Okegawa et al., 2007). Blanco et al. (2013) also indicated that enhancement of PSI-CET caused a variegated leaf phenotype. Both A. immutans and the 35Spro:PGR5 lines accumulated phytoene, indicating a block in carotenoid biosynthesis at the phytoene desaturase step (Kambakam et al., 2016). In this study, the pgr5 mutation complemented the disturbed chloroplast development in trx m124-2 (Figure 1). We suggest that one of the physiological functions of Trx m–dependent downregulation of the PGR5/PGRL1-dependent pathway is similar to the role of PTOX, which is maintenance of the redox state of the PQ pool during early chloroplast development.
Trx m Regulates the PGR5/PGRL1-Dependent Pathway
The deficiency of Trx m caused the induction of high NPQ at low light intensities (Figure 2). A recent study indicated that activity of zeaxanthin epoxidase was impaired in the Trx m–deficient mutant, where a high proton motive force was also reported (Da et al., 2018). This may be related to the high NPQ phenotype (Niyogi et al., 1998). Also, in the Arabidopsis ntrc mutants where the activation of the Calvin–Benson cycle enzymes was suppressed, high NPQ was induced (Naranjo et al., 2016). Several mutants, defective in the Calvin–Benson cycle enzymes, were reported to induce high NPQ (Jin et al., 2008; Livingston et al., 2010). The Arabidopsis high cyclic electron flow1 (hcef1) mutant, defective in FBPase, was demonstrated to induce high NPQ due to the enhancement of NDH-dependent PSI-CET (Livingston et al., 2010). The NDH-dependent pathway was also stimulated in a mutant impaired in plastid fructose-1,6-bisP aldolase activity (Gotoh et al., 2010). In crr2 trx m124-2, however, high NPQ was still induced at low light intensities (Supplemental Figure 2), indicating that this high NPQ did not depend on the NDH-dependent pathway. By contrast, the high NPQ phenotype was completely suppressed in pgr5 trx m124-2 (Figure 2), suggesting that high NPQ in trx m124-2 was caused by the enhancement of the PGR5/PGRL1-dependent PSI-CET. In fact, the 35Spro:PGR5 lines also exhibited a high NPQ phenotype (Okegawa et al., 2007). NPQ is essential for photoprotection, but high NPQ under low light conditions limits plant growth (Roach and Krieger-Liszkay, 2012). To avoid the growth defects, NPQ is regulated by various mechanisms (Ruban, 2016). We propose the possibility that the amplitude of NPQ is appropriately maintained also by Trx m–dependent regulation of the PGR5/PGRL1-dependent pathway.
Trx m4 Directly Downregulates the PGR5/PGRL1-Dependent Pathway via an Interaction with PGRL1
We showed that Trx m4 formed a disulfide complex with PGRL1 in vivo (Figure 6). Formation of this complex was observed also in the in vitro assay using ruptured chloroplasts and purified Trx m proteins (Supplemental Figure 7). Consistent with the suppression effect of Trxs on Fd-dependent PQ reduction (Figures 5), Trxs m1 and m2 also formed complexes with PGRL1 in vitro (Supplemental Figure 7). On the basis of these observation, we propose that formation of Trx m-PGRL1 complex suppresses PGR5/PGRL1-dependent PQ reduction.
In contrast to the in vitro experiments (Supplemental Figure 7), in planta, Trx m4 formed a complex with PGRL1, but Trxs m1 and m2 did not (Figure 6; Supplemental Figure 5A). This discrepancy could be explained by comparison of deduced amino acid sequences and the midpoint redox potential (Em) of Trx m proteins. Compared to Trx m4, Trxs m1 and m2 show a higher sequence identity of amino acids in the mature protein region (Trxs m1 and m2, 81%; Trxs m1 and m4, 57%; and Trxs m2 and m4, 53%), and Trxs m1 and m2 have the same Em value (–335 mV), but Trx m4 has a higher Em value (–312 mV; Yoshida and Hisabori, 2017). Because of these properties of Trx m isoforms, Trxs m1, m2, and m4 may be able to interact with PGRL1 in vitro, but in planta, Trxs m1 and m2 may prefer to interact with other Trx target proteins, because there are many Trx targets other than PGRL1 in the chloroplasts. Thormählen et al. (2017) suggested that a main target of Trxs m1 and m2 is NADP-MDH from mutant analysis. Therefore, it is probably necessary to consider the competition between the Trx targets in vivo. Another possibility could be that the Trx m1-PGRL1 or Trx m2-PGRL1 complexes are more unstable than the Trx m4-PGRL1 complex, which would result in dissociation of these complexes during protein extraction from leaves. In fact, Trx m1 was shown to interact with PGRL1 in planta by a biomolecular fluorescence complementation test (Nikkanen et al., 2018). Since the growth defects were only evident in the trx m124-2 triple mutant, and not the trx m4 single or double mutants (Okegawa and Motohashi, 2015), Trxs m1 and m2 may interact with PGRL1 also in vivo, especially in the trx m4 mutant background.
Analysis of the PGRL1 Cys variants identified that Cys-123 of PGRL1 is the target residue for Trx m4 binding (Figures 7D and 7E). These results suggest that the PGR5/PGRL1-dependent pathway is not regulated by Trx m in the C123S and C22SC123S lines. The C123S and C22SC123S lines induced slightly higher NPQ than the wild type at light intensities less than 50 µmol photons m–2 s–1 (Figure 7C), but did not show the marked growth defects observed in trx m124-2. This is probably because the C123S and C22SC123S lines had lower PGR5 than the wild type, although a higher amount of PGRL1 accumulated (Figure 7A). More than 30 transgenic lines were analyzed, but transgenic plants of the C123S and C22SC123S lines accumulating the same level of PGR5 as the wild type could not be found. Cys-123 may also be required for the stability of PGR5, although either Cys-212/215 or Cys-240/243 was suggested to interact with PGR5 (Hertle et al., 2013). The detailed role of Cys-212/215 or Cys-240/243 in PGRL1 remains unclear, since transgenic plants carrying these Cys variants of PGRL1 has not been analyzed in vivo. We do not exclude the possibility that these Cys residues also may contribute to the redox regulation of PGRL1 and the PGR5/PGRL1-dependent pathway.
Cys-22 and Cys-123 also appear to be involved in the homodimerization of PGRL1, since the C22SC123S lines did not form PGRL1 dimers (Figure 7E; Supplemental Figure 6E). The C22SC123S PGRL1 variant complemented the pgrl1 mutant phenotype (Figures 7B and 7C), suggesting that PGRL1 dimer is dispensable for the function of PGRL1, at least under the studied conditions. In the wild type, the PGRL1 dimer was detected via chloroplast isolation (Supplemental Figure 7), but it was missing from the samples isolated directly from leaves (Figure 6). By contrast, the dimer could be detected in leaves of the 35Spro:PGRL1A, C22S, and C123S lines (Figure 7E). Thus, formation of a PGRL1 dimer is likely to be transient and prone to dissociation during extraction from leaves. A similar case was also reported for the thylakoid-associated protein kinase STN7. Although STN7 dimer was undetectable in vivo, STN7 dimerization was shown to regulate STN7 activity (Shapiguzov et al., 2016). Hertle et al. (2013) proposed that Trx m monomerized PGRL1 dimer and promoted Fd-dependent PQ reductase activity. Although the function of PGRL1 dimer in the regulation of PSI-CET remains unclear, our results indicate that reduced Trx m4 directly downregulated the PGR5/PGRL1-dependent pathway by forming a heterodimer with PGRL1.
Physiological Function of Trx m4–Dependent Regulation of the PGR5/PGRL1-Dependent Pathway
Compared to Trx m4-PGRL1 complex, monomers of PGRL1 were more abundant (Figures 6 and 8). It is unclear what the physiological significance of Trx m4–dependent regulation of a small part of PGRL1 is. PGR5 has been proposed to be associated with PGRL1 and required for Fd oxidation in the PGR5/PGRL1-dependent PSI-CET (Hertle et al., 2013). PGR5 is considerably less than PGRL1 (0.09 versus 0.70 mmol/mol chlorophyll; Hertle et al., 2013). Similarly, Trx m4 was also less than PGRL1 (0.03 mmol/mol chlorophyll; Okegawa and Motohashi, 2015). Based on the stoichiometry of these proteins, the formation of Trx m4-PGRL1 may sufficiently contribute to the downregulation of the PGR5/PGRL1-dependent pathway in vivo. In fact, analysis of Fd-dependent PQ reduction in ruptured chloroplasts showed that only part of PGRL1 formed a complex with Trx m4 (Supplemental Figure 7), but Fd-dependent PQ reduction activity was substantially suppressed (Figure 5A).
The Trx m4-PGRL1 complex was transiently dissociated during the induction of photosynthesis but formed again within 30 min after illumination (Figure 8A; Supplemental Figure 8A). These results suggest that PGR5/PGRL1-dependent PSI-CET is enhanced during the induction of photosynthesis. This is supported by previous studies, which demonstrated that PSI-CET operates efficiently during the induction of photosynthesis (Heber and Walker, 1992; Joliot and Joliot, 2002). Furthermore, the dynamics of the Trx m4-PGRL1 complex during the induction of photosynthesis were consistent with the time course of the induction and relaxation of NPQ (Supplemental Figure 8). After a shift from dark to light, NPQ was transiently induced and was relaxed within several minutes (Supplemental Figure 8B). Although relaxation of ∆pH via ATP synthase in response to activation of the Calvin–Benson cycle mainly affects the relaxation of NPQ (Joliot and Joliot, 2006), the regulation of the PGR5/PGRL1-dependent pathway by Trx m4 might also contribute to the dynamics of NPQ during the induction of photosynthesis to some extent.
Typical chloroplast-localized Trxs reduce the disulfide bonds of target proteins, so the observation that Trx m4 forms a complex with PGRL1 seems unusual, raising questions on dissociation and reformation of this complex. The chloroplastic atypical Trx (ACHT1) was also demonstrated to form a complex with target proteins in vivo (Dangoor et al., 2012). ACHT1 functions as an oxidative sensor in the chloroplast, by forming a complex with 2-Cys peroxiredoxin, which detoxifies hydrogen peroxide. Formation of the ACHT1-2-Cys peroxiredoxin complex was linked to changes in the photosynthetic production of peroxides. We propose a possible model for the regulation of PGRL1 protein by Trx m4 (Supplemental Figure 10). After a shift from dark to light, a disulfide bond between Trx m4 and PGRL1 is cleaved by the reducing equivalents from the photosynthetic electron transport chain, resulting in transient dissociation of the Trx m4-PGRL1 complex during the induction of photosynthesis. Since electron transport from FTR to Trx m4 is more efficient than to other Trx m isoforms (Yoshida and Hisabori, 2017), Trx m4 may have an advantage in the regulation of PGRL1 to activate the PGR5/PGRL1-dependent pathway, soon after a shift from dark to light. However, when the stromal redox state reaches equilibrium (during the steady state photosynthesis), the photochemically reduced Trx m4 could again reduce PGRL1, and form the complex. Indeed, the Trx m4-PGRL1 complex accumulated stably during the steady state photosynthesis, as well as in the dark (Figure 8B). The Em of PGRL1 (–270 mV at pH 7.0; Hertle et al., 2013) is similar to that of spinach (Spinacia oleracea) Trx m (–282 mV at pH 7.0; Motohashi and Hisabori, 2006). During the induction of photosynthesis, the Em values for PGRL1 and Trx m4 are expected to change, due to the dynamic changes in the stromal pH and the redox state of the stroma. As such, the inversion of the redox potential values between PGRL1 and Trx m4 may determine dissociation and association of the Trx m4-PGRL1 complex.
In the dark, the Trx m4-PGRL1 complex accumulated consistently (Figure 8), suggesting that the PGR5/PGRL1-dependent pathway is downregulated in the dark. This is consistent with measurement results of PSI-CET in the dark. The afterglow luminescence signal and the post-illumination increase in chlorophyll fluorescence (which are measured in the dark) are used to probe NDH-dependent PSI-CET (Shikanai et al., 1998; Havaux et al., 2005). However, these methods are not applicable for measuring PGR5/PGRL1-dependent PSI-CET, because these techniques cannot detect its activity in Arabidopsis (Hashimoto et al., 2003; Havaux et al., 2005). Taken together with our results that Trx m4 formed a complex with PGRL1 in the dark (Figure 8), these observations suggest that the PGR5/PGRL1-dependent pathway may need to be downregulated in the dark.
The Trx m4-PGRL1 complex could also be observed during the steady state of photosynthesis, regardless of the light intensities (Figure 8B). Under the same conditions, the reduction rate of thiol enzymes was significantly higher in pgr5 trx m124-2 than in trx m124-2 (Figure 3), indicating that the activity of thiol enzymes was promoted in pgr5-defective trx m124-2. The FTR-dependent Trx system, as well as the PGR5/PGRL1-dependent pathway, accepts electrons from Fd (Hanke and Mulo, 2013). Other than the above-mentioned two processes, many enzymes depend on Fd for reducing power in the chloroplast, such as Fd-NADPH reductase (FNR), sulfite reductase, nitrite reductase, and Glu synthase (Mondal and Bruce, 2018). Therefore, the regulation of electron partitioning from Fd is important for efficient photosynthesis and other metabolic pathways. Although studies on the structural analysis of interactions between Fd and its partner enzymes have progressively accumulated, the mechanism of electron distribution from Fd to these diverse processes is largely unknown. Since the pgr5 mutation caused a Trx-dependent activation of thiol enzymes in the trx m124-2 mutant background, the Fd-dependent Trx system may compete with PSI-CET pathways over the oxidation of Fd in the wild type. Trx m4 may support other Trxs in accepting electrons from Fd via FTR and activating their target enzymes through downregulation of the PGR5/PGRL1-dependent pathway. From these results, we suggest the possibility that Trx m4 fine-tunes electron partitioning from Fd to distinct processes, including the Fd-Trx system, by regulating the PGR5/PGRL1-dependent pathway. Recently, Guan et al. (2018) demonstrated that a newly identified Fd homolog with an extra C terminus, AtFdC1, interacted with FTR, PGR5, and PGRL1B, but not with FNR, which is different from other leaf-type Fds. Analysis of AtFdC1 may provide insight into the function of Trx m4 in the electron partitioning from Fd. Further analysis is required to elucidate the physiological function of Trx m4–dependent regulation of PGR5/PGRL1-dependent PSI-CET.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type. The trx m124-2 (Okegawa and Motohashi, 2015) and pgrl1ab (Hertle et al., 2013) mutants are in the Columbia-0 background, whereas the crr2-2 (Hashimoto et al., 2003) and pgr5 (Munekage et al., 2002) mutants are in the Columbia gl1 background. Plants were either grown in soil or in Petri dishes containing Murashige and Skoog (MS) medium with 1.0% (w/v) agar and 1.0% (w/v) Suc, and grown in growth chambers (50 µmol photons m–2 s–1 of white light, continuous light, 23°C, and 55% humidity) for 3 to 4 weeks. The seeds sown on agar were surface sterilized by soaking them in 5% (v/v) sodium hypochlorite for 15 min. For analysis of fresh weight, chlorophyll content, and electron microscopy, 3-week-old plants were used. For other analyses, 4-week-old-plants were used.
To generate quadruple mutants, the trx m124-2 triple mutant was crossed either with the crr2-2 or pgr5 mutant, and the crr2-2 trx m124-2 and pgr5 trx m124-2 mutants were selected from the F2 plants. The presence of T-DNA insertions and the mutations were confirmed by PCR (for primers, see Supplemental Table 3).
The T-DNA lines mentioned in the article were obtained from the Salk Institute Genomic Analysis Laboratory under the stock numbers SALK_087118 (trx m1; Laugier et al., 2013) and SALK_023810 (trx m4; Laugier et al., 2013) and the Nottingham Arabidopsis Stock Centre under the stock number SAIL_100_E02 (trx m2-2; Okegawa and Motohashi, 2015). A schematic diagram of the structure of the T-DNA lines is provided in Supplemental Figure 11.
Vector Construction and Transformation of A. thaliana
All primers used in this study are listed in Supplemental Table 3. The coding region of PGPL1A excluding the transit peptide sequence was amplified by PCR with specific primers. Primers for all site-directed mutagenesis were designed using the PrimerX on-line software (http://www.bioinformatics.org/primerx), and all site-directed mutagenesis was performed using the SLiP-method (Motohashi, 2017). DNA fragments harboring the wild type and mutagenized PGRL1A coding region sequences were cloned into the binary vector pART27 (Gleave, 1992) and introduced via Agrobacterium tumefaciens C58 into pgrl1ab (DalCorso et al., 2008) using the floral dip procedure (Clough and Bent, 1998). Transgenic seedlings were selected based on their resistance to kanamycin on selection medium (MS + 1.0% [w/v] Suc + 50 mg/L kanamycin). More than 20 transgenic plants were isolated for each line. The T2 generation was used for analysis.
Analysis of Chlorophyll Content
Leaves (30 mg fresh weight) were harvested from 3-week-old seedlings grown on MS plates and were immediately powdered by grinding in liquid nitrogen. Chlorophyll was extracted in 80% (v/v) acetone and collected by centrifugation at 15,000g for 5 min at 4°C. The residue was re-extracted with 80% (v/v) acetone and centrifuged once again (15,000g for 5 min at 4°C). Chlorophyll content was determined by spectrophotometry as described by Porra et al. (1989).
Isolation of Intact Chloroplasts
Leaves of 4-week-old plants were homogenized by a Polytron PT 10-35 GT homogenizer (Kinematica) in 20 mM Tricine-NaOH, pH 8.4, containing 400 mM sorbitol, 5 mM MgCl2, 5 mM MnCl2, 2 mM EDTA, 10 mM NaHCO3, 0.5% (w/v) BSA, and 5 mM ascorbate. After centrifugation at 3000g for 5 min at 4°C, the pellet was gently resuspended in 50 mM HEPES-KOH, pH 7.6, containing 400 mM sorbitol, 5 mM MgCl2, and 2.5 mM EDTA. Isolated intact chloroplasts were suspended in 25 mM HEPES-KOH, pH 7.6, containing 3 mM MgCl2. The insoluble fraction containing thylakoids and envelopes was separated from the stroma fraction by centrifugation at 10,000g for 3 min at 4°C. The thylakoid membrane proteins and stromal proteins were solubilized in SDS-PAGE sample buffer (62.5 mM Tris-HCl, pH 6.8, containing 2% [w/v] SDS, 10% [v/v] glycerol, and 0.0005% [w/v] bromophenol blue) with 5% (v/v) 2-mercaptoethanol.
SDS-PAGE and Immunoblot Analysis
Proteins were separated by SDS-PAGE, using the conventional Laemmli (Tris-Gly) system (Laemmli, 1970) or using the Tris-tricine buffer system (for PGR5 detection; Schägger and von Jagow, 1987), and transferred onto polyvinylidene difluoride membranes by using a semidry blotting apparatus. Polyclonal antibodies against PsbO (AS05 092) and PsaD (AS09 461) were obtained from Agrisera. Immunoblot signal was visualized using the Immobilon western chemiluminescent horseradish peroxidase substrate (EMD Millipore) or ECL Plus protein gel blotting detection kit (GE Healthcare; Whitehead et al., 1983). The chemiluminescence was detected with a LAS-3000 UV mini lumino-image analyzer (Fujifilm) and analyzed by the Multi Gauge 3.1 software (Fujifilm).
Preparation of Proteins and Antibodies
All recombinant Trx proteins in this study were purified as previously described by Motohashi and Okegawa (2014) and Okegawa and Motohashi (2015). Specific antibodies against Trx isoforms, SBPase, FBPase, NADP-MDH, PetC, and ATPC1 were prepared as previously described by Okegawa and Motohashi (2015). Polyclonal anti-RbcS was raised in rabbits, against the synthetic peptide NSPGYYDGRYWTMWKLPLFGC. Polyclonal anti-PGRL1 was raised in rabbits, against the synthetic peptide CTEQSGPVGGDNVDSN, located at the N terminus of PGRL1A. To remove nonspecific bands, PGRL1A peptide antibodies were prepared by affinity purification using the immobilized synthetic peptide of PGRL1A (TEQSGPVGGDNVDSNVL) on EAH-Sepharose 4B medium (GE Healthcare). Polyclonal anti-PGR5 was raised in rabbits, against the PGR5 recombinant protein. The four tandem repeats of the mature form of PGR5 (amino acids 61 to 133) synthesized by gBlocks Gene Fragments (Integrated DNA Technologies; Supplemental Table 4) were cloned into the pET23c vector. Expression of the PGR5 recombinant protein in Escherichia coli BL21-CodonPlus (DE3)-RIL strain was induced by 0.5 mM isopropyl β-d-thiogalactopyranoside at an OD600 of 0.8. Next, E. coli cells were grown for 3 h at 37°C and harvested by centrifugation at 5000g for 10 min at 4°C. The expressed four tandem repeat PGR5 protein (32 kD) obtained as the inclusion bodies was washed with a buffer containing 2% (w/v) Triton X-100 as previously described by Okegawa et al. (2016) and further washed with 25 mM Tris-HCl, pH 7.5, containing 8 M urea and 2 mM EDTA. The washed inclusion bodies were solubilized in 25 mM Tris-HCl, pH 7.5, containing 6 M guanidinium chloride. The purity of the proteins was checked by SDS-PAGE, and this protein was used as antigen.
Electron Microscopy
TEM analysis was conducted by the Tokai Electron Microscopy service, using chemical fixation. For TEM, Arabidopsis rosette leaves from 3-week-old plants grown on MS plates were cut into 1 × 2-mm pieces and fixed in 2% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.05 M cacodylic acid buffer, pH 7.4, at 4°C overnight. After this fixation, the samples were postfixed in 2% (v/v) osmium tetroxide, in the same buffer, at 4°C for 3 h. Samples were further dehydrated with graded ethanol solutions (50, 70, 90, and 100% [v/v]). Ethanol was then replaced for propylene oxide (PO), and the samples were infiltrated with a 70:30 mixture of PO:resin (Quetol 651; Nissin EM) for 1 h, after which the PO was volatilized overnight. Next, the resin was polymerized for 48 h at 60°C. Ultrathin sections (80 nm) were cut with an ultramicrotome (Ultracut UCT; Leica) and stained with 2% (w/v) uranyl acetate for 15 min. Sections were examined using a transmission electron microscope (JEM-1400Plus; JEOL) at 100 kV.
Analysis of the in Vivo Chlorophyll Fluorescence
Chlorophyll fluorescence was measured with a Mini-pulse amplitude modulation portable chlorophyll fluorometer (Walz). Minimum fluorescence (Fo) was obtained from the open PSII reaction centers in the dark-adapted state, by a nonactinic measuring light (650 nm; 0.05 to 0.1 µmol photons m–2 s–1). A saturating pulse (SP) of white light (800 ms; 3000 µmol photons m–2 s–1) was applied to determine the maximum fluorescence with closed PSII centers in the dark-adapted state (Fm) and during illumination with actinic light (AL; Fm′). The steady state fluorescence level (Fs) was recorded during AL illumination. PSII quantum yield (ФPSII) was calculated as (Fm′ – Fs)/Fm′ (Genty et al., 1989). The relative ETR through PSII was calculated as ФPSII × light intensity (µmol photons m–2 s–1). NPQ was calculated as (Fm – Fm′)/Fm′. The dependence of fluorescence parameters on the light intensity was analyzed by increasing AL intensity in a stepwise manner, every 2 min after the application of SP (15 to 955 µmol photons m–2 s–1). The NPQ induction was measured at a light intensity of 62 µmol photons m–2 s–1 for 20 min and NPQ recovery in the dark was recorded for 8 min.
In Vivo Photo-Reduction of the Thiol Enzymes
Photo-reduction of the thiol enzymes in seedlings was determined using AMS as previously described by Okegawa and Motohashi (2015), with minor modifications. In a light intensity-dependent experiment (Figure 3), seedlings were dark incubated (for 8 h) and then illuminated at three different light intensities (50, 200, or 800 µmol photons m–2 s–1) for 1 h each. In a time-dependent experiment during the induction of photosynthesis (Figure 4), seedlings were dark adapted for 8 h and exposed to light (200 µmol photons m–2 s–1) for up to 30 min. Samples were then collected at the indicated conditions and frozen in liquid nitrogen. Frozen samples were ground by tungsten beads (5-mm diameter) and incubated with 125 mM Tris-HCl, pH 6.8, containing 10 mM AMS (Thermo Fisher Scientific), 10 mM EDTA, 4% (w/v) SDS, 8 M urea, and 1% (v/v) plant protease inhibitor (Sigma-Aldrich) for 2 h at 25°C. Hydrophilic proteins were separated from the lipid fraction by the addition of an equal volume of chloroform, and the aqueous portion was collected by centrifugation (at 15,000g for 10 min at 4°C). Proteins were solubilized in 2× SDS-PAGE sample buffer. The protein samples were separated by nonreducing SDS-PAGE (10 or 15% [w/v] acrylamide) and detected by immunoblotting. The reduction level of the proteins was quantified using Multi Gauge 3.1 software and is presented as the ratio of the reduced per the total protein.
In Vitro Fd-Dependent PQ Reduction Assay
The in vitro Fd-dependent PQ reduction assay was performed as described previously, with minor modifications, by Okegawa et al. (2008). Intact chloroplasts (20 µg of chlorophyll mL–1) were osmotically ruptured in 50 mM HEPES-NaOH, pH 8.0, containing 7 mM MgCl2, 1 mM MnCl2, 2 mM EDTA, 30 mM KCl, and 0.25 mM KH2PO4, for each assay. As electron donors, 5 µM spinach (Spinacia oleracea) Fd (Sigma-Aldrich) and 0.25 mM NADPH (Oriental kobo) were used. In this assay system, NADPH is essential as an electron donor to Fd via FNR (Miyake and Asada, 1994). Increases in the chlorophyll fluorescence—through the addition of NADPH and Fd, under weak light (650 nm; 1.0 µmol photons m–2 s–1)—were monitored by a Mini-pulse amplitude modulation portable chlorophyll fluorometer. At this light intensity, the fluorescence level predominantly reflects the PQ reduction by electron transport from Fd, not by PSII photochemistry (Munekage et al., 2002) The fluorescence levels were standardized by the Fm levels and monitored by the application of SP. To reduce or oxidize the purified recombinant Trxs, Trxs were pretreated with 100 µM DTT or 10 µM CuCl2 at 25°C for 30 min; 5.0 µM of the reduced or oxidized Trxs was added to the ruptured chloroplasts before the measurement.
Isolation of Trx m–Interacting Proteins and Detection of the Trx m4-PGRL1 Complex
For this analysis, 4-week-old seedlings were harvested in the dark period and after 1-h illumination with growth light (50 µmol photons m–2 s–1; Figures 6 and 7) or at the indicated time or light conditions (Figure 8). Samples were frozen in liquid nitrogen and treated in the same way as for in vivo photo-reduction of the thiol enzymes (Figures 3 and 4). AMS was added to block the free thiols. The protein samples were separated by nonreducing SDS-PAGE (10 or 15% [w/v] acrylamide) and detected by immunoblot analysis using Trx m4 and PGRL1 antibodies.
Statistical Analysis
Calculations were performed on more than three independent biological replicates (see figure legends). One-way ANOVA with Tukey–Kramer multiple comparison test was used to determine significant differences among the materials tested (P < 0.05). Statistical data are provided in Supplemental Tables 5 and 6.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Trx m1 (AT1G03680); Trx m2 (AT4G03520); Trx m4 (AT3G15360); CRR2 (AT3G46790); PGR5 (AT2G05620); PGRL1A (AT4G22890); PGRL1B (AT4G11960).
Supplemental Data
Supplemental Figure 1. Phenotypes of the pgrl1ab trx m124-2 mutant.
Supplemental Figure 2. Light intensity dependence of NPQ in WT, crr2-2, pgr5, trx m124-2 and crr2-2 trx m124-2 mutants.
Supplemental Figure 3. Activity of Arabidopsis chloroplastic Trxs to reduce disulfide bonds.
Supplemental Figure 4. Fd-dependent PQ reduction assay in the ruptured chloroplasts.
Supplemental Figure 5. Identification of Trx m target proteins.
Supplemental Figure 6. Characterization of the 35Spro:PGRL1A Cys variant lines.
Supplemental Figure 7. Complex formation between the Trxs and PGRL1, in ruptured chloroplasts.
Supplemental Figure 8. The dynamics of the Trx m4-PGRL1 complex and NPQ during the induction of photosynthesis.
Supplemental Figure 9. Accumulation of photosynthesis-related proteins during the de-etiolation of etioplasts into chloroplasts.
Supplemental Figure 10. A hypothetical model for Trx m4-mediated regulation of PGRL1.
Supplemental Figure 11. Schematic diagram of the structure of Trx m T-DNA mutants.
Supplemental Table 1. Growth phenotype of the wild type, trx m124-2, crr2-2 trx m124-2 and pgr5 trx m124-2 mutants grown under short day and medium light conditions.
Supplemental Table 2. Fresh weight of the wild type, trx m124-1, crr2-2 trx m124-1 and pgr5 trx m124-1 mutants.
Supplemental Table 3. Primers used in this study.
Supplemental Table 4. DNA sequence of the four tandem repeats of the mature form of PGR5 (amino acids 61 to 133) synthesized by gBlocks Gene Fragments.
Supplemental Table 5. Parameters for the statistical analyses in Figure 1B.
Supplemental Table 6. Parameters for the statistical analyses in Figure 1C.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Author Contributions
Y.O. and K.M. designed the research; Y.O. performed the experiments; Y.O and K.M. analyzed the data; and Y.O and K.M. wrote the article.
Acknowledgements
We are grateful to Toshiharu Shikanai (Kyoto University) for the fruitful discussion and critical reading of the manuscript. We are also grateful to Hiroshi Yamamoto (Kyoto University) for suggesting the method of PGR5 antibody production. We thank Dario Leister (Ludwig-Maximilians University) for providing the pgrl1ab mutant seeds and Tsuyoshi Endo (Kyoto University) for providing NDF antibody. This work was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Young Scientists (B) (grant JP16K18573 to Y.O.) and for Scientific Research on Innovative Areas (grants JP17H05730 and JP19H04733 to Y.O.) and the Ministry of Education, Culture, Sports, Science and Technology Supported Program for the Strategic Research Foundation at Private Universities (grant S1511023 to K.M.)
References
- Arnon D.I., Allen M.B., Whatley F.R.(1954). Photosynthesis by isolated chloroplasts. Nature 174: 394–396. [DOI] [PubMed] [Google Scholar]
- Blanco N.E., Ceccoli R.D., Vía M.V., Voss I., Segretin M.E., Bravo-Almonacid F.F., Melzer M., Hajirezaei M.R., Scheibe R., Hanke G.T.(2013). Expression of the minor isoform pea ferredoxin in tobacco alters photosynthetic electron partitioning and enhances cyclic electron flow. Plant Physiol. 161: 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton C., Nandha B., Johnson G.N., Joliot P., Finazzi G.(2006). Redox modulation of cyclic electron flow around photosystem I in C3 plants. Biochemistry 45: 13465–13475. [DOI] [PubMed] [Google Scholar]
- Buchanan B.B.(2016). The path to thioredoxin and redox regulation in chloroplasts. Annu. Rev. Plant Biol. 67: 1–24. [DOI] [PubMed] [Google Scholar]
- Carol P., Stevenson D., Bisanz C., Breitenbach J., Sandmann G., Mache R., Coupland G., Kuntz M.(1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejudo F.J., Ojeda V., Delgado-Requerey V., González M., Pérez-Ruiz J.M.(2019). Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 10: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Courteille A., Vesa S., Sanz-Barrio R., Cazalé A.C., Becuwe-Linka N., Farran I., Havaux M., Rey P., Rumeau D.(2013). Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol. 161: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Q., Sun T., Wang M., Jin H., Li M., Feng D., Wang J., Wang H.B., Liu B.(2018). M-Type thioredoxins are involved in the xanthophyll cycle and proton motive force to alter NPQ under low-light conditions in Arabidopsis. Plant Cell Rep. 37: 279–291. [DOI] [PubMed] [Google Scholar]
- DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., Leister D.(2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285. [DOI] [PubMed] [Google Scholar]
- Dangoor I., Peled-Zehavi H., Wittenberg G., Danon A.(2012). A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24: 1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P., Fernie A.R.(2014). Metabolic control of redox and redox control of metabolism in plants. Antioxid. Redox Signal. 21: 1389–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.M., Baker N.R.(1989). The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92. [Google Scholar]
- Gleave A.P.(1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gotoh E., Matsumoto M., Ogawa K., Kobayashi Y., Tsuyama M.(2010). A qualitative analysis of the regulation of cyclic electron flow around photosystem I from the post-illumination chlorophyll fluorescence transient in Arabidopsis: A new platform for the in vivo investigation of the chloroplast redox state. Photosynth. Res. 103: 111–123. [DOI] [PubMed] [Google Scholar]
- Guan X., Chen S., Voon C.P., Wong K.B., Tikkanen M., Lim B.L.(2018). FdC1 and leaf-type ferredoxins channel electrons from photosystem I to different downstream electron acceptors. Front. Plant Sci. 9: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke G., Mulo P.(2013). Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 36: 1071–1084. [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T.(2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36: 541–549. [DOI] [PubMed] [Google Scholar]
- Havaux M., Rumeau D., Ducruet J.M.(2005). Probing the FQR and NDH activities involved in cyclic electron transport around photosystem I by the ‘afterglow’ luminescence. Biochim. Biophys. Acta 1709: 203–213. [DOI] [PubMed] [Google Scholar]
- Heber U., Walker D.(1992). Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol. 100: 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertle A.P., Blunder T., Wunder T., Pesaresi P., Pribil M., Armbruster U., Leister D.(2013). PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 49: 511–523. [DOI] [PubMed] [Google Scholar]
- Horton P., Ruban A.V., Walters R.G.(1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 655–684. [DOI] [PubMed] [Google Scholar]
- Jin S.-H., Wang D., Zhu F.-Y., Li X.-Q., Sun J.-W., Jiang D.A.(2008). Up-regulation of cyclic electron flow and down-regulation of linear electron flow in antisense-rca mutant rice. Photosynthetica 46: 506–510. [Google Scholar]
- Joët T., Cournac L., Horvath E.M., Medgyesy P., Peltier G.(2001). Increased sensitivity of photosynthesis to antimycin A induced by inactivation of the chloroplast ndhB gene. Evidence for a participation of the NADH-dehydrogenase complex to cyclic electron flow around photosystem I. Plant Physiol. 125: 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Johnson G.N.(2011). Regulation of cyclic and linear electron flow in higher plants. Proc. Natl. Acad. Sci. USA 108: 13317–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Joliot A.(2002). Cyclic electron transfer in plant leaf. Proc. Natl. Acad. Sci. USA 99: 10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Joliot A.(2006). Cyclic electron flow in C3 plants. Biochim. Biophys. Acta 1757: 362–368. [DOI] [PubMed] [Google Scholar]
- Kambakam S., Bhattacharjee U., Petrich J., Rodermel S.(2016). PTOX mediates novel pathways of electron transport in etioplasts of Arabidopsis. Mol. Plant 9: 1240–1259. [DOI] [PubMed] [Google Scholar]
- Kanervo E., Singh M., Suorsa M., Paakkarinen V., Aro E., Battchikova N., Aro E.M.(2008). Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol. 49: 396–410. [DOI] [PubMed] [Google Scholar]
- Krause G.H., Weis E.(1991). Chlorophyll fluorescence and photosynthesis - The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 313–349. [Google Scholar]
- Laemmli U.K.(1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Laugier E., Tarrago L., Courteille A., Innocenti G., Eymery F., Rumeau D., Issakidis-Bourguet E., Rey P.(2013). Involvement of thioredoxin y2 in the preservation of leaf methionine sulfoxide reductase capacity and growth under high light. Plant Cell Environ. 36: 670–682. [DOI] [PubMed] [Google Scholar]
- Livingston A.K., Cruz J.A., Kohzuma K., Dhingra A., Kramer D.M.(2010). An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake C., Asada K.(1994). Ferredoxin-dependent photoreduction of the monodehydroascorbate radical in spinach thylakoids. Plant Cell Physiol. 35: 539–549. [Google Scholar]
- Mondal J., Bruce B.D.(2018). Ferredoxin: The central hub connecting photosystem I to cellular metabolism. Photosynthetica 56: 279–293. [Google Scholar]
- Motohashi K.(2017). Seamless ligation cloning extract (SLiCE) method using cell lysates from laboratory Escherichia coli strains and its application to SLiP site-directed mutagenesis. Methods Mol. Biol. 1498: 349–357. [DOI] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T.(2006). HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281: 35039–35047. [DOI] [PubMed] [Google Scholar]
- Motohashi K., Okegawa Y.(2014). Method for enhancement of plant redox-related protein expression and its application for in vitro reduction of chloroplastic thioredoxins. Protein Expr. Purif. 101: 152–156. [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T.(2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582. [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T.(2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371. [DOI] [PubMed] [Google Scholar]
- Naranjo B., Mignée C., Krieger-Liszkay A., Hornero-Méndez D., Gallardo-Guerrero L., Cejudo F.J., Lindahl M.(2016). The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ. 39: 804–822. [DOI] [PubMed] [Google Scholar]
- Nikkanen L., Toivola J., Trotta A., Diaz M.G., Tikkanen M., Aro E.M., Rintamäki E.(2018). Regulation of cyclic electron flow by chloroplast NADPH-dependent thioredoxin system. Plant Direct 2: e00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K., Grossman A.R., Björkman O.(1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y., Kagawa Y., Kobayashi Y., Shikanai T.(2008). Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol. 49: 825–834. [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Kobayashi Y., Shikanai T.(2010). Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J. 63: 458–468. [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Koshino M., Okushima T., Motohashi K.(2016). Application of preparative disk gel electrophoresis for antigen purification from inclusion bodies. Protein Expr. Purif. 118: 77–82. [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Long T.A., Iwano M., Takayama S., Kobayashi Y., Covert S.F., Shikanai T.(2007). A balanced PGR5 level is required for chloroplast development and optimum operation of cyclic electron transport around photosystem I. Plant Cell Physiol. 48: 1462–1471. [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Motohashi K.(2015). Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. Plant J. 84: 900–913. [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz J.M., Spínola M.C., Kirchsteiger K., Moreno J., Sahrawy M., Cejudo F.J.(2006). Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöscher M., Reisinger V., Eichacker L.A.(2011). Proteomic comparison of etioplast and chloroplast protein complexes. J. Proteomics 74: 1256–1265. [DOI] [PubMed] [Google Scholar]
- Porra R., Thompson W., Kriedemann P.(1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Roach T., Krieger-Liszkay A.(2012). The role of the PsbS protein in the protection of photosystems I and II against high light in Arabidopsis thaliana. Biochim. Biophys. Acta 1817: 2158–2165. [DOI] [PubMed] [Google Scholar]
- Ruban A.V.(2016). Nonphotochemical chlorophyll fluorescence quenching: Mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 170: 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G.(1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379. [DOI] [PubMed] [Google Scholar]
- Serrato A.J., Pérez-Ruiz J.M., Spínola M.C., Cejudo F.J.(2004). A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279: 43821–43827. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A., Chai X., Fucile G., Longoni P., Zhang L., Rochaix J.D.(2016). Activation of the Stt7/STN7 kinase through dynamic interactions with the cytochrome b6f complex. Plant Physiol. 171: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A.(1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95: 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Järvi S., Grieco M., Nurmi M., Pietrzykowska M., Rantala M., Kangasjärvi S., Paakkarinen V., Tikkanen M., Jansson S., Aro E.M.(2012). PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24: 2934–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa K., Tsujimoto H.Y., Arnon D.I.(1963). Role of chloroplast ferredoxin in the energy conversion process of photosynthesis. Proc. Natl. Acad. Sci. USA 49: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira Y., Okegawa Y., Sugimoto K., Abe M., Miyoshi H., Shikanai T.(2013). Antimycin A-like molecules inhibit cyclic electron transport around photosystem I in ruptured chloroplasts. FEBS Open Bio 3: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen I., Zupok A., Rescher J., Leger J., Weissenberger S., Groysman J., Orwat A., Chatel-Innocenti G., Issakidis-Bourguet E., Armbruster U., Geigenberger P.(2017). Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol. Plant 10: 168–182. [DOI] [PubMed] [Google Scholar]
- Whitehead T., Thopre G., Carter T., Groucutt C., Kricka L.(1983). Enhanced luminescence procedure for sensitive determination of peroxidase-labelled conjugates in immunoassay. Nature 305: 158–159. [Google Scholar]
- Wu D., Wright D.A., Wetzel C., Voytas D.F., Rodermel S.(1999). The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Peng L., Fukao Y., Shikanai T.(2011). An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Shikanai T.(2019). PGR5-dependent cyclic electron flow protects photosystem I under fluctuating light at donor and acceptor sides. Plant Physiol. 179: 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Hisabori T.(2016). Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. USA 113: E3967–E3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Hisabori T.(2017). Distinct electron transfer from ferredoxin-thioredoxin reductase to multiple thioredoxin isoforms in chloroplasts. Biochem. J. 474: 1347–1360. [DOI] [PubMed] [Google Scholar]