SPIKE1, a DOCK family guanine nucleotide exchange factor, interacts with and activates the GTPase LjROP6 to mediate the polar progression of infection threads in Lotus japonicus.
Abstract
In legumes, rhizobia attach to root hair tips and secrete nodulation factor to activate rhizobial infection and nodule organogenesis. Endosymbiotic rhizobia enter nodule primordia via a specialized transcellular compartment known as the infection thread (IT). The IT elongates by polar tip growth, following the path of the migrating nucleus along and within the root hair cell. Rho-family ROP GTPases are known to regulate the polarized growth of cells, but their role in regulating polarized IT growth is poorly understood. Here, we show that LjSPK1, a DOCK family guanine nucleotide exchange factor (GEF), interacts with three type I ROP GTPases. Genetic analyses showed that these three ROP GTPases are involved in root hair development, but only LjROP6 is required for IT formation after rhizobia inoculation. Misdirected ITs formed in the root hairs of Ljspk1 and Ljrop6 mutants. We show that LjSPK1 functions as a GEF that activates LjROP6. LjROP6 enhanced the plasma membrane localization LjSPK1 in Nicotiana benthamiana leaf cells and Lotus japonicus root hairs, and LjSPK1 and LjROP6 interact at the plasma membrane. Taken together, these results shed light on how the LjROP6-LjSPK1 module mediates the polarized growth of ITs in L. japonicus.
INTRODUCTION
To form symbiotic root nodules, legumes and rhizobia initiate their symbiotic interaction via a molecular dialog. The host legume secretes flavonoid compounds that function as signals sensed by rhizosphere rhizobia, and the expression of nodulation (Nod) genes is induced for the biosynthesis and secretion of nodulation factors (NF)s. NFs are lipochito-oligosaccharide molecular signals that are perceived by NF receptors. NFs initiate a series of plant signaling and developmental events, including root hair deformation, membrane depolarization, intracellular calcium oscillations, and cortex cell division. These events lead to the formation of the nodule primordium (Ehrhardt et al., 1992; Downie and Walker, 1999; Timmers et al., 1999; Cullimore et al., 2001; Esseling et al., 2003; Oldroyd and Downie, 2008).
In temperate legumes such as Lotus japonicus and Medicago truncatula, rhizobia attach to the host’s root hair tips and redirect root hair growth to entrap the rhizobia within an infection chamber (Esseling et al., 2003; Murray, 2011; Fournier et al., 2015). Within the root hair chamber, an infection pocket filled with bacteria forms from a tubular invagination of the cell wall and membrane; this structure is known as an infection thread (IT). The IT extends from the infection chamber down through the root hairs to the root cortex, where a nodule primordium is produced (Fournier et al., 2008; Fournier et al., 2015). The bacteria grow, divide, and fill the IT. The bacteria are then released from the IT into the nodule primordium via an unwalled droplet (Brewin, 2004). This leads to the formation of a nitrogen-fixing root nodule that provides the proper microenvironment for nitrogen fixation by rhizobia and nutrient exchange between the two partners (Jones et al., 2007; Oldroyd, 2013; Roy et al., 2020).
Genetic studies of L. japonicus and M. truncatula have revealed several genes that are required for IT formation, including LjNAP/MtRIT1, LjPIR, LjSCARN, and LjARPC1. These genes encode components of the WAVE/SCAR-ARP2/3 complex, which is required for actin nucleation and cytoskeletal rearrangement. LjNPL/MtNPL encode a legume-specific pectate lyase involved in cell wall remodeling. Mutant analyses revealed some genes that are required for rhizobial infection, but their precise biological/biochemical functions are unknown. These genes include LjRINRK1, encoding an atypical leucine-rich repeat receptor-like kinase; LjCERBERUS/MtLIN, encoding a U-box and WD40-repeat domain protein; MtVAPYRIN, encoding a VAP/MSP and ankyrin-repeats domain protein; MtRPG, encoding a coiled-coil protein; and MtCBS1, encoding a DUF21 and cystathionine-β-synthase domain protein (Arrighi et al., 2008; Kiss et al., 2009; Yano et al., 2009; Yokota et al., 2009; Miyahara et al., 2010; Murray et al., 2011; Hossain et al., 2012; Xie et al., 2012; Qiu et al., 2015; Sinharoy et al., 2016; Li et al., 2019; Liu et al., 2019a).
The IT initiates and extends from the root hair tip to the nodule primordium. This process is directional, indicating that ITs show polarized growth. The polarized growth of pollen tubes and root hairs requires a strong calcium gradient at the tip, a polarized actin cytoskeleton, tip-directed vesicle trafficking, exocytosis, and signaling by ROP (Rho of plants) GTPases (Samaj et al., 2006; Kost, 2008; Craddock and Yang, 2012; Craddock et al., 2012). The LIN-VAPYRIN-Exo70H4 protein complex in M. truncatula localizes to the tips of elongated pre-ITs, suggesting that an exocytosis process may be involved in the polarized growth of ITs (Liu et al., 2019b). Type I ROP GTPases (including ROP1 to ROP8) play important roles in root hair and pollen tube tip growth in Arabidopsis (Arabidopsis thaliana; Kost et al., 1999; Li et al., 1999; Jones et al., 2002; Eklund et al., 2010; Craddock et al., 2012). The constitutively active (CA) forms of AtROP4 and AtROP6 do not function in the polarized growth of root hairs, nor do they form a localized Ca2+ gradient at the root hair tip (Molendijk et al., 2001). Compared to the wild type, plants expressing CA AtROP2 produce additional and misplaced root hairs on the cell surface as well as longer root hairs, whereas plants expressing dominant-negative (DN) forms of AtROP2 produce shortened wavy root hair (Jones et al., 2002).
MtROP10 is associated with the NF receptor MtNFP and is involved in regulating the polarized growth of root hairs and NF-induced root hair deformation in M. truncatula (Lei et al., 2015). The roles of ROP genes in M. truncatula have been clarified to some extent by knockdown RNA interference (RNAi) studies. Compared to the wild type, MtROP8 RNAi transgenic plants show larger numbers of infection events and increased nodulation in response to inoculation, and MtROP9 RNAi plants show enhanced mycorrhizal and early hyphal root colonization but reduced rhizobial infection (Kiirika et al., 2012; Wang et al., 2014). In L. japonicus, the Nod factor receptor LjNFR5 interacts with LjROP6 to regulate IT formation and nodulation (Ke et al., 2012), and LjROP6 interacts with LjCHC1, a central component in clathrin-mediated endocytosis, to regulate rhizobial infection and nodule formation (Wang et al., 2015). These findings demonstrate that LjROP6 plays an essential role in regulating legume-rhizobium symbiosis. However, it is unknown whether ROP GTPase functions in the polarized growth of ITs.
ROP GTPases act as molecular switches in various signaling pathways by cycling between the GTP-bound active and GDP-bound inactive forms (Yang, 2002). The shuttling between the active and inactive forms of ROP GTPases is regulated by three types of factors: RhoGEF (guanine nucleotide exchange factor), which controls the transition of these GTPase from the inactive (GDP-bound) to active (GTP-bound) form; RhoGAP (GTPase-activating protein), which accelerates GTP hydrolysis; and RhoGDI (guanine nucleotide dissociation inhibitor), which inhibits GDP release so that ROP GTPases remain in their inactive state (Yang, 2002; Kost, 2008). The M. truncatula genome contains 10 genes encoding plant Rop nucleotide exchanger-type RopGEFs. MtRopGEF2 affects the cytosolic Ca2+ gradient and the subcellular architecture of root hairs and is required for root hair development (Riely et al., 2011).
In addition to plant Rop nucleotide exchanger-type RopGEFs, a single DOCK family GEF, SPIKE1 (SPK1), is also present in plants. In Arabidopsis, spk1 seedlings show a lethal phenotype and display serious defects in the polarized growth of cotyledons and leaf epidermal cells (Qiu et al., 2002). The AtSPK1 DOCK homology region 2 (DHR2) domain binds to ROPs to facilitate nucleotide exchange and generates signals to activate two heteromeric complexes, WAVE/SCAR and ARP2/3, to control actin polymerization (Basu et al., 2008). AtSPK1 is involved in activating the AtROP6-AtRIC1 pathway, which regulates the auxin-mediated internalization of AtPIN2 (Lin et al., 2012). In rice (Oryza sativa), OsSPK1 interacts with OsPit, an R protein against rice blast fungus, and regulates the activation of OsRac1 to trigger the immune response (Wang et al., 2018).
Although ITs show polar growth, it is unknown whether a ROP GTPase regulates this process and, if so, how this ROP GTPase is activated. Here, we show that LjSPK1 interacts with LjROP6 and activates its ROP GTPase activity. Some ITs were found to be misdirected in the root hairs of the Ljspk1 and Ljrop6 mutants after rhizobial infection. Our results suggest that an LjROP6-LjSPK1 signaling module guides the direction of IT growth during the early stage of rhizobial infection in L. japonicus.
RESULTS
Expression Pattern of SPK1 in L. japonicus
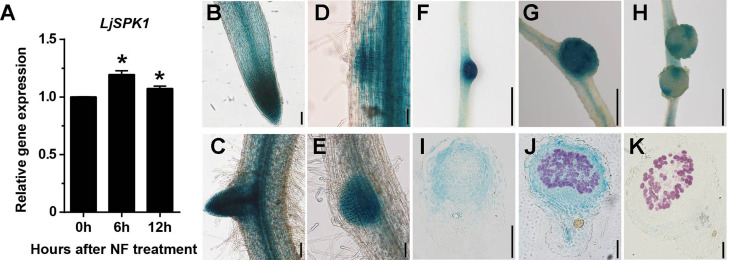
SPK1 is a DOCK family GEF that activates ROP GTPases (Basu et al., 2008). Using BLAST tools, we searched for the SPK1 gene in the L. japonicus genome. This search identified LjSPK1 (LotjaGi3g1v0299000), which encodes an 1829-amino acid protein showing 79.2% identity with AtSPK1. Phylogenetic analysis revealed that SPK1 is conserved across eukaryotes, usually as a single copy gene, with one exception being soybean (Glycine max), which has undergone a recent whole-genome duplication (Schmutz et al., 2010; Supplemental Figure 1A). Sequence analyses revealed three conserved domains in LjSPK1: DHR1 (amino acids 446 to 726); DHR2 (amino acids 1299 to 1829), which is the GEF catalytic domain; and DHR3 (amino acids 140 to 287; Supplemental Figure 1B). To investigate the transcriptional pattern of LjSPK1 during nodulation, we monitored the transcript levels of LjSPK1 after NF treatment by RT-qPCR. LjSPK1 transcript levels slightly increased at 6 and 12 h after NF inoculation versus the 0 h control (Figure 1A).
Figure 1.
LjSPK1 Is Induced by Nod Factors and Is Specifically Expressed in Young Nodules.
(A) RT-qPCR analysis of LjSPK1 transcript levels in roots of wild-type L. japonicus (Gifu) after inoculation with purified Nod factor (0 h, 6 h, 12 h). Expression is relative to that in mock-treated samples (0 h) and normalized to that of Lotus Ubiquitin. Mean and SD were derived from two biological replicates. Statistical significance (*P < 0.05) was evaluated by Student’s t test.
(B) and (C) pLjSPK1:GUS expression pattern in wild-type roots without rhizobial inoculation.
(D) to (H) pLjSPK1:GUS expression pattern in wild-type roots after inoculation with M. loti R7A expressing lacZ. GUS activity was detected in nodule primordia and young nodules (D) to (G) and in vascular bundles of mature nodules (H).
(I) to (K) Section of nodules showing that pLjSPK1:GUS expression in all cell layers in young nodules (I) and (J), but not in mature nodules’ nitrogen-fixation zone (K).
Transgenic roots were costained with X-Gluc and magenta-Gal to visualize pLjSPK1:GUS expression (in blue) and M. loti (in purple), respectively. Bars = 100 μm in (B) to (H) and 1 mm in (I) to (K).
We analyzed the spatial expression pattern of LjSPK1 by monitoring β-glucuronidase (GUS) expression from the GUS gene driven by the LjSPK1 promoter (pLjSPK1:GUS) in L. japonicus transgenic hairy roots. In the absence of rhizobial inoculation, we detected GUS signals in root vascular tissues and lateral roots (Figures 1B and 1C). However, after inoculation with Mesorhizobium loti R7A/lacZ, GUS signals were present in the dividing cortex region (Figure 1D), nodule primordia (Figure 1E), bumps (Figure 1F), young nodules (Figure 1G), and vascular bundles of mature nodules (Figure 1H). Light microscopy analyses of sections of these nodules revealed GUS signals in bump and young nodule cells (Figures 1I and 1J), but not in the nitrogen-fixation zone of mature nodules (Figure 1K). These expression patterns suggest that LjSPK1 might be involved in root nodule symbiosis in L. japonicus.
LjSPK1 Is Required for Polarized Growth in Plants and Rhizobial Infection
To explore the roles of LjSPK1 in root nodule symbiosis, we obtained two LORE1 insertion lines (30,019,873 and 30,051,253) in which LjSPK1 had insertions at amino acid positions 273 and 910 from the start codon, respectively. These lines were designated as Ljspk1-1 (30,019,873) and Ljspk1-2 (30,051,253; Supplemental Figure 1B). RT-qPCR analysis revealed that endogenous LjSPK1 transcript levels were significantly reduced in both Ljspk1 lines (Supplemental Figure 1C). For both Ljspk1 alleles, the homozygotes were nearly completely sterile, so we could only use seedlings obtained from the segregation of heterozygous plants for phenotypic analyses. Soil-grown Ljspk1 mutants survived but showed some growth defects, including severe dwarfism of adult plants (Supplemental Figure 1D), few and smaller flowers with an abnormal petal shape (Supplemental Figure 1E), and smaller, dark-green leaves (Supplemental Figures 1F and 1H). Wild-type plants formed elongated, filamentous trichomes on the abaxial midribs of leaves, whereas the Ljspk1 mutants formed fewer and smaller trichomes (Supplemental Figure 1H). Scanning electron microscopy of epidermal cells on the dorsal leaves of wild-type (Gifu) and Ljspk1 mutants revealed that wild-type pavement cells had a clear neck and lobe, whereas those on Ljspk1 leaves were nearly round (Supplemental Figures 1I and 1J). The phenotypes of these Ljspk1 mutants were similar to those reported for the Arabidopsis spk1-1 mutant (Qiu et al., 2002). These results suggest that LjSPK1 is involved in the polarized growth of cells in L. japonicus.
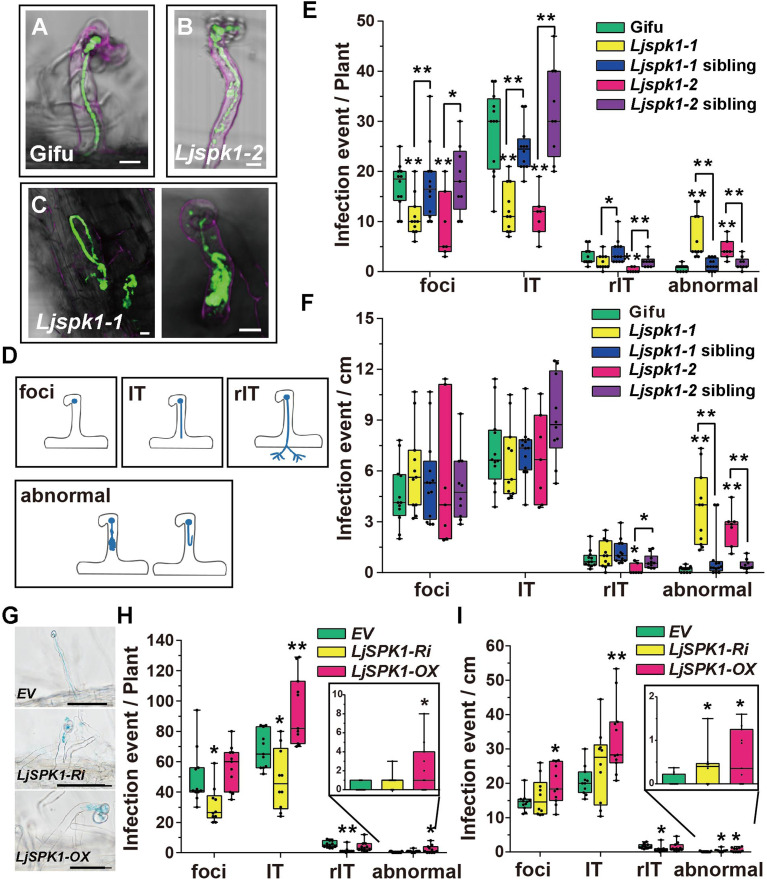
We analyzed the infection and nodulation phenotypes of the Ljspk1 mutants following inoculation with M. loti R7A constitutively expressing GFP or lacZ. Wild-type (Gifu) and Ljspk1 wild-type siblings (sibling) produced normal elongated ITs in curled root hairs (Figure 2A; Supplemental Figures 2A, 2B, and 2D). In the Ljspk1 mutants, infection initiated normally from a curled root hair, but some ITs showed anomalous growth patterns, including the formation of sac-like structures or depolarized loop shapes inside the root hairs (Figures 2B to 2D; Supplemental Figures 2C and 2E). We counted the total number of infection events in the Ljspk1 mutants 1 week after inoculation. The number of infection events including infection foci (foci), ITs in root hairs (IT), and ITs extending into cortex cells (rIT) was significantly lower in the Ljspk1 mutants than in the wild type. In addition, there were significantly more abnormal ITs (abnormal in Figure 2) in the Ljspk1 mutants than in the wild type (Figure 2E).
Figure 2.
ITs in Ljspk1 Mutants Are Abnormal Compared with Wild-Type L. japonicus.
(A) to (D) Normal elongating ITs in the wild type (Gifu) (A) and typical abnormal infection events in the Ljspk1 mutants (B) and (C) at 1 week after inoculation with M. loti R7A/GFP. Roots were counterstained with propidium iodide before observation. Green fluorescence shows normal infection foci and a normal IT in a curled root hair in the wild type, whereas Ljspk1 mutants show abnormal infection processes, such as sac-like or looped ITs (B) and (C). Bars = 10 μm. (D) A cartoon diagram of the infection events. Foci, infection foci; IT, IT in an epidermis cell; rIT, IT extending into a cortex cell; abnormal, abnormal IT in root hairs.
(E) and (F) Boxplots representing the number of infection events in wild-type, Ljspk1 siblings, and Ljspk1 mutants. Total number of infection events per plant (E) and number of infection events per centimeter root (F) were scored at 1 week after inoculation with M. loti R7A/lacZ. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, Student’s t test, comparison between the wild type and mutants).
(G) to (I) IT phenotypes (G) and infection events ([H] and [I]) in wild-type L. japonicus hairy roots expressing control plasmid (EV), LjSPK1-Ri, or LjSPK1-OX. Total number of infection events per plant (H) and number of infection events per centimeter root (I) scored 1 week after inoculation with M. loti R7A/lacZ. Abnormal infection events are enlarged in the inset in (H) and (I). Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, Student’s t test, comparison between EV control and experimental group). Bars = 50 μm.
For each boxplot, the center line in the box shows the median; the box limits are the upper and lower quartiles; the whiskers represent the maximum and minimum values.
Because the Ljspk1 mutants had shorter primary roots than the wild type (Supplemental Figures 2F and 2I), we calculated the number of infection events per centimeter (cm). The number of infection foci and ITs per-cm root did not differ significantly between the wild type and the Ljspk1 mutants except that Ljspk1-2 had fewer rITs (Figure 2F). However, both Ljspk1 mutants had more abnormal infection events than the wild type and Ljspk1 siblings (Figure 2F). These results suggest that the infection events were not significantly affected, but the polarized growth of ITs was markedly affected in the Ljspk1 mutants. At 2 weeks after inoculation, the wild type had produced abundant pink mature nodules, while the Ljspk1 mutants had produced fewer nodules per plant (Supplemental Figures 2G, 2J, and 2K). Light microscopy of semithin sections of nodules stained with toluidine-blue revealed that Ljspk1-1 nodules had fewer infected cells than the wild type and Ljspk1-1 siblings’ nodules (Supplemental Figure 2H).
Next, we explored the role of LjSPK1 by generating LjSPK1-overexpressing lines (LjSPK1-OX) and lines with knocked-down LjSPK1 expression (LjSPK1-Ri). The LjSPK1-knockdown lines were generated by introducing an RNAi vector and the LjSPK1-OX lines were generated by introducing the LjSPK1 cDNA driven by the L. japonicus Ubiquitin promoter into wild-type L. japonicus hairy roots. Compared with empty vector (EV) control, the LjSPK1-Ri lines had markedly fewer infection events, but the LjSPK1-OX lines had increased numbers of infection events, including infection foci and ITs (Figures 2G and 2H). The LjSPK1-Ri lines had shorter roots than the EV control (Supplemental Figure 2L). Analysis of the infection events revealed that both the LjSPK1-Ri and LjSPK1-OX lines had more abnormal infection events per cm of root than the EV control (Figure 2I). The phenotype of LjSPK1-Ri was similar to that of the Ljspk1 LORE1 insertion mutants. RT-qPCR analysis revealed that LjSPK1 transcript levels were significantly lower in the LjSPK1-Ri lines and significantly higher in the LjSPK1-OX lines compared to the control (Supplemental Figure 2M). Together, these results demonstrate that LjSPK1 is required for the rhizobial infection process and is involved in the polarized growth of ITs in L. japonicus.
SPK1 Interacts with Three L. japonicus ROP GTPases in Yeast Cells, but Only LjROP6 Is Required for Polarized Growth of ITs
AtSPK1 is a DOCK-family GEF protein, and DHR2 is the GEF catalytic domain that facilitates the nucleotide exchange activity of ROP GTPases (Basu et al., 2008). We were interested in determining which ROP GTPase is activated by LjSPK1 to regulate rhizobial infection in L. japonicus. BLAST searches of the L. japonicus genome revealed 10 ROP GTPases. We conducted phylogenetic analysis of ROP GTPases in Arabidopsis, L. japonicus, and M. truncatula (Supplemental Figure 3A). Because AtSPK1 interacts and facilitate nucleotide exchange with type I ROP GTPase, and most ROP GTPases are membrane-localized proteins, we used the DUAL membrane yeast two-hybrid system to determine which type I ROP GTPase interacts with the LjSPK1 DHR2 domain (named SPK1-DHR2). Of the four type I and one type II ROP GTPases examined, three type I ROP GTPases (LjROP1, LjROP3, and LjROP6) interacted with the LjSPK1 DHR2 domain in yeast (Saccharomyces cerevisiae) cells (Supplemental Figure 3B). No interaction was detected between LjSPK1 DHR2 and LjROP5 or LjROP10 in yeast cells (Supplemental Figure 3B).
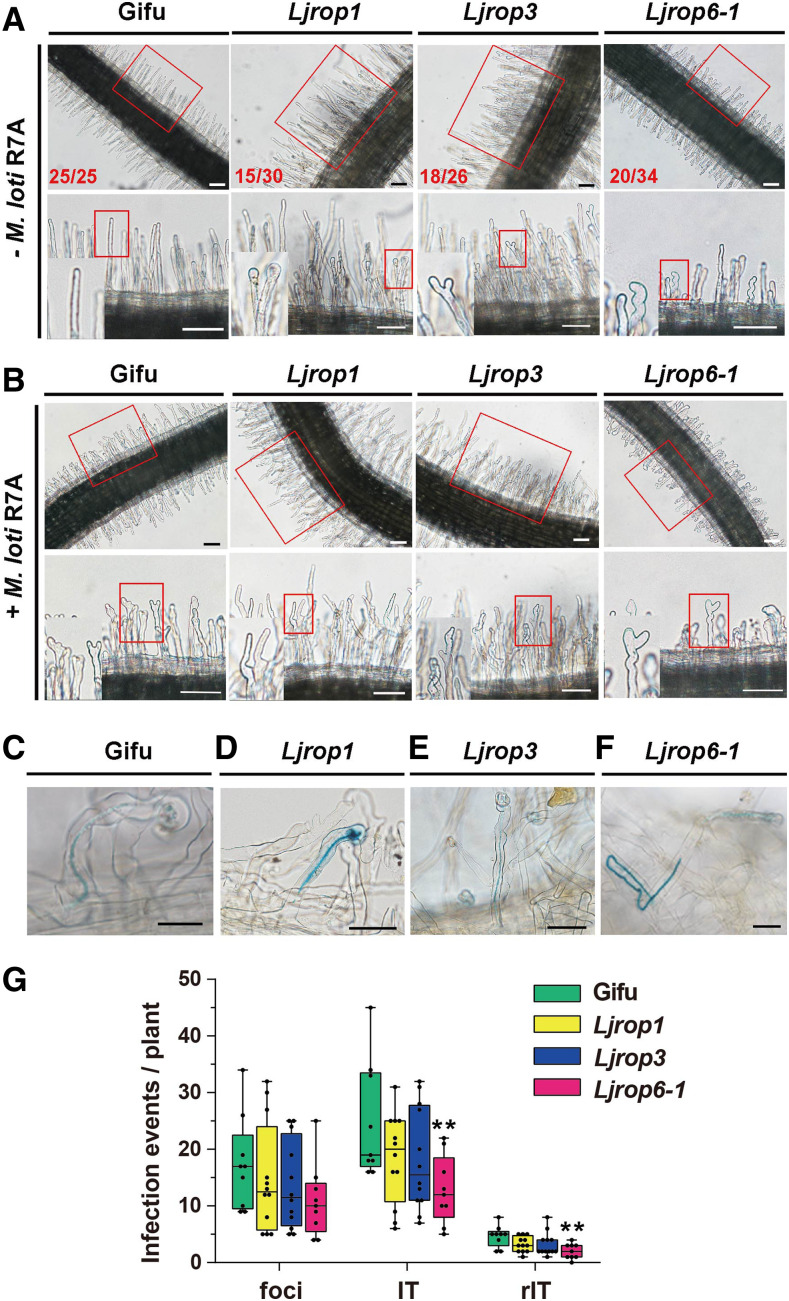
To explore whether these three type I LjROPs function in rhizobial infection, we obtained their LORE1 insertion mutants (30,000,786 with an insertion at 83 bp in LjROP1; 30,000,537 with an insertion at 67 bp before the start codon in LjROP3; and 30,031,226 with an insertion at 258 bp in LjROP6). We then obtained homozygotes of these insertion mutants. RT-qPCR analysis confirmed that endogenous LjROP transcript levels were significantly reduced in all three Ljrop mutants (Supplemental Figure 3C). We analyzed the root hair and infection phenotypes of the mutants. First, we observed the root hairs of the mutants at 2 d after germination with or without rhizobial inoculation. In the absence of rhizobial inoculation, the roots of wild-type plants formed straight root hairs, but more than half of the plants in lines Ljrop1 (15 out of 30), Ljrop3 (18 out of 26), and Ljrop6-1 (20 out of 34) formed branched root hairs or root hairs with swollen tips (Figure 3A). At 18 h after M. loti R7A/lacZ inoculation, both the wild type and Ljrop mutants displayed rhizobia-induced root hair deformation in the infection zone, with no distinguishable differences between these lines (Figure 3B). These observations suggest that all three LjROP GTPases play roles in root hair development and respond normally to rhizobia.
Figure 3.
Root Hair and IT Phenotypes of Ljrop Mutants.
(A) Light micrographs of root hairs in wild-type (Gifu) and Ljrop mutants at 2 d after germination. The numbers in the lower left corners show the number of plants with the type of root hair shown in the image out of the total number of plants observed. Bars = 100 μm.
(B) Light micrograph of M. loti R7A-induced root hair deformation in the infection zone of wild-type and Ljrop mutants at 18 h after M. loti R7A inoculation. Bars = 100 μm.
(A) and (B) Insets (bottom) are enlargements of the areas in red boxes (top).
(C) to (F) IT phenotypes of wild-type (C) and Ljrop mutants ([D] to [F]) at 5 d after inoculation with M. loti R7A/LacZ. Bars = 50 μm.
(G) Boxplot representing the number of infection events on roots of wild-type and Ljrop mutant roots at 5 d after inoculation with M. loti R7A/lacZ. The center line in the box shows the median; the box limits are the upper and lower quartiles; the whiskers represent the maximum and minimum values. Asterisks indicate a significant difference (**P < 0.01, Student’s t test, comparison between wild-type and Ljrop mutants).
We further analyzed IT formation and infection events after rhizobial inoculation. There were no differences in IT formation or infection events between the wild type and Ljrop1 or Ljrop3, but there were fewer infection events in Ljrop6-1 than in the wild type (Figure 3G). Interestingly, Ljrop6-1 contained some misdirected ITs in the root hairs, which were similar to, but more severe than, those in Ljspk1 (Figure 3F and described in detail below). However, this phenotype was not observed in the wild type, Ljrop1, or Ljrop3 plants (Figures 3C to 3E). To analyze whether these LjROPs play functionally redundant roles during rhizobial infection, we crossed Ljrop1 × Ljrop3 and Ljrop1 × Ljrop6-1 to obtain the Ljrop1 Ljrop3 and Ljrop1 Ljrop6-1 double mutants. We analyzed the infection events in these mutants after inoculation with M. loti R7A/LacZ. Compared with wild-type plants, Ljrop1 Ljrop3 plants had more infection foci but no misdirected ITs, and Ljrop1 Ljrop6-1 plants produced approximately the same number of misdirected ITs as Ljrop6-1 (Supplemental Figure 3D). These results indicate that ROP1, ROP3, and ROP6 affect the polarized growth of root hairs in L. japonicus, but only LjROP6 is required for the polarized growth of ITs in root hairs. The results also indicate that the misdirection of ITs in Ljrop6-1 is not caused by a deficiency in root hair development.
LjROP6 Is Required for the Polarized Growth of ITs in L. japonicus
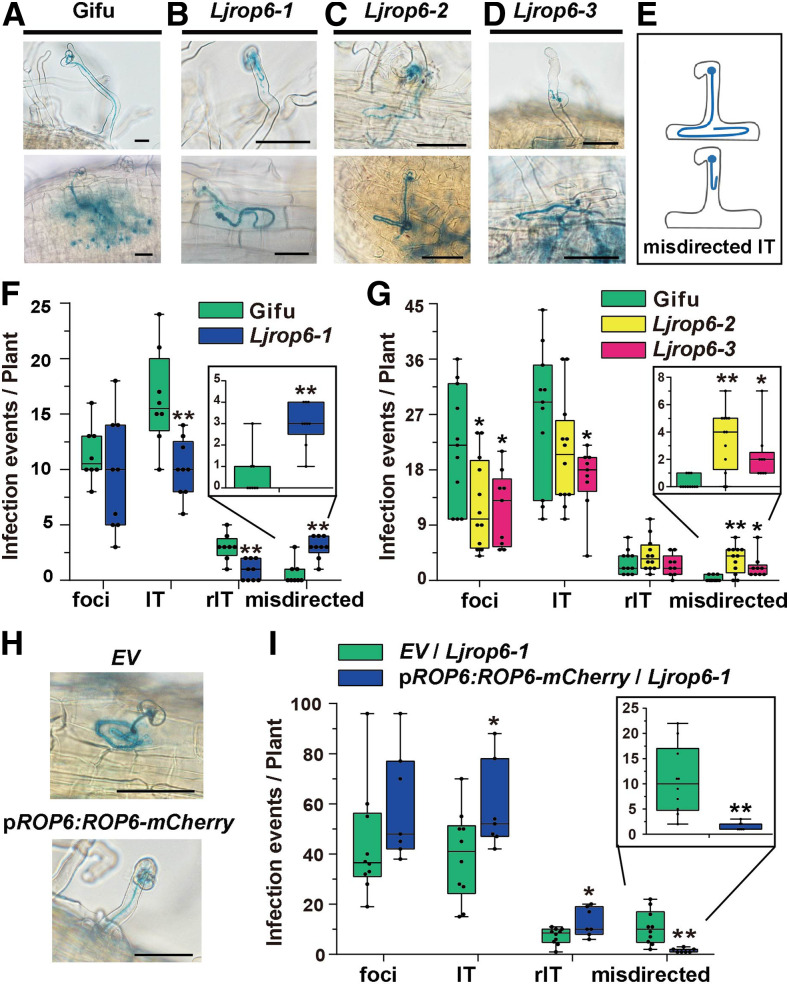
To confirm the notion that ROP6 is required for the polarized growth of ITs in root hairs, we obtained two more LORE1 insertion lines: 30,142,846 (Ljrop6-2) and 30,103,232 (Ljrop6-3), with insertions at 505 bp after and 114 bp before the LjROP6 start codon, respectively (Supplemental Figure 4A). RT-qPCR analysis of homozygous Ljrop6-2 and Ljrop6-3 plants showed that LjROP6 transcript levels were significantly lower in these plants than in wild-type plants (Supplemental Figure 4E).
We observed the root hair phenotypes of the Ljrop6-2 and Ljrop6-3 mutants at 2 d after germination. Both before and after rhizobial inoculation, the root hair phenotypes and responses to rhizobia of the Ljrop6-2 and Ljrop6-3 mutants were similar to those of Ljrop6-1 (Supplemental Figures 5A and 5B). We analyzed rhizobial symbiosis in these three Ljrop6 mutants after M. loti R7A/LacZ inoculation. After rhizobial infection, the three Ljrop6 mutants produced normal ITs like those in the wild type (Figure 4A). However, as noted above, some ITs in the Ljrop6 mutants were misdirected or tangled in the root epidermal cells (Figures 4B to 4E), suggesting that the ITs became misdirected as they elongated from the infection chamber down to the cortex nodule primordium. Compared to the wild type, the Ljrop6 mutants had significantly fewer infection events and formed more misdirected ITs at 5 d after inoculation with M. loti R7A/lacZ (Figures 4F and 4G). At 2 weeks after M. loti R7A/lacZ inoculation, the Ljrop6 mutants and wild-type plants produced normal mature pink nodules (Supplemental Figure 4B). The total nodule number was not significantly different between the Ljrop6 mutants and wild-type plants (Supplemental Figures 4C and 4D).
Figure 4.
Disorientated IT Phenotypes of Ljrop6 Mutants and Complementation of IT Phenotype via Hairy Root Transformation of Ljrop6-1.
(A) to (E) Normal IT phenotypes of the wild-type (A) and typical misdirected infection events in all three Ljrop6 mutants in (B) to (D). Bars = 50 μm. (E) A cartoon diagram of the misdirected ITs. Misdirected indicates misdirected or looped IT in root hairs or epidermis cell.
(F) and (G) Boxplots representing the number of infection events in wild-type L. japonicus (Gifu) and Ljrop6 mutants at 5 d after inoculation with M. loti R7A/lacZ. Misdirected infection events are enlarged in the inset. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, Student’s t test, comparison between wild-type and Ljrop6 mutants).
(H) and (I) IT phenotype (H) and infection events (I) of Ljrop6-1 hairy roots expressing control plasmid (EV) and pLjROP6:LjROP6-mCherry. ITs were stained with X-Gal and scored 5 d after inoculation with M. loti R7A/lacZ. Misdirected infection events are enlarged in the inset. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, Student’s t test, comparison between EV and LjROP6). Bars = 50 μm.
For each boxplot, the center line in the box shows the median; the box limits are the upper and lower quartiles; the whiskers represent the maximum and minimum values.
To further confirm the notion that LjROP6 is required for the polarized growth of ITs, we generated transgenic hairy roots in the Ljrop6-1 background using the LjROP6 native promoter to drive the expression of LjROP6 cDNA fused with mCherry (pLjROP6:LjROP6-mCherry). The disorientated ITs in Ljrop6-1 were rescued by complementation with ROP6-mCherry (Figure 4H). Statistical analyses showed that the complemented group (pLjROP6:LjROP6-mCherry/Ljrop6-1) had more ITs and rITs and fewer misdirected ITs compared with the EV control at the same time point (Figure 4I). LjROP6 transcript levels were significantly higher in Ljrop6-1 hairy roots than the control (Supplemental Figure 4F). Together, these observations confirm the notion that LjROP6 is required for the polarized growth of ITs in L. japonicus.
LjSPK1 Physically Interacts with LjROP6 In Planta and Activates Its GTPase Activity In Vitro
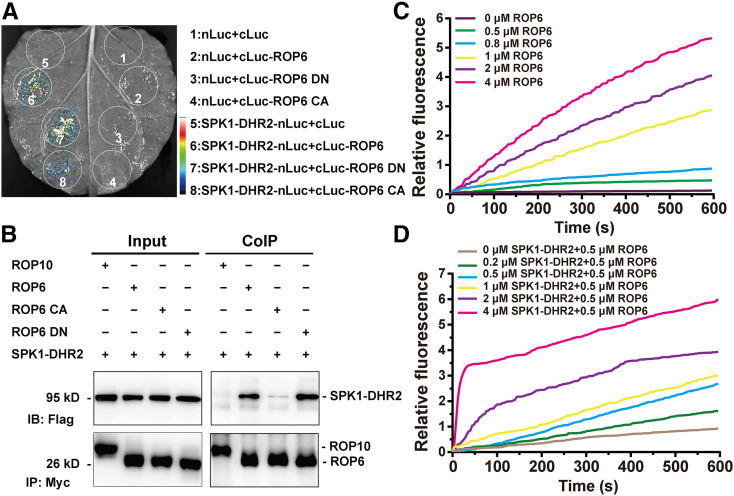
As described above, LjROP6 interacted with LjSPK1 in yeast cells. We further confirmed their interaction in Nicotiana benthamiana leaf pavement cells using split-luciferase complementation and coimmunoprecipitation (co-IP) assays. LjROP6 interacted with the LjSPK1-DHR2 catalytic domain in split-luciferase complementation (Figure 5A) and co-IP assays (Figure 5B). However, we did not detect coprecipitation of LjSPK1 with LjROP10, a type II ROP GTPase, in our co-IP assays (Figure 5B). The activation of ROP GTPases relies on GDP and GTP exchange, and this cycle requires GEFs to facilitate the dissociation of GDP (Kost, 2008). We therefore investigated the ability of LjSPK1 to interact with LjROP6 in the DN (D121A) or CA (G15V) form. LjSPK1-DHR2 interacted more strongly with the LjROP6 DN form than with the LjROP6 CA form in co-IP assays (Figure 5B). These results suggest that LjSPK1 preferentially interacts with the inactive form, but not the active form, of LjROP6 to activate the nucleotide exchange activity of LjROP6.
Figure 5.
LjSPK1 Physically Interacts with LjROP6 and Activates Its GTPase Activity In Vitro.
(A) Luciferase biomolecular complementation assays showing the interaction between LjSPK1-DHR2 and LjROP6, LjROP6 CA, or LjROP6 DN in N. benthamiana leaf cells. The indicated constructs were transiently coexpressed in N. benthamiana leaves, and luciferase complementation imaging was captured 2 d after agroinfiltration. nLuc, N-terminal fragment of firefly luciferase; cLuc, C-terminal fragment of firefly luciferase. Fluorescence signal intensity is indicated.
(B) Co-IP assay showing the interaction between LjSPK1-DHR2 and LjROP6 in N. benthamiana leaves. The indicated constructs were coexpressed in N. benthamiana leaves. The co-IP assay was performed using anti-Myc antibody, and the proteins were detected by immunoblot analysis with anti-Flag and anti-Myc antibodies. LjSPK1-DHR2 interacted more strongly with LjROP6 DN than with LjROP6 CA. No interaction was detected between LjSPK1-DHR2 and LjROP10, a type II ROP GTPase.
(C) Time course of intrinsic nucleotide exchange in LjROP6. The intrinsic guanine nucleotide exchange rates of LjROP6 increased with increasing reaction time and increasing concentration of LjROP6. The assays contained 1 μM mant-GTP and the indicated concentration of GDP-LjROP6.
(D) LjSPK1-DHR2 shows concentration-dependent GEF activity toward LjROP6. The assays contained 1 μM mant-GTP, 0.5 μM GDP-LjROP6, and the indicated concentrations of LjSPK1-DHR2.
Results are a representative of three independent assays with similar results.
The LjSPK1 DHR2 domain is known to activate small GTPases (Basu et al. 2008; Wang et al. 2018). To examine whether LjSPK1 facilitates the guanine nucleotide exchange activity of LjROP6, we tested the GEF activity of the LjSPK1 DHR2 domain in vitro using a fluorescence spectroscopy-based procedure (Gu et al., 2006; Wang et al., 2017). First, we expressed and purified glutathione S-transferase-tagged LjSPK1-DHR2 and unlabeled-GDP His-tagged LjROP6 in Escherichia coli. To start the nucleotide exchange reaction, we added fluorescently labeled N-methylanthraniloyl (mant)-GTP and unlabeled-GDP LjROP6 to the reaction buffer and recorded the fluorescence values for 600 s. The intrinsic guanine nucleotide exchange rate of LjROP6 increased with increasing reaction time and with increasing concentrations of LjROP6 (Figure 5C). Next, to investigate the effect of LjSPK1 on the guanine nucleotide exchange rate of LjROP6, we added LjSPK1-DHR2 protein to reaction buffer containing 0.5 μM unlabeled-GDP LjROP6 protein and 1 μM mant-GTP before recording the change in fluorescence intensity. The LjSPK1 GEF activity toward LjROP6 increased with increasing LjSPK1-DHR2 concentrations (Figure 5D). These results demonstrate that LjROP6 has intrinsic guanine nucleotide exchange activity, which is enhanced by SPK1.
LjROP6 Colocalizes and Interacts with LjSPK1 in the Plasma Membrane
LjROP6 was shown to be localized to the plasma membrane (PM) and cytoplasm (Ke et al., 2012), but SPK1 is localized to the endoplasmic reticulum (ER; Zhang et al., 2010; Wang et al., 2018). To validate the subcellular localization of LjROP6 and confirm its interaction with LjSPK1, we analyzed the subcellular localization of LjROP6 in N. benthamiana leaf cells and L. japonicus roots. We coexpressed GFP-LjROP6 or LjROP6-mCherry under the control of the L. japonicus Ubiquitin promoter in N. benthamiana leaf cells. In these analyses, LjROP6 localized to the PM, regardless of whether it was tagged with a fluorescent marker at its N terminus or C terminus (Supplemental Figures 6A to 6C). We then expressed LjROP6-mCherry in wild-type L. japonicus hairy roots and monitored its localization before and after inoculation with M. loti R7A/mTag. Signals from mCherry were observed at the PM in root hairs without rhizobia (Supplemental Figure 6D) and in elongating or curled root hairs after inoculation with M. loti R7A (Supplemental Figure 6E). The LjROP6-mCherry fusion protein displayed a similar subcellular localization pattern regardless of whether it was expressed under the control of the Ubiquitin promoter (Supplemental Figures 6D and 6E) or the LjROP6 native promoter (Supplemental Figure 6F).
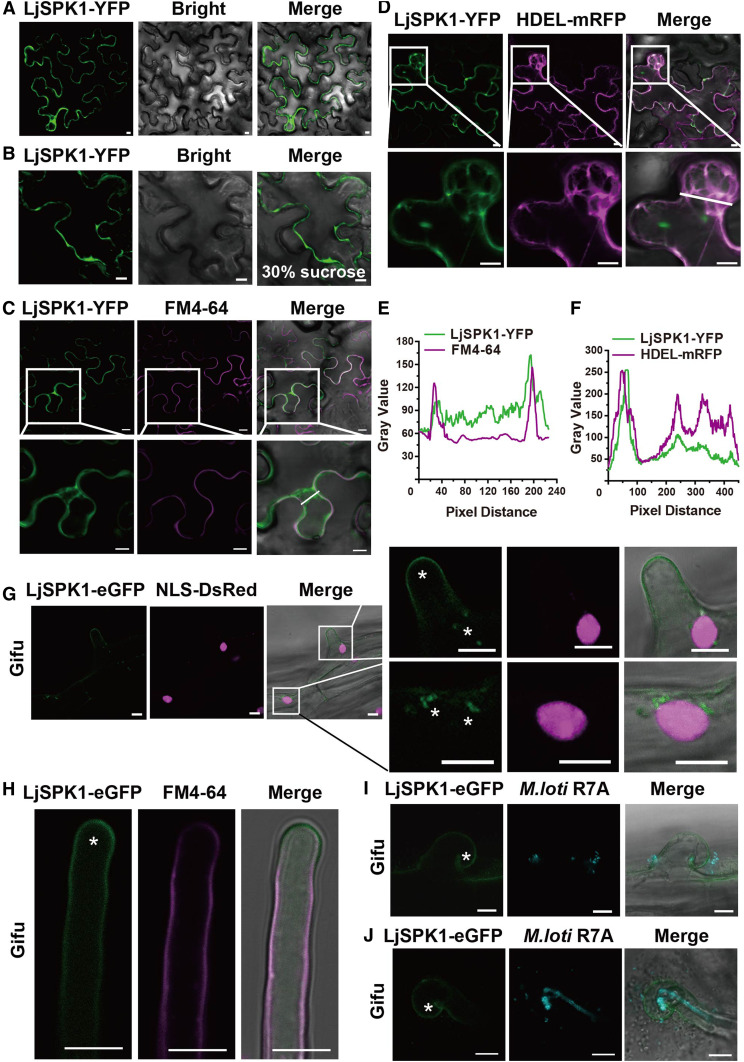
Next, we analyzed the subcellular localization of LjSPK1. We transformed N. benthamiana leaves with Agrobacterium carrying LjSPK1-yellow fluorescent protein (YFP) and observed the localization of LjSPK1 by monitoring the fluorescence of YFP by laser scanning confocal microscopy. LjSPK1 showed a punctate distribution close to the PM or cytoplasm (Figure 6A). Following plasmolysis using 30% (w/v) Suc, LjSPK1-YFP was not associated with the cell wall (Figure 6B). To examine whether LjSPK1 localizes to the PM, we stained the N. benthamiana leaves with the PM localization marker FM4-64. LjSPK1 showed almost no colocalization with FM4-64 (Figures 6C and 6E). AtSPK1 and OsSPK1 are localized to subdomains of the ER (Zhang et al., 2010; Wang et al., 2018). To determine whether this is also the case for LjSPK1, we coexpressed LjSPK1-YFP with the ER marker HDEL-mRFP in N. benthamiana leaves. LjSPK1 colocalized with the ER marker (Figures 6D and 6F). These findings suggest that LjSPK1 is associated with subdomains of the ER. We then explored the subcellular distribution of LjSPK1-eGFP in L. japonicus hairy roots. LjSPK1-eGFP displayed PM and punctate distribution close to the nucleus in L. japonicus root hairs (Figure 6G), and LjSPK1-eGFP also colocalized with the PM marker FM4-64 (Figure 6H). LjSPK1-GFP expressed in Ljrop6-1 hairy roots also showed a punctate distribution close to the nucleus (Supplemental Figure 7A), similar to that of LjSPK1-eGFP expressed in the wild type. After rhizobia inoculation, LjSPK1-eGFP was detected in the PM of root hairs with infection foci or elongated ITs (Figures 6I and 6J).
Figure 6.
Subcellular Localization of LjSPK1 in N. benthamiana Leaves and Wild-Type L. japonicus Hairy Roots.
(A) to (F) Confocal images of LjSPK1-YFP expressed in N. benthamiana leaf cells. (A) and (B) Subcellular localization of LjSPK1-YFP in N. benthamiana leaves (A) and after plasmolysis via 30% (w/v) Suc treatment (B). (C) LjSPK1-YFP (green) expressed in N. benthamiana leaves and stained with the PM marker FM4-64 dye (magenta), showing that SPK1 does not merge with the PM marker. (D) LjSPK1-YFP (green) and ER marker HDEL-mRFP (magenta) were coexpressed in N. benthamiana leaves. Image shows merging of LjSPK1-YFP and HDEL-mRFP fluorescence. These genes were driven by the CaMV 35S promoter. (E) and (F) Intensity profiles of LjSPK1 and FM4-64 or HDEL-mRFP. Plots show fluorescence intensities of LjSPK1-YFP (green) and FM4-64 or HDEL-mRFP (magenta) in regions of interest (insets in [C] and [D]). (C) and (D) Insets (bottom) are enlargements of the areas in white boxes (top). Bars = 10 μm.
(G) to (J) Live cell confocal images of LjSPK1-eGFP expressed in root hairs before (G) and (H) and after (I) and (J) M. loti R7A/mTag inoculation in wild-type L. japonicus hairy roots. (G) LjSPK1-eGFP (green) fluorescence was detected in L. japonicus hairy roots in the PM and puncta in the vicinity of the nucleus. NLS-DsRed (magenta; nuclear marker) was used as a transgenic marker. (H) LjSPK1-eGFP (green) was detected in L. japonicus hairy root PM and stained with the PM marker FM4-64 dye (magenta). (I) and (J) LjSPK1-eGFP (green) in the PM of curled root hair (I) or with elongated IT (J) after M. loti R7A/mTag inoculation. IT is indicated by M. loti R7A/mTag (cyan). Asterisks indicate GFP fluorescence. Expression of LjSPK1 was driven by the L. japonicus Ubiquitin gene promoter. Insets (right) are enlargements of the areas in white boxes (left). Bars = 10 μm.
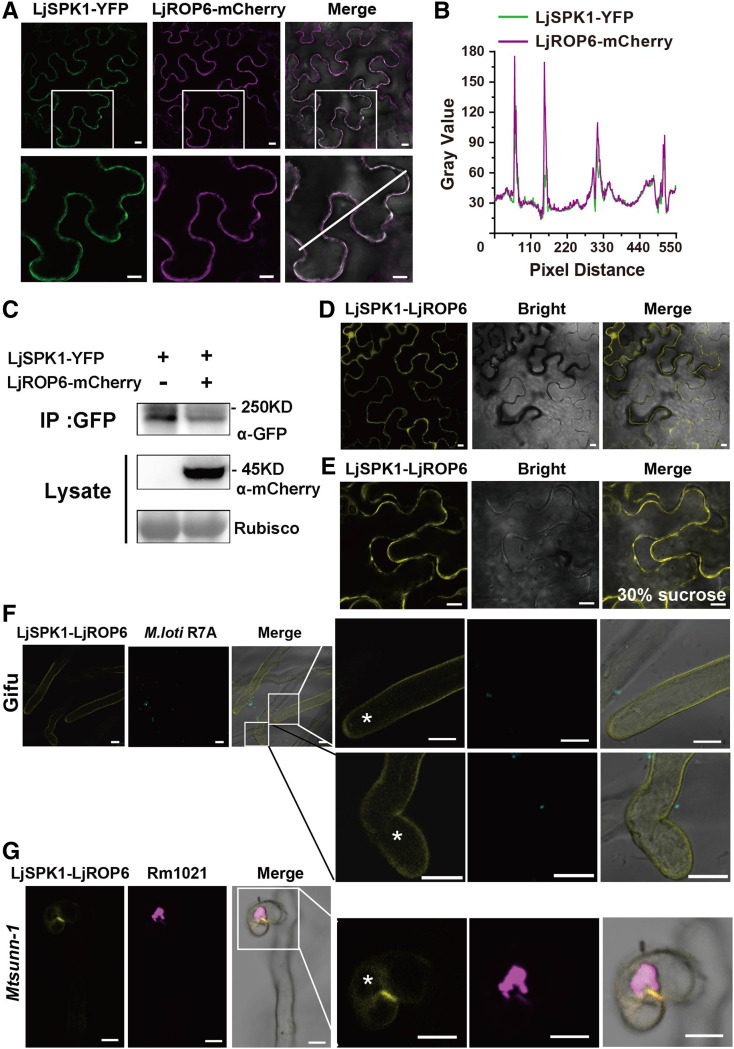
We then coexpressed LjSPK1-YFP and LjROP6-mCherry in N. benthamiana leaf pavement cells. In contrast to the punctate localization of LjSPK1-YFP when expressed alone, LjSPK1-YFP coexpressed with LjROP6-mCherry showed reduced punctate distribution, and most LjSPK1-YFP signals were localized in patches on the PM (Figures 7A to 7C). This observation suggests that LjROP6 promotes the PM localization of LjSPK1.
Figure 7.
LjROP6 Colocalizes and Interacts with LjSPK1 at the PM in N. benthamiana Leaves and Hairy Roots in Legumes.
(A) to (C) LjSPK1-YFP (green) and LjROP6-mCherry (magenta) were coexpressed in N. benthamiana leaves, showing that LjROP6 promotes LjSPK1 distribution in the PM (A). Insets (bottom) are enlargements of the areas in white boxes (top). (B) Intensity profiles of LjSPK1-YFP and LjROP6-mCherry. Plots show fluorescence intensities of LjSPK1-YFP (green) and LjROP6-mCherry (magenta) in regions of interest (insets in [A]). (C) Immunoblots showing protein levels of LjSPK1 and LjROP6 in N. benthamiana leaves. Total protein was extracted and analyzed by immunoblotting with anti-GFP and anti-mCherry antibodies. Rubisco was used as the loading control. Bars = 10 μm.
(D) to (E) BiFC assay of LjSPK1-cVenus and LjROP6-nVenus expressed in N. benthamiana leaves, before (D) and after (E) plasmolysis with 30% (w/v) Suc treatment. Split Venus fluorescent was detected in the PM. Bars = 10 μm.
(F) to (H) BiFC assay of LjSPK1-cVenus and LjROP6-nVenus in wild-type L. japonicus hairy roots (F) or M. truncatula sunn-1 hairy roots (G) 5 d after rhizobial inoculation. Asterisks represent the Venus fluorescence and indicate that LjROP6 interacts with LjSPK1 in the root hair PM (F) or infection foci (G). Cyan and magenta fluorescence represent M. loti R7A/mTag (cyan, [F]) or Sm1021/mCherry (magenta, G), respectively. Insets (right) are enlargements of the areas in white boxes (left). Bars = 10 μm.
We evaluated the interaction between LjSPK1 and LjROP6 in planta via bimolecular fluorescence complementation (BiFC) assays, with C-terminal and N-terminal split-Venus fragments fused to LjSPK1 and LjROP6, respectively, driven by the L. japonicus or Arabidopsis Ubiquitin promoter. When this construct (pLjUb:LjSPK1-cVenus/pAtUb:LjROP6-nVenus) was agro-infiltrated into N. benthamiana leaf cells, Venus fluorescence was detected in the PM and in some punctate structures adjacent to the PM (Figure 7D). This distribution was confirmed by plasmolysis experiments using 30% (w/v) Suc (Figure 7E). These results confirm the notion that LjSPK1 interacts with LjROP6 at the PM. We then introduced this construct into wild-type L. japonicus hairy roots and monitored the localization of Venus fluorescence signals after inoculation with M. loti R7A/mTag. Venus fluorescence signals were detected in the PM of deformed root hairs (Figure 7F) as well as in infection foci (Supplemental Figure 7B). We also expressed these constructs in the M. truncatula sunn-1 mutant, which had many more infection events than the wild type. Venus fluorescence was observed in the infection chamber of the root hairs at the site of the initiating IT (Figure 7G; Supplemental Figure 7C).
Polarized Growth of Root Hairs and Rhizobial Infection Are Affected in LjROP6 Overexpression, CA, and DN Lines
To explore the role of LjROP6 in root hair development and IT formation, we generated LjROP6 overexpression, LjROP6 CA, and LjROP6 DN hairy roots with each construct under the control of the L. japonicus Ubiquitin promoter. Green fluorescent protein (GFP) signals were visualized in the transformed roots. In the EV control, root hairs showed normal tip growth, and the root hairs grew straight and away from the primary root axis (n = 30) (Supplemental Figures 8A and 8B). However, in hairy roots overexpressing LjROP6, root hairs were morphologically similar to those of the control in that they displayed normal tip growth, but approximately one-third of the hairy roots formed shorter root hairs (25/82; Supplemental Figures 8A to 8C). Surprisingly, all LjROP6 CA (42/42) and LjROP6 DN (51/51) lines formed completely depolarized ballooning root hairs on transgenic roots (Supplemental Figures 8A to 8C).
We analyzed the infection events in the LjROP6 overexpression, LjROP6 CA, and LjROP6 DN transgenic hairy roots after M. loti R7A/lacZ inoculation. The LjROP6 overexpression line formed elongated ITs similar to those of the EV control but had a few infection foci in uncurled root hairs and formed some disorientated ITs (Supplemental Figures 8D and 8E). The number of infection events (foci and ITs) in hairy roots appeared to be slightly higher in the LjROP6 overexpression line than in the EV control, but this difference was not significant (Supplemental Figure 8H). In the hairy roots of LjROP6 CA and LjROP6 DN, most ITs were abnormally short and formed without root hair curling (Supplemental Figures 8F and 8G). In addition, the number of infection events was significantly reduced in LjROP6 CA and LjROP6 DN hairy roots versus the EV control (Supplemental Figure 8H). LjROP6 transcript levels in hairy roots were higher in the LjROP6 overexpression, LjROP6 CA, and LjROP6 DN lines than in the EV control (Supplemental Figure 8I). These observations demonstrate that LjROP6 homeostasis is essential for its function in root hair development and IT formation.
We investigated the functions of LjROP1 and LjROP3 in root hair development by expressing LjROP1 or LjROP3 cDNA under the control of the LjUb promoter in wild-type L. japonicus hairy roots and analyzing their CA and DN forms. In all of these transgenic lines, the hairy roots formed shorter root hairs than the EV control (Supplemental Figure 9), which is similar to the phenotype of LjROP6-overexpressing L. japonicus hairy roots.
Actin Filaments Are Disordered in the Root Hairs of Ljrop6-1 Compared with Wild-Type L. japonicus
The actin cytoskeleton is important for root hair tip growth. To determine whether LjROP6 regulates the arrangement of actin filaments to influence the polarized growth of root hair tips, we used Alexa Fluor 488-conjugated phalloidin to stain wild-type and Ljrop6-1 roots. As expected, the root hairs of wild-type plants predominantly displayed the characteristic arrangement of actin filaments in long cables aligned longitudinally (Supplemental Figure 10A). However, 70% of Ljrop6-1 root hairs had fewer longitudinally aligned actin filaments and significantly more transversely oriented ones compared to the wild type (Supplemental Figure 10B). Most of the short swollen or medium-length root hairs of Ljrop6-1 showed disordered or web-like arrangements of actin filaments (Supplemental Figure 10C). These results indicate that LjROP6 affects the arrangement of actin filaments in root hairs to regulate their development.
DISCUSSION
Rhizobial ITs elongate following the migration of the nucleus along and within the root hairs via polar tip growth. ROP GTPases are key components required for the polarized growth of cells during processes such as pollen tube elongation and root hair development (Zheng and Yang, 2000; Samaj et al., 2006). LjROP6 interacts with the NF receptor LjNFR5 to mediate nodulation, and LjROP6 associates with LjCHC1 (Ke et al., 2012; Wang et al., 2015). The CHC1-hub domain, a dominant effector of clathrin-mediated endocytosis, impairs rhizobial infection (Wang et al., 2015). Here, we demonstrated that LjROP6 is required for the polarized growth of ITs in root hairs. GTPases act as molecular switches that fluctuate between an inactive GDP-bound form and an active GTP-bound form. We found that the DOCK-family GEF protein LjSPK1 interacts with, and activates, LjROP6. We also found that LjROP6 promotes the localization of LjSPK1 to the PM and that this may be important for its function. Together, our results show that the DOCK-family GEF LjSPK1 activates LjROP6 to mediate the polarized growth of ITs, and they provide evidence that early NF signaling is connected to morphological changes associated with rhizobial infection.
Rhizobial invasion into host plants via the IT requires an actively growing root hair. Inside the root hair, an infection chamber forms and elongates to form the IT, the tubular structure that guides bacteria toward cortical cells (Fournier et al., 2008, 2015). ROP GTPases play important roles in the polarized growth of cells. ROPs are polarized into single, discrete PM domains in tip-growing cells such as pollen tubes and root hairs (Lin et al., 1996; Molendijk et al., 2001). The NF receptor LjNFR5 interacts with LjROP6 to mediate IT formation and nodulation (Ke et al., 2012). In this study, we identified three LORE1 insertion mutants of Ljrop6. All three mutants were able to form normal ITs after rhizobial inoculation. However, compared to the wild type, the mutants had fewer infection events (foci and ITs) and formed many more disoriented ITs in root hairs. This observation suggests that LjROP6 mediates the polarized growth of ITs in root hairs.
Disoriented ITs have also been observed in other mutants, such as mutants of the cytokine receptor gene LjLhk1 (Murray et al., 2007) and topoisomerase VI (TOPO6A) subunit LjSUNERGOS1 or LjVAG1 (Suzaki et al., 2014; Yoon et al., 2014). These mutants exhibit misdirected ITs and highly defective nodule organogenesis, which in turn affects nodule primordium formation. However, in the Ljrop6 mutants observed in this study, the ITs formed loops and showed disoriented growth in root epidermis cells, but nodule formation was not affected. This finding suggests that the LjROP6-mediated polar elongation of ITs is not associated with nodule primordium formation. The Golgi/trans-Golgi-localized Rab GTPase PvRabA2 is required for IT progression and the maintenance of membrane integrity in common bean (Phaseolus vulgaris; Blanco et al., 2009; Dalla et al., 2017). In M. truncatula, the LIN-VPY-Exo70H4 complex shows a polar distribution in pre-ITs, suggesting that this complex mediates the polarized growth of ITs via exocytosis (Liu et al., 2019b). In the future, it would be interesting to determine whether LjROP6 interacts with this complex to guide the polarized growth of ITs.
ROP GTPases regulate cell growth and shape by organizing the actin filaments that drive cytoplasmic streaming and vesicle trafficking in the cytoskeleton. These GTPases also affect the arrangement of cortical membrane-associated microtubules, which guide cell wall deposition via cellulose synthase complexes (Fu et al., 2005; Endler and Persson, 2011; Tominaga and Ito, 2015). ROP GTPase activity is regulated by GAPs, GEFs, and RhoGDIs (Yang, 2002; Kost, 2008). SPK1 is a DOCK-family GEF that is conserved in animals and plants (Meller et al., 2005). In Arabidopsis, AtSPK1 has been implicated in activating ROP signaling to regulate actin polymerization via WAVE and ARP2/3 complexes in leaves (Qiu et al., 2002; Basu et al., 2008). Similarly, in rice, OsPit interacts with OsSPK1 to activate OsRAC1 and trigger immune responses (Wang et al., 2018). These results indicate that LjSPK1 interacts with and activates LjROP6 and that both LjSPK1 and LjROP6 are involved in the polarized growth of ITs following rhizobial infection. The formation of ITs requires several components of the WAVE/SCAR-ARP2/3 complex, including NAP, PIR, SCARN, and ARPC1 (Yokota et al., 2009; Hossain et al., 2012; Qiu et al., 2015). It will be important to investigate whether the LjSPK1-LjROP6 module interacts with the WAVE/SCAR-ARP2/3 complex to regulate actin rearrangement and mediate IT formation.
Exocytosis and the polar inhibition of clathrin-dependent endocytosis regulate the polarized growth of cells in both animals and plants. Studies of ROPs/RACs have revealed that these processes are important targets of Rho-GTPase signaling (Craddock and Yang, 2012; Craddock et al., 2012; Feiguelman et al., 2018). For example, the yeast exocyst subunit SEC3 is a direct effector of the Rho GTPase Cdc42. In Arabidopsis, the ROP effector ICR1/RIP1 (INTERACTOR OF CONSTITUTIVE ACTIVE ROP1) interacts with the Arabidopsis SEC3 homolog and is required for the polar localization of PIN1 (Lavy et al., 2007; Hazak et al., 2010). SPK1 is required for the auxin-activated ROP6-RIC1 pathway to inhibit the internalization of PIN2 by stabilizing actin microfilaments. These processes affect auxin distribution and lateral root development (Lin et al., 2012). In L. japonicus, LjROP6 interacts with LjCHC1 and is involved in rhizobial infection (Wang et al., 2015). Interestingly, LjROP6 alone localizes to the PM but shows a punctate distribution on the PM when it is coexpressed with LjCHC1 (Wang et al., 2015) or LjSPK1, as demonstrated in the BiFC assay between LjSPK1 and LjROP6 in N. benthamiana leaves in this study. Further studies are needed to investigate whether LjSPK1-activated LjROP6 can activate other downstream effectors such as RICs or RIPs. Such interactions could control the polarized growth of rhizobial ITs via effects on the organization of actin microfilaments, auxin distribution, and exocytosis and endocytosis pathways.
AtSPK1 is localized to subdomains of the ER (Zhang et al., 2010), and OsSPK1 (Wang et al., 2018) and LjSPK1 (this study) show a punctate distribution and colocalize with an ER marker. However, coexpressing OsSPK1-OsRAC1 or LjSPK1-LjROP6 in N. benthamiana leaves enhanced the PM localization of SPK1, suggesting that OsSPK1-OsRAC1 or LjSPK1-LjROP6 interact at the PM (Wang et al., 2018). In this study, LjSPK1 alone was distributed at the PM and in punctate spots around the nucleus in L. japonicus, but it interacted with LjROP6 in the PM and infection pocket in curled root hairs of L. japonicus infected with M. loti. In rice, OsRacGEF1 is transported from the ER to the PM with OsCERK1 via a vesicle trafficking pathway (Akamatsu et al., 2013). It would be interesting to explore which pathway is involved in the translocation of LjSPK1 from the ER to the PM.
GEFs are activated by PM-bound receptor-like kinases (Akamatsu et al., 2013; Liao et al., 2017). In mammals, the kinase Akt and the protein phosphatase PP2A interact with the DHR2 domain of DOCK6. DOCK6 is phosphorylated by Akt and dephosphorylated by PP2A at Ser1194; the phosphorylation status of DOCK6 determines its ability to promote axon growth (Miyamoto et al., 2013). A phosphoproteomic study of root nodule symbiosis in M. truncatula revealed that two phosphoisoforms of DOCK-family proteins, MtSPK1 and DOCK7, displayed lower phosphorylation levels upon NF treatment in nfp and dmi3 than in the wild type. Those findings suggest that rhizobia infection may affect the phosphorylation status of MtSPK1 (Rose et al., 2012). Thus, it will be important to investigate whether there is a kinase that affects the phosphorylation status of LjSPK1 to regulate its activity toward LjROP6 during the establishment of symbiosis.
ROP GTPases play an essential role in controlling the polarized growth of pollen tubes and root hairs. In Arabidopsis, AtSPK1 functions as a GEF that interacts with and activates a series of ROP GTPases (Basu et al., 2008). Our results show that LjSPK1 interacts with three type I ROP GTPases in L. japonicus. All three LjROP GTPases are required for the polarized growth of root hairs. The Ljrop mutants in this study formed branched and swollen root hairs, while lines overexpressing the LjROP GTPases and their CA or DN forms produced short, ballooning root hairs. These phenomena are not consistent with the previous finding that plants expressing the DN and CA forms of ROP showed different phenotypes (Molendijk et al., 2001; Jones et al., 2002; Lei et al., 2015). This discrepancy can be explained by the notion that ROP GTPases function as molecular switches. Because the homeostasis of their activity is strictly regulated, either stronger or weaker activity will affect their function. Although LjSPK1 can interact with three ROP GTPases, only one of them, LjROP6, is required for the polar elongation of ITs in root hairs. Perhaps LjROP6 interacts with LjNFR5 to transduce NF signaling to mediate the progression and elongation of ITs. Interestingly, in a study of M. truncatula ROP10, only the MtROP10 CA form showed defects in the polarized growth of root hairs; its infection phenotype was very similar to those of the LjROP6 CA and LjROP6 DN forms. However, although MtROP10 was able to interact with the NF receptor MtNFP and it was required for root hair deformation, it was not required for the polarized growth of ITs (Lei et al., 2015). Therefore, these and previous findings suggest that different ROP GTPases that are associated with NF signaling mediate different infection processes.
Prenylation is required for the membrane attachment and function of type I ROP GTPases, and LjROP6 is a type I ROP GTPase. When we examined the subcellular localization of LjROP6 in N. benthamiana, the placement of the fluorescent protein at the N terminus or C terminus of this protein did not affect its PM localization. Moreover, in the complementation assay, LjROP6 tagged with mCherry at its C terminus fully rescued the infection phenotype of Ljrop6-1. These results suggest that the prenylation of LjROP6 has minor effects on its function, which are consistent with the results of another study in Arabidopsis (Sorek et al., 2011).
The penetration of bacteria into legume roots is a key step in the specific recognition of compatible rhizobia during the formation and progression of ITs. These and previous findings provide genetic evidence that the molecular machinery associated with the LjNFR5-LjROP6-LjSPK1 module is involved in the vesicle trafficking and cytoskeleton rearrangements required for the polarized growth of ITs. Thus, we suggest that the LjNFR5-LjROP6-LjSPK1 module has been co-opted to participate in some events in rhizobial infection, particularly the polarized growth of ITs (Supplemental Figures 11A and 11B). Our results also show that LjSPK1 can activate other type I ROP GTPases such as LjROP1 and LjROP3 and that these ROP GTPases are required for the growth of the root hair tip (Supplemental Figure 11C).
METHODS
Biological Materials and Strains
Lotus japonicus accession Gifu B-129 was used in this study. The Ljrop and Ljspk1 mutants were obtained from the LORE1 transposon insertion library at Lotus Base (https://lotus.au.dk; Fukai et al., 2012; Urbański et al., 2012). The mutants were genotyped by PCR and sequenced using the primers shown in Supplemental Data Set 1.
The rhizobium strain Mesorhizobium loti R7A carrying pXLGD4 (lacZ), pMP2444 (GFP), or mTag was used for infection and the nodulation assays. The Agrobacterium rhizogenes strain AR1193 was used for hairy root transformation of L. japonicus and Medicago truncatula roots, and the Agrobacterium tumefaciens strain EHA105 was used for transient expression in N. benthamiana. Plasmids were transformed into Escherichia coli DH10B or DH5α for cloning or into E. coli Rosetta (DE3) for protein expression. The yeast (Saccharomyces cerevisiae) strain NMY51 was used for the DUAL membrane yeast two-hybrid system.
Cloning, DNA Manipulation, and Plasmid Construction
The coding sequences (CDS) of the LjROPs and LjSPK1 were amplified from the Gifu cDNA library by PCR amplification; the CA and DN forms of LjROPs were generated with a Hieff Mut site-directed mutagenesis kit (Yeasen Biotechnology) as per the manufacturer’s instructions.
For expression analysis of LjSPK1 in L. japonicus hairy roots, the LjSPK1 promoter (2 kb upstream of the start codon) was amplified from Gifu genomic DNA by PCR. The PCR product was inserted into pDONR207 by BP reaction (Invitrogen), and combined into pKGWFS7 to generate the pLjSPK1:GUS construct by LR reaction (Invitrogen).
For the LjSPK1 RNAi construct, the CDS fragment of LjSPK1 was amplified by PCR. The PCR product was inserted into pDONR207 by BP reaction (Invitrogen) and combined into pUB-GWS-GFP to generate the LjSPK1-Ri construct by LR reaction (Invitrogen).
To overexpress LjSPK1 or LjROPs in L. japonicus hairy roots, the LjSPK1, LjROPs, LsROPs CA, and LjROPs DN CDS were transferred from X-pDONR207 into pUB-GW-GFP by LR reaction to generate the LjSPK1-OX, LjROPs-OX, LjROPs CA, or DN constructs, respectively.
For the DUAL membrane yeast two-hybrid system, the PCR products and pCCW-STE or pDSL-Nx were digested with Sfi1, and the LjROPs and LjSPK1-DHR2 were inserted into pCCW-STE and pDSL-Nx, respectively, using T4 DNA ligase (Takara).
To measure guanine nucleotide exchange activity, the LjROP6 and LjSPK1-DHR2 PCR products were recombined into pDONR207 by BP reaction (Invitrogen). LjROP6 pDONR207 and LjSPK1-DHR2 pDONR207 were recombined into pHGWA with a His-tag or pGGWA with a glutathione S-transferase-tag by LR reaction and transformed into E. coli Rosetta for protein expression.
For the luciferase biomolecular complementation assays in N. benthamiana, the PCR products and luciferase vector pCambia1300-35S-nLuc or pCambia1300-35S-cLuc were digested with Kpn1 and Sal1, and LjSPK1-DHR2 and LjROP6 were inserted into pCambia1300-X-nLuc or pCambia1300-cLuc-X using T4 DNA ligase.
For co-IP assays in N. benthamiana, the PCR products (LjROP6, LjROP6 CA, LjROP6 DN, or LjROP10) were inserted into destination vector pCambia1305-35S-Myc following Kpn1 and Sal1 digestion. The LjSPK1-DHR2 PCR products were inserted into destination vector pUB-GFP/X-FLAG, which was modified from pUB-GFP (Maekawa et al., 2008; Li et al., 2019), following Kpn1 and Asc1 digestion.
To determine the subcellular localization of LjROP6 in L. japonicus leaves and L. japonicus hairy roots, LjROP6 pDONR207 was recombined into pK7WGF2 to generate GFP-LjROP6 by LR reaction. LjROP6 PCR products were inserted into pUB-GFP/X-mCherry (this vector was modified from pUB-GFP; Li et al., 2019) following Xba1 and Kpn1 digestion using T4 DNA ligase to generate LjROP6-mCherry.
For the complementation assay via hairy root transformation of Ljrop6-1, the LjROP6 promoter (1.8 kb upstream of the start codon) was amplified from Gifu genomic DNA by PCR amplification. The PCR products and destination vector pUB-GFP were digested with Pst1 and Xba1, and the LjROP6 promoter was inserted into pUB-GFP using T4 DNA ligase to generate pLjROP6-GFP. pLjROP6-GFP and pUB-GFP/LjROP6-mCherry were digested with Xba1 and Asc1, and LjROP6-mCherry was inserted into pLjROP6-GFP using T4 DNA ligase to generate pLjROP6:LjROP6-mCherry. This vector was also used to determine the subcellular localization of LjROP6 in L. japonicus transformed hairy roots.
To determine the subcellular localization of LjSPK1 in N. benthamiana leaves, the LjSPK1 PCR products were inserted into pCambia1300-35S-YFP following Sma1 and Spe1 digestion to generate LjSPK1-YFP. For LjSPK1 localization and BiFC analysis in N. benthamiana leaves, L. japonicus, or M. truncatula hairy roots, the LjSPK1-eGFP and BiFC constructs were generated by Golden Gate cloning (Weber et al., 2011). The LjSPK1 CDS was synthesized in the level 0 vector pL0V-SC3 (Shanghai Xitubio Biotechnology). This vector and the EC16570 vector were digested with Bsa1 to generate LjSPK1-eGFP as the level 1 construct. This level 1 LjSPK1-eGFP was assembled into EC50507 (https://www.ensa.ac.uk/), adding p35S:NLS-DsRed as a transgenic marker, to generate the level 2 construct LjSPK1-eGFP. The synthesized LjSPK1 CDS in vector pL0V-SC3 was assembled into EC12850 to generate LjSPK1-cVenus; the LjROP6 PCR product was inserted into pL0V-SC3 following Bbs1 digestion, and this construct was assembled into EC12849 to generate LjROP6-nVenus. Finally, these constructs were assembled into EC50507, adding p35S:DsRed as a transgenic marker, to generate the BiFC construct pLjUb:LjSPK1-cVerns/pAtUb:LjROP6-nVenus.
All PCR amplification was performed using high-fidelity DNA polymerase KOD Plus (Toyobo) or MAX (Vazyme), and all constructs were confirmed by DNA sequencing. The constructs in destination vectors were introduced into A. rhizogenes AR1193 for hairy root transformation in L. japonicus or M. truncatula or into A. tumefaciens strain EHA105 for transient expression in N. benthamiana by electroporation. All primers are listed in Supplemental Data Set 1, and all constructs are listed in the Supplemental Table.
Analysis of Plant Growth, Rhizobial Inoculation, Infection, Nodulation, Root Hairs, and Actin Rearrangement
L. japonicus seeds were scarified, surface sterilized in 10% (v/v) sodium hypochlorite for 7 min, and rinsed five times with sterile water. The sterilized seeds were imbibed in water, transferred to 0.8% (v/v) water agar plates, and grown in 22°C in the dark for 3 to 4 d for germination. The seedlings were transferred to a mixture of perlite and vermiculite (1:1), cultivated in a growth chamber under a 16 h/8 h light (250 µmol/m2/s)/dark cycle, and inoculated with rhizobia at 5 to 7 d after transfer. The infection phenotypes and events were determined at the indicated time points by laser scanning confocal microscopy (FV1000, Olympus) using GFP-marked M. loti R7A or light microscopy (ECLIPSE Ni, Nikon) after staining the roots with 5-bromo-4-chloro-3-indolylbeta-D-galacto-pyranoside (X-Gal).
To prepare nodule sections, the nodules were fixed in glutaraldehyde (2.5% [v/v]), embedded in Technovit 7100 resin (Kulzer) according to the manufacturer’s instructions, and cut into 5- to 10-mm transverse sections with a microtome (RM2265, Leica Microsystems). Root nodule sections were costained with toluidine blue or magenta-Gal to visualize GUS or M. loti.
To analyze the root hair phenotypes of the Ljrop mutants, the seedlings were transferred to glass slides containing 1 mL liquid Fahraeus medium and incubated overnight. The seedlings were inoculated by adding fresh Fahraeus medium with or without M. loti R7A (OD600nm∼0.01) and incubated in the dark for ∼18 h before analysis. The root hairs were observed and imaged under a light microscope (Nikon ECLIPSE Ni). For observation root hairs in L. japonicus hairy roots, transformed hairy roots of the LjROPs overexpression, LjROPs CA, and LjROPs DN lines were scored by GFP fluorescence using a Nikon SMZ1500 microscope, and then root hairs were observed and imaged under a light microscope (Nikon ECLIPSE Ni). The length of root hairs was measured using ImageJ. Five root hair cells were measured per transformed root, and at least 10 transformed roots were scored.
Phalloidin staining was done as previously described by Yokota et al. (2009), and actin was observed by laser scanning confocal microscopy (FV1000, Olympus).
All assays were validated in at least three independent experiments.
Analysis of Promoter:GUS, RNAi, Overexpression, and Complementation in L. japonicus Transformed Hairy Roots
The indicated constructs in A. rhizogenes AR1193 were introduced into L. japonicus Gifu or Ljrop6-1 roots on 1/2 B5 medium via hairy root transformation. After 2 weeks, the transformed hairy roots were scored by GFP fluorescence, as observed under a Nikon SMZ1500 microscope. The transformed chimeric plants were transferred to pots filled with a mixture of vermiculite and perlite (1:1) and inoculated with M. loti R7A/lacZ at 5 to 7 days after transfer. For pLjSPK1:GUS in L. japonicus hairy roots, the hairy roots were stained with both magenta-Gal and X-Gluc to visualize M. loti R7A and gene expression at the indicated time points. For the LjSPK1-Ri, LjSPK1-OX, and LjROP6 complementation assays, the transgenic roots were stained with X-Gal, and the infection events were counted under an ECLIPSE Ni light microscope (Nikon) at 5 or 7 days after inoculation. At least seven plants were analyzed per experiment. The number of nodules was counted at 2 or 3 weeks after inoculation with M. loti R7A/lacZ using at least nine plants at each time point. These assays were repeated in three individual experiments.
RNA Extraction and RT-qPCR
Total RNA was extracted from plant roots with TRIpure reagent (Aidlab Biotechnologies) according to the manufacturer’s instruction and quantified using a Nanodrop 2000 instrument (Thermo Scientific). Reverse transcription of cDNA was performed using TransScript one-step gDNA removal and cDNA synthesis SuperMix (TransGen Biotech). RT-PCR was conducted using SYBR green real-time PCR master mix (Toyobo), and the products were detected using the ABI StepOnePlus PCR system (Thermo Fisher Scientific). Amplification conditions consisted of an initial denaturation step at 95°C for 20 s, 40 cycles at 95°C for 5 s and 60°C for 30 s, followed by a melting curve stage of 95°C for 15 s, 60°C for 60 s and 95°C for 10 s. Ubiquitin (Lj5g3v2060710.1) was used as an internal control, and RT-qPCR data for each sample were normalized to the respective Ubiquitin expression level. Standard errors and statistical significance based on three biological replicates were calculated using the 2−ΔΔCt method. The primers used are shown in Supplemental Data Set 1.
Scanning Electron Microscopy
The leaves of the Ljspk1 mutants were fixed in formalin-aceto-alcohol solution (5% [v/v] formalin, 5% [v/v] acetic acid and 45% [v/v] alcohol) overnight at 4°C and dehydrated in in an ethanol series (30%, 50%, 70%, 80%, 90%, 95%, and 100% [v/v] ethanol) for 5 min per step at room temperature. The leaves were dried using a carbon dioxide critical point drier (LEICA CPD300), transferred onto a copper mount for ion sputtering, and imaged under a scanning electron microscope (JSM-6360LV, JEOL) using 1 to 5 kV accelerating voltage.
Phylogenetic Analysis
The Arabidopsis, M. truncatula, and L. japonicus protein sequences were obtained from the genome databases at TAIR, M. truncatula Mt4.0, and L. japonicus miyakogusa.jp website version 3.0. Soybean (G. max), common bean (P. vulgaris), and pea (Pisum sativum) SPK1 protein sequences were obtained from Legume Information System (https://www.legumeinfo.org/). Rice (Oryza sativa) OsSPK1, Homo sapiens DOCK180, nematode (Caenorhabditis elegans) CED-5, fruit fly (Drosophila melanogaster) MYOBLAST CITY (MBC), yeast Ylr422wp, and slime mold (Dictyostelium discoideum) DocA protein sequences were obtained from the National Center for Biotechnology Information. All protein sequences were aligned using ClustalW, and the phylogenetic tree was built using the maximum likelihood method in MEGA7. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a Jones Taylor Thornton model and selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths representing the number of substitutions per site. Sequence alignments of SPK1 proteins and ROP GTPases are shown in Supplemental Files 1 and 2, respectively.
DUAL Membrane Yeast Two-Hybrid Assays
The yeast strain NMY51 was transformed with the constructs in destination vectors as per manufacturer’s instructions using lithium acetate transformation (Yeast Protocols Handbook PT3024-1, Clontech). The transformants were grown on synthetic defined medium (0.67% yeast nitrogen base, 2% Glc, 2% Bacto-agar and amino acid mix) without the appropriate auxotrophic markers and in the presence of 3-amino-1,2,4-triazole at different concentrations. These assays were repeated three times.
Luciferase Biomolecular Complementation Assays
cLuc-LjROP6, cLuc-LjROP6 CA, or cLuc-LjROP6 DN was coexpressed with LjSPK1-DHR2-nLuc in N. benthamiana leaves via agroinfiltration along with p19, which inhibits gene silencing. The transformed plants were grown in a growth chamber. After 2 to 3 days, Luc images were captured by a charged coupled device camera (Tanon) after spraying the leaves with 1 mM luciferin. All images were acquired using the same exposure settings. Each interaction group was validated in at least three replicates, and three independent experiments were performed.
BiFC Assays
The LjSPK1 and LjROP6 BiFC construct (pLjUb:LjSPK1-cVenus/pAtUb:LjROP6-nVenus) was expressed in N. benthamiana leaves by agroinfiltration, along with p19. The transformed plants were grown in a growth chamber, and 2 to 3 d later, the images were captured by laser scanning confocal microscopy (TCS SP8, Leica). Leaves were immersed in 30% (w/v) Suc for 30 min for the plasmolysis experiment. The LjSPK1-LjROP6 BiFC construct was also expressed in wild-type L. japonicus or M. truncatula sunn-1 by hairy root transformation. The transgenic hairy roots were scored based on NLS-DsRed marker and inoculated with M. loti R7A/mTag or Rm1021/mCherry (OD600 ∼0.01). The images were analyzed at 5 to 7 d after inoculation. The filter sets for excitation (ex) and emission (em) were as follows: Venus (ex/em, 514 nm/524 to 545 nm), mCherry/DsRed (ex/em, 561 nm/600 to 630 nm) and mTag (ex/em, 405 nm/415 to 454 nm). All BiFC experiments were repeated twice, and at least five leaves or roots were analyzed each time.
Subcellular Localization in L. japonicus Transformed Hairy Roots and N. benthamiana Leaves
To determine the subcellular localization of LjSPK1 and LjROP6 in L. japonicus hairy roots, the indicated constructs in A. tumefaciens strain AR1193 were expressed in L. japonicus hairy roots, and the transgenic roots were inoculated by M. loti R7A/mTag. The subcellular localization of the proteins was observed by laser scanning confocal microscopy (TCS SP8, Leica). The filter sets for ex and em were as follows: GFP (ex/em, 488 nm/505 to 550 nm), FM4-64/mCherry/DsRed (ex/em, 561 nm/600 to 630 nm) and mTag (ex/em, 405 nm/415 to 454 nm).
To observe the subcellular localization of LjSPK1 and LjROP6 in N. benthamiana leaves, constructs in A. tumefaciens strain EHA105 were expressed in N. benthamiana leaves by agroinfiltration along with p19. Agrobacterium cells were grown overnight, collected, resuspended in buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone), adjusted to OD600 (∼0.4), and incubated at 28°C for 2 h before infiltration. Three or four leaves were selected for injection. Following injection, the plants were kept in a growth chamber under a 16 h/8 h light (250 μmol/m2/s)/dark cycle. After 2 to 3 d, the leaves were observed by confocal microscopy (Olympus FV1000 or Leica TCS SP8). Leaves were immersed in 30% (w/v) Suc for 30 min for plasmolysis or FM4-64 solution for colocalization with PM. The filter sets were as follows: GFP (ex/em, 488 nm/505 to 550 nm), YFP (ex/em, 514 nm/524 to 545 nm), FM4-64/mRFP/mCherry (ex/em, 561 nm/600 to 630 nm). Gray values were analyzed using ImageJ. We used GFP beads (SMART, CAT.SA070001) for immunoprecipitation, and GFP antibody (Abmart, CAT.M20004) and mCherry antibody (AffinitY, CAT.T0090) for immunoblot analysis. These assays were validated with three independent experiments. All protein subcellular localization assays were repeated in at least three independent experiments.
Protein Extraction and Co-IP Assays
Constructs in different combinations (ROP6-Myc, ROP6 CA-Myc, ROP6 DN-Myc, ROP10-Myc, and SPK1-Flag) were transformed into N. benthamiana leaves by agroinfiltration along with p19. The plants were grown in a growth room for 48 h under a 16 h/8 h light (250 μmol/m2/s)/dark cycle. Approximately 0.3 g leaf tissue was harvested, and leaf proteins were extracted with 1 mL extraction buffer (50 mM TRIS-MES, pH 8.0, 0.5 M Suc, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail CompleteMini tablets). The mixture was centrifuged at 10,000g at 4°C for 15 min, and the supernatant was collected for co-IP assays. We used Myc beads (Abmart, CAT.M20012L) for immunoprecipitation and Flag antibody (HuiOu Biotech, CAT.HOA012FL01) and Myc antibody (Abmart, CAT.M2000M) for immunoblotting. These assays were validated with three independent experiments.
Guanine Nucleotide Exchange Activity Assay
To investigate the guanine nucleotide exchange activity of ROP6 and determine how it is affected by SPK1, the constructs in destination vectors described above were introduced into E. coli Rosetta cells by electroporation. The guanine nucleotide exchange assay was essentially performed as described previously, with small modifications by Gu et al. (2006) and Wang et al. (2017). The guanine nucleotide exchange activity of ROP6 at different concentrations and the effect of SPK1 on this activity were determined by monitoring fluorescence using a Varioskan Flash (Thermo Fisher Scientific). The changes in fluorescence intensity were recorded every 4 s for 600 s (ex/em 360 nm/440 nm). This assay was repeated three times.
Statistical Analyses
Statistical significance was calculated by two-tailed Student’s t test (*P < 0.05, **P < 0.01) and error bars indicate SD. Vertical box plots were generated using GraphPad Prism 6.0 software. For each box plot, the line in the box represents the median; the boxes show the upper and lower quartiles; the whiskers represent the maximum and minimum values. One-way ANOVA followed by Tukey’s multiple comparisons test was used to determine the differences. Values of P < 0.05 were considered to be statistically significant. The results of statistical analyses are shown in Supplemental Data Set 2.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers MT701563 for LjSPK1 and the Lotus Base Gifu v1.2 (https://lotus.au.dk/) for LotjaGi3g1v0299000. The LjROP GTPase sequences can be found in miyakogusa.jp v3.0 (http://www.kazusa.or.jp/lotus/) under the following accession numbers: LjROP1 (Lj1g3v3331690), LjROP3 (Lj2g3v1670990), LjROP5 (Lj3g3v3453430), LjROP6 (Lj0g3v0167719), and LjROP10 (Lj1g3v0415230).
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of DOCK family GEF proteins and growth phenotypes of Ljspk1 mutants.
Supplemental Figure 2. Symbiotic phenotypes of wild type (Gifu) and Ljspk1 mutants.
Supplemental Figure 3. Interaction between LjSPK1 and L. japonicus ROP GTPases in yeast cells, and the transcription levels and infection events of Ljrop mutants.
Supplemental Figure 4. LjROP6 transcript levels and nodulation phenotypes of Ljrop6 mutants.
Supplemental Figure 5. Root hair phenotypes of Ljrop6-2 and Ljrop6-3 mutants.
Supplemental Figure 6. LjROP6 protein subcellular localization in N. benthamiana epidermal leaf cells and wild-type L. japonicus hairy roots.
Supplemental Figure 7. LjSPK1 subcellular localization in Ljrop6-1 and BiFC assay of LjSPK1 and LjROP6 interactions in L. japonicus wild-type and Mtsunn-1 hairy roots.
Supplemental Figure 8. Root hairs growth and rhizobial infection in LjROP6 overexpression, LjROP6 CA, and LjROP6 DN lines.
Supplemental Figure 9. Root hair phenotypes of LjROP1/3 overexpression, CA, and DN lines.
Supplemental Figure 10. Actin filaments are disordered in Ljrop6-1 root hairs compared with wild type.
Supplemental Figure 11. A proposed model of LjSPK1-LjROP function in rhizobial IT formation and polarized growth of root hairs.
Supplemental Table. Constructs used in this study.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. ANOVA and Student’s t test results for the data shown in the figures.
Supplemental File 1. Sequence alignment of SPK1 proteins.
Supplemental File 2. Sequence alignment of ROP GTPase proteins.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Phil Pool (University of Oxford, United Kingdom) for kindly providing us with the M. loti R7A/mTag strain, Jeremy Murray (CAS Center for Excellence in Molecular Plant Sciences [CEMPS], Chinese Academy of Sciences, China) for sharing the Golden Gate vectors, and Yoji Kawano (Shanghai Center for Plant Stress Biology, CAS, China) and Zhao Liu (Hebei University of Chinese Medicine) for suggestions about GEF enzyme activity. We thank Yongfei Wang (CEMPS, CAS, China) for helpful discussions on this study. This work was funded by the National Key R&D Program of China (grant 2016YFA0500500), the Strategic Priority Research Program of Chinese Academy of Sciences (grant XDB27040208), and the International Partnership Program of CAS (grant 153D31KYSB20160074).
AUTHOR CONTRIBUTIONS
J.L. and F.X. designed the experiments; J.L. performed most of the experiments; M.X.L. and L.P.Q. isolated the rop6-1 and spk1-1 mutants, respectively; J.L. and F.X. analyzed the data and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Akamatsu A., Wong H.L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., Shimamoto K.(2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476. [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., Godfroy O., de Billy F., Saurat O., Jauneau A., Gough C.(2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. USA 105: 9817–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F.A., Meschini E.P., Zanetti M.E., Aguilar O.M.(2009). A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21: 2797–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., Le J., Zakharova T., Mallery E.L., Szymanski D.B.(2008). A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc. Natl. Acad. Sci. USA 105: 4044–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin N.J.(2004). Plant cell wall remodelling in the rhizobium–legume symbiosis. Crit. Rev. Plant Sci. 23: 293–316. [Google Scholar]
- Craddock C., Lavagi I., Yang Z.(2012). New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 22: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C., Yang Z.(2012). Endocytic signaling in leaves and roots: Same rules different players. Front Plant Sci 3: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore J.V., Ranjeva R., Bono J.J.(2001). Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 6: 24–30. [DOI] [PubMed] [Google Scholar]
- Dalla V. V., Traubenik S., Rivero C., Aguilar O.M., Zanetti M.E., Blanco F.A.(2017). The monomeric GTPase RabA2 is required for progression and maintenance of membrane integrity of infection threads during root nodule symbiosis. Plant Mol. Biol. 93: 549–562. [DOI] [PubMed] [Google Scholar]
- Downie J.A., Walker S.A.(1999). Plant responses to nodulation factors. Curr. Opin. Plant Biol. 2: 483–489. [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Atkinson E.M., Long S.R.(1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000. [DOI] [PubMed] [Google Scholar]
- Eklund D.M., Svensson E.M., Kost B.(2010). Physcomitrella patens: A model to investigate the role of RAC/ROP GTPase signalling in tip growth. J. Exp. Bot. 61: 1917–1937. [DOI] [PubMed] [Google Scholar]
- Endler A., Persson S.(2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4: 199–211. [DOI] [PubMed] [Google Scholar]
- Esseling J.J., Lhuissier F.G., Emons A.M.(2003). Nod factor-induced root hair curling: Continuous polar growth towards the point of nod factor application. Plant Physiol. 132: 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguelman G., Fu Y., Yalovsky S.(2018). ROP GTPases structure-function and signaling pathways. Plant Physiol. 176: 57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J., Teillet A., Chabaud M., Ivanov S., Genre A., Limpens E., de Carvalho-Niebel F., Barker D.G.(2015). Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167: 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J., Timmers A.C., Sieberer B.J., Jauneau A., Chabaud M., Barker D.G.(2008). Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148: 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z.(2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700. [DOI] [PubMed] [Google Scholar]
- Fukai E., Soyano T., Umehara Y., Nakayama S., Hirakawa H., Tabata S., Sato S., Hayashi M.(2012). Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 69: 720–730. [DOI] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z.(2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O., Bloch D., Poraty L., Sternberg H., Zhang J., Friml J., Yalovsky S.(2010). A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol. 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.S., Liao J., James E.K., Sato S., Tabata S., Jurkiewicz A., Madsen L.H., Stougaard J., Ross L., Szczyglowski K.(2012). Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol. 160: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.M., Kobayashi H., Davies B.W., Taga M.E., Walker G.C.(2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Shen J.J., Fu Y., Li H., Yang Z., Grierson C.S.(2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D., Fang Q., Chen C., Zhu H., Chen T., Chang X., Yuan S., Kang H., Ma L., Hong Z., Zhang Z.(2012). The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 159: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E., Oláh B., Kaló P., Morales M., Heckmann A.B., Borbola A., Lózsa A., Kontár K., Middleton P., Downie J.A., Oldroyd G.E., Endre G.(2009). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiirika L.M., Bergmann H.F., Schikowsky C., Wimmer D., Korte J., Schmitz U., Niehaus K., Colditz F.(2012). Silencing of the Rac1 GTPase MtROP9 in Medicago truncatula stimulates early mycorrhizal and oomycete root colonizations but negatively affects rhizobial infection. Plant Physiol. 159: 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B.(2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 18: 119–127. [DOI] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N.H.(1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Bloch D., Hazak O., Gutman I., Poraty L., Sorek N., Sternberg H., Yalovsky S.(2007). A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17: 947–952. [DOI] [PubMed] [Google Scholar]
- Lei M.J., et al. (2015). The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 27: 806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z.(1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2019). Atypical receptor kinase RINRK1 required for rhizobial infection but not nodule development in Lotus japonicus. Plant Physiol. 181: 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Tang R., Zhang X., Luan S., Yu F.(2017). FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58: 1143–1150. [DOI] [PubMed] [Google Scholar]
- Lin D., et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wang Y., Zhu J.K., Yang Z.(1996). Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.W., et al. (2019a). NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 179: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.W., et al. (2019b). A protein complex required for polar growth of rhizobial infection threads. Nat. Commun. 10: 2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M.(2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382. [DOI] [PubMed] [Google Scholar]
- Meller N., Merlot S., Guda C.(2005). CZH proteins: A new family of Rho-GEFs. J. Cell Sci. 118: 4937–4946. [DOI] [PubMed] [Google Scholar]
- Miyahara A., Richens J., Starker C., Morieri G., Smith L., Long S., Downie J.A., Oldroyd G.E.(2010). Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol. Plant Microbe Interact. 23: 1553–1562. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Torii T., Yamamori N., Ogata T., Tanoue A., Yamauchi J.(2013). Akt and PP2A reciprocally regulate the guanine nucleotide exchange factor Dock6 to control axon growth of sensory neurons. Sci. Signal. 6: ra15. [DOI] [PubMed] [Google Scholar]
- Molendijk A.J., Bischoff F., Rajendrakumar C.S., Friml J., Braun M., Gilroy S., Palme K.(2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20: 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D.(2011). Invasion by invitation: Rhizobial infection in legumes. Mol. Plant Microbe Interact. 24: 631–639. [DOI] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K.(2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E.(2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A.(2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Jilk R., Marks M.D., Szymanski D.B.(2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Lin J.S., Xu J., Sato S., Parniske M., Wang T.L., Downie J.A., Xie F.(2015). SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PLoS Genet. 11: e1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely B.K., He H., Venkateshwaran M., Sarma B., Schraiber J., Ané J.M., Cook D.R.(2011). Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 65: 230–243. [DOI] [PubMed] [Google Scholar]
- Rose C.M., et al. (2012). Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11: 724–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Liu W., Nandety R.S., Crook A., Mysore K.S., Pislariu C.I., Frugoli J., Dickstein R., Udvardi M.K.(2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32: 15–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J., Müller J., Beck M., Böhm N., Menzel D.(2006). Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 11: 594–600. [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Sinharoy S., Liu C., Breakspear A., Guan D., Shailes S., Nakashima J., Zhang S., Wen J., Torres-Jerez I., Oldroyd G., Murray J.D., Udvardi M.K.(2016). A Medicago truncatula cystathionine-β-synthase-like domain-containing protein is required for rhizobial infection and symbiotic nitrogen fixation. Plant Physiol. 170: 2204–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N., Gutman O., Bar E., Abu-Abied M., Feng X., Running M.P., Lewinsohn E., Ori N., Sadot E., Henis Y.I., Yalovsky S.(2011). Differential effects of prenylation and s-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 155: 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ito M., Yoro E., Sato S., Hirakawa H., Takeda N., Kawaguchi M.(2014). Endoreduplication-mediated initiation of symbiotic organ development in Lotus japonicus. Development 141: 2441–2445. [DOI] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G.(1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tominaga M., Ito K.(2015). The molecular mechanism and physiological role of cytoplasmic streaming. Curr. Opin. Plant Biol. 27: 104–110. [DOI] [PubMed] [Google Scholar]
- Urbański D.F., Małolepszy A., Stougaard J., Andersen S.U.(2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 69: 731–741. [DOI] [PubMed] [Google Scholar]
- Wang C., Zhu M., Duan L., Yu H., Chang X., Li L., Kang H., Feng Y., Zhu H., Hong Z., Zhang Z.(2015). Lotus japonicus clathrin heavy Chain1 is associated with Rho-Like GTPase ROP6 and involved in nodule formation. Plant Physiol. 167: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Lei M., Chen A., Wang R., Li G., Wang Y.(2014). MtROP8 is involved in root hair development and the establishment of symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti. Chin. Sci. Bull. 59: 4289–4297. [Google Scholar]
- Wang Q., Li Y., Ishikawa K., Kosami K.I., Uno K., Nagawa S., Tan L., Du J., Shimamoto K., Kawano Y.(2018). Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. USA 115: E11551–E11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Liu Z., Bao L.J., Zhang S.S., Zhang C.G., Li X., Li H.X., Zhang X.L., Bones A.M., Yang Z.B., Chen Y.L.(2017). The RopGEF2–ROP7/ROP2 pathway activated by phyB suppresses red light-induced stomatal opening. Plant Physiol. 174: 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S.(2011). A modular cloning system for standardized assembly of mutltigene constructs. PLos One 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Murray J.D., Kim J., Heckmann A.B., Edwards A., Oldroyd G.E., Downie J.A.(2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. USA 109: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.(2002). Small GTPases: Versatile signaling switches in plants. Plant Cell 14: S375–S388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2009). CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J. 60: 168–180. [DOI] [PubMed] [Google Scholar]
- Yokota K., et al. (2009). Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.J., Hossain M.S., Held M., Hou H., Kehl M., Tromas A., Sato S., Tabata S., Andersen S.U., Stougaard J., Ross L., Szczyglowski K.(2014). Lotus japonicus SUNERGOS1 encodes a predicted subunit A of a DNA topoisomerase VI that is required for nodule differentiation and accommodation of rhizobial infection. Plant J. 78: 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Kotchoni S.O., Samuels A.L., Szymanski D.B.(2010). SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr. Biol. 20: 2144–2149. [DOI] [PubMed] [Google Scholar]
- Zheng Z.L., Yang Z.(2000). The Rrop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 5: 298–303. [DOI] [PubMed] [Google Scholar]