Abstract
The oxylipins, 5S,12S-dihydroxy-6E,8Z,10E,14Z-eicosatetraenoic acid (5S,12S-diHETE) and 5S,15S-dihydroxy-6E,8Z,11Z,13E-eicosatetraenoic acid (5S,15S-diHETE), have been identified in cell exudates and have chemotactic activity toward eosinophils and neutrophils. Their biosynthesis has been proposed to occur by sequential oxidations of arachidonic acid (AA) by lipoxygenase enzymes, specifically through oxidation of AA by h5-LOX followed by h12-LOX, h15-LOX-1, or h15-LOX-2. In this work, h15-LOX-1 demonstrates altered positional specificity when reacting with 5S-HETE, producing 90% 5S,12S-diHETE, instead of 5S,15S-diHETE, with kinetics 5-fold greater than that of h12-LOX. This is consistent with previous work in which h15-LOX-1 reacts with 7S-HDHA, producing the noncanonical, DHA-derived, specialized pro-resolving mediator, 7S,14S-diHDHA. It is also determined that oxygenation of 5S-HETE by h15-LOX-2 produces 5S,15S-diHETE and its biosynthetic kcat/KM flux is 2-fold greater than that of h15-LOX-1, suggesting that h15-LOX-2 may have a greater role in lipoxin biosynthesis than previously thought. In addition, it is shown that oxygenation of 12S-HETE and 15S-HETE by h5-LOX is kinetically slow, suggesting that the first step in the in vitro biosynthesis of both 5S,12S-diHETE and 5S,15S-diHETE is the production of 5S-HETE.
The process of inflammation plays an important role in tissue homeostasis and is tightly regulated through the balance of pro-inflammatory and pro-resolving signals. In the initial stage of inflammation, damaged tissues produce pro-inflammatory signaling molecules, such as leukotrienes, that recruit neutrophils and macrophages to sites of injury.1 During the resolution stage of inflammation, cells generate specialized pro-resolving mediators (SPMs), such as the lipoxins,2 which limit inflammation by antagonizing leukotriene-mediated pro-inflammatory processes3–5 and enhancing phagocytosis of apoptotic neutrophils by macrophages.6 Without proper resolution, chronic inflammation can contribute to a variety of diseases, such as diabetes and cancer, to name a few.7–9
Lipoxins and leukotrienes are oxylipins produced by lipoxygenase-catalyzed oxygenation of arachidonic acid (AA).10 Lipoxygenases are non-heme iron enzymes that catalyze the stereospecific hydroperoxidation of AA, and other polyunsaturated fatty acids (PUFAs).11
Humans have one 12S-LOX isozyme and two 15S-LOX isozymes (Table 1). Human platelet 12-lipoxygenase (h12-LOX or ALOX12) is expressed in platelets and pancreatic islets of Langerhans;12 human reticulocyte 15-lipoxygenase-1 (h15-LOX-1 or ALOX15) is expressed in reticulocytes and macrophages,13,14 and human epithelial 15-lipoxygenase-2 (h15-LOX-2 or ALOX15B) is expressed in macrophages, neutrophils, epithelium, and prostate tissue.15–17 When reacting with AA, h12-LOX produces only 12S-hydroperoxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (12S-HpETE), h15-LOX-1 produces mostly 15S-hydroperoxy-5Z,8Z,11Z,13E-eicosatetraenoic acid (15S-HpETE), along with a minor amount of 12S-HpETE, while h15-LOX-2 produces only 15S-HpETE.18 These LOX isozymes are also postulated to oxygenate 5S-HpETE to produce 5S,12S-6E,8Z,10E,14Z-dihydroxyeicosatetraenoic acid (5S,12S-diHpETE)19–21 and 5S,15S-dihydroperoxy-6E,8Z,11Z,13E-eicosatetraenoic acid (5S,15S-diHpETE),22 which are reduced in the cell and are the focus of this work (Figure 1).
Table 1.
Common Protein Names for Human LOX Genes
| LOX human gene | LOX human protein |
|---|---|
| ALOX5 | h5-LOX |
| ALOX12 | h12-LOX |
| ALOX15 | h15-LOX-1, 12/15-LOX |
| ALOX15b | h15-LOX-2 |
Figure 1.
5S,12S-diHETE and 5S,15S-diHETE are the reduced forms of the LOX products, the di-HpETEs. The structures are artificially drawn in a straight line for the purpose of comparison.
Lipoxin production requires a hydroperoxide precursor that is dehydrated to form an epoxide. Formation of lipoxin A4 (LXA4) requires a hydroperoxide at C5, while formation of lipoxin B4 (LXB4) requires a hydroperoxide at C15. 5S,15S-diHpETE can serve as an intermediate in the biosynthesis of both LXA4 and LXB4 because it contains hydroperoxides at both C5 and C15 and can be converted to an epoxide by either h12-LOX or h15-LOX-1.23–26 5S,15S-diHETE, the form of 5S,15S-diHpETE reduced by glutathione peroxidase, is found in macrophages and PMNs,24,27 and its concentration is elevated in PMNs isolated from patients with asthma28 and rheumatoid arthritis.29 5S,15S-diHETE functions as an eosinophil chemoattractant,30 potentiates neutrophil degranulation,31 and inhibits platelet aggregation.23 5S,15S-diHpETE is proposed to be formed by two lipoxygenase-catalyzed dioxygenation reactions and can be produced from either 5S-HpETE or 15S-HpETE, as the starting reagent.22,24,32–34 It was postulated that h5-LOX and an h15-LOX isozyme can carry out this biosynthesis; however, the order of the reaction and the relative efficiency of these two pathways have not been examined in vitro.
5S,12S-diHETE is an isomer of leukotriene B4 (LTB4) and appears to be biosynthesized in a manner similar to that of 5S,15S-diHETE, through sequential LOX oxygenations, followed by reduction. It is produced by both platelets and neutrophils34–36 and manifests at elevated levels in patients with psoriasis.37 5S,12S-diHETE can be produced at larger amounts than LTB4 in cells38 but has lower chemotactic activity toward PMNs compared to LTB4.39–41 On the basis of cell studies, 5S,12S-diHETE is proposed to be formed through two sequential lipoxygenase-catalyzed oxygenations,19–21 with either h5-LOX reacting with 12S-HETE or h12-LOX reacting with 5S-HETE,19,20,42 followed by reduction. However, the relative efficiency of these two pathways is unknown, and there has been no investigation into a possible role of h15-LOX-1 in the formation of 5S,12S-diHETE, although it is biosynthetically feasible due to its ability to oxidize at the C12 position (vide supra).
In general, it is postulated that AA binds to the active site of both h12-LOX43 and h15-LOX-1,44 methyl-end first, with amino acid residues, 417 and 418, defining the bottom of the pocket for both LOXs.45,46 If the volumes of these two residues are enlarged by mutagenesis in h12-LOX, the percentage of 15S-HpETE production increases due to shallower positioning of AA into the active site.43,47 If the volumes of these two residues are decreased by mutagenesis in h15-LOX-1, the percentage of 12-HpETE production increases due to the deeper positioning of AA into the active site.48 This binding model for both h12-LOX and h15-LOX-1 is termed the “volume hypothesis”, where the depth of penetration of the fatty acid [i.e., AA or docosahexaenoic acid (DHA)] defines the position of the abstracted hydrogen atom and subsequently the position of oxygenation. However, if the substrate is the oxylipin of DHA, 7S-hydroperoxydocosahexaenoic acid (7S-HpDHA), this “volume” binding model is not maintained49 and h15-LOX-1 has altered positional specificity.50 Upon reaction with DHA, h15-LOX-1 primarily abstracts a hydrogen atom from the ω−8 carbon (C15) and oxygenates C17; however, upon reaction with 7S-HpDHA, h15-LOX-1 abstracts primarily from the ω−11 carbon (C12) and oxygenates on C14.
This shift in product profile raises questions about the role of h15-LOX-1 in the biosynthesis of 5S,12S-diHpETE and 5S,15S-diHpETE. If 5S-HpETE binds in a manner similar to that of 7S-HpDHA, then the presumed biosynthetic pathways of 5S,12S-diHpETE and 5S,15S-diHpETE could be redefined. In addition, the contribution of h15-LOX-2 to the formation of lipoxin intermediates has been largely overlooked because it reacts more slowly with AA compared to h15-LOX-1 and was initially thought to have limited tissue expression.15 More recent studies indicate that h15-LOX-2 reacts as well with 7S-HpDHA, producing >98% of 7S,17S-diHpDHA,50 and is expressed in primary macrophages at levels higher than those of both h15-LOX-1 and h12-LOX,17,51 suggesting a possible greater role for h15-LOX-2 in the biosynthesis of resolvin D5 (RvD5 or 7S,17S-diHDHA).
In this work, we have determined that the positional specificity of h15-LOX-1 and h15-LOX-2 with 7S-HpDHA does extend to the analogous AA-derived oxylipin, 5S-HpETE. In addition, we have measured the relative rates of h5-LOX, h12-LOX, h15-LOX-1, and h15-LOX-2 in the in vitro biosynthesis of 5S,15S-diHpETE and 5S,12S-diHpETE and have proposed possible in vivo biosynthetic pathways that could have implications for the therapeutic effect of LOX isozyme inhibition.
METHODS
Expression and Purification of h15-LOX-1, h15-LOX-2, h12-LOX, and h5-LOX.
Overexpression and purification of wild-type (wt) h15-LOX-1 (Uniprot entry P16050), h12-LOX (Uniprot entry P18054), h5-LOX (Uniprot entry P09917), and h15-LOX-2 (Uniprot entry O15296) were performed as previously described.52–54 The purity of h15-LOX-1, h15-LOX-2, and h12-LOX was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to be >90%. The metal content was assessed on a Finnigan inductively coupled plasma mass spectrometer (ICP-MS), via comparison with an iron standard solution. Cobalt-EDTA was used as an internal standard. The wt h5-LOX used in the kinetics of this work was not purified due to a dramatic loss of activity and was prepared as an ammonium sulfate precipitate. The amount of h5-LOX contained in the ammonium sulfate pellet was estimated to be approximately 1% of total protein by comparing values obtained by a Bradford assay and quantitative SDS-PAGE using a His-tagged purified Stable h5-LOX mutant as a positive control (Figure S1). The stable h5-LOX mutant was expressed in Rosetta 2 cells (Novagen) transformed with the pET14b-Stable-h5-LOX plasmid (a gift from M. Newcomer of Louisiana State University, Baton Rouge, LA), grown in Terrific Broth containing 34 μg mL−1 chloramphenicol and 100 μg mL−1 ampicillin at 37 °C for 3.5 h, and then held at 20 °C for an additional 26 h. Cells were pelleted and resuspended in 50 mM Tris (pH 8.0), 500 mM NaCl, 20 mM imidazole with 1 μM Pepstatin, 100 μM PMSF, and DNaseI (2 Kunitz/g) (Sigma). The cells were lysed in a French pressure cell and centrifuged at 40000g for 20 min. The cell lysate was applied to a Ni(II)-IDA Sepharose column and eluted with 50 mM Tris (pH 8.0), 500 mM NaCl, and 200 mM imidazole. The final product was stored at −80 °C with 10% glycerol. The kcat and kcat/KM values determined for our ammonium sulfate-precipitated h5-LOX are similar to values seen from the purified stable h5-LOX mutant55 when accounting for addition of ATP to buffers in this work, which induces a 5-fold increase in h5-LOX activity.56 Due to the difficulty determining the iron content of crude ammonium sulfate-precipitated extracted enzyme, the metalation of h5-LOX was assumed to be 100%, indicating that the estimated kinetic parameters presented in this work are lower limits.
Production and Isolation of Oxylipins.
5(S)-Hydroperoxyeicosatetraenoic acid (5S-HpETE) was synthesized by the reaction of AA (40 μM) with ~200 mg of ammonium sulfate-precipitated h5-LOX. The reaction was carried out for 1 h in 1000 mL of 25 mM HEPES (pH 7.5) containing 50 mM NaCl, 100 μM EDTA, and 200 μM ATP. The reaction was quenched with 0.5% glacial acetic acid, and the mixture was extracted three times with ⅓ volume of dichloromethane and evaporated to dryness under nitrogen. The products were purified isocratically via high performance liquid chromatography (HPLC) on a Higgins Haisil Semipreparative (5 μm, 250 mm × 10 mm) C18 column with a 50:50 ratio of 99.9% acetonitrile with 0.1% acetic acid and 99.9% water with 0.1% acetic acid. The isolated products were assessed to be >95% pure by LC-MS/MS. 5(S)-Hydroxyeicosatetraenoic acid (5S-HETE) was synthesized the same as for 5S-HpETE; however, trimethylphosphite was added as a reductant prior to HPLC. 12S-HpETE was synthesized by reaction of AA (25–50 μM) with h12-LOX and was run for 30 min in 500 mL of 25 mM HEPES (pH 8.0). 15(S)-Hydroperoxyeicosatetraenoic acid (15S-HpETE) was synthesized by reaction of AA (25–50 μM) with soybean 15-LOX and was run for 30 min in 500 mL of 50 mM sodium borate (pH 9.0). 15(S)-Hydroxyeicosatetraenoic acid (15S-HETE) and 12(S)-hydroxyeicosatetraenoic acid (12S-HETE) were synthesized the same as for 15S-HpETE and 12S-HpETE, but with trimethylphosphite added as a reductant prior to HPLC. The products were extracted and purified isocratically via HPLC, as described above for 5S-HpETE.
Steady-State Kinetics of h5LOX, h12-LOX, h15-LOX-1, and h15-LOX-2.
Steady-state kinetic reactions were performed at ambient temperature (25 °C) in a 1 cm path length quartz cuvette and monitored on a PerkinElmer Lambda 45 ultraviolet-visible (UV/Vis) spectrophotometer. Reaction mixtures contained 2 mL of either 25 mM HEPES (pH 7.5) (h15-LOX-1 and h15-LOX-2), 25 mM HEPES, (pH 8.0) (h12-LOX), or 25 mM HEPES (pH 7.5), 50 mM NaCl, 100 μM EDTA, and 200 μM ATP (h5-LOX). Substrate concentrations were varied from 0.25 to 10 μM for AA or from 0.5 to 40 μM for 5S-HETE and 5S-HpETE. Reactions were initiated by the addition of either h5-LOX (~300–900 nM final concentrations), h12-LOX (~300–900 nM final concentrations), h15-LOX-1 (~200–600 nM final concentrations), or h15-LOX-2 (~300–700 nM final concentrations) and monitored on a PerkinElmer Lambda 45 UV/Vis spectrophotometer. Product formation was determined by the increase in absorbance at 234 nm for 5S-HETE (ε234 = 27000 M−1 cm−1), 270 nm for 5S,12S-diHETE (ε270 = 40000 M−1 cm−1), and 254 nm for 5S,15S-diHETE (ε254 = 21500 M−1 cm−1).24 It should be noted that 5S,15S-diHDHA has an absorbance maximum of 245 nm; however, due to overlap with the substrate peak at 234 nm, formation of this product was measured at 254 nm using an extinction coefficient of 21900 M−1 cm−1 to isolate it from the substrate absorption band.57 KaleidaGraph (Synergy) was used to fit kcat and kcat/KM to the Michaelis-Menten equation for the calculation of kinetic parameters.
Product Analysis of Reactions of LOX with AA, 5S-HETE, and 5S-HpETE.
Reactions were carried out with stirring at ambient temperature in the buffers indicated above. Reaction mixtures with AA contained 10 μM substrate and a final concentration of either h5-LOX (~300 nM), h12-LOX (~300 nM), h15-LOX-1 (~200 nM), or h15-LOX-2 (~350 nM). Those with 5S-HETE and 5S-HpETE contained 10 μM substrate and a final concentration of either h12-LOX (950 nM), h15-LOX-1 (550 nM), or h15-LOX-2 (700 nM). Reaction mixtures with 15S-HETE and 15S-HpETE, 12S-HETE, and 12S-HpETE contained 20 μM substrate and 1100 nM h5-LOX. Reactions were monitored via an UV/Vis spectrophotometer and quenched at 50% turnover with 0.5% glacial acetic acid. Each quenched reaction mixture was extracted with DCM and reduced with trimethylphosphite. The samples were then evaporated under a stream of nitrogen to dryness and reconstituted in 50 μL of a 90% acetonitrile/10% water mixture containing 0.1% formic acid and 3 μM 13-HODE as an internal standard. Control reactions without enzymes were also conducted and used for background subtraction, ensuring oxylipin degradation products were removed from analysis. Reactions were analyzed via LC-MS/MS using a Synergi 4 μm Hydro-RP 80 Å C18 LC column with a polar end cap (150 mm × 2 mm). Mobile phase solvent A consisted of 99.9% water and 0.1% formic acid, and solvent B consisted of 99.9% acetonitrile and 0.1% formic acid. Analysis was carried out over 60 min using an isocratic 50:50 A:B system for 0–30 min followed by a gradient from 50:50 A:B to 75:25 A:B, from 30 to 60 min. The chromatography system was coupled to a Thermo-Electron LTQ LC-MS/MS instrument for mass analysis. All analyses were performed in negative ionization mode at the normal resolution setting. MS2 was performed in a targeted manner with a mass list containing the following m/z ratios: 319.4 ± 0.5 (HETEs), 335.4 ± 0.5 (diHETEs), and 351.4 ± 0.5 (triHETEs). Products were identified by matching retention times and fragmentation patterns to those of known standards, or in the cases in which MS standards were not available, structures were deduced from comparison with known and theoretical fragments. It should be noted that the rates of product formation as seen by UV detection mirrored the area of the products detected by MS, with only trace amounts of unidentified products. MS/MS fragments used to identify 5,12-diHETE included m/z 317, 299, 273, 203, 195, and 141. MS/MS fragments used to identify 5,12-diHETE included m/z 317, 299, 273, 235, 177, and 163.
Chiral Chromatography.
5S,12S-diHETE and 5S,15S-diHETE were synthesized and isolated via HPLC as described above. Purified 5S,12S-diHETE and 5S,15S-diHETE were analyzed via LC-MS/MS using a Chirapak AD-RH 2.1 mm × 150 mm, 5 μm chiral column coupled to a Thermo electron LTQ instrument. Retention times and fragmentations were compared to those of 5S,12S-diHETE, 5S,15S-diHETE, LTB4, and 6-trans-12-epi-LTB4 standards purchased from Cayman Chemicals (Ann Arbor, MI). Analysis was carried out using a gradient from 40:60 to 70:30 A:B, over 25 min. Mobile phase solvent A consisted of 99.9% acetonitrile and 0.1% formic acid, and solvent B consisted of 99.9% water and 0.1% formic acid. MS/MS conditions were the same as described above. The standards were purchased from Cayman Chemicals.
Molecular Modeling.
A structure of h15-LOX-1 (ALOX15) is not available in the Protein Data Bank (PDB). Therefore, we constructed a homology model of h15-LOX-1 from its sequence (Uniprot entry P16050) using the substrate mimetic inhibitor-bound, high-resolution structure of porcine 12-LOX (PDB entry 3rde). The h15-LOX-1 sequence is 86% identical to the porcine 12-LOX sequence, with both being considered ALOX15 genes. The homology model was constructed using Prime (version 5.4, Schrodinger Inc.). During the modeling step, we retained the co-crystallized inhibitor, metal ion (Fe3+), and a hydroxide ion that coordinated the metal ion from the homologue structure. The model was subsequently energy minimized using the Protein Preparation Wizard module of Maestro (version 11.8, Schrodinger Inc.). During this step, hydrogen atoms were added to the protein, co-crystallized ligand, and the hydroxide ion. Hydrogen atoms of the titratable residues were adjusted, and side chains optimized so they could make better hydrogen bonding interactions. The structure was finally minimized such that the heavy atoms did not move beyond 0.3 Å from their starting positions. Oxylipins were modeled using the Edit/Build panel of Maestro; their energy was minimized using LigPrep software (Schrodinger Suite 2018–4, Schrodinger Inc.), and they were docked to the h15-LOX-1 model using the docking software Glide (version 8.1, Schrodinger Inc.) with the extra-precision (XP) docking score. We used the co-crystallized ligand coordinates to define the binding pocket. During the ligand docking step, the protein was initially kept rigid, but despite extensive ligand conformation sampling, no low-energy docking pose was identified for the ligands. Therefore, we used InducedFit docking (Schrodinger Inc.), in which residues in the active site of the protein and ligand are treated flexibly, with docking and side chain optimization steps being iterated until a converged low-energy docking pose was obtained.
Oxylipin Titration into Human Platelets.
The University of Michigan Institutional Review Board approved all research involving human volunteers. Washed platelets were isolated from human whole blood via serial centrifugation and adjusted to 3.0 × 108 platelets/mL in Tyrode’s buffer (10 mM HEPES, 12 mM NaHCO3, 127 mM NaCl, 5 mM KCl, 0.5 mM NaH2PO4, 1 mM MgCl2, and 5 mM glucose), as previously published.58 Platelets (250 μL at a density of 3.0 × 108 platelets/mL) were dispensed into glass cuvettes and incubated with the indicated 5S-HETE at 1, 10, and 15 μM for 10 min at 37 °C. Oxylipin-treated platelets were stimulated with 0.25 μg/mL collagen (Chrono-log), under stirring conditions (1100 rpm) at 37 °C, in a Chrono-log model 700D lumi-aggregometer, and platelet aggregation was recorded for 6 min.
To determine if 5S-HETE was enzymatically converted to another chemical ex vivo, 1 mL of platelets (1.0 × 109 platelets/mL) was incubated with 10 μM 5S-HETE or vehicle (DMSO) for 10 min at 37 °C and then pelleted by centrifugation at 1000g for 2 min. The supernatant was transferred to a fresh tube and snap-frozen. Oxylipins were extracted and analyzed via LC-MS/MS, as described previously.9 The m/z transitions for 5S-HETE, 5S,12S-diHETE, and 5S,15S-diHETE were 319.4 → 205.4, 335.4 → 195.4, and 351.4 → 235.4, respectively.
RESULTS
Biosynthesis of 5S,15S-diHpETE.
The two pathways by which 5S,15S-diHpETE might be synthesized depend on the order of the LOX isozyme reaction (Scheme 1, pathways 1 and 2). These two pathways were therefore investigated to compare the relative ability of h5-LOX, h15-LOX-1, and h15-LOX-2 to form the pathway intermediates in vitro.
Scheme 1.
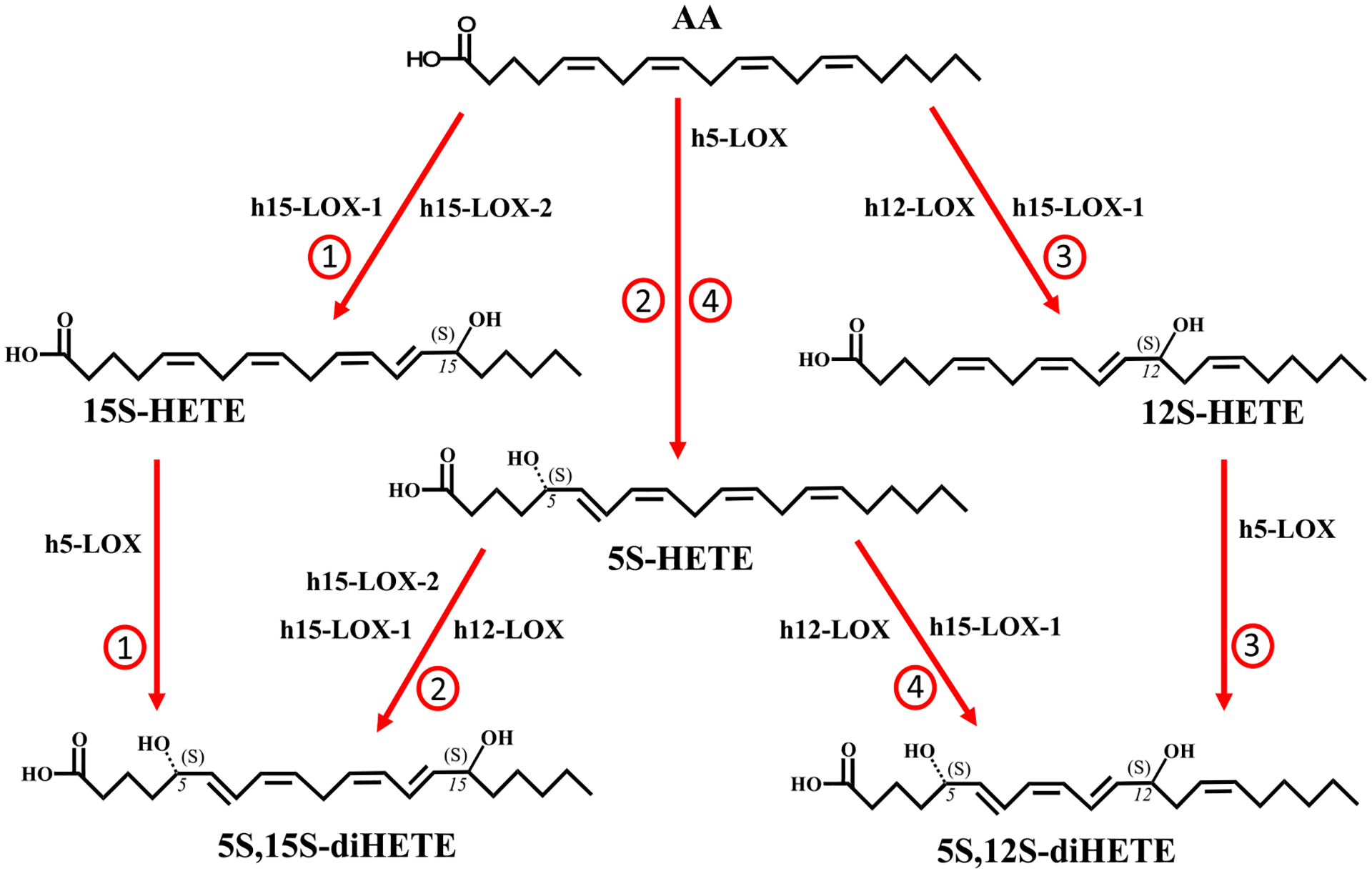
Possible Biosynthetic Pathways of 5S,12S-diHETE and 5S,15S-diHETEa
aIt should be noted that LOXs initially produce hydroperoxides that are subsequently reduced by glutathione peroxidases. The reduced products have been presented for the sake of simplicity.
Pathway 1: AA and h15-LOX-1 or h15-LOX-2 to 15S-HpETE, and Then h5-LOX to 5S,15S-diHpETE (Scheme 1).
The first possible route that may contribute to the formation of 5S,15S-diHpETE involves oxidation of AA by h15-LOX-1 or h15-LOX-2 to form 15S-HpETE. Therefore, the steady-state kinetics and product profiles were previously measured; h15-LOX-1 was found to have a kcat with AA of 10 s−1, while the kcat/KM was s−1 2.1 μM−1 (Table 2).50 The major product of the reaction was 15S-HpETE (84%), with 12S-HpETE comprising the remaining 16% (Table 3), as previously observed.47 The kcat for AA with h15-LOX-2 was measured to be 0.96 s−1, while the kcat/KM was found to be 0.19 s−1 μM−1 (Table 2).50 The reaction of h15-LOX-2 with AA produced 100% 15S-HpETE, as previously observed.15,18 Multiplying the kcat/KM values by product percentages allows calculation of the biosynthetic flux. Comparing these biosynthetic flux data indicates that h15-LOX-1 (2.1 s−1 μM−1 × 0.84 = 1.8) is 10-fold more efficient than h15-LOX-2 (0.19 s−1 μM−1 × 1 = 0.19) in producing 15S-HpETE (Table 4).
Table 2.
Steady-State Kinetic Values of h5-LOX, h12-LOX, h15-LOX-1, and h15-LOX-2 with Substrates That Serve as Intermediates in the Biosynthesis of 5S,12S-diHpETE and 5S,15S-diHpETE
| enzyme | substrate | kcat (s−1) | kcat/KM (s−1 μM−1) | KM (μM) |
|---|---|---|---|---|
| h5-LOX | AAa | 1.2 ± 0.4 | 0.35 ± 0.04 | 3.3 ± 0.9 |
| h12-LOX | AAb | 9.7 ± 1 | 5.8 ± 0.4 | 1.7 ± 0.3 |
| h12-LOX | 5S-HETE | 0.088 ± 0.006 | 0.051 ± 0.01 | 1.7 ± 0.4 |
| h12-LOX | 5S-HpETE | 0.15 ± 0.01 | 0.15 ± 0.02 | 1.0 ± 0.2 |
| h15-LOX-1 | AAb | 10 ± 1 | 2.1 ± 0.3 | 5.0 ± 0.3 |
| h15-LOX-1 | 5S-HETE | 1.1 ± 0.4 | 0.23 ± 0.03 | 4.9 ± 0.6 |
| h15-LOX-1 | 5S-HpETE | 1.8 ± 0.2 | 0.23 ± 0.04 | 7.8 ± 2 |
| h15-LOX-2 | AAb | 0.96 ± 0.09 | 0.19 ± 0.02 | 5.0 ± 0.2 |
| h15-LOX-2 | 5S-HETE | 2.1 ± 0.2 | 0.072 ± 0.008 | 29 ± 2 |
| h15-LOX-2 | 5S-HpETE | 2.0 ± 0.2 | 0.035 ± 0.004 | 57 ± 3 |
Table 3.
Distribution of Products Created by Reaction of h5-LOX, h12-LOX, h15-LOX-1, and h15-LOX-2 with Substrates That Serve as Intermediates in the Biosynthesis of 5S,12S-diHpETE and 5S,15S-diHpETEa
| enzyme | substrate | C5 product (%) | C12 product (%) | C15 product (%) |
|---|---|---|---|---|
| h5-LOX | AAb | 100 | – | – |
| 12S-HpETE | 100 | – | – | |
| 15S-HETE | 100 | – | – | |
| 15S-HpETE | 100 | – | – | |
| h12-LOX | AA | – | 100 | – |
| 5S-HETE | – | 87 ± 4 | 13 ± 4 | |
| 5S-HpETE | – | 90 ± 3 | 10 ± 3 | |
| h15-LOX-1 | AA | – | 16 ± 5 | 84 ± 5 |
| 5S-HETE | – | 86 ± 6 | 14 ± 6 | |
| 5S-HpETE | – | 61 ± 6 | 39 ± 6 | |
| h15-LOX-2 | AA | – | – | 100 |
| 5S-HETE | – | – | 100 | |
| 5S-HpETE | – | – | 100 |
The product label (i.e., C5 product) refers to the carbon that is oxygenated on the substrate.
The epoxide product from 5S-HpETE was observed at low levels and considered a derivative of 5S-HpETE.
Table 4.
Biosynthetic Flux of Reactions Leading to the Production of 5S,12S-diHETE and 5S,15S-diHETEa
| enzyme | substrate | kcat/KM (s−1 μM−1)c | 5S,12S-diHETE flux | 5S,15S-diHETE flux |
|---|---|---|---|---|
| h5-LOX | AAb | 0.35 | 0.35 | 0.35 |
| h12-LOX | AA | 5.8 | 5.8 | – |
| h12-LOX | 5S-HETE | 0.051 | 0.044 | 0.0066 |
| h12-LOX | 5S-HpETE | 0.15 | 0.14 | 0.015 |
| h15-LOX-1 | AA | 2.1 | 0.34 | 1.8 |
| h15-LOX-1 | 5S-HETE | 0.23 | 0.20 | 0.032 |
| h15-LOX-1 | 5S-HpETE | 0.23 | 0.14 | 0.089 |
| h15-LOX-2 | AA | 0.19 | – | 0.19 |
| h15-LOX-2 | 5S-HETE | 0.072 | – | 0.072 |
| h15-LOX-2 | 5S-HpETE | 0.035 | – | 0.035 |
Biosynthetic flux is calculated by multiplying each kcat/KM value (s−1 μM–1) by the percentage of total product from each reaction that serves as the intermediate in the synthesis of either 5S,12S-diHETE or 5S,15S-diHETE.
The AA flux values are also listed as they produce intermediates in the biosynthetic pathway, such as 5S-HpETE, 12S-HpETE, and 15S-HpETE.
kcat/KM error values are listed in Table 2.
The 15S-HpETE formed by h15-LOX-1 or h15-LOX-2 can then react with h5-LOX to form 5S,15S-diHpETE. The ability of h5-LOX to carry out this reaction was assessed using UV-vis spectroscopy, but no reaction was observed at either 234, 270, or 302 nm. However, upon analysis of multiple time points with LC-MS/MS, a small amount of product was observed from both 15S-HETE and 15S-HpETE, consisting entirely of 5S,15S-diHpETE. Although kcat and KM values could not be obtained for these reactions due to their slow rates, the V10 values for 15S-HETE and 15S-HpETE were determined to be 0.0013 and 0.002 mol s−1 mol−1 at 10 μM substrate, respectively (Table 5). Compared with the rate of h5-LOX with AA at 10 μM, these values for 15S-HETE and 15S-HpETE catalysis with h5-LOX are 130- and 85-fold slower, respectively.
Table 5.
V10 Values (moles per second per mole) of h5-LOX with 10 μM Substrate (i.e., AA, 12S-HETE, 12S-HpETE, 15S-HETE, and 15S-HpETE)a
| enzyme | substrate | V10 (mol s−1 mol−1) |
|---|---|---|
| h5-LOX | AA | 0.17 ± 0.05 |
| h5-LOX | 15S-HETE | 0.0013 ± 0.0004 |
| h5-LOX | 15S-HpETE | 0.0020 ± 0.001 |
| h5-LOX | 12S-HETE | 0.00050 ± 0.0003 |
| h5-LOX | 12S-HpETE | 0.0030 ± 0.001 |
V10 is defined as the moles of product produced per second per mole of enzyme (moles per second per mole) at 10 μM substrate.
Pathway 2: AA and h5-LOX to 5S-HpETE, Then h15-LOX-1, h15-LOX-2, or h12-LOX to 5S,15S-diHpETE (Scheme 1).
The second route of formation of 5S,15S-diHpETE begins with the oxidation of AA by h5-LOX to form 5S-HpETE. The kinetics and product profile of this reaction determined a k of 1.2 s−1 cat and a kcat/KM of 0.35 s−1 μM−1 (Table 2) and produced 98% 5S-HpETE and its 5,6-epoxide (Table 3), with trace amounts of other oxylipins. These values are consistent with those obtained from a purified stable h5-LOX mutant55 when accounting for addition of ATP to buffers in this work, which induces a 5-fold increase in h5-LOX activity.56 It should be noted that these values are from protein estimates of the crude preparation utilizing a SDS-PAGE assay, and its iron occupancy was assumed to be 100%.
5S-HpETE generated by h5-LOX can further react with a h15-LOX isozyme to form 5S,15S-diHpETE. The kcat of 5S-HpETE with h15-LOX-1 was 1.8 s−1, and the kcat/KM was 0.23 s−1 μM−1 (Table 2). The major product of the reaction was 5S,12S-diHpETE (61% of the total), with 5S,15S-diHpETE comprising only 39% (Table 3). The kcat of 5S-HETE with h15-LOX-1 was 1.1 s−1, and the kcat/KM was 0.23 s−1 μM−1 (Table 2); however, the majority of the product produced was 5S,12S-diHpETE (86% of the total), with 5S,15S-diHpETE comprising only 14% (Table 3). This unusual result was previously observed in the reaction of h15-LOX-1 with 7S-HpDHA to produce the noncanonical ω−9 product, 7S,14S-diHpDHA.50 In contrast, h15-LOX-2 and 5S-HETE displayed a k of 2.1 s−1 cat and a kcat/KM of 0.072 s−1 μM−1 (Table 2), producing only the canonical product, 5S,15S-diHpETE. The biosynthetic flux values of these two reactions indicate that h15-LOX-2 (0.072 s−1 μM−1 × 1 = 0.072) is more efficient than h15-LOX-1 (0.23 s−1 μM−1 × 0.14 = 0.032) in producing 5S,15S-HpETE (Table 4).
Biosynthesis of 5S,12S-diHETE.
In addition to identification of 5S,15S-diHETE in biological samples, 5S,12S-diHETE has also been observed,34–36 indicating its biological importance. Therefore, each of the two possible biosynthetic routes of the oxidized form, 5S,12S-diHpETE (Scheme 1, pathways 3 and 4), was investigated to compare their relative efficiency in vitro.
Pathway 3: AA and h12-LOX or h15-LOX-1 to 12S-HpETE, Then h5-LOX to 5S,12S-diHpETE (Scheme 1).
The first possible route for generating 5S,12S-diHpETE begins when h12-LOX oxidizes AA to form 12S-HpETE. The kcat for AA with h12-LOX was shown to be 9.7 s−1; the kcat/KM was found to be 5.8 s−1 μM−1 (Table 2),50 and the reaction produced 100% 12S-HpETE (Table 3), consistent with previously published values.59 Along with h12-LOX, h15-LOX-1 also contributes to the formation of 12S-HpETE, producing 14% 12S-HpETE and 86% 15S-HpETE, with a kcat of 10 s−1 and a kcat/KM of 2.1 s−1 μM−1 (Table 2), similar to previous work.47,60,61 Multiplying their kcat/KM values by their product percentages results in biosynthetic flux values of 5.8 for h12-LOX and 0.34 for h15-LOX-1, indicating that h12-LOX is a 16-fold more efficient in producing 12S-HpETE than h15-LOX-1 (Table 4).
Once 12S-HpETE is produced by h12-LOX or h15-LOX-1, it may react further with h5-LOX to form 5S,12S-diHpETE. The ability of h5-LOX to carry out this reaction was investigated; however, no reaction was observed at either 234, 270, or 302 nm, using UV-vis spectroscopy. The more sensitive LC-MS/MS method (vide supra) detected small amounts of only 5S,12S-diHpETE from 12S-HETE and 12s-HpETE with V10 values of 0.0005 and 0.003 mol s−1 mol−1, respectively (Table 5). Compared with that of AA, the values for 12S-HETE and 12S-HpETE reactivity are 340- and 57-fold slower, respectively.
Pathway 4: AA and h5-LOX to 5S-HpETE, Then h12-LOX and h15-LOX-1 to 5S,12S-diHpETE (Scheme 1).
As reported above, the reaction of h5-LOX and AA was found to have a kcat of 1.2 s−1 and a kcat/KM of 0.35 s1 μM−1 cat, with the major product of the reaction being 5S-HpETE, which can subsequently be a substrate for h12-LOX. Investigating the kinetic and product profile of h12-LOX reacting with 5S-HpETE revealed that 87% of the product made was 5S,12S-diHpETE, while 13% was 5S,15S-diHpETE (Table 3). The kcat of h12-LOX with 5S-HETE was 0.088 s−1, and the kcat/KM was 0.051 s−1 μM−1, which are 110- and 114-fold slower than with AA, respectively (Table 2). In comparing the biosynthetic flux values of h12-LOX and h15-LOX-1, we found h12-LOX to have a flux 5-fold lower than that of h15-LOX-1 in producing 5S,12S-diHpETE (0.051 s−1 μM−1 × 0.87 = 0.044 and 0.23 s−1 μM−1 × 0.85 = 0.20 s−1 μM−1, respectively) (Table 4). This dramatic decrease in the catalytic activity of h12-LOX with 5S-HpETE indicates that h15-LOX-1 may be involved in a novel biosynthetic pathway for producing 5S,12S-diHpETE.
Chiral Chromatography.
When reacting with polyunsaturated fatty acids (PUFAs), h5-LOX, h12-LOX, h15-LOX-1, and h15-LOX-2 produce an S-configured hydroperoxide with an E,Z-conjugated double bond.62,63 Therefore, it was assumed that the doubly oxygenated products of the work presented here, 5S,12S-diHpETE and 5S,15S-diHpETE, had similar regio- and stereospecificity, as previously seen with the 7S-HpDHA LOX products.50 This assumption was confirmed when 5S,12S-diHpETE and 5S,15S-diHpETE were analyzed via LC-MS/MS using a reverse-phase chiral C18 column, demonstrating retention times and fragmentations identical to those of known standards (Figures S2–S4). As controls, an LTB4 standard eluted at 6.6 min, while a 6-trans-12-epi-LTB4 standard eluted at 11.6 min, demonstrating adequate separation of these isomers.
To assess adequate separation of 5R,12S-diHpETE from 5S,12S-diHpETE, h15-LOX-1 was reacted with both 5R-HETE and 5S-HETE and reduced to the alcohol, resulting in two peaks with retention times of 4.2 and 5.5 min, respectively. Similarly, adequate separation of 5R,15S-diHETE from 5S,15S-diHETE was assessed by reacting h15-LOX-2 with both 5R-HETE and 5S-HETE, resulting in two peaks with retention times of 17.5 and 27.8 min, respectively (Figures S3 and S4).
To further differentiate the double bond geometry of the various products, the UV spectra of the products were compared to those of known standards. The 5,12-diHETE isomers contain a central peak at ~270 nm, flanked by a shoulder at ~260 nm that is of greater intensity than the shoulder at ~280 nm, indicating an EZE configuration.64 Parenthetically, products with EEZ double bond geometry contain shoulders at ~260 and ~280 nm with approximately equal intensity; however, products with EEE geometry have a shoulder with slightly higher intensity at 280 nm compared to that at 260 nm.65–67
Finally, it was confirmed that the products were the diHpETE products by two methods. First, enzymatic products were still observed when the substrate was 5-HETE. An epoxide can be formed only by reacting 5-HpETE with LOX, so the fact that product is formed with 5-HETE is consistent with oxygenation on C12 or C15, depending on the LOX used. Second, the product from 5-HETE was extracted with and without a reductant, and a shift in retention times was observed. This is due to the fact that the hydroperoxide and alcohol have distinct retention times during HPLC, so when the reductant reduces the hydroperoxide to the alcohol, a shift in the retention time is observed. If the products were from hydrolysis of the epoxide, then no hydroperoxide would be formed and thus no shift in retention time would be observed.
5S-HETE Docking to h15-LOX-1 and h15-LOX-2.
5S-HETE was docked against the active site of h15-LOX-1 and h15-LOX-2 (Figure 2), which demonstrated a methyl-end first U-shaped binding mode in both proteins. The carboxylate group of 5S-HETE makes a hydrogen bond with the Gln595 residue of h15-LOX-1, and the hydroxyl group makes a hydrogen bond with the backbone carbonyl oxygen atom of the Ile399 residue (Figure 2A). The hydrophobic tail of 5S-HETE is buried deep in the hydrophobic pocket created by Phe352, Ile417, and Ile592. This docking mode is consistent with our previous docking pose with 7S-HDHA; however, the carboxylic acid of 7S-HDHA hydrogen bonds with Arg402, due to its increased length relative to that of 5S-HETE.50 It is interesting to note that Arg402 was implicated in AA binding, but it is not predicted in our docking model.68
Figure 2.
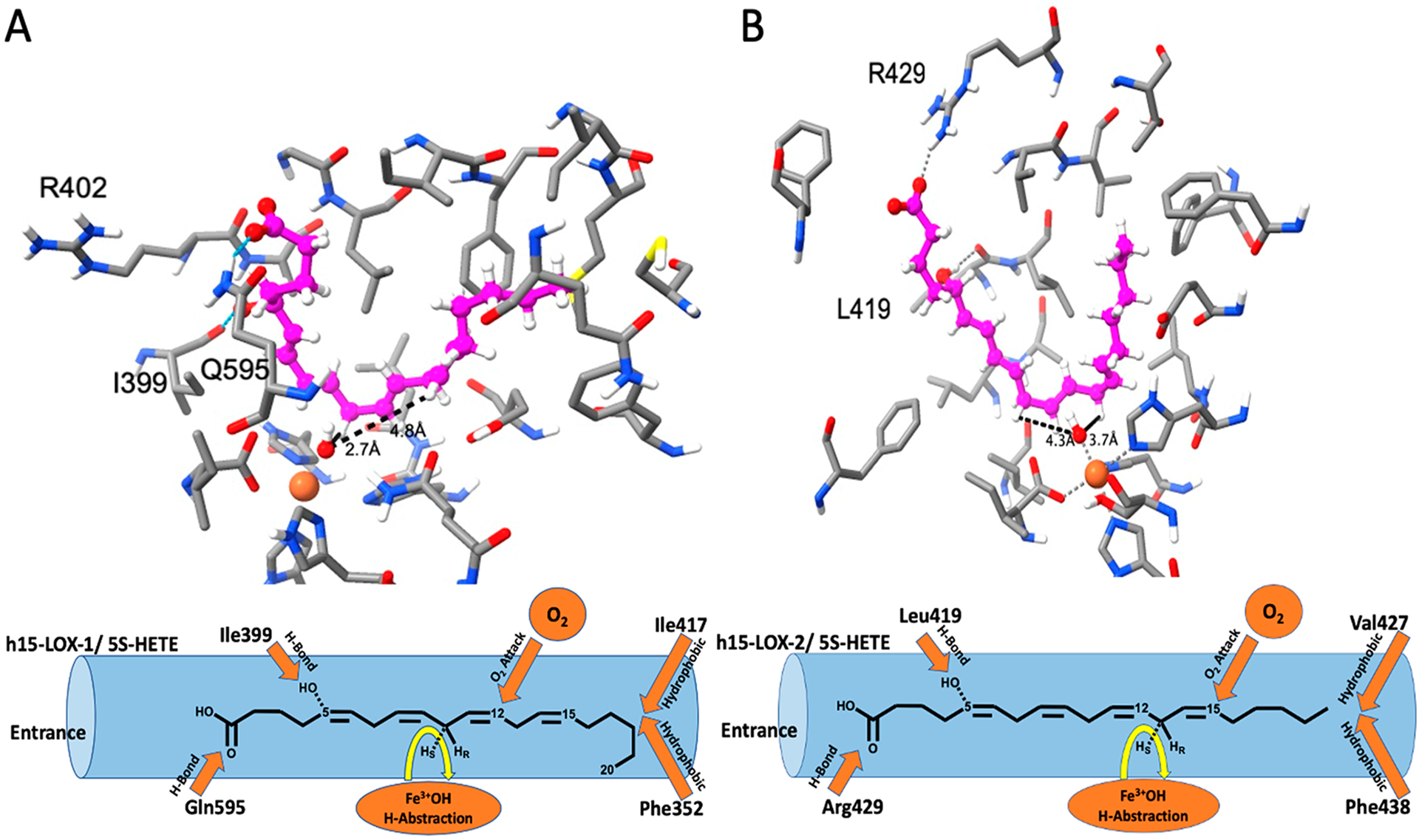
InducedFit docking pose of 5S-HETE against (A) h15-LOX-1 and (B) h15-LOX-2. 5S-HETE, metal ion (Fe3+), and hydroxide ion are shown in ball-and-stick representation, and protein residues in the active site are shown in stick representation. Carbon atoms of 5s-HETE and proteins are colored magenta and gray, respectively, oxygen, hydrogen, nitrogen, and Fe3+ ion are colored red, white, blue, and orange, respectively. The distances between the hydroxide oxygen atom and the pro-S hydrogen of C10 and C13 reactive carbons are also shown. In addition, we have also labeled residues that form hydrogen bonds to the substrate. Below each docking structure is a cartoon.
In the case of h15-LOX-2, the hydrophobic tail of 5S-HETE is buried deeply in the hydrophobic pocket created by residues Phe365, Leu420, Ile421, Val427, Phe438, and Leu607, with the carboxylate group forming a hydrogen bond with Arg429. An additional hydrogen bond interaction is observed between the hydroxyl group of 5S-HETE and the backbone carbonyl oxygen of residue Leu419 (Figure 2B). This pose is also consistent with the previous model proposed for 7S-HDHA with the exception of the carboxylic acid interaction.50 The carboxylate of 7S-HDHA forms a salt bridge with Arg429, whereas the carboxylate of 5S-HETE forms a single hydrogen bond with Arg429 due to its reduced length.
Distances between pro-S hydrogen atoms on the abstracted carbons, C10 and C13, and the reactive hydroxide oxygen atom in the active site are shown in Figure 2 and in Table 6. In the case of h15-LOX-1, the pro-S hydrogen of C10 is 2.7 Å from the hydroxide oxygen atom, while the pro-S hydrogen of C13 is 4.8 Å from the hydroxide oxygen atom, supporting the experimental results where h15-LOX-1 produces 5S,12S-diHETE. However, in the case of h15-LOX-2, the pro-S hydrogen atom of C13 is 3.7 Å from the hydroxide oxygen atom, whereas the pro-S hydrogen atom of C10 is 4.3 Å from the hydroxide oxygen atom, supporting the experimental results where h15-LOX-2 produces 5S,15S-diHETE. Similar differences in abstraction distances were also observed with 7S-HDHA docking poses.50
Table 6.
Distances between the Hydroxide Oxygen Atom and the pro-S Hydrogens of the Reactive Carbons (C10 and C13) of 5S-HETE against h15-LOX-1 and h15-LOX-2
| enzyme | hydroxide oxygen-pro-S C10 hydrogen distance (Å) | hydroxide oxygen-pro-S C13 hydrogen distance (Å) |
|---|---|---|
| h15-LOX-1 | 2.7 | 4.8 |
| h15-LOX-2 | 4.3 | 3.7 |
Platelet Reactivity toward 5S-HETE.
The presence of 5S,12S-diHETE in vivo has been assumed to result from the activity of h5-LOX and h12-LOX; however, our current work (vide supra) indicates that 5S-HETE is a poor substrate for h12-LOX in vitro. To assess the ability of h12-LOX to react with 5S-HETE in vivo, thrombin-activated platelets were incubated with 5S-HETE and analyzed for the presence of 5S-HETE metabolites using LC-MS/MS. Unreacted 5S-HETE was detectable in the samples; however, no 5S-HETE metabolites were detectable down to 2 ng/mL. 12S-HETE was detectable from endogenous AA, indicating that the platelets contained active h12-LOX. The lack of any detectable 5S,12S-diHETE is consistent with the in vitro finding that 5S-HETE is a poor substrate for h12-LOX and suggests that another enzyme, such as h15-LOX-1, may be the source of 5S,12S-diHETE observed in vivo. It should be noted that 5S,12S-diHETE was previously observed when 5S-HETE was added to platelets; however, the conditions of the previous work were distinct from our conditions.19 We are currently further investigating these confounding results to understand the details of the in vivo activity of h12-LOX in platelets.
To determine the platelet activity of 5S,12S-diHETE, it was also titrated into activated platelets and found to have low potency, 50 ± 20% aggregation at 10 μM (Figure 3). This is in contrast to the case for 12S-HETrE, which demonstrated 5 ± 2% aggregation at 10 μM, and 5S,15S-diHETE, which previously demonstrated 0 ± 2% aggregation at 10 μM.23 Despite the low potency of 5S,12S-diHETE in reducing the level of platelet aggregation, the fact that it is observed at high levels in biological samples34–38 suggests that 5S,12S-diHETE may have another, as yet unidentified, role in other biological processes.
Figure 3.
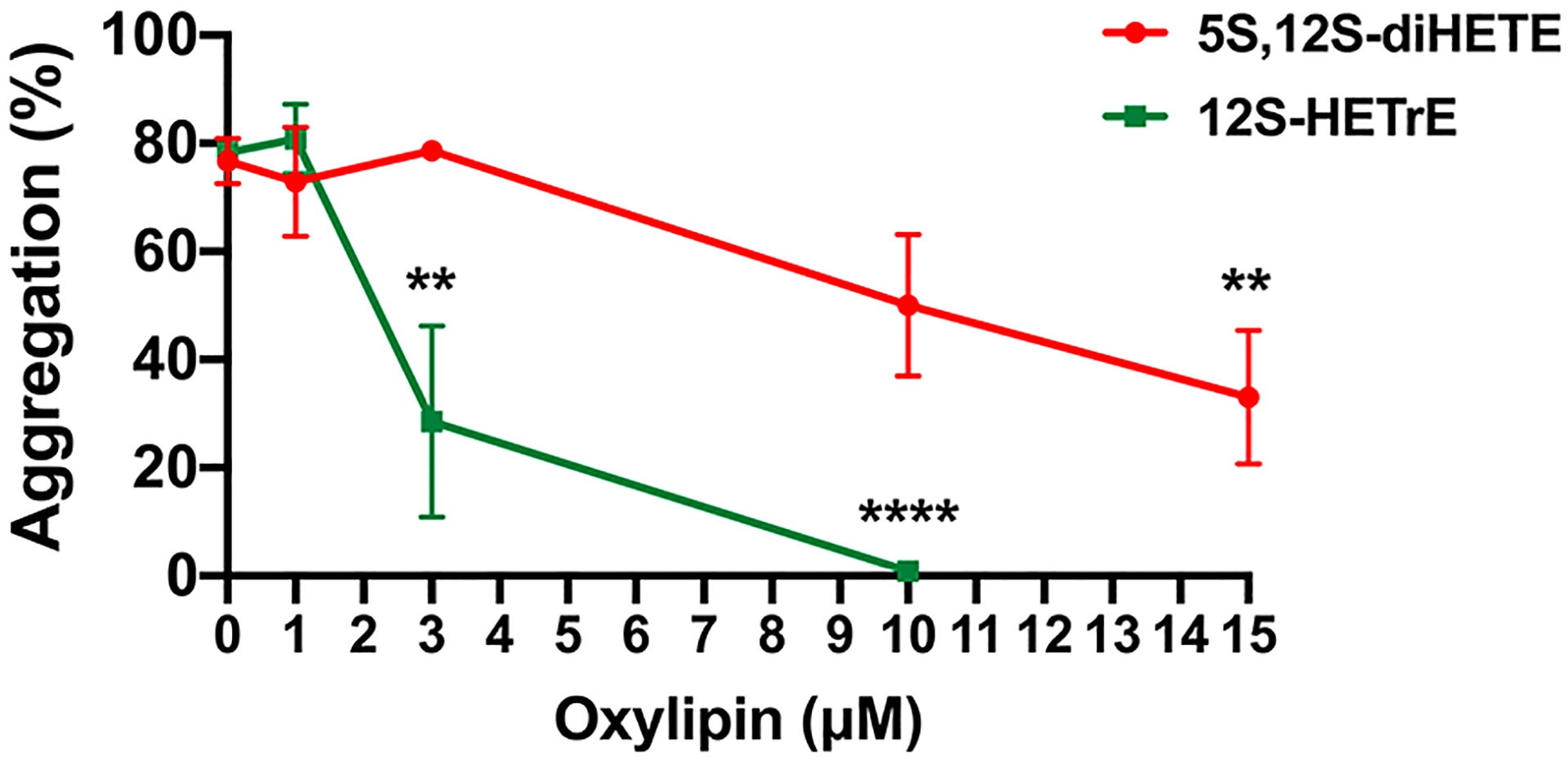
Effect of 5S,12S-diHETE on the aggregation of activated platelets. Washed human platelets were treated with vehicle (DMSO) or 5S,12S-diHETE or 12S-HETrE in half-log increments ranging from 1 to 10 μM for 10 min and then stimulated with collagen (0.25 μg/mL) in an aggregometer. Data represent the mean ± the standard error of the mean of six independent experiments. Statistical analysis was performed using one-way analysis of variance with Dunnett’s test comparing the aggregation of platelets treated with each concentration of oxylipin to the aggregation of vehicle-treated platelets. **P < 0.01; ***P < 0.001.
DISCUSSION
Biosynthesis of 5S,15S-diHpETE.
There are two pathways for synthesizing 5S,15S-diHpETE. Pathway 1 involves the formation of 15S-HpETE by either h15-LOX-1 or h15-LOX-2, followed by reaction with h5-LOX to form 5S,15S-diHpETE. Formation of 15S-HpETE occurs rapidly through oxidation of AA by h15-LOX-1, with kcat and kcat/KM values that are 15- and 12-fold greater than that of h15-LOX-2, respectively. However, once formed, 15S-HpETE and its reduced form, 15S-HETE, are comparatively poor substrates for h5-LOX. This is consistent with previous work indicating that h5-LOX reacts poorly with oxygenated fatty acids.50,57 The slow reaction of h5-LOX with 15S-HpETE makes this a rate-limiting step and indicates that it is a relatively poor mechanistic pathway for producing 5S,15S-diHpETE in vitro.
In pathway 2, 5S-HpETE is first formed through oxygenation of AA by h5-LOX and is subsequently oxidized by an h15-LOX isozyme to form 5S,15S-diHpETE. h5-LOX reacts rapidly with AA to generate a 100% yield of 5S-HpETE. This is in contrast to the reaction of h5-LOX and DHA, which produces multiple mono-oxylipins, including 7S-HpDHA, 14S-HpDHA, and 17S-HpDHA,50 and implicates the added length and unsaturation of DHA in the decrease in the product selectivity of h5-LOX. After 5S-HpETE is formed, the second step in pathway 2 is the formation of 5S,15S-diHpETE, which has been assumed to be performed by h15-LOX-1. This is a reasonable assumption, given that h15-LOX-1 primarily oxygenates C15 of AA, and is markedly faster at reacting with fatty acids than h15-LOX-2.61 However, the role of h15-LOX-1 in this pathway may be less significant, given that h15-LOX-1 shows an altered positional specificity with 7S-HpDHA, primarily oxygenating the ω−9 carbon, C14, instead of the expected ω−6 carbon, C17.50 This altered positional specificity is confirmed in the current work, where h15-LOX-1 is shown to primarily oxygenate at the ω−9 carbon, C12, when reacting with 5S-HpETE. This is a significant result as it demonstrates that the altered positional specificity seen with 7S-HpDHA also occurs with 5S-HpETE and indicates this is a new canonical reaction for h15-LOX-1. A model was previously developed for the binding of 7S-HDHA to h15-LOX-1, where the C7 alcohol hydrogen bonded with the backbone carbonyl of Ile399, thus pushing the methyl end of 7S-HDHA farther into the active site, allowing for the generation of 7S,14S-diHDHA. A similar docking procedure was performed with 5S-HETE, and a similar binding mode was observed between the C5 alcohol hydrogen and the backbone carbonyl of Ile399. This supports the hypothesis of the methyl-end binding registration, because both C7 of 7S-HDHA and C5 of 5S-HETE are in the ω−16 position and hence are positioned to interact with the backbone of Ile399. It should also be noted that Ile399 is conserved in mouse and porcine Alox15 proteins. Considering that these gene products produce more 12-HpETE than 15-HpETE, it is interesting to speculate that they would also produce 5,12-diHpETE if 5-HpETE were the substrate. We are currently testing this hypothesis.
With h15-LOX-1 primarily generating 5S,12S-diHpETE from 5S-HpETE, this indicates that h15-LOX-2 may play a larger role in making 5S,15S-diHpETE and its reduced counterpart, 5S,15S-diHETE, than previously thought (Scheme 2). This work supports this hypothesis, with h15-LOX-2 producing 5S,15S-diHpETE with a biosynthetic flux that is 2-fold greater than that of h15-LOX-1, despite the fact that h15-LOX-1 has 10-fold higher activity with AA than does h15-LOX-2 (Scheme 2). Therefore, the most kinetically favorable in vitro route in producing 5S,15S-diHpETE is the formation of 5S-HpETE by h5-LOX, followed by oxygenation at C15 by h15-LOX-2 or h15-LOX-1 (Scheme 2). This unrecognized role of h15-LOX-2 in the synthesis of 5S,15S-diHpETE is mirrored by previous work showing that h15-LOX-2 is more efficient than h15-LOX-1 in forming RvD5, due to the altered positional specificity of h15-LOX-1 with 7S-HpDHA and reinforces our hypothesis of a greater role of h15-LOX-2 in the biosynthesis of oxylipins.50
Scheme 2.
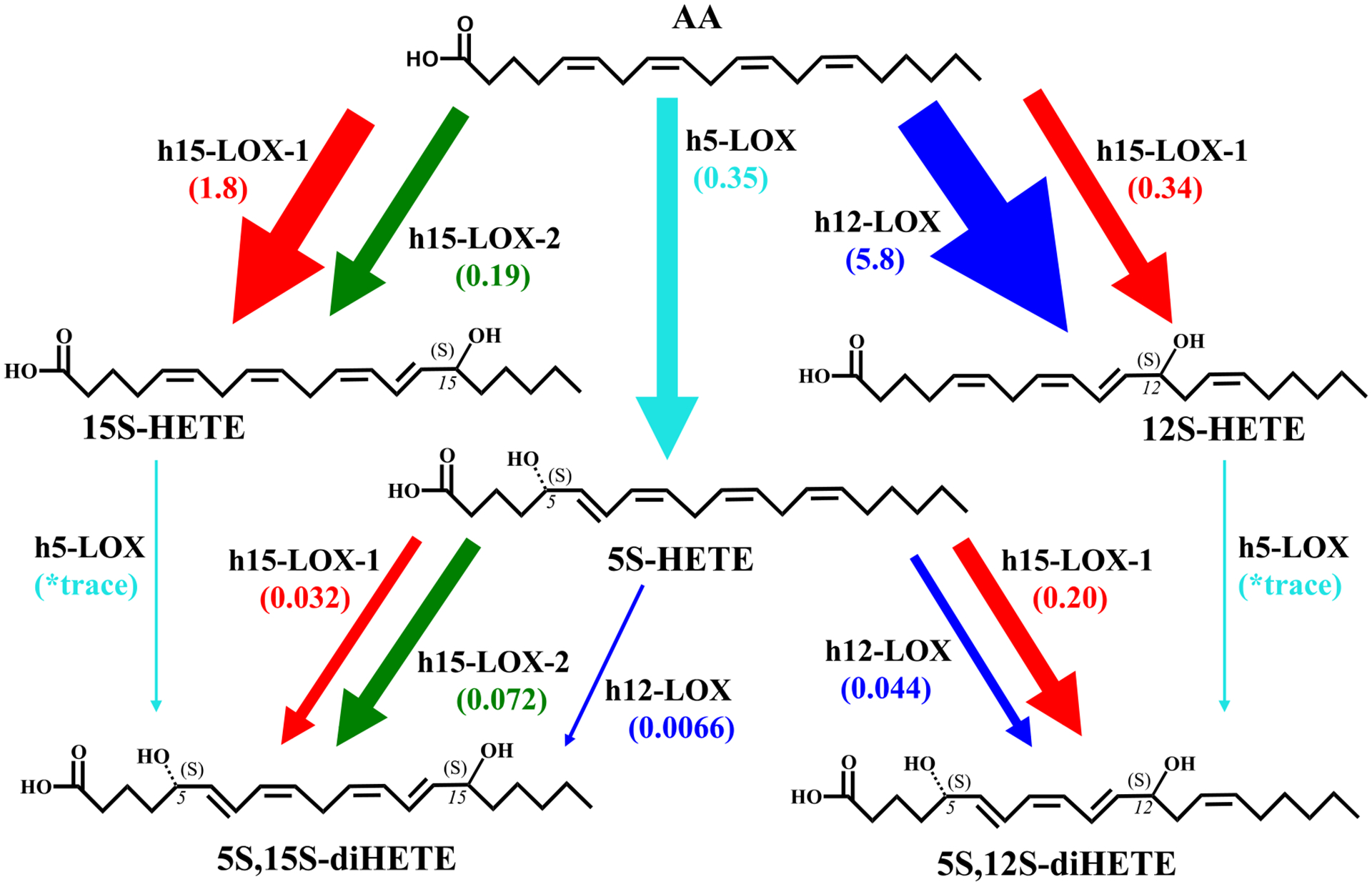
5S,12S-diHETE and 5S,15S-diHETE Biosynthesis Pathwaysa
aThe total flux values for the reactions of h5-LOX, h12-LOX, h15-LOX-1, and h15-LOX-2 with AA, 12S-HETE, 15S-HETE, and 5S-HETE are compared by multiplying kcat/KM values (s–1 μM–1) by the percentage of total product from each reaction that serves as the intermediate in the synthesis of 5S,12S-diHETE or 5S,15S-diHETE. An asterisk indicates that flux values are not reported for h5-LOX due to its low reactivity and its inability to manifest a kcat/KM value. It should be noted that LOXs initially produce hydroperoxides that are subsequently reduced by glutathione and other cellular peroxidases. Only reduced products are shown for the sake of simplicity.
If we compare the LOX biosynthetic V10 flux values between AA and DHA and their derivatives, we observe some illuminating similarities and differences. The V10, defined as the moles of product produced per second per mole of enzyme at 10 μM substrate, of h5-LOX with AA is comparable to that of DHA at 10 μM (0.17 and 0.14 mol s−1 mol−1, respectively);50 however, given the lower percentage of 7S-HpDHA produced (52%), the flux value is 2-fold lower for DHA. Upon comparison of 5S,15S-diHpETE and 7S,17S-diHpDHA production, the kcat/KM flux value for 7S,17S-diHpDHA is 48-fold greater for h12-LOX (0.32 vs 0.0066), 1.6-fold greater for h15-LOX-1 (0.051 vs 0.032), and 2-fold greater for h15-LOX-2 (0.15 vs 0.072).50 These in vitro data suggest that at low substrate concentrations, LOX isozymes will preferentially produce 7S,17S-diHpDHA over 5S,15S-diHpETE, which could have biological implications depending on the concentration of specific LOX isozymes and specific fatty acids/oxylipins. It should be noted that kcat/KM flux values are valid in vivo only if the cellular concentration of the oxylipin is below the KM value.
Biosynthesis of 5S,12S-diHETE.
As discussed previously, the reduced form of 5S,12S-diHpETE, 5S,12S-diHETE, has been observed in lipidomic analysis and is thus potentially a significant biomolecule with respect to inflammation. There are two main pathways for producing 5S,12S-diHpETE, with the first, pathway 3, involving the formation of 12S-HpETE by either h12-LOX or h15-LOX-1, followed by reaction with h5-LOX to form 5S,12S-diHpETE. Although 12S-HpETE is formed rapidly by h12-LOX, the second step in this pathway, reaction of h5-LOX with 12S-HpETE, occurs relatively slowly. Similar to that seen with other mono-oxylipins, the reaction with h5-LOX and 12S-HETE occurs at a rate that is >300-fold lower than its rate with AA, making it the rate-limiting step of this pathway, for the in vitro biosynthetic pathway. It should be emphasized that in the cell, protein expression will also affect the rate-limiting step of a particular biosynthetic pathway.
However, pathway 4, involving formation of 5S,12S-diHpETE from 5S-HpETE, is not as rate restricted. The reaction of h5-LOX with AA occurs rapidly producing 5S-HpETE (vide supra). The second step with h12-LOX produces the expected 5S,12S-diHpETE product; however, 5S-HETE is a relatively poor substrate for h12-LOX, with a kcat and a kcat/KM >100-fold lower than those of the reaction with AA. In contrast, 5S-HETE is a relatively better substrate for h15-LOX-1, with its kcat/KM value being ~4-fold greater than that of h12-LOX, despite the fact that h12-LOX is more efficient in reacting with AA than is h15-LOX-1. The comparatively high kinetic values of h15-LOX-1 with 5S-HETE combined with its altered product profile make its biosynthetic kcat/KM flux almost 5-fold greater at producing 5S,12S-diHpETE than is h12-LOX. This result is consistent with previous in vitro results, where h15-LOX-1 is 20-fold more efficient than h12-LOX at producing lipoxins from 5,15-diHpETE,23,69 and indicates a negative bias against oxylipin substrates for h12-LOX relative to h15-LOX-1. In addition, these results suggest that h15-LOX-1 could be a source of the 5S,12S-diHpETE detected in biological samples, where 5S-HpETE is the available substrate.
It should be noted that h12-LOX shows a small shift in product specificity while reacting with 5S-HpETE, producing 13% 5S,15S-diHpETE instead of the expected 100% 5S,12S-diHpETE. A similar shift was previously observed when h12-LOX reacted with 7S-HDHA to produce 7S,17S-diHpDHA,50 both being the product of ω−8 hydrogen atom abstraction. These results indicate that both h12-LOX and h15-LOX-1 have altered product profiles when reacting with oxylipins, indicating caution should be shown when considering biosynthetic pathways for specific oxylipin production.
Biological Consequences.
5S,15S-diHpETE is an oxylipin that is produced in high abundance in many inflammatory conditions, including asthma and rheumatoid arthritis.29 It has been suggested that while the reduced form is observed, a hydroperoxide intermediate is necessary at C5 for formation of LXA4 and at C15 for formation of LXB4. This is because the hydroperoxide is required for the critical dehydration step by a LOX isozyme to generate the epoxide intermediate, which is then hydrolyzed to the corresponding lipoxin.18,23 5S,15S-diHpETE contains hydroperoxides at both C5 and C15 and can serve as an intermediate for the production of either lipoxin A4 (LXA4) and lipoxin B4 (LXB4). Previously, it was shown that h15-LOX-1 converted 5S,15S-diHpETE to LXB4 in vitro, while h15-LOX-2 was unable to do so,18 implying that h15-LOX-2 is not involved in the biosynthesis of LXB4. However, increases in h15-LOX-2 concentrations were observed to coincide with increases in lipoxin levels in airway epithelial cells.70 These data, combined with our in vitro data, may indicate that both 15-LOX isozymes are involved in LXB4 biosynthesis in vivo, with h15-LOX-2 oxygenating C15 of 5S-HETE or 5S-HpETE and h15-LOX-1 performing the dehydration step to produce the epoxide LXB4 intermediate. This is a remarkable hypothesis because the presumption has been that h15-LOX-1 and not h15-LOX-2 is the isozyme producing 5S,15S-diHpETE, due to its greater enzymatic activity in producing 15S-HpETE from AA. In the in vitro work presented here, h15-LOX-1 is not as efficient in producing 5S,15S-diHpETE from 5S-HpETE, but rather it has a greater propensity to synthesize 5S,12S-diHpETE. h15-LOX-2, however, does react well with 5S-HpETE, producing only 5S,15S-diHpETE. If we consider our previous work in the biosynthesis of lipoxins,23 we can propose the most efficient in vitro biosynthetic pathway for lipoxin production (Scheme 3). The first in vitro biosynthetic step is the production of 5S-HpETE by h5-LOX, followed by catalysis with h15-LOX-2 to produce 5S,15S-diHpETE. At this point, h15-LOX-1 produces the epoxide precursor of LXB4,23 which is subsequently converted to LXB4 by a hydrolase. This proposed in vitro biosynthetic pathway suggests that the role of h15-LOX-2 in producing LXB4 in vivo may be larger than previously suspected. In vivo work supports this hypothesis, where upregulation of h15-LOX-2 in dendritic cells is linked to increased 5S,15S-diHETE production71 and keratinocytes expressed h15-LOX-2 and produced 5S,15S-diHETE when incubated with 5S-HETE.15–17 We are currently investigating this further by applying our LOX specific inhibitors to human cell lines to confirm the role of particular biosynthetic pathways in 5S,12S-diHETE and 5S,15S-diHETE production.
Scheme 3.
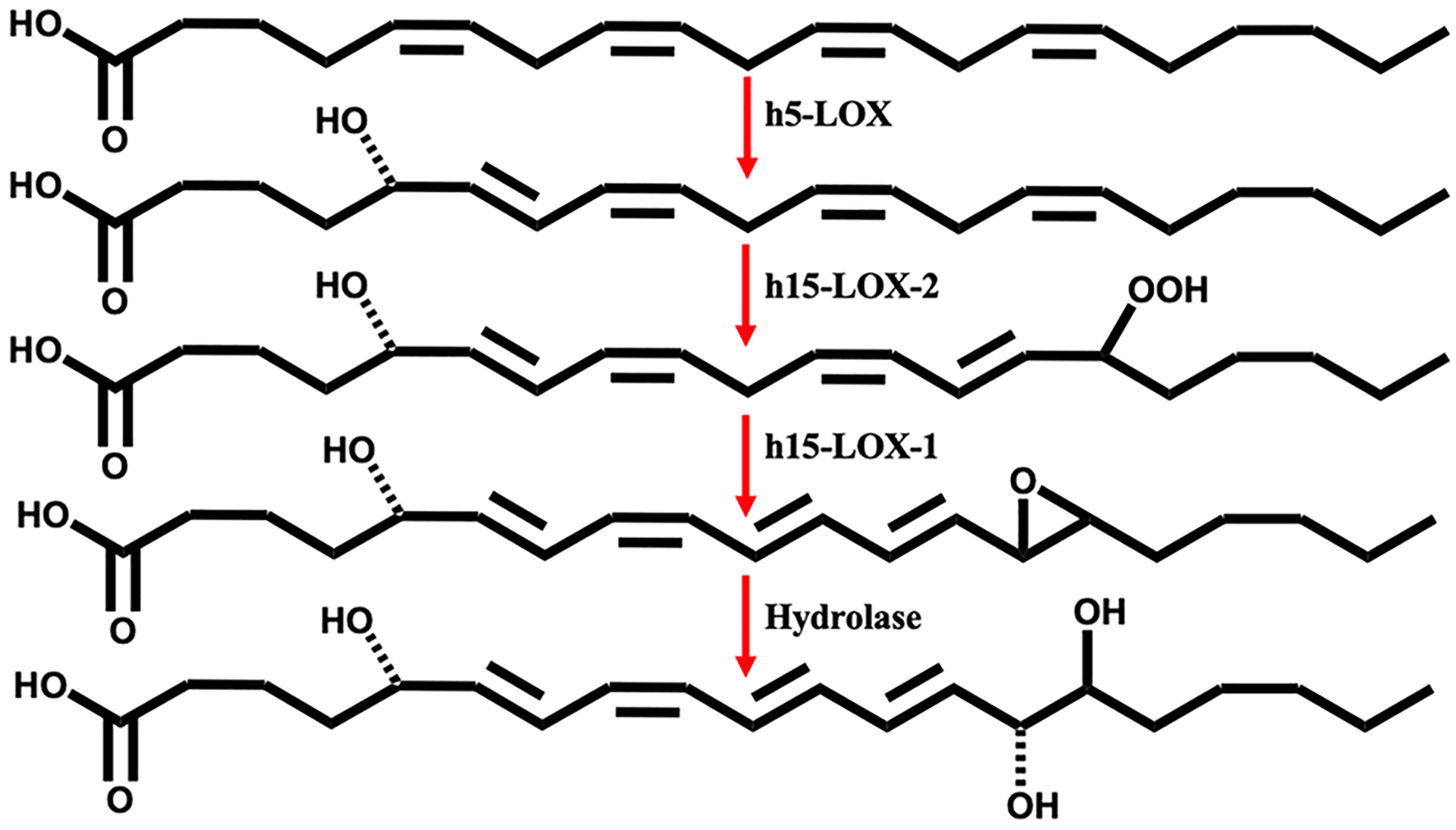
Proposed In Vitro Biosynthetic Pathway for LXB4
If h15-LOX-2 does have a greater role in lipoxin biosynthesis in vivo, then this may affect the predicted involvement of specific cells in lipoxin formation.22,24,32–34 For example, h15-LOX-2 is expressed at high levels in macrophages,17,51 and because macrophages also express h5-LOX and h15-LOX-1,13,72 they could contain all of the enzymes necessary to produce lipoxins on their own. This hypothesis is supported by the fact that the level of expression of h5-LOX is elevated in pro-inflammatory M1 macrophages,73 while h15-LOX-1 expression is upregulated in the pro-resolving M2 sub-type.71,73,74 Therefore, lipoxin production may occur with changes in macrophage LOX expression, without the need for a transcellular mechanism of synthesis. Nevertheless, the proposed in vitro biosynthetic routes (vide supra) should be considered with caution when extrapolating to in vivo systems. Although h5-LOX is a poor biocatalyst with oxylipins in vitro (vide supra), its activity can be altered by protein expression and the presence of FLAP,75 calcium,24 and hydroperoxides in the cell.76–78 In addition, 15-LOX isozymes are known to have altered catalysis with allosteric effector molecules in vitro, which could also be present in vivo.61,79 Therefore, we are currently utilizing specific LOX inhibitors in primary cells to determine which LOX isozyme plays a role in specific oxylipin biosynthetic pathways based on enzymatic activity and protein expression.
CONCLUSION
In conclusion, the altered positional specificity of h15-LOX-1 extends beyond 7S-HpDHA to 5S-HpETE. Therefore, the in vivo biosynthesis of dioxylipins, such as 5S,15S-diHpETE and 5S,12S-diHpETE, can occur by routes that may not be obvious when extrapolating from LOX activity with AA. In particular, h15-LOX-2 may possibly make a larger contribution to the biosynthesis of lipoxins and other oxylipins, which was previously attributed solely to h15-LOX-1, and therefore, interpretation of in vivo data should be carefully evaluated with the current results in mind.
Supplementary Material
Funding
National Institutes of Health (NIH) Grants R01 GM105671 (M.H. and T.R.H.), R01 HL11405 (M.H. and T.R.H.), R35 GM131835 (M.H. and T.R.H.), and K99HL136784 (B.E.T.).
ABBREVIATIONS
- AA
arachidonic acid
- DHA
docosahexaenoic acid
- PUFA
polyunsaturated fatty acid
- SPM
specialized pro-resolving mediator
- 5S-HETE
5S-hydroxy-6E,8Z,11Z,14Z-eicosatetraenoic acid
- 5S-HpETE
5S-hydroperoxy-6E,8Z,11Z,14Z-eicosatetraenoic acid
- 12S-HETE
12S-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid
- 12S-HpETE
12S-hydroperoxy-5Z,8Z,10E,14Z-eicosatetraenoic acid
- 15S-HETE
15S-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid
- 15S-HpETE
15S-hydroperoxy-5Z,8Z,11Z,13E-eicosatetraenoic acid
- 5S,12S-diHETE
5S,12S-dihydroxy-6E,8Z,10E,14Z-eicosatetraenoic acid
- 5S,12S-diHpETE
5S,12S-dihydroperoxy-6E,8Z,10E,14Z-eicosatetraenoic acid
- 5S,15S-diHETE
5S,15S-dihydroxy-6E,8Z,11Z,13E-eicosatetraenoic acid
- 5S,15S-diHpETE
5S,15S-dihydroperoxy-6E,8Z,11Z,13E-eicosatetraenoic acid
- 7S-HDHA
7S-hydroperoxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid
- 7S-HpDHA
7S-hydroperoxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid
- 14S-HpDHA
14S-hydroperoxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid
- 17S-HpDHA
17S-hydroperoxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid
- 7S,14S-diHDHA
7S,14S-dihydroxy-4Z,8E,10Z,12E,16Z,19Z-docosahexaenoic acid
- 7S,17S-diHDHA
7S,17S-dihydroxy-4Z,8E,10Z,13Z,15E,19Z-docosahexaenoic acid
- 13-HODE
13S-hydroxy-octadecadienoic acid; RvD5, Resolvin D5
- LTB4
leukotriene B4
- LXB4
lipoxin B4
- LXA4
lipoxin A4
- LOX
lipoxygenase
- h15-LOX-1 or ALOX15
human reticulocyte 15-lipoxygenase-1
- h5-LOX or ALOX5
human 5-lipoxygenase
- h12-LOX or ALOX12
human platelet 12-lipoxygenase
- h15-LOX-2 or ALOX15B
human epithelial 15-lipoxygenase-2
- PMN
polymorphonuclear leukocyte
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00622.
h5-LOX SDS-PAGE and MS characterization of oxylipin standards (PDF)
The authors declare the following competing financial interest(s): M.P.J. is a consultant to and shareholder of Schrodinger LLC, which licenses the software used in this work.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00622
Contributor Information
Chakrapani Kalyanaraman, Department of Pharmaceutical Chemistry, School of Pharmacy, University of California, San Francisco, San Francisco, California 94158, United States.
Michael Holinstat, Department of Pharmacology, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States.
Matthew P. Jacobson, Department of Pharmaceutical Chemistry, School of Pharmacy, University of California, San Francisco, San Francisco, California 94158, United States.
Theodore R. Holman, Department of Chemistry and Biochemistry, University of California, Santa Cruz, Santa Cruz, California 95064, United States.
REFERENCES
- (1).Tabas I, and Glass CK (2013) Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Serhan CN, Hamberg M, and Samuelsson B (1984) Trihydroxytetraenes: a novel series of compounds formed from arachidonic acid in human leukocytes. Biochem. Biophys. Res. Commun 118, 943–949. [DOI] [PubMed] [Google Scholar]
- (3).Badr KF, DeBoer DK, Schwartzberg M, and Serhan CN (1989) Lipoxin A4 antagonizes cellular and in vivo actions of leukotriene D4 in rat glomerular mesangial cells: evidence for competition at a common receptor. Proc. Natl. Acad. Sci. U. S. A 86, 3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hedqvist P, Raud J, Palmertz U, Haeggstrom J, Nicolaou KC, and Dahlen SE (1989) Lipoxin A4 inhibits leukotriene B4-induced inflammation in the hamster cheek pouch. Acta Physiol. Scand 137, 571–572. [DOI] [PubMed] [Google Scholar]
- (5).Mangino MJ, Brounts L, Harms B, and Heise C (2006) Lipoxin biosynthesis in inflammatory bowel disease. Prostaglandins Other Lipid Mediators 79, 84–92. [DOI] [PubMed] [Google Scholar]
- (6).Serhan CN (2010) Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? Am. J. Pathol 177, 1576–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nathan C, and Ding A (2010) Nonresolving inflammation. Cell 140, 871–882. [DOI] [PubMed] [Google Scholar]
- (8).Weaver JR, Holman TR, Imai Y, Jadhav A, Kenyon V, Maloney DJ, Nadler JL, Rai G, Simeonov A, and Taylor-Fishwick DA (2012) Integration of pro-inflammatory cytokines, 12-lipoxygenase and NOX-1 in pancreatic islet beta cell dysfunction. Mol. Cell. Endocrinol 358, 88–95. [DOI] [PubMed] [Google Scholar]
- (9).Cole BK, Lieb DC, Dobrian AD, and Nadler JL (2013) 12- and 15-lipoxygenases in adipose tissue inflammation. Prostaglandins Other Lipid Mediators 104–105, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Serhan CN, Chiang N, and Van Dyke TE (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 8, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O’Donnell VB, Kuhn H, and Walther M (2010) Molecular enzymology of lipoxygenases. Arch. Biochem. Biophys 503, 161–174. [DOI] [PubMed] [Google Scholar]
- (12).Kuhn H, Banthiya S, and van Leyen K (2015) Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1851, 308–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Colakoglu M, Tuncer S, and Banerjee S (2018) Emerging cellular functions of the lipid metabolizing enzyme 15-Lipoxygenase-1. Cell Proliferation 51, No. e12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ford-Hutchinson AW (1991) Arachidonate 15-lipoxygenase; characteristics and potential biological significance. Eicosanoids 4, 65–74. [PubMed] [Google Scholar]
- (15).Brash AR, Boeglin WE, and Chang MS (1997) Discovery of a second 15S-lipoxygenase in humans. Proc. Natl. Acad. Sci. U. S. A 94, 6148–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jisaka M, Kim RB, Boeglin WE, Nanney LB, and Brash AR (1997) Molecular cloning and functional expression of a phorbol ester-inducible 8S-lipoxygenase from mouse skin. J. Biol. Chem 272, 24410–24416. [DOI] [PubMed] [Google Scholar]
- (17).Hulten LM, Olson FJ, Aberg H, Carlsson J, Karlstrom L, Boren J, Fagerberg B, and Wiklund O (2010) 15-Lipoxygenase-2 is expressed in macrophages in human carotid plaques and regulated by hypoxia-inducible factor-1alpha. Eur. J. Clin. Invest 40, 11–17. [DOI] [PubMed] [Google Scholar]
- (18).Green AR, Barbour S, Horn T, Carlos J, Raskatov JA, and Holman TR (2016) Strict Regiospecificity of Human Epithelial 15-Lipoxygenase-2 Delineates Its Transcellular Synthesis Potential. Biochemistry 55, 2832–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Borgeat P, Fruteau de Laclos B, Picard S, Drapeau J, Vallerand P, and Corey EJ (1982) Studies on the mechanism of formation of the 5S, 12S-dihydroxy-6,8,10,14(E, Z, E, Z)-icosatetraenoic acid in leukocytes. Prostaglandins 23, 713–724. [DOI] [PubMed] [Google Scholar]
- (20).Serhan CN, Broekman MJ, Korchak HM, Marcus AJ, and Weissman G (1982) Endogenous phospholipid metabolism in stimulated neutrophils differential activation by FMLP and PMA. Biochem. Biophys. Res. Commun 107, 951–958. [DOI] [PubMed] [Google Scholar]
- (21).Borgeat P, Nadeau M, Salari H, Poubelle P, and Fruteau de Laclos B (1985) Leukotrienes: biosynthesis, metabolism, and analysis. Adv. Lipid Res 21, 47–77. [DOI] [PubMed] [Google Scholar]
- (22).Chavis C, Vachier I, Chanez P, Bousquet J, and Godard P (1996) 5(S),15(S)-dihydroxyeicosatetraenoic acid and lipoxin generation in human polymorphonuclear cells: dual specificity of 5-lipoxygenase towards endogenous and exogenous precursors. J. Exp. Med 183, 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Green AR, Freedman C, Tena J, Tourdot BE, Liu B, Holinstat M, and Holman TR (2018) 5 S,15 S-Dihydroperox-yeicosatetraenoic Acid (5,15-diHpETE) as a Lipoxin Intermediate: Reactivity and Kinetics with Human Leukocyte 5-Lipoxygenase, Platelet 12-Lipoxygenase, and Reticulocyte 15-Lipoxygenase-1. Biochemistry 57, 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Maas RL, Turk J, Oates JA, and Brash AR (1982) Formation of a novel dihydroxy acid from arachidonic acid by lipoxygenase-catalyzed double oxygenation in rat mononuclear cells and human leukocytes. J. Biol. Chem 257, 7056–7067. [PubMed] [Google Scholar]
- (25).Kuhn H, Wiesner R, Alder L, Fitzsimmons BJ, Rokach J, and Brash AR (1987) Formation of lipoxin B by the pure reticulocyte lipoxygenase via sequential oxygenation of the substrate. Eur. J. Biochem 169, 593–601. [DOI] [PubMed] [Google Scholar]
- (26).Ueda N, Yokoyama C, Yamamoto S, Fitzsimmons BJ, Rokach J, Oates JA, and Brash AR (1987) Lipoxin synthesis by arachidonate 12-lipoxygenase purified from porcine leukocytes. Biochem. Biophys. Res. Commun 149, 1063–1069. [DOI] [PubMed] [Google Scholar]
- (27).Dalli J, and Serhan CN (2012) Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood 120, e60–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chavis C, Chanez P, Vachier I, Bousquet J, Michel FB, and Godard P (1995) 5–15-diHETE and lipoxins generated by neutrophils from endogenous arachidonic acid as asthma biomarkers. Biochem. Biophys. Res. Commun 207, 273–279. [DOI] [PubMed] [Google Scholar]
- (29).Thomas E, Leroux JL, Blotman F, and Chavis C (1995) Conversion of endogenous arachidonic acid to 5,15-diHETE and lipoxins by polymorphonuclear cells from patients with rheumatoid arthritis. Inflammation Res. 44, 121–124. [DOI] [PubMed] [Google Scholar]
- (30).Morita E, Schroder JM, and Christophers E (1990) Identification of a novel and highly potent eosinophil chemotactic lipid in human eosinophils treated with arachidonic acid. J. Immunol 144, 1893–1900. [PubMed] [Google Scholar]
- (31).O’Flaherty JT, and Thomas MJ (1985) Effect of 15-lipoxygenase-derived arachidonate metabolites on human neutrophil degranulation. Prostaglandins, Leukotrienes Med. 17, 199–212. [DOI] [PubMed] [Google Scholar]
- (32).Green FA (1990) Transformations of 5-HETE by activated keratinocyte 15-lipoxygenase and the activation mechanism. Lipids 25, 618–623. [DOI] [PubMed] [Google Scholar]
- (33).McDonald PP, McColl SR, Naccache PH, and Borgeat P (1992) Activation of the human neutrophil 5-lipoxygenase by leukotriene B4. British journal of pharmacology 107, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Serhan CN (1989) On the relationship between leukotriene and lipoxin production by human neutrophils: evidence for differential metabolism of 15-HETE and 5-HETE. Biochim. Biophys. Acta, Lipids Lipid Metab 1004, 158–168. [DOI] [PubMed] [Google Scholar]
- (35).Yoshimoto T, Miyamoto Y, Ochi K, and Yamamoto S (1982) Arachidonate 12-lipoxygenase of porcine leukocyte with activity for 5-hydroxyeicosatetraenoic acid. Biochim. Biophys. Acta, Lipids Lipid Metab 713, 638–646. [PubMed] [Google Scholar]
- (36).Marcus AJ, Safier LB, Ullman HL, Broekman MJ, Islam N, Oglesby TD, Gorman RR, and Ward JW (1985) Inhibition of platelet function in thrombosis. Circulation 72, 698–701. [DOI] [PubMed] [Google Scholar]
- (37).Sorokin AV, Norris PC, English JT, Dey AK, Chaturvedi A, Baumer Y, Silverman J, Playford MP, Serhan CN, and Mehta NN (2018) Identification of proresolving and inflammatory lipid mediators in human psoriasis. J. Clin. Lipidol 12, 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Borgeat P, Picard S, Vallerand P, and Sirois P (1981) Transformation of arachidonic acid in leukocytes. Isolation and structural analysis of a novel dihydroxy derivative. Prostaglandins Med. 6, 557–570. [DOI] [PubMed] [Google Scholar]
- (39).Larfars G, Lantoine F, Devynck MA, Palmblad J, and Gyllenhammar H (1999) Activation of nitric oxide release and oxidative metabolism by leukotrienes B4, C4, and D4 in human polymorphonuclear leukocytes. Blood 93, 1399–1405. [PubMed] [Google Scholar]
- (40).Valles J, Santos MT, Marcus AJ, Safier LB, Broekman MJ, Islam N, Ullman HL, and Aznar J (1993) Downregulation of human platelet reactivity by neutrophils. Participation of lipoxygenase derivatives and adhesive proteins. J. Clin. Invest 92, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sirois P, Roy S, Beraldin G, Valler P, and Borgeat P (1982) Comparative activity of natural mono-, di- and tri-hydroxy derivatives of arachidonic acid on guinea-pig lungs: myotropic effect and stimulation of cyclooxygenase activity. Prostaglandins 24, 405–418. [DOI] [PubMed] [Google Scholar]
- (42).Maas RL, Ingram CD, Taber DF, Oates JA, and Brash AR (1982) Stereospecific removal of the DR hydrogen atom at the 10-carbon of arachidonic acid in the biosynthesis of leukotriene A4 by human leukocytes. J. Biol. Chem 257, 13515–13519. [PubMed] [Google Scholar]
- (43).Suzuki H, Kishimoto K, Yoshimoto T, Yamamoto S, Kanai F, Ebina Y, Miyatake A, and Tanabe T (1994) Site-directed mutagenesis studies on the iron-binding domain and the determinant for the substrate oxygenation site of porcine leukocyte arachidonate 12-lipoxygenase. Biochim. Biophys. Acta, Lipids Lipid Metab 1210, 308–316. [DOI] [PubMed] [Google Scholar]
- (44).Gan Q-F, Browner MF, Sloane DL, and Sigal E (1996) Defining the Arachidonic Acid Binding Site of Human 15-Lipoxygenase: Molecular Modeling and Mutagenesis. J. Biol. Chem 271, 25412–25418. [DOI] [PubMed] [Google Scholar]
- (45).Sloane DL, Leung R, Barnett J, Craik CS, and Sigal E (1995) Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: the side-chain geometry of amino acids 417 and 418 determine positional specificity. Protein Eng., Des. Sel 8, 275–282. [DOI] [PubMed] [Google Scholar]
- (46).Borngraber S, Kuban RJ, Anton M, and Kuhn H (1996) Phenylalanine 353 is a primary determinant for the positional specificity of mammalian 15-lipoxygenases. J. Mol. Biol 264, 1145–1153. [DOI] [PubMed] [Google Scholar]
- (47).Vogel R, Jansen C, Roffeis J, Reddanna P, Forsell P, Claesson HE, Kuhn H, and Walther M (2010) Applicability of the triad concept for the positional specificity of mammalian lipoxygenases. J. Biol. Chem 285, 5369–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sloane DL, Leung R, Craik CS, and Sigal E (1991) A Primary determinant for Lipoxygenase positional specificity. Nature 354, 149–152. [DOI] [PubMed] [Google Scholar]
- (49).Kuhn H, Walther M, and Kuban RJ (2002) Mammalian arachidonate 15-lipoxygenases: Structure, function, and biological implications. Prostaglandins Other Lipid Mediators 68–69, 263–290. [DOI] [PubMed] [Google Scholar]
- (50).Perry SC, Kalyanaraman C, Tourdot BE, Conrad WS, Akinkugbe O, Freedman JC, Holinstat M, Jacobson MP, and Holman TR (2020) 15-Lipoxygenase-1 biosynthesis of 7S,14S-diHDHA implicates 15-Lipoxygenase-2 in biosynthesis of resolvin D5. J. Lipid Res 61, 1087–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wuest SJ, Crucet M, Gemperle C, Loretz C, and Hersberger M (2012) Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis 225, 121–127. [DOI] [PubMed] [Google Scholar]
- (52).Amagata T, Whitman S, Johnson TA, Stessman CC, Loo CP, Lobkovsky E, Clardy J, Crews P, and Holman TR (2003) Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and 15-soybean lipoxygenases. J. Nat. Prod 66, 230–235. [DOI] [PubMed] [Google Scholar]
- (53).Robinson SJ, Hoobler EK, Riener M, Loveridge ST, Tenney K, Valeriote FA, Holman TR, and Crews P (2009) Using enzyme assays to evaluate the structure and bioactivity of sponge-derived meroterpenes. J. Nat. Prod 72, 1857–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jameson JB, Kantz A, Schultz L, Kalyanaraman C, Jacobson MP, Maloney DJ, Jadhav A, Simeonov A, and Holman TR (2014) A High Throughput Screen Identifies Potent and Selective Inhibitors to Human Epithelial 15-Lipoxygenase-2. PLoS One 9, No. e104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, and Newcomer ME (2011) The structure of human 5-lipoxygenase. Science 331, 217–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Smyrniotis CJ, Barbour SR, Xia Z, Hixon MS, and Holman TR (2014) ATP allosterically activates the human 5-lipoxygenase molecular mechanism of arachidonic acid and 5(S)-hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic acid. Biochemistry 53, 4407–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Green AR, Freedman C, Tena J, Tourdot BE, Liu B, Holinstat M, and Holman TR (2018) 5 S,15 S-Dihydroperox-yeicosatetraenoic Acid (5,15-diHpETE) as a Lipoxin Intermediate: Reactivity and Kinetics with Human Leukocyte 5-Lipoxygenase, Platelet 12-Lipoxygenase, and Reticulocyte 15-Lipoxygenase-1. Biochemistry 57, 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Adili R, Tourdot BE, Mast K, Yeung J, Freedman JC, Green A, Luci DK, Jadhav A, Simeonov A, Maloney DJ, Holman TR, and Holinstat M (2017) First Selective 12-LOX Inhibitor, ML355, Impairs Thrombus Formation and Vessel Occlusion In Vivo With Minimal Effects on Hemostasis. Arterioscler., Thromb., Vasc. Biol 37, 1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Ikei KN, Yeung J, Apopa PL, Ceja J, Vesci J, Holman TR, and Holinstat M (2012) Investigations of human platelet-type 12-lipoxygenase: role of lipoxygenase products in platelet activation. J. Lipid Res 53, 2546–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Armstrong M, van Hoorebeke C, Horn T, Deschamps J, Freedman JC, Kalyanaraman C, Jacobson MP, and Holman T (2016) Human 15-LOX-1 active site mutations alter inhibitor binding and decrease potency. Bioorg. Med. Chem 24, 5380–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wecksler AT, Kenyon V, Deschamps JD, and Holman TR (2008) Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry 47, 7364–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Borgeat P, Hamberg M, and Samuelsson B (1976) Transformation of arachidonic acid and homo-gamma-linolenic acid by rabbit polymorphonuclear leukocytes. Monohydroxy acids from novel lipoxygenases. J. Biol. Chem 251, 7816–7820. [PubMed] [Google Scholar]
- (63).Hamberg M, and Samuelsson B (1974) Prostaglandin endoperoxides. Novel transformations of arachidonic acid in human platelets. Proc. Natl. Acad. Sci. U. S. A 71, 3400–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Balas L, Guichardant M, Durand T, and Lagarde M (2014) Confusion between protectin D1 (PD1) and its isomer protectin DX (PDX). An overview on the dihydroxy-docosatrienes described to date. Biochimie 99, 1–7. [DOI] [PubMed] [Google Scholar]
- (65).Chen P, Fenet B, Michaud S, Tomczyk N, Vericel E, Lagarde M, and Guichardant M (2009) Full characterization of PDX, a neuroprotectin/protectin D1 isomer, which inhibits blood platelet aggregation. FEBS Lett 583, 3478–3484. [DOI] [PubMed] [Google Scholar]
- (66).Aursnes M, Tungen JE, Vik A, Colas R, Cheng CY, Dalli J, Serhan CN, and Hansen TV (2014) Total synthesis of the lipid mediator PD1n-3 DPA: configurational assignments and anti-inflammatory and pro-resolving actions. J. Nat. Prod 77, 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Tungen JE, Aursnes M, Vik A, Ramon S, Colas RA, Dalli J, Serhan CN, and Hansen TV (2014) Synthesis and anti-inflammatory and pro-resolving activities of 22-OH-PD1, a mono-hydroxylated metabolite of protectin D1. J. Nat. Prod 77, 2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Gan Q-F, Browner MF, Sloane DL, and Sigal E (1996) Defining the arachidonic acid binding site of human 15-lipoxygenase. J. Biol. Chem 271, 25412–25418. [DOI] [PubMed] [Google Scholar]
- (69).Sheppard KA, Greenberg SM, Funk CD, Romano M, and Serhan CN (1992) Lipoxin generation by human megakaryocyte-induced 12-lipoxygenase. Biochim. Biophys. Acta, Mol. Cell Res 1133, 223–234. [DOI] [PubMed] [Google Scholar]
- (70).Ringholz FC, Buchanan PJ, Clarke DT, Millar RG, McDermott M, Linnane B, Harvey BJ, McNally P, and Urbach V (2014) Reduced 15-lipoxygenase 2 and lipoxin A4/leukotriene B4 ratio in children with cystic fibrosis. Eur. Respir. J 44, 394–404. [DOI] [PubMed] [Google Scholar]
- (71).Spanbroek R, Hildner M, Kohler A, Muller A, Zintl F, Kuhn H, Radmark O, Samuelsson B, and Habenicht AJ (2001) IL-4 determines eicosanoid formation in dendritic cells by down-regulation of 5-lipoxygenase and up-regulation of 15-lipoxygenase 1 expression. Proc. Natl. Acad. Sci. U. S. A 98, 5152–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Abrial C, Grassin-Delyle S, Salvator H, Brollo M, Naline E, and Devillier P (2015) 15-Lipoxygenases regulate the production of chemokines in human lung macrophages. British journal of pharmacology 172, 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Werner M, Jordan PM, Romp E, Czapka A, Rao Z, Kretzer C, Koeberle A, Garscha U, Pace S, Claesson HE, Serhan CN, Werz O, and Gerstmeier J (2019) Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome. FASEB J 33, 6140–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Profita M, Sala A, Siena L, Henson PM, Murphy RC, Paterno A, Bonanno A, Riccobono L, Mirabella A, Bonsignore G, and Vignola AM (2002) Leukotriene B4 production in human mononuclear phagocytes is modulated by interleukin-4-induced 15-lipoxygenase. J. Pharmacol. Exp. Ther 300, 868–875. [DOI] [PubMed] [Google Scholar]
- (75).Peters-Golden M, and Brock TG (2003) 5-lipoxygenase and FLAP. Prostaglandins, Leukotrienes Essent. Fatty Acids 69, 99–109. [DOI] [PubMed] [Google Scholar]
- (76).Hatzelmann A, Schatz M, and Ullrich V (1989) Involvement of glutathione peroxidase activity in the stimulation of 5-lipoxygenase activity by glutathione-depleting agents in human polymorphonuclear leukocytes. Eur. J. Biochem 180, 527–533. [DOI] [PubMed] [Google Scholar]
- (77).Weitzel F, and Wendel A (1993) Selenoenzymes regulate the activity of leukocyte 5-lipoxygenase via the peroxide tone. J. Biol. Chem 268, 6288–6292. [PubMed] [Google Scholar]
- (78).Petrich K, Ludwig P, Kuhn H, and Schewe T (1996) The suppression of 5-lipoxygenation of arachidonic acid in human polymorphonuclear leucocytes by the 15-lipoxygenase product (15S)-hydroxy-(5Z,8Z,11Z,13E)-eicosatetraenoic acid: structure-activity relationship and mechanism of action. Biochem. J 314 (Part 3), 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Wecksler AT, Kenyon V, Garcia NK, Deschamps JD, van der Donk WA, and Holman TR (2009) Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry 48, 8721–8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


