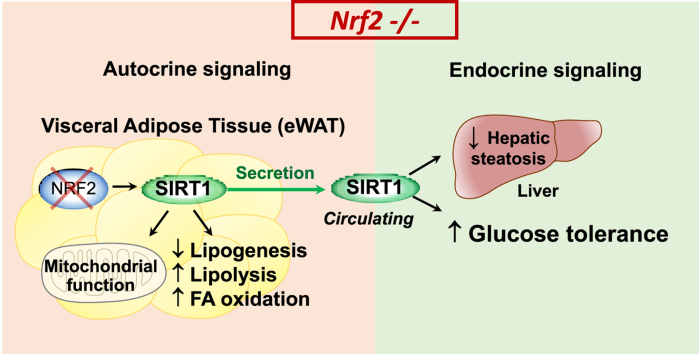
Abstract
Obesity is associated with metabolic dysregulation characterized by insulin resistance and glucose intolerance. Nuclear factor E2-related factor (Nrf2) is a critical regulator of the stress response and Nrf2-deficient mice (Nrf2−/−) are protected against high fat diet (HFD)-induced metabolic derangement. We searched for factors that could underline this favorable phenotype and found that Nrf2−/− mice exhibit higher circulating levels of sirtuin 1 (Sirt1), a key player in cellular homeostasis and energy metabolism, compared to wild-type mice. Increased Sirt1 levels in Nrf2−/− mice were found not only in animals under standard diet but also following HFD. Interestingly, we report here that the visceral adipose tissue (eWAT) is the sole source of increased Sirt1 protein in plasma. eWAT and other fat depots displayed enhanced adipocytes lipolysis, increased fatty acid oxidation and glycolysis, suggesting autocrine and endocrine actions of Sirt1 in this model. We further demonstrate that removal of eWAT completely abolishes the increase in circulating Sirt1 and that this procedure suppresses the beneficial effect of Nrf2 deficiency on glucose tolerance, but not insulin sensitivity, following a HFD regime. Thus, in contrast to many other stressful conditions where Nrf2 deficiency exacerbates damage, our study indicates that up-regulation of Sirt1 levels specifically in the visceral adipose tissue of Nrf2−/− mice is a key adaptive mechanism that mitigates glucose intolerance induced by nutritional stress.
Keywords: Nrf2, Sirtuin 1, Obesity, White adipose tissue, Glucose metabolism
Graphical abstract
Highlights
-
•
Nrf2−/− mice are protected against weight gain and metabolic dysfunction under high fat diet (HFD).
-
•
Nrf2−/− mice display increased production of sirtuin 1 exclusively in visceral white adipose tissue (eWAT).
-
•
Removal of eWAT prior to HFD abolishes the increase in sirtuin 1 and impairs glucose tolerance.
-
•
A strong link exists between Nrf2 deficiency, SIRT1 in eWAT and regulation of glucose homeostasis.
1. Introduction
Obesity and associated metabolic dysfunction constitute a great threat for global health and the study of mechanisms implicated in these conditions may help to identify potential therapeutic targets. The white adipose tissue (WAT) is crucial in obesity due its fat storing capacity as well as its endocrine function, promoted by the secretion of several factors and hormones that influence whole body metabolism [1]. For example, WAT releases leptin and adiponectin to control appetite and insulin sensitivity, and produces inflammatory factors such as interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) that are involved in the metabolic complications of obesity. These and other observations led to a shift in the perception of WAT from that of an inert tissue to that of a highly plastic organ that can be manipulated to provide beneficial effects in obesity. In line with this, we have recently demonstrated that a switch in adipose tissue metabolism towards glycolysis induced by a carbon monoxide-releasing molecule (CORM-401) reduces body weight gain and improves insulin resistance in HFD-fed mice [2]. In addition, stimulation of brown-like features in WAT, based on the idea that brown fat augments energy expenditure as opposed to the energy storage function of WAT, has been reported. Treatment with fibroblast growth factor 21 (FGF21), irisin or the overexpression of sirtuin 1 (Sirt1), were shown to confer ‘browning’ characteristics to WAT and exert beneficial effects on insulin and glucose tolerance [3]. In particular, Sirt1 is a NAD+-dependent protein deacetylase that plays a crucial role in controlling cellular metabolism as well as the energy status of the organism, and has been shown to protect mice against HFD-induced metabolic dysfunction [4]. Additional key roles of Sirt1 include inhibition of adipogenesis and increased free fatty acids utilization in WAT [5], while in the liver Sirt1 enhances gluconeogenesis and inhibits glycolysis via forkhead box protein O1 (FOXO1) [6]. Overexpression of Sirt1 also ameliorates hepatic steatosis and glucose intolerance in obese mice by inducing FGF21 [7].
Previous studies have highlighted a crosstalk between Sirt1 and nuclear factor erythroid 2-related factor 2 (Nrf2), the transcription factor that activates the response to stress conditions by regulating the expression of a variety of detoxifying and antioxidant genes including NAD(P)H-quinone oxidoreductase 1 (NQO1), glutathione S-transferases (GSTs) and heme oxygenase-1 (HO-1) [[8], [9], [10], [11]]. Besides its well-described cytoprotective activities against tissue damage triggered by oxidative stress and inflammation [[12], [13], [14]], the possible implications of Nrf2 in the regulation of metabolism started to emerge only recently [15]. However, whether Nrf2 plays a protective or deleterious role in obesity is unclear as divergent data have been published so far. In fact, pharmacologic induction of Nrf2 by the synthetic triterpenoid CDDO-imidazolide prevents body weight gain, adipogenesis and hepatic lipid accumulation in mice [16]. Activation of Nrf2 also improves leptin and insulin resistance provoked by hypothalamic oxidative stress [17] and impedes the onsets of HFD-induced diabetes [18]. In addition, genetic deletion of Nrf2 leads to hepatic insulin resistance via the activation of NF-κB [19]. In contrast, complete or adipose-specific Nrf2 ablation protects mice against HFD-induced obesity and insulin resistance; these effects are associated with impaired adipogenesis mediated by the nuclear receptor PPARγ in adipose tissue [20,21] and by increased production of FGF21, which regulates glucose intake [22,23]. These discrepancies may be due to the origin of the metabolic disorders or the experimental models used to study the impact of Nrf2 in obesity, emphasizing the need to investigate further the exact role of Nrf2 and its downstream effectors in metabolism.
In the present study, we extensively examined the response of Nrf2 deficient mice to HFD-induced obesity. We demonstrate that Nrf2 deficient mice (Nrf2−/−) exhibit significantly elevated basal plasma levels of Sirt1 compared to wild type (WT) mice. Higher circulating Sirt1 appear to derive most exclusively from the epididymal WAT (eWAT). By comparing wild type (WT) and Nrf2−/− animals we reveal that Nrf2 deficient mice under HFD maintain an increased production of Sirt1, are resistant to weight gain and metabolic derangement and display enhanced adipocytes lipolysis together with increased bioenergetic metabolism. Finally, we show that removal of eWAT (eWATectomy) before subjecting mice to HFD suppresses the increase in circulating Sirt1 and the beneficial effect of Nrf2 deficiency on glucose tolerance after a HFD regime. Our study demonstrates that increased eWAT-derived Sirt1 is a critical downstream adaptation of Nrf2 deficiency that mitigates glucose intolerance induced by obesity.
2. Materials and methods
2.1. Animals
Wild type (WT) C57/BL6J mice (8 weeks old) were purchased from Janvier Labs (France). Nrf2−/− mice (8 weeks old), which had been backcrossed with C57/BL6J mice and kept as homozygous, were purchased from the TAAM laboratory (Orléans, France). To take in consideration that mice were from different suppliers, even if from the same genetic background, mice from both groups were received at the age of 4 weeks, put into quarantine for a 4 weeks period in order to acclimatize them to our animal facility. Thus, the same environment and nutritional states were applied prior to the beginning of our protocols. Mice were housed under controlled conditions of temperature (21 ± 1°C), hygrometry (60 ± 10%) and lighting (12 h per day). Animals were acclimatized in the laboratory for one week before the start of the experiments. Mice were fed either a standard diet SD (A04, SAFE Diet, Augy, France) or a HFD (60 kcal % fat, Brogaarden, DK). All animals received care according to institutional guidelines, and all experiments were approved by the Institutional Ethics committee (licence number 16–090). During follow-up, animals underwent body weight, food intake, metabolic and echocardiography assessments. For tissue collection, mice were euthanatized 6 h after fasting and organs and blood were collected and processed for further evaluation.
2.2. Fasting blood glucose, glucose and insulin tolerance tests
Whole-body glucose tolerance and insulin sensitivity were assessed in all groups at weeks 12th and 13th by intraperitoneal glucose (GTT) and insulin (ITT) tolerance tests, respectively. First, blood was collected via tail clip to measure fasting blood glucose (Caresens® N, DinnoSanteTM). Then, mice received glucose (1.5 g/kg) or insulin (0.3 UI/kg) in solution by intraperitoneal injection and blood glucose was measured at 15, 30, 60, 90 and 120 min after the injection. The HOmeostasis Model Assessment of Insulin Resistance (HOMA-IR) adjusted to rodents was calculated as ([glucose (mg/dl)/18] x [insulin (ng/ml)/0.0347])/108.24 as reported [24]. The area under the curve (AUC) for the glucose excursion was calculated using Graph Prism.
2.3. Body composition
Body composition (fat mass, free water, and total water) using magnetic resonance imaging (EchoMRI-900TM, Echo Medical System, Houston, TX) as previously described [25,26]. Conscious mice were placed in a thin-walled plastic cylinder with a plastic insert that functioned to limit the movement while the mice were in the EchoMRI instrument. Body composition analysis was measured in mice exposed to a low-intensity electromagnetic field and the amount of fat and lean mass detected was expressed as a percentage of total body weight.
2.4. Removal of visceral adipose tissue (eWATectomy)
A total of 10 male WT C57/BL6J mice (WT-eWAT) and 10 male Nrf2−/− C57/BL6J mice (Nrf2-eWAT), 10 weeks of age and weighing approximately 25–30 g, were used for adipose tissue removal performed by epididymal fat resection (eWATectomy). Before eWATectomy, mice underwent baseline assessment of metabolic functions (glucose tolerance and insulin tolerance test). Mice were anesthetized with inhaled 3% isoflurane and a 1-cm single abdominal midline incision was made. Bilateral epididymal fat pads were lifted from the peritoneal cavity onto a sterilized and humidified surgical drape, dissected with an electronic scalpel, and removed without damaging the testicular blood supply. The sham operation was performed in the same manner without fat pads removal. The abdominal peritoneum was closed with prolyne sutures and the skin was closed with silk sutures (Angiotech, Canada). After eWATectomy, mice were subjected to a 8 week HFD regime. GTT and ITT tests were performed at week 7 and mice sacrificed at the end of the protocol for tissue analysis.
2.5. Preparation of eWAT-conditioned medium (eCM)
Epididymal white adipose tissue (eWAT, 0.1 g) was collected and kept at room temperature in a 24-well plate with 1 ml/well of Dulbecco's modified Eagle's medium DMEM (Life Technologies, France). The tissue was minced into ~1 mm3 pieces and incubated for 1 h at 37°C and 5% CO2 prior to transferring it into a new plate with fresh DMEM medium containing 4.5 g glucose, 2 mM glutamine, 1% free fatty acid bovine serum albumin, 1% antibiotic and antimycotic solution (Sigma Aldrich). eWAT-conditioned medium (eCM) was collected 24 h after incubation and stored at −80°C for further analysis.
2.6. Plasma and eCM analysis
Enzyme-linked immunosorbent assay (ELISA) kits were used to measure insulin (ALPCO Diagnostics, Salem, NH), FGF21 (R&D Systems, Minneapolis, MN) and Sirt1 (E-EL-M0350, Cliniscience). Lactate, alanine transaminase (ALT), aspartate transaminase (AST) were measured in plasma samples using a Cobas 8000 analyser (Roche, Indianapolis, USA). Cytokines (IL-1β, IL-10 and IL-6) and leptin, adiponectin and resistin were measured using Mesoscale Multiplex and Single-plex plates, respectively (Mesoscale Discovery, Gaithersburg, USA).
2.7. Analysis of plasma lipids and metabolites
Following an 8 h fast, a blood sample was obtained via the orbital sinus under isoflurane anesthesia for analysis of plasma lipids and metabolites. Nuclear magnetic resonance NMR spectroscopy analysis of plasma samples was performed using a 14.0 T Bruker magnet equipped with a Bruker AV-III console operating at 600.13 MHz. All spectra were acquired in 3 mm NMR tubes using a Bruker 5 mm QCI cryogenically cooled NMR probe. Plasma samples were prepared and analyzed according to the Bruker In-Vitro Diagnostics research (IVDr) protocol. Sample preparation consisted of combining 50 μl of plasma with 150 μl of buffer supplied by Bruker Biospin specifically for the IVDr protocol. For 1D 1H NMR, data was acquired using the 1D-NOE experiment, which filters NMR signals associated with broad line widths arising from proteins that might be present in plasma samples that adversely affect spectral quality. Experiment conditions included: sample temperature of 310K, 96k data points, 20 ppm sweep width, a recycle delay of 4 s, a mixing time of 150 ms, and 32 scans. Lipoprotein subclass analysis was performed using regression analysis of the NMR data which is done automatically as part of the IVDr platform as previously described [27].
2.8. Analysis of mRNA expression
After an initial extraction step by mixing Extract-All (Eurobio, France) and chloroform to samples, total RNA purification was performed with a column extraction Kit (RNeasy Mini®, Qiagen, Germany). Double-strand cDNA was synthesized from total RNA with the High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA). Quantitative real-time PCR (qPCR) was performed in a StepOnePlus Real-Time PCR System using commercially available TaqMan primer-probe sets (Life Technologies, Carlsbad, CA). Gene expression was assessed by the comparative CT (ΔΔCT) method with β-actin as the reference gene.
2.9. Western blot analysis
Snap-frozen eWAT samples (200 mg) were lysed in cell lysis buffer (Cell Signaling, Danvers, MA France) supplemented with 1% phenylmethylsulfonyl fluoride (PMSF). Protein samples were resolved on 12% bis-Tris gels followed by transfer to nitrocellulose membrane. Antibodies for AMPK (#2532S), AMPK-P (#2535), Foxo1 (#2880) were from Cell Signaling and antibodies for β-actin (sc-47778) were from Santa Cruz. Bands were visualized by enhanced chemiluminescence and quantified using ImageJ software.
2.10. Histology and immunohistochemistry
Fresh visceral adipose tissue (eWAT) was fixed in 10% phosphate-buffered formalin overnight. Paraffin wax sections of 5 μm were processed for immunostaining. Hematoxylin-eosin (H&E) stained images from mice were analyzed using Adiposoft software. Perilipin antibodies (#9349) were obtained from Cell Signaling, Sirt1 antibodies (GTX17532) were from GeneTex Inc. and CD68 antibodies (ab125512) were from Abcam. Electron microscopy (EM) investigation was performed on eWAT sections fixed with glutaraldehyde and stained with, osmium tetroxyde. Primary adipocytes from eWAT samples were stained with Mitotracker green (M7514) from Invitrogen. 3D-Z stack of images were acquired using confocal microscope Zeiss LSM510 and analysis was performed using Fiji software.
2.11. Pre-adipocytes isolation and culture
Pre-adipocytes were isolated from eWAT obtained from WT or Nrf2−/− mice. Fat pads were placed in Hanks Balanced Salt Solution (HBSS) medium (Life Technologies) under a tissue culture hood, minced into ~1 mm3 pieces and transferred into fresh digestion medium containing 70% HBSS, 30% Bovine Serum Albumin (Sigma Aldrich) and 1 mg/mL collagenase type II (Life Technologies). Samples were incubated at 37°C for 1 h under shaking (120 rpm). The resulting material was filtered through 100 μm mesh and flushed with stop digestion medium collagenase-free. Adipocytes, pre-adipocytes and stromavascular components were separated by centrifugation at 1500 rpm for 5 min at room temperature. The pre-adipocytes were collected and transferred into a new tube with 1:1 v/v DMEM growth media containing 10% new born calf serum, 1% penicillin/streptomycin and 2.4 nM insulin. Pre-adipocytes were centrifuged at 1500 rpm for 5 min at room temperature, medium was removed and the pre-adipocyte pellet was washed with 10 mL growth medium. This washing step was repeated twice. The pre-adipocytes pellet was resuspended, plated in 12 well plate and cultured for 5-6 days for differentiation at 37°C in 5% CO2.
2.12. Preparation of stromal and adipocyte fractions
Stromal cells and mature adipocytes were isolated from epididymal adipose tissue from WT and NRF2−/− male mice. Adipose tissue was harvested, weighed, finely minced and digested in HBSS containing 1 mg/mL collagenase type II (Sigma) for approximately 40 min at 37 °C with gentle shaking. The digested material was passed through a 100 μm filter (Falcon-352360), washed with HBSS+10%FBS+P/S and spun at 300 g × 5min. The medium was removed, floating mature adipocytes and stromal vascular fraction (SVF) were collected and washed again. Isolation of mRNA was performed by RNeasy Lipid Tissue Mini Kit (Quiagen). Adiponectin (ADIPOQ), a late marker of adipocyte differentiation, was used to confirm the purity of the fractions.
2.13. 3T3-L1 cell culture
3T3-L1 murine pre-adipocytes (reference 088SP-L1-F) were purchased from the ZenBio company (NC, USA) and cultured in an atmosphere of 5% CO2 at 37 °C using DMEM supplemented with 10% newborn calf serum. For adipocyte differentiation, cells were stimulated with 3T3-L1 differentiation medium containing IBMX (500 μM), dexamethasone (250 nM) and insulin (175 nM) for 2 days after cells reached confluency. The medium was changed to DMEM containing 10% FBS and insulin (175 nM) after 2 days and adipocytes were then kept into DMEM containing only 10% FBS. Prior to the experiments, adipocytes were subjected to serum deprivation for 16 h with DMEM supplemented with 0.5% FBS.
2.14. siRNA treatment
siRNA for Nrf2 (reference L-040766-00-0005) was purchased from Horizon Discovery LTD company (Cambridge, UK) and siRNA treatment was performed according to manufacturer's instructions.
2.15. Cellular bioenergetic analysis using the Seahorse Bioscience XF analyzer
Bioenergetic profiles of the adipocytes were determined using a Seahorse Bioscience XF24 Analyzer (Billerica, MA, USA) that provides real-time measurements of oxygen consumption rate (OCR), indicative of mitochondrial respiration, and extracellular acidification rate (ECAR), an index of glycolysis as previously described by our group [2].
2.16. Determination of ATP, NAD+ and NADH
NAD+ and NADH levels were determined in frozen adipose tissue samples using an enzymatic method (EnzyChrom, BioAssays Systems, Hayward, CA) according to manufacturer's instructions. Intracellular ATP levels were measured at the end of the experiments with the XF Analyzer using an ATPLiteTM Bioluminescence Assay Kit (PerkinElmer, Courtaboeuf, France) according to manufacturer's instructions.
2.17. Statistical analysis
Data are expressed as mean values ± standard error of the mean (SEM). Statistical analysis was performed by one or two-way analysis of variance (ANOVA) with Fisher multiple comparison test. The result were considered significant at p-value<0.05.
3. Results
3.1. Nrf2-deficient mice exhibit enhanced Sirt1 levels originating from WAT
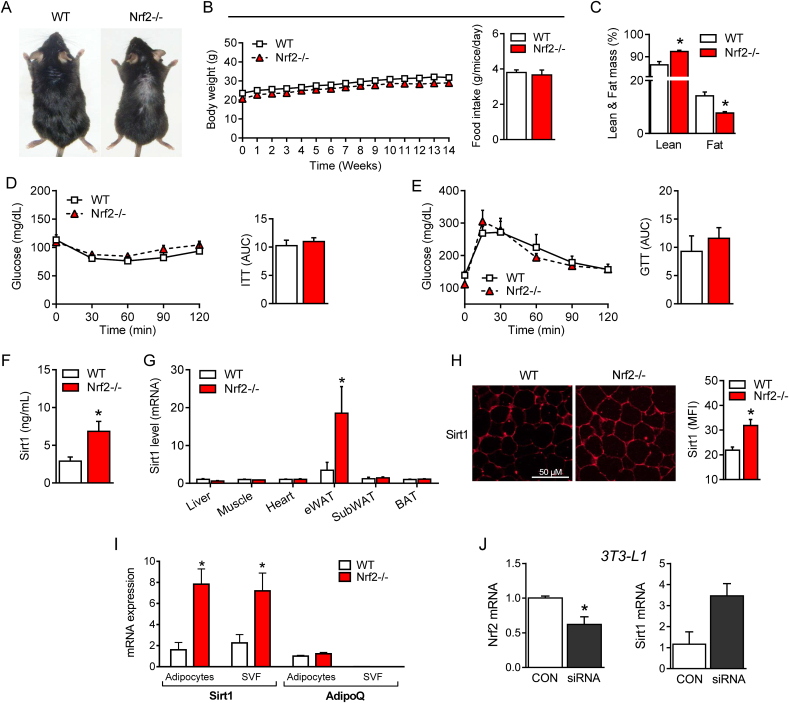
A representative photograph of 22 week-old mice reveals that the Nrf2−/− mouse is slightly smaller than its WT counterpart (Fig. 1A). When mice were fed a standard diet (SD) for 14 weeks, we did not observe a significant difference in the changes of total body weight and food intake between the two groups (Fig. 1B). Nevertheless, Nrf2−/− mice remained slightly smaller and displayed significantly higher total lean mass and lower fat mass compared to WT mice (Fig. 1C). Interestingly, the plasma levels of the main adipose tissue cytokines leptin, adiponectin and resistin were also markedly lower in Nrf2−/− than in WT mice (Table 1). We did not find any difference in glucose and insulin metabolism assessed by the glucose (GTT) and insulin (ITT) tolerance tests, respectively (Fig. 1D and 1E). Similarly, plasma levels of glucose, insulin and lipids (triglycerides, non-esterified fatty acid, cholesterol, apolipoproteins, low and high-density lipoproteins (LDL and HDL, respectively)) were all comparable between WT and Nrf2−/− mice (Table 1). However, among the various parameters measured, we noticed that plasma Sirt1 levels were strongly enhanced in Nrf2−/− mice compared to their WT counterparts (Fig. 1F). To identify the source of Sirt1, we examined the expression of the Sirt1 gene by qPCR in various organs. Interestingly, and compared to WT mice, we detected a significant up-regulation of Sirt1 mRNA expression only in eWAT of Nrf2−/− mice, while no difference was observed in liver, muscle, heart, subcutaneous white (SubWAT) and brown adipose tissues (BAT) (Fig. 1G). Immunofluorescence staining revealed that the enhanced Sirt1 gene expression in eWAT of Nrf2−/− mice coincided with an increase of Sirt1 protein levels (Fig. 1H). Notably, the marked increase in Sirt1 mRNA expression in eWAT of Nrf2−/− mice was originating from both the adipocyte and stromal vascular fraction (SVF) as indicated by the similar level of Sirt1 in these two type of cellular components (Fig. 1I). To confirm further that an increase in Sirt1 expression is due to the absence of Nrf2, we cultured 3T3-L1 fully differentiated adipocytes and silenced the Nrf2 gene by siRNA transfection. As shown in Fig. 1J, we found that ablating Nrf2 in adipocytes causes an increased Sirt1 gene expression. These data clearly show that Nrf2 deficiency results in a distinctive adaptation characterized by increased circulating levels of Sirt1, which most likely originates from enhanced secretion of Sirt1 from eWAT.
Fig. 1.
Metabolic adaptation of Nrf2-deficient mice fed a standard diet (SD) is associated with enhanced circulating Sirt1 levels. (A) Representative photographs of 22-week-old wild-type (WT) and Nrf2-deficient (Nrf2−/−) mice. (B) Changes in body weight (BW) in WT and Nrf2−/− mice during a 14-week SD regime. (C) Lean and fat mass measured by indirect calorimetry in WT and Nrf2−/− mice after 14 weeks of SD. (D) Insulin tolerance test (ITT) and (E) glucose tolerance test (GTT) in WT and Nrf2−/− mice after 14 weeks of SD. (F) Plasma levels of sirtuin 1 (Sirt1) protein and (G) Sirt1 mRNA gene expression in different organs of WT and Nrf2-/-mice after 14 weeks of SD. (H) Immunofluorescence staining for Sirt1 protein in eWAT of WT and Nrf2−/− mice and quantification assessed by mean of fluorescence intensity (MFI). (I) Sirt1 and adiponectin (AdipoQ) mRNA expression in adipocytes and stromal vascular fraction (SVF) separated from eWAT of WT and Nrf2−/− mice. (J) Nrf2 and Sirt1 mRNA expression in 3T3-L1 cells transfected with Nrf2 siRNA. Values represent the mean ± SEM of n = 8–9 mice per group. *p < 0.05 vs. WT mice (white bars).
Table 1.
Plasma biochemistry analysis of WT and Nrf2−/− mice on standard (SD) or high fat diet (HFD).
| SD |
HFD |
|||
|---|---|---|---|---|
| WT | Nrf2−/− | WT | Nrf2−/− | |
| Glucose (mg/dL) | 139.3 ± 11.5 | 111.3 ± 8.4 | 231.1 ± 12.9* | 149.4 ± 7.9# |
| Insulin (ng/mL) | 0.4 ± 0.1 | 0.3 ± 0.1 | 1.2 ± 0.2* | 0.6 ± 0.1 |
| HOMA-IR | 0.8 ± 0.2 | 0.6 ± 0.2 | 2.8 ± 0.6* | 1.3 ± 0.4 |
| Triglycerides (mg/dL) | 143.1 ± 4.8 | 142.6 ± 4.4 | 137.6 ± 4.2 | 147.5 ± 6.3 |
| NEFA (mmol/L) | 0.7 ± 0.03 | 0.7 ± 0.12 | 0.5 ± 0.06 | 0.7 ± 0.06 |
| Cholesterol (mg/dL) | 45.2 ± 2.2 | 45.3 ± 2.2 | 107.8 ± 4.9* | 68.3 ± 5.0*# |
| LDL (mg/dL) | 15.8 ± 1.4 | 14.9 ± 1.0 | 53.2 ± 2.5* | 30.2 ± 2.4*# |
| HDL (mg/dL) | 34.3 ± 1.3 | 31.8 ± 1.2 | 53.6 ± 1.7* | 43.6 ± 2.5*# |
| LDL:HDL (arbitrary units) | 0.4 ± 0.03 | 0.5 ± 0.02 | 1.0 ± 0.03* | 0.7 ± 0.03*# |
| ApoA1 (mg/dL) | 65.7 ± 1.7 | 65.5 ± 1.9 | 89.6 ± 2.4* | 81.3 ± 3.2*# |
| ApoA2 (mg/dL) | 13.7 ± 0.9 | 15.5 ± 0.9 | 12.1 ± 1.1 | 20.8 ± 1.2*# |
| ApoB100 (mg/mL) | 30.4 ± 0.8 | 31.3 ± 0.8 | 51.5 ± 1.9* | 39.2 ± 1.4*# |
| Leptin (ng/mL) | 2.1 ± 0.7 | 0.6 ± 0.2* | 57.9 ± 14.9* | 18.0 ± 2.5 |
| Adiponectin (μg/mL) | 7.8 ± 0.9 | 5.3 ± 0.4* | 12.3 ± 0.7* | 14.2 ± 0.8*# |
| Resistin (ng/mL) | 0.9 ± 0.09 | 0.5 ± 0.05* | 0.5 ± 0.07* | 0.5 ± 0.04* |
| β-hydroxybutyrate (mmol/L) | 0.4 ± 0.05 | 0.4 ± 0.04 | 0.5 ± 0.04 | 0.5 ± 0.04 |
| FGF21 (ng/mL) | 0.64 ± 0.2 | 0.78 ± 0.17 | 1.4 ± 0.17* | 1.98 ± 0.36*# |
Apo: apolipoprotein; HDL: high densitiy lipoprotein; HOMA-IR. Homeostasic model assessment of insulin resistance; LDL: low density lipoprotein; NEFA: Non-esterified fatty acid; FGF21: fibroblast growth factor 21. Results are shown as mean ± SEM. *p < 0.05 vs. SD WT group; #p < 0.05 vs. HFD WT group.
3.2. Nrf2 deficiency enhances lipolysis and fatty acid oxidation in adipose tissue
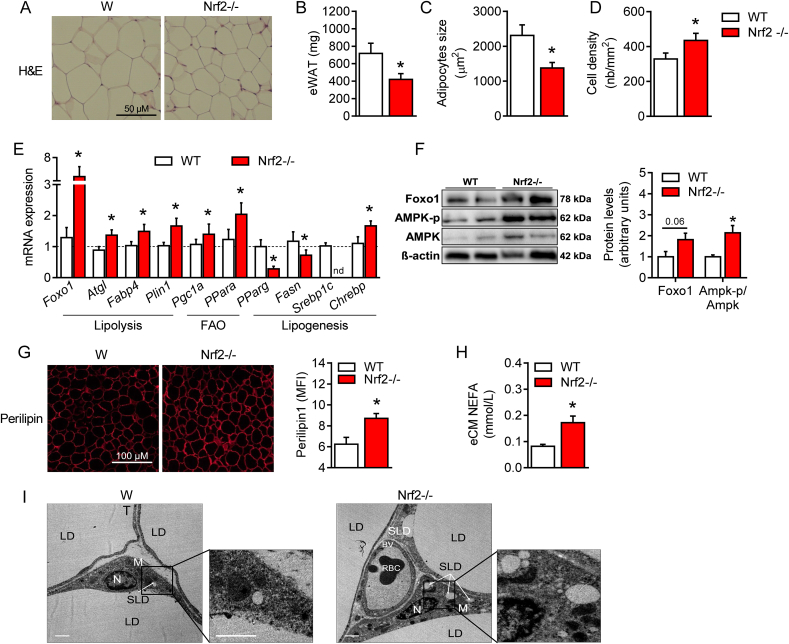
Because Sirt1 controls lipid metabolism in adipocytes [5,28], we further investigated the impact of Nrf2 deficiency on lipolysis, fatty acid oxidation and lipogenesis. We first noticed that the amount of eWAT in Nrf2-deficient mice is significantly reduced, with much smaller and denser adipocytes compared to WT mice (Fig. 2A–D). BAT and SubWAT of Nrf2−/− mice were also smaller and exhibited a reduced adipocyte size than in WT (Supplementary Figs. 1 and 2), even in the absence of increased Sirt1 gene levels (Fig. 1G). Secondly, the expression of several genes involved in lipolysis and fatty acid oxidation in eWAT of Nrf2−/− mice was significantly higher compared to WT. These included forkhead box protein O1 (Foxo1), adipose triglyceride lipase (Atgl), fatty acid binding protein 4 (Fabp4) and perilipin (Plin1), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) and peroxisome proliferator-activated receptor alpha (PPARα) (Fig. 2E). In contrast, lipogenic genes such as peroxisome proliferator-activated receptor gamma (PPARγ), fatty acid synthase (Fasn) and sterol regulatory element-binding transcription factor 1 (Srebp1c) in eWAT of Nrf2−/− were all lower than in WT mice (Fig. 2E). A similar profile for adipocytes lipolytic and lipogenic gene expressions was found in SubWAT and for some genes (i.e. PGC1α) also in BAT (Supplementary Figs. 1 and 2). We also show that increased Fox1 gene expression correlated with up-regulation of FOXO1 protein in eWAT of Nrf2−/− mice in association with enhanced AMPK and phosphorylated-AMPK proteins (Fig. 2F). Since FOXO1, AMPK and phosphorylated-AMPK are downstream targets of Sirt1, these data suggest a Sirt1-driven metabolic adaptation of eWAT resulting from Nrf2-deficiency. Moreover, we found a significant increase in perilipin, a protein known to promote hydrolysis of lipid droplets in adipose tissue (Fig. 2G), and higher levels of non-esterified free fatty acid (NEFA) secreted in the conditioned media from eWAT of Nrf2−/− mice compared to WT (Fig. 2H). Finally, we demonstrated by electron microscopy that eWAT from Nrf2−/− mice contained more lipid droplets than eWAT from WT animals (Fig. 2I). Altogether, these data establish that: 1) Nrf2 deficiency modifies adipose tissue metabolism, driving it towards fatty acids mobilization and elimination; 2) this modification is associated with an increased Sirt1 production, as demonstrated by activation of Sirt1 downstream targets in eWAT and consistent with the recognized role of Sirt1 in fatty acid utilization [28]; 3) in addition to autocrine signaling on eWAT, it is likely that Sirt1 secreted by eWAT and circulating in blood also affects BAT and SubWAT fatty acid metabolism in an endocrine manner.
Fig. 2.
Nrf2 deficiency is characterized by enhanced lipolysis and fatty acid oxidation in adipose tissue. (A) Hematoxylin and eosin (H&E) staining of visceral adipose tissue (eWAT) showing differences in size and cell density from WT and Nrf2−/− mice. (B-D) eWAT weight, adipocytes size and cell density based on the H&E pictures. (E) mRNA expression of genes involved in lipolysis, fatty acid oxidation and lipogenesis in eWAT of WT and Nrf2−/− mice. (F) Western blot analysis of Foxo1, AMPK-p, AMPK and βactin expression levels from eWAT samples. (G) Representative photographs showing perilipin staining in eWAT and its quantification assessed by mean fluorescence intensity (MFI). (H) Non-esterified fatty acid (NEFA) levels released from eWAT conditioned medium (eCM). (I) Electron microscopy of eWAT from WT and Nrf2−/− mice showing: Nucleus (N), Lipid Droplets (LD), Small Lipid Droplets (SLD), Mitochondria (M), Blood Vessel (BV) and Red Blood Cell (RBC). Values represent the mean ± SEM. n = 8–9 mice per group. *p < 0.05 vs. WT mice (white bars).
3.3. Nrf2 deficiency enhances bioenergetic metabolism in adipocytes
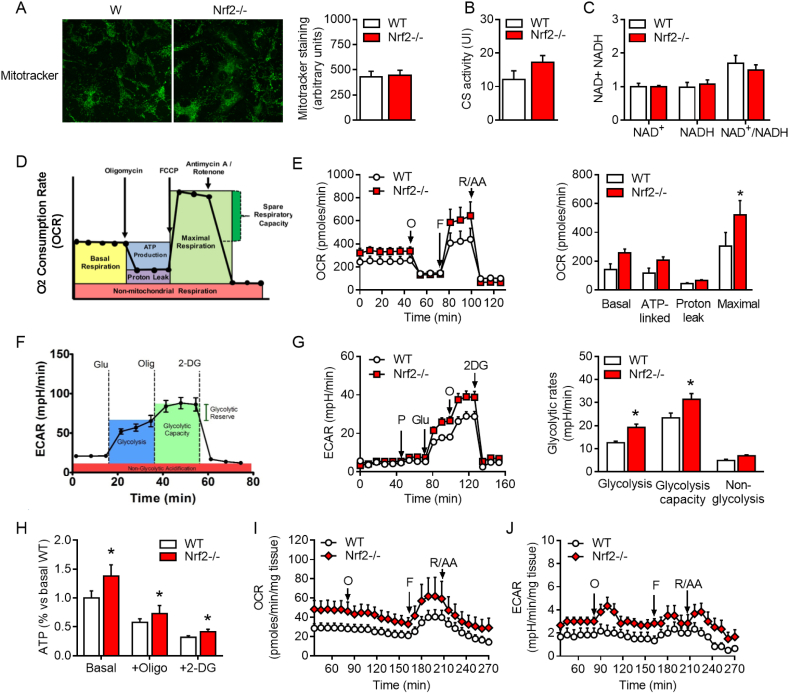
Because of the enhanced fatty acid metabolism in eWAT, we next assessed the impact of Nrf2 deficiency on bioenergetics in adipocytes and adipose tissue punches isolated from Nrf2−/− and WT mice. Mitochondrial content, and NAD+/NADH ratio were not significantly different between the two groups, although citrate synthase activity tended to increase (Fig. 3A, 3B and 3C). Using the Seahorse analyzer (Fig. 3D and 3F), we also found that Nrf2−/− adipocytes displayed a slightly higher basal and ATP-linked oxygen consumption rate (OCR), a significant increase in maximal respiration (Fig. 3E) as well as higher glycolysis and glycolytic capacity compared to WT adipocytes (Fig. 3G). Concomitantly, Nrf2−/− adipocytes exhibited higher ATP levels compared to WT adipocytes (Fig. 3H). The increased ATP originated from both oxidative phosphorylation, as evidenced by the experiments conducted in the presence of the glycolysis inhibitor 2-deoxyglucose, and glycolysis, revealed by the experiments performed in the presence of the ATP synthesis inhibitor oligomycin (Fig. 3H). Importantly, a similar increase in mitochondrial oxygen consumption and glycolytic activity was found in ex vivo adipose tissue punches collected from Nrf2−/− compared to WT eWAT (Fig. 3I and 3J). These results indicate that adipocytes from Nrf2−/− mice are more metabolically active in vitro and ex vivo. Accordingly, Nrf2−/− mice displayed increased plasma levels of both citric acid and lactic acid compared with WT mice under a SD regime (Table 2), confirming that Nrf2 deficiency is characterized by an overall increase in energetic metabolism in adipocytes.
Fig. 3.
Mitochondrial bioenergetics and glucose metabolism in Nrf2-deficient adipocytes. (A) Immunofluorescence staining for Mitotracker in primary adipocytes isolated from eWAT tissue collected from WT or Nrf2−/− mice. (B-C) Citrate synthase (CS) activity and NAD+/NADH ratio in eWAT tissue from WT and Nrf2−/− mice. (D-E) Oxygen consumption rate (OCR) was measured by a MitoStress assay (Seahorse Analyzer) performed on primary adipocytes with a sequential addition of oligomycin (O), FCCP, and rotenone/antimycin A (R/AA). (F-G) Extracellular acidification rate (ECAR) was measured by a Glycolytic assay (Seahorse Analyzer) performed on primary adipocytes with a sequential addition of pyruvate (P), Glucose (Glu), Oligomycin (O) and 2-deoxyglucose (2-DG). (H) ATP content in primary adipocytes isolated from eWAT of WT and Nrf2−/− mice. (I-J) OCR and ECAR measured by the Seahorse Analyzer on punches of eWAT collected from WT and Nrf2−/− mice. Values represent the mean ± SEM of n = 5–6 mice per group. *p < 0.05 vs. WT mice.
Table 2.
Plasma energy metabolites in WT and Nrf2−/− mice on standard (SD) or high-fat diet (HFD).
| SD |
HFD |
|||
|---|---|---|---|---|
| WT | Nrf2−/− | WT | Nrf2−/− | |
| Keto Acids | ||||
| Pyruvic acid (mmol/L) | 0.047 ± 0.003 | 0.056 ± 0.009 | 0.052 ± 0.003 | 0.071 ± 0.007 |
| Acetic acid (mmol/L) | 0.040 ± 0.003 | 0.089 ± 0.011* | 0.038 ± 0.003* | 0.045 ± 0.006 |
| Carboxylic Acids | ||||
| Citric acid (mmol/L) | 0.07 ± 0.005 | 0.14 ± 0.004* | 0.08 ± 0.007 | 0.11 ± 0.005*# |
| Formic acid (mmol/L) | 0.027 ± 0.002 | 0.029 ± 0.002 | 0.023 ± 0.001 | 0.028 ± 0.003 |
| Lactic acid (mmol/L) | 1.5 ± 0.14 | 2.2 ± 0.015* | 1.6 ± 0.08 | 2.1 ± 0.12*# |
| Amino Acids | ||||
| Alanine (nmol/L) | 0.11 ± 0.005 | 0.16 ± 0.007* | 0.14 ± 0.005* | 0.14 ± 0.009 |
| Glutamine (nmol/L) | 0.15 ± 0.006 | 0.25 ± 0.01* | 0.23 ± 0.01* | 0.18 ± 0.01# |
| Histidine (nmol/L) | 0.02 ± 0.004 | 0.03 ± 0.002 | 0.03 ± 0.002 | 0.03 ± 0.002* |
| Leucine (nmol/L) | 0.03 ± 0.002 | 0.03 ± 0.003 | 0.03 ± 0.001 | 0.04 ± 0.002 |
| Valine (nmol/L) | 0.05 ± 0.003 | 0.06 ± 0.003 | 0.06 ± 0.003 | 0.05 ± 0.003 |
Results are shown as mean ± SEM. *p < 0.05 vs. SD WT group; #p < 0.05 vs. HFD WT group.
3.4. Improved glucose metabolism and lipid profile in Nrf2-deficient mice fed a high fat diet (HFD)
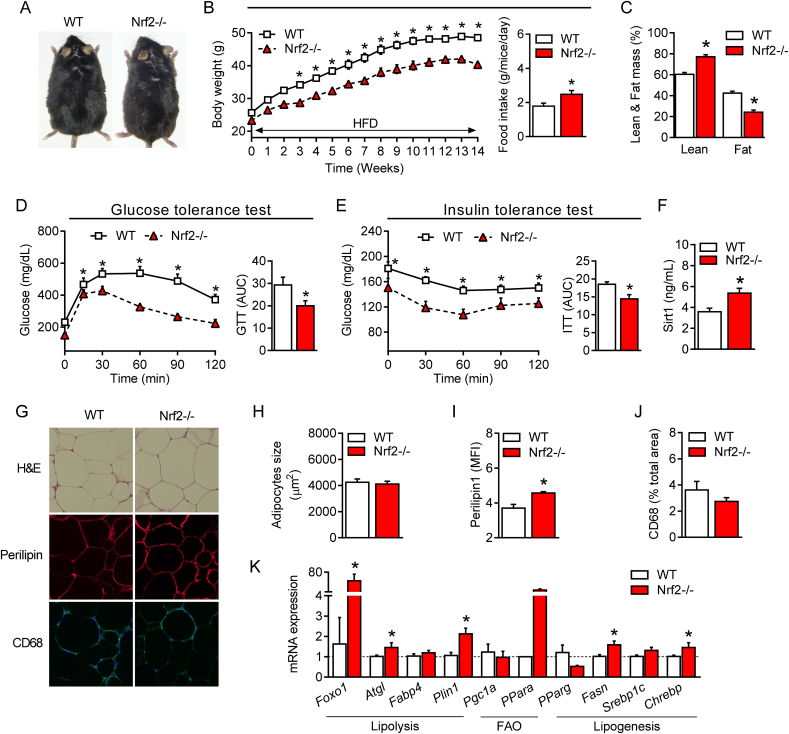
We next asked whether the beneficial changes in glucose and lipid metabolism promoted by Nrf2 deficiency protect against obesity. For this, WT and Nrf2−/− mice were subjected to a HFD regime for 14 weeks as previously described by our group [2]. As shown in Fig. 4, at the end of this protocol Nrf2−/− mice displayed a lower body weight despite having a higher food intake (Fig. 4A and 4B) compared to WT mice. Nrf2−/− mice also had a higher lean mass and a lower fat mass (Fig. 4C) as well as improved glucose metabolism and insulin tolerance compared to their WT counterpart (Fig. 4D and 4E), confirming previously published data [20]. In line with these findings, Nrf2−/− mice under HFD showed reduced levels of plasma glycemia and insulin after fasting and improved HOMA-IR (Table 1). Although plasma triglyceride and NEFA levels remained unchanged, HDL, LDL, ApoA1, ApoA2 and ApoB100 were all significantly reduced in Nrf2−/− mice after HFD (Table 1). As previously reported [22], plasma levels and the hepatic gene expression of FGF21 were significantly higher in Nrf2−/− mice (Table 1 and Supplementary Fig. 3G) together with increased circulating lactic acid and citric acid levels (Table 2). Interestingly, and similarly to mice fed a SD, Nrf2−/− obese mice maintained significantly higher levels of plasma Sirt1 compared to WT obese mice (Fig. 4F). No difference in the size of adipocytes was observed between the two groups (Fig. 4G and 4H) but the visceral adipose tissue of Nrf2−/− mice still displayed a different profile compared to WT. This included increased perilipin content (Fig. 4G and 4I) and an elevated expression of genes involved in lipolysis and fatty acid oxidation (Fig. 4K). Moreover, a slight, non significant, decrease in macrophage infiltration, which is known to contribute to adipose tissue low-grade inflammation during obesity, was observed in Nrf2−/− mice (Fig. 4G and 4J). Of note, the metabolic protection afforded by Nrf2 deficiency against HFD was evident not only in the adipose tissue but also in the liver, where our analysis revealed reduced hepatic injury (ALT/AST) and steatosis and decreased expression of certain genes involved in lipogenesis (Supplementary Figs. 3A–C). These data support the concept that Nrf2 deficiency counteracts the metabolic derangements caused by HFD resulting in improved phenotype of the adipose tissue and other organs such as the liver.
Fig. 4.
Improved glucose metabolism, insulin sensitivity and lipid profile in obese Nrf2-deficient mice. (A) Representative photographs of 22-week-old wild-type (WT) and Nrf2-deficient (Nrf2−/−) mice fed a HFD for 14 weeks. (B) Changes in body weight (BW) in WT and Nrf2−/− mice during a 14-week HFD regime. (C) Percentage of lean and fat mass and (D) plasma levels of Sirtuin 1 (Sirt1) protein in obese WT and Nrf2−/− mice. (E) Glucose tolerance test (GTT) and (F) insulin tolerance test (ITT) in WT and Nrf2−/− mice after 14 weeks under HFD. (G) Hematoxylin and eosin (H&E) analysis and immunofluorescence staining for perilipin and CD68 of visceral adipose tissue (eWAT) collected from obese WT and Nrf2-/-mice. (H, I and J) Quantification of adipocyte size, perilipin and CD68 in eWAT of obese WT and Nrf2−/− mice. (K) mRNA gene expression of genes involved in lipid metabolism in eWAT of obese WT and Nrf2−/− mice. . Values represent the mean ± SEM of n = 8–9 mice per group. *p < 0.05 vs. WT mice. MFI: mean fluorescence intensity.
3.5. eWAT removal suppresses the increase in Sirt1 and impairs glucose tolerance in Nrf2-deficient obese mice
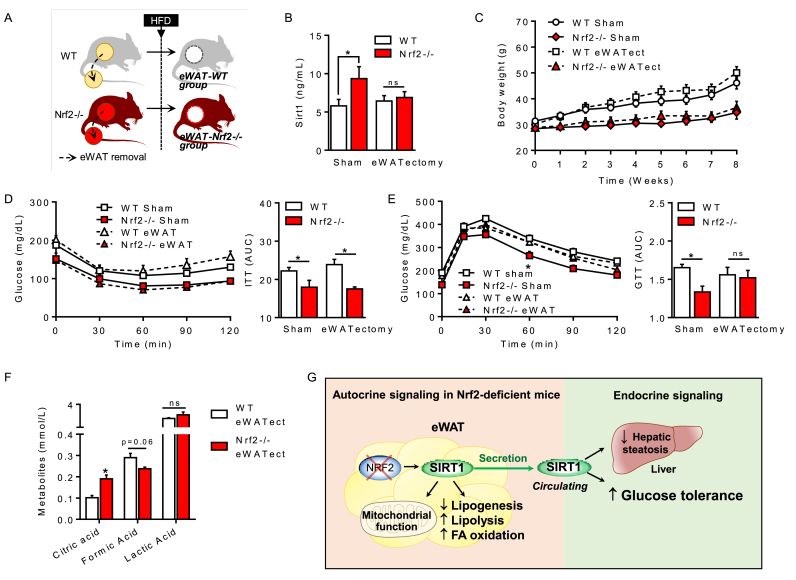
Since eWAT of Nrf2−/− mice is the only tissue expressing high Sirt1 levels, we hypothesized that eWAT-derived Sirt1 is responsible for protecting Nrf2−/− mice against the metabolic disruption caused by HFD. To test this hypothesis, we directly removed eWAT from WT and Nrf2−/− mice (eWATectomy) prior to subjecting them to HFD for 8 weeks (see schematic diagram in Fig. 5A). Plasma levels of Sirt1, body weight and metabolic parameters were assessed at the end of the protocol. Notably, we found that removal of fat completely abolished the increase in circulating Sirt1 characteristic of Nrf2 deficiency to levels comparable to sham-operated WT mice (Fig. 5B), unequivocally demonstrating that eWAT is the main source of Sirt1 in Nrf2−/− mice. Importantly, Nrf2−/− mice subjected to eWATectomy became glucose intolerant in a manner similar to WT mice under HFD (Fig. 5E) but retained their enhanced insulin sensitivity in both sham and eWATectomy conditions (Fig. 5D). Moreover, eWATectomy did not affect body weight gain induced by HFD since Nrf2−/− mice still exhibited lower body weight gain compared to WT (Fig. 5C). We also found that lactic acid, which was augmented in Nrf2−/− mice both under SD and HFD regimes (Table 2), was normalized by eWATectomy to WT levels (Fig. 5F) while circulating citric acid remained elevated. Circulating lipids were still similar in Nrf2-deficient animals before and after eWATectomy (data not shown). Taken together, these results indicate that Sirt1 produced by eWAT is a critical factor in preserving improved glucose metabolism in Nrf2−/− mice during obesity.
Fig. 5.
Removal of adipose tissue abolishes the increase in Sirt1 and suppresses the improved glucose metabolism in Nrf2−/−obese mice. (A) Schematic representation of the visceral (epididymal) white adipose tissue removal (eWATectomy) protocol in WT and Nrf2−/− mice. Sham groups underwent the same surgical procedure without fat removal. After eWATectomy, WT and Nrf2−/− mice were subjected to an 8-week HFD regime before analysis of samples. (B) Plasma levels of Sirt1 protein in obese WT and Nrf2−/− mice after eWATectomy. (C) Changes in body weight of WT and Nrf2−/− mice without (sham) or with eWATectomy during the 8 week HFD regime. (D) Insulin tolerance test (ITT) and (E) glucose tolerance test (GTT) were performed in WT and Nrf2−/− obese mice after eWATectomy at week 7. (F) Plasma levels of different metabolites in WT and Nrf2−/− obese mice following eWATectomy. (G) Proposed mechanisms underlying the protective effects of Nrf2 deletion on glucose and lipid metabolism. Values represent the mean ± SEM of n = 10 mice per group. *p < 0.05 indicate statistical difference vs. respective control groups.
4. Discussion
Our study reveals for the first time that Nrf2 deficiency in lean and obese mice is characterized by increased systemic levels of Sirt1. We establish the visceral adipose tissue as the sole source of high Sirt1 protein expression that leads to increased Sirt1 plasma levels. We further demonstrate that removal of eWAT in Nrf2−/− mice restores circulating Sirt1 to levels of WT mice. Interestingly, we show that Nrf2−/− mice are protected from insulin and glucose intolerance when subjected to HFD but that, following eWATectomy, insulin sensitivity is maintained while glucose tolerance is completely lost. Our study identifies Sirt1 as a downstream regulator participating to the beneficial effects of Nrf2 deficiency on glucose homeostasis in mice during nutritional stress induced by HFD.
We first observed that lean Nrf2−/− mice exhibited higher lean mass and lower fat mass than WT mice without significant changes in whole body weight. This difference in body composition was maintained under HFD but with the important distinction that Nrf2−/− mice gained less body weight compared to WT obese mice. This is consistent with previous studies reporting a protective role of Nrf2 deficiency in obese mice through FGF21 overexpression after challenge with HFD [22]. However, in that study and ours, lean Nrf2−/− mice did not display increased FGF21 levels and it was unclear why Nrf2 deficiency caused a different body composition even under standard diet. We therefore looked for other plasma metabolic markers that could explain this difference between the genotypes and discovered that Sirt1 was significantly higher in the circulation of lean Nrf2−/− mice. By evaluating its gene expression in different organs, we demonstrated that in Nrf2−/− mice, the visceral adipose tissue was the only tissue where Sirt1 was elevated compared to WT. We fully validated this finding by showing that this increase was not detected in the liver, muscle, heart, SubWAT and BAT and that circulating Sirt1 returned to WT levels when eWATectomy was carried out in Nrf2−/− mice. Interestingly, Sirt1 levels remained higher also after HFD, supporting its contribution to the beneficial metabolic adaptation of Nrf2−/− mice to an obesogenic regime.
It is intriguing that Sirt1 levels increased exclusively in the adipose tissue in response to whole mouse Nrf2 deficiency and a detailed investigation is required to understand the underlying mechanisms. Nevertheless, we speculate that Nrf2 suppresses Sirt1 expression, as indicated also by our preliminary findings showing that silencing Nrf2 with siRNA in 3T3-L1 pre-adipocytes is directly associated with up-regulation of Sirt1 mRNA. Whether this is due to direct control of Sirt1 gene expression by the Nrf2 transcription factor or because Nrf2 deficiency results in higher oxidative stress that in turn may stimulate Sirt1 [29] is unknown. A very recent report showed that HFD induced marked oxidative stress in adipose tissue with concomitant increase in Nrf2 [30], suggesting that oxidative stress may be a participating factor. These authors also reported that Nrf2 activation promoted lipid accumulation and lipogenesis in adipocytes during obesity [30], supporting our current results on the beneficial role of Nrf2 deficiency against HFD-mediated lipid dysregulation.
Sirt1 is known to be a key metabolic sensor in various metabolic tissues [31]. Sirt1 promotes lipolysis via FOXO1 and adipose triglyceride lipase (ATGL)-dependent mechanisms [28] and stimulates fat utilization in white adipose tissue by repressing PPARγ [5]. Consistent with this, our results demonstrated that Nrf2 deficiency led to decreased adipocyte size, increased expression of the lipolysis-related genes Foxo1, Atgl, Fabp4, Perilipin1 and decreased expression of the adipogenesis gene PPARγ in both lean and obese mice. A higher lypolysis activity was corroborated by electronic microscopy imaging of visceral adipose tissue showing more numerous lipid droplets in Nrf2−/− than WT mice, thus indicating a more metabolically active adipose tissue in Nrf2−/− animals. This effect was accompanied by enhanced mitochondrial oxidative and glycolytic activities in primary adipocytes in vitro and in ex vivo adipose tissue. Although a previous study on embryonic fibroblast cells from Nrf2-deficient mice showed an increased oxygen consumption compared to WT cells [32], other reports have demonstrated a decrease in oxygen consumption in cells lacking Nrf2 [33] but these studies were performed on cell types different from adipocytes. Our main findings on gene expression data in different adipose tissue depots, phenotypic and plasma analysis of circulating HDL and LDL as well as apoproteins levels and bioenergetic profile of eWAT strongly support a positive impact of Nrf2 deficiency on systemic metabolism that becomes highly relevant during HFD conditions.
Our work highlights that Sirt1 activation is a crucial adaptive mechanism following the lack of Nrf2, adding to previous investigations which identified increased expression of FGF21 as contributing factor to the improved metabolic phenotype of Nrf2−/− mice under HFD [22]. We also found that plasma levels of FGF21 were elevated in Nrf2−/− mice after HFD and we suggest here that Sirt1 produced by the adipose tissue and FGF21 originating from the liver act in a concerted manner to ameliorate energy homeostasis of Nrf2 deficient animals under HFD. Recent studies report that Sirt1 promotes FGF21 signaling in oxytocin neurons and stimulates oxytocin transcription through Nrf2, contributing to the homeostatic regulation of macronutrient-based diet selection in mice [34,35]. Thus, we cannot exclude that Sirt1 overproduction in eWAT of Nrf2−/− mice may also affect FGF21 levels via an endocrine action on the liver.
Our findings on improved insulin-resistance and glucose intolerance in murine Nrf2 deficiency after HFD are in line with previous studies [[19], [20], [21], [22]]. Calorimetry assessments have also shown that Nrf2−/− mice under HFD display a significant increase in energy expenditure compared to WT mice [30,32]. This, combined with our novel data on Sirt1 overexpression derived exclusively from eWAT, prompted us to determine the specific role of Sirt1 on metabolism in Nrf2−/− mice. Because we observed a reduction of Sirt1 to WT levels after eWATectomy in Nrf2-deficient mice, we asked ourselves: what would be the metabolic consequences of subjecting Nrf2−/− mice lacking eWAT to HFD? Our results clearly demonstrate that following the HFD regime, Nrf2−/− mice still exhibit improved insulin sensitivity but lose the protection against glucose intolerance, narrowing down the unique impact of Sirt1 on glucose metabolism in these conditions. Plasma lactic acid also returned to WT levels after eWATectomy, substantiating the role of eWAT-derived Sirt1 on glucose metabolism in Nrf2−/− mice and consistent with a study demonstrating the importance of Sirt1 in glucose tolerance but not insulin resistance [4]. In contrast, circulating citric acid remained elevated, indicating that other players, originating from tissues different from eWAT, contribute to metabolic adaptation in Nrf2−/− mice. These additional factors (including FGF21) likely explain why Nrf2 deficient animals are still protected from HFD-induced weight gain and insulin resistance even after eWATectomy.
5. Conclusions
In conclusion, our study advances our understanding on the role Nrf2 plays in modulating energetic metabolism and identifies novel pathways that are controlled by this master regulator of the stress response. This is the first study reporting a link between Nrf2 deficiency and Sirt1 expression in visceral adipose tissue and their beneficial implications in the regulation of glucose metabolism. The use of Nrf2 inhibitors alone or in combination with other drugs could be an effective therapeutic strategy to counteract the metabolic dysfunction and glucose deregulation that characterize diseases such as obesity and type 2 diabetes.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by a public grant overseen by the French National Research Agency (ANR) as part of the second “Investissements d'Avenir” programme (CARMMA Project, reference: ANR‐15‐RHUS‐0003). The NMR instrumentation was supported in part by an NIH grant (S10 RR025677) and Vanderbilt University matching funds. The authors thank Florine Weber and Philippe Druelle for help in the animal facility platform; Xavier Decrouy, Christelle Gandolphe and Wilfried Verbecq-Morlot from the histology platform; Alain Schmitt from the electronic microscopy platform of Institut Cochin; Dr. Costin Radu for kindly showing the surgical procedure to remove adipose tissue (eWATectomy) and Stéphane Moutereau at Henri Mondor Hospital for blood analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101805.
Contributor Information
Laura Braud, Email: laura.braud@inserm.fr.
Roberta Foresti, Email: roberta.foresti@inserm.fr.
Roberto Motterlini, Email: roberto.motterlini@inserm.fr.
Abbreviations
- ALT
alanine transaminase
- AST
aspartate transaminase
- DMEM
Dulbecco's Modified Eagle Medium
- DPBS
Dulbecco Phosphate Buffer Solution
- ECAR
extracellular acidification rate
- eWAT
epididymal adipose tissue
- FBS
fetal bovine serum
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- GTT
glucose tolerance test
- HBSS
Hanks Balanced Salt Solution
- HFD
high fat diet
- ITT
insulin tolerance test
- OCR
oxygen consumption rate
- SD
standard diet
- SIRT 1
sirtuin-1
Authors’ contribution
L.B., R.F. and R.M. designed the study, performed experiments, collected and analyzed data and wrote the manuscript. L.B. and M.P. performed metabolic tests and their analysis. D.F.S. and D.E.S. performed metabolomic analysis of keto acids, carboxylic acids and amino acids as well as analysis of plasma lipids. S.M. performed RNA and protein extractions, ELISA and immunostaining. G.D. read the manuscript. R.M. is the guarantor of this work and, as such, had full access to all the data in this study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1. Effects of Nrf2 deficiency on brown adipose tissue in lean and obese mice. Mice received a standard diet (SD) or a high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of brown adipose tissue (BAT) in WT and Nrf2-/- mice. (B) Hematoxylin and eosin (H&E) staining of BAT in mice. (C-D) Percentage of lipid and mRNA expression of genes involved in metabolism in BAT of WT and Nrf2-/- mice. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM, n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.
Supplementary Figure 2. Effects of Nrf2 deficiency on subcutaneous adipose tissue in lean and obese mice. Mice received a standard diet (SD) or a high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of the subcutaneous adipose tissue (SubWAT) in WT and Nrf2-/- mice. (B) Hematoxylin and eosin (H&E) staining of subWAT in WT and Nrf2-/- mice. (C-D) Quantification of adipocyte size and mRNA expression of genes involved in metabolism in subWAT of WT and Nrf2-/- mice. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM of n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.
Supplementary Figure 3. Effects of Nrf2 deficiency on hepatic function and metabolic profile in lean and obese mice. Mice received a standard diet (SD) or high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of liver in WT and Nrf2-/- mice. (B-D) Plasma levels of aspartate amino transferase (AST), alanine amino transferase (ALAT) and the ratio AST/ALAT in WT and Nrf2-/- mice. (E-F) Hematoxylin and eosin (H&E) staining of liver samples and percentage of hepatic lipid droplets in WT and Nrf2-/- mice. (G) mRNA expression of hepatic genes involved in metabolism. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM of n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.
References
- 1.Ronti T., Lupattelli G., Mannarino E. The endocrine function of adipose tissue: an update. Clin. Endocrinol. (Oxf.). 2006;64:355–365. doi: 10.1111/j.1365-2265.2006.02474.x. [DOI] [PubMed] [Google Scholar]
- 2.Braud L., Pini M., Muchova L., Manin S., Kitagishi H., Sawaki D., Czibik G., Ternacle J., Derumeaux G., Foresti R., Motterlini R. Carbon monoxide–induced metabolic switch in adipocytes improves insulin resistance in obese mice. JCI Insight. 2018;3 doi: 10.1172/jci.insight.123485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y., Rosenbaum M., Zhao Y., Gu W., Farmer S.R., Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfluger P.T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., de Oliveira R.M., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Wong K., Giles A., Jiang J., Lee J.W., Adams A.C., Kharitonenkov A., Yang Q., Gao B., Guarente L., Zang M. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology. 2014;146:539–549. doi: 10.1053/j.gastro.2013.10.059. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Zhao G., Chen L., Ding Y., Lian J., Hong G., Lu Z. Resveratrol protects mice from paraquat-induced lung injury: the important role of SIRT1 and NRF2 antioxidant pathways. Mol. Med. Rep. 2016;13:1833–1838. doi: 10.3892/mmr.2015.4710. [DOI] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakabayashi N., Dinkova-Kostova A.T., Holtzclaw W.D., Kang M.-I., Kobayashi A., Yamamoto M., Kensler T.W., Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balogun E., Hoque M., Gong P., Killeen E., Green C.J., Foresti R., Alam J., Motterlini R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 2003;371:887–895. doi: 10.1042/bj20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.-L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 13.Keleku-Lukwete N., Suzuki M., Otsuki A., Tsuchida K., Katayama S., Hayashi M., Naganuma E., Moriguchi T., Tanabe O., Engel J.D., Imaizumi M., Yamamoto M. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:12169–12174. doi: 10.1073/pnas.1509158112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Shin S., Wakabayashi J., Yates M.S., Wakabayashi N., Dolan P.M., Aja S., Liby K.T., Sporn M.B., Yamamoto M., Kensler T.W. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur. J. Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagishita Y., Uruno A., Fukutomi T., Saito R., Saigusa D., Pi J., Fukamizu A., Sugiyama F., Takahashi S., Yamamoto M. Nrf2 improves leptin and insulin resistance provoked by hypothalamic oxidative stress. Cell Rep. 2017;18:2030–2044. doi: 10.1016/j.celrep.2017.01.064. [DOI] [PubMed] [Google Scholar]
- 18.Uruno A., Furusawa Y., Yagishita Y., Fukutomi T., Muramatsu H., Negishi T., Sugawara A., Kensler T.W., Yamamoto M. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol. Cell Biol. 2013;33:2996–3010. doi: 10.1128/MCB.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Dou W., Ni Z., Wen Q., Zhang R., Qin M., Wang X., Tang H., Cao Y., Wang J., Zhao S. Deletion of Nrf2 leads to hepatic insulin resistance via the activation of NF-κB in mice fed a high-fat diet. Mol. Med. Rep. 2016;14:1323–1331. doi: 10.3892/mmr.2016.5393. [DOI] [PubMed] [Google Scholar]
- 20.Pi J., Leung L., Xue P., Wang W., Hou Y., Liu D., Yehuda-Shnaidman E., Lee C., Lau J., Kurtz T.W., Chan J.Y. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Dasuri K., Fernandez-Kim S.-O., Bruce-Keller A.J., Keller J.N. Adipose-specific ablation of Nrf2 transiently delayed high-fat diet-induced obesity by altering glucose, lipid and energy metabolism of male mice. Am. J. Transl. Res. 2016;8:5309–5319. [PMC free article] [PubMed] [Google Scholar]
- 22.Chartoumpekis D.V., Ziros P.G., Psyrogiannis A.I., Papavassiliou A.G., Kyriazopoulou V.E., Sykiotis G.P., Habeos I.G. Nrf2 represses FGF21 during long-term high-fat diet-induced obesity in mice. Diabetes. 2011;60:2465–2473. doi: 10.2337/db11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y.-K.J., Wu K.C., Liu J., Klaassen C.D. Nrf2 deficiency improves glucose tolerance in mice fed a high-fat diet. Toxicol. Appl. Pharmacol. 2012;264:305–314. doi: 10.1016/j.taap.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S., Muniyappa R., Yan X., Chen H., Yue L.Q., Hong E.-G., Kim J.K., Quon M.J. Comparison between surrogate indexes of insulin sensitivity and resistance and hyperinsulinemic euglycemic clamp estimates in mice. Am. J. Physiol. Endocrinol. Metab. 2008;294:E261–E270. doi: 10.1152/ajpendo.00676.2007. [DOI] [PubMed] [Google Scholar]
- 25.Hosick P.A., AlAmodi A.A., Hankins M.W., Stec D.E. Chronic treatment with a carbon monoxide releasing molecule reverses dietary induced obesity in mice. Adipocyte. 2016;5:1–10. doi: 10.1080/21623945.2015.1038443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinds Terry D., Jr., Hosick Peter A., Chen Shujuan, Tukey Robert H., Hankins Michael W., Nestor-Kalinoski Andrea, Stec David E. Mice with hyperbilirubinemia due to Gilbert's syndrome polymorphism are resistant to hepatic steatosis by decreased serine 73 phosphorylation of PPARα. Am. J. Physiol. Endocrinol. Metab. 2017;312:E244–E252. doi: 10.1152/ajpendo.00396.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen M., Dyrby M., Toubro S., Engelsen S.B., Nørgaard L., Pedersen H.T., Dyerberg J. Quantification of lipoprotein subclasses by proton nuclear magnetic resonance-based partial least-squares regression models. Clin. Chem. 2005;51:1457–1461. doi: 10.1373/clinchem.2004.046748. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarti P., English T., Karki S., Qiang L., Tao R., Kim J., Luo Z., Farmer S.R., Kandror K.V. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J. Lipid Res. 2011;52:1693–1701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salminen A., Kaarniranta K., Kauppinen A. Crosstalk between oxidative stress and SIRT1: impact on the aging process. Int. J. Mol. Sci. 2013;14:3834–3859. doi: 10.3390/ijms14023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun X., Li X., Jia H., Wang H., Shui G., Qin Y., Shu X., Wang Y., Dong J., Liu G., Li X. Nuclear factor E2-related factor 2 mediates oxidative stress-induced lipid accumulation in adipocytes by increasing adipogenesis and decreasing lipolysis. Antioxidants Redox Signal. 2019 doi: 10.1089/ars.2019.7769. [DOI] [PubMed] [Google Scholar]
- 31.Li X. SIRT1 and energy metabolism. Acta Biochim. Biophys. Sin. 2013;45:51–60. doi: 10.1093/abbs/gms108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider K., Valdez J., Nguyen J., Vawter M., Galke B., Kurtz T.W., Chan J.Y. Increased energy expenditure, Ucp1 expression, and resistance to diet-induced obesity in mice lacking nuclear factor-erythroid-2-related transcription factor-2 (Nrf2) J. Biol. Chem. 2016;291:7754–7766. doi: 10.1074/jbc.M115.673756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinkova-Kostova A.T., Abramov A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015;88:179–188. doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki T., Kitamura T. Roles of FoxO1 and Sirt1 in the central regulation of food intake. Endocr. J. 2010;57:939–946. doi: 10.1507/endocrj.k10e-320. [DOI] [PubMed] [Google Scholar]
- 35.Matsui S., Sasaki T., Kohno D., Yaku K., Inutsuka A., Yokota-Hashimoto H., Kikuchi O., Suga T., Kobayashi M., Yamanaka A., Harada A., Nakagawa T., Onaka T., Kitamura T. Neuronal SIRT1 regulates macronutrient-based diet selection through FGF21 and oxytocin signalling in mice. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-07033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Effects of Nrf2 deficiency on brown adipose tissue in lean and obese mice. Mice received a standard diet (SD) or a high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of brown adipose tissue (BAT) in WT and Nrf2-/- mice. (B) Hematoxylin and eosin (H&E) staining of BAT in mice. (C-D) Percentage of lipid and mRNA expression of genes involved in metabolism in BAT of WT and Nrf2-/- mice. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM, n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.
Supplementary Figure 2. Effects of Nrf2 deficiency on subcutaneous adipose tissue in lean and obese mice. Mice received a standard diet (SD) or a high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of the subcutaneous adipose tissue (SubWAT) in WT and Nrf2-/- mice. (B) Hematoxylin and eosin (H&E) staining of subWAT in WT and Nrf2-/- mice. (C-D) Quantification of adipocyte size and mRNA expression of genes involved in metabolism in subWAT of WT and Nrf2-/- mice. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM of n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.
Supplementary Figure 3. Effects of Nrf2 deficiency on hepatic function and metabolic profile in lean and obese mice. Mice received a standard diet (SD) or high fat diet (HFD) for 14 weeks and sacrificed at the end of the experiments. (A) Weight of liver in WT and Nrf2-/- mice. (B-D) Plasma levels of aspartate amino transferase (AST), alanine amino transferase (ALAT) and the ratio AST/ALAT in WT and Nrf2-/- mice. (E-F) Hematoxylin and eosin (H&E) staining of liver samples and percentage of hepatic lipid droplets in WT and Nrf2-/- mice. (G) mRNA expression of hepatic genes involved in metabolism. mRNA expression was determined by real-time PCR and normalized to β-actin. Values represent the mean ± SEM of n=6-8 per group. *p<0.05 vs. control group (SD) and #p<0.05 vs. HFD group.