Rice lipid binding protein EPAD1 is a male meiocyte-derived determinant that controls grass-specific pollen exine pattern formation by maintaining primexine integrity.
Abstract
The highly variable and species-specific pollen surface patterns are formed by sporopollenin accumulation. The template for sporopollenin deposition and polymerization is the primexine that appears on the tetrad surface, but the mechanism(s) by which primexine guides exine patterning remain elusive. Here, we report that the Poaceae-specific EXINE PATTERN DESIGNER 1 (EPAD1), which encodes a nonspecific lipid transfer protein, is required for primexine integrity and pollen exine patterning in rice (Oryza sativa). Disruption of EPAD1 leads to abnormal exine pattern and complete male sterility, although sporopollenin biosynthesis is unaffected. EPAD1 is specifically expressed in male meiocytes, indicating that reproductive cells exert genetic control over exine patterning. EPAD1 possesses an N-terminal signal peptide and three redundant glycosylphosphatidylinositol (GPI)-anchor sites at its C terminus, segments required for its function and localization to the microspore plasma membrane. In vitro assays indicate that EPAD1 can bind phospholipids. We propose that plasma membrane lipids bound by EPAD1 may be involved in recruiting and arranging regulatory proteins in the primexine to drive correct exine deposition. Our results demonstrate that EPAD1 is a meiocyte-derived determinant that controls primexine patterning in rice, and its orthologs may play a conserved role in the formation of grass-specific exine pattern elements.
INTRODUCTION
Pollen is the male gametophyte of flowering plants and is enclosed within a multi-layered cell wall whose structure directs successful pollination and provides physical and chemical resistance against environmental stresses (Edlund et al., 2004; Jiang et al., 2013; Shi et al., 2015). Patterning of the pollen wall surface exhibits vast morphological diversity between species and constitutes an important feature of plant taxonomic classification (Blackmore and Barnes, 1990; Scott, 1994; Ariizumi and Toriyama, 2011). However, its structure and development also share common features across plant taxa. The outer wall (exine) is composed of a flat bottom layer (nexine) and an upper sexine layer, further divided into column-like baculae and roof-like tectum layers. The unpatterned inner wall (intine) is deposited between the microspore plasma membrane (PM) and nexine. After formation of the exine layers, the pollen coat (also known as tryphine or pollenkitt) fills the spaces between the baculae (Shi et al., 2015). While the general process of pollen wall development is well conserved in plants, the factors that regulate species-specific wall patterning remain unknown.

Pollen wall patterning, which affects inter- and intra-species recognition by the pistil, is largely determined by the exine structure (Wang and Dobritsa, 2018). Exine is composed predominantly of sporopollenin, a stable and durable biopolymer, whose deposition and polymerization are key determinants of the exine wall pattern. Sporopollenin precursors are synthesized and transported from the tapetum, the innermost layer of the anther wall (Quilichini et al., 2010; Choi et al., 2011; Dou et al., 2011; Quilichini et al., 2014). Genetic and biochemical analyses suggest that aliphatic units and phenylpropanoid derivatives are the major constituents of sporopollenin (Quilichini et al., 2015; Shi et al., 2015). Recently, aliphatic-polyketide-derived polyvinyl alcohol units, 7-O-p-coumaroylated C16 aliphatic units, and minor amounts of naringenin were found in sporopollenin of pitch pine (Pinus rigida; Li et al., 2019). In addition, phenylpropanoid derivatives including p-hydroxybenzoate, p-coumarate, ferulate, and lignin guaiacyl units were detected in lily (Lilium brownii) and Japanese cedar (Cryptomeria) sporopollenin (Xue et al., 2020). Interestingly, Xue et al. (2020) described land plants with a wide range of sporopollenin compositions.
Exine development starts with the establishment of a thin microfibrillar matrix (primexine) in the tetrad microspores between the callose wall and microspore PM. Primexine composition is similar to the primary cell wall, containing cellulose, pectin, and xylan, as well as arabinogalactan-proteins and lipoproteins (Dickinson and Heslop-Harrison, 1968; Li et al., 2017; Suzuki et al., 2017). Primexine is thought to serve as the template for exine deposition and subsequent assembly of sporopollenin into the initial precursors of tectum and baculae known as protectum and probaculae. In many species, regular undulation of the microspore PM is observed at this stage (Takahashi, 1989; Takahashi and Skvarla, 1991; Fitzgerald and Knox, 1995; Paxson-Sowders et al., 2001). It has been proposed that this PM undulation is caused by patterned deposition of materials in the primexine, called “spacers” (Fitzgerald and Knox, 1995), and that PM undulation is accompanied by deposition of probaculae on top of the membrane protrusions. However, while pollen grains of the grass species sorghum (Sorghum bicolor) and maize (Zea mays) generally follow a similar process of pollen wall formation, they do not exhibit PM undulation (Skvarla and Larson, 1966; Christensen et al., 1972).
Comparative phylogenomics and biochemical experiments have revealed that the genes that regulate sporopollenin biosynthesis and primexine formation are highly conserved across land plants (Scott, 1994; Ariizumi and Toriyama, 2011; Wallace et al., 2011; Jiang et al., 2013; Lou et al., 2014; Quilichini et al., 2015; Shi et al., 2015). Although many molecular factors required for primexine formation have been identified, the biological functions of most remain unknown. Several Arabidopsis (Arabidopsis thaliana) mutants exhibit abnormal, delayed, or absent primexine formation, for example mutations affecting the calcium binding protein DEFECTIVE IN EXINE FORMATION 1 (DEX1; Paxson-Sowders et al., 2001), the plastidic integral membrane protein NO EXINE FORMATION 1 (NEF1; Ariizumi et al., 2004), the sugar transporter RUPTURED POLLEN GRAIN 1 (RPG1, also named SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERS 8, or SWEET8) and its paralog RPG2 (Guan et al., 2008; Sun et al., 2013), the PM-localized protein NO PRIMEXINE AND PLASMA MEMBRANE UNDULATION (NPU; Chang et al., 2012), and the de novo DNA methyltransferase Exine Formation Defect (EFD; Hu et al., 2014). Polysaccharide biosynthetic components, such as the β-1,3-galactosyltransferase UNEQUAL PATTERN OF EXINE1/KAONASHI4 (UPEX1/KNS4) and the putative xylosyl transferases SPONGY2/ IRREGULAR XYLEM9-LIKE (SPG2/IRX9L) and its paralog IRX14L, are also required for primexine formation (Dobritsa et al., 2011; Li et al., 2017; Suzuki et al., 2017). In rice (Oryza sativa), mutation in OsDEX1 impairs callose degradation and primexine formation (Yu et al., 2016), while DEFECTIVE POLLEN WALL 3 (DPW3) encodes a membrane alpha integrin-like protein whose loss of function affects callose deposition and primexine formation, resulting in pollen abortion (Mondol et al., 2020). Most of these genes are exclusively or predominantly expressed in the tapetum, indicating that the tapetum plays an essential role, not only in producing sporopollenin precursors, but also in controlling sporopollenin deposition and assembly (Ariizumi and Toriyama, 2011; Quilichini et al., 2015). Pollen mother cells (PMCs) have also been implicated in exine patterning (Sheldon and Dickinson,1983), but there is little evidence for their direct involvement in regulating exine patterning.
Nonspecific lipid transfer proteins (nsLTPs) are small, basic proteins, characterized by eight conserved Cys residues (Boutrot et al., 2008). nsLTPs are involved in a wide range of biological functions: several preferentially accumulate in the tapetum with proposed roles in pollen wall formation. Arabidopsis type III LTPs are specifically expressed in the tapetum, and their encoded proteins have been proposed to function as exine precursor distributers and exine constituents (Huang et al., 2013). In rice, OsC6 encodes an LTP synthesized in the tapetum that has been implicated in sporopollenin precursor translocation (Zhang et al., 2010). Recently, the wheat (Triticum aestivum) Male Sterility 1 (Ms1) gene, whose mutation leads to severe defects in pollen development, was shown to encode a phospholipid binding nsLTP, although its function and subcellular location are not clear. One study indicated that TaMs1 is a PM-localized protein expressed in the tapetum and PMCs and suggested that TaMs1 plays a role in sporopollenin precursor delivery (Kouidri et al., 2018). A contradictory report showed that TaMs1 specifically accumulates in male meiocytes and localized to plastidic and mitochondrial membranes, and suggested that TaMs1 is required for microgametogenesis (Wang et al., 2017).
In this study, we describe the grass-specific nsLTP EXINE PATTERN DESIGNER 1 (EPAD1) in rice, which affects pollen exine patterning by controlling primexine integrity, that is, the continuity and homogeneity of the primexine, and probaculae distribution. EPAD1 is exclusively expressed in male meiocytes, and the epad1 male-sterile mutant shows altered pollen exine patterning. EPAD1 possesses an N-terminal signal peptide and redundant C-terminal GPI-anchor attachment sites that are required for its function and location to the microspore PM at the tetrad stage. With its GPI-anchor, EPAD1 belongs to the rice type G nsLTP subclade and is the ortholog of TaMs1. EPAD1 can bind a range of phospholipids in vitro, and we propose that EPAD1 itself and/or the lipids carried by EPAD1 are required to maintain primexine integrity. Our data provide substantial molecular evidence that the rice PMC plays an essential role in the species-specific exine pattern formation.
RESULTS
epad1 Flowers Are Male-Sterile
We identified the male-sterile mutant exine pattern designer 1 (epad1) from a rice mutant library (Chen et al., 2006). epad1 plants exhibited normal vegetative development, and normal panicle and spikelet morphology (Figures 1A to 1C). However, when compared to the wild type, epad1 flowers had smaller, paler anthers that failed to produce viable pollen (Figures 1C to 1F). When epad1 flowers were pollinated with wild-type pollen, all F1 plants were fertile and their F2 progeny showed an approximate 3:1 ratio for phenotype segregation (fertile:sterile = 80:24, χ2 = 0.21, P > 0.05), indicating that the epad1 mutation specifically affects male, but not female, reproduction and is monogenic and recessive.
Figure 1.
epad1 Plants Are Male-Sterile.
(A) Wild-type (WT) rice (japonica cv 9522) and epad1 plants after heading. Bar = 15 cm.
(B) Panicles showing fertile pollen grains in the wild type and sterile pollen grains in epad1 plants. Bar = 5 cm.
(C) Spikelets after removal of the palea and lemma, showing normal epad1 floral organs, with smaller, paler anthers than the wild type. gl, glume; pi, pistil; st, stamen. Bar = 1 mm.
(D) Mature wild-type and epad1 anthers. Bar = 1 mm.
(E) and (F) Staining with Lugol’s iodine solution of wild-type (E) and epad1 (F) mature pollen grains. Viable pollens are stained dark blue. Bars = 50 µm.
(G) to (N) Wild-type ([G] to [J]) and epad1 ([K] to [N]) anthers at stage 8b (tetrad; [G] and [K]); stage 9 (early microspore; [H] and [L]); stage 10 (vacuolated microspore; [I] and [M]); and stage 12 (mature pollen; [J] and [N]. E, epidermis; En, endothecium; M, middle layer; T, tapetum; Tds, tetrads; Msp, microspore; P, pollen; Dsp, degenerated microspore. At least six biological replicates for epad1 and the wild type were used for semi-thin transverse section analysis of each stage (stages 8–12). Representative images are shown. Bars = 25 μm.
We used semi-thin transverse sections to analyze developmental defects of epad1 anthers. We observed no detectable differences during meiosis or the early stages of anther development before stage 10 (stages labeled according to Zhang and Wilson, 2009 and Zhang et al., 2011) when compared to the wild type (Figures 1G to 1L; Supplemental Figure 1). Similarly, callose deposition appeared normal in epad1 microsporocytes and tetrads during meiosis (Supplemental Figure 2). However, at stage 10, epad1 microspores were irregularly shaped and less vacuolated than wild-type microspores (Figures 1I and 1M). By stage 12, when wild-type microspores had undergone mitosis and generated starch-filled mature pollen grains (Figure 1J), epad1 microspores had degenerated (Figure 1N). The epad1 phenotype was restricted to the microspores, as we observed no obvious differences in anther cell wall development (Figures 1I to 1J and 1M to 1N).
The epad1 Mutation Disrupts Primexine Integrity
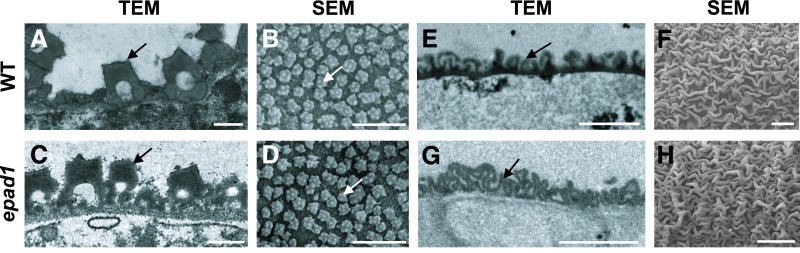
We next used transmission electron microscopy (TEM) to examine developmental defects in epad1 microspores more closely. In wild-type microspores, a continuous layer of primexine matrix was deposited between the callose layer and the microspore PM soon after meiotic cell division (Figure 2A). Probaculae then accumulated on the primexine at regular intervals at the late tetrad stage (Figures 2B and 2M). As the callose wall degenerates at stage 9, a thin membrane-like structure became discernible and the probaculae continued to increase in size (Figures 2C and 2M). Later during stage 9, a triple-layered structure, comprising the protectum, fibrillar material, and the foot layer, formed outside the microspore PM. This structure functions as a base for pollen exine assembly. Probaculae serve as assembly units to direct sporopollenin accumulation and polymerization on and between them (Figures 2G and 2M). At the vacuolated microspore stage (stage 10), a two-layered pollen exine, including the tectum layer, foot layer, and the baculae perpendicular to them, became evident, with a faint demarcation line between the layers, and pronounced microchannels formed in both the tectum and foot layers (Figures 2H and 2M). At stage 12, the mature pollen grains developed a well-organized wall with a dense accumulation of sporopollenin (Figures 2I and 2M).
Figure 2.
Integrity of the Primexine Layer Is Disrupted in epad1 Pollen.
Pollen exine of wild-type (WT; [A] to [C] and [G] to [I]) and epad1 ([D] to [F] and [J] to [L]) microspores at different developmental stages. Msp, microspore; Ca, callose; PM, plasma membrane; PE, primexine; PB, probaculae; ML, membrane-like structure; PT, protectum; FL, foot layer; FM, fibrillar material; DL, demarcation line; Mch, microchannel; Te, tectum; Ba, baculae; Ex, exine. At least ten biological replicates for epad1 and the wild type were used for TEM analysis of each stage (stages 8–12). Representative images are shown. Bars = 0.2 µm in ([A] to [G] and [J]), and 0.5 µm in ([H], [I], [K] and [L]).
(M) Model showing typical structural features in wild-type and epad1 pollen exine at different stages. Darker green colors in stages 8b to 12 echo the darker structures observed in ([B], [C], [E] to [L]), indicating the increasing accumulation of sporopollenin on exine structures.
Meiosis successfully completed in the epad1 mutant and the newly formed tetrad was normal (Supplemental Figure 3). However, we also detected phenotypic deviations from the wild type at stage 8b during microspore development. Indeed, the initial deposition of the primexine layer was broken and discontinuous (Figures 2D and 2M), resulting in the subsequent irregular accumulation of probaculae (Figures 2E and 2M). At stage 9, as in the wild type, the callose wall disappeared and a membrane-like structure became visible in epad1 microspores, but probaculae deposition remained irregular (Figures 2F and 2M). No triple-layered structure formed during stage 9 in epad1 (Figures 2J and 2M), hindering the formation of a continuous tectum and foot layers at stage 10; however, microchannel formation was not affected (Figure 2K). In epad1, baculae randomly clustered into irregular rings, resulting in a fused tectum and foot layer (Figures 2K and 2M). At stage 12, we observed thickened elliptical clumps of exine in epad1 microscopes (Figures 2L and 2M).
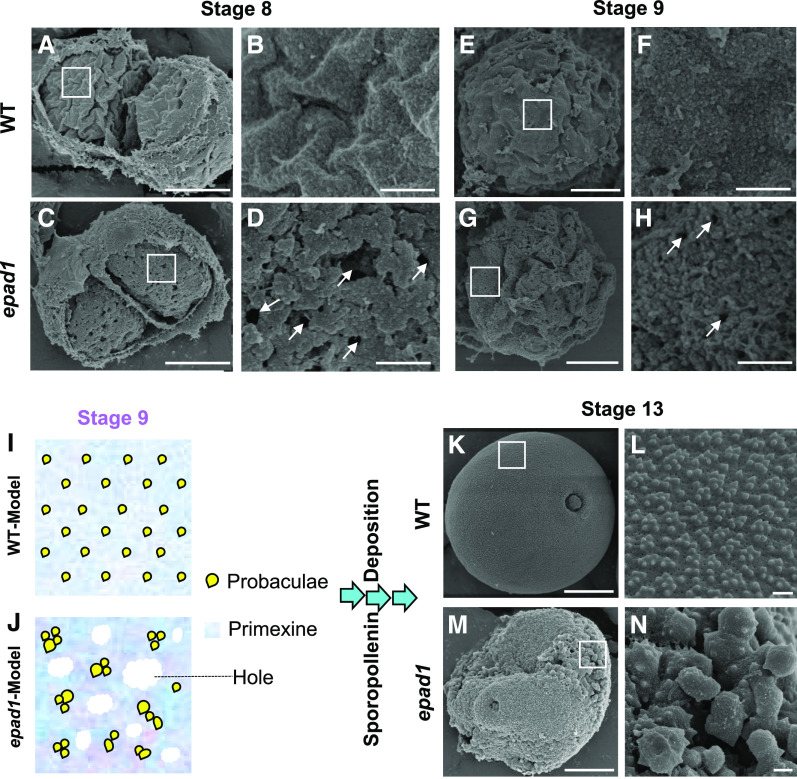
Scanning electron microscopy (SEM) validated the altered primexine and exine patterning seen in epad1 microspores. At stage 8, wild-type primexine was wrinkled and covered the whole surface of tetrad microspores uniformly, with probaculae densely but evenly distributed (Figures 3A and 3B). Regular assembly of exine components persisted during development, and spherical pollen grains displayed an elaborate surface pattern at maturity (Figures 3E, 3F, 3K, and 3L). However, in epad1 microspores, primexine coverage was disrupted at stage 8, with cavities and irregularly-clumped probaculae that became more pronounced by stage 9 (Figures 3C, 3D, 3G, and 3H). At stage 13, epad1 pollen grains collapsed and carried large sporopollenin-like aggregates on their surface (Figures 3M and 3N).
Figure 3.
The Exine Pattern Is Altered in epad1 Pollen.
Wild-type (WT; [A], [E], and [K]) and epad1 ([C], [G], and [M]) pollen grains and the enlarged view of the wild-type ([B], [F], and [L]) and epad1 ([D], [H], and [N]) pollen surface. Arrows indicate holes in the primexine of epad1 microspores. At least ten biological replicates for epad1 and the wild type were used for scanning electron microscopy analysis of each stage (stages 8–13). Representative images are shown. Bars = 10 µm in ([A], [C], [E], [G], [K], and [M]) and 0.5 µm in ([B], [D], [F], [H], [L], and [N]).
(I) and (J) Schematic illustration of primexine integrity and probacula distribution in wild-type (I) and epad1 (J) microspores.
We also examined the effects of the epad1 mutation on anther lipidic structures (Shi et al., 2015) during pollen development. Ubisch bodies, composed of sporopollenin, are spherical structures produced on the inner surface of the tapetum, while a spaghetti-like cuticle layer, consisting of cutin and wax, forms on the outside of the anther. Cutin shares some aliphatic precursors with sporopollenin (Shi et al., 2015; Xu et al., 2016), and rice mutants in sporopollenin biosynthetic genes always show abnormalities in the cuticle layer and Ubisch body formation (Shi et al., 2011; Liu et al., 2017). Surprisingly, although the epad1 mutation caused severe defects in pollen exine formation, the anther cuticle and Ubisch body looked similar to the wild type (Figure 4), suggesting that EPAD1 is not required for biosynthesis and allocation of sporopollenin precursors. Together, these observations indicate that EPAD1 specifically affects pollen exine patterning by disrupting primexine integrity, rather than sporopollenin production or deposition.
Figure 4.
Ubisch Body and Cuticle Formation Is Not Affected in epad1 Anthers.
(A) and (C) TEM observation of Ubisch bodies (arrows) on the inner surface of the wild-type (WT; A) and epad1 (C) anther wall. Bars = 0.5 µm.
(B) and (D) Scanning electron microscopy (SEM) observation of Ubisch bodies (arrows) on the inner surface of the wild-type (B) and epad1 (D) anther wall. Bars = 2.5 µm.
(E) and (G) TEM images of the cuticle layer (arrows) on the wild-type (E) and epad1 (G) anther surface. Bars = 5 µm.
(F) and (H) Scanning electron microscopy images of the cuticle layer on the wild-type (F) and epad1 (H) anther surface. Bars = 5 µm.
At least six biological replicates for epad1 and the wild type were used for Ubisch body and cuticle TEM and scanning electron microscopy analysis. Representative images are shown.
EPAD1 Encodes a Grass-Specific Type G nsLTP
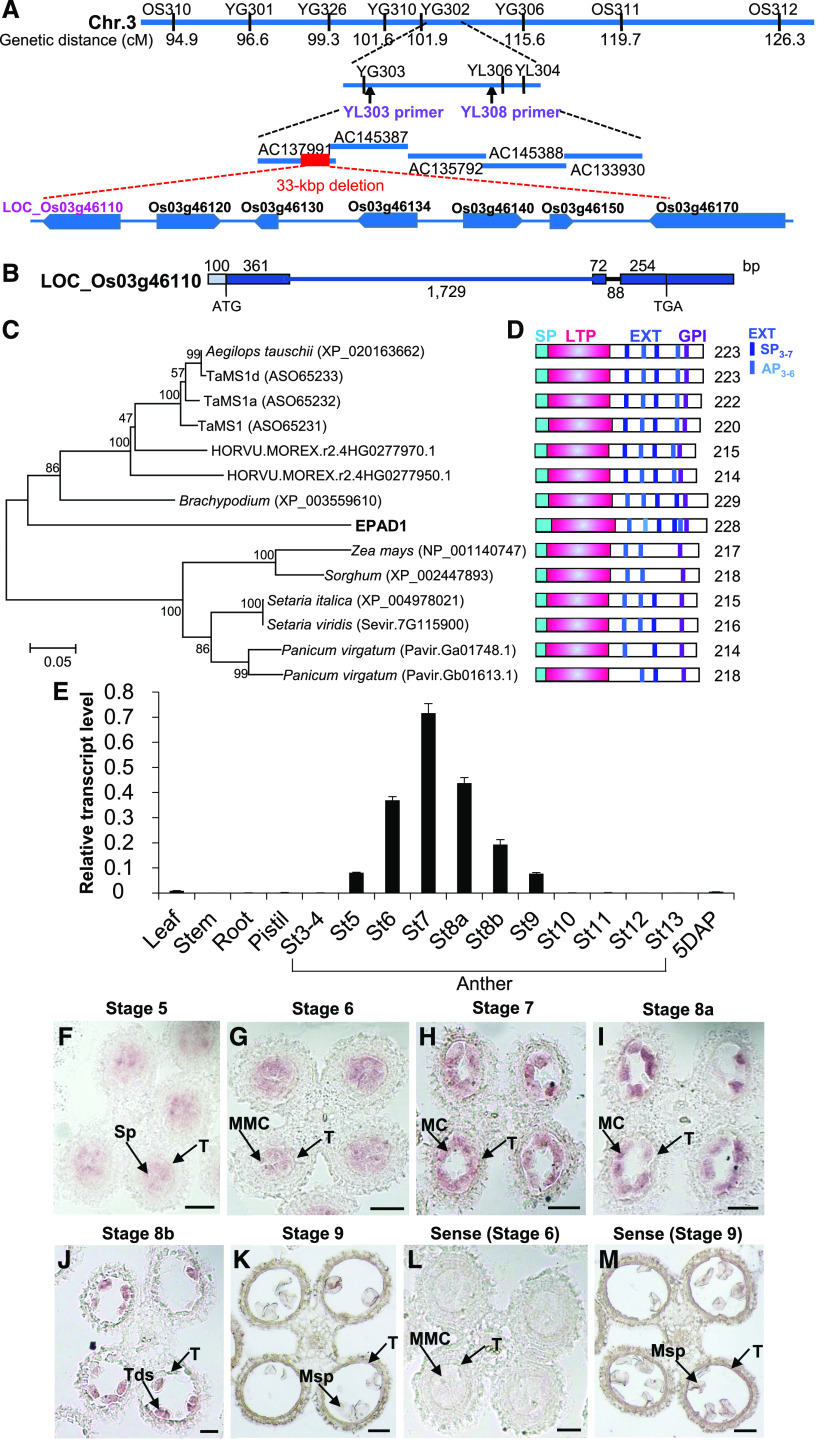
We applied a map-based cloning approach to identify EPAD1, using an F2 progeny from a cross between epad1 and a wild-type indica cultivar. We initially mapped EPAD1 between the two Insertion-Deletion (InDel) molecular markers OS310 and OS311 on chromosome 3, an interval that we further narrowed down to a 51-kbp region between indel markers YL303 and YL308 (Figure 5A). Sequence analysis revealed a 33-kbp deletion in the epad1 background, covering seven genes that were predicted to encode two LTP family proteins (Os03g46110 and Os03g46150), two F-box proteins (Os03g46120 and Os03g46140), and three retrotransposons (Os03g46130, Os03g46134, and Os03g46170; Figure 5A).
Figure 5.
Map-Based Cloning and Expression of EPAD1.
(A) Fine mapping of the EPAD1 gene. Names and positions of the molecular markers and primers are shown. The red dashed line shows the 33 kbp deleted region in epad1 plants. cM, centimorgan.
(B) Schematic representation of the gene organization of EPAD1 (Os03g46110). Boxes indicate exons and intervening lines indicate introns. Numbers indicate sequence length, in bp.
(C) Phylogenetic tree of EPAD1 and its orthologs from other species (Supplemental File 2). Scale bar indicates number of expected changes per amino acid residue.
(D) Schematic representation of functional domains in EPAD1 and its orthologs. The predicted signal peptides (SP), LTP domain, extensin-like motif (EXT), and GPI anchor are indicated as cyan, pink, blue, and purple boxes, respectively. Numbers indicate sequence length (amino acids).
(E) RT-qPCR analysis of relative EPAD1 transcript levels in various tissues. St, anther stage; 5 DAP, seeds 5 d after pollination. Error bars indicate SDs from three biological replicates.
(F) to (M) mRNA in situ hybridization for EPAD1 in stage 5 (F), stage 6 (G), stage 7 (H), stage 8a (I), stage 8b (J), and stage 9 (K) anthers. Sp, sporogenous cell; MMC, microspore mother cell; MC, meiotic cell; Tds, tetrads; Msp, microspore; T, tapetum. Sense probe was used as negative control ([L] and [M]). At least six biological replicates were used for anti-sense and sense probes signal analysis at each stage (stages 5–9). Representative images are shown. Bars = 20 µm.
We selected two genes for complementation tests based on their predicted gene functions and expression patterns. Constructs carrying wild-type genomic sequences for Os03g46110 or Os03g46120 were transformed into epad1 homozygous plants. Only those transformed with the Os03g46110 fragment were restored to normal fertility (Supplemental Figure 4), demonstrating that Os03g46110 is EPAD1.
The gene structure of EPAD1 — three exons and two introns — was confirmed by sequencing the EPAD1 cDNA and comparing it with the genomic sequence (Figure 5B). The predicted EPAD1 protein consists of 228 amino acids, with a predicted 23 amino acid signal peptide for secretion, and a putative LTP domain at the N terminus (Figures 5C and 5D). The LTP domain contains eight highly conserved Cys residues at positions 40, 53, 62, 63, 78, 80, 105, and 119, which are critical for the LTP structure (Douliez et al., 2000). In addition, EPAD1 possesses an extensin-like motif at its C terminus that is not present in canonical nsLTPs, followed by three predicted GPI-anchor attachment sites (Ser residues 204, 205, and 206), which is characteristic of type-G nsLTPs (Edstam et al., 2011).
To understand the evolutionary context of EPAD1 in plants, we used the full-length EPAD1 cDNA to search sequence databases. We identified EPAD1 orthologs only in grasses (Figure 5C), with the wheat ortholog TaMs1 being essential for male fertility (Tucker et al., 2017; Wang et al., 2017). Multiple sequence alignment revealed that the eight-Cys motif in the LTP domain (C-X12-C-X8-CC-X14-CXC-X24-C-X13-C) is highly conserved, whereas the extensin-like motif with the S/A-P3-7-SP3 sequence (Johnson et al., 2017) and GPI-anchor region diverge between EPAD1 orthologs (Supplemental Figure 5). In addition, we compared the LTP domains of EPAD1 and known rice nsLTPs, including OsC6 (Boutrot et al., 2008; Zhang et al., 2010; Edstam et al., 2011). A phylogenetic tree based on the eight-Cys residue motif confirmed that EPAD1 clustered with type-G nsLTPs (Supplemental Figure 6), most of which are GPI-anchored proteins.
The absence of EPAD1 orthologs in nongrass species is in stark contrast with other genes involved in pollen exine patterning, including those involved in callose degradation, primexine formation and sporopollenin assembly such as DEX1, NEF1, RPG1, NPU, and EFD (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Guan et al., 2008; Chang et al., 2012; Sun et al., 2013; Hu et al., 2014). This observation suggests that the EPAD1 gene was acquired relatively recently in plant evolutionary history, and that EPAD1 and its orthologs represent a divergent nsLTP clade present only in grasses.
EPAD1 Is Specifically Expressed in the PMC and Young Microspore
We performed an RT-qPCR analysis to examine whether EPAD1 expression correlated with our phenotypic observations. Indeed, we detected EPAD1 transcripts in anthers from stage 5 to stage 9, but not in other organs (Figure 5E). RNA in situ hybridization analysis showed that EPAD1 expression was first observed in the sporogenous cell (stage 5; Figure 5F). EPAD1 was highly expressed in the PMCs and tetrads, but not detectable in young microspores (Figures 5G to 5K). EPAD1 orthologs may have a conserved function in pollen development: in silico analysis of microarray or transcriptome deep-sequencing (RNA-seq) data revealed the flower-specific expression of EPAD1 orthologs in purple false broom (Brachypodium distachyon) and barley (Hordeum vulgare; Supplemental Figures 7A and 7B), and high expression in panicle, floret, and meiotic tassel in Panicoideae subfamily plants, including sorghum, maize, green foxtail (Setaria viridis), and switchgrass (Panicum virgatum; Supplemental Figures 7C to 7F).
EPAD1 Localizes to the Plasma Membrane
To elucidate the subcellular localization of EPAD1, we transiently expressed a GFP-tagged EPAD1 fusion protein in rice protoplasts and onion (Allium cepa) epidermal cells under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. We detected a clear GFP signal at the PM, as it closely colocalized with the PM marker CD3-1007-mCherry (Figures 6A to 6F). To distinguish whether the signal was in the PM or/and cell wall, we treated onion cells with 30% (w/v) Suc solution to induce plasmolysis. The GFP signal was associated with the PM indicative of PM localization of the protein (Figures 6G to 6I).
Figure 6.
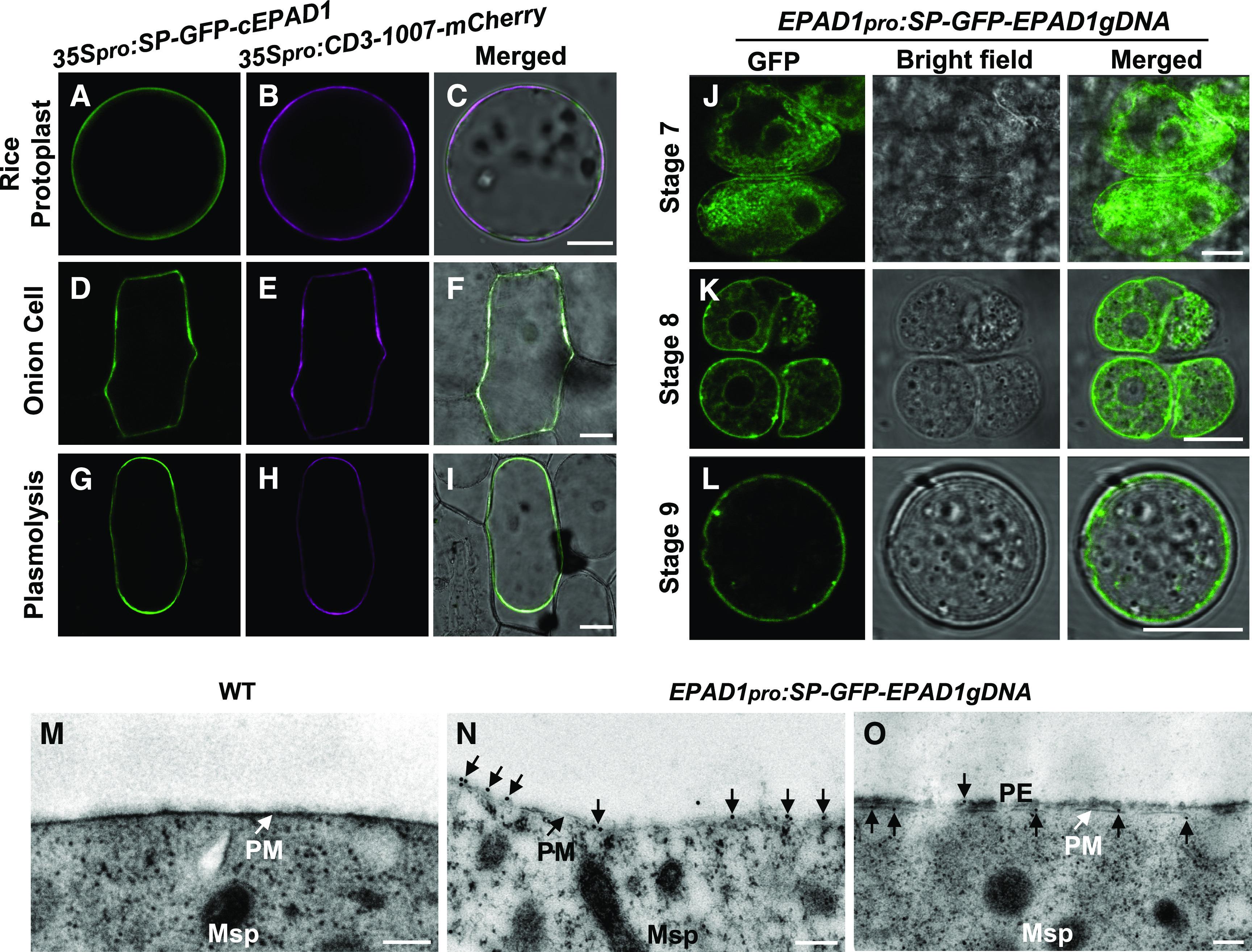
The Subcellular Localization of EPAD1.
(A) to (C) Representative confocal images of rice protoplast cells transiently expressing GFP-tagged EPAD1 (A), the plasma membrane marker CD3-1007-mCherry (B), and merged images (C). Bars = 10 µm.
(D) to (I) Representative confocal images of onion epidermal cells transiently expressing GFP-tagged EPAD1 (D, G), the plasma membrane marker CD3-1007-mCherry ([E] and [H]), and merged images ([F] and [I]), before ([D] to [F]) and after ([G] to [I]) plasmolysis. Bars = 50 µm.
(J) to (L) Meiocytes expressing EPAD1pro:SP-GFP-EPAD1gDNA at stage 7 (J), stage 8 (K), and early stage 9 (L).
From left to right in each row is GFP fluorescence, bright field, and the merged image, respectively ([J] to [L]). Bars = 10 µm.
(M) to (O) Immunogold GFP-labeled TEM section of wild type (WT; [M]) and transgenic line expressing EPAD1pro:SP-GFP-EPAD1gDNA ([N] and [O]) shows that EPAD1 accumulates on the plasma membrane of microspores during early and late stage 8b. Msp, microspore; PM, plasma membrane; PE, primexine. Bars = 0.2 µm.
At least fifteen biological replicates of protoplasts or microspores were used for subcellular localization analysis. Representative images are shown.
To confirm these results in rice anthers in vivo, we introduced the GFP-tagged EPAD1 construct, under the control of its native promoter, in which GFP was put between the signal peptide and the LTP domain (EPAD1pro:SP-GFP-EPAD1gDNA) into epad1 plants. The construct was functional, as it partially rescued the male fertility of epad1 (Supplemental Figures 8J to 8L). We detected GFP signals exclusively in the PMCs and young microspores (stages 7–9; Supplemental Figures 8A to 8C). Male meiocytes displayed a strong cytosolic GFP signal during meiosis (Figure 6J), after which the signal gathered at the PM of tetrads (Figure 6K) and young microspores (Figure 6L). TEM of immuno-gold labeled GFP confirmed that EPAD1 localized to the PM at the tetrad stage (stage 8; Figures 6N to 6O). There was no signal detected in wild-type microspores (Figure 6M).
The Signal Peptide and GPI-Anchor Are Required for EPAD1 Localization and Function
GPI-anchored proteins are known to be targeted to detergent-resistant membrane areas (liquid-ordered nanodomains) in the outer leaflet of the PM (Mayor and Riezman, 2004; Yeats et al., 2018). To analyze whether the N-terminal signal peptide (SP) and C-terminal GPI-anchor of EPAD1 affect its PM localization, we expressed GFP-tagged deletion constructs lacking either the SP (GFP-cEPAD1ΔSP) or the GPI motif (SP-GFP-cEPAD1ΔGPI) in Arabidopsis protoplasts. Both truncated proteins predominantly localized to the cytosol, similar to the GFP control (Supplemental Figures 9A to 9C), suggesting that both N- and C-terminal domains are required for the PM localization of EPAD1.
We used additional EPAD1 permutation constructs, placing GFP at either end of the EPAD1 genomic fragment — EPAD1gDNA-GFP and GFP-EPAD1gDNA, respectively — to further assess the biological importance of the N- and C-terminal regions. These constructs were placed under the control of the EPAD1 promoter and introduced into epad1 plants. Ubiquitous GFP signal distribution was observed in the meiocyte cytosol at stage 7 with all EPAD1 constructs (Supplemental Figures 8A, 8D, and 8G). However, by stages 8 and 9, GFP signals of the GFP-terminal constructs remained cytosolic while the SP-GFP-EPAD1gDNA construct clearly labeled the PM (Figures 6K and 6L; Supplemental Figures 8B to 8C, 8E to 8F, and 8H to 8I). In addition, the two GFP-terminal constructs did not complement the male-sterile phenotype of epad1 (Supplemental Figures 8M to 8R), suggesting that changes to the N- or C-terminal sequences of EPAD1 affect both subcellular localization and function of EPAD1 in vivo.
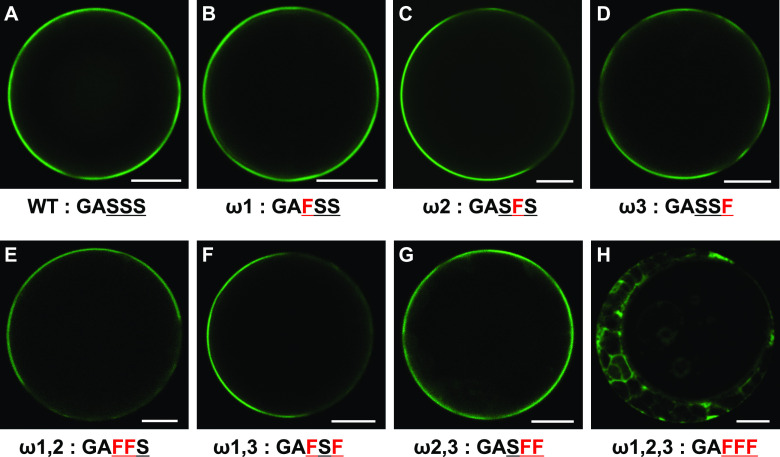
To define the exact GPI-anchor attachment site (the ω-site), we mutated three predicted ω-site Ser residues (amino acids 204–206), either alone or in combinations in the backbone of the SP-GFP-cEPAD1 construct. None of the constructs carrying one- (ω1; ω2; ω3) or two-amino acid mutations (ω1,2; ω1,3; ω2,3) showed any change in protein PM localization compared with the wild-type protein (Figures 7A to 7G). However, the three-amino acid mutation (ω1,2,3) prevented PM localization and confined EPAD1 accumulation to the cytoplasm (Figure 7H), indicating that these Ser residues act as redundant GPI-anchor attachment sites in EPAD1. Complementation analysis using mutant constructs of EPAD1 genomic DNA fragments carrying one- (ω2), two- (ω1,3; ω2,3) or three-amino acid (ω1,2,3) mutations confirmed that EPAD1 function was not affected by modifying one or two of the ω-sites, whereas EPAD1 function was abolished by changing all three ω-sites (Supplemental Figure 10). Thus, the Ser residues are critical for EPAD1 PM localization and function, likely by facilitating GPI-anchoring of the protein to attach properly to the membrane.
Figure 7.
Analysis of the EPAD1 GPI-Anchor Sites.
Representative GFP fluorescence images of Arabidopsis protoplast cells expressing 35Spro:SP-GFP-cEPAD1 wild type (WT; A), 35Spro:SP-GFP-cEPAD1 ω1 (B), 35Spro:SP-GFP-cEPAD1 ω2 (C), 35Spro:SP-GFP-cEPAD1 ω3 (D), 35Spro:SP-GFP-cEPAD1 ω1,2 (E), 35Spro:SP-GFP-cEPAD1 ω1,3 (F), 35Spro:SP-GFP-cEPAD1 ω2,3 (G), 35Spro:SP-GFP-cEPAD1 ω1,2,3 (H). Amino acid sequence under each panel indicates the position of the mutation in the GPI-anchor attachment sequence in red. Underlined residues are predicted GPI-anchor attachment sites. At least ten randomly chosen protoplasts were used for GFP signal analysis. Representative images are shown. Bars = 10 µm.
EPAD1 Binds to Phospholipids in vitro
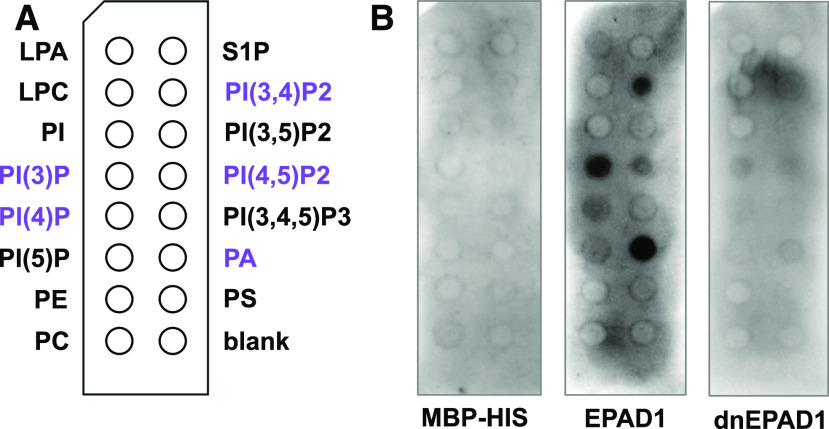
Plant nsLTPs can bind or transfer a broad spectrum of hydrophobic aliphatic molecules including fatty acids, fatty acyl-CoA, hydroxylated fatty acid, phospholipids, glycolipids, and cutin monomers (Carvalho and Gomes, 2007; Liu et al., 2015). To characterize whether EPAD1 has lipid binding capacity, we purified a recombinant EPAD1 protein without the SP and GPI-anchor segments and used it in protein–lipid overlay assays. Maltose binding protein (MBP), which does not bind lipids, was used as the negative control. The recombinant EPAD1 protein bound to several phospholipids, including PA and several phosphatidylinositols (PIs) such as PI(3)P, PI(4)P, PI(3,4)P, PI(4,5)P2 (Figure 8). The lipid binding activity of EPAD1 was dependent on protein structure: when disulfide bonds were disrupted by Tris-2-carboxyethy1-phosphine, binding of EPAD1 to phospholipids was abolished (Figure 8). These results indicate that EPAD1 can bind several phospholipids that are found in microspores (Lee et al., 2018).
Figure 8.
Lipid Binding Ability of EPAD1.
(A) Schematic diagram of the layout of phospholipid spots on membranes incubated in the presence of different EPAD1 proteins. Purple labels indicate spots bound by EPAD1.
(B) Representative results of lipid-protein interaction assay for MBP-HIS (negative control), MBP-EPAD1-HIS (EPAD1), and denatured MBP-EPAD1-HIS (dnEPAD1) recombinant protein, respectively.
LPA, lysophosphatidic acid; LPC, lysophosphocholine; PI, phosphatidylinositol; PI(3)P, phosphatidylinositol-3-phosphate; PI(4)P, phosphatidylinositol-4-phosphate; PI(5)P, phosphatidylinositol-5-phosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PI(3,4)P2, phosphatidylinositol-3,4-bisphosphate; PI(3,5)P2, phosphatidylinositol-3,5bisphosphate; PI(4,5)P2, phosphatidylinositol-4,5-bisphosphate; PI(3,4,5)P3, phosphatidylinositol-3,4,5-triphosphate; PA, phosphatidic acid; PS, phosphatidylserine.
DISCUSSION
EPAD1 Is Specifically Involved in Primexine Patterning in Rice
Primexine is thought to provide a template to guide sporopollenin deposition, polymerization and pattern formation in the developing pollen cell wall (Figure 9A; Xu et al., 2016); however, the mechanisms by which this occurs are largely unknown. Here, we describe EPAD1, a type-G nsLTP that plays a critical role in maintaining primexine integrity to direct exine patterning in the monocot crop rice.
Figure 9.
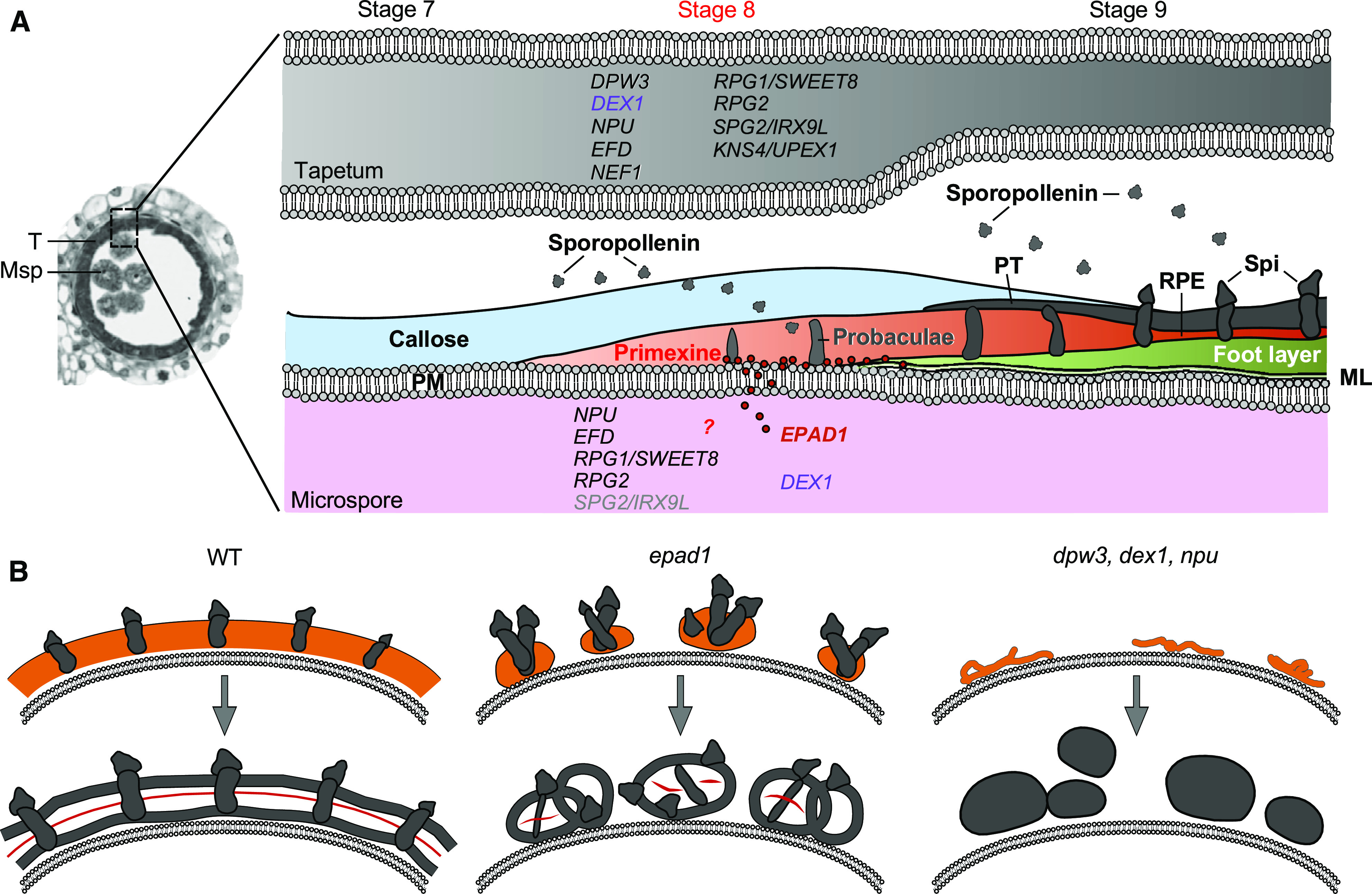
Schematic Model of EPAD1 Function in Pollen Exine Patterning.
(A) The stages of pollen exine patterning and related genes in the tapetum and microspore. EPAD1 function is required in the microspore for maintenance of primexine homogeneity during stage 8.
PM, plasma membrane; Spi, spinule; PT, protectum; RPE, remnant of primexine; ML, membrane-like structure.
(B) Representative defects in epad1 and comparison to other pollen exine pattern defective mutants. Orange and dark gray colors represent primexine and sporopollenin-derived pollen exine, respectively. Red line represents primexine remnants. Wild-type (WT) microspore forms uniform primexine at stage 8, which defines sporopollenin accumulation. The epad1 mutation affects pollen exine patterning, in contrast to previously reported mutants such as dpw3, dex1, npu, which affect sporopollenin accumulation.
Current dogma proposes three major functions for primexine. First, primexine acts like a glue to adhere sporopollenin precursors to the microspore surface. Second, primexine templates polymerization of sporopollenin precursors into specific assembly units, the probaculae and protectum. Consistent with previous reports on grass pollen wall formation (Skvarla and Larson, 1966; Christensen et al., 1972), we did not observe regular microspore PM undulation at the tetrad stage (Figures 2A, 2B, and 6M to 6O). These results are also consistent with a recent report (Radja et al., 2019) in which species whose pollen grains are smooth and featureless do not always undergo PM undulation during early pollen wall development, indicating that these species may exploit a different strategy to position the probaculae. Finally, primexine drives localization of the probaculae and protectum, which comprise the base of global exine pattern formation. Most Arabidopsis and rice primexine mutants have no or a greatly reduced primexine layer (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Chang et al., 2012; Yu et al., 2016; Mondol et al., 2020). In these mutants, amorphous sporopollenin-like aggregates cannot be, or are rarely, deposited onto the microspore (Figure 9B, the right panel). By contrast, EPAD1 does not affect the formation of primexine, but specifically affects its deposition pattern and integrity (Figure 9B, epad1). In addition, callose wall development and degradation were normal in the epad1 PMCs (Supplemental Figure 2), which differs from most known primexine-defective mutants (Guan et al., 2008; Ma et al., 2013; Sun et al., 2013; Yu et al., 2016). Furthermore, in contrast to the mutant phenotypes in sporopollenin-related genes (Zhang et al., 2010; Shi et al., 2011; Liu et al., 2017), the cuticle and Ubisch body development were normal in epad1 anthers, indicating that abnormalities in epad1 pollen exine are not caused by defects in sporopollenin biosynthesis or translocation. These results suggest that EPAD1 is specifically involved in controlling primexine deposition, which subsequently influences the location of assembly units and final exine pattern (Figure 9B, epad1).
EPAD1 Is a Male Meiocyte-Derived Determinant of Exine Patterning
Factors that influence primexine formation are typically derived from the sporophytic tapetum (Figure 9A). However, the PMCs may also contribute to exine patterning — for example, the primexine-related genes DEX1, NPU, RPG1/SWEET8, and RPG2 are expressed in both the tapetum and PMCs (Guan et al., 2008; Ma et al., 2013; Sun et al., 2013; Yu et al., 2016) — but so far no PMC-specific proteins have been found to play a role in primexine patterning. We show that EPAD1 is expressed exclusively in the PMCs and tetrads (Figures 5F to 5J). These results are consistent with exine pattern determinants being present in PMCs during prophase I (Heslop-Harrison, 1971; Sheldon and Dickinson, 1983). Our results thus indicate that the deposition and assembly of sporopollenin is regulated, not only by tapetal cells, but also by the microspore (Figure 9A).
Recent investigations on the pollen aperture, where little or no primexine and sporopollenin is deposited, have shown that two key aperture development regulators, INAPERTURATE POLLEN 1 (INP1) and DEFECTIVE IN APERTURE FORMATION 1 (OsDAF1), are similarly expressed (Dobritsa et al., 2018; Zhang et al., 2020). During anther development, both genes are expressed specifically in the PMCs and their daughter cells, microspores during meiosis to the tetrad stage. The pollen aperture first appears at young microspore stage, indicating that although the genes are transcribed in PMCs, their corresponding proteins do not function until the completion of meiosis. Like EPAD1 (Figures 6J to 6L), INP1 and OsDAF1 are evenly distributed in the PMC cytoplasm, but then re-locate to their functional location at the tetrad stage (Zhang et al., 2020). Thus, PMC-derived determinants of pollen morphology may be produced and prepared at the diploid cell stage, but only take action at the haploid cell stage.
EPAD1 Maintains Primexine Integrity and Affects Probaculae Localization
The exact component(s) that drive sporopollenin adhesion within primexine remain elusive. Sporopollenin acceptor particles (SAPs) might work as the initial sites for sporopollenin polymer accumulation in primexine (El-Ghazaly and Jensen, 1986; Skvarla and Rowley, 1987; Rowley and Claugher, 1991; Xu et al., 2016). In epad1, the assembly units form normally, suggesting that SAP-like components are present and functional, although their arrangement on the cell wall is disturbed. Moreover, when compared to the evenly distributed probaculae in wild-type primexine (Figures 2B and 2C), the regions covered by primexine contain more probaculae in epad1 (Figures 2E and 2F, Figure 2M, late stage 8b and early stage 9), indicating that primexine is not simply spatially interrupted, but is more densely decorated and contains more SAP-like components. EPAD1 may therefore be required to define SAP location at the tetrad stage, when it localizes to the PM (Figures 6 and 7; Supplemental Figures 8 to 10). These findings have important implications for how primexine determines positions of exine assembly units in a precise pattern.
As a lipid binding protein anchored to the microspore PM, EPAD1 may define PM features that affect the distribution of proteins such as SAPs. The signal peptide and GPI-anchor sites are indispensable for its PM location and function (Supplemental Figures 8D to 8I, 8M to 8R, and 9), and mutation of the putative GPI-anchor sites renders the EPAD1 protein nonfunctional (Supplemental Figure 10G to 10H). EPAD1 displays PI binding activity in vitro, as does its wheat ortholog TaMs1 where four amino acid changes in the TaMs1 LTP domain (C52Y, V55D, D60N, and C94Y) totally abolished this function (Tucker et al., 2017; Wang et al., 2017). PIs, enriched in specific PM domains, play crucial roles in directing cell polarity and vesicular trafficking in plants, and act as docking sites or substrates to recruit peripheral membrane proteins (Heilmann and Heilmann, 2013; Stanislas et al., 2015; Barbosa et al., 2016; Lee et al., 2018). PIs also influence PM biophysical properties directly through their polyanionic head groups or in combination with other lipids (Heilmann, 2016; Gerth et al., 2017), indicating that PIs may stabilize PM nanodomains via multiple mechanisms. Two PI phosphates, PI(4)P and PI(4,5)P2, are enriched in the PM underneath the aperture site (Lee et al., 2018), where minimal sporopollenin deposition occurs. Although PIs normally locate to the inner surface of the PM, some reports indicate that they also present in the outer leaflet of the PM (Kale et al., 2010; Boss and Im, 2012). Therefore, EPAD1 may bind to specific PI-rich PM domains that can facilitate recruitment of proteins, such as SAPs, involved in primexine development. Alternatively, EPAD1 may provide docking sites for PI molecules in the PM, which go on to recruit primexine-regulatory proteins. A third possibility is that the lipid molecules carried by EPAD1 are required to maintain the homogeneity of primexine components. As new tools become available, examination of the in vivo composition and distribution of PIs or other lipids in the wild-type and epad1 PM and primexine will shed light on the specific mechanisms of EPAD1 function.
EPAD1 possesses a hydrophilic extensin-like motif at its C terminus. In canonical cellulosic cell walls, extensins interact with acidic pectin to create a template for the orderly assembly of the nascent cell wall (Cannon et al., 2008; Lamport et al., 2011), echoing the proposed function of EPAD1. In addition, both the LTP-domain and C-terminal (extensin-like domain) of TaMs1 are required for its lipid binding capability (Wang et al., 2017). How this motif is required for EPAD1 function is yet unclear, but it is possible that EPAD1 is stabilized in primexine by cross-linking with pectin via the extensin-like motif. Another possibility is that EPAD1 may maintain primexine homogeneity by extensin-pectin cross-linking.
EPAD1 and its Orthologs may Represent Grass-Specific Exine Patterning Determinants
Primexine-regulating genes have widely distributed orthologs in flowering plants (Xu et al., 2016; Mondol et al., 2020), and are therefore unlikely to be involved in species-specific exine patterning. However, EPAD1 and its orthologs (Tucker et al., 2017; Wang et al., 2017) are only found in the grasses, suggesting that they may play a role in defining species-specific exine pattern.
EPAD1 clusters into the type-G nsLTP subclade in a genome-wide analysis of rice nsLTPs (Supplemental Figure 6). Type-D and -G nsLTPs are found in all land plants, representing the earliest nsLTPs (Edstam, et al., 2011). Comparative studies of LTP genes within the Poaceae, including rice, wheat, and sorghum, suggest that independent duplication events promoted an increasingly functional diversity during speciation (Jang et al., 2007, 2008; Wang et al., 2010, 2012). EPAD1 and its orthologs appear to have evolved together with the emergence of the Poaceae, and we hypothesize that the common EPAD1 ancestor arose via fusion of two quite distinct domains that evolved independently in angiosperms: the hydrophobic, negatively-charged LTP domain containing the N-terminal signal peptide, and the hydrophilic, positively-charged extensin-like domain containing the GPI-anchor region (Supplemental Figure 5; Carvalho et al., 2007; Wei and Zhong, 2014; Wang et al., 2017).
Orthologs of EPAD1 may play conserved roles in grass pollen development, as evidenced by similarities in their protein sequences, such as the conserved eight-Cys LTP domain, preferential expression in young spikelets (Figure 5; Supplemental Figures 5 and 7), and similar pollen wall phenotypes in the Tams1 and epad1 mutants (Tucker et al., 2017). Sequence differences exist, particularly in the variable C-terminal domain: while EPAD1 possesses three redundant GPI-anchor attachment sites (Figure 7), TaMs1 only has one potential GPI-anchor attachment Ser residue (Tucker et al., 2017; Wang et al., 2017). Although grass pollen grains display similar surface ornamentation patterns, the size and density of sculptural elements vary slightly between species (Mander et al., 2013). Whether other EPAD1 orthologs have roles in exine patterning, and whether the variable C terminus causes minor differences in pollen wall pattern, remain to be seen.
Interestingly, the exine structure of epad1 pollen resembles that of pointed centrolepis (Centrolepis aristata, Centrolepidaceae), where the tectum and foot-layer tie together to form circular structures separated at regular intervals (Figure 2L; Rowley and Dunbar, 1996). Centrolepidaceae are close relatives of grasses, but originated later than the Poaceae (Bouchenak-Khelladi et al., 2014; Luo et al., 2015). The pollen exine pattern of C. aristata may have arisen through the loss of genes, such as EPAD1 ortholog(s), during evolution. Our findings have significantly extended our understanding of pollen exine patterning via an evolutionarily diversified role for nsLTPs. Further investigations on EPAD1 orthologs in other grass species will help to unravel the mechanisms by which exine patterning is imprinted onto the PM via male meiocyte-derived determinants.
METHODS
Plant Materials
All rice plants (Oryza sativa) were grown in the paddy field of Shanghai Jiao Tong University, located in Shanghai (31.03°N, 121.45°E), China. The epad1 mutation was isolated from an existing rice mutant library in Oryza sativa ssp. japonica cv 9522 that was created by irradiation with 60Co γ-rays (Chen et al., 2006). The F2 mapping population was generated from a cross between epad1 and cv Guang Lu Ai 4 (ssp. indica). Arabidopsis (Arabidopsis thaliana) plants from the Columbia-0 (Col‐0) accession were grown in growth chambers under a photoperiod of 16-h white light (Philips, YZ36RR25, cool daylight, TLD 36W/865)/8-h darkness at 23°C.
Characterization of Mutant Phenotypes
We documented phenotypes of whole plants and panicles by taking photographs with an E995 digital camera (Nikon); dissected spikelets, flowers, and anthers were photographed with an M205A microscope (Leica). Pollen viability was analyzed under a Eclipse 80i microscope (Nikon) after pollen grains were released and immersed into Lugol’s iodine solution (2% [w/v] potassium iodide and 0.2% [w/v] iodine in water). Anthers from different developmental stages, as defined by Zhang and Wilson (2009) and Zhang et al. (2011), were collected based on spikelet and anther length. We performed 4′,6-diamidino-2-phenylindole staining of chromosomes, callose staining of microspores, and semi-thin cross-section, scanning electron microscopy, and TEM of developing anthers as described by Dong et al. (2005), Li et al. (2006), Zhang et al. (2011), and Xu et al. (2016), and the results examined with a Eclipse Ni-E Microscope (Nikon; 4′,6-diamidino-2-phenylindole and callose), Eclipse 80i microscope (Nikon), S-4800 scanning electron microscopy (Hitachi), and G2 spirit Biotwin TEM (Technai), respectively.
Generation of Localization and Complementation Constructs
For functional complementation analysis of the epad1 mutant, we cloned genomic fragments for EPAD1 containing a 2712-bp upstream region (promoter + 5′ untranslated region), 2504-bp genic region (EPAD1gDNA), and 1098-bp downstream region (EPAD1pro:EPAD1gDNA), and for OsFBOX100 containing 3379-bp of upstream sequence, 2444-bp genic region, and 994-bp downstream sequence (OsFBOX100pro:OsFBOX100gDNA) into the binary vector pCAMBIA1301 by In-Fusion cloning (In-Fusion HD Cloning Kit; Takara PT5162-1).
To confirm the gene structure of EPAD1, we PCR-amplified the EPAD1 coding sequence with the EPAD1-F and EPAD1-R primer pair. The PCR product was sequenced and compared with the EPAD1 genomic sequence. The cDNA used above was synthesized using total RNA isolated from meiosis-stage rice flowers (see RNA Extraction and RT-qPCR section below).
The 35Spro:SP-GFP-cEPAD1 construct was generated by fusing the coding region for the EPAD1 N-terminal signal sequence, GFP fragment, and the remaining 603-bp region of EPAD1 cDNA sequence (cEPAD1) into the SpeI and BstEII restriction sites in pCAMBIA1301:GFP by In-Fusion cloning. The plasma membrane marker construct, 35Spro:CD3-1007-mCherry, was kindly provided by Chikuang Wen (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences).
The EPAD1pro:EPAD1gDNA-GFP construct was generated by fusing the EPAD1 genomic fragment containing the 2712-bp upstream sequence and 2501-bp genic region without the stop codon into pCAMBIA1301:GFP by In-Fusion cloning. We also inserted a PCR-amplified GFP sequence fragment with NcoI sites as linker sequences into EPAD1 genomic DNA in front of the N-terminal signal sequence by digesting EPAD1pro:EPAD1gDNA with NcoI to yield EPAD1pro:GFP-EPAD1gDNA. To generate EPAD1pro:SP-GFP-EPAD1gDNA, we fused the EPAD1 genomic fragment containing the 2712-bp upstream sequence and 84-bp encoding the N-terminal signal sequence of EPAD1 with an amplified GFP sequence fragment into the BamHI and BstEII restriction sites in pCAMBIA1301:GFP, generating EPAD1pro:Sig-GFP, into which we subsequently inserted the remaining 2420 bp EPAD1 genic region with In-Fusion cloning.
Each plasmid was transformed into Escherichia coli strain DH5α and verified by sequencing. All constructs were introduced into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 for transformation of rice calli induced from young panicles of homozygous epad1 plants. All primers used are listed in the Supplemental Table with relevant restriction enzyme sites underlined.
Sequence and Phylogenetic Analysis
Full-length EPAD1 cDNA was used as the query for its closest relatives in published databases including the National Center for Biotechnology Information, The Arabidopsis Information Resource, Phytozome, and Gramene. We used GPI-SOM (http://genomics.unibe.ch/cgi-bin/gpi.cgi) and GPI prediction server (http://mendel.imp.ac.at/gpi/gpi_server.html) to predict sites of posttranslational addition of a GPI-anchor (Eisenhaber et al., 1999; Pierleoni et al., 2008). SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) was used to predict signal peptide sequence for cleavage, and Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) were used for LTP domain prediction. We analyzed the characteristics of the extensin-like domain according to Johnson et al. (2017). ExPASy ProtParam tool (https://web.expasy.org/protparam/) was used to analysis the LTP domain and extensin-like domain physical and chemical parameters.
Protein sequences were aligned using ClustalX, with a gap extension penalty and gap opening penalty set to 0.01 and 10, respectively. The phylogenetic tree (Figure 5C) was constructed based on multiple alignments of complete protein sequences. The rice nsLTP phylogeny (Supplemental Figure 6) was constructed using protein sequences of the eight-Cys motif (C…C…CC…CXC…C…C); the GPI-anchor and the link to the anchor were excluded. Based on the eight-Cys motif sequences, a neighbor-joining tree was constructed with pairwise deletion, with a bootstrap value of 1000 permutations by MEGA software (Tamura et al., 2013). The information on rice nsLTPs was obtained from Edstam, et al. (2011) and Wei and Zhong (2014).
RNA Extraction and RT-qPCR
We isolated total RNA from rice tissues with the TIANGEN RNAprep Pure Kit for plants, according to the manufacturer’s protocol. After treatment with DNase I, 1 μg RNA was used to synthesize oligo(dT)-primed first-strand cDNAs using PrimeScript reverse transcriptase (Takara). RT-qPCR analyses were performed on a CFX96 (Bio-Rad) instrument using SYBR Green Master mix (QIAGEN), according to the manufacturer’s instructions. Transcript levels were expressed relative to ACTIN (LOC_Os03g50885) housekeeping gene. All reactions were performed with three biological replicates.
We obtained expression levels of Bradi1g13030 from Brachypodium distachyon, Sobic.006g089700 from Sorghum bicolor, Setaria viridis (Sevir.7G115900.1), and Pavir.Gb01613.1 and Pavir.Ga01748.1 from Panicum virgatum from Phytozome. RNA sequencing data for HORVU.MOREX.r2.4HG0277970.1 and HORVU.MOREX.r2.4HG0277950.1 from Hordeum vulgare, and GRMZM2G151021 from Zea mays obtained from Barley RTD (https://ics.hutton.ac.uk/barleyrtd/index.html) and MaizeGDB (https://maizegdb.org/).
in situ Hybridization
RNA in situ hybridizations were performed according to Li et al. (2006). We used the EPAD1 cDNA fragment as template to generate the sense and antisense probes, which were amplified by primers listed in Supplemental Table with fused SP6 and T7 promoters, respectively. The in vitro RNA transcription and labeling was performed using the SP6/T7 DIG RNA labeling kit (Roche), following manufacturer’s protocols. An E600 microscope (Nikon) was used to examine and photograph the results of the assay.
Subcellular Localization of EPAD1
Protoplasts from both Arabidopsis and rice, and onion epidermal cell were used for the transient expression assay. We isolated Arabidopsis and rice protoplasts from mesophyll cells of 20-d-old Arabidopsis leaves and the etiolated hypocotyl of 12-d-old rice plants, respectively, and subjected them to polyethylene glycol-mediated transformation as described by Bart et al. (2006). The onion epidermal cell bombardment experiment was performed using a helium biolistic device (Bio-Rad PDS-1000) as described by Tan et al. (2012). Plasmolysis was induced with 30% (w/v) Suc solution for 10 min.
We performed site-directed mutagenesis on the SP-GFP-cEPAD1 (ω1), SP-GFP-cEPAD1 (ω2), SP-GFP-cEPAD1 (ω3), SP-GFP-cEPAD1 (ω1,2), SP-GFP-cEPAD1 (ω2,3), SP-GFP-cEPAD1 (ω1,3), and SP-GFP-cEPAD1 (ω1,2,3) constructs at single or multiple Ser residues acting as putative GPI-anchor ω sites (Figure 7; Supplemental Figure 5). All Ser residues were replaced by Phe, and all constructs were cloned into pCAMBIA1301. The EPAD1pro:EPAD1gDNA (ω2), EPAD1pro:EPAD1gDNA (ω2,3), EPAD1pro:EPAD1gDNA (ω1,3), and EPAD1pro:EPAD1gDNA (1,2,3) constructs were similarly modified by site-directed mutagenesis at putative GPI-anchor ω sites in the EPAD1 genomic DNA fragment and used for complementation assays. All mutated constructs were made by Sangon Biotech company (Shanghai, China).
The 35Spro:GFP-cEPAD1/ΔSP and 35Spro:SP-GFP-cEPAD1/ΔGPI constructs were made as translational fusions between GFP and the EPAD1 cDNA fragment lacking either the SP or the GPI motif and inserted into the XhoI and EcoRI restriction sites in pGREEN (kindly provided by Hao Yu, National University of Singapore, Singapore).
We collected fluorescence and bright field images on a laser scanning confocal microscope (Leica TCS SP5). GFP fluorescence was imaged at the excitation wavelength of 488 nm and an emission wavelength of 505 to 530 nm. mCherry and chlorophyll autofluorescence were imaged at an emission wavelength of 600 to 700 nm.
For immunological detection of GFP, we fixed and embedded rice anthers from the wild type and from EPAD1pro:SP-GFP-EPAD1gDNA transgenic plants at stages 7 and 8 by high-pressure freezing and the freeze-substitution method (Zhang et al., 2016). Briefly, fresh anthers were processed using a Leica EM HPM100 high pressure freezer with 1-headecene (Sigma-Aldrich) as a cryoprotectant. Then samples were freeze-substituted in 0.25% (w/v) glutaraldehyde, 0.1% (w/v) uranyl acetate and 8,2,2-dimethoypropane (Sigma-Aldrich) in acetone, and embedded in LR White resin (London Resin Company). Sections of 70 nm were deposited on nickel grids and labeled as previously described by Wilson and Bacic (2012). In brief, sections were treated with 0.1 mol/L Gly for 10 min, blocked in 1% (w/v) BSA for 30 min, followed by hybridization using a 1:200 dilution of rabbit polyclonal anti-GFP antibody (Life Technologies, A6455) and a 1:40 dilution of goat-anti-rabbit conjugated to 18 nm gold particles (ImmunoResearch Inc). After post-staining with uranyl acetate and Reynolds lead citrate, samples were examined with a G2 spirit Biotwin TEM (Tecnai).
Recombinant Protein Purification and Protein-Lipid Overlay Assay
We amplified the EPAD1 coding sequence missing both the signal peptide and the C terminus using primers MBP-EPAD1-HIS-F and MBP-EPAD1-HIS-R (Supplemental Table). The PCR product was inserted into the EcoRI and SalI restriction sites of the pMAL-C2X expression vector to generate MBP-EPAD1-HIS. The negative control, pMAL-C2X, encodes a His-tagged maltose binding protein (MBP-HIS). Both constructs (MBP-EPAD1-HIS or MBP-HIS) were introduced into E. coli expression strain BL21 and the resulting colonies were grown in Luria-Bertani (LB) medium with 100 mg/L ampicillin at 37°C until OD600 reached 0.6 to 0.8, at which point the temperature was reduced to 16°C. Isopropyl β-D-thiogalactoside was added to a final concentration of 0.2 mM, and the cultures were maintained overnight. The cells were pelleted by centrifugation (4000 rpm, 40 min, at 4°C) and resuspended in Ni+-column balance buffer (300 mM NaCl, 25 mM Tris, 20 mM imidazole, pH 8) containing 0.5mM phenylmethanesulfonyl fluoride. After sonication, the lysates were centrifuged (13,000 rpm, 40 min, at 4°C) and protein was purified on a Ni2+-chelating Sepharose column (Thermo Fisher Scientific) and eluted from the column by the addition of 300 mM imidazole, according to manufacturer’s instructions. The purified proteins were concentrated with ultrafiltration tubes to remove imidazole. Protein purity and integrity were checked by SDS-PAGE gel, and the purified recombinant proteins were stored at –80°C in 25 mM Tris buffer, pH 7.5, containing 20% (v/v) glycerol and 20 mM MgCl2. MBP-EPAD1-HIS protein was denatured by adding 10 mM Tris-2-carboxyethy1-phosphine for 20 min at room temperature.
We performed the protein-lipid overlay assay with phosphoinositide phosphate strips (P-6001, Echelon Biosciences) according to manufacturer’s instructions (Strip Array v9) with modifications. The lipid membranes were blocked in 1% (w/v) albumin (Sigma-Aldrich) in Tris Buffered Saline with Tween-20 (TBST; 10 mM Tris-HCl pH 8.0, 140 mM NaCl, 0.1% [v/v] Tween-20) for 3 h at room temperature. Recombinant protein was added to a final concentration of 10 μg/mL and incubated overnight at 4°C, followed by three washes with TBST, 10 min each time. We used an anti-MBP antibody (New England Biolabs), diluted to 1:3,000 in blocking buffer for incubation at room temperature for 2 h, followed by three washes with TBST. The lipid membranes were then incubated with anti-mouse horseradish peroxidase antibody (Abmart), diluted 1:3,000 in blocking buffer, for 1 h at room temperature. After three washes in TBST, the signals were detected by enhanced chemiluminescence detection (Thermo Fisher) on a ChemiDocTM MP imaging system (Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/Gramene data libraries under the following accession numbers: EPAD1 (Os03g46110). Accession numbers for the sequences used in the phylogenic analysis are listed in Supplemental Files 1 and 3, and raw format of phylogentic trees are provided in Supplemental Files 2 and 4.
Supplemental Data Files
Supplemental Figure 1. Meiotic chromosome behavior in wild-type and epad1 male meiocytes.
Supplemental Figure 2. Callose deposition in wild-type and epad1 anthers.
Supplemental Figure 3. Newly formed tetrads are normal in epad1 plants.
Supplemental Figure 4. Phenotype of the flowers, anthers, and pollen grains in wild type, epad1, and transgenic complementation line carrying EPAD1pro:EPAD1gDNA.
Supplemental Figure 5. Alignment of the predicted EPAD1 protein sequence and orthologs.
Supplemental Figure 6. Unrooted phylogenetic relationship of rice nsLTPs.
Supplemental Figure 7. Expression of EPAD1 orthologs.
Supplemental Figure 8. Expression of EPAD1 in the anther and complementation test of EPAD1pro:SP-GFP-EPAD1gDNA, EPAD1pro:EPAD1gDNA-GFP and EPAD1pro:GFP-EPAD1gDNA in epad1 lines.
Supplemental Figure 9. Both the signal peptide and GPI-anchor are required for the plasma membrane localization of EPAD1.
Supplemental Figure 10. Phenotype of the flower and pollen grains in epad1 lines expressing EPAD1 with mutated GPI-anchor ω sites.
Supplemental Table. Primers used in this study.
Supplemental File 1. MEGA 6 alignment of EPAD1 and orthologs for Figure 5C.
Supplemental File 2. Newick Format of the phylogenetic tree from Figure 5C.
Supplemental File 3. MEGA 6 alignment of rice nsLTPs shown in Supplemental Figure 6.
Supplemental File 4. Newick format of the phylogenetic tree from Supplemental Figure 6.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Zhijin Luo, Mingjiao Chen, and Zibo Chen (Shanghai Jiao Tong University) for mutant screening and generation of F2 populations for mapping; Yu Kong and Xu Wang (Electron Microscopy Facilities of Neuroscience, Chinese Academy of Science) for assistance with EM sample preparation and analysis; Natalie Betts (School of Agriculture, Food and Wine, University of Adelaide) for editing; and Hongyan Yao (State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200438, China) for assisting protein–lipid overlay assays. This work was supported by funds from the National Key Technologies Research and Development Program of China (grant 2016YFD0100902); the National Natural Science Foundation of China (NSFC; grants U19A2031 and 31670309); and the China Innovative Research Team, Ministry of Education, and the Program of Introducing Talents of Discipline to Universities (grants 111 Project and B14016). Funding was also provided by ARC FT and DP (grants DP190101941 and FT160100218 to S.P.); Villum Investigator (grant 25915 to S.P.); a Novo Nordisk Laureate grant (NNF19OC0056076; to S.P.); and SJTU JiRLMDS Joint Research Fund (grant MDS-JF-2019A05 to S.P. and L.W.).
AUTHOR CONTRIBUTIONS
D.Z. and W.L. designed the experiments; H.L., Y.-J.K., and L.Y. carried out most of the experiments; H.L., Y.-J.K., and W.L. wrote the article; Z.L., J.Z., H.S., and G.H. assisted with experiments; M.C. conducted rice transformation; and S.P. helped with data analyses and editing.
Footnotes
Articles can be viewed without a subscription.
References
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K.(2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K.(2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Shikata H., Zourelidou M., Heilmann M., Heilmann I., Schwechheimer C.(2016). Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143: 4687–4700. [DOI] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S., Barnes S.H.(1990). Pollen wall development in angiosperms In Microspores evolution and ontogeny, Blackmore S., and Knox R.B., eds (London, The United Kingdom: Academic Press; ), pp. 173–192. [Google Scholar]
- Boss W.F., Im Y.J.(2012). Phosphoinositide signaling. Annu. Rev. Plant Biol. 63: 409–429. [DOI] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y., Muasya A.M., Linder H.P.(2014). A revised evolutionary history of poales: Origins and diversification. Bot. J. Linn. Soc. 175: 4–16. [Google Scholar]
- Boutrot F., Chantret N., Gautier M.F.(2008). Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M.C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B.L., Chen L., Lamport D.T.A., Chen Y., Kieliszewski M.J.(2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Ade.O., Gomes V.M.(2007). Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Chang H.S., Zhang C., Chang Y.H., Zhu J., Xu X.F., Shi Z.H., Zhang X.L., Xu L., Huang H., Zhang S., Yang Z.N.(2012). No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chu H., Yuan Z., Pan A., Liang W., Huang H., Shen M., Zhang D., Chen L.(2006). Isolation and genetic analysis for rice mutants treated with 60 Co -Ray. J. Xiamen Univ. 45: 82–85. [Google Scholar]
- Choi H., Jin J.Y., Choi S., Hwang J.U., Kim Y.Y., Suh M.C., Lee Y.(2011). An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 65: 181–193. [DOI] [PubMed] [Google Scholar]
- Christensen J.E., Horner H.T., Lersten N.R.(1972). Pollen wall and tapetal orbicular wall development in sorghum bicolor (gramineae). Am. J. Bot. 59: 43–58. [Google Scholar]
- Dickinson H.G., Heslop-Harrison J.(1968). Common mode of deposition for the sporopollenin of sexine and nexine. Nature 220: 926–927. [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Geanconteri A., Shrestha J., Carlson A., Kooyers N., Coerper D., Urbanczyk-Wochniak E., Bench B.J., Sumner L.W., Swanson R., Preuss D.(2011). A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157: 947–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Kirkpatrick A.B., Reeder S.H., Li P., Owen H.A.(2018). Pollen aperture factor inp1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiol. 176: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P.S.(2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328. [DOI] [PubMed] [Google Scholar]
- Dou X.Y., Yang K.Z., Zhang Y., Wang W., Liu X.L., Chen L.Q., Zhang X.Q., Ye D.(2011). WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J. Integr. Plant Biol. 53: 74–88. [DOI] [PubMed] [Google Scholar]
- Douliez J.P., Michon T., Elmorjani K., Marion D.(2000). Mini review: Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 32: 1–20. [Google Scholar]
- Edlund A.F., Swanson R., Preuss D.(2004). Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 16 (Suppl): S84–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstam M.M., Viitanen L., Salminen T.A., Edqvist J.(2011). Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 4: 947–964. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Bork P., Eisenhaber F.(1999). Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292: 741–758. [DOI] [PubMed] [Google Scholar]
- El-Ghazaly G., Jensen W.(1986). Studies of the development of wheat (Triticum aestivum) pollen: Formation of the pollen aperture. Can. J. Bot. 64: 1–29. [Google Scholar]
- Fitzgerald M.A., Knox R.B.(1995). Initiation of primexine in freeze-substituted microspores of Brassica campestris. Sex. Plant Reprod. 8: 99–104. [Google Scholar]
- Gerth K., Lin F., Menzel W., Krishnamoorthy P., Stenzel I., Heilmann M., Heilmann I.(2017). Guilt by association: A phenotype-based view of the plant phosphoinositide network. Annu. Rev. Plant Biol. 68: 349–374. [DOI] [PubMed] [Google Scholar]
- Guan Y.F., Huang X.Y., Zhu J., Gao J.F., Zhang H.X., Yang Z.N.(2008). RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147: 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I.(2016). Phosphoinositide signaling in plant development. Development 143: 2044–2055. [DOI] [PubMed] [Google Scholar]
- Heilmann M., Heilmann I.(2013). Arranged marriage in lipid signalling? The limited choices of PtdIns(4,5)P2 in finding the right partner. Plant Biol. 15: 789–797. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J.(1971). Wall pattern formation in angiosperm microsporogenesis. Symp. Soc. Exp. Biol. 25: 277–300. [PubMed] [Google Scholar]
- Hu J., Wang Z., Zhang L., Sun M.X.(2014). The Arabidopsis Exine Formation Defect (EFD) gene is required for primexine patterning and is critical for pollen fertility. New Phytol. 203: 140–154. [DOI] [PubMed] [Google Scholar]
- Huang M.D., Chen T.L., Huang A.H.(2013). Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiol. 163: 1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.S., Jung J.H., Yim W.C., Lee B.M., Seo Y.W., Kim W.(2007). Divergence of genes encoding non-specific lipid transfer proteins in the poaceae family. Mol. Cells 24: 215–223. [PubMed] [Google Scholar]
- Jang C.S., Yim W.C., Moon J.C., Hung J.H., Lee T.G., Lim S.D., Cho S.H., Lee K.K., Kim W., Seo Y.W., Lee B.M.(2008). Evolution of non-specific lipid transfer protein (nsLTP) genes in the Poaceae family: Their duplication and diversity. Mol. Genet. Genomics 279: 481–497. [DOI] [PubMed] [Google Scholar]
- Jiang J., Zhang Z., Cao J., Weber A.(2013). Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 15: 249–263. [DOI] [PubMed] [Google Scholar]
- Johnson K.L., Cassin A.M., Lonsdale A., Bacic A., Doblin M.S., Schultz C.J.(2017). A motif and amino acid bias bioinformatics pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol. 174: 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295. [DOI] [PubMed] [Google Scholar]
- Kouidri A., Baumann U., Okada T., Baes M., Tucker E.J., Whitford R.(2018). Wheat TaMs1 is a glycosylphosphatidylinositol-anchored lipid transfer protein necessary for pollen development. BMC Plant Biol. 18: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D.T.A., Kieliszewski M.J., Chen Y., Cannon M.C.(2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Weber Z.T., Zourelidou M., Hofmeister B.T., Schmitz R.J., Schwechheimer C., Dobritsa A.A.(2018). Arabidopsis protein kinase d6pkl3 is involved in the formation of distinct plasma membrane aperture domains on the pollen surface. Plant Cell 30: 2038–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.S., Phyo P., Jacobowitz J., Hong M., Weng J.K.(2019). The molecular structure of plant sporopollenin. Nat. Plants 5: 41–46. [DOI] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.L., Liu Y., Douglas C.J.(2017). Role of glycosyltransferases in pollen wall primexine formation and exine patterning. Plant Physiol. 173: 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Zhang X., Lu C., Zeng X., Li Y., Fu D., Wu G.(2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66: 5663–5681. [DOI] [PubMed] [Google Scholar]
- Liu Z., Lin S., Shi J., Yu J., Zhu L., Yang X., Zhang D., Liang W.(2017). Rice No Pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J. 91: 263–277. [DOI] [PubMed] [Google Scholar]
- Lou Y., Zhu J., Yang Z.(2014). Molecular cell biology of pollen walls In Applied Plant Cell Biology, Nick P., and Opatrny Z., eds (Berlin, Germany: Springer; ), pp. 179–205. [Google Scholar]
- Luo Y., Lu L., Wortley A.H., Li D.Z., Wang H., Blackmore S.(2015). Evolution of angiosperm pollen. 3. monocots. Ann. Mo. Bot. Gard. 101: 406–455. [Google Scholar]
- Ma L.J., Yang Z.N., Zhang S.(2013). Dex1, a plasma membrane-localized protein, functions in microspore development by affecting cals5 expression in Arabidopsis thaliana. Chin. Sci. Bull. 58: 2855–2861. [Google Scholar]
- Mander L., Li M., Mio W., Fowlkes C.C., Punyasena S.W.(2013). Classification of grass pollen through the quantitative analysis of surface ornamentation and texture. Proc. Biol. Sci. 280: 20131905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Riezman H.(2004). Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5: 110–120. [DOI] [PubMed] [Google Scholar]
- Mondol P.C., Xu D., Duan L., Shi J., Wang C., Chen X., Chen M., Hu J., Liang W., Zhang D.(2020). Defective Pollen Wall 3 (DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice. New Phytol. 225: 807–822. [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders D.M., Dodrill C.H., Owen H.A., Makaroff C.A.(2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127: 1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A., Martelli P.L., Casadio R.(2008). PredGPI: A GPI-anchor predictor. BMC Bioinformatics 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J.(2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Grienenberger E., Douglas C.J.(2015). The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113: 170–182. [DOI] [PubMed] [Google Scholar]
- Quilichini T.D., Samuels A.L., Douglas C.J.(2014). ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 26: 4483–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radja A., Horsley E.M., Lavrentovich M.O., Sweeney A.M.(2019). Pollen cell wall patterns form from modulated phases. Cell 176: 856–868.e10. [DOI] [PubMed] [Google Scholar]
- Rowley J.R., Dunbar A.(1996). Pollen development in Centrolepis aristata (Centrolepidaceae). Grana 35: 1–15. [Google Scholar]
- Rowley J.R., Claugher D.(1991). Receptor-independent sporopollenin. Bot. Acta 104: 316–323. [Google Scholar]
- Scott R.J.(1994). Pollen exine: The sporopollenin enigma and the physics of pattern In Molecular and cellular aspects of plant reproduction, Scott R.J., and Stead M.A., eds (London: Cambridge University Press; ), pp. 49–81. [Google Scholar]
- Sheldon J.M., Dickinson H.G.(1983). Determination of patterning in the pollen wall of Lilium henryi. J. Cell Sci. 63: 191–208. [DOI] [PubMed] [Google Scholar]
- Shi J., Cui M., Yang L., Kim Y.J., Zhang D.(2015). Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20: 741–753. [DOI] [PubMed] [Google Scholar]
- Shi J., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvarla J.J., Larson D.A.(1966). Fine structural studies of zea mays pollen. I: Cell membranes and exine ontogeny. Am. J. Bot. 53: 1112–1125. [Google Scholar]
- Skvarla J.J., Rowley J.R.(1987). Ontogeny of pollen in Poinciana (Leguminosae). I. Development of exine template. Rev. Palaeobot. Palynol. 50: 239–311. [Google Scholar]
- Stanislas T., Hüser A., Barbosa I.C.R., Kiefer C.S., Brackmann K., Pietra S., Gustavsson A., Zourelidou M., Schwechheimer C., Grebe M.(2015). Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat. Plants 1: 15162. [DOI] [PubMed] [Google Scholar]
- Sun M.X., Huang X.Y., Yang J., Guan Y.F., Yang Z.N.(2013). Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 26: 83–91. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Narciso J.O., Zeng W., van de Meene A., Yasutomi M., Takemura S., Lampugnani E.R., Doblin M.S., Bacic A., Ishiguro S.(2017). KNS4/UPEX1: A type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173: 183–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M.(1989). Pattern determination of the exine in Caesalpinia japonica (Leguminosae: Caesalpinioideae). Am. J. Bot. 76: 1615–1626. [Google Scholar]
- Takahashi M., Skvarla J.J.(1991). Exine pattern formation by plasma membrane in Bougainvillea spectabilis Willd. (Nyctaginaceae). Am. J. Bot. 78: 1063–1069. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Liang W., Hu J., Zhang D.(2012). MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 22: 1127–1137. [DOI] [PubMed] [Google Scholar]
- Tucker E.J., et al. (2017). Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat. Commun. 8: 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S., Fleming A., Wellman C.H., Beerling D.J.(2011). Evolutionary development of the plant and spore wall. AoB Plants 2011: plr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Hwang S.G., Karuppanapandian T., Liu A., Kim W., Jang C.S.(2012). Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res. 19: 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Kwon H.J., Yim W.C., Lim S.D., Moon J.C., Lee B.M., Seo Y.W., Kim W., Jang C.S.(2010). Expressional diversity of wheat nsLTP genes: Evidence of subfunctionalization via cis-regulatory divergence. Genetica 138: 843–852. [DOI] [PubMed] [Google Scholar]
- Wang R., Dobritsa A.A.(2018). Exine and aperture patterns on the pollen surface: their formation and roles in plant reproduction. Annual Plant Reviews 1: 1–40. [Google Scholar]
- Wang Z., Li J., Chen S., Heng Y., Chen Z., Yang J., Zhou K., Pei J., He H., Deng X.W., Ma L.(2017). Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. USA 114: 12614–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Zhong X. (2014). Non-specific lipid transfer proteins in maize. BMC Plant Biol. 14: 281–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Bacic A.(2012). Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat. Protoc. 7: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Xu T., Zhang C., Zhou Q., Yang Z.N.(2016). Pollen wall pattern in Arabidopsis. Sci. Bull. (Beijing) 11: 1–6. [Google Scholar]
- Xue J.S., et al. (2020). Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Mol. Plant S1674-2052–2056. [DOI] [PubMed] [Google Scholar]
- Yeats T.H., Bacic A., Johnson K.L.(2018). Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J. Integr. Plant Biol. 60: 649–669. [DOI] [PubMed] [Google Scholar]
- Yu J., et al. (2016). A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiol. 172: 1772–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D.(2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Luo X., Zhu L.(2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38: 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang D.B., Wilson Z.A.(2009). Stamen specification and anther 1 development in rice. Chin. Sci. Bull. 54: 2342–2353. [Google Scholar]
- Zhang X., Zhao G., Tan Q., Yuan H., Betts N., Zhu L., Zhang D., Liang W.(2020). Rice pollen aperture formation is regulated by the interplay between OsINP1 and OsDAF1. Nat. Plants 6: 394–403. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2016). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7: 11656. [DOI] [PMC free article] [PubMed] [Google Scholar]