JANUS connects RNA polymerase II (Pol II) with PLT1 to promote the autotranscription of PLT1 while GIF1 titrates JANUS away from Pol II by competitive binding.
Abstract
Plant roots are sustained through meristem activity at the root tip. Two transcriptional pathways, one mediated by PLETHORAs (PLTs) and the other by SHORTROOT and SCARECROW, play major roles in root meristem development. The role of PLTs during root meristem development requires a concentration gradient, which is not only contributed by posttranslational regulation such as growth dilution and intercellular movement but also likely by a largely unknown fine-tuned transcriptional regulatory mechanism. We report here that Arabidopsis (Arabidopsis thaliana) JANUS positively regulates PLT1 expression in the root meristem by recruiting RNA polymerase II (Pol II) to PLT1 and by interacting with PLT1. JANUS-dependent recruitment of Pol II is inhibited through the competitive binding of JANUS by GRF-INTERACTING FACTOR1 (GIF1)/ANGUSTIFOLIA3, a transcriptional cofactor that negatively regulates PLT1 expression. Finally, GIF1 and JANUS, the antagonistic regulators of PLT1, both depend on Arabidopsis IMPORTIN β4 for their nuclear accumulation. The combination of an importin and its two antagonistic cargos in PLT1 transcription may have logistic benefits in fine-tuning the transcription of PLT1 in root meristem.
INTRODUCTION
The activity of the meristem at the root tip sustains plant root growth. The root meristem is organized into well-defined domains, like the stem cell niche and transit-amplifying cells produced by root stem cells (Dolan et al., 1993; Watt and Hogan, 2000; Weigel and Jürgens, 2002; Laux, 2003; Aida et al., 2004). The root stem cell niche consists of the quiescent center (QC) and surrounding stem cells. Stem cells are pluripotent cells whose progeny normally generate defined yet flexible cell types, depending on their position (Xu et al., 2006; Scheres, 2007; Sena et al., 2009), while QC cells are mitotically inactive but are critical for maintaining and replenishing stem cells (Dolan et al., 1993; van den Berg et al., 1995; Cruz-Ramírez et al., 2013).
Two major transcriptional routes specify the QC cell fate and the adjacent stem cells: PLETHORA family members (PLTs) and SHORTROOT/SCARECROW (SCR; Wysocka-Diller et al., 2000; Sabatini et al., 2003; Aida et al., 2004; Heidstra et al., 2004; Galinha et al., 2007; Scheres, 2007; Heidstra and Sabatini, 2014). Mutations in either pathway cause the collapse of the QC and the disorganization of stem cells, thereby compromising root growth and meristem development (Petricka et al., 2012; Heyman et al., 2014; Shimotohno et al., 2018). The establishment and maintenance of a PLT concentration gradient is critical for root development, such that higher PLT levels promote stem cell maintenance while lower PLT levels promote the mitotic activity of stem cell daughters (Galinha et al., 2007; Mahönen et al., 2014). A further reduction in PLT levels promotes cell differentiation (Mahönen et al., 2014). The PLT gradient controls cell fate by the spatial induction of genes required for cell proliferation and the repression of genes involved in cell differentiation (Santuari et al., 2016). Moreover, PLT targets include major patterning genes and components of an autoregulatory feedback loop, thus reenforcing their key roles in organ development (Santuari et al., 2016). Growth dilution and intercellular movement both contribute to the establishment of the PLT gradient (Mahönen et al., 2014). However, it is not fully understood how transcriptional regulation influences this protein gradient.
A recent study demonstrated that the transcriptional coactivator GRF-INTERACTING FACTOR1 (GIF1)/ANGUSTIFOLIA3 negatively regulates PLT1 expression, likely by interacting with transcription factors such as GROWTH REGULATORY FACTORS (GRFs) in the root meristem (Rodriguez et al., 2015; Ercoli et al., 2018). We recently demonstrated that the Arabidopsis (Arabidopsis thaliana) importin, IMPORTIN β4 (IMB4), mediates the nuclear accumulation of GIF1 (Liu et al., 2019). However, in contrast to the longer roots seen in gif1 mutants (Ercoli et al., 2018), null mutants for IMB4 showed reduced root growth (Liu et al., 2019), suggesting a complexity in their interaction. Here, we report that IMB4 mediates the nuclear accumulation of another transcriptional regulator of PLT1, JANUS, in addition to that of GIF1. JANUS positively regulates the expression of PLT1 in the root meristem by recruiting RNA polymerase II (Pol II). Interestingly, GIF1 inhibits the binding of Pol II with JANUS through competitive binding with the RNA recognition motif II (RRM2) domain of JANUS. Furthermore, JANUS directly interacts with PLT1 via its C terminus and is required for the autotranscriptional regulation of PLT1. Thus, GIF1 and JANUS, whose nuclear accumulation is controlled by IMB4, antagonistically mediate root meristematic activity. Such a tripartite module may fine-tune the expression of PLT1 to control root meristem homeostasis.
RESULTS AND DISCUSSION
Functional Loss of IMB4 Compromises Root Meristem Activity
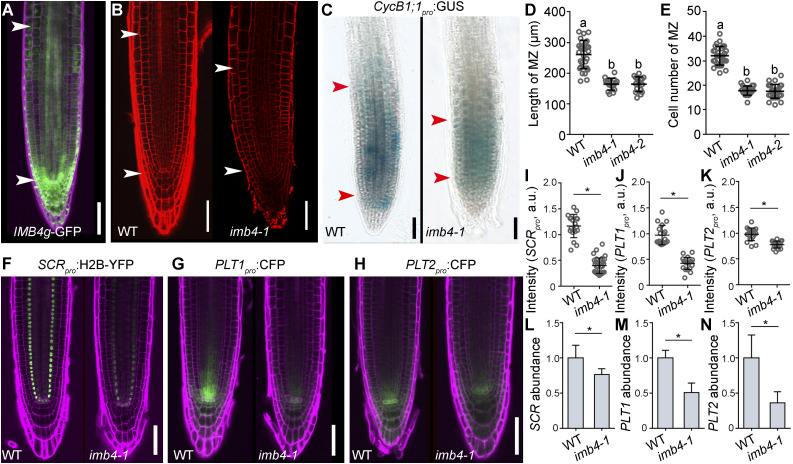
The nuclear accumulation of GIF1 in ovules depends on IMB4 (Liu et al., 2019). Indeed, imb4-1, a null mutation of IMB4, shows defective ovule development comparable to that of gif1 (Liu et al., 2019). We determined that root apical meristem (RAM) cells in imb4-1 exhibit a significant reduction of nuclear GIF1 accumulation compared with the wild type (Supplemental Figure 1) even though IMB4 is highly expressed in roots (Figure 1A), based on confocal laser scanning microscopy (CLSM) of the IMB4g-GFP construct in transgenic plants (Liu et al., 2019). Thus, we expected a longer RAM in the imb4-1 mutant, as seen in gif1 or higher-order mutants of GIF family members (Ercoli et al., 2018). However, imb4-1 instead showed a significantly reduced root meristem (Figures 1B and 1D), which resulted from a reduced activity in cell division (Figures 1C and 1E). The genes PLT1, PLT2, and SCR, which are critical for root meristem development (Wysocka-Diller et al., 2000; Sabatini et al., 2003; Aida et al., 2004; Heidstra et al., 2004; Galinha et al., 2007; Scheres, 2007; Petricka et al., 2012; Heidstra and Sabatini, 2014; Heyman et al., 2014; Shimotohno et al., 2018), were all transcriptionally downregulated in the imb4-1 mutant relative to the wild type, based on the analysis of their respective reporter lines in which the expression of Cyan Fluorescent Protein (CFP) or Yellow Fluorescent Protein (YFP) is driven by their native promoters (Figures 1F to 1K): PLT1pro:CFP (Chen et al., 2011), PLT2pro:CFP (Chen et al., 2011), or SCRpro:H2B-YFP (Heidstra et al., 2004). RT-qPCR independently validated these results (Figures 1L to 1N), thus suggesting that IMB4 plays a positive role in root meristem development.
Figure 1.
Functional Loss of IMB4 Compromises Root Meristem Activity.
(A) Representative CLSM image of an IMB4g-GFP seedling root. The root was stained with propidium iodide (PI; magenta).
(B) Representative CLSM images of PI-stained wild-type and imb4-1 roots.
(C) CycB1;1pro:GUS activity in wild-type and imb4-1 roots.
The region of the RAM is indicated by two arrowheads in (A) to (C).
(D) and (E) Length (D) and cell number (E) of the root meristem zone (MZ). Values are means ± sd (n > 16). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
(F) to (K) Representative CSLM images ([F] to [H]) and fluorescence intensities ([I] to [K]) of SCRpro:H2B-YFP ([F] and [I]), PLT1pro:CFP ([G] and [J]), and PLT2pro:CFP ([H] and [K]) in wild-type and imb4-1 roots. a.u. indicates arbitrary units. Roots were stained with PI (magenta). Values are means ± sd (n > 16; [J] and [K]) or means ± se (n = 10; [I]). Fluorescence of the nuclei from 10 roots was measured for (I); fluorescence of the stem cell niche from over 16 roots was measured for (J) and (K).
(L) to (N) Relative transcript abundance for SCR (L), PLT1 (M), and PLT2 (N) 5 d after germination (DAG) in wild-type and imb4-1 seedlings, as determined by RT-qPCR.
Asterisks in (I) to (N) indicate significant differences (t test, P < 0.05). Bars in (A) to (C) and (F) to (H) = 50 μm.
IMB4 Mediates the Nuclear Accumulation of JANUS
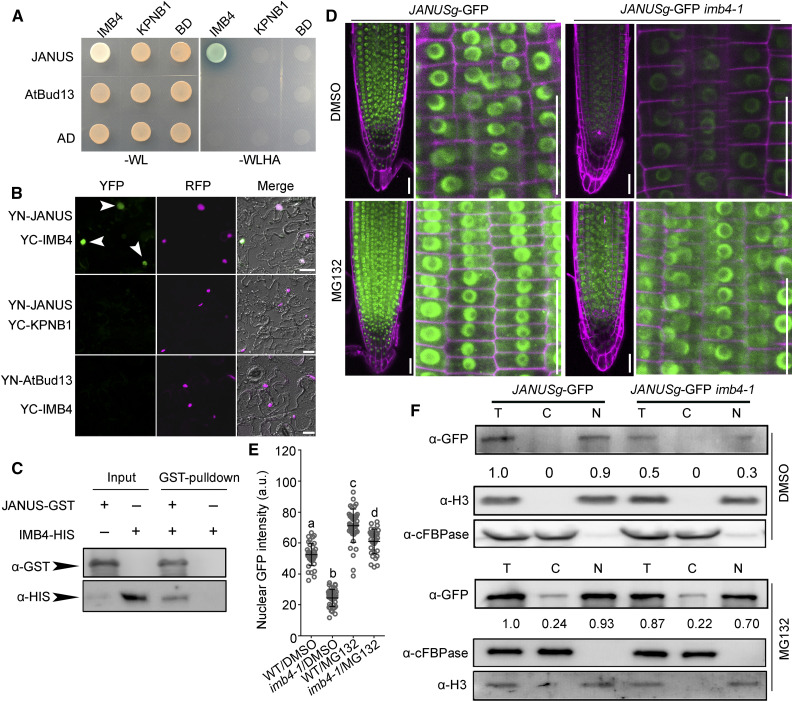
The fact that imb4-1 differed from gif1, a mutant in one of its cargos (Liu et al., 2019) during root meristem development, suggested that other IMB4 cargos might play positive roles in this process. By screening the Arabidopsis Protein Interactome Database (Li et al., 2011), we focused on JANUS out of a few potential IMB4 interactors. JANUS is a putative component of the spliceosome (Xiong et al., 2019a; Xiong and Li, 2020) that was recently demonstrated to recruit Pol II for the transcription of WUSCHEL-RELATED HOMEOBOX2 (WOX2) and PIN-FORMED7 (PIN7) during embryogenesis (Xiong et al., 2019a). Indeed, JANUS interacted with IMB4 by yeast two-hybrid (Y2H; Figure 2A), by bimolecular fluorescence complementation (BiFC; Figure 2B), and by in vitro pull-down (Figure 2C) assays.
Figure 2.
IMB4 Mediates the Nuclear Accumulation of JANUS.
(A) and (B) Y2H (A) and BiFC (B) assays demonstrating the specific interaction between JANUS and IMB4. KPNB1, another Arabidopsis importin β (Luo et al., 2013), was used as a negative control for IMB4, while AtBud13, another component of the spliceosome (Xiong et al., 2019b), was used as a negative control for JANUS. Yeast colonies bearing both the BD and AD vectors were selected on synthetic minimal medium lacking Trp and Leu (–WL); positive interaction was assessed on synthetic minimal medium lacking Trp, Leu, His, and Ade (–WLHA). Bars in (B) = 50 μm.
(C) In vitro pull-down assay. Results are representative of three biological replicates.
(D) and (E) Representative CLSM images (D) and GFP intensities (E) of RAM from JANUSg-GFP in the wild type and JANUSg-GFP in imb4-1 upon DMSO or MG132 treatment. a.u. indicates arbitrary units. Roots were stained with PI (magenta). Images were taken from seedling roots at 5 DAG treated with DMSO or 100 μM MG132 for 2 h. Values are means ± se (n = 20). Fluorescence of the nuclei in the root meristem zone was measured. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). Bars in (D) = 50 μm.
(F) Cell fractionation and immunoblot analysis. Anti-GFP antibody was used in immunoblots to detect JANUS-GFP. Numbers at the bottom are the values of JANUS-GFP in the nuclear fraction (N) or cytoplasmic fraction (C) relative to that of the total protein fraction (T), which was set to 1. Anti-histone H3 and anti-cFBPase were used to indicate nuclear and cytoplasmic fractions, respectively.
To determine how IMB4 might regulate JANUS, we analyzed GFP distribution in the root meristem of the wild type or the imb4-1 mutant expressing the JANUSg-GFP construct by CLSM. JANUSg-GFP rescues the embryo lethality normally seen in janus mutants (Xiong et al., 2019a), indicating that the JANUS-GFP fusion protein is functional. We detected strong GFP signal in the nuclei of wild-type cells but not in imb4-1 (Figures 2D and 2E), although JANUSg-GFP was expressed at a comparable level in the wild type and imb4-1 (Supplemental Figure 2B), suggesting the IMB4-dependent nuclear accumulation of JANUS. Indeed, inhibiting 26S proteasome-mediated degradation with the proteasome inhibitor MG132 substantially increased the nuclear intensity of the JANUS-GFP fusion protein in both the wild type and imb4-1 (Figures 2D and 2E), indicating that IMB4 protects JANUS against 26S proteasome-mediated degradation. In addition, GFP signal appeared in the cytoplasm in both the wild type and imb4-1 upon MG132 treatment (Figures 2D and 2E), suggesting that JANUS undergoes rapid turnover in the cytoplasm. By employing cell fractionation and immunoblot analyses, we confirmed that IMB4 positively regulates the nuclear accumulation of JANUS by preventing 26S proteasome-mediated degradation (Figure 2F). Ivermectin, an inhibitor of nuclear import, significantly inhibited the nuclear accumulation of JANUS-GFP (Supplemental Figure 3), further supporting a dynamic import of JANUS to the nucleus in RAM.
Because JANUS is essential for embryogenesis (Xiong et al., 2019a), it was surprising that the imb4 mutant would be viable (Liu et al., 2019). A close examination of the expression pattern associated with the IMB4g-GFP construct (Liu et al., 2019) showed that IMB4 is expressed late during embryogenesis—that is, in embryos at the cotyledon stage (Supplemental Figure 2A). In addition, by examining JANUS-GFP distribution in developing embryos, we determined that the nuclear accumulation of JANUS-GFP was comparable between the wild type and the imb4-1 mutant expressing the JANUSg-GFP construct during early embryogenesis. However, GFP signal from the JANUSg-GFP transgene was significantly reduced in imb4-1 embryos at the cotyledon stage compared with wild-type embryos (Supplemental Figures 2C and 2D). These results suggest that nuclear accumulation of JANUS during early embryogenesis is regulated by factors other than IMB4.
JANUS Positively Regulates Root Meristem Activity
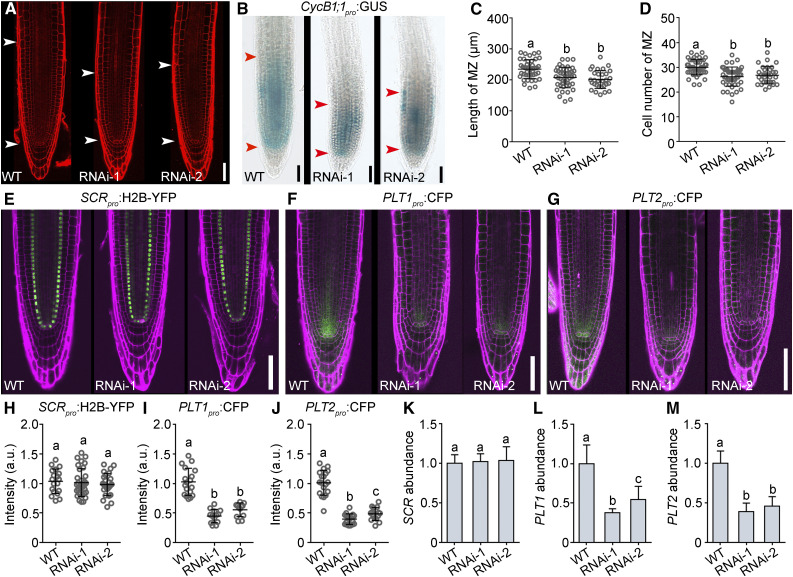
To examine the potential role of JANUS in root meristem development, we examined transgenic roots expressing 35Spro:JANUS-RNAi to downregulate JANUS transcripts by RNAi interference (RNAi; Xiong et al., 2019a), since JANUS loss of function results in embryo lethality (Xiong et al., 2019a). Downregulating JANUS indeed caused a severe reduction in root meristem development (Figure 3A). 35Spro:JANUS-RNAi plants showed a significantly shorter root meristem (Figure 3C), which was due to reduced cell division (Figures 3B and 3D). By analyzing fluorescence intensity of SCRpro:H2B-YFP (Figure 3E), PLT1pro:CFP (Figure 3F), or PLT2pro:CFP (Figure 3G) in JANUS-RNAi lines, we determined that the transcriptional activity at PLT1 (Figure 3I) and PLT2 (Figure 3J), but not SCR (Figure 3H), was significantly downregulated by JANUS-RNAi, which we confirmed by RT-qPCR in wild-type and JANUS-RNAi seedlings (Figures 3K to 3M).
Figure 3.
JANUS Positively Regulates Root Meristem Activity.
(A) Representative CLSM image of PI-stained roots from the wild type and two 35Spro:JANUS-RNAi lines (RNAi-1 and RNAi-2).
(B) CycB1;1pro:GUS activity in roots from the wild type and two 35Spro:JANUS-RNAi lines.
The region of the RAM is indicated by two arrowheads in (A) and (B).
(C) and (D) Length (C) and cell number (D) of the root meristem zone (MZ). Values are means ± sd (n > 33).
(E) to (J) Representative CSLM images ([E] to [G]) and fluorescence intensities ([H] to [J]) of SCRpro:H2B-YFP ([E] and [H]), PLT1pro:CFP ([F] and [I]), and PLT2pro:CFP ([G] and [J]) in the wild type and two 35Spro:JANUS-RNAi lines. a.u. indicates arbitrary units. Roots were stained with PI (magenta). Values are means ± sd (n > 12) for (I) and (J) and means ± se (n = 27) for (H). Fluorescence of the stem cell niche from more than 12 roots was measured for (I) and (J); fluorescence of nuclei from 27 roots was measured for (H).
(K) to (M) Relative transcript abundance of SCR (K), PLT1 (L), and PLT2 (M) at 5 DAG in the wild type and two lines of 35Spro:JANUS-RNAi by RT-qPCR. Values are means ± se (n = 3).
Different letters in (C), (D), and (H) to (M) indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05). Bars in (A), (B), and (E) to (G) = 50 μm.
Because all three genes are expressed during embryogenesis (Wysocka-Diller et al., 2000; Aida et al., 2004; Xiong et al., 2019a) and since JANUS is essential for embryogenesis (Xiong et al., 2019a), we examined developing janus embryos expressing the reporters SCRpro:H2B-YFP (Supplemental Figure 4A), PLT1pro:CFP (Supplemental Figure 4B), or PLT2pro:CFP (Supplemental Figure 4C) by CLSM. We obtained results in embryos that were very similar to those seen in the root meristem—that is, the transcriptional activity of PLT1 and PLT2 but not SCR was substantially downregulated in janus embryos (Supplemental Figure 4). These results suggested that JANUS positively regulates root meristem development, possibly by transcriptional regulation of PLTs. In addition, JANUS accounts for a specific subset of downstream events mediated by IMB4 during RAM development, since the transcriptional activity of SCR was compromised in imb4-1 (Figure 1) but not in JANUS-RNAi lines (Figure 3).
JANUS Mediates Pol II-Dependent PLT1 Transcription
JANUS was previously demonstrated to recruit Pol II for the transcription of specific genes (Xiong et al., 2019a; Xiong and Li, 2020), despite its lack of obvious DNA binding domains (Xiong et al., 2019a; Xiong and Li, 2020). To test whether the function of JANUS in root meristem was through Pol II, we performed the following experiments. First, we examined whether the noncatalytic subunit of RNA-dependent RNA polymerase II, IV, and V NRPB10, a JANUS-interacting Pol II component (Xiong et al., 2019a), participated in root meristem development. NRPB10 is highly expressed in the root meristem (Supplemental Figure 5A), and its loss of function resulted in a significant reduction in root meristem length (Supplemental Figures 5B, 5C, and 5E) and in meristem cell number (Supplemental Figure 5F), suggesting that NRPB10 is a key component of root meristem development. Second, the transcriptional activity at PLT1 was significantly reduced in the nrpb10 mutant, based on CFP intensity at the stem cell niche of the PLT1pro:CFP reporter expressed in the nrpb10 background and on RT-PCR (Supplemental Figures 5D, 5G, and 5H). These results suggest that Pol II positively regulates root meristem development by mediating the transcription of key regulatory genes.
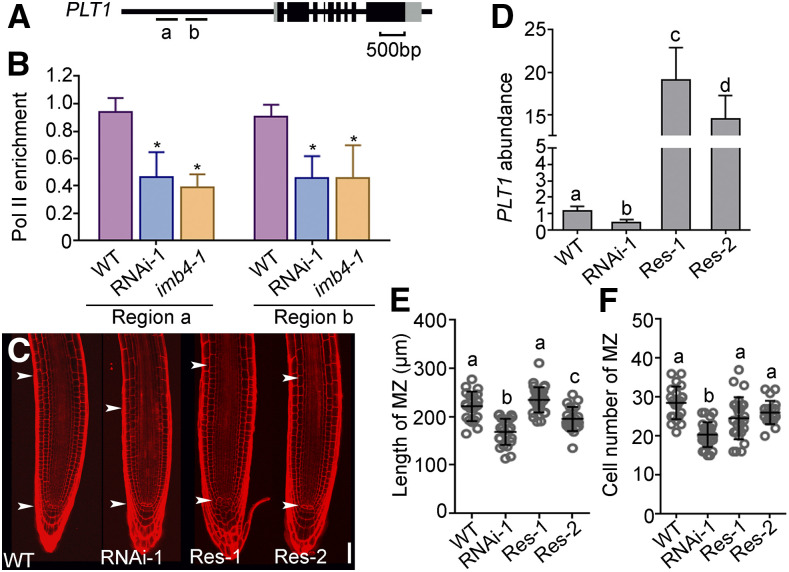
Because JANUS specifically recruits Pol II for the transcription of WOX2 and PIN7 during embryogenesis (Xiong et al., 2019a), we suspected that the role of Pol II in the transcription of PLT1 might also depend on JANUS. To test this hypothesis, we examined Pol II occupancy at the PLT1 promoter by chromatin immunoprecipitation (ChIP) assays (Figure 4A). Indeed, Pol II occupancy was significantly reduced at the promoter regions of PLT1 by the downregulation of JANUS in JANUS-RNAi lines or in the imb4-1 mutant (Figure 4B), indicating that JANUS mediates PLT1 transcription during root meristem development by recruiting Pol II to the PLT1 promoter. To provide further evidence that reduced PLT1 levels resulted from reduced RAM size in JANUS-RNAi lines, we introduced UBQ10pro:GFP-PLT1 into the JANUS-RNAi background. As expected, the ectopic expression of PLT1 partially rescued the reduced RAM length of JANUS-RNAi lines (Figures 4C, 4E, and 4F), consistent with a significant increase in PLT1 abundance seen by RT-qPCR (Figure 4D). These results suggest that a reduction in PLT1 transcript levels is a major cause for RAM size reduction in JANUS-RNAi.
Figure 4.
JANUS Mediates Pol II-Dependent PLT1 Transcription.
(A) Schematic illustration of the PLT1 genomic locus. Letters (a and b) indicate target regions in ChIP-PCR.
(B) Quantification of ChIP-PCR. Pol II enrichment at PLT1 is presented as the ratio of PLT1 (Pol II/input) to ACTIN (Pol II/input). Results are means ± se (n = 3). Asterisks indicate significant differences from the wild type (t test, P < 0.05).
(C) Representative PI-stained roots from the wild type, 35Spro:JANUS-RNAi-1, and two 35Spro:JANUS-RNAi UBQ10pro:GFP-PLT1 lines (Res-1 and Res-2). The region of the RAM is indicated by two arrowheads. Bar = 50 μm.
(D) Relative transcript abundance of PLT1 in the wild type, 35Spro:JANUS-RNAi-1, and two lines of 35Spro:JANUS-RNAi-1 UBQ10pro:GFP-PLT1 (Res-1 and Res-2). Values are means ± se (n = 3). RNAs were extracted from seedlings at 5 DAG.
(E) and (F) Length (E) and cell number (F) of the root meristem zone (MZ). Values are means ± sd (n > 17).
Different letters in (D) to (F) indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
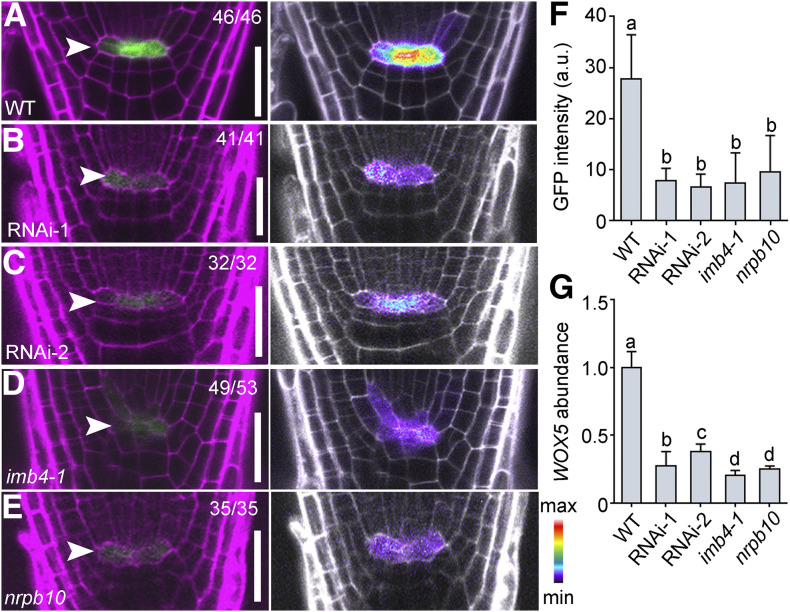
PLT1 was reported to positively regulate the expression of WOX5 (Shimotohno et al., 2018), whose activity defines the QC (Sarkar et al., 2007). We hypothesized that a significant reduction in PLT1 abundance in JANUS-RNAi lines would result in reduced WOX5 expression. To test this hypothesis, we introduced the WOX5pro:GFP reporter (Blilou et al., 2005) into JANUS-RNAi lines as well as in the imb4-1 and nrpb10 mutants, both of which were compromised in PLT1 expression. By examining GFP intensity at the QC of WOX5pro:GFP in the wild type (Figure 5A), two WOX5pro:GFP lines in JANUS-RNAi (Figures 5B and 5C), WOX5pro:GFP in imb4-1 (Figure 5D), and WOX5pro:GFP in nrpb10 (Figure 5E), we determined that WOX5 transcriptional activity was significantly reduced in the JANUS-RNAi, imb4-1, and nrpb10 backgrounds (Figure 5F). RT-qPCR analysis of seedlings from the various genetic materials confirmed a reduction of WOX5 transcript abundance in the JANUS-RNAi, imb4-1, and nrpb10 backgrounds (Figure 5G). These results further support the JANUS-dependent transcription of PLT1.
Figure 5.
WOX5 Expression Depends on IMB4, JANUS, and Pol II.
(A) to (E) Representative CLSM images of the QC from PI-stained 5-DAG seedling roots expressing the WOX5pro:GFP reporter in the Col-0 wild type, two 35Spro:JANUS-RNAi lines (RNAi-1 and RNAi-2), imb4-1, and nrpb10. Arrowheads point to QC cells. Left, the merge of the RFP (PI; magenta) and GFP channels; right, the GFP channel displayed in pseudocolors, with intensity scale at right bottom. Bars = 50 μm.
(F) and (G) GFP intensity (F) and WOX5 abundance (G) from the WOX5pro:GFP reporter in the Col-0 wild type, two 35Spro:JANUS-RNAi lines (RNAi-1 and RNAi-2), imb4-1, and nrpb10 in the QC cells. a.u. indicates arbitrary units. Values are means ± sd (n > 30) in (F) and means ± se (n = 3) in (G). RNAs were extracted from seedlings at 5 DAG. Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
JANUS Is Required for Autotranscriptional Regulation of PLT1 by Interacting with PLT1
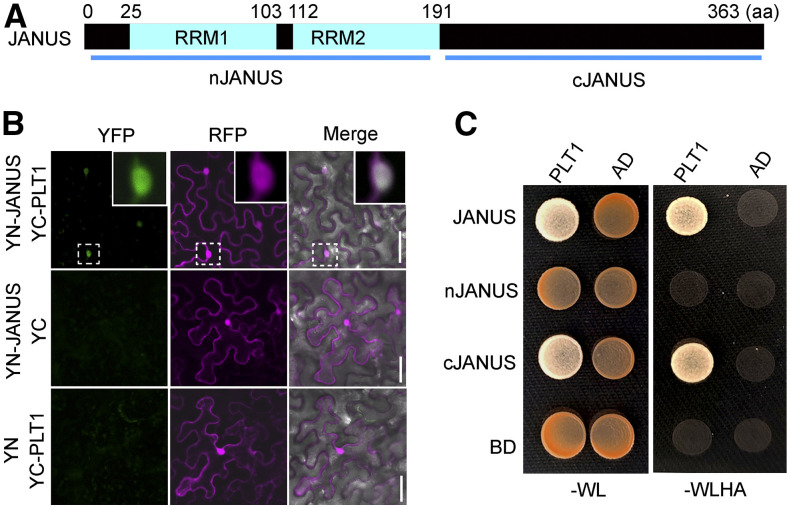
Because JANUS contains no discernible DNA binding domains (Xiong et al., 2019a; Xiong and Li, 2020), we wondered how JANUS might influence PLT1 transcription in the root meristem, as suggested here, as well as WOX2 and PIN7 transcription during embryogenesis (Xiong et al., 2019a). To solve this issue, we performed a Y2H screen using JANUS as the bait. Surprisingly, we identified PLT1 among potential JANUS interactors and confirmed this result by targeted Y2H (Figure 6C) and BiFC (Figure 6B) assays. Upon close examination, we determined that PLT1 interacts with the C terminus of JANUS (Figure 6C) while Pol II components interact with the N-terminal domain of JANUS (Xiong et al., 2019a).
Figure 6.
JANUS Interacts with PLT1.
(A) Schematic illustration of JANUS domain organization. RRM indicates RNA-recognition motif; nJANUS indicates the N-terminal domain of JANUS including both RRMs; cJANUS indicates the C-terminal domain of JANUS. aa, amino acids.
(B) A representative BiFC assay showing that JANUS interacts with PLT1 in the nucleus. From left to right: the YFP channel, the RFP channel for RFP expressed from 35Spro:RFP (magenta), and merges of the YFP, RFP, and transmission channels. Insets are closeups of the dotted squares in the top panels. Bars = 50 μm.
(C) A representative Y2H assay showing that the C terminus of JANUS interacts with PLT1. Yeast colonies transformed with both BD and AD vectors were grown on synthetic minimal medium lacking Trp and Leu (–WL). Positive interactions were determined by growth on synthetic minimal medium lacking Trp, Leu, His, and Ade (–WLHA). Results are representative of three biological replicates.
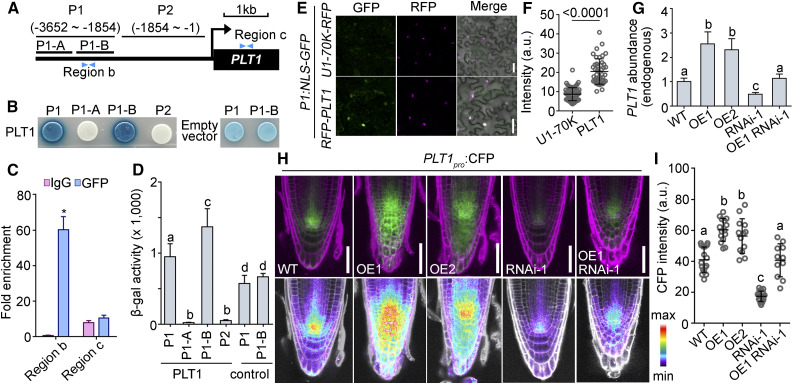
PLT2 was reported to regulate its own transcription (Durgaprasad et al., 2019) and PLT1 was hinted to have a similar autotranscriptional regulatory role (Xu et al., 2020). We first tested whether PLT1 was able to regulate its own transcription. We introduced the PLT1pro:CFP reporter into a wild-type line already carrying UBQ10pro:GFP-PLT1 (PLT1-OE) and in two PLT loss-of-function mutants, the plt1-4 single mutant and the plt1-4 plt2-2 double mutant (Aida et al., 2004). By examining CFP intensity at the stem cell niche as a proxy for endogenous PLT1 transcriptional activity, we determined that PLT1-OE enhanced, whereas PLT loss of function reduced, PLT1 transcriptional activity (Supplemental Figures 6A to 6F). We validated these results by RT-qPCR (Supplemental Figure 6G). Indeed, PLT1 bound to its own promoter sequences based on yeast one-hybrid (Y1H; Figures 7A, 7B, and 7D), ChIP (Figure 7C), and in vitro transcriptional (Figures 7E and 7F) assays. Thus, we conclude that PLT1 positively regulates its own transcription.
Figure 7.
Autotranscriptional Regulation of PLT1 in RAM Requires JANUS.
(A) Schematic illustration of the promoter and 5′ untranslated region of PLT1. P1-A spans from –3652 to –2809 bp; P1-B spans from –2809 to –1854 bp; region b spans from –2047 to –1919 bp; region c spans from 726 to 1103 bp.
(B) Representative Y1H assay. Empty vector used as the negative control shows background activation to the P1 and P1-B regions.
(C) Quantification of ChIP-PCR. Fold enrichment at PLT1 is presented as the ratio of PLT1 (ChIP/input) to ACTIN (ChIP/input). Results are means ± se (n = 3). Seedlings at 5 DAG were used for the assay. The asterisk indicates a significant difference from the wild type (t test, P < 0.05).
(D) β-Galactosidase activity from Y1H assays. Control indicates empty vector. Values are means ± sd (n = 7). Different letters indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
(E) and (F) Representative CLSM images (E) and GFP intensities (F) of leaf epidermal cells cotransformed with P1pro:NLS-GFP and 35Spro:RFP‐PLT1 (magenta) or U1pro-70K-RFP (a nuclear protein as a control; magenta). a.u. indicates arbitrary units. Results are means ± sd (n > 30). The P value is shown on top of the bar graph (t test). Bars = 50 μm.
(G) to (I) Endogenous PLT1 abundance (G), CLSM images (H), and CFP intensities (I) of the PLT1pro:CFP reporter in the wild type Col-0, two UBQ10pro:GFP-PLT1 lines (OE1 and OE2), 35Spro:JANUS-RNAi (RNAi-1), and 35Spro:JANUS-RNAi UBQ10pro:GFP-PLT1 (OE1 RNAi-1). The same PLT1pro:CFP transgene was used in all genetic backgrounds. Images in the top row are merges of the CFP and RFP channels (PI; magenta); images in the bottom row are the CFP channel in pseudocolors, for which intensities cover the full range of measured values within each data set (maximum to minimum). Values in (G) are means ± se (n = 3). RNAs were extracted from seedlings at 5 DAG. Values in (I) are means ± sd (n > 20). Fluorescence of the stem cell niche was measured. Different letters indicate significantly different groups (Tukey’s multiple comparisons test, P < 0.05). Bars = 50 μm.
Because JANUS interacted with PLT1 and Pol II through nonoverlapping domains, we suspected that JANUS might be critical for the autotranscriptional regulation of PLT1 by recruiting Pol II. To test whether JANUS was required for the autotranscriptional regulation of PLT1, we examined CFP fluorescence from the PLT1pro:CFP reporter introduced in the wild type as well as in PLT1-OE, JANUS-RNAi, and PLT-OE JANUS-RNAi plants. Indeed, the enhanced transcription of endogenous PLT1 by ectopic PLT1 was significantly reduced by the downregulation of JANUS, based on CFP intensity (Figures 7H and 7I) and RT-qPCR (Figure 7G). Notably, overexpression of PLT1 did activate the expression of endogenous PLT1 in JANUS-RNAi plants, albeit to a lesser extent than in the wild type (Figures 7H and 7I). If the autoregulation of PLT1 depended solely on JANUS, the overexpression of PLT1 would be expected to have no or very limited effects on the expression of endogenous PLT1 in the absence of JANUS. However, there is still residual JANUS present in the JANUS-RNAi plants, which may contribute to this upregulation. Alternatively, the autoregulation of PLT1 may not be solely dependent on JANUS. Regardless, we also observed JANUS-dependent PLT1 autoactivation during the formation of the embryonic root pole (Supplemental Figure 7), supporting the idea that JANUS interacts with PLT1 to participate in the autotranscriptional regulation of PLT1.
GIF1 Competitively Binds JANUS Away from Pol II to Regulate PLT1 Transcription
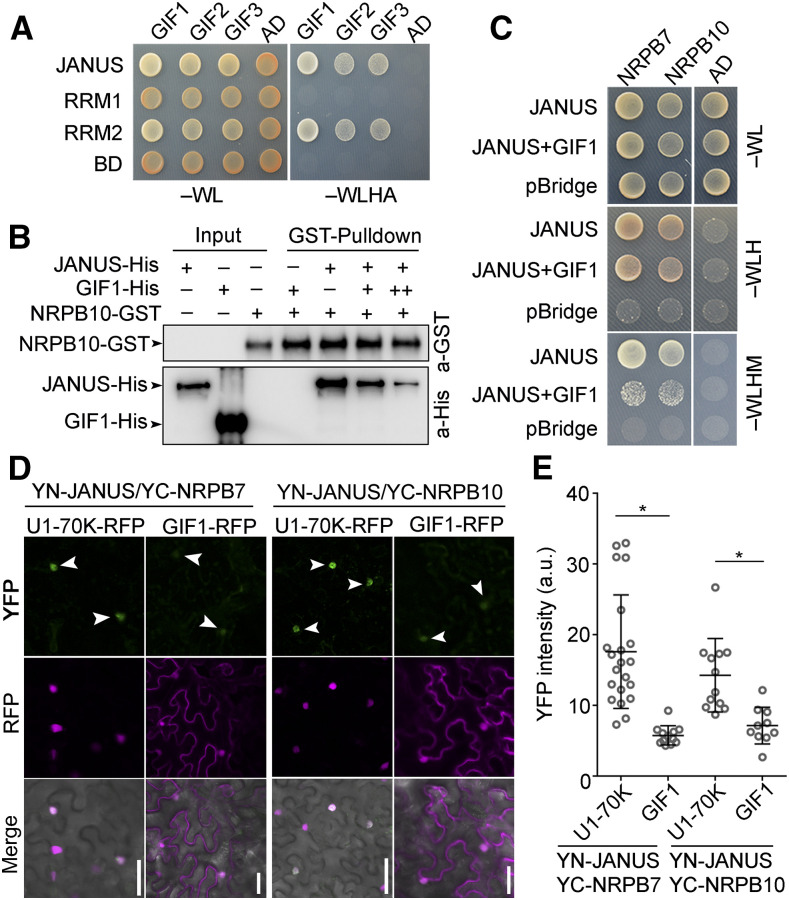
It was unexpected that although both GIF1 and JANUS are regulated by IMB4 for their nuclear accumulation, they play antagonistic roles in root meristem development. In fact, GIF1 appeared in the Arabidopsis Protein Interactome Database as another JANUS interactor (Li et al., 2011). First, we validated the interaction between GIF1 and JANUS by Y2H (Figure 8A) and by BiFC (Supplemental Figure 8) assays. Notably, GIF1 interacted with the N-terminal RRM2 of JANUS (Figure 8A), the same domain responsible for its interaction with Pol II (Xiong et al., 2019a). We thus considered the possibility that GIF1 may interact with the RRM2 domain of JANUS to competitively prevent the JANUS-Pol II interaction, which would explain the antagonistic roles of GIF1 and JANUS during root meristem development. To test this hypothesis, we performed in vitro pull-down (Figure 8B), yeast three-hybrid (Y3H; Figure 8C), and BiFC (Figures 8D and 8E) assays. By all experimental approaches, we determined that GIF1 inhibits the interaction between JANUS and Pol II (Figures 8B to 8E).
Figure 8.
GIF1 Competitively Binds JANUS Away from Pol II.
(A) Y2H assay showing that the RRM2 domain of JANUS interacts with GIFs. Positive interactions were determined by growth on synthetic minimal medium lacking Trp, Leu, His, and Ade (–WLHA). Results are representative of three biological replicates.
(B) In vitro pull-down showing that GIF1 interacts with JANUS to inhibit the interaction between JANUS and the Pol II component NRPB10. Experiments were performed three times with similar results.
(C) Representative Y3H assay showing that GIF1 inhibits the interaction between JANUS and its interacting Pol II components NRPB7 and NRPB10.
(D) and (E) Representative CLSM images (D) and YFP intensities (E) of BiFC assays showing that the positive interaction between YN-JANUS and YC-NRPB7 or YC-NRPB10 is reduced by coexpressing GIF1-RFP but not U1-70K-RFP (magenta), a nuclear marker protein. Arrowheads point at the YFP-positive nuclei. a.u. represents arbitrary units. Values in (E) are means ± sd (n > 11). Asterisks indicate significant differences (t test, P < 0.05). Bars in (D) = 50 μm.
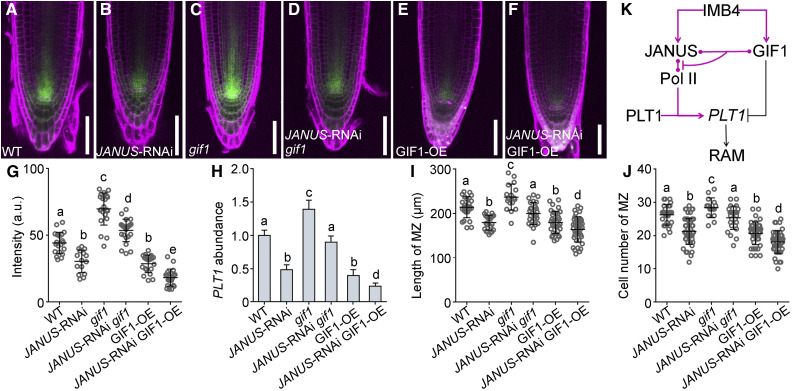
To provide genetic evidence supporting the antagonistic roles of GIF1 and JANUS in the root meristem, we introduced the GIF1 loss-of-function mutation gif1 or the gain-of-function UBQ10pro:GFP-GIF1 (Liu et al., 2019) into JANUS-RNAi plants and examined root meristem development of the resulting genetic combinations. Downregulating JANUS in the gif1 mutant caused a reduction of the root meristem, as expected based on their opposite phenotypes, rendering it comparable to that of the wild type (Figures 9I and 9J; Supplemental Figure 9). Consistent with the measured root meristem length, PLT1 abundance returned to wild-type levels in JANUS-RNAi gif1 plants (Figure 9H). By contrast, increasing GIF1 expression in JANUS-RNAi plants caused a further reduction in root meristem length (Figures 9I and 9J; Supplemental Figure 9) and an additional reduction in PLT1 abundance (Figure 9H). To determine whether the antagonistic roles of GIF1 and JANUS in root meristem development were mediated by PLT1 transcription, we examined PLT1 transcriptional activity by examining the activity of the PLT1pro:CFP reporter in the wild type (Figure 9A) and in JANUS-RNAi (Figure 9B), gif1 (Figure 9C), JANUS-RNAi gif1 (Figure 9D), GIF1-OE (Figure 9E), and JANUS-RNAi GIF1-OE (Figure 9F) plants. A careful examination of the CFP signal (Figure 9G) and RT-qPCR (Figure 9H) confirmed the antagonistic roles of GIF1 and JANUS on PLT1 transcription. These results demonstrate that JANUS and GIF1 play antagonistic roles in root meristem development, likely through PLT1 transcription.
Figure 9.
GIF1 and JANUS Antagonistically Mediate the Transcription of PLT1 and Root Meristem Development.
(A) to (F) Representative CLSM images of PI-stained roots expressing the PLT1pro:CFP reporter in wild-type Col-0 (A), 35Spro:JANUS‐RNAi (RNAi-1; [B]), gif1 (C), 35Spro:JANUS‐RNAi gif1 (JANUS-RNAi gif1; [D]), UBQ10pro:GFP-GIF1 (GIF1-OE; [E]), and 35Spro:JANUS‐RNAi UBQ10pro:GFP-GIF1 (JANUS-RNAi GIF1-OE; [F]). Images are merges of the CFP channel (green) and the RFP channel (magenta). Bars = 50 μm.
(G) CFP intensity at the stem cell niche. a.u. indicates arbitrary units. Values are means ± sd (n > 20).
(H) Relative transcript abundance of PLT1. Results are means ± se (n = 3). RNAs were extracted from seedlings at 5 DAG.
(I) and (J) Length (I) and cell number (J) of the root meristem zone (MZ). Values are means ± sd (n > 16).
Different letters in (G) to (J) indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, P < 0.05).
(K) A working model showing that two cargos regulated by IMB4 perform antagonistic roles in PLT1 expression and thus root meristem development.
In summary, we showed here that JANUS and GIF1 antagonistically regulate PLT1 transcription (Figure 9K). JANUS positively mediated the autotranscriptional regulation of PLT1 by binding to both Pol II and PLT1, a role similar to that of subunits of the mediator complex (Dolan and Chapple, 2017). Because JANUS is essential for the autotranscriptional activation of PLT1, JANUS-dependent PLT1 transcription is critical to amplify differential PLT1 levels established by mitotic dilution and cell-to-cell movement (Mahönen et al., 2014). GIF1, a negative transcriptional regulator of PLTs (Ercoli et al., 2018), competitively interacted with JANUS to inhibit its Pol II recruitment. Transcription of PLT1 was thus spatiotemporally regulated depending on the GIF1-JANUS interaction and JANUS-dependent autotranscriptional regulation (Figure 9K). The antagonistic PLT regulators GIF1 and JANUS both relied on the same importin for their nuclear accumulation (Figure 9K). That two or more cargos in the same pathway are regulated by the same importin may have logistical benefits adopted during evolution.
METHODS
Plant Materials and Growth Conditions
We used Arabidopsis (Arabidopsis thaliana) accession Col‐0 as the wild type for all experiments. T‐DNA insertion mutants in JANUS (SALK_008993; janus; Xiong et al., 2019a), NRPB10 (SALK_114301; nrpb10; Xiong et al., 2019a), IMB4 (SALK_049564; imb4-1; Liu et al., 2019), GIF1 (SALK_150407; gif1; Lee et al., 2014), and plt1-4 plt2-2 (Aida et al., 2004) have been described previously. Marker lines carrying PLT1pro:CFP (Chen et al., 2011), PLT2pro:CFP (Chen et al., 2011), ProWOX5:GFP (Blilou et al., 2005), SCRpro:H2B-YFP (Heidstra et al., 2004), and CycB1;1pro:GUS (Chen et al., 2011) have been described. We crossed these marker lines into janus/+, 35Spro:JANUS‐RNAi, imb4-1, gif1, plt1-4, plt1-4 plt2-2, nrpb10, UBQ10pro:GFP-GIF1, and 35Spro:JANUS‐RNAi UBQ10pro:GFP-GIF1. Transgenic plants including IMB4g-GFP, UBQ10pro:GFP-GIF1, 35Spro:JANUS‐RNAi, JANUSg-GFP, and NRPB10g-GFP were previously described by Liu et al. (2019) and Xiong et al. (2019a). Seedlings and plants were grown in a growth chamber at 20 ± 2°C under long‐day conditions (16 h of light/8 h of dark). For growth on plates, we surface-sterilized seeds with 2.6% (v/v) sodium hypochlorite for 10 min, rinsed with sterile water four times, and sowed seeds on half-strength Murashige and Skoog medium. Plates were incubated at 4°C for 2 d and then transferred to a growth chamber at 20 ± 2°C in long-day conditions with 120 to 140 µmol of light (Philips F96T8/TL841/HO/PLUS 86-W bulbs).
DNA Manipulation
We generated all constructs using Gateway technology (Invitrogen) unless otherwise specified. Entry clones were generated in the pENTR/D/TOPO vector (Invitrogen). We used the destination vectors pJG4-5 and pLacZi2u for Y1H (Lin et al., 2007), pDEST‐GBKT7 and pDEST‐GADT7 (Invitrogen) for Y2H, pET30a and pGEX4T-1 for pull-down assays, pBridge (Clontech) for Y3H, and pSITE::cEYFP‐C1 and pSITE::nEYFP‐C1 (Martin et al., 2009) for BiFC. Constructs used in Y2H assays, including IMB4-BD, JANUS-AD, JANUS-BD, AtBud13-AD, GIF1-AD, GIF2-AD, GIF3-AD, RRM1-BD, RRM2-BD, NRPB7-AD, NRPB10-AD, and NRPB11-AD, were previously described by Liu et al. (2019) and Xiong et al. (2019a, 2019b). Constructs used in BIFC assays, including YN-JANUS, YN-AtBud13, YC-IMB4, YC-GIF1, YC-GIF2, YC-GIF3, YC-NRPB7, and YC-NRPB10, were were previously described by Liu et al. (2019) and Xiong et al. (2019a, 2019b). For YC-KPNB1 or PLT1 overexpression, we amplified the full-length coding sequences of KPNB1 and PLT1 by PCR with the primer pairs ZY7164/ZY7165 and ZY7197/ZY7198, respectively. The entry vectors were used in LR reactions with the destination vector pSITE::cEYFP-C1 (Martin et al., 2009), UBQ10pro:GFP-GW, or 35Spro:RFP-GW (Wang et al., 2017) to generate YC-KPNB1, UBQ10pro:GFP‐PLT1, or 35Spro:RFP‐PLT1, respectively.
We performed all PCR amplifications with Phusion hot‐start high‐fidelity DNA polymerase (Thermo Fisher Scientific) with the annealing temperature and extension times recommended by the manufacturer. All entry vectors were sequenced. All primers are listed in the Supplemental Table.
PCR, RNA Extraction, RT‐PCR, and qPCR
We performed genotyping PCR to verify the identity of janus, imb4-1, gif1, and nrpb10 mutants using the following primers: ZY7860/ZY7861 for JANUS and ZY1/ZY7861 for janus; ZY4342/ZY4343 for IMB4 and ZY1/ZY4342 for imb4-1; ZY6811/ZY6812 for GIF1 and ZY1/ZY6812 for gif1; and ZY7892/ZY7893 for NRPB10 and ZY1/ZY7893 for nrpb10.
For qPCR analyses, we isolated total RNAs from seedlings 5 DAG using a Qiagen RNeasy plant mini kit according to the manufacturer’s instructions with on-column DNase I digestion (Invitrogen). We initiated first-strand cDNA synthesis with oligo(dT) oligonucleotides and SuperScript III reverse transcriptase. We performed RT‐qPCR on a Bio‐Rad CFX96 real‐time system using SYBR Green real‐time PCR master mix (Toyobo) as described by Zhou et al. (2013). GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE and ACTIN2 were used as reference genes for qPCR. Primers used are ZY9590/ZY9591 for PLT1, ZY9592/ZY9593 for PLT2, ZY9594/ZY9595 for SCR, ZY11346/ZY11347 for the endogenous PLT1, WOX5-F/WOX5-R for WOX5, and GFP-F/ZY7642 for ectopic JANUS. All experiments were repeated in three biological replicates with similar results, and each biological replicate consisted of three technical replicates. All primers are listed in the Supplemental Table.
Y1H Assay
We inserted the P1, P1-A, P1-B, and P2 sequences of the PLT1 promoter into the pLacZi2u vector after double digestion with KpnI and SalI. We inserted the full‐length PLT1 coding sequence into the pJG4-5 vector (Lin et al., 2007) after double digestion with XhoI and EcoRI. We cotransformed the two plasmids into the yeast (Saccharomyces cerevisiae) strain EGY48. We plated cells onto selective medium (without Ura and Trp) to select transformants. We then grew positive colonies on selective medium without Ura and Trp, supplemented with 20% galactose, 20% raffinose, 50 mL of buffer salts (1.95 g of Na2HPO4 and 1.855 g of NaH2PO4·2H2O), and X‐Gal (5-Bromo-4-chloro-3-indolyl β-D-galactoside).
Y2H, Y3H, and BiFC Assays
We performed Y2H assays as described by Xiong et al. (2019a). We cloned the full-length coding sequence for NRPB7 and NRPB10 into the pGADT7 vector. For pBridge-JANUS-GIF1, we cloned the coding sequence of JANUS into the MCS I site of the pBridge vector (Clontech) fused to the GAL4 DNA binding domain, and the coding sequence of GIF1 was cloned into the MCS II site of the pBridge vector to generate the “bridge” protein in the absence of Met. Y3H assays were based on the MATCHMAKER GAL4 Two-Hybrid System (Clontech). We cotransformed constructs into yeast strain Y2HGold (Clontech). The presence of the transgenes was confirmed by growth onto synthetic solid medium lacking Trp and Leu. We spotted positive yeast colonies onto plates containing synthetic solid medium lacking Trp, Leu, and His to assess the JANUS-Pol II interaction without the expression of GIF1. We used plates containing synthetic solid medium lacking Trp, Leu, His, and Met to induce GIF1 expression. Interactions were observed after 3 d of incubation at 30°C. BiFC assays were performed as described by Xiong et al. (2019a).
Protein Purification and in Vitro Pull-Downs
For the purification of recombinant proteins in in vitro pull-down assays, we introduced pET30a-JANUS, GST-GIF1, and GST-NRPB10 vectors into BL21 competent cells (DE3). We grew BL21 cells at 37°C in Luria-Bertani medium in the presence of antibiotics (100 μg/mL kanamycin) to an OD600 of 0.6 to 0.8. We induced protein accumulation by adding IPTG to a final concentration of 0.5 mM. Cells were further incubated in a shaker set to 90 rpm overnight at 16°C before pellet collection by centrifugation at 12,000 rpm for 10 min at 4°C. We resuspended cell pellets in lysis buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 2 mM PMSF) and lysed cells by sonication on ice with seven pulses of 15-s on/off at 30 mW amplitude. We collected the supernatant by centrifugation at 16,000g for 30 min at 4°C to completely remove cell debris. We purified the 6× His-JANUS, GST-GIF1, or GST-NRPB10 recombinant protein with Ni-NTA column or glutathione agarose beads (Zhang et al., 2017).
We performed in vitro pull-down assays as described by Wang et al. (2016). We incubated ∼1 μg each of GST or GST-NRPB10 protein to 25 μL of glutathione-Sepharose beads for 1 h at 4°C. We then incubated bound beads with 1 μg of 6× His-JANUS protein for another 1 h at 4°C. After three washes with 1× PBS buffer, we eluted the beads with 40 μL of 10 mM glutathione in 50 mM Tris (pH 8.0), mixed with a loading buffer, and then boiled at 100°C for 10 min. We loaded ∼15 μL of each boiled sample for SDS-PAGE and immunoblot analysis with an anti-His antibody (Beyotime; 1:1000 dilution) and an anti-GST antibody (Beyotime; 1:1000 dilution). For secondary antibodies, we used a goat anti-mouse IgG horseradish peroxidase (Beyotime; 1:1000 dilution) and a goat anti-rabbit IgG horseradish peroxidase (Beyotime; 1:1000 dilution).
For pull-down assays to test whether a high concentration of GIF1 affects the JANUS-NRPB10 interaction, we added 1 µg of purified GST-NRPB10 and 6× His-JANUS to each sample. We added purified GST-GIF1 according to the concentration gradient. We then used the glutathione-Sepharose beads to pull down proteins. The sequential procedures and buffers were the same as those described above.
Fluorescence Imaging and Quantification
We captured all fluorescence images on a Zeiss LSM880 laser scanning microscope with a 20 or 40/1.3 oil objective. Fluorescence of GFP, YFP, mCherry, and PI staining was captured using the following excitation/emission settings: 488 nm/505 to 550 nm for GFP, 458 nm/463 to 500 nm for CFP, 514 nm/530 to 590 nm for YFP, and 561 nm/600 to 650 nm for mCherry and PI staining. Image processing was performed with the LSM image-processing software (Zeiss). We imaged embryos or roots of 5-DAG seedlings from the SCRpro:H2B-YFP, PLT1pro:CFP, PLT2pro:CFP, or JANUSg-GFP plants by CLSM. We defined regions of interest (ROIs) as the nuclei for SCRpro:H2B-YFP and JANUSg-GFP in embryogenesis or upon ivermectin treatment or in BiFC assays as in Figure 8. We defined ROIs as the entire stem cell niche for PLT1pro:CFP and PLT2pro:CFP. The ROI was defined as the entire embryonic root pole for PLT1pro:CFP during late embryogenesis. We quantified fluorescence intensity using 40 to 60 cells from 15 to 20 roots for each genetic background or each treatment with ImageJ. We determined statistical significance by Student’s t test or one-way ANOVA (Tukey’s multiple comparisons test).
Cell Fractionation and Immunoblot Analysis
We performed cell fractionation as described by Wierzbicki et al. (2008) with slight modifications. Specifically, we ground 2 g of seedlings into powder in liquid nitrogen, suspended the resulting powder in 20 mL of Honda buffer (20 mM HEPES-KOH, pH 7.4, 0.44 M Suc, 1.25% [v/v] Ficoll, 2.5% [w/v] Dextran T40, 10 mM MgCl2, 0.5% [v/v] Triton X-100, 5 mM DTT, 1 mM PMSF, and 1% [w/v] plant protease inhibitors) before filtering through two layers of Miracloth and centrifugation at 5000g for 20 min. We collected the supernatant as the cytosolic fraction. We washed the pellets five times, each time with 1 mL of Honda buffer. We obtained total nuclear proteins by adding 3 volumes of 1% (w/v) SDS directly to the nucleus pellet, followed by boiling at 95°C for 10 min. We mixed equal volumes of each fraction with loading buffer, boiled, gel-separated, and subjected them to immunoblot analysis. Immunolabeling of GFP-JANUS was performed using an anti-GFP antibody (Beyotime; 1:1000 dilution). Anti-histone H3 (Beyotime; 1:1000 dilution) and anti-cFBPase (Agrisera; 1:5000 dilution) were used as the nuclear and cytoplasmic markers, respectively. Quantification of total proteins was performed using Gel Image System 4.1.2 (Bio-Tanon).
ChIP
We performed ChIP assays as described by Saleh et al. (2008). We harvested 2 g of wild‐type, imb4-1, and 35Spro:JANUS‐RNAi seedlings at 12 DAG in cross‐linking buffer (0.4 M sucrose, 10 mM Tris‐HCl, pH 8.0, 1 mM PMSF, 1 mM EDTA, and 1% formaldehyde) for 10 min under vacuum infiltration and then halted in 2 M Gly. After the addition of 5 μg of anti-Pol II antibodies (Abcam) to the chromatin and incubation at 4°C overnight, we added agarose beads conjugated with protein A and protein G and incubated at 4°C for 2.5 h. After reverse cross‐linking, we eluted DNA and dissolved it in 20 μL of water. We diluted the immunoprecipitated DNA with sterile water before quantification by real‐time PCR. Real‐time PCR data were normalized to ACTIN. We express the enrichment of Pol II at the PLT1 genomic locus as the ratio of (Pol II PLT1/input PLT1) to (Pol II ACTIN/input ACTIN). For PLT1 autoactivation, we performed ChIP assays using anti-GFP antibodies on 5-d-old wild-type or 35Spro:GFP‐PLT1 seedlings. The enrichment of PLT1 at its genomic locus is given as the ratio of (ChIP PLT1/input PLT1) to (ChIP ACTIN/input ACTIN). Primers used are ZY9923/ZY9924 for PLT1 fragment a, ZY9925/ZY9926 for PLT1 fragment b, and ZY9590/ZY9591 for PLT1 fragment c. Primers are listed in the Supplemental Table.
Ivermectin Treatment
We incubated JANUSg-GFP seedlings at 5 DAG in liquid half-strength Murashige and Skoog medium supplemented with DMSO or 5 to 15 μM ivermectin (Sigma-Aldrich PHR1380-1G; predissolved in DMSO) for 8 h before imaging.
Accession Numbers
Sequence data in this article can be found in The Arabidopsis Information Resource under the following accession numbers: At2g18510 (JANUS), At5g28640 (GIF1), At1g01160 (GIF2), At4g00850 (GIF3), At4g27640 (IMB4), At5g59180 (NRPB7), At1g11475 (NRPB10), At3g20840 (PLT1), At1g51190 (PLT2), At3g54220 (SCR), At3g11260 (WOX5), At1g31870 (AtBud13), and At5g53480 (KPNB1).
Supplemental Data
Supplemental Figure 1. IMB4 mediates nuclear accumulation of GIF1 in RAM.
Supplemental Figure 2. Nuclear accumulation of JANUS in early embryogenesis is independent of IMB4.
Supplemental Figure 3. Ivermectin inhibits the nuclear accumulation of JANUS.
Supplemental Figure 4. JANUS positively regulates embryonic root meristem activity.
Supplemental Figure 5. Pol II positively regulates root meristem activity and PLT1 transcription.
Supplemental Figure 6. Self-transcriptional regulation of PLT1.
Supplemental Figure 7. Auto-transcriptional regulation of PLT1 in late embryogenesis requires JANUS.
Supplemental Figure 8. GIFs interact with JANUS.
Supplemental Figure 9. GIF1 and JANUS antagonistically mediate RAM.
Supplemental Table. Oligos used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the ABRC for mutant seeds, Ben Scheres for SCRpro:H2B-YFP, Qian Chen for the PLT1pro:CFP and PLT2pro:CFP marker lines, Xu-Gang Li for the plt1-4 and plt1-4 plt2-2 mutants, and Xian Sheng Zhang for the CycB1;1pro:GUS and WOX5pro:GFP lines. This work was supported by the National Natural Science Foundation of China (grants 31771558 and 31970332 to S.L.).
AUTHOR CONTRIBUTIONS
F.X. conceptualized the study. F.X., S.L., and Y.Z. created the methodology. F.X., B.-K.Z., H.-H.L., G.W., J.-H.W., and Y.-N.W. performed the research. F.X. wrote the original draft of the article, and S.L. and Y.Z. were responsible for the editing and review. S.L. and Y.Z. supervised the study. Funding was acquired by S.L.
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B.(2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B.(2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A., Díaz-Triviño S., Wachsman G., Du Y., Arteága-Vázquez M., Zhang H., Benjamins R., Blilou I., Neef A.B., Chandler V., Scheres B.(2013). A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 11: e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B.(1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Dolan W.L., Chapple C.(2017). Conservation and divergence of mediator structure and function: Insights from plants. Plant Cell Physiol. 58: 4–21. [DOI] [PubMed] [Google Scholar]
- Durgaprasad K., Roy M.V., Venugopal M A., Kareem A., Raj K., Willemsen V., Mähönen A.P., Scheres B., Prasad K.(2019). Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep. 29: 453–463.e3. [DOI] [PubMed] [Google Scholar]
- Ercoli M.F., Ferela A., Debernardi J.M., Perrone A.P., Rodriguez R.E., Palatnik J.F.(2018). GIF transcriptional coregulators control root meristem homeostasis. Plant Cell 30: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B.(2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Sabatini S.(2014). Plant and animal stem cells: Similar yet different. Nat. Rev. Mol. Cell Biol. 15: 301–312. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B.(2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18: 1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Kumpf R.P., De Veylder L.(2014). A quiescent path to plant longevity. Trends Cell Biol. 24: 443–448. [DOI] [PubMed] [Google Scholar]
- Laux T.(2003). The stem cell concept in plants: A matter of debate. Cell 113: 281–283. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Wynn A.N., Franks R.G., Hwang Y.S., Lim J., Kim J.H.(2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386: 12–24. [DOI] [PubMed] [Google Scholar]
- Li P., Zang W., Li Y., Xu F., Wang J., Shi T.(2011). AtPID: The overall hierarchical functional protein interaction network interface and analytic platform for Arabidopsis. Nucleic Acids Res. 39: D1130–D1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H.(2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.H., Xiong F., Duan C.Y., Wu Y.N., Zhang Y., Li S.(2019). Importin β4 mediates nuclear import of GRF-interacting factors to control ovule development in Arabidopsis. Plant Physiol. 179: 1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Wang Z., Ji H., Fang H., Wang S., Tian L., Li X.(2013). An Arabidopsis homolog of importin β1 is required for ABA response and drought tolerance. Plant J. 75: 377–389. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Ten Tusscher K., Siligato R., Smetana O., Díaz-Triviño S., Salojärvi J., Wachsman G., Prasad K., Heidstra R., Scheres B.(2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M.(2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Winter C.M., Benfey P.N.(2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Ercoli M.F., Debernardi J.M., Breakfield N.W., Mecchia M.A., Sabatini M., Cools T., De Veylder L., Benfey P.N., Palatnik J.F.(2015). MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27: 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B.(2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z.(2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Santuari L., et al. (2016). The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima M., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T.(2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Scheres B.(2007). Stem-cell niches: Nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8: 345–354. [DOI] [PubMed] [Google Scholar]
- Sena G., Wang X., Liu H.Y., Hofhuis H., Birnbaum K.D.(2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Heidstra R., Blilou I., Scheres B.(2018). Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 32: 1085–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hage W., Weisbeek P., Scheres B.(1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- Wang J.G., Feng C., Liu H.H., Feng Q.N., Li S., Zhang Y.(2017). AP1G mediates vacuolar acidification during synergid-controlled pollen tube reception. Proc. Natl. Acad. Sci. USA 114: E4877–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liang L., Xue Y., Jia P.F., Chen W., Zhang M.X., Wang Y.C., Li H.J., Yang W.C.(2016). A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244. [DOI] [PubMed] [Google Scholar]
- Watt F.M., Hogan B.L.(2000). Out of Eden: Stem cells and their niches. Science 287: 1427–1430. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jürgens G.(2002). Stem cells that make stems. Nature 415: 751–754. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S.(2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N.(2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603.10631180 [Google Scholar]
- Xiong F., Li S.(2020). SF3b4: A versatile player in eukaryotic cells. Front. Cell Dev. Biol. 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Liu H.H., Duan C.Y., Zhang B.K., Wei G., Zhang Y., Li S.(2019a). Arabidopsis JANUS regulates embryonic pattern formation through Pol II-mediated transcription of WOX2 and PIN7. iScience 19: 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Ren J.J., Yu Q., Wang Y.Y., Kong L.J., Otegui M.S., Wang X.L.(2019b). AtBUD13 affects pre-mRNA splicing and is essential for embryo development in Arabidopsis. Plant J. 98: 714–726. [DOI] [PubMed] [Google Scholar]
- Xu J., Hofhuis H., Heidstra R., Sauer M., Friml J., Scheres B.(2006). A molecular framework for plant regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- Xu M., Gu X., Liang N., Bian X., Wang H., Qin Y., Pi L., Wu S.(2020). Intersected functional zone of transcriptional regulators patterns stemness within stem cell niche of root apical meristem. J. Integr. Plant Biol. 62: 897–911. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tucker E., Hermann M., Laux T.(2017). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40: 264–277.e4. [DOI] [PubMed] [Google Scholar]
- Zhou L.Z., Li S., Feng Q.N., Zhang Y.L., Zhao X., Zeng Y.L., Wang H., Jiang L., Zhang Y.(2013). PROTEIN S-ACYL TRANSFERASE10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]