Hydrogen sulfide negatively regulates the progress of autophagy through persulfide modification of the Arabidopsis AtATG4a protease.
Abstract
Hydrogen sulfide is a signaling molecule that regulates essential processes in plants, such as autophagy. In Arabidopsis (Arabidopsis thaliana), hydrogen sulfide negatively regulates autophagy independently of reactive oxygen species via an unknown mechanism. Comparative and quantitative proteomic analysis was used to detect abscisic acid-triggered persulfidation that reveals a main role in the control of autophagy mediated by the autophagy-related (ATG) Cys protease AtATG4a. This protease undergoes specific persulfidation of Cys170 that is a part of the characteristic catalytic Cys-His-Asp triad of Cys proteases. Regulation of the ATG4 activity by persulfidation was tested in a heterologous assay using the Chlamydomonas reinhardtii CrATG8 protein as a substrate. Sulfide significantly and reversibly inactivates AtATG4a. The biological significance of the reversible inhibition of the ATG4 by sulfide is supported by the results obtained in Arabidopsis leaves under basal and autophagy-activating conditions. A significant increase in the overall ATG4 proteolytic activity in Arabidopsis was detected under nitrogen starvation and osmotic stress and can be inhibited by sulfide. Therefore, the data strongly suggest that the negative regulation of autophagy by sulfide is mediated by specific persulfidation of the ATG4 protease.
INTRODUCTION
Hydrogen sulfide (H2S) is currently recognized as a signaling molecule. In plant systems, H2S is considered to be as important as NO and H2O2 and regulates essential processes of plant performance (García-Mata and Lamattina, 2013; Calderwood and Kopriva, 2014; Jin and Pei, 2015; Gotor et al., 2017). Sulfide mediates tolerance against a range of plant stresses from heavy metal toxicity to salinity and drought to enhance plant viability (Gotor et al., 2019). Sulfide regulates critical processes, including autophagy (Álvarez et al., 2012; Gotor et al., 2013, 2015; Laureano-Marín et al., 2016a, 2016b) and the abscisic acid (ABA)-dependent stomatal movement (Jin et al., 2013; Scuffi et al., 2014, 2018; Papanatsiou et al., 2015).

Despite increasing evidence of the biological function of H2S, there is a considerable lack of information on the mechanism of action of H2S in particular physiological processes. The mechanism of action must be related to chemical reactivity of H2S with other molecules. H2S was suggested to coordinate the metal centers of metalloproteins (Vitvitsky et al., 2018) or act as a reductant of reactive oxygen species (Zaffagnini et al., 2019). A third mechanism of action of H2S is based on its ability to modify the thiol group (−SH) of the Cys residues in target proteins to form a persulfide group (−SSH) resulting in functional changes in the protein structure, activity, or subcellular localizations (Aroca et al., 2015, 2017b). This posttranslational modification is called persulfidation (known previously as S-sulfhydration) and has been initially demonstrated in the mammalian (Mustafa et al., 2009; Paul and Snyder, 2012) and plant systems (Aroca et al., 2015, 2017a) using specific labeling methods.
Our previous investigations in Arabidopsis (Arabidopsis thaliana) demonstrated that hydrogen sulfide functions as a signaling molecule in the cytosol that negatively regulates autophagy (Álvarez et al., 2012; Gotor et al., 2013; Romero et al., 2013), which is a highly conserved process involving digestion of the cell contents for recycling and maintenance of cell homeostasis. Autophagy occurs at the basal levels in eukaryotic cells and is induced by internal or external perturbations. In plants, autophagy is involved in development, immune response, and senescence and is induced by stress conditions, including nutrient limitation and other abiotic stresses (Liu and Bassham, 2012; Pérez-Pérez et al., 2012; Masclaux-Daubresse et al., 2017; Üstün et al., 2017; Marshall and Vierstra, 2018). This catabolic process is characterized by de novo synthesis of autophagosomes in the cytosol, in which cytoplasmic materials to be recycled are sequestrated, transported, and released into the plant vacuole. Various receptors have been described to assist with specific cargo recruitment and degradation via selective autophagy. The core autophagy machinery is highly conserved in all studied eukaryotes, and involved proteins are referred to as autophagy-related (ATG). These ATG proteins include the ATG8 and ATG12 ubiquitin-like conjugation systems that catalyze the covalent attachment of ATG8 to the phospholipid phosphatidylethanolamine (PE), which is an essential adduct for the formation of autophagosomes (Mizushima et al., 2011). Before this conjugation, the C-terminal extension of newly synthetized ATG8 has to be cleaved by the Cys-type protease ATG4 to expose a highly conserved Gly, which is necessary for conjugation to PE. Additionally, ATG4 functions as a deconjugating enzyme that cleaves the amide bond between ATG8 and PE allowing the recycling of free ATG8 (Nair et al., 2012; Nakatogawa et al., 2012; Yu et al., 2012).
The role of sulfide as a repressor of autophagy is independent of nutrient conditions and specific tissues because sulfide inhibits autophagy in leaves under dark-induced carbon starvation (Álvarez et al., 2012) or in roots under nitrogen deprivation (Laureano-Marín et al., 2016b), and both conditions are unrelated to sulfur metabolism. A study aiming to decipher the mechanism of action of H2S showed that it is independent of the formation of reactive oxygen species, such as hydrogen peroxide or superoxide anions, and therefore H2S does not serve as a reducer in the regulation of autophagy (Laureano-Marín et al., 2016b). Interestingly, a comparative and quantitative proteomic analysis was performed to detect endogenous persulfidated proteins; the results indicated that at least 10% of the entire Arabidopsis proteome undergoes persulfidation under physiological conditions, suggesting a widespread distribution of this posttranslational modification (Aroca et al., 2017a). Furthermore, persulfidation of various components of the ABA signaling pathway has been recently described as a specific mechanism of action by which H2S controls the guard cell ABA signaling (Chen et al., 2020; Shen et al., 2020).
In this study, a comparative and quantitative proteomic analysis was used to detect persulfidated proteins in the leaves of Arabidopsis exogenously treated with ABA. Interestingly, comparison of the untreated and ABA-treated samples indicated that AtATG4a was the protein with the highest difference in the persulfidation level. We then sought to determine whether persulfidation is the mechanism of the regulation of autophagy by sulfide in Arabidopsis under the stress conditions and to ascertain whether persulfidation regulates the activity of ATG4. To this aim, an enzymatic assay using Chlamydomonas reinhardtii CrATG8 as ATG4 substrate was developed. In-depth analysis of ATG4 proteolytic activity was performed in vitro and in a cell-free system using total extracts from Arabidopsis under basal or autophagy-activating conditions, including nitrogen limitation or osmotic stress. The results indicate that ATG4 is a specific target of persulfidation.
RESULTS
Autophagy Induction by ABA-Triggered Persulfide Modification Is Mediated by AtATG4a Regulation by Hydrogen Sulfide in Arabidopsis
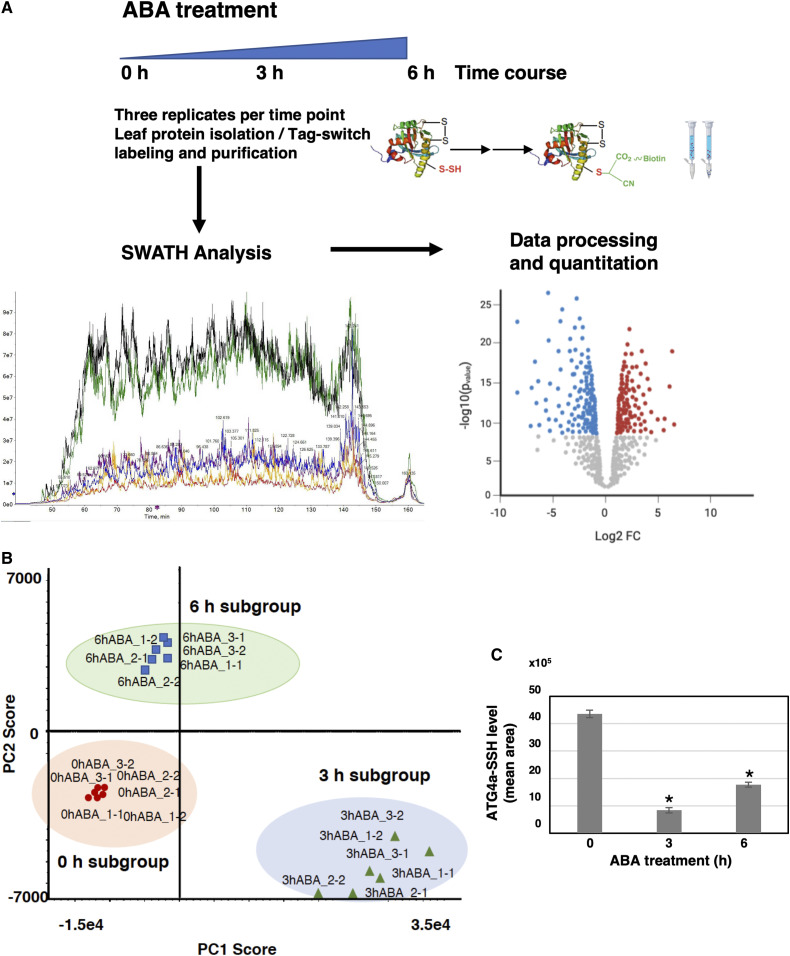
ABA can trigger changes in hydrogen sulfide level and protein persulfide modification in the guard cells to modulate stomatal closure (Shen et al., 2020). To assess whether ABA also regulates protein persulfidation levels in the mesophyll cells, a sequential window acquisition of all theoretical spectra-mass spectrometry (SWATH-MS) quantitative approach was combined with the tag-switch method to measure protein persulfidation (Aroca et al., 2017a). Protein samples from three biological replicates (independent pools) of leaf tissue treated with ABA for 0 h (control sample), 3 h, and 6 h were isolated and subjected to the tag-switch procedure (Figure 1A). The proteins eluted from the streptavidin beads were digested, and the peptide solutions analyzed in two sequential steps: a shotgun data-dependent acquisition approach to generate the spectral library and SWATH acquisition by the data-independent acquisition (DIA) method. In the first step, integration of the nine data sets resulted in identification of a total of 10,329 peptides (1% false discovery rate [FDR] and 90% confidence) and 1434 unique proteins (1% FDR) that were used as a spectral library (Supplemental Data Set 1). In the second step, to quantify protein levels using SWATH acquisition, the same six biological samples were analyzed in two technical replicates by the DIA method using the liquid chromatography (LC) gradient and LC-MS equipment employed in generation of the spectral library.
Figure 1.
Proteomic Analysis of Protein Persulfidation in Response to ABA in Mesophyll Cells.
(A) Workflow of ABA leaf treatments followed by tag-switch protein labeling with CN-biotin, protein purification, and quantitative SWATH analysis of eluted proteins.
(B) Principal component analysis representation plot of the 18 samples after SWATH analysis. Score for PC1 (70%) versus PC2 (9.9%), Pareto scaling.
(C) Level of persulfidated AtATG4a after ABA treatments. Values represent the mean peak areas of extracted ion chromatogram of identified AtATG4a peptides. *P < 0.05.
Therefore, six data sets were generated from the control and ABA-treated (for 3 and 6 h) samples to yield a total of 18 data sets used for the quantitative analysis. The fragment spectra were extracted for the 18 runs, and 33,887 ion transitions, 4871 peptides, and 1157 proteins were quantified. Principal component analysis of the protein sample subgroups revealed significant reproducible data between the replicates and differences between the ABA treatments (Figure 1B).
Comparison between the control (0 h) and the 3 h ABA treatment samples (0 h versus 3 h ABA; Supplemental Data Set 2) showed that 192 proteins were more abundant in the control samples with the fold change > 1.5 (P < 0.05), and 242 proteins were less abundant with the fold change < 0.66 (P < 0.05; Supplemental Data Set 3). Higher abundance of a protein in the control samples than that in the samples prepared after 3 h ABA treatment means that the protein is more persulfidated in the control because the tag-switch labeling recovers more protein from the streptavidin column. Lower abundance in the control means that a protein is more persulfidated in the 3 h ABA samples.
A total of 192 proteins with reduced persulfidation level after 3 h of ABA treatment were analyzed based on their assigned functions and classified into 32 functional groups using the MapMan nomenclature (Supplemental Table 1; Thimm et al., 2004; Klie and Nikoloski, 2012). The most numerous set corresponded to the general protein group (bin 29; Supplemental Table 2), which included 18.2% of the total identified proteins with 35 total elements, 29 of which are involved in protein amino acid activation (eight elements, tRNA ligases), protein synthesis (12 elements) and protein degradation (nine elements). The latter group included the Cys-type protease AtATG4a involved in autophagy, which showed the highest persulfidation change (5.20-fold change), and the S1 RNA BINDING RIBOSOMAL PROTEIN 1 (3.69-fold change) that regulates seedling growth in the presence ABA or under abiotic stress conditions (Gu et al., 2015). Additional proteins involved in the regulatory process and with a reduction in their persulfidation levels included MAPK kinase 6 (MPK6, AT2G43790), Tyr phosphatase 1 (PTP1, AT1G71860), Gly-rich protein 8 (GRP8, AT4G39260), and 9-cis-epoxycarotenoid dioxygenase 4 (NCED4, AT4G19170).
The quantitative analysis of the control and the 6-h ABA treatment samples (Supplemental Data Set 4) showed a reduction in the number of regulated proteins; only 120 proteins were more abundant in the control versus the ABA treatment samples with the fold change > 1.5 (P < 0.05), and 198 proteins were less abundant with the fold change < 0.66 (P < 0.05; Supplemental Data Set 5).
Comparison of differentially regulated proteins at 3 and 6 h of ABA treatment showed that 42% of the proteins are regulated under both conditions and had different levels of persulfidation. The Cys protease AtATG4a showed a reduction in the level of persulfidation down to only 2.45-fold after 6 h of ABA treatment compared to that under the control conditions; this level was higher than that in the 3-h ABA treatment samples (Figure 1C). Therefore, persulfidation level of AtATG4a was transiently reduced after a short ABA treatment, and this reduction was very significant compared to the untreated control.
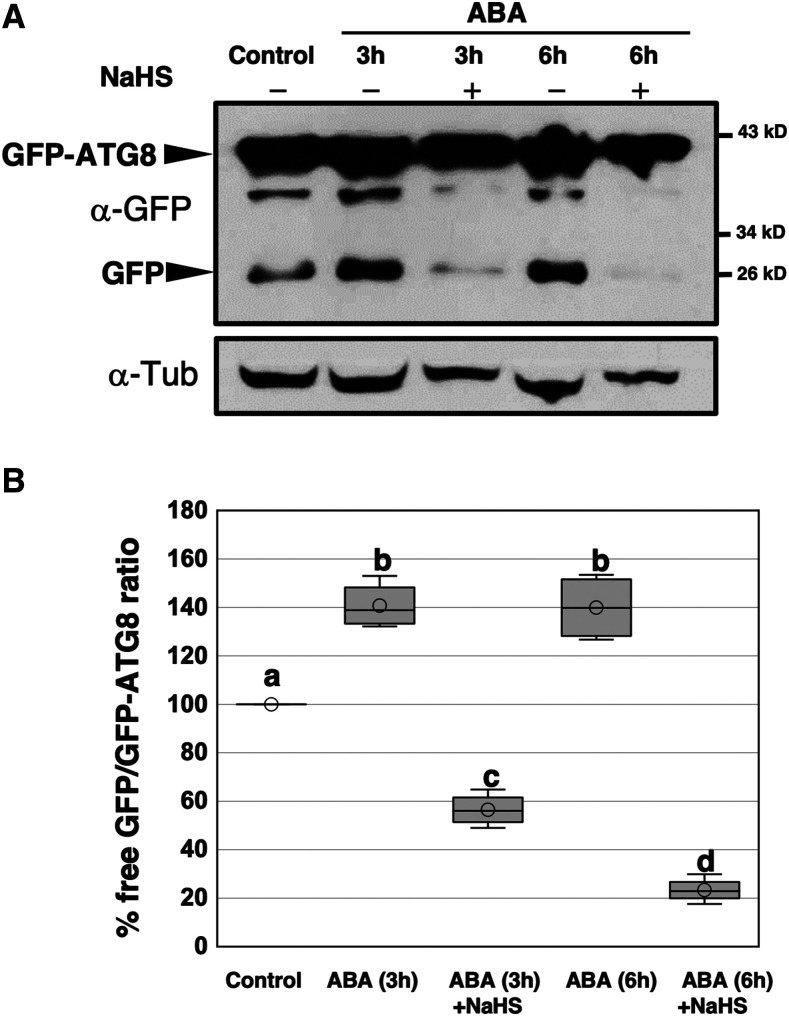
The proteomics data suggest that AtATG4a protease is differentially persulfidated depending on the treatment conditions; this difference may be related to the progress of autophagy. To test this assumption, we analyzed the regulation of autophagy by ABA treatments and the effects of sulfide under these conditions. Arabidopsis seedlings expressing GFP-ATG8e fusion protein (Xiong et al., 2007) were treated with 50 μM ABA for 3 and 6 h and subjected to additional treatment of 200 μM NaHS for 1 h. Total protein extracts were obtained, and immunoblot analysis was performed using anti-GFP antibodies to detect free GFP and the GFP-ATG8e fusion protein (Figure 2A). A clear increase in the free GFP protein level in ABA-treated seedlings was detected compared with the control and a significant reduction in the GFP accumulation after the additional sulfide treatment was observed resulting in protein levels lower than those detected in the control. Quantification of the ratio-free GFP/GFP-ATG8 under each condition showed that ABA induced the autophagic flux and that this ABA-induced autophagy was repressed by NaHS (Figure 2B). These findings and the proteomic data suggest that sulfide is a negative regulator of bulk autophagy independently of the condition used to induce this catabolic process, including at least nutrient limitation (Álvarez et al., 2012; Laureano-Marín et al., 2016b) and ABA-dependent stress (this study). Furthermore, persulfidation appears to be the mechanism of action of sulfide and the AtATG4a protease may be one of the specific targets.
Figure 2.
Effect of Sulfide on Autophagy Induced by ABA Treatment.
(A) Immunoblot analysis of GFP-ATG8e fusion protein. One-week-old seedlings expressing GFP-ATG8e fusion protein were transferred to liquid MS media and were not treated (control) or treated with 50 μM ABA for 3 and 6 h and then 200 μM NaHS for 1 h. Total protein extracts were prepared and subjected to SDS-PAGE and immunoblot analysis with anti-GFP antibodies. Anti-tubulin antibodies were used as the protein-loading control.
(B) Quantification of the free GFP/GFP-ATG8 ratio. For each condition, the levels of free GFP and GFP-ATG8e fusion protein were quantified in a less exposed blot shown in Supplemental Figure 6. The value of 100% was assigned to the free GFP/GFP-ATG8 ratio of the control sample. Data are from three independent experiments and evaluated by two-factor ANOVA. Same letters indicate no significant differences. P < 0.05.
Persulfidation of AtATG4a at Cys170
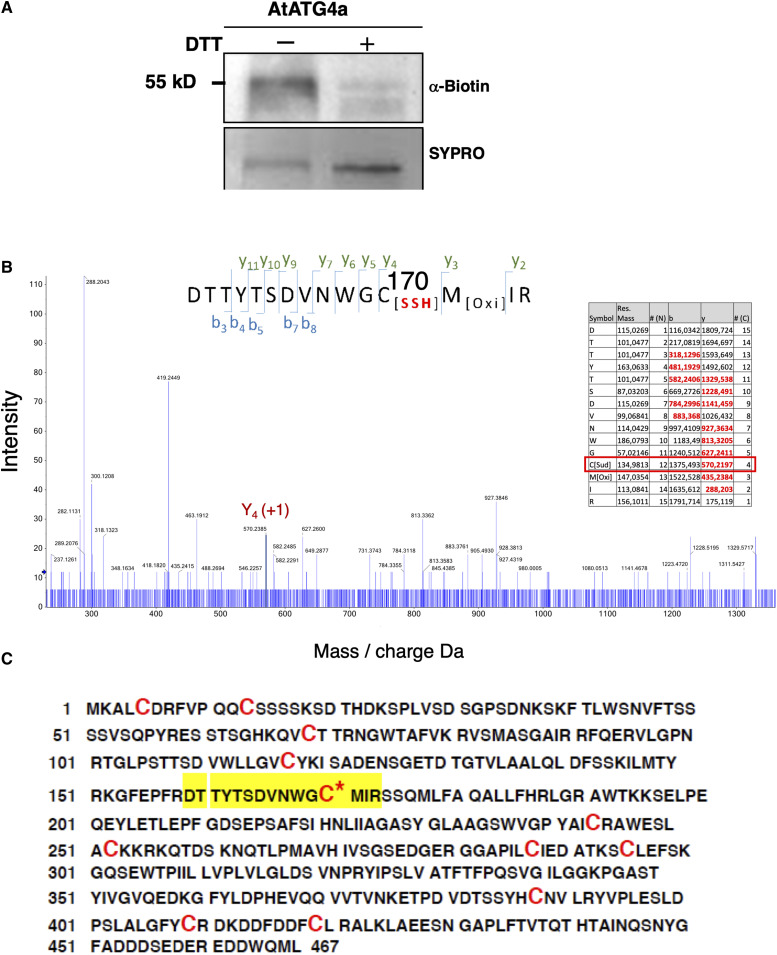
To demonstrate the persulfidation-based modification of Cys residues in AtATG4a, recombinant protein was purified, subjected to the tag-switch procedure, and analyzed by immunoblotting using anti-biotin antibodies (Aroca et al., 2017a). A biotin-labeled protein band corresponding to AtATG4a was clearly detected by the antibody. Moreover, when AtATG4a was pretreated with DTT to reduce the persulfide residues, the biotin-labeled protein bands were not detected (Figure 3A). These results clearly indicate that AtATG4a undergoes persulfidation in vitro. To identify the Cys residue that can be modified by persulfidation, recombinant AtATG4a was analyzed by LC-tandem mass spectrometry (MS/MS). The protein was digested with trypsin under nonreducing conditions in the absence of alkylating agents to avoid the reduction or modification of the persulfide residues. The digested peptides were analyzed to detect a 32 D molecular mass increase in the fragmentation spectrum. The identified peptides included DTTYTSDVNWGCMIR that showed a persulfidation modification of Cys170 (Figure 3B). AtATG4a protein was identified with a sequence coverage of 97%, and no other persulfidated peptides were detected despite the presence of another 11 Cys residues in the primary structure (Figure 3C). Cys170 is located in the highly conserved catalytic site of ATG4 proteins from various organisms (Satoo et al., 2009; Pérez-Pérez et al., 2014, 2016) suggesting that the modification by persulfidation may have an important impact on the AtATG4a proteolytic activity and biological function.
Figure 3.
Persulfidation of Arabidopsis AtATG4a.
(A) Immunoblot analysis of persulfidated AtATG4a. Purified recombinant AtATG4a was treated in the absence (−) or in the presence of 50 mM DTT (+) for 30 min at 4°C, dialyzed, and subjected to the tag-switch labeling as described in the Methods. Then, the proteins were subjected to immunoblot analysis using anti-biotin antibodies. Sypro Ruby fluorescent staining is shown as the protein loading control.
(B) Analysis of AtATG4a using mass spectrometry. LC-MS/MS analysis of a tryptic peptide of AtATG4a containing Cys170. The table inside the spectrum contains the predicted ion types for the modified peptide, and the ions detected in the spectrum are highlighted in red.
(C) AtATG4a protein sequence identified with 97% coverage. The peptide containing persulfidated Cys170 is highlighted in yellow, and all the Cys residues are red.
In Vitro Processing of CrATG8 by AtATG4a
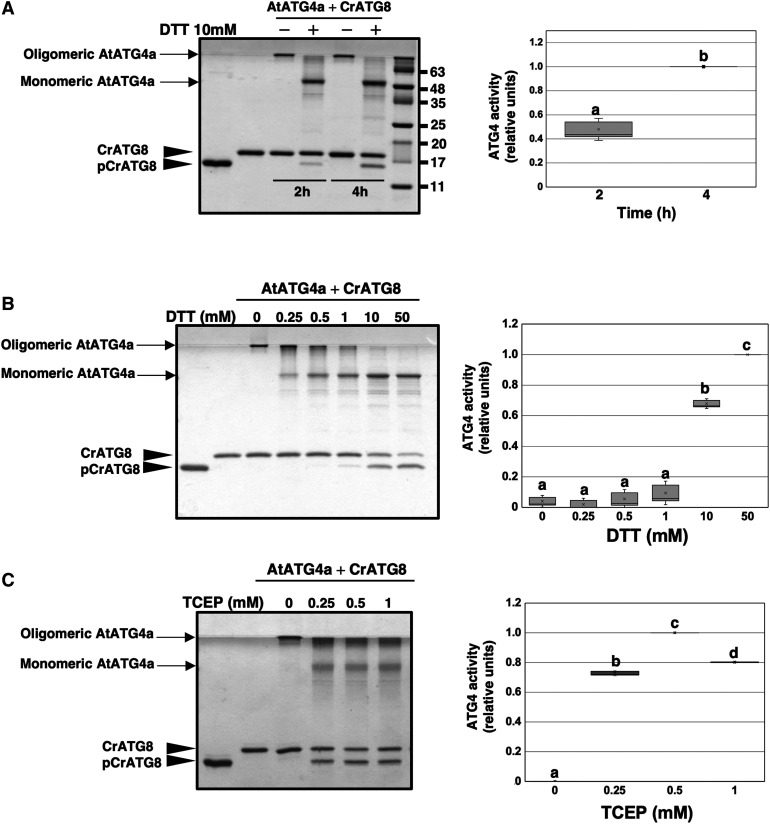
To determine whether modification of AtATG4 by persulfidation has an effect on its biological activity, an in vitro assay using CrATG8 from C. reinhardtii as a substrate was established (Pérez-Pérez et al., 2010). CrATG8 was previously used to monitor the activity of yeast (Saccharomyces cerevisiae) ATG4 (Pérez-Pérez et al., 2010, 2014). ATG4 processes ATG8 at a conserved Gly residue located at the C terminus of the protein (Kirisako et al., 2000). Arabidopsis contains nine different ATG8 isoforms, none of which has more than five amino acid residues after the conserved Gly (Doelling et al., 2002). In contrast, CrATG8 harbors a 14-amino acid extension after the Gly, and CrATG8 processing can be easily monitored by Coomassie Blue-stained SDS-PAGE due to differences in mobility between the unprocessed and processed CrATG8 forms (Supplemental Figure 1). When purified AtATG4a was incubated with CrATG8 in the presence of DTT, the processed protein with the same mobility as a truncated form lacking the last 14 amino acids (CrATG8G120, referred to as pCrATG8) was detected (Figure 4A). Therefore, AtATG4a was active and was able to process CrATG8 at its C-terminal Gly validating the ATG4 processing activity assay. The results indicated that AtATG4a activity was increased in a time-dependent manner and required a reducing agent to adopt the monomeric form required for the activity (Figure 4A), as described previously in other systems (Pérez-Pérez et al., 2010).
Figure 4.
Arabidopsis ATG4a Cleaves Chlamydomonas ATG8.
ATG4 activity of the recombinant AtATG4a was assayed by monitoring the cleavage of CrATG8 from the unprocessed (CrATG8) to processed (pCrATG8) forms (indicated by arrowheads) using SDS-PAGE followed by Coomassie Blue staining and quantification of protein band intensities. The ATG4 activity (relative units) was determined as the ratio of the band intensity of the processed CrATG8 to the sum of the intensities of the unprocessed and processed CrATG8. Activity value of 1 corresponds to the maximum. Processed pCrATG8 (lane 1) and unprocessed CrATG8 (lane 2) were loaded as controls. Representative images are shown. Data are from three independent experiments and evaluated by two-factor ANOVA. Same letters indicate no significant differences. P < 0.05.
(A) Effect of incubation time. AtATG4a was incubated with CrATG8 in the absence or in the presence of 10 mM DTT for the indicated times.
(B) Effect of DTT concentration. AtATG4a was incubated with CrATG8 in the absence or in the presence of increasing concentrations of DTT for 4 h.
(C) Effect of TCEP concentration. AtATG4a was incubated with CrATG8 in the absence or in the presence of increasing concentrations of TCEP for 4 h.
To analyze the effect of a reducing agent on AtATG4a activity, the in vitro assay in the presence or in the absence of different DTT or tris(2-carboxyethyl)phosphine (TCEP) concentrations was performed. In the absence of DTT and TCEP, recombinant AtATG4a was in an oligomeric form that was retained in the upper part of the acrylamide gel. Increasing concentrations of the reducing agent enhanced the monomerization of recombinant AtATG4a and consequently the processing of CrATG8 by AtATG4a (Figures 4B and 4C). Therefore, properties of Arabidopsis ATG4a were similar to those of the Chlamydomonas and yeast ATG4 (Pérez-Pérez et al., 2014, 2016). Interestingly, TCEP was more efficient than DTT in the activation of CrATG8 cleavage, showing the maximum level of ATG4 activity at 0.5 mM TCEP while two orders of magnitude higher DTT concentration was required for optimal activity (Figures 4B and 4C).
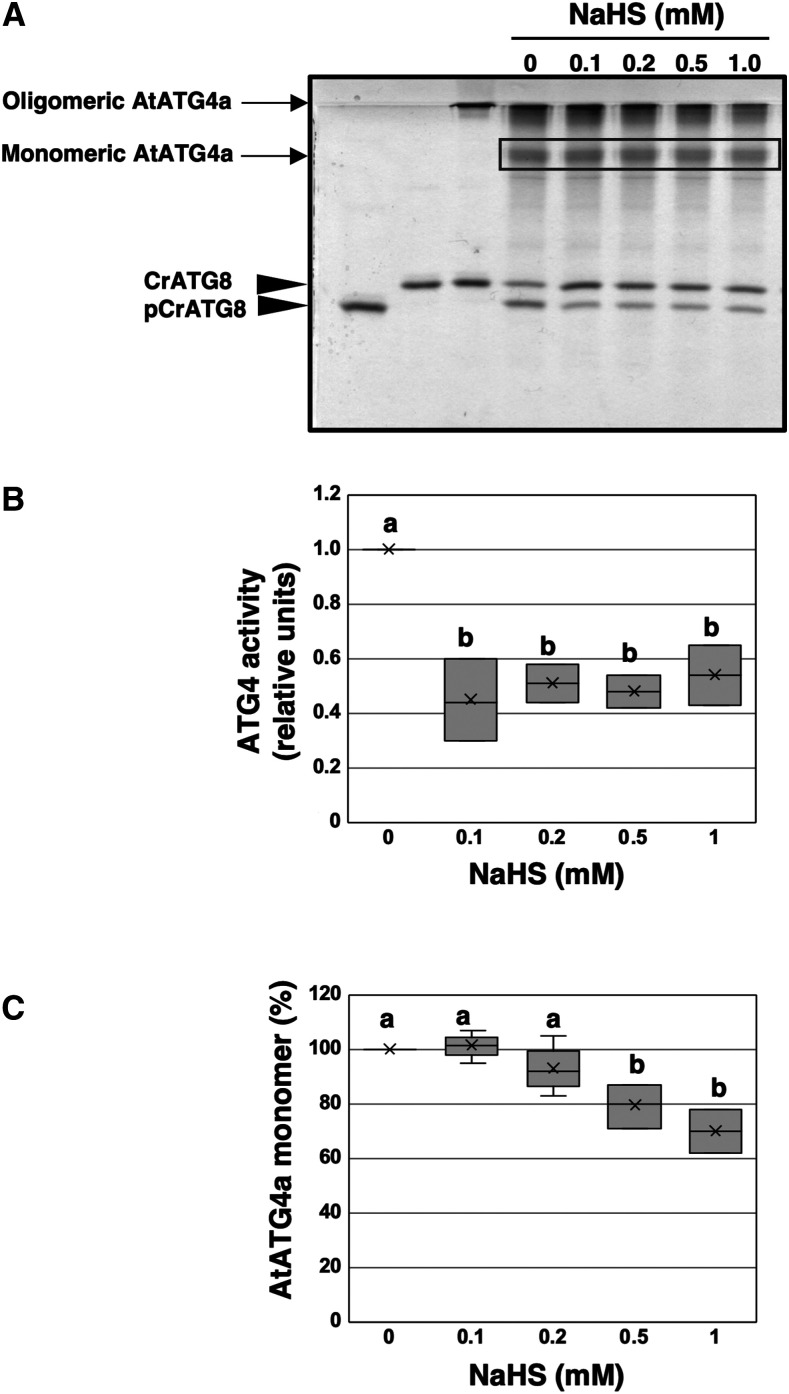
Sulfide Inhibits the Proteolytic Activity of AtATG4a
A suitable ATG4 enzyme activity assay was developed and used to study whether persulfidation plays a role in the regulation of this activity. Thus, purified recombinant AtATG4a was pretreated with TCEP to produce an active enzyme and then incubated with increasing concentrations of NaHS as a sulfide donor. The ATG4 activity-dependent CrATG8 processing was determined using the Coomassie-stained gel method (Figure 5A). An increase in the accumulation of the unprocessed CrATG8 form was detected when AtATG4a was incubated in the presence of NaHS at a concentration as low as 0.1 mM. ATG4 activity was determined as the ratio of the band intensity of the processed CrATG8 to the sum of the intensities of the unprocessed and processed CrATG8; the data indicate that 0.1 mM NaHS significantly inhibits the ATG4 activity to ∼40% of the activity in the absence of sulfide (Figure 5B). Interestingly, analysis of the aggregation state of AtATG4a in the presence of the sulfide donor demonstrated that NaHS has different effects on monomerization and activity of AtATG4a (Figure 5C). Sulfide did not promote the oligomerization of the active monomeric AtATG4a to the same extent as it inhibited ATG4 proteolytic activity under the conditions used in the assays. Concentrations of NaHS from 0.1 to 0.5 mM did not significantly increase the abundance of high molecular weight and inactive oligomers of AtATG4a (Figure 5C). These results indicate that sulfide donor inhibits AtATG4a activity but does not directly influence the aggregation state of the protein in contrast to the effect of DTT or thioredoxin on yeast and C. reinhardtii ATG4 proteins (Pérez-Pérez et al., 2014, 2016).
Figure 5.
Effect of NaHS on AtATG4a Enzyme Activity.
(A) AtATG4a was incubated with 0.5 mM TCEP for 2 h and treated in the absence or in the presence of indicated concentrations of NaHS for 1 h. Then, CrATG8 was added to the incubation mixture, and ATG4 activity was monitored after 4 h using Coomassie-stained gels as described in Methods. All procedures were performed at 25°C. Lanes 1 and 2, unprocessed and processed CrATG8, respectively. Lane 3, AtATG4a incubated with CrATG8 in the absence of TCEP and of NaHS. Lanes 4 to 8, TCEP-pretreated reduced AtATG4a incubated with CrATG8 in the presence of increasing concentrations of NaHS (from 0 to 1 mM). A representative image is shown.
(B) Quantification of ATG4 activity (relative units) determined as the ratio of the band intensity of the processed CrATG8 to the sum of the intensities of the unprocessed and processed CrATG8. A value of 1 corresponds to AtATG4a in the absence of NaHS (lane 4).
(C) Quantification of the protein band intensity corresponding to the monomeric AtATG4a form marked by a rectangle in (A). A value of 100% corresponds to AtATG4a in the absence of NaHS (lane 4).
Data are from three independent experiments and evaluated by two-factor ANOVA. Same letters indicate no significant differences. P < 0.05.
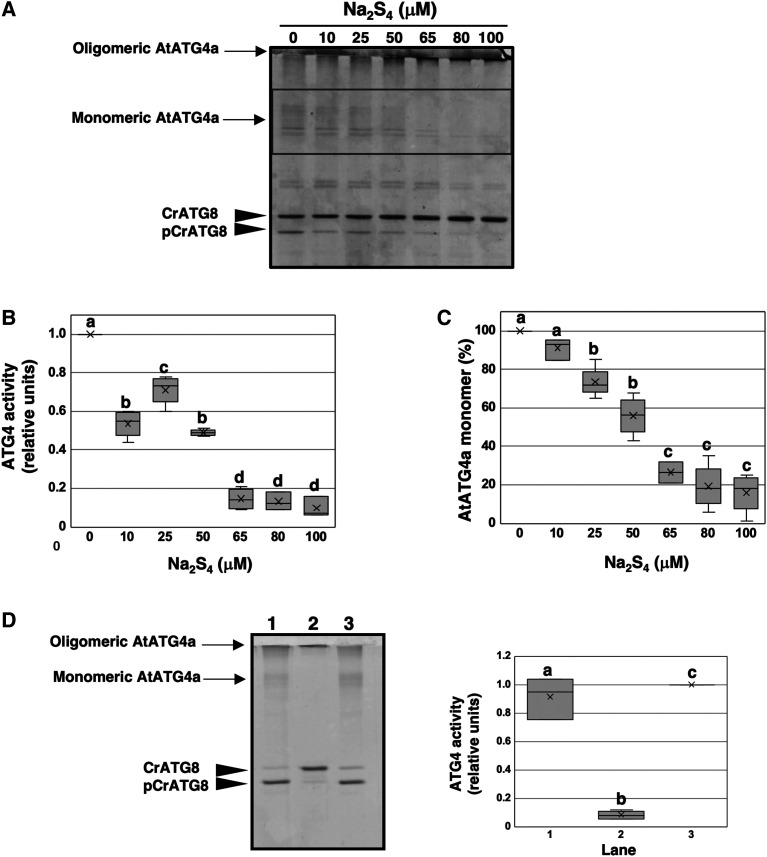
Recent studies have questioned whether NaHS can be the sulfurating molecule instead of the proposed polysulfide and persulfide molecules. These molecules contain sulfane sulfur, the form of sulfur (S0) with the ability to reversibly attach to other sulfur atoms. Most of the reported biological activity associated with sulfide may be due to persulfides, which are considered responsible for intracellular signal transduction through persulfidation in vivo (Toohey, 2011; Ida et al., 2014; Mishanina et al., 2015). Thus, assays similar to those described above were performed using various concentrations of polysulfide Na2S4 used as a sulfur donor. Our results indicated that polysulfide inactivates AtATG4a more efficiently than NaHS (Figures 6A and 6B). Concentrations of Na2S4 three orders of magnitude (10 to 50 μM) lower than those of NaHS were necessary to achieve a similar inhibition, and complete inactivation of the enzyme was observed at 100 μM Na2S4. Furthermore, polysulfides were more active in promoting the aggregation of AtATG4a, and the differences in the effects on activity and oligomerization were not as pronounced as those observed with NaHS (Figure 6C).
Figure 6.
Effect of Polysulfides on AtATG4a Enzyme Activity.
(A) AtATG4a was incubated with 0.5 mM TCEP for 2 h and subsequently treated in the absence or in the presence of indicated concentrations of Na2S4 for 1 h. Then, CrATG8 was added to the incubation mixture, and ATG4 activity was monitored after 4 h using Coomassie-stained gels as described in Methods. All procedures were performed at 25°C. A representative image is shown.
(B) Quantification of ATG4 activity (relative units) determined as the ratio of the band intensity of the processed CrATG8 to the sum of the intensities of the unprocessed and processed CrATG8. A value of 1 corresponds to AtATG4a treated in the absence of Na2S4 (lane 1).
(C) Quantification of the protein band intensity corresponding to the monomeric AtATG4a form marked by a rectangle in (A). A value of 100% corresponds to AtATG4a treated in the absence of Na2S4 (lane 1).
(D) Reversibility of the effect. AtATG4a was incubated with 0.25 mM TCEP for 2 h (lane 1) and treated with 100 μM Na2S4 for 1 h (lane 2) and 1 mM TCEP for 1 h (lane 3). ATG4 activity was monitored using Coomassie-stained gels. The experiment was performed at least three times and a representative image used for the quantification of the activity is shown.
Data are from three independent experiments and evaluated by two-factor ANOVA. Same letters indicate no significant differences. P < 0.05.
Collectively, our results indicate that sulfide can inhibit the ATG4 proteolytic activity and that this inhibition is independent on the ATG4 aggregation state, at least at low concentrations of sulfurating species. Our results also suggest that this inhibitory effect is mediated by specific persulfidation of the catalytic Cys170 residue. Site-directed mutagenesis of this catalytic Cys inactivates ATG4 activity in all tested systems and conditions (Scherz-Shouval et al., 2007; Shu et al., 2010; Pérez-Pérez et al., 2014, 2016), and therefore the mutant enzyme cannot be used to test the inhibitory effect of sulfide.
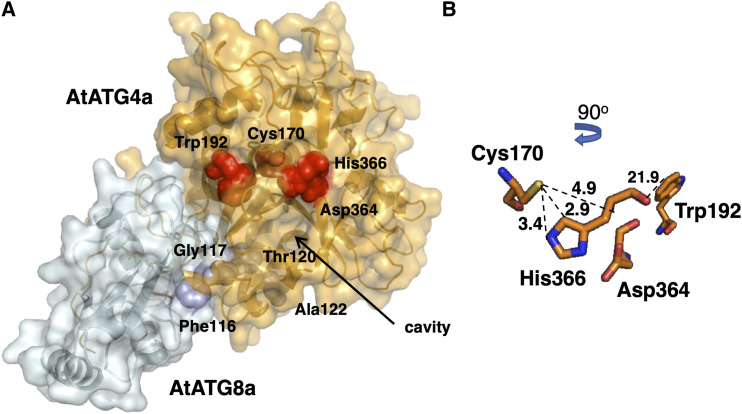
To examine the impact of posttranslational modification of Cys170 by persulfidation on the interaction between AtATG4a and its substrate AtATG8a, 3D homology modeling was performed using the structure of the Homo sapiens HsATG4b-LC3 complex (Satoo et al., 2009). AtATG4a shares up to 33.4% sequence identity with HsAtg4B (E-value 7.3 × 10−90) and AtATG8a shares up to 59.4% sequence identity with HsLC3 (E-value 1.8 × 10−40) with conserved residues covering the whole sequence. Persulfidation of Cys170 in the AtATG4a-AtATG8a protein complex caused conformational changes and intramolecular rearrangements of the catalytic site (Figure 7A) influencing substrate recognition. Addition of the −SH group to Cys170 induced an unfavorable steric effect particularly affecting the His366 residue since the imidazole Cδ2 and Nε2 atoms are 2.9 Å and 3.4 Å from the S atom of Cys170, respectively. The additional sulfur and hydrogen atoms have a covalent radius of 1.02 Å and 0.37 Å, respectively (Figure 7B). Therefore, this Cys modification can inhibit the activity of the plant AtATG4a protein.
Figure 7.
Predicted Structure of the AtATG4a-AtATG8a Complex.
(A) 3D modeling of the AtATG4a-AtATG8a complex based on the structure of the HsAtg4B-LC3 protein complex (PDB ID: 2Z0E). The AtATG8 protein sequence (Q8LEM4 in UniProtkB) corresponds to the splice variant 1. Surface representation of the protein complex and the equivalent residues surrounding the catalytic cavity Cys170, Trp192, Asp364, and His366 in AtATG4a (red) and Phe116, Gly117, Thr120, and Ala122 in AtATG8a (blue) are shown in the structural models.
(B) Zoomed view of the putative conformation of the active site showing the spheres corresponding to the position and distance (Å) of catalytic residues Cys170, Trp192, Asp364, and His366 in AtATG4a.
Interestingly, the simulation of surface electrostatic potential reveals the differences between HsATG4b-LC3 and AtATG4a-AtATG8a protein complexes; specifically, the electrostatic potential surface around the catalytic site of the human complex is more electronegative compared to that of the Arabidopsis complex (Supplemental Figure 2). However, Cys persulfidation does not perturb the charge, since the additional −SH group is a neutral contribution. A closer look at the catalytic cavity region suggests that it is somewhat smaller and less exposed to the solvent in Arabidopsis compared to the human homologue likely hampering the accessibility of the substrate. Overall, these data suggest that inhibition of the processing activity of Arabidopsis AtATG4a may be due to a conformational change of the catalytic site induced by persulfidation.
Endogenous Arabidopsis ATG4 Processes CrATG8 for Conjugation
Our results indicate that sulfide can regulate ATG4 proteolytic activity through persulfidation of a specific Cys residue. This conclusion is based on in vitro experiments. If sulfide has a biological role in the control of ATG4 activity in living plants, the inhibitory effect of sulfide should be reversible. To explore this hypothesis, active AtATG4a was inhibited by a high concentration of Na2S4 to promote complete inactivation and to determine the extent of reversion by reduction of all persulfide residues with TCEP. The reactivation of AtATG4a was monitored as the ratio of the protein band intensities of unprocessed and processed CrATG8 (Figure 6D). The results clearly confirm that the inhibition of ATG4 proteolytic activity by sulfide is reversible, suggesting that it may play a role in the control of ATG4 in vivo.
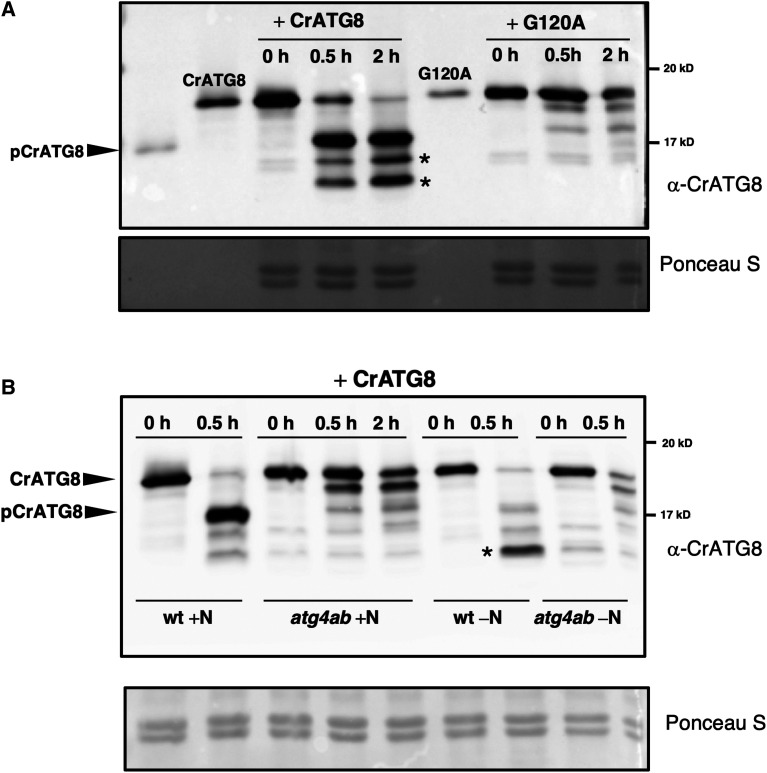
To determine whether sulfide regulation of ATG4 also occurs in vivo, ATG4 enzyme activity was detected in Arabidopsis leaves using the addition of exogenous CrATG8 to leaf protein extracts. Immunoblot analysis using antibodies against CrATG8 indicated that CrATG8 was efficiently processed by incubation with Arabidopsis leaf protein extracts (Figure 8A, right). To confirm the specificity of the CrATG8 cleavage assay, Arabidopsis leaf protein extracts were incubated with a mutant of CrATG8 with conserved Gly120 replaced by Ala (G120A). This CrATG8 mutant is not processed by ATG4 (Pérez-Pérez et al., 2010, 2016). When G120A was used as the substrate, the processed form was not detected, demonstrating the specificity of the endogenous ATG4 activity in Arabidopsis (Figure 8A, left). Moreover, in addition to full-length and processed CrATG8, the antibodies specifically recognized other bands with faster mobility than pCrATG8 (Figure 8, asterisks). These bands were exclusively detected when wild-type CrATG8, but not the G120A mutant, was used in the assay and they apparently correspond to the conjugated form of CrATG8 protein, as demonstrated previously in Chlamydomonas (Pérez-Pérez et al., 2010). Incubation of the Arabidopsis protein extract with the processed form of the CrATG8 (pCrATG8), which does not require cleavage by ATG4 and can be conjugated to PE (Supplemental Figure 3), confirmed that the faster mobility bands correspond to lipidated forms. The antibodies detected mainly an intense protein band that was progressively accumulated with incubation time and fully lipidated at the shortest time of incubation with the extracts under autophagy-activating conditions induced by nitrogen deficiency (Supplemental Figure 3). Therefore, our data demonstrated that the Arabidopsis protein extracts contain all the active proteins required for the conjugation of ATG8 and efficiently recognize CrATG8, thus validating the results of the ATG4 activity assay in the cell-free total extract.
Figure 8.
Cleavage and Conjugation of Chlamydomonas ATG8 by Arabidopsis Proteins.
(A) ATG4 proteolytic activity in wild-type Arabidopsis leaves. Arabidopsis protein extracts prepared from leaves of wild-type seedlings grown for 11 d on MS medium were incubated with CrATG8 or site-directed mutant G120A proteins at 25°C for the indicated times, and ATG4 activity was monitored as the cleavage of the ATG8 forms to the processed (pCrATG8) forms by immunoblotting analysis with anti-CrATG8. Processed pCrATG8 (lane 1), unprocessed CrATG8 (lane 2), and site-directed mutant G120A (lane 6) were loaded as controls.
(B) ATG4 proteolytic activity in the Arabidopsis atg4ab mutant. Arabidopsis protein extracts were prepared from the leaves of wild-type and atg4ab double mutant seedlings grown for 7 d on the MS medium and transferred to the same medium (+N) or to a nitrogen-deficient medium (−N) for additional 4 d. The protein extracts were incubated with CrATG8 at 25°C, and ATG4 activity was monitored after 0, 0.5, or 2 h as indicated by immunoblotting analysis with anti-CrATG8.
The arrowheads show the unprocessed CrATG8 and processed pCrATG8 protein bands, and the asterisks indicate faster-mobility protein bands. Ponceau staining is shown as the protein loading control of the Arabidopsis extract.
To confirm the conclusion that Arabidopsis ATG4 proteins catalyze the processing of CrATG8 in the cell-free total extract assay, the ATG4 enzyme activity of the Arabidopsis protein extracts prepared from the atg4ab double mutant seedlings was assayed (Supplemental Figure 4; Yoshimoto et al., 2004; Chung et al., 2010). When CrATG8 was incubated with the Arabidopsis atg4ab protein extract, the processed form of CrATG8 was not detected even after extended incubation. Similarly, when protein extracts were prepared from nitrogen-limited seedlings, the immunoblot analysis revealed a prominent protein band corresponding to the lipidated form of CrATG8 only in the presence of protein extracts form wild-type plants but not from the atg4ab mutant (Figure 8B).
Therefore, our data show that endogenous Arabidopsis ATG4 proteins recognize the cleavage site of the Chlamydomonas ATG8 substrate, which is efficiently processed in the cell-free enzymatic assay.
Sulfide Reversibly Inhibits the ATG4 Activity in Arabidopsis Seedlings
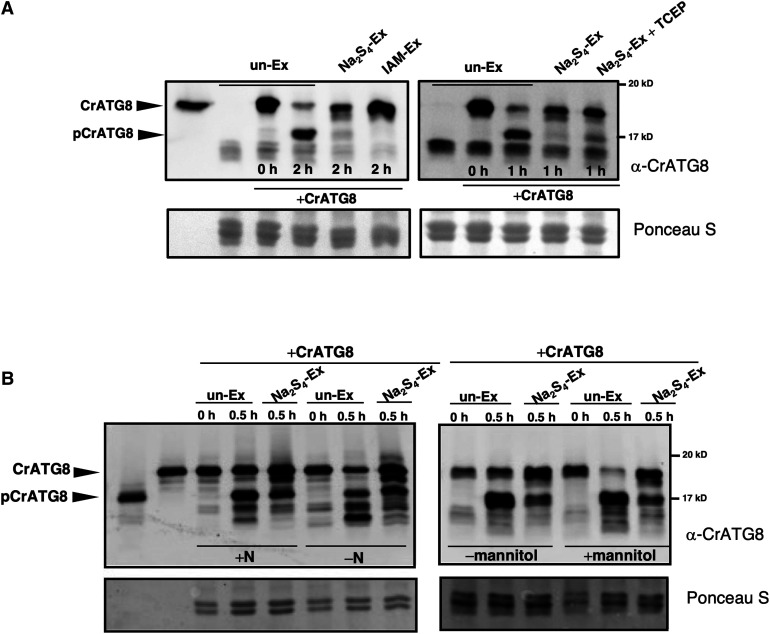
The effect of a sulfurating species on endogenous Arabidopsis ATG4 proteins was investigated to confirm the results obtained in in vitro analysis. Leaf protein extracts were treated with polysulfide, and CrATG8-processing activity was compared with that in the untreated extract. A pronounced decrease in the ATG4 activity was observed when the Arabidopsis extract was pretreated with Na2S4 (Figure 9A). Additionally, treatment of the Arabidopsis leaf protein extract with the alkylating agent iodoacetamide inhibited ATG4 activity as expected because ATG4 is a Cys protease and its activity is dependent on the catalytic Cys; this effect was similar to the effect of polysulfide (Figure 9A, left). Thus, these findings strongly suggest that sulfide negatively regulates ATG4 activity in vivo, at least the cleavage activity of the C-terminal extension of ATG8. Reversibility of the inhibitory effect of polysulfide was also analyzed. When the polysulfide-incubated Arabidopsis extract was treated with TCEP as a reducing agent to reduce the persulfide-modified Cys residues in the protein extract, a significant difference in the activity was detected compared with that in the polysulfide-treated extract (Figure 9A, right). When the extract was incubated with Na2S4, the processed CrATG8 form was barely detected; however, incubation with TCEP significantly increased the abundance of this band, suggesting that sulfide may inhibit Arabidopsis ATG4 activity in a reversible manner.
Figure 9.
Sulfide Inhibits the Endogenous Proteolytic Activity of Arabidopsis ATG4.
(A) Effect of polysulfides on endogenous enzyme activity of Arabidopsis ATG4. Arabidopsis protein extracts (Ex) were prepared from the leaves of seedlings grown for 11 d on the MS medium. The extracts were treated in the absence (un-Ex) and in the presence of 200 μM Na2S4 (Na2S4-Ex) or 20 mM iodoacetamide (IAM-Ex) for 30 min, or in the presence of 200 μM Na2S4 for 30 min and 1 mM TCEP for 30 min (Na2S4-Ex + TCEP). Then, CrATG8 was added to the incubation mixture, and ATG4 proteolytic activity was monitored. Lane 1, unprocessed CrATG8.
(B) Sulfide reverts the endogenous enzyme activity of Arabidopsis ATG4 under autophagy-induced conditions. Arabidopsis protein extracts were prepared from the leaves of seedlings grown for 7 d on the MS medium and transferred to the same medium (+N) or to a nitrogen-deficient medium (−N; left); or transferred to the same medium (−mannitol) or to same medium containing 300 mM mannitol (+mannitol; right) for additional 4 d. The extracts were treated in the absence (un-Ex) or in the presence of 200 μM Na2S4 for 30 min (Na2S4-EX); CrATG8 was added to the incubation mixture, and ATG4 activity was monitored. Lane 1, processed pCrATG8, and lane 2, unprocessed CrATG8.
The ATG4 activity was monitored at the indicated times by immunoblotting analysis with anti-CrATG8. All procedures were performed at 25°C. Ponceau staining is shown as the protein loading control.
To characterize the inhibition of ATG4 proteolytic activity by sulfide, the effect of polysulfide on the ATG4 activity was assayed under basal and autophagy-inducing conditions. Two different established conditions of autophagy induction were tested: nitrogen deficiency, which has been extensively characterized previously by Doelling et al. (2002), Hanaoka et al. (2002), Xiong et al. (2005), Phillips et al. (2008), Guiboileau et al. (2013), and Laureano-Marín et al. (2016b), and osmotic stress (Liu et al., 2009) imposed by the addition of mannitol to the growth medium, which induces the ABA-dependent signaling pathway. The processed form of CrATG8 was detected in total extracts under the control and autophagy-activating conditions, and sulfide inhibited endogenous ATG4 activity under both conditions (Figure 9B). The faster-mobility protein band corresponding to the lipidated form of CrATG8 was more prominent under nitrogen limitation than osmotic stress; however, sulfide treatment decreased the accumulation of the conjugated forms of CrATG8 under both conditions.
DISCUSSION
Accumulating evidence emphasizes the importance of autophagy in plant growth and development. This catabolic process is highly dynamic and occurs at the basal levels to maintain cellular homeostasis during growth; however, autophagy is fine-tuned to adjust plant metabolism to internal and external perturbations. Various regulators of autophagy have been identified in plant systems, such as the energy sensor Snf1-related protein kinase 1 (SnRK1), the kinase Target of Rapamycin (TOR), the TOR downstream substrate ATG1 kinase complex, and the endoplasmic reticulum stress sensor inositol-requiring enzyme-1 (IRE1; Marshall and Vierstra, 2018; Soto-Burgos et al., 2018).
Other regulators of autophagy, such as hydrogen sulfide, have been identified, although the details of the molecular mechanism of action of these regulators remain poorly understood. In the animal systems, interactions of sulfide with autophagy have been described in various pathologies; depending on the disease, sulfide can either suppress or activate autophagy. In all cases, hydrogen sulfide has protective effects (Sen, 2017; Wang et al., 2017; Wu et al., 2018). Despite substantial progress, the exact mechanism of autophagy regulation by sulfide in mammals remains unknown. In plants, particularly in Arabidopsis, the interplay between sulfide and autophagy has been shown, and progress in understanding of the mechanism has been obtained. Hydrogen sulfide generated in the cytosol functions as a signaling molecule negatively regulating autophagy independently of the nutritional conditions. Furthermore, sulfide was shown to repress autophagy via a mechanism that is independent of redox conditions (Álvarez et al., 2012; Laureano-Marín et al., 2016b).
Sulfur and autophagy are also linked by the mechanism of Arabidopsis sensing of the sulfur-containing amino acid Cys. Two different pathways have been identified for sensing of its carbon/nitrogen precursor and its sulfur precursor. The sulfur precursor is transduced to TOR by downregulation of Glc metabolism; therefore, sulfide increases Glc levels and TOR kinase activity, downregulating autophagy (Dong et al., 2017). This study demonstrated that cytosolic sulfide is not the signal responsible for the regulation but does not exclude chloroplast sulfide as the signaling molecule. In fact, a proteomic study showed that other sulfurating species in addition to the cytosolic sulfide can be responsible for regulation of diverse biological processes (Aroca et al., 2017a).
The data of this study emphasize sulfide regulation of autophagy. Our findings indicate that ABA activates autophagy and hydrogen sulfide downregulates this catabolic process. A link between autophagy and ABA was previously described through the Arabidopsis multistress regulator tryptophan-rich sensory protein-related (TSPO). ABA induces TSPO as a heme scavenger, which binds the excessive or deleterious heme and then is targeted for degradation through autophagy (Vanhee and Batoko, 2011; Vanhee et al., 2011). Other connections between autophagy and ABA signaling via TOR have been described. Under nonstressed conditions, TOR phosphorylation of ABA receptors leads to inactivation of SnRK2 kinases and disrupts ABA signaling. Under abiotic stress conditions, ABA activates SnRK2 that phosphorylates RAPTOR, resulting in the inhibition of TOR and consequential activation of autophagy (Wang et al., 2018).
Previous studies have shown that the main mechanism of action of sulfide involves persulfidation of reactive Cys residues of the target proteins resulting in changes in enzyme structure, activity, and subcellular localization demonstrated in several target plant proteins (Aroca et al., 2015, 2017b, 2018). A proteomic analysis performed on mature leaves from Arabidopsis plants grown under nonstress conditions revealed that a high proportion of the whole Arabidopsis proteome may undergo persulfidation under the basal conditions (Aroca et al., 2017a). In this study, a comparative and quantitative proteomic analysis has been performed on ABA-treated leaves to determine whether persulfidation mechanism is involved in sulfide regulation of the ABA signaling pathways. Significant differences in the levels of persulfidation were observed after ABA treatment compared to that under the control conditions. Surprisingly, the protein with the lowest level of persulfidation after 3 h ABA treatment was identified as Cys-type protease AtATG4a (5.20-fold change, P < 0.05, control versus 3 h ABA), and even after 6 h of ABA treatment, the reduction in the level of persulfidation remained very significant (2.46-fold change, P < 0.05, control versus 6 h ABA). These data suggest that AtATG4a may be a target of persulfidation and that this posttranslational modification may be the molecular mechanism mediating sulfide regulation of autophagy. A detailed analysis of the ABA-triggered changes in the persulfidation proteome will be performed in a future study. Our results demonstrated that ATG4 proteolytic activity in Arabidopsis is reversibly regulated by sulfide, and this regulation effectively controls the progression of autophagy. Thus, our findings contribute to the understanding of the mechanism of regulation of autophagy by sulfide in the plant systems and add another level of regulation to this process.
The AtATG4a protease contains a specific site of persulfidation detected by the tag-switch procedure and confirmed by mass spectrometry. The specifically persulfidated Cys residue is Cys170, which is a part of the characteristic catalytic triad Cys-His-Asp of Cys proteases (Sugawara et al., 2005). Comparison of the amino acid sequences of ATG4s from various biological systems indicated low similarity, although the amino acids required for the Cys protease activity, including the catalytic Cys170, are highly conserved. Interestingly, Cys170 is the only Cys residue that is conserved in all known ATG4s (Supplemental Figure 5; Pérez-Pérez et al., 2014, 2016; Seo et al., 2016). In contrast, redox regulation of specific Cys residues has been detected in ATG4 from human, yeast, and Chlamydomonas. However, these residues are not conserved in various organisms, and therefore the details of the regulatory mechanisms may be different. In humans, HsATG4a and HsATG4b proteases are the targets of reversible oxidation by H2O2, and the Cys residue Cys81 located close to the catalytic Cys residue (Cys77, analogous to Cys170 in Arabidopsis) was shown to be critical for this regulation (Scherz-Shouval et al., 2007). Recently, Cys292 and Cys361 have been shown to be HsATG4b sites essential for reversible oxidative modification (Zheng et al., 2020). Redox regulation of the yeast ScATG4 protein is due to the formation of a disulfide bond between the noncatalytic residues Cys338 and Cys394, which is thioredoxin dependent (Pérez-Pérez et al., 2014). In Chlamydomonas reinhardtii, CrATG4 activity depends on the formation of a single disulfide bond regulated by the NADPH/thioredoxin system; however, only Cys400, which is the equivalent to Cys338 in yeast, has been demonstrated to be required for redox regulation of the algal CrATG4 enzyme (Pérez-Pérez et al., 2016). In Arabidopsis, the activity of AtATG4a and AtATG4b is reversibly inhibited in vitro by reactive oxygen species (Woo et al., 2014), although redox regulation of the critical Cys residues was not reported. Additionally, other posttranslational modifications of Cys residues of ATG4 proteases have been described, such as S-nitrosylation of HsATG4b at Cys189 and Cys292, though these two residues are not conserved in the HsATG4a amino acid sequence (Li et al., 2017). Therefore, persulfidation of the catalytic Cys residue of Cys proteases ATG4 can be a posttranslational modification conserved in various biological systems.
Because the target residue of persulfidation involves the active site, an effect of the sulfide donor molecules on the enzymatic activity of AtATG4a was anticipated. In fact, a 3D modeling analysis predicted that the addition of a sulfur atom to the −SH group of Cys170 can cause unfavorable effects on the catalytic site of AtATG4a that should affect substrate recognition and impair the enzyme activity. To test this hypothesis, a heterologous activity assay was developed using CrATG8 as a substrate similar to assays previously reported by Pérez-Pérez et al. (2014) and Seo et al. (2016). Our results indicate that the plant protease is functional and can process the algal substrate.
Our findings demonstrate that sulfide plays a specific role in the regulation of ATG4 enzymatic activity. The sulfide donor molecules used in this study, such as hydrosulfide and tetrasulfide, significantly inactivate AtATG4a cleavage activity even at relatively low concentrations, polysulfide being the most efficient inhibitor. The chemical mechanism of the reaction of the thiol groups with the sulfide molecule remains a matter of debate. H2S cannot directly react directly with thiols, and the Cys group must be partially oxidized (converted to disulfide, glutathiolated, S-nitrosylated, or to sulfenic acid) prior to sulfide attack. Alternatively, certain chemical studies have demonstrated that sulfane sulfur (S0) of the polysulfide molecule is responsible for the production of persulfide during interaction with the thiol group (Toohey, 2011; Kimura, 2015; Mishanina et al., 2015). This phenomenon can explain why polysulfide is a more potent inhibitor of ATG4 activity than hydrosulfide. Moreover, the inhibitory effect of low concentrations of sulfide on AtATG4a activity compatible with the in vivo conditions is reversible by a reducing agent, suggesting a biological role of this effect. Interestingly, an enzymatic mechanism of reversing persulfidation by the thioredoxin system has been demonstrated in animal systems (Wedmann et al., 2016; Doka et al., 2020). Thus, it is plausible that the thioredoxin system also functions in plants to modulate the activity of AtATG4.
The biological significance of sulfide regulation of autophagy through reversible inhibition of ATG4 protease is reinforced by the assays in Arabidopsis leaves under basal and autophagy-inducing conditions. Our experimental system was shown to be suitable for specific assay of Arabidopsis ATG4 processing activity by using various experimental approaches. Endogenous plant ATG4 Cys proteases specifically recognize the Gly120 cleavage site in the substrate from green algae based on the experiment with the G120A mutant of the CrATG8 substrate. The processed form was not detected when the Arabidopsis protein extracts were deficient in ATG4a and ATG4b enzymes, demonstrating the specificity of our experimental system. The endogenous Arabidopsis ATG4 proteins mimic the effect of sulfurating molecules on AtATG4a in vitro, including significant inhibition by the sulfide donor and reversal by a reducing agent. A significant increase in the overall ATG4 protease activity in Arabidopsis was detected under autophagy-inducing conditions, including nitrogen starvation and osmotic stress. Thus, a correlation was detected between the progress of autophagy and the ATG4 enzymatic activity estimated using the heterologous assay method. Additionally, the inhibitory effect of sulfide on the protease activity was observed under the conditions of induced autophagy. Overall, our findings suggest that negative regulation of the progress of autophagy by sulfide is mediated by specific persulfidation of Cys protease ATG4. However, additional targets of sulfide cannot be ruled out and further analysis is needed.
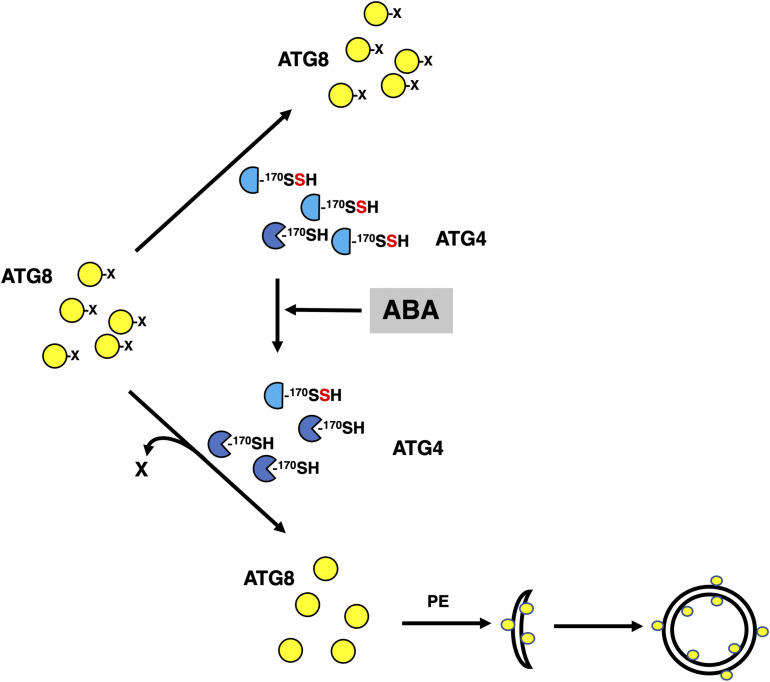
In conclusion, our data suggest a new level of regulation of ATG4 activity by sulfide. Based on our findings, we propose that intracellular sulfide maintains high levels of persulfidation of the ATG4 pool under basal conditions, resulting in the inhibition of ATG4 proteolytic activity. ATG4 inhibition limits the formation of ATG8-PE adducts and consequently de novo synthesis of autophagosomes. An increase in the intracellular level of ABA transiently reduces the level of persulfidation of the ATG4 population and then activates the protease activity of the enzyme to process ATG8 that can be further lipidated (Figure 10).
Figure 10.
Graphical Model of ABA-Triggered Induction of Autophagy Mediated by Posttranslational Modification of ATG4.
Under basal conditions, intracellular sulfide maintains high levels of persulfidation of the ATG4 pool, which inhibits the proteolytic activity of the enzyme for ATG8 C-terminal processing. An increase in the intracellular level of ABA transiently decreases the level of persulfidation of the ATG4 population activating the protease activity of the enzyme and the processing of ATG8 that can be further lipidated to progress autophagy. Yellow circles represent ATG8 protein with or without the processed C terminus (represented as X). Blue semicircles represent persulfidated ATG4 at the thiol group of Cys170 residue. Blue Pacman symbols represent ATG4 with reduced thiol group of Cys170 residue. The conjugation process of ATG8 with PE, and autophagosome initiation and closure are also shown.
METHODS
Plant Material, Treatments, and Protein Extraction
Plant growth conditions were 16 h of light (120 μE m−2 s−1) at 22°C and 8 h of dark at 20°C. The Arabidopsis (Arabidopsis thaliana) wild-type and the atg4ab mutant (Nottingham Arabidopsis Stock Center) seeds were sown on Murashige and Skoog (MS) solid medium containing 0.8% (w/v) agar, synchronized at 4°C for 2 d, and incubated vertically in a growth chamber (LUMILUX cool white bulbs). For exposure to the nitrogen-starvation conditions, nitrogen-deficient MS medium was prepared by replacing nitrate salts with chloride salts. For mannitol treatment, 300 mM was added to the MS medium. One-week-old seedlings were transferred to nitrogen-deficient or mannitol-containing MS solid medium for an additional 4 d of growth.
For proteomic analysis, wild-type Arabidopsis plants were grown in soil for 30 d and then sprayed with water (control conditions) or 50 μM ABA for 3 and 6 h. To assess the autophagic activity, 1-week-old Arabidopsis seedlings overexpressing GFP-ATG8e (Xiong et al., 2007) were transferred to the MS liquid medium and treated with 50 μM ABA for 3 and 6 h and with 200 μM NaHS for 1 h.
Arabidopsis material (200 mg) was ground in liquid nitrogen with 400 μL of extraction buffer (100 mM Tris-HCl, pH 7.5, 400 mM Suc, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride) using a mortar and pestle. After centrifugation at 500g for 10 min at 4°C, the supernatant was used as the Arabidopsis protein extract.
Persulfidated Protein Quantitation by Label-free SWATH-MS Acquisition and Analysis
Protein samples from three biological replicates (independent pools) of leaf tissues treated with ABA for 0 h (control sample), 3 h, and 6 h were isolated, and 1 mg of protein per sample was used for the tag-switch labeling for enrichment of persulfidated proteins as described by Aroca et al. (2017a). After elution from the streptavidin beads, the proteins were precipitated by trichloroacetic acid/acetone. Precipitated samples were resuspended in 50 mM ammonium bicarbonate with 0.2% (v/v) Rapigest (Waters) for protein determination. Protein (50 µg) was digested with trypsin as previously described by García et al. (2019), and the SWATH-MS analysis was performed at the Proteomic Facility of the Institute of Plant Biochemistry and Photosynthesis (Seville, Spain). A data-dependent acquisition approach using nano-LC-MS/MS was initially performed to generate the SWATH-MS spectral library as described by García et al. (2019).
The peptide and protein identifications were performed using ProteinPilot software (version 5.0.1, Sciex) with the Paragon algorithm. The search was conducted against a Uniprot proteome (June 2017 release), and the corresponding reversed entries and common contaminants were assembled in the FASTA format using ProteinPilot software version 5.0.1 (AB Sciex) with the Paragon algorithm. Samples were input as unlabeled with no special factors, trypsin digested, and methylsulfonyl benzothiazole alkylated. The automatically generated report in ProteinPilot was manually inspected for FDR cut-off proteins, and only proteins identified at FDR ≤1% were considered for output and subsequent analysis.
For relative quantification using SWATH analysis, the same samples used to generate the spectral library were analyzed using a DIA method. Each sample (2 μL) was analyzed using the SWATH-MS acquisition method on the LC-MS equipment with the LC gradient as described. The method consisted of repeated acquisition cycles of time-of-flight (TOF) MS/MS scans (230 to 1500 m/z, 60 ms acquisition time) of 60 overlapping sequential precursor isolation windows of variable width (1 m/z overlap) covering the 400 to 1250 m/z mass range from a previous TOF MS scan (400 to 1250 m/z, 50 ms acquisition time) for each cycle. The total cycle time was 3.7 s.
Autocalibration of the equipment and chromatographic conditions were controlled by an injection of a standard of digested β-galactosidase from Escherichia coli between the replicates.
SWATH MS spectra alignment was performed with the PeakView 2.2 (Sciex) software with the MicroApp SWATH 2.0 using the reference spectral library generated as described above. Two DIA raw files for each biological replicate were loaded in unison using the following parameters: 10 peptides, seven transitions, peptide confidence > 99%, 1% FDR including shared peptides, and extracted-ion chromatogram width set at 0.05 Da. After data processing, three distinct files were exported for subsequent quantification. The processed mrkvw files containing protein information from PeakView were loaded into MarkerView (version 1.2.1, AB Sciex) for normalization of protein intensity (peak area) for all runs using the built-in total ion intensity sum plug-in. Log2 transformation was performed prior to statistical analysis. A histogram plot was used to check the normality of distribution of each technical replicate. Mean values of protein expression were used for calculation of fold change. Proteins with adjusted P < 0.05 and fold change ≥ 1.5 were considered differentially expressed.
The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the identifier PXD019802.
Expression of AtATG4a in E. coli
Total RNA was extracted from wild-type Arabidopsis leaves using an RNeasy plant mini kit (Qiagen) and reverse transcribed using an oligo (dT) primer and a SuperScript first-strand synthesis system for RT-PCR (Invitrogen). Subsequently, a 1404-bp sequence encoding the full-length AtATG4a (At2g44140) protein was amplified by PCR using the primers ATG4-F, CACCATGAAGGCTTTATGTGA, and ATG4-R, ATGACTGGCAAATGCTCTGA and the proofreading Platinum Pfx DNA polymerase (Invitrogen). The PCR conditions were as follows: a denaturation cycle of 2 min at 94°C followed by 30 amplification cycles of 15 s at 94°C, 30 s at 57°C, and 1 min at 68°C. The amplified cDNA was then ligated into the pENTR/D-TOPO vector using the pENTR directional TOPO cloning kit (Invitrogen) according to the manufacturer’s instructions. Positive clones were identified by PCR and selected for plasmid DNA isolation. The AtATG4a cDNA was then cloned into the expression vector pDEST17 using an E. coli expression system with gateway technology (Invitrogen), which generates a fusion protein with an N-terminal 6× His tag that was confirmed by sequencing; the expression was induced with L-arabinose in BL21-AI E. coli cells.
Purification of the Recombinant AtATG4a Protein
The 6× His-tagged recombinant protein was isolated from 200 mL of BL21-AI E. coli cells that were cultured at 37°C to an optical density of 0.5 at 600 nm and induced with 0.2% (w/v) L-arabinose for 2.5 h at 37°C. Prior to purification, His-tagged AtATG4a was solubilized with 6 M urea because the recombinant protein was contained in the inclusion bodies. Then, the protein was purified from the soluble fraction by nickel affinity chromatography using an Invitrogen Ni-NTA purification system (Thermo Fisher Scientific), according to the manufacturer’s instructions. The purified protein was concentrated and desalted using 10-kD cutoff pore size centrifugal filter units (Millipore). The purity of the protein was confirmed by SDS-PAGE using 12% (w/v) polyacrylamide gels stained by Coomassie Blue.
Detection of Persulfidation on the Recombinant AtATG4a
An untreated aliquot of the purified recombinant AtATG4a and another aliquot pretreated with 50 mM DTT for 30 min at 4°C to reduce all persulfide groups were precipitated with acetone for 20 min at −20°C and centrifuged at a maximum speed for 20 min at 4°C. After acetone removal, the proteins were resuspended in 50 mM Tris-HCl (pH 8) buffer supplemented with 2.5% (w/v) SDS and subjected to the tag-switch procedure as previously described by Aroca et al. (2017a). The cyano-biotinylated proteins were then detected using an immunoblot assay as follows. The CN-biotinylated proteins were separated using nonreducing SDS-PAGE through 12% (w/v) polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Bio-Rad) according to the manufacturer’s instructions. The anti-biotin (Abcam, catalog no. ab191354) and secondary antibodies (Bio-Rad, catalog no.170-6515) were diluted 1:500,000 and 1:100,000, respectively, and enhanced chemiluminescent select protein gel blotting detection reaction (GE Healthcare) was used to detect the proteins using horseradish peroxidase-conjugated anti-rabbit secondary antibodies. For protein loading control, the membranes before immunodetection were stained with SYPRO Ruby (Invitrogen) to detect all protein bands.
Identification of Persulfidated Cys Residues of Recombinant AtATG4a Using Mass Spectrometry
Recombinant AtATG4a was separated using nonreducing SDS-PAGE through 12% (w/v) polyacrylamide gels, and the band corresponding to AtATG4a was manually excised from Coomassie-stained gels. Gel plugs were washed twice using 50 mM ammonium bicarbonate and acetonitrile and dried under a steam of nitrogen. Then, proteomics-grade trypsin (Sigma-Aldrich) at a final concentration of 16 ng/μL in 25% (v/v) acetonitrile/50 mM ammonium bicarbonate solution was added; the samples were digested at 37°C for 5 h. The reaction was stopped by adding 50% (v/v) acetonitrile/0.5% (v/v) trifluoroacetic acid for peptide extraction. The eluted tryptic peptides were dried by speed-vacuum centrifugation and resuspended in 6 μL of 0.1% (v/v) formic acid in water. Digested peptides were subjected to nanoliquid chromatography electrospray ionization MS/MS analysis using a nanoliquid chromatography system (ExcionLC AD, Sciex) coupled to a TripleTOF 5600+ mass spectrometer (Sciex) with a spray ionization source. Mass spectrometry and MS/MS data of individual samples were processed using the Analyst TF 1.5.1 software (Sciex). Peptide mass tolerance was set to 25 μD D−1 and 0.05 D for fragment masses, and only one or two missed cleavages were allowed. Peptides with an individual Mr search score ≥ 20 were considered correctly identified.
In Vitro ATG4 Enzyme Activity Assay
The typical reaction mixture contained 5 μM recombinant AtATG4a, 5 μM CrATG8 (Pérez-Pérez et al., 2010), and 1 mM EDTA in Tris-buffered saline (50 mM Trizma base, 138 mM NaCl, and 27 mM KCl at pH 8). When indicated, AtATG4a was incubated in the presence of DTT, TCEP, NaHS, Na2S4, or iodoacetamide (IAM) alone or in combination at the indicated times and concentrations. The reaction mixtures were incubated at 25°C, and the reaction was stopped by the addition of β-mercaptoethanol-free Laemmli sample buffer followed by 5 min boiling. The proteins were resolved using nonreducing SDS-PAGE through 15% (w/v) polyacrylamide gels and stained with Coomassie Brilliant Blue (Sigma-Aldrich). The gels were scanned with a GS-800 densitometer (Bio-Rad), and the signals corresponding to the unprocessed and processed CrATG8 forms were quantified with the Quantity One software (Bio-Rad). The ATG4 activity (in arbitrary units) was calculated as the ratio of the band intensity of the processed CrATG8 to the sum of the intensities of the unprocessed and processed CrATG8. An activity value of 1 corresponds to the maximum value.
Assay of Endogenous ATG4 Enzyme Activity in Cell-free Total Extract
The in vivo assay of ATG4 activity in cell-free total extract was performed in a typical reaction mixture containing 40 μg of leaf or 20 μg of root protein extract and 0.05 μM purified unprocessed CrATG8, processed pCrATG8, or Gly-to-Ala mutant protein (G120A). When required, a sulfur donor (Na2S4), a reducing agent (TCEP), or an alkylating agent (IAM) was added at the indicated concentrations. The reaction mixture was incubated at 25°C for the indicated time, stopped by addition of β-mercaptoethanol-free Laemmli sample buffer, and boiled for 5 min. Then, proteins were separated using nonreducing SDS-PAGE through 15% (w/v) polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad) as previously described by Pérez-Pérez et al. (2010). Anti-CrATG8 (Pérez-Pérez et al., 2010) and secondary antibodies were diluted 1:3000 and 1:10,000, respectively. An enhanced chemiluminescent select protein gel blotting detection reaction (GE Healthcare) was used to detect the proteins. For protein loading control, the membranes before immunodetection were stained with Ponceau S (Sigma-Aldrich) to detect all protein bands.
Protein Modeling
3D homology modeling was driven by Modeler (Sali and Blundell, 1993) using the structure of the Homo sapiens Atg4B-LC3 complex (PDB ID: 2Z0E; Sato et al., 2009) as a template. Molecular crystal X-ray structures and structural model of Arabidopsis AtATG4a-AtATG8a complex were inspected, analyzed, and plotted with PyMol 1.4.1 (Schrodinger). Surface electrostatic potentials were calculated and visualized using the PyMol 1.4.1 software.
Accession Numbers
The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD019802. Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: AtATG4a (At2g44140), AtATG4b (At3g59950), AtATG8a (At4g21980), and CrATG8 (Cre16.g689650.t1.1).
Supplemental Data
Supplemental Figure 1. C termini of the Chlamydomonas ATG8 proteins and nine Arabidopsis ATG8 proteins.
Supplemental Figure 2. Simulation of surface electrostatic potential distribution in the HsATG4b-LC3 protein complex (PDB ID: 2Z0E) and AtATG4a-AtATG8a 3D structural model.
Supplemental Figure 3. Conjugation of Chlamydomonas ATG8 processed by ATG4s proteases from Arabidopsis.
Supplemental Figure 4. Phenotypes of wild-type and atg4ab double mutant seedlings under the basal and induced autophagy conditions.
Supplemental Figure 5. Alignment of amino acid sequences of ATG4 from various sources.
Supplemental Figure 6. Lower exposure of the GFP blot shown in Figure 2.
Supplemental Table 1. Classification of the proteins with persulfidation level reduced after 3 h ABA treatment.
Supplemental Table 2. Protein list of the elements included in the bin of “proteins” with persulfidation level reduced after 3 h ABA treatment.
Supplemental Data Set 1. Spectral library.
Supplemental Data Set 2. Proteins quantified using SWATH acquisition method in the control sample versus 3 h ABA treatment sample.
Supplemental Data Set 3. Proteins with different abundance in the control sample compared to that in the 3 h ABA treatment sample at P <0.05.
Supplemental Data Set 4. Proteins quantified using SWATH acquisition method in the control sample versus 6 h ABA treatment sample.
Supplemental Data Set 5. Proteins with different abundance in the control sample compared to that in the 6 h ABA treatment sample at P <0.05.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Rocío Rodríguez for the proteomics services. This work was supported in part by the European Regional Development Fund through the Ministerio de Economía y Competitividad and the Agencia Estatal de Investigación (grant nos. BFU2015-68216-P and PGC2018-099048-B-I00 to J.L.C.; grant nos. BIO2015-74432-JIN and PID2019-110080GB-I00 to M.E.P.-P.; grant nos. BIO2016-76633-P and PID2019-109785GB-I00; and Junta de Andalucía grant no. P18-RT-3154 to C.G.)., the Marie Skłodowska-Curie Grant Agreement (grant no. 834120 to Á.A.), Government of Aragón-FEDER (grant no. E35_17R to I.Y.), and the Ministerio de Economia y Competitividad through the program of “Formación de Personal Investigador” (to A.M.L.-M. and A.J.-F.).
AUTHOR CONTRIBUTIONS
Á.A., A.M.L.-M., M.E.P.-P., I.Y., A.J.-F., and I.M. performed research and analyzed the data; J.L.C. and L.C.R. designed the research and analyzed the data; C.G. designed the research, analyzed the data, and wrote the article; all authors contributed to the discussion.
References
- Álvarez C., García I., Moreno I., Pérez-Pérez M.E., Crespo J.L., Romero L.C., Gotor C.(2012). Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24: 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Benito J.M., Gotor C., Romero L.C.(2017a). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68: 4915–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Gotor C., Romero L.C.(2018). Hydrogen sulfide signaling in plants: Emerging roles of protein persulfidation. Front Plant Sci 9: 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Schneider M., Scheibe R., Gotor C., Romero L.C.(2017b). Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 58: 983–992. [DOI] [PubMed] [Google Scholar]
- Aroca Á., Serna A., Gotor C., Romero L.C.(2015). S-sulfhydration: A Cys posttranslational modification in plant systems. Plant Physiol. 168: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood A., Kopriva S.(2014). Hydrogen sulfide in plants: From dissipation of excess sulfur to signaling molecule. Nitric Oxide 41: 72–78. [DOI] [PubMed] [Google Scholar]
- Chen S., Jia H., Wang X., Shi C., Wang X., Ma P., Wang J., Ren M., Li J.(2020). Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 13: 732–744. [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D.(2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D.(2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Doka E., et al. (2020). Control of protein function through oxidation and reduction of persulfidated states. Sci Adv. 6: eaax8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., et al. (2017). Sulfur availability regulates plant growth via glucose-TOR signaling. Nat. Commun. 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- García I, Arenas-Alfonseca L, Moreno I, Gotor C, Romero L C(2019). HCN regulates cellular processes through posttranslational modification of proteins by S-cyanylation. Plant Physiol 179: 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C., Lamattina L.(2013). Gasotransmitters are emerging as new guard cell signaling molecules and regulators of leaf gas exchange. Plant Sci. 201-202: 66–73. [DOI] [PubMed] [Google Scholar]
- Gotor C., García I., Aroca Á., Laureano-Marín A.M., Arenas-Alfonseca L., Jurado-Flores A., Moreno I., Romero L.C.(2019). Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 70: 4251–4265. [DOI] [PubMed] [Google Scholar]
- Gotor C., García I., Crespo J.L., Romero L.C.(2013). Sulfide as a signaling molecule in autophagy. Autophagy 9: 609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C., Laureano-Marín A.M., Arenas-Alfonseca L., Moreno I., Aroca Á., García I., Romero L.C.(2017). Advances in Plant Sulfur Metabolism and Signaling In Botany, Cánovas F.M., Lüttge U., and Matyssek R., eds, Volume 78 (Cham, Switzerland: Springer International Publishing; ), pp. 45–66. [Google Scholar]
- Gotor C., Laureano-Marín A.M., Moreno I., Aroca Á., García I., Romero L.C.(2015). Signaling in the plant cytosol: Cysteine or sulfide? Amino Acids 47: 2155–2164. [DOI] [PubMed] [Google Scholar]
- Gu L., Jung H.J., Kim B.M., Xu T., Lee K., Kim Y.O., Kang H.(2015). A chloroplast-localized S1 domain-containing protein SRRP1 plays a role in Arabidopsis seedling growth in the presence of ABA. J. Plant Physiol. 189: 34–41. [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Avila-Ospina L., Yoshimoto K., Soulay F., Azzopardi M., Marmagne A., Lothier J., Masclaux-Daubresse C.(2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199: 683–694. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y.(2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T., et al. (2014). Reactive Cys persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 111: 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Pei Y.(2015). Physiological implications of hydrogen sulfide in plants: Pleasant exploration behind its unpleasant odour. Oxid. Med. Cell. Longev. 2015: 397502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Xue S., Luo Y., Tian B., Fang H., Li H., Pei Y.(2013). Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol. Biochem. 62: 41–46. [DOI] [PubMed] [Google Scholar]
- Kimura H.(2015). Hydrogen sulfide and polysulfides as signaling molecules. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 91: 131–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y.(2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie S., Nikoloski Z.(2012). The choice between MapMan and Gene Ontology for automated gene function prediction in plant science. Front. Genet. 3: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureano-Marín A.M., Moreno I., Aroca Á., García I., Romero L.C., Gotor C.(2016a). Regulation of autophagy by hydrogen sulfide In Gasotransmitters in Plants: The Rise of a New Paradigm in Cell Signaling, Lamattina L., and García-Mata C., eds (Cham, Switzerland: Springer International Publishing; ), pp. 53–75. [Google Scholar]
- Laureano-Marín A.M., Moreno I., Romero L.C., Gotor C.(2016b). Negative regulation of autophagy by sulfide is independent of reactive oxygen species. Plant Physiol. 171: 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Y., Wang L., Wang P., Xue Y., Li X., Qiao X., Zhang X., Xu T., Liu G., Li P., Chen C.(2017). Autophagy impairment mediated by S-nitrosation of ATG4B leads to neurotoxicity in response to hyperglycemia. Autophagy 13: 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C.(2012). Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C.(2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Vierstra R.D.(2018). Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69: 173–208. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Chen Q., Havé M.(2017). Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 39: 8–17. [DOI] [PubMed] [Google Scholar]
- Mishanina T.V., Libiad M., Banerjee R.(2015). Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y.(2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27: 107–132. [DOI] [PubMed] [Google Scholar]
- Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K., Barrow R.K., Yang G., Wang R., Snyder S.H.(2009). H2S signals through protein S-sulfhydration. Sci. Signal. 2: ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Yen W.-L., Mari M., Cao Y., Xie Z., Baba M., Reggiori F., Klionsky D.J.(2012). A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 8: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ishii J., Asai E., Ohsumi Y.(2012). Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 8: 177–186. [DOI] [PubMed] [Google Scholar]
- Papanatsiou M., Scuffi D., Blatt M.R., García-Mata C.(2015). Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiol. 168: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B.D., Snyder S.H.(2012). H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13: 499–507. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Florencio F.J., Crespo J.L.(2010). Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152: 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Lemaire S.D., Crespo J.L.(2012). Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 160: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Lemaire S.D., Crespo J.L.(2016). Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172: 2219–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Zaffagnini M., Marchand C.H., Crespo J.L., Lemaire S.D.(2014). The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10: 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D.(2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero L.C., García I., Gotor C.(2013). L-cysteine Desulfhydrase 1 modulates the generation of the signaling molecule sulfide in plant cytosol. Plant Signal. Behav. 8: e24007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L.(1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815. [DOI] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D.B., Uozumi N.(2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424: 439–448. [DOI] [PubMed] [Google Scholar]
- Satoo K., Noda N.N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F.(2009). The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28: 1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z.(2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26: 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuffi D., Álvarez C., Laspina N., Gotor C., Lamattina L., García-Mata C.(2014). Hydrogen sulfide generated by L-Cys desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol. 166: 2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuffi D., Nietzel T., Di Fino L.M., Meyer A.J., Lamattina L., Schwarzländer M., Laxalt A.M., García-Mata C.(2018). Hydrogen sulfide increases production of NADPH oxidase-dependent hydrogen peroxide and phospholipase D-derived phosphatidic acid in guard cell signaling. Plant Physiol. 176: 2532–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N.(2017). Functional and molecular insights of hydrogen sulfide signaling and protein sulfhydration. J. Mol. Biol. 429: 543–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E., Woo J., Park E., Bertolani S.J., Siegel J.B., Choi D., Dinesh-Kumar S.P.(2016). Comparative analyses of ubiquitin-like ATG8 and Cys protease ATG4 autophagy genes in the plant lineage and cross-kingdom processing of ATG8 by ATG4. Autophagy 12: 2054–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., et al. (2020). Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 32: 1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C.W., Drag M., Bekes M., Zhai D., Salvesen G.S., Reed J.C.(2010). Synthetic substrates for measuring activity of autophagy proteases: Autophagins (Atg4). Autophagy 6: 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Burgos J., Zhuang X., Jiang L., Bassham D.C.(2018). Dynamics of autophagosome formation. Plant Physiol. 176: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F.(2005). Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 280: 40058–40065. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M.(2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Toohey J.I.(2011). Sulfur signaling: Is the agent sulfide or sulfane? Anal. Biochem. 413: 1–7. [DOI] [PubMed] [Google Scholar]
- Üstün S., Hafrén A., Hofius D.(2017). Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40: 122–130. [DOI] [PubMed] [Google Scholar]
- Vanhee C., Batoko H.(2011). Autophagy involvement in responses to abscisic acid by plant cells. Autophagy 7: 655–656. [DOI] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H.(2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitvitsky V., Miljkovic J.L., Bostelaar T., Adhikari B., Yadav P.K., Steiger A.K., Torregrossa R., Pluth M.D., Whiteman M., Banerjee R., Filipovic M.R.(2018). Cytochrome c reduction by H2S potentiates sulfide signaling. ACS Chem. Biol. 13: 2300–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J A, Csordas A, del-Toro N, Dianes J A, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q W, Wang R, et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleaic Acids Res 44: D447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tang W., Zhu Y.Z.(2017). An update on AMPK in hydrogen sulfide pharmacology. Front. Pharmacol. 8: 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69: 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedmann R., et al. (2016). Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci. (Camb.) 7: 3414–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J., Park E., Dinesh-Kumar S.P.(2014). Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 Cys proteases. Proc. Natl. Acad. Sci. USA 111: 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wang H., Teng T., Duan S., Ji A., Li Y.(2018). Hydrogen sulfide and autophagy: A double edged sword. Pharmacol. Res. 131: 120–127. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C.(2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C.(2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y.(2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.-Q., Ni T., Hong B., Wang H.-Y., Jiang F.-J., Zou S., Chen Y., Zheng X.-L., Klionsky D.J., Liang Y., Xie Z.(2012). Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Fermani S., Marchand C.H., Costa A., Sparla F., Rouhier N., Geigenberger P., Lemaire S.D., Trost P.(2019). Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 31: 155–210. [DOI] [PubMed] [Google Scholar]
- Zheng X., Yang Z., Gu Q., Xia F., Fu Y., Liu P., Yin X.M., Li M.(2020). The protease activity of human ATG4B is regulated by reversible oxidative modification. Autophagy 16: 1838–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]