Gihana et al. demonstrate that Cdc14 phosphatase activity, released from the nucleolus through the mitotic exit network, terminates the asymmetric localization of Cdc42 at the end of mitosis in budding yeast. Redistribution of Cdc42 activity between the bud and mother preserves normal cell size and mitotic spindle position.
Abstract
During G1 in budding yeast, the Cdc42 GTPase establishes a polar front, along which actin is recruited to direct secretion for bud formation. Cdc42 localizes at the bud cortex and then redistributes between mother and daughter in anaphase. The molecular mechanisms that terminate Cdc42 bud-localized activity during mitosis are poorly understood. We demonstrate that the activity of the Cdc14 phosphatase, released through the mitotic exit network, is required for Cdc42 redistribution between mother and bud. Induced Cdc14 nucleolar release results in premature Cdc42 redistribution between mother and bud. Inhibition of Cdc14 causes persistence of Cdc42 bud localization, which perturbs normal cell size and spindle positioning. Bem3, a Cdc42 GAP, binds Cdc14 and is dephosphorylated at late anaphase in a Cdc14-dependent manner. We propose that Cdc14 dephosphorylates and activates Bem3 to allow Cdc42 inactivation and redistribution. Our results uncover a mechanism through which Cdc14 regulates the spatiotemporal activity of Cdc42 to maintain normal cell size at cytokinesis.
Introduction
Chromosome partitioning is coordinated with cell surface growth to ensure that, at every division cycle, daughter cells reach appropriate and consistent size to accommodate the nucleus and other organelles. In polarized cells, asymmetric cell growth is directed by a mechanism whereby an activated GTPase (Cdc42, Rac, or Rop) accumulates at a local cortical domain on the plasma membrane to define a polar front (Chiou et al., 2017; Olayioye et al., 2019). The cytoskeleton is subsequently guided along the GTPase front and directs secretion, resulting in cell surface growth targeted to the polarity site. The spatiotemporal activity of polarity factors affects both the pattern and magnitude of cell surface growth (Bi and Park, 2012; Gulli et al., 2000; Howell and Lew, 2012; Knaus et al., 2007; Lew and Reed, 1993; Okada et al., 2013; Pruyne and Bretscher, 2000; Sopko et al., 2007; Woods and Lew, 2019).
In budding yeast, growth is first polarized to one site to allow the initiation of bud formation in G1 (Woods and Lew, 2019). Activated Cdc42 localizes to one presumptive bud site, where it recruits the septin and the actin cytoskeleton. Following polarization of GTP-Cdc42 at the presumptive site, actin filaments are oriented along the polar front, and cortical actin patches become concentrated at the site (Fig. 1 A; Adams et al., 1990; Engqvist-Goldstein and Drubin, 2003; Lew and Reed, 1993; Rodal et al., 2005; Valdez-Taubas and Pelham, 2003; Young et al., 2004). Cdc42 activity is regulated by one guanine exchange factor, Cdc24, and four GTPase-activating proteins (GAPs), Rga1, Rga2, Bem2, and Bem3 (Johnson, 1999; Marquitz et al., 2002; Smith et al., 2002; Zheng et al., 1993, 1994). Cdks, associated with G1 cyclins, promote Cdc42 activation and bud formation by inhibiting Cdc42 GAPs and the nuclear sequestration of Cdc24 (Gulli et al., 2000; Knaus et al., 2007; Sopko et al., 2007).
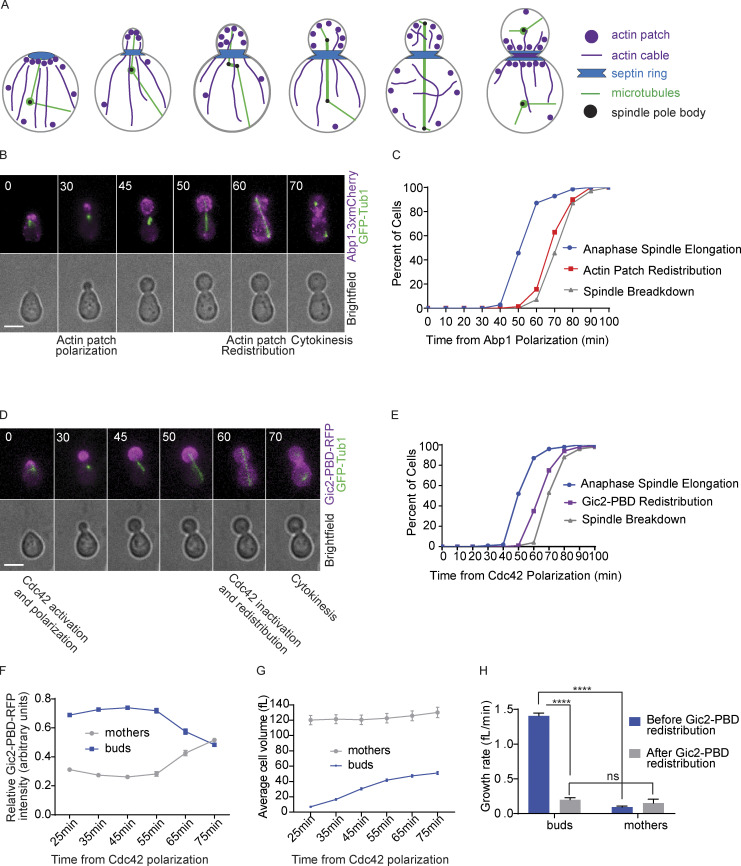
Figure 1.
Cdc42 is inactivated at late anaphase to preserve normal cell size. (A) Cartoon of actin filament and actin patch localization throughout the budding yeast cell cycle. (B) Representative time-lapse images showing the localization of actin patches by monitoring Abp1-3xmCherry during mitosis in a wild-type cell. Numbers indicate time in minutes from the initial polarization of actin patches. Cells were synchronized in G1 with α factor and released. Scale bar, 5 µm. (C) Timing of anaphase onset, actin patch redistribution, and spindle breakdown (n = 70 cells from three experiments). (D) Representative time-lapse images showing localization of a GTP-Cdc42 biosensor (Gic2-PBD-RFP) during mitosis in a wild-type cell. Numbers indicate time in minutes from the initial polarization of GTP-Cdc42 in G1. Cells were synchronized in G1 with α factor and then released. Scale bar, 5 µm. (E) Timing of anaphase onset, Cdc42 redistribution, and spindle breakdown (n = 100 cells from three experiments). (F) Gic2-PBD-RFP fluorescence intensity (n = 30 cells from three experiments for each time point indicated). Average intensity from each cell compartment was normalized over the total intensity in the cell, and the average ratio from 30 cells is plotted for each time point. Error bars represent SEM. (G) Cell volume of buds and mothers (n = 30 cells from three experiments for each time point indicated; average ± SEM). (H) Growth rate before and after Cdc42 redistribution (n = 100 cells from three experiments; average ± SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, paired t test). ns, not statistically significant.
The mechanism of Cdc42 polarity establishment and the initiation of bud formation in G1 have been thoroughly investigated, but less is known about how bud growth is regulated throughout S, G2, and M phases. In these phases, the bud continues to grow but switches from a polarized tip growth to an isotropic growth (Adams and Pringle, 1984; Farkaš et al., 1974; Kilmartin and Adams, 1984; Lew and Reed, 1993; Tkacz and Lampen, 1972). Cdc42 remains concentrated at the bud cortex, directing secretory vesicles to the growth site through actin-dependent and independent mechanisms (Bi and Park, 2012; Howell and Lew, 2012). At the end of mitosis, the actin patches and Cdc42 redistribute between the bud and the mother before accumulating at the bud neck at cytokinesis. The molecular mechanisms through which Cdc42 is redistributed at the end of mitosis are not fully understood.
Here, we investigated the spatiotemporal activity of Cdc42 in S and M phases of the cell cycle in budding yeast. We found that cells arrested at S or M phases maintain bud-localized Cdc42 activity; the bud continues to grow, and the spindles become mispositioned into the bud. We show that the redistribution of Cdc42 between mother and bud at late anaphase requires the nucleolar release of the Cdc14 phosphatase through the mitotic exit network (MEN). We identified one substrate of Cdc14, the Cdc42 GAP Bem3. Because GAPs are inactivated by phosphorylation by Cdk, dephosphorylation of Bem3 by Cdc14 likely leads to the activation of the GAP, suggesting a mechanism for Cdc42 inactivation. We conclude that the regulation of the spatiotemporal activity of Cdc42 by Cdc14 is important to preserve cell size at cytokinesis and for normal positioning of the mitotic spindle.
Results
Cdc42 inactivation at late anaphase maintains cell size
Previous studies showed that, in mitosis, actin patches redistribute between mother and daughter cells before localizing to the bud neck at cytokinesis (Fig. 1 A; Adams and Pringle, 1984; Kilmartin and Adams, 1984). We investigated the timing of the redistribution of the actin patches relative to the elongation and the breakdown of the mitotic spindle. We used time-lapse microscopy to monitor cells expressing Abp1 fused to three tandem copies of mCherry (Abp1-mCherry). The use of time-lapse microscopy allows us to continually track the cells and differentiate between mother and bud. Abp1 binds to actin patches, which are cortical actin structures made of short and branched actin filaments (Drubin et al., 1988). The cells also expressed GFP-TUB1 to monitor the microtubule spindle, anaphase onset, and spindle breakdown (Carminati and Stearns, 1997). Cells were synchronized in G1 by treatment with α factor mating pheromone and then released into the cell cycle (Breeden, 1997). We observed that actin patches redistributed between the bud and mother compartments at late anaphase before anaphase spindle breakdown (Fig. 1, B and C). From their initial polarization in G1, actin patches remained restricted to the bud for 70 ± 8 min (mean ± SD).
To determine how the timing of the actin patch redistribution is coordinated with Cdc42 activity, we monitored the localization of active GTP-bound Cdc42 using a biosensor consisting of the Gic2 p21-binding domain (PBD) bound to RFP (Gulli et al., 2000; Okada et al., 2013). As expected from previous studies, GTP-Cdc42 polarized and accumulated at the presumptive bud site in G1 (Fig. 1 D; Gulli et al., 2000; Okada et al., 2013; Pringle et al., 1995). As the cell cycle progressed, GTP-Cdc42 remained concentrated in the bud, and intriguingly, when the spindle reached full length but before spindle breakdown and cytokinesis, the GTP-Cdc42 biosensor redistributed between the bud and the mother (Fig. 1, D and E). From the initial polarization in G1, GTP-Cdc42 remained concentrated in the bud for an average of 66 ± 9 min (mean ± SD) in wild-type cells (Fig. 1, E and F). Similar results were obtained in asynchronous cells that were not treated with α factor (Fig. S1, A–C). The redistribution of the biosensor fluorescence signal that we observed likely indicates the inactivation of Cdc42, as previous work showed that Cdc42 is inhibited at the end of mitosis to allow cytokinesis to occur (Atkins et al., 2013). As Cdc42 is inactivated, the biosensor molecules redistribute between the bud and the mother (Fig. 1 F). However, we cannot exclude the possibility that active GTP-Cdc42 is redistributed between mother and bud. These results reveal that the redistribution of Cdc42 from the bud occurs just before actin patch redistribution and before spindle breakdown, thus providing further insight into the order of events at mitosis exit.
Figure S1.
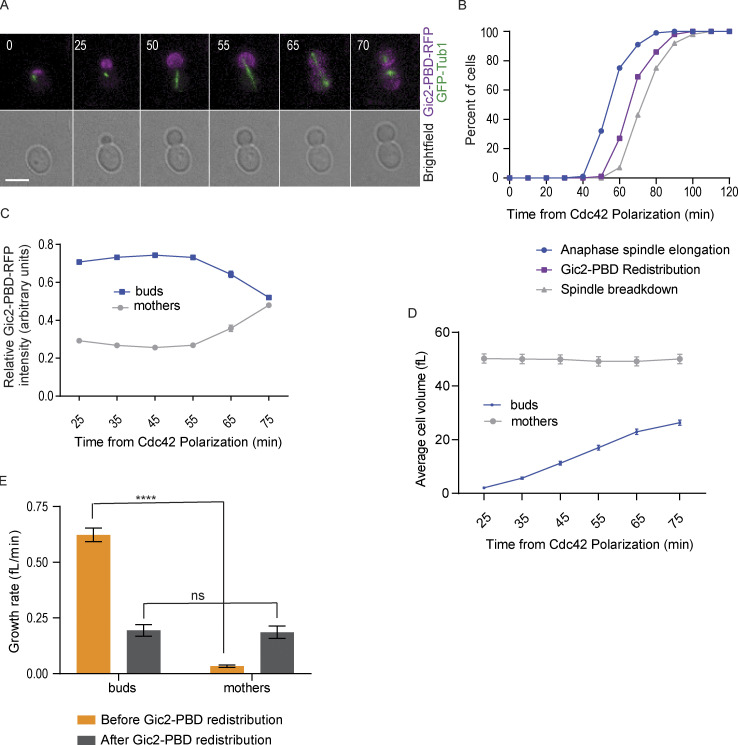
Localization of GTP-Cdc42 in asynchronously dividing cells. (A) Representative time-lapse images showing the localization of the GTP-Cdc42 biosensor (Gic2-PBD-RFP) during mitosis in a wild-type cell without α factor treatment. Numbers indicate time in minutes from the initial polarization of Cdc42 in G1. Scale bar, 5 µm. (B) Timing of anaphase onset, Cdc42 redistribution, and spindle breakdown in wild-type cells without α factor synchronization (n = 100 cells from three experiments). (C) Gic2-PBD fluorescence intensity measurements over time (n = 30 cells for each time point indicated from three experiments). Average intensity from each cell compartment was normalized to the total intensity in the cell, and the average ratio from 30 cells was plotted for each time point. Error bars indicate SEM. (D) Cell volume of buds and mothers (n = 30 cells for each time point from three experiments; average ± SEM). (E) Growth rate before and after Cdc42 redistribution (n = 54 cells from three experiments; average ± SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, paired t test). ns, not statistically significant.
In budded cells, the concentration of active Cdc42 in the bud targets growth to the bud cortex (Bi and Park, 2012; Howell and Lew, 2012; Woods and Lew, 2019). As a result, during mitosis, the buds grow faster than the mother cells (Leitao and Kellogg, 2017). We hypothesized that the inactivation of Cdc42 would result in an equal growth rate in the mother and the bud before cytokinesis. To test this hypothesis, we measured both the volume and growth rate of the bud and the mother compartments before and after redistribution of the GTP-Cdc42 biosensor. To measure the growth rate, we divided the total change in volume by the change in time. Before GTP-Cdc42 redistribution, for each cell analyzed, volume and time were measured starting from 25 min after the initial polarization of GTP-Cdc42 until the time of GTP-Cdc42 biosensor redistribution. The same measurements were done from the GTP-Cdc42 redistribution time until the time of cytokinesis. As previously shown, cell growth was restricted to the bud before anaphase (Fig. 1, G and H; Leitao and Kellogg, 2017). In contrast, after redistribution of the GTP-Cdc42 biosensor, the growth rate of the buds decreased, and the average growth rate of the buds and the mothers became similar. Cells without α factor treatment gave similar outcomes (Fig. S1, D and E). These results further suggest that Cdc42 is inactivated with the redistribution, and this inactivation balances the growth rate between the daughter and mother cell before cytokinesis and before the start of a new cell cycle.
GTP-Cdc42 remains restricted to the bud during S and M phase cell cycle arrest
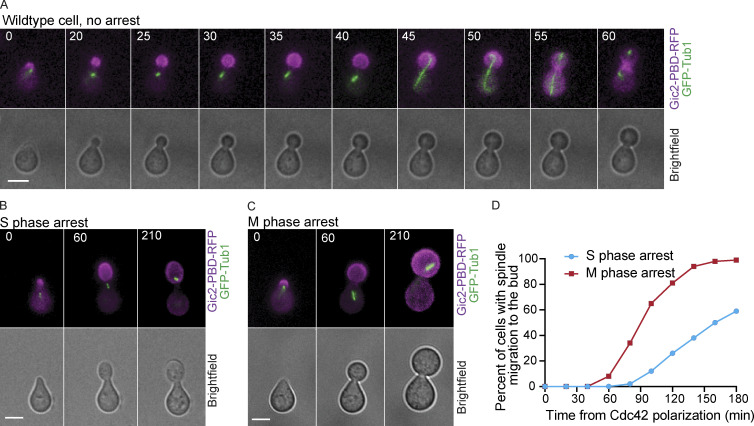
When cell cycle progression is blocked, cells are able to continue to grow in size (Demidenko and Blagosklonny, 2008; Johnston et al., 1977; Neurohr et al., 2019). However, it remains unclear how cell cycle arrest affects the localization of GTP-Cdc42 in budded cells that have concentrated GTP-Cdc42 in the bud. Using the GTP-Cdc42 biosensor, we assessed the localization of GTP-Cdc42 and size of the mothers and the buds in cells arrested in S and M phases following a release from a G1 synchronization. Cells were arrested in S phase with hydroxyurea treatment and in M phase with depletion of CDC20, an activator of the anaphase-promoting complex. We observed that GTP-Cdc42 remained concentrated in the buds in the arrested cells (Fig. 2, A–D). These results demonstrate that active Cdc42 can remain concentrated in a subcellular compartment when cell cycle progression is blocked before cytokinesis.
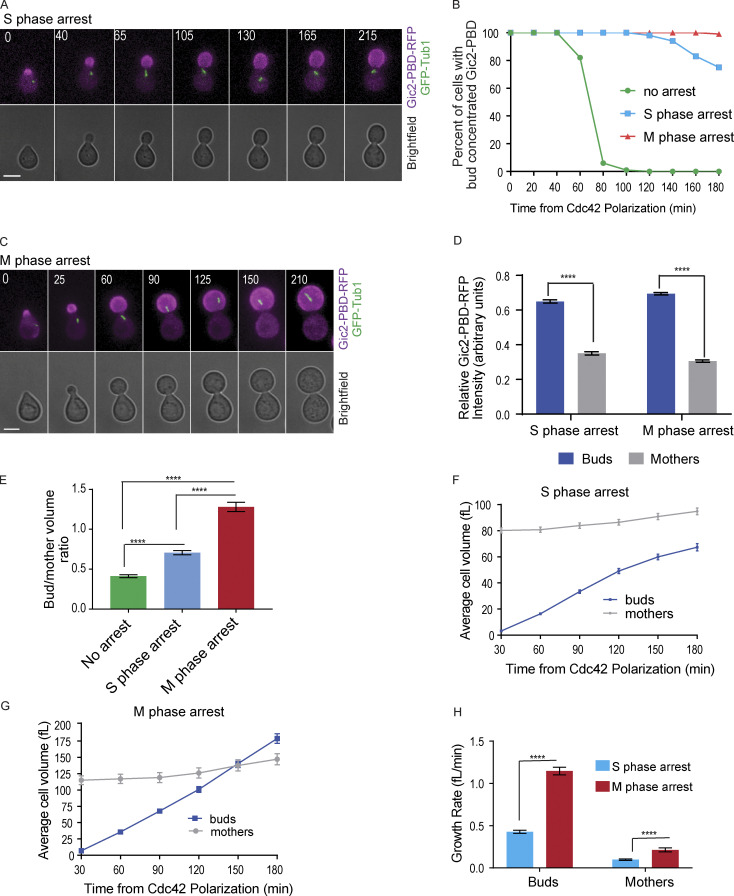
Figure 2.
GTP-Cdc42 remains restricted to the bud during S and M phase cell cycle arrest. (A) Representative time-lapse images of a cell arrested in S phase with hydroxyurea. Numbers indicate time in minutes from the initial Cdc42 polarization. Cells were synchronized in G1 with α factor. Scale bar, 5 µm. (B) GTP-Cdc42 concentration in the buds of cells that are not arrested, cells in S phase arrest, and cells in M phase arrest (n = 100 cells from three experiments). (C) Representative time-lapse images of a cell arrested in metaphase by CDC20 depletion. Numbers indicate time in minutes from the initial Cdc42 polarization. Cells were synchronized in G1 with α factor. Scale bar, 5 µm. (D) Graph of relative Gic2-PBD-RFP fluorescence intensity. The average intensity from each cell compartment was normalized to the total intensity in the cell ± SEM. Intensity was measured at 2 h from α factor release (n = 50 cells from three experiments). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test). (E) Bud/mother volume ratio measurement in arrested cells, calculated at 180 min from α factor release. For cells that were not arrested, the ratio was calculated at cytokinesis (n = 30 from three experiments for each; average ± SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney test). (F) Volume of cells arrested in S phase (n = 30 cells from three assays for each time point; average ± SEM). (G) Volume of cells arrested in M phase (n = 30 cells from three assays for every time point; average ± SEM). (H) Growth rate of cells arrested in S and in M phase (n = 30 cells from three experiments; average ± SEM). The growth rate was measured between 30 min and 180 min. Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney test).
The cell size ratio between the buds and the mother compartments was perturbed in cells arrested in S or M phase for 3 h after initial Cdc42 polarization in G1 (Fig. 2, E–G). Without cell cycle arrest, the buds reached, on average, 40% of the size of the mothers by the time of cytokinesis (Fig. 2 E). The buds of the cells that were arrested in S phase reached 70% of the average size of the mothers (Fig. 2, E and F). Strikingly, in M phase arrest, the buds attained a size that was 130% of the average size of the mothers (Fig. 2, E and G). Thus, the M phase arrest led to buds that, on average, grew even larger than their mothers, therefore reversing the characteristic cell size ratio between the buds and the mothers (Fig. 2 G). Similar results were obtained in cells arrested in M phase with nocodazole treatment (Fig. S2, A–D). We also noticed that cells arrested in M phase grew larger and faster compared with the cells arrested in S phase with hydroxyurea (Fig. 2, E and H). The data suggest that, when the cell cycle is arrested, persistence of GTP-Cdc42 in the bud can perturb the normal size ratio between the mother and daughter cell compartments.
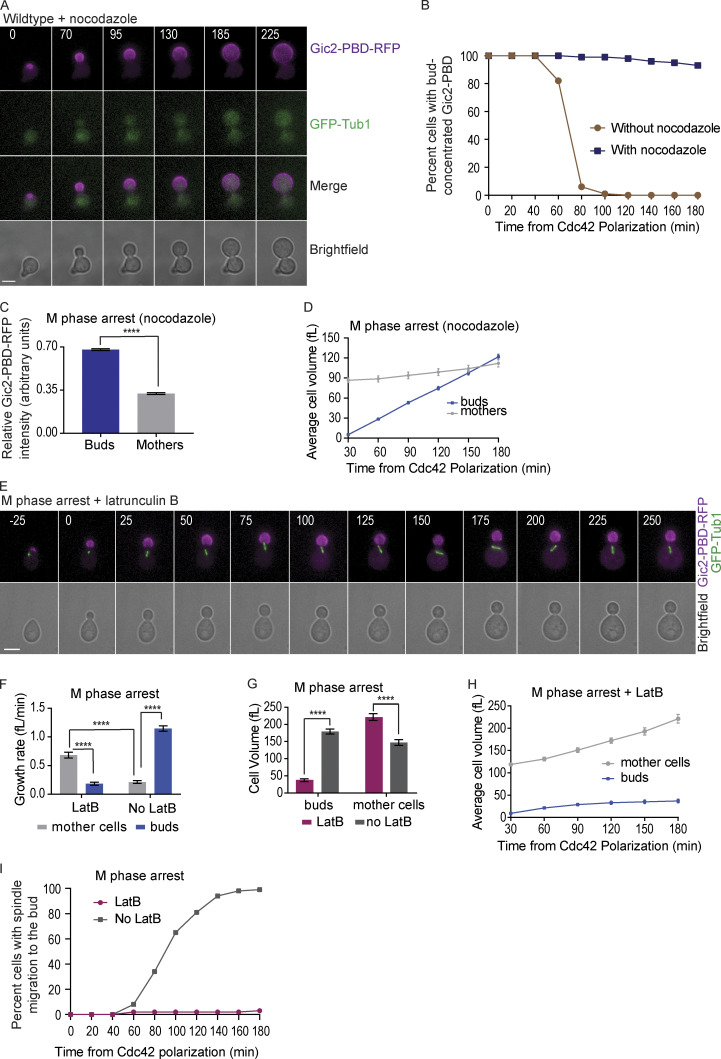
Figure S2.
GTP-Cdc42 localization and cell size in cells treated with nocodazole or latrunculin. (A) Representative time-lapse image showing the localization of Gic2-PBD during mitosis in a wild-type cell treated with nocodazole. Numbers indicate time in minutes from the initial polarization of Cdc42 in G1. Cells were synchronized in G1 with α factor and released into medium containing nocodazole. Scale bar: 5 µm. (B) Percent of wild-type cells without nocodazole and with nocodazole with Gic2-PBD bud concentration over time (n = 100 cells from three experiments for each background). (C) Gic2-PBD-RFP fluorescence intensity in cells treated with nocodazole for two hours. Average intensity from each cell compartment was normalized to the total intensity in the cell (n = 50 cells from three experiments; average ± SEM). Asterisks indicate statistically significant differences (Mann-Whitney t test; P < 0.0001). (D) Cell volume of buds and mothers of cells treated with nocodazole (n = 30 cells from three experiments for each time point indicated; average ± SEM). (E) Representative time-lapse image showing the localization of Gic2-PBD in a cell treated with LatB. Numbers indicate time in minutes from the initial polarization of Cdc42 in G1. Scale bar, 5 µm. For E–I, cells were synchronized in G1 with α factor, released into the cell cycle and arrested in M phase by CDC20 depletion. (F) Growth rate of buds versus mothers with and without LatB treatment (n = 30 cells from three experiments; average ± SEM). The growth rate was measured between 0 and 180 min, where time 0 is the time of LatB addition. Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney test). (G) Cell volume of mothers and buds with and without LatB treatment (n = 30 cells for every time point indicated from three experiments; average ± SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test). (H) Average cell volume after 3 h of LatB treatment (n = 30 from three experiments; average ± SEM). (I) Percentage of cells with spindle migration to the bud with or without LatB treatment (n = 100 cells for each category from three experiments).
Because active and polarized Cdc42 guides the actin cytoskeleton, which in turn directs secretion and cell growth (Pruyne and Bretscher, 2000), we tested if the excessive bud growth in cells arrested in M phase was actin dependent. We synchronized cells in G1, released them into the cell cycle, and arrested them at metaphase with the depletion of CDC20. To the budded and metaphase-arrested cells, we added latrunculin B (LatB), a drug that disrupts actin polymerization (Spector et al., 1983). We found that actin depolymerization prevented the reverse distribution of cell size between the bud and mother cells. The growth rate of the buds was decreased in the presence of LatB, and the buds remained smaller than the mothers during M phase arrest (Fig. S2, E–H). Surprisingly, in the absence of actin polymerization in cells arrested in M phase, the mother compartments grew more and faster than their buds (Fig. S2, F and H). The mothers of cells arrested at M phase with latrunculin also grew larger than mothers of cells arrested in M phase without latrunculin (Fig. S2 G). The results demonstrate that, by targeting secretory vesicles to the buds, polymerized actin not only promotes the growth of the buds but also maintains the normal growth rate of the mothers to prevent abnormally large mothers.
Prolonged bud localization of Cdc42 activity perturbs normal positioning of the mitotic spindle, cell cycle timing, bud site selection, and bud morphology
In budding yeast, the actin cytoskeleton helps position the nucleus, and therefore the mitotic spindle, at the bud neck for division of the chromosomes between the mother and daughter (Fig. 1 A; Miller et al., 1999; Miller and Rose, 1998; Palmer et al., 1992; Theesfeld et al., 1999). A microtubule motor protein, Kar9, connects cytoplasmic microtubules to an actin motor protein, which binds actin filaments oriented along the mother–bud axis (Beach et al., 2000; Theesfeld et al., 2003; Yin et al., 2000). We hypothesized that persistent GTP-Cdc42 bud localization in cells arrested at S or M phase could result in a mislocalization of the spindle due to prolonged forces that pull the spindle along the actin filaments and into the daughter cell. We therefore monitored spindle position in cells arrested in S and M phases.
As expected from previous work, in wild-type cells that are not arrested, the mitotic spindle remained in the mother, positioned at or close to the mother–bud neck, until anaphase onset, when the spindle elongated across the bud neck and into the bud (Fig. 3 A; Kusch et al., 2002). In cells arrested in S or M phase, the entire spindle migrated into the buds in 59% and 99% of cells, respectively (Fig. 3, B–D). When we treated cells arrested in M phase with LatB, the spindles did not migrate to the bud. Instead, the spindle remained positioned at or near the bud neck (Fig. S2, E and I), likely because spindle positioning can also occur through actin independent pathways (Carminati and Stearns, 1997; Eshel et al., 1993; Kusch et al., 2002; Li et al., 1993; Moore et al., 2009). These results suggest that prolonged GTP-Cdc42 bud localization leads to persistent actin filaments pointing toward the mother–bud axis, causing spindle mispositioning.
Figure 3.
Aberrant spindle position in cells arrested with GTP-Cdc42 concentrated in the bud. (A) Representative time-lapse images showing normal spindle position in a wild-type cell. Numbers indicate time in minutes from the initial polarization of Cdc42 in G1. Scale bar, 5 µm. (B and C) Representative images showing spindle migration to the bud in cells arrested in S phase (B) and M phase (C). Scale bars, 5 µm. (D) Percentage of cells in which the spindle migrated in the bud (n = 100 cells from three experiments).
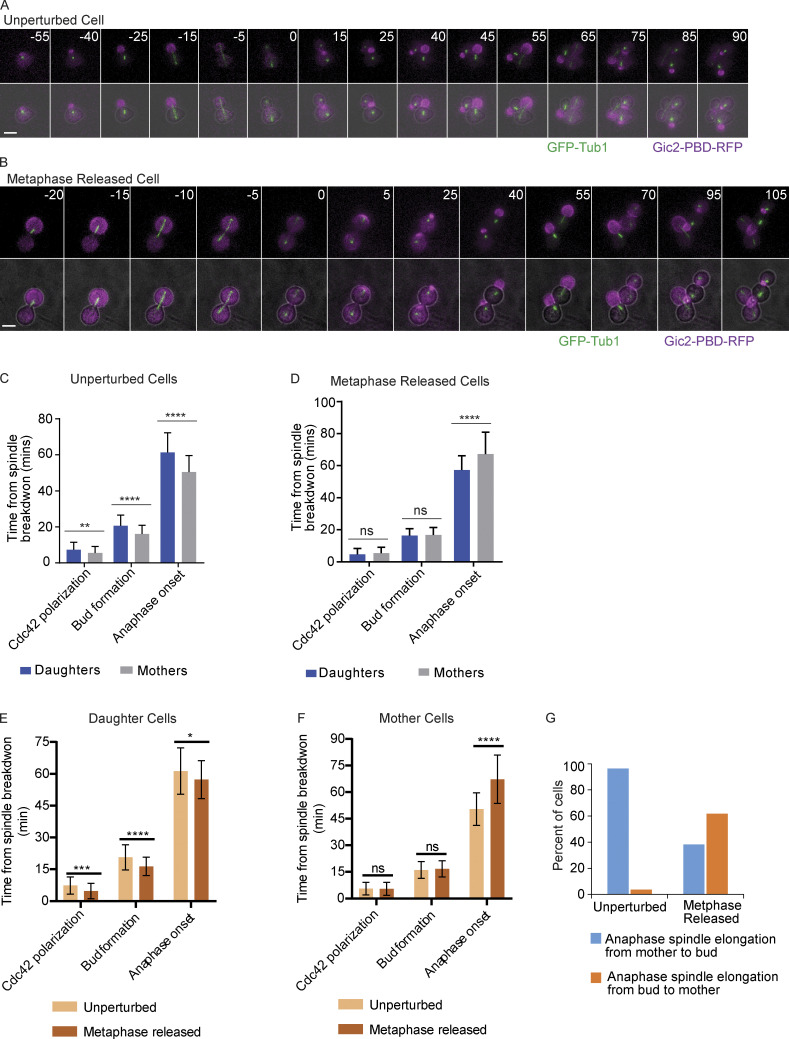
Previous work has shown that in budding yeast, cell size is negatively correlated with the timing of cell cycle entry (Hartwell and Unger, 1977; Wheals, 1982). The daughter cells, which are smaller at birth than their mother cells, are delayed in cell cycle reentry after cytokinesis. We considered that the perturbation of cell size ratio that we observed between the mother and bud compartments could disrupt the timing of normal cell cycle processes (Fig. 2). Therefore, we compared the timing of Cdc42 polarization, bud formation, and anaphase onset in unperturbed cells (with a normal mother to daughter size ratio; Fig. S3 A) to cells that were originally arrested at metaphase with prolonged localization of Cdc42 activity to the bud (metaphase-released cells with buds larger than the mothers; Fig. S3 B). These cells were initially depleted of Cdc20 to arrest them at metaphase for 3 h but then induced to express CDC20 to exit metaphase. We measured the time from anaphase spindle disassembly to Cdc42 polarization, bud formation, and anaphase onset in the next cell cycle. As expected, in unperturbed cells, the mother was faster than the daughter for each of the analyzed events (Fig. S3 C). In contrast, in the metaphase-released cells, Cdc42 polarization and bud formation occurred with similar timing in both mother and daughter cells (Fig. S3 D). In contrast to the unperturbed cell cycle, anaphase onset was faster in the daughter than in mother cells in metaphase-released cells.
Figure S3.
Altered cell cycle timing and anaphase spindle orientation after release from a prolonged metaphase arrest. (A) Representative time-lapse images showing an unperturbed control cell released from α factor. Numbers indicate time in minutes, with time 0 indicating the time of spindle breakdown in the first division cycle. Scale bar, 5 µm. (B) Representative time-lapse images showing a cell that was first released from α factor, then underwent a 3-h metaphase arrest, and then was released from the metaphase arrest. In this cell, the anaphase spindle elongates from the mother into the bud. Time 0 represents the time of anaphase spindle breakdown after metaphase release. Scale bar, 5 µm. (C) The timing of Cdc42 polarization, bud formation, and anaphase onset measured from the first spindle breakdown (time 0) after α factor release of unperturbed cells (n = 55 cells from two experiments). Error bars indicate SD. ****, P < 0.0001; **, P = 0.006 (Mann-Whitney t test). (D) The timing of Cdc42 polarization, bud formation, and anaphase onset measured from spindle breakdown (time 0) after the release from a 3-h metaphase arrest. Cells have MET3prCDC20, which allows a metaphase block with Cdc20 depletion in methionine-containing medium. The release occurs when methionine is removed from the medium. Error bars indicate SD (n = 55 cells from two experiments). ****, P < 0.0001 (Mann-Whitney t test). ns, not statistically significant. (E) The timing of Cdc42 polarization, bud formation, and anaphase onset measured from the first spindle breakdown (time 0) after α factor release (unperturbed) or metaphase release (metaphase released) daughter cells. Error bars indicate SD (n = 55 cells from two experiments). ****, P < 0.0001; ***, P = 0.002; *, P < 0.0273 (Mann-Whitney t test). (F) The timing of Cdc42 polarization, bud formation, and anaphase onset measured from the first spindle breakdown (time 0) after α factor release (unperturbed) or metaphase release (metaphase released) mother cells. Error bars indicate SD (n = 55 cells from two experiments). ****, P < 0.0001; ns, not statistically significant (Mann-Whitney t test). (G) Orientation of anaphase spindle elongation in unperturbed cells and cells released from a 3-h metaphase arrest (n = 55 cells from two experiments).
We also compared the timing of Cdc42 polarization, bud formation, and anaphase onset in the daughters of unperturbed cells to the daughters of metaphase-released cells and the timing of the same cell cycle events in the mothers of unperturbed cells versus the mothers of metaphase-released cells. The metaphase-released daughters undergo Cdc42 polarization, budding, and anaphase onset faster than the unperturbed daughters (Fig. S3 E). Interestingly, the metaphase-released mothers had a delayed anaphase onset compared with the unperturbed mothers (Fig. S3 F). As shown in Fig. 3, the mitotic spindle of the cells arrested in metaphase migrate to the buds. We assessed the orientation of spindle elongation during anaphase in cells released from the metaphase arrest. In the unperturbed cells, the spindle was found in the mother compartment and then elongated into the bud in 96% of cells (Fig. S3, A and G). In contrast, in the metaphase-rescued cells, which often had the spindle located in the bud compartment, 62% of the cells underwent an anaphase spindle elongation from the bud into the mother, a reverse order of the normal process (Fig. S3, B and G). Overall, these results show that the reversed cell size ratio between the bud and the mother alters the normal timing of cell cycle events. In addition, the data reveal a reversed elongation of spindle during anaphase in cells released from a metaphase arrest.
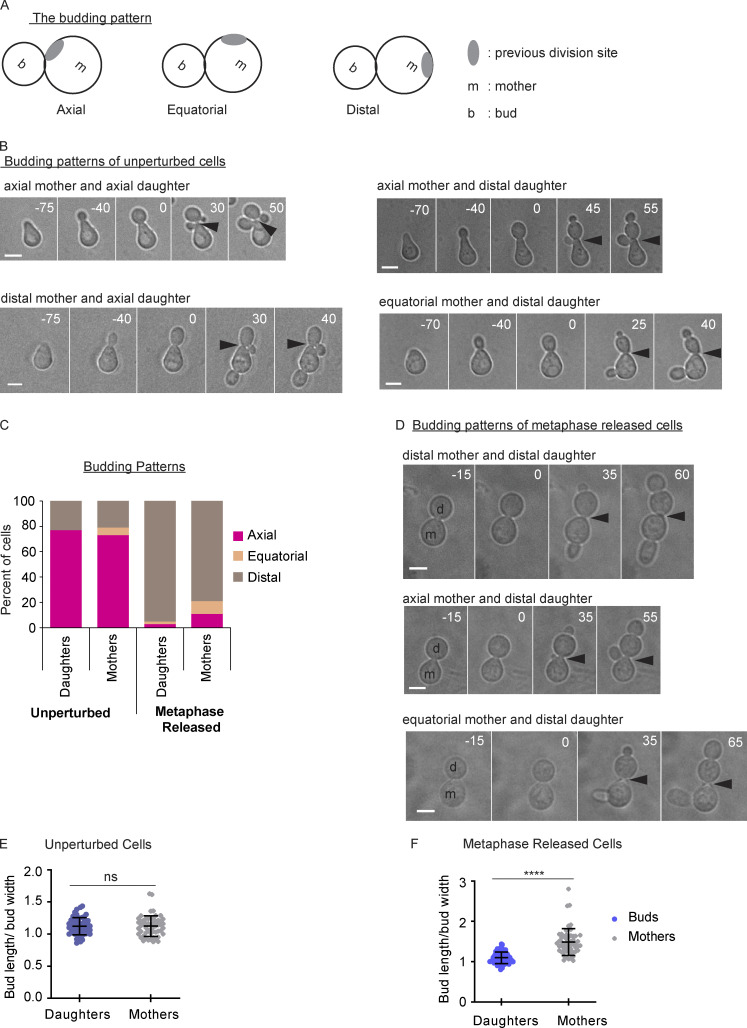
We next asked if the pattern of bud site selection was different in the unperturbed cells compared with the cells released from a prolonged metaphase arrest. Budding yeast cells choose a site of polarized growth in a nonrandom pattern. Haploid cells typically bud in an axial pattern in which mother and daughter cells bud at a location adjacent to the previous bud site (Fig. S4 A; Bi and Park, 2012). We released the unperturbed cells from an α factor arrest, allowed them to undergo one division cycle, and then examined the position of the bud in both the mother and original daughter cells in the second division cycle. By following the cells with time-lapse microscopy, we can identify the original bud site and the new bud site. Most unperturbed cells display the expected pattern with new buds adjacent to the original bud site in both the mother and daughters (Fig. S4, B and C). However, some unperturbed cells have mothers or daughters with distal buds. Infrequently, the cells bud in an equatorial pattern. Cells released from the metaphase arrest have an altered budding pattern. In most of the metaphase-released cells, both the mother and daughter cells bud at a site distal from their previous bud site (Fig. S4, C and D). A small fraction of daughter and mother cells bud at equatorial or axial sites.
Figure S4.
Altered budding pattern after prolonged metaphase arrest. (A) A cartoon showing budding patterns. (B) Representative images of budding patterns in unperturbed cell cycle. Cells were released from α factor. Numbers indicate time in minutes, with time 0 indicating the time of spindle breakdown in the first division cycle after α factor release. Scale bars, 5 µm. Arrowheads indicate the previous division site. (C) Distribution of budding patterns in an unperturbed cell cycle and from cells released from a metaphase arrest (n = 100 cells from two experiments for each treatment). (D) Representative images of budding patterns in metaphase-released cells. Cells were first released from α factor and arrested at metaphase by depleting Cdc20 using a methionine-repressible promoter. After 3 h, cells were released from the metaphase arrest. Numbers indicate time in minutes, with time 0 showing the time of spindle breakdown in the first division cycle after α factor release. Scale bars, 5 µm. Arrowheads indicate the previous division site. (E) Measurements of the bud length divided by the bud width in both the mother and daughter cells (buds) in an unperturbed cell cycle. Error bars indicate SD (n = 55 cells from two experiments). ns, not statistically significant (Mann-Whitney t test). (F) Measurements of the bud lengths divided by the bud width from both the mother and daughter cells (buds) in cells released from a metaphase arrest (n = 55 cells from two experiments). Error bars indicate SD. ****, P < 0.0001 (Mann-Whitney t test).
We also noticed that in the cells released from a metaphase arrest, the mother cells often produced elongated buds (Fig. S4 D). To quantify this phenotype, we measured the ratio of bud length (the length from the bud neck to the bud tip) to bud width. The buds from unperturbed mother and daughter cells had a similar length (Fig. S4 E). In contrast, in metaphase-released cells, the buds from the mother cells were longer than the buds from the daughter cells (Fig. S4 F). These results demonstrate that the mother cells from a prolonged metaphase arrest, which have a delayed anaphase onset, also form elongated buds. Overall, these results suggest that a prolonged metaphase arrest with bud-localized Cdc42 activity causes defects in spindle positioning, cell cycle timing, bud site selection, and bud morphology.
The MEN promotes Cdc42 redistribution
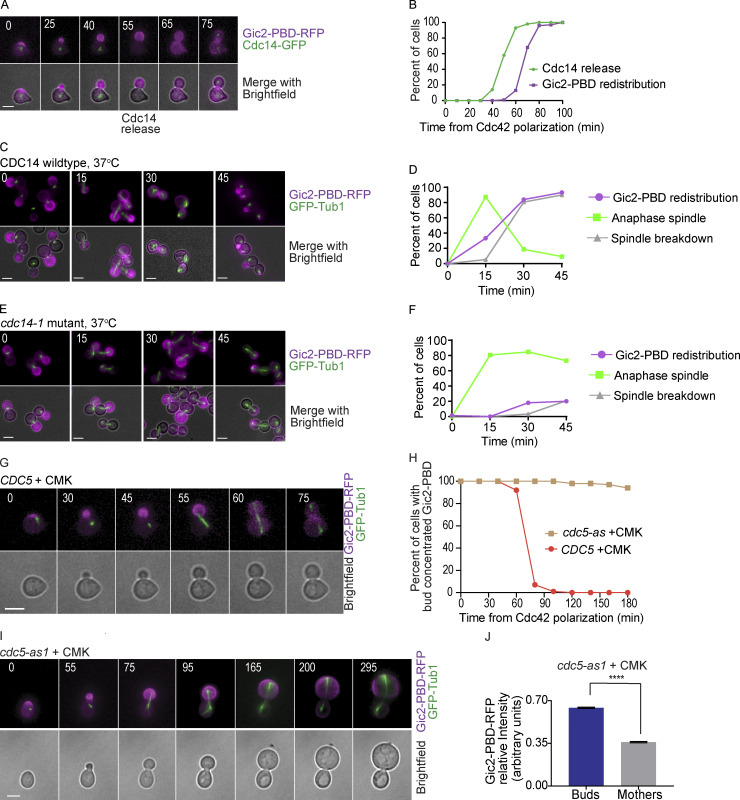
The observation that the GTP-Cdc42 biosensor redistributed consistently at late anaphase before spindle breakdown and cytokinesis led to the hypothesis that Cdc14 is involved in Cdc42 redistribution. The Cdc14 phosphatase is sequestered in the nucleolus until it is released, first in early anaphase through the Cdc14 early anaphase release (FEAR) pathway and then in a sustained release through the MEN (Pereira et al., 2002; Shou et al., 1999; Stegmeier and Amon, 2004; Stegmeier et al., 2002; Traverso et al., 2001; Visintin et al., 1998; Yoshida et al., 2002). Once released, Cdc14 reverses Cdk phosphorylation and promotes cyclin degradation, resulting in spindle breakdown and cytokinesis. To investigate the timing of Cdc14 release and Cdc42 activity, we imaged live cells expressing the GTP-Cdc42 biosensor and Cdc14-GFP. As shown in Fig. 4, A and B, the release of Cdc14 from the nucleolus preceded the redistribution of the GTP-Cdc42 biosensor by 10–20 min.
Figure 4.
Cdc14 activity is required for Gic2-PBD redistribution. (A) Representative time-lapse images showing Cdc14 nucleolar release and Gic2-PBD redistribution in a wild-type cell released from a G1 α factor arrest. Numbers indicate time in minutes, counted from the time of initial Cdc42 polarization. Scale bar, 5 µm. (B) Timing of Cdc14 nucleolar release and Gic2-PBD-RFP redistribution in wild-type cells released from a G1 α factor synchronization (n = 100 cells from three experiments). (C) Representative images showing Gic2-PBD localization in wild-type cells at 37°C. Scale bars, 5 µm. Cells were released from a G1 α factor block and then arrested at metaphase by depletion of Cdc20 using a methionine-repressible MET3 promoter. Cells were incubated in methionine medium for 2 h at 25°C and then released in media lacking methionine at 37°C. Numbers indicate time in minutes, counted from the transfer to methionine-free medium. (D) The timing of progression through anaphase, Gic2-PBD redistribution, and spindle breakdown in wild-type cells released from a metaphase arrest at 37°C. Time was counted from the transfer to methionine-free medium. n = 150 cells from three experiments. (E) Representative images showing Gic2-PBD localization in cdc14-1 mutant cells released from metaphase arrest at 37°C. Cells were treated as in C. Numbers indicate time in minutes, counted from the transfer to methionine-free medium. Scale bars, 5 µm. (F) The timing of progression through anaphase, Gic2-PBD redistribution, and spindle breakdown in cdc14-1 mutant cells released from a metaphase arrest at 37°C. Scale bar, 5 µm. n = 150 cells from three experiments. (G) Representative time-lapse images showing the mitotic spindle and Gic2-PBD in a wild-type cell treated with CMK. Numbers indicate time in minutes from the initial Cdc42 polarization in G1. Scale bar, 5 µm. (H) The percentage of wild-type and cdc5-as1 cells treated with CMK with bud-concentrated Gic2-PBD. For each background, n = 100 cells from three experiments. (I) Representative time-lapse images showing the mitotic spindle and the Gic2-PBD biosensor in a cdc5-as1 cell treated with the CMK inhibitor. Numbers indicate time in minutes from the initial Cdc42 polarization in G1. Scale bar, 5 µm. (J) Gic2-PBD-RFP fluorescence intensity in cdc5-as1 cells treated with CMK for 3 h. Average intensity in each cell compartment was normalized to the total intensity in the cell (n = 50 cells from three experiments; error bars indicate SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test).
To test our hypothesis that Cdc14 nucleolar release is required for Cdc42 redistribution, we used a temperature-sensitive CDC14 allele, cdc14-1, to conditionally inactivate Cdc14. Cells were released from a metaphase arrest at the restrictive temperature (37°C) and imaged at 0-, 15-, 30-, and 45-min time points. Cells with wild-type CDC14 grown at the restrictive temperature underwent anaphase spindle elongation before GTP-Cdc42 biosensor redistribution, as expected (Fig. 4, C and D). cdc14-1 cells grown at 37°C enter anaphase with similar timing to wild-type cells; however, the anaphase spindles do not break down and the GTP-Cdc42 biosensor remains concentrated in the bud in the majority of the cells (Fig. 4, E and F). We noted that a small fraction of cdc14-1 cells break through the temperature arrest and undergo spindle breakdown and Gic2-PBD redistribution, probably due to the leakiness of the temperature-sensitive mutation.
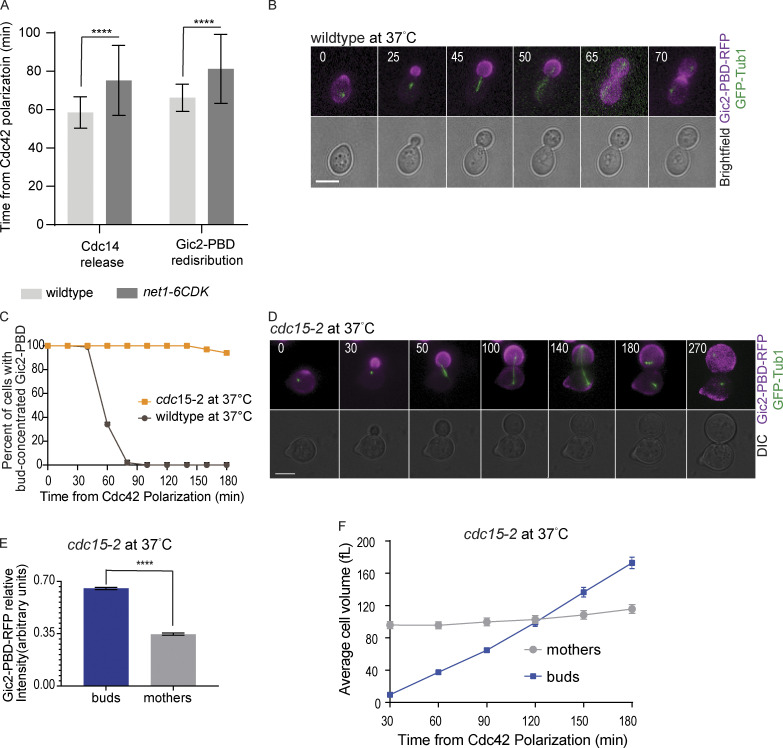
We also tested the redistribution of Cdc42 in cells expressing a net1-6cdk mutant. Net1 inhibits Cdc14 activity by anchoring the phosphatase in the nucleolus (Visintin et al., 1999). Cdk-dependent phosphorylation of Net1 releases Cdc14 through the FEAR pathway (Azzam et al., 2004). The net1-6cdk mutant cannot be phosphorylated by Cdk, thus delaying Cdc14 release (Azzam et al., 2004; Liang and Wang, 2007). We found that the redistribution of Cdc42 is delayed in the cells in which net1-6cdk is the only source of Net1 (Fig. S5 A).
Figure S5.
Gic2-PBD localization in cdc15-2 mutant cells. (A) The timing of Cdc14 release and Gic2-PBD redistribution in wild-type and net1-6cdk cells. The time was measured from the initial GTP-Cdc42 polarization (n = 30 cells). Error bars indicate SD. Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test). (B) Representative time-lapse images showing GFP-Tub1 and Gic2-PBD in a wild-type cell incubated at 37°C. Numbers indicate time in minutes from the initial GTP-Cdc42 polarization in G1. Scale bar, 5 µm. (C) Percentage of cells with Gic2-PBD concentrated in the buds of wild-type and cdc15-2 cells incubated at 37°C (n = 100 cells for each background from three experiments). (D) Representative time-lapse images showing GFP-Tub1 and the Gic2-PBD in a cdc15-2 cell incubated at 37°C (restrictive temperature). Numbers indicate time in minutes from the initial GTP-Cdc42 polarization in G1. Scale bar, 5 µm. (E) Gic2-PBD-RFP fluorescence intensity in cdc15-2 cells incubated at 37°C for 3 h. Average intensity in each cell compartment was normalized to the total intensity in the cell (n = 50 cells from three experiments; average ± SEM). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test). (F) Average cell volume in cdc15-2 cells incubated at 37°C (n = 30 cells for each time point from three experiments; average ± SEM). DIC, differential interference contrast.
For further confirmation, we tested cdc15-2 cells, a temperature-sensitive mutant of the Cdc15 kinase involved in the MEN pathway but not involved in FEAR (Jaspersen et al., 1998; Lee et al., 2001; Menssen et al., 2001; Shou et al., 1999; Visintin et al., 1999). At the restrictive temperature, cdc15-2 cells arrest in late anaphase. Inhibition of Cdc15 blocks full Cdc14 release from the nucleolus, with a minor amount of Cdc14 released through the FEAR pathway (Stegmeier et al., 2002). Wild-type cells released from an α factor arrest at the restrictive temperature redistributed Cdc42 with normal timing (Fig. S5, B and C). In agreement with our observations in the cdc14-1 mutants, the majority of cdc15-2 cells released from an α factor arrest at the restrictive temperature have GTP-Cdc42 largely concentrated in the bud (Fig. S5, C–E). The buds of the cdc15-2 arrested cells also grew larger than their mothers (Fig. S5, D and F). These results suggest that release of Cdc14 through the MEN pathway is important for the full redistribution of Cdc42.
Furthermore, in a complementary approach, we blocked Cdc14 nucleolar release by inhibiting Cdc5, the budding yeast polo kinase. Cdc5 is essential for Cdc14 nucleolar release, with roles in both the FEAR and the MEN pathways (Geymonat et al., 2003; Hu and Elledge, 2002; Hu et al., 2001; Lee et al., 2001; Visintin et al., 2003). We inhibited Cdc5 kinase activity using a cdc5-as1 allele, which can be catalytically inactivated by a chloromethylketone (CMK) inhibitor (Snead et al., 2007). Synchronous cells were released from G1 in the presence of CMK. The cells also expressed GFP-Tub1 to mark the mitotic spindle and monitor cell cycle progression. Wild-type CDC5 control cells treated with CMK redistributed the GTP-Cdc42 biosensor at a time comparable to untreated cells (Fig. 4, G and H). cdc5-as1 cells treated with CMK had an average delay to anaphase spindle elongation of only 10 min compared with the wild-type cells treated with CMK, but cdc5-as1 cells arrested with long anaphase spindles, indicating a lack of Cdc14 release, as previously reported (Fig. 4 I; Snead et al., 2007). Importantly, the GTP-Cdc42 biosensor remained concentrated in the buds in cdc5-as1 cells treated with CMK (Fig. 4, H–J). These results support our conclusion that Cdc14 nucleolar release is important for the redistribution of Cdc42.
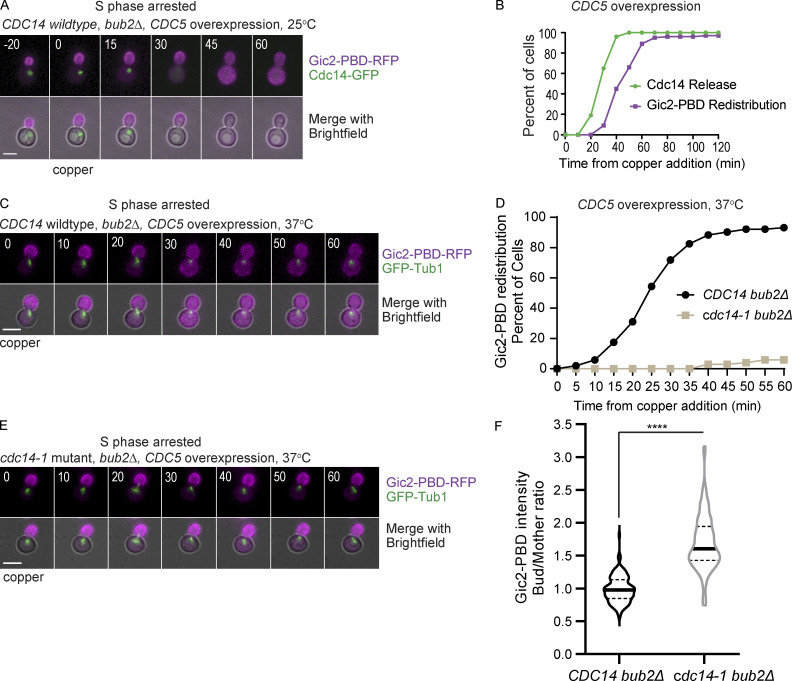
We predicted that if Cdc14 release was responsible for Cdc42 redistribution, then premature release of Cdc14 would cause the redistribution of the GTP-Cdc42 biosensor before anaphase. Previous work showed that overexpressing CDC5 in bub2Δ mutant cells can cause Cdc14 nucleolar release in cells arrested in S phase (Visintin et al., 2003). Bub2 is part of a complex that inhibits Cdc14 release to prevent untimely exit from mitosis (Pereira et al., 2000). We integrated an extra copy of CDC5 under control of a copper-inducible CUP1 promotor in bub2Δ cells. We arrested the cells in S phase with hydroxyurea and, after the cells formed buds, added copper to overexpress CDC5. Within 1 h, bub2Δ cells had released Cdc14. Remarkably, 97% of the cells that released Cdc14 also redistributed the GTP-Cdc42 biosensor (Fig. 5, A and B).
Figure 5.
Induced Cdc14 release from the nucleolus is sufficient to redistribute GTP-Cdc42 between the mother and bud. (A) Representative time-lapse images showing Cdc14-GFP and Gic2-PBD in a bub2Δ cell arrested at S phase with hydroxyurea and overexpressing CDC5 from a copper-inducible promotor. Numbers indicate time in minutes, with 0 indicating the time of copper addition. Scale bar, 5 µm. (B) Timing of Cdc14 release and Gic2-PBD biosensor redistribution in bub2Δ cells arrested at S phase with hydroxyurea and overexpressing CDC5 (n = 100 cells from three experiments). (C) Representative time-lapse images showing Gic2-PBD redistribution in S phase in bub2Δ mutant cells overexpressing Cdc5 from a copper-inducible promotor. Numbers indicate time in minutes, counted from the time of copper addition. Scale bar, 5 µm. (D) Timing of Gic2-PBD redistribution in bub2Δ and in bub2Δ cdc14-1 cells overexpressing Cdc5 from a copper-inducible promotor and arrested in S phase at 37°C (n = 100 cells from three experiments for each genotype). (E) Representative time-lapse images showing Gic2-PBD redistribution in S phase–arrested bub2Δ cdc14-1 cells overexpressing Cdc5. Numbers indicate time in minutes, counted from the time of copper addition. Scale bar, 5 µm. (F) Gic2-PBD intensity of bub2Δ and bub2Δ cdc14-1 cells overexpressing Cdc5 from a copper-inducible promotor and arrested in S phase at 37°C. The intensity was measured at 60 min after copper addition (n = 100 cells for each genotype from three experiment). Asterisks indicate statistically significant differences (****, P < 0.0001, Mann-Whitney t test).
The results suggest that Cdc5 overexpression in S phase–arrested cells induces release of Cdc14 from the nucleolus and causes the redistribution of Cdc42. However, we cannot exclude the possibility that Cdc5 could have other substrates that cause Cdc42 redistribution. In fact, a previous study implicated Cdc5 in the inactivation of Cdc42 before cytokinesis (Atkins et al., 2013). To distinguish between these two possibilities, we asked if Cdc5 overexpression could cause Cdc42 redistribution in cdc14-1 bub2Δ cells. We arrested cells at S phase, shifted the temperature to 37°C, and induced CDC5 expression with the addition of copper. Cdc42 redistributed as expected in the CDC14 bub2Δ control cells overexpressing CDC5 at 37°C (Fig. 5, C and D). In contrast, GTP-Cdc42 remained concentrated in the bud in cdc14-1 bub2Δ cells overexpressing CDC5 at 37°C (Fig. 5, D and E). We measured the Gic2-PBD fluorescence intensity in the mothers and buds 60 min after CDC5 overexpression. In cdc14-1 bub2Δ cells, as opposed to CDC14 bub2Δ cells, the GTP-Cdc42 biosensor median fluorescence intensity remains higher in the buds than in the mothers, suggesting that Cdc14 inhibition impaired the redistribution of the biosensor molecules between the bud and the mother cells (Fig. 5 F). We conclude that Cdc14 release is necessary and sufficient to terminate the asymmetric localization of Cdc42 activity to the buds.
Dephosphorylation of Cdc42 GAP Bem3 is dependent on Cdc14
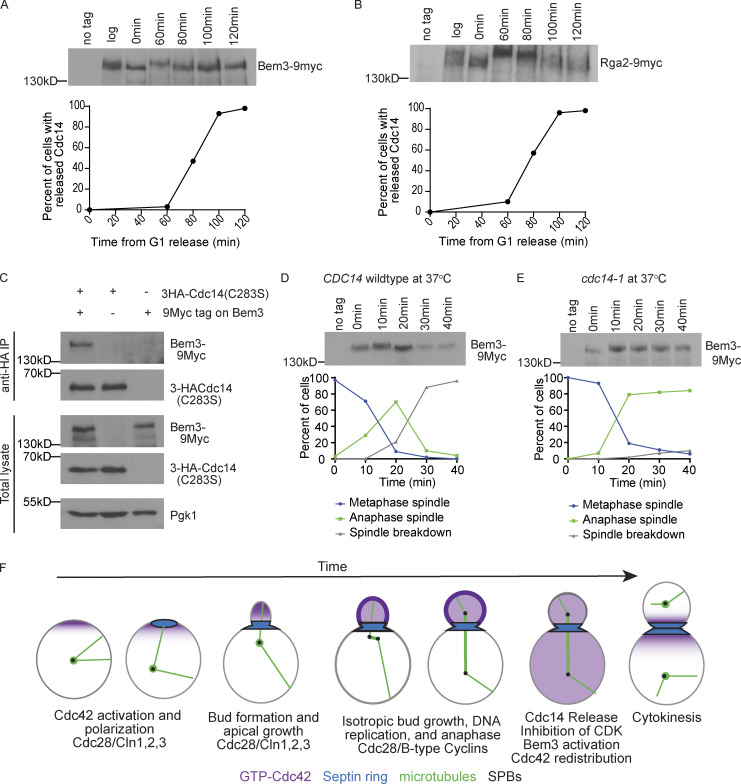
Our results demonstrate that the nucleolar release of Cdc14 results in Cdc42 redistribution between the mother and the bud. As a potential mechanism of action, we considered that Cdc14 could dephosphorylate Cdc42 regulators. Previous work showed that phosphorylation of Cdc42 GAPs in G1 results in their inactivation, enhancing Cdc42 activation and polarization (Knaus et al., 2007; Sopko et al., 2007). We hypothesized that Cdc14 could dephosphorylate and activate a GAP to inactivate Cdc42. Previous work predicted that Bem3 and Rga2 are potential substrates of Cdc14 based on a Cdc14 binding consensus sequence (Eissler et al., 2014). Therefore, we asked if these proteins were dephosphorylated at the time of Cdc14 release. We released cells from an α factor arrest and harvested proteins at different time points through mitosis and cytokinesis. As expected, Bem3-9Myc underwent a band shift after entering into the cell cycle that we are interpreting as phosphorylation (Fig. 6 A; comparison of t = 0 to t = 60). At 80 min, Bem3-9Myc underwent a faster migrating band shift, which correlates with the time of Cdc14 release (Fig. 6 A). Rga2 underwent a modest band shift at 80 min with a further shift at 100 min (Fig. 6 B). Previous data showed that overexpression of Bem3, but not Rga2, is highly toxic to the cell due to Bem3’s GAP activity, causing a failure in bud emergence and actin polarization (Knaus et al., 2007; Sopko et al., 2007). Therefore, we considered that the dephosphorylation and activation of Bem3 at the end of mitosis would be important for inactivation and redistribution of Cdc42. We focused our studies on Bem3 as a potential substrate of Cdc14.
Figure 6.
Cdc14 regulates Bem3 phosphorylation. (A) Western blot of Bem3-9myc in wild-type cells during a synchronous mitosis. Cells were released from α factor and aliquots were taken at indicated times. Alpha factor was added back at 80 min to prevent the cells from starting a new cell cycle. A graph of the corresponding timing of Cdc14 nucleolar release is shown below (n = 100 cells per time point). (B) Western blot images of Rga2-9myc in wild-type cells undergoing synchronous mitosis. A graph of the corresponding timing of Cdc14 release is shown below (n = 100 cells per time point). (C) Western blots of coimmunoprecipitated 3HA-Cdc14(C283S) and Bem3-9Myc. Pgk1 serves as a loading control. (D) Western blot of Bem3-9myc in wild-type cells released from a metaphase arrest at 37°C. Cells were released from α factor into a metaphase arrest by depleting Cdc20 using a methionine-repressible promoter. Cells were incubated in methionine medium for 2 h at 25°C, and then washed and released in media lacking methionine to resume cell cycle progression at 37°C. Aliquots were taken at indicated times. A graph of cell cycle progression is shown below. Cell cycle progression was assessed by monitoring the mitotic spindle with GFP-Tub1 (n = 100 cells per time point). (E) Western blot of Bem3-9myc in cdc14-1 cells released from a metaphase arrest at 37°C (n = 100 cells per time point). Cells were processed as in D. (F) Proposed model of GTP-Cdc42 (magenta) polarity and bud localization during the cell cycle with microtubules (green), spindle pole bodies (SPBs; black), and septins (blue).
We next asked if Bem3 interacts with Cdc14. Because phosphatases usually interact transiently with their substrates, we expressed an epitope-tagged Cdc14 trap allele in which the catalytic cysteine was changed to a serine (3HA-Cdc14 [C283S]; Powers et al., 2017). The C283S mutation prevents Cdc14 catalytic activity but allows formation of stable interactions with substrates (Blanchetot et al., 2005; Taylor et al., 1997). We found that immunoprecipitated 3HA-Cdc14(C283S) pulled down Bem3-9Myc (Fig. 6 C). This result demonstrates that Bem3 and Cdc14 interact in vivo.
We considered that if Bem3 is a substrate of Cdc14, Bem3 should not be dephosphorylated in the cdc14-1 mutants. To investigate Bem3 phosphorylation in anaphase and mitosis exit, we arrested cells at metaphase through Cdc20 depletion, released them from the arrest, and isolated protein at 10-min time points for 40 min. We found that Bem3 underwent a band upshift at early anaphase in both cdc14-1 and CDC14 control cells at 37°C (Fig. 6 D; t = 20). This upshift correlates in timing with results from a previous study showing that Cdc42 activity increases in early anaphase just before it decreases (Atkins et al., 2013). In late anaphase, Bem3 displays a band downshift in CDC14 control cells, suggesting dephosphorylation of Bem3. In contrast, in cdc14-1 cells, Bem3 did not undergo a band downshift, suggesting that Bem3 remained phosphorylated (Fig. 6 E). To our knowledge, this is the first demonstration of the Cdc14-dependent dephosphorylation of a Cdc42 GAP at the end of mitosis. Because previous work has shown that the phosphorylation of Cdc42 GAPs by Cdk is inhibitory (Knaus et al., 2007; Sopko et al., 2007), we presume that dephosphorylation at the end of mitosis would activate the GAPs to inhibit Cdc42. We propose that Bem3 is a substrate of Cdc14 and that the dephosphorylation of Bem3 by Cdc14 may activate Bem3, resulting in inactivation and redistribution of Cdc42 (Fig. 6 F).
Discussion
The mitotic exit pathway promotes Cdc42 redistribution
Our results demonstrate that Cdc14 release from the nucleolus, through the MEN, is required for Cdc42 redistribution at the end of anaphase. The redistribution of Cdc42 between the mother and bud occurs shortly after Cdc14 release from the nucleolus (Fig. 4, A and B). Inactivation of Cdc14, by growing cdc14-1 cells at the restrictive temperature, maintains Cdc42 localization in the bud (Fig. 4, E and F). Furthermore, preventing Cdc14 release by inhibiting polo kinase, Cdc5, or by inactivating Cdc15, also maintained GTP-Cdc42 bud localization (Fig. 4, H–J; and Fig. S5, C–E). Forced premature release of Cdc14 in S phase–arrested cells led to a premature redistribution of Cdc42 (Fig. 5). As Cdc14 release reverses Cdk substrate phosphorylation, we searched for potential substrates of Cdc14 that regulate Cdc42 activity. We found that the dephosphorylation of the Cdc42 GAP Bem3 depends on Cdc14 activity. Previous studies have shown that the GAPs are phosphorylated in G1 and that this phosphorylation inactivates them (Gulli et al., 2000; Knaus et al., 2007; Sopko et al., 2007). This leads us to propose the model that dephosphorylation of Bem3 results in its activation, causing the inactivation and redistribution of Cdc42 (Fig. 6 F).
Interestingly, Cdc42 bud localization is also required for the normal function of the MEN. Lte1, a component of the MEN signaling pathway, is a guanine nucleotide exchange factor for the Tem1 GTPase. Tem1 serves as an upstream activator of the MEN signaling pathway, ultimately leading to release of Cdc14 from its inhibitor in the nucleolus (reviewed in Scarfone and Piatti, 2015). Lte1 becomes asymmetrically localized to the bud cortex in S phase, and this asymmetric localization requires Cdc42 activity (Bardin et al., 2000; Höfken and Schiebel, 2002; Pereira et al., 2000; Seshan et al., 2002). Tem1 is localized to the old spindle pole body that enters the bud when the cells undergo anaphase spindle elongation. The asymmetry of Lte1 localization ensures that the MEN is activated only once chromosomes have been partitioned into the bud. Our work showing that the MEN is required for Cdc42 redistribution and inactivation implies a negative feedback loop in which bud-localized Cdc42 promotes its own inactivation by recruiting MEN components.
Maintenance of cell size to preserve cellular fitness
While cell size varies across cell types, every cell type has a characteristic size that is preserved during cell division. Chromosome segregation is coordinated with cell growth to ensure that cells retain their size. In budding yeast, buds are characteristically smaller than their mothers at the end of mitosis. Here, we show that budding yeast cells inactivate Cdc42 at late anaphase to redistribute growth between the bud and the mother compartments (Fig. 1). By the time budding yeast cells reach cytokinesis, the bud size is, on average, 40% of the mother size (Fig. 2 E). We find that arresting the cell cycle before spindle breakdown keeps Cdc42 asymmetrically localized and active in the buds, which leads to abnormally large buds and causes spindle position defects (Fig. 2, B–E). After 3 h of metaphase arrest, the buds grew larger than their mothers, reversing the normal size relationship between the buds and their mothers (Fig. 2 G).
Perturbation of the size ratio between the mother and the bud could be consequential to the cells. Cells released from a prolonged metaphase arrest have alterations in the pattern of bud site selection and the timing of the next cell cycle when compared with unperturbed cells (Figs. S3 and S4). For example, Cdc42 polarization and bud formation occurred at a similar time in both the buds and the mothers in metaphase-released cells (Fig. S3). In an unperturbed cell cycle, the daughter cell is delayed in bud initiation due to the asymmetric nuclear localization of a transcription factor Ace2, driving a bud-specific transcriptional program (Di Talia et al., 2009; Laabs et al., 2003; Mazanka et al., 2008; Parnell and Stillman, 2008). Ace2 activity contributes to the bud-specific delay in cell cycle reentry after cytokinesis (Laabs et al., 2003). Intriguingly, a recent study reported that prolonged metaphase delay is sufficient to disrupt Ace2 asymmetry (Herrero et al., 2020), thus providing a potential account for the altered timing of budding that we observe in cells released from a prolonged metaphase arrest: the disruption of Ace2 asymmetry allows the daughter cells to reenter the cell cycle at the same time as the mother cells after cytokinesis. Interestingly, in the metaphase-released cells, the mothers underwent anaphase later than the daughters (Fig. S3, B and D). Whether this phenotype is due to Ace2 asymmetric localization needs to be further investigated. Nonetheless, the finding provokes an untested presumption that prolonged metaphase arrest may deplete the mother cells of certain determinants that contribute to timely anaphase entry.
What is the importance of a smaller size of the buds compared with their mothers at every division? Recent work has shown that as cells divide, they increase in size, and the size increase contributes to aging and reduces cellular fitness by diluting the cytoplasm (Neurohr et al., 2019). Therefore, cells that are born smaller have an advantage of a longer replicative lifespan; they can undergo more divisions before they reach a critical size at which their size starts to impede their function. Thus, having a mechanism in place to inactivate Cdc42 after chromosome segregation ensures that the bud maintains a smaller size than the mother to enhance cellular fitness and delay cellular aging.
Materials and methods
Strains, plasmids, and manipulations
Gene deletion and tagging were done using a standard PCR-based transformation method (Longtine et al., 1998; Janke et al., 2004; Sheff and Thorn, 2004). Plasmids containing GFP-TUB1, GIC2-PBD(W23A)-RFP, and CUP1prCDC5 were integrated into the genome. Yeast strains used in this study are derivatives of W303 with the following basal genetic background: ade2-1, his3-11,15 leu2-3,112 trp1-1, ura3-1, and can1-100.
The following strains were used in this study: LY4667: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3; LY4146: MATa, PTUB1-GFP-TUB1:LEU2, ABP1-3xmCherry:HphMX; LY5339: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, PMET-3HA-CDC20:TRP1 (at the CDC20 locus); LY7677: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, PMET-3HA-CDC20:TRP1 (at the CDC20 locus), cdc41-1; LY4593: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, cdc5-L158G:NAT; LY4686: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, cdc15-2; LY4894: MATa, CDC14-GFP:KanMX, GIC2PBD-(W23A)-RFP:URA3:HIS3, PCUP1-CDC5:TRP1; LY4677: MATa, CDC14-GFP:KanMX, GIC2PBD-(W23A)-RFP:URA3:HIS3; LY6191: MATa, CDC14-GFP:KanMX, GIC2PBD-(W23A)-RFP:URA3:HIS3, PCUP1-CDC5:TRP1, bub2::hphNT1; LY6335: MATa, CDC14-GFP:KanMX, GIC2PBD-(W23A)-RFP:URA3:HIS3, bub2::hphNT1; LY6300: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, PCUP1-CDC5:TRP1, bub2::hphNT1; LY7752: MATa, PTUB1-GFP-TUB1:LEU2, GIC2PBD-(W23A)-RFP:URA3:HIS3, PCUP1-CDC5:TRP1, bub2::hphNT1, cdc14-1; LY4674: MATa, Bem3-9Myc:KanMX; LY6853: MATa, Bem3-9Myc:KanMX, Cdc14-GFP:His; LY7723: MATa, Bem3-9Myc:KanMX, PTUB1-GFP-TUB1:LEU2, PMET-3HA-CDC20:TRP1 (at the CDC20 locus); LY7725: MATa, Bem3-9Myc:KanMX, PTUB1-GFP-TUB1:LEU2, PMET-3HA-CDC20:TRP1 (at the CDC20 locus), cdc14-1; LY4672: MATa, Rga2-9Myc:KanMX; LY6854: MATa, Rga2-9Myc:KanMX, Cdc14-GFP:HIS; LY4942: MATa, net1::HIS5, net1-6Cdk-TEV-9myc:TRP1Cdc14-GFP:KanMX, GIC2PBD-(W23A)-RFP:URA3:HIS3.
Growth conditions
The yeast growth media used in our experiments were synthetic complete (SC) medium (0.67% bacto-yeast nitrogen base without amino acids, 0.2% dropout mix with all amino acids, and 2% glucose), YPD medium (2% yeast extract, 1% peptone, and 2% glucose), or YP medium (2% yeast extract and 1% peptone) supplemented with 2% raffinose or 2% galactose. It is specified below where each medium type was used.
Cell synchronization
Cells were synchronized in G1 with 10 µM α factor (Zymo Research; Y1001) for 2 h in SC or YPD medium at 25°C with 175 rpm shaking.
Cell cycle arrest
S phase arrest was achieved by synchronizing the cells in G1 with α factor and then releasing them into SC medium with 200 µM hydroxyurea (Sigma; H8726). For cells harboring the CUP1prCDC5 construct and their respective controls, hydroxyurea was added to asynchronous cultures, since adding the drug to α factor–synchronized cells was slightly toxic. 100 µM copper sulfate (Sigma; 209198) was added after to cells after 2 h in hydroxyurea. To achieve arrest in M phase, cells were synchronized in G1 and released into cell cycle without CDC20, an activator of the anaphase-promoting complex. CDC20 was under a methionine-repressible MET3 promotor. Therefore, cells were grown and synchronized in medium lacking methionine but released into medium with methionine to block Cdc20 expression.
Microscope image acquisition, processing, and volume measurements
Three microscope systems were used for this study. cdc15-2 and their counterpart wild-type controls were loaded in coverslip-mounted chamber containing liquid SC medium; the coverslip was coated with concanavalin A (Sigma; 11028–71-0) to stabilize the cells during imaging. Imaging was performed on the Applied Precision Delta Vision personal Delta Vision (pDV) controlled by the SoftWoRx software and equipped with a CoolSNAP_HQ2/HQ2-ICX285 camera using a 60× oil objective (u-Plan S-APO N, 1.4 NA) at room temperature. cdc14-1 strains and corresponding controls were imaged on the pDV with the incubator temperature set to 37°C. All other time-lapse live images were acquired on a Nikon Ti-E inverted microscope controlled by the NIS-Elements software and equipped with Cool-SNAPHQ2 CCD camera and a 60× oil objective (PlanAPO VC, 1.4 NA), using the ONIX microfluidics system (Millipore) or in coverslip chambers containing SC medium at room temperature. During time-lapse imaging, the time interval was 5 min, and images were acquired for 6–8 h. For Z scan, 5–11 Z plans were acquired, with a step of 1–1.5 µm. Time-lapse live images were acquired in five Z steps with a 1-µm step on the pDV microscope or 1.2-µm step on the Nikon Ti-E microscope. All static images were acquired using a Nikon Ti-E2 microscope controlled by the NIS-Elements software equipped with a Photometrics Prime 95B 25-mm camera and a 100× oil objective (Plan Apo λ, 1.45 NA) at room temperature. Cells incubated in SC medium were collected and imaged on a slide immediately without fixation. The fluorochromes used were GFP fused to TUB1, RFP fused to the Gic2 PBD domain, and mCherry fused to Abp1.
Maximum intensity projections were performed to obtain the final fluorescence images presented in this article. To measure Gic2-PBD-RFP fluorescence intensity, sum projections were used instead. Imaging processing and fluorescence intensity measurements were done using ImageJ. Cell volume was calculated using the BudJ plugin in ImageJ (Ferrezuelo et al., 2012).
Western blotting
For Western blot experiments, cells were grown in YPD medium. A 5-ml culture was started and grown overnight to saturation at 30°C on a roller drum. In the morning, the cells were diluted 1/20 and grown to log phase (OD600 = 1) by incubating in YPD at 25°C for 5 h with 175 rpm shaking. Cells were then synchronized in G1 with α factor for 2 h, washed YPD medium, and released in fresh medium. For each time point indicated, 1.5 ml of the culture was harvested in a 1.5-ml screw-cap tube containing ∼100 µl of 0.5-mm glass beads, spun down for 30 s at 7,000 rpm on a tabletop centrifuge, and snap frozen in liquid nitrogen. At 80 min after releasing from α factor, the cells were treated with α factor again to prevent them from starting a new division cycle. The frozen cells were lysed in a buffer (65 mM Tris-HCl, pH 6.8, 50 mM NaF, 100 mM sodium β-glycerophosphate, 3% sodium dodecyl sulfate, 10% glycerol, 1 mM PMSF, 5% β-mercaptoethanol, and one tablet of protease inhibitor; Thermo Fisher Scientific; A32963) and a very small amount of bromophenol blue by vigorously shaking three times for 30 s, with a 30-s break on ice between shaking rounds. The cells extracts were boiled at 95–100°C for 5 min; 10 µl were loaded per well during gel electrophoresis. The gel was run for 2.5 h at room temperature and constant 150 V. Proteins were transferred onto a polyvinylidene difluoride membrane for 1.5 h at 4°C and constant 180 mAmps.
Blocking and antibody incubation were done in 5% milk in a TBS/T buffer (50 mM Trizma base, 150 mM NaCl, and 0.1% Tween, pH 7.4). The primary antibody used was an anti-myc 9E10 mouse monoclonal (Roche; 11 667 149 001); it was incubated overnight at 4°C with gentle shaking. The secondary antibody used was a sheep anti-mouse IgG horseradish peroxidase-linked (GE Healthcare; NA9310V); it was incubated at room temperature for 1 h with gentle shaking. Antibodies were washed three times for 5 min in TBS/T buffer. The polyvinylidene difluoride membrane was exposed to chemiluminescent reagents (Thermo Fisher Scientific; 34095) for 5 min and exposed to autoradiography films (MidSci) for 1–10 min in the dark. The films were developed on a Konica Minolta SRX-101A processor. Films were scanned on a C5X25A Deskjet HP printer, and the scanned images were cropped in ImageJ for final presentation.
Immunoprecipitation
Cells were grown and processed as previously reported (Powers et al., 2017). Briefly, a 5-ml starter culture was incubated overnight at 30°C on a roller drum. Cells were grown in SC medium lacking leucine to maintain the plasmid carrying the cdc14 substrate trapping allele. The allele was expressed from GAL1 promoter. The next morning, the overnight culture was diluted into 50 ml YPR (2% yeast extract, 1% peptone, and 2% raffinose) and grown at 30°C with 200 rpm shaking until OD600= 0.8. The culture was then divided into two 25-ml batches, and to one batch, 20 ml fresh YP medium (2% yeast extract and 1% peptone) and 5 ml of 20% galactose were added. To the other batch, 20 ml fresh YP medium and 5 ml of 20% glucose were added. The two cultures were incubated at 30°C for 2 h. The cells were then pelleted in 50-ml conical tubes, and the pellet was washed once with 1 ml water. The pellet was then transferred to 1.5-ml screw-cap tubes containing ∼100 µl of 0.5-mm glass beads and snap frozen in liquid nitrogen after a quick spin and removal of residual supernatant. Cells were lysed at 4°C by vigorous shaking for 15 min. The lysis buffer contained 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.1% Triton X-100, 5 mM EDTA, 20 mM sodium fluoride, and 20 mM β-glycerophosphate. Before lysing the cells, the lysis buffer was supplemented with 10 µM leupeptin (Sigma; L2884) and 1 µM pepstatin (Sigma; P4265). PMSF was added to the lysate at a final concentration of 1 mM, and then the lysate was cleared by centrifugation at 14,000 rcf for 20 min at 4°C. The cleared lysate was incubated with anti-HA affinity resin (Abcam; ab270603) for 2 h at 4°C with gentle rocking. The resin was collected by centrifugation at 8,000 rcf for 30 s at 4°C and washed four times with 1 ml lysis buffer. The resin was then suspended in 40 μl sample buffer and boiled at 95–100°C for 5 min. The boiled samples were centrifuged at 8,000 rcf for 30 s, and the supernatant was collected and mixed with 0.5% β-mercaptoethanol before submission to SDS-PAGE electrophoresis. The primary antibody used was the 12CA5 mouse monoclonal against HA (Sigma; 11583816001). The secondary antibody used was a sheep anti-mouse IgG horseradish peroxidase-linked (GE Healthcare; NA9310V). Western blotting and electrophoresis were performed as described above (see Western blotting).
Primers
The following primers were used: Bem3 C-terminal tagging: 5′-TGAGGCATCTCCAAGTCTTTAC-3′, 5′-CTTCTTTATCTCAGCTCTTCGACC-3′, 5′-AAGAAGAGGAAAGGGAGAAAGTAGATATACATATTCCTCAGGTTCGTACGCTGCAGG-3′, and 5′-TGGCAACGTTATATTTCTACAATTTTAGACCATCAGATATCATCGATGAATTCGAGC-3′; Rga2 C-terminal tagging: 5′-GAGAGGCTACCATTAAACAACGAC-3′, 5′-CAAGAAGAAGAGGAGACGCAATAC-3′, 5′-TGGAATTTATACTTGGAAACTATAGAGACATATTTAAGCAAGCACGTACGCTGCAGG-3′, and 5′-TCCTATGTTTATTTAACTTTTGCAAATCTGTATTAGATATCATCGATGAATTCGAGCTCG-3′; Cdc14 C-terminal tagging: 5′-GAAGCCTAGTGAGGCCATTG-3′, 5′-CCATCAGGAGGAGAAGATTGT-3′.; BUB2 deletion: 5′-AGGTAAAAGAAACAACAGACTTTTAAACTTGTTAACTTTTGCATGCGTACGC-3′, 5′-TGCAGGTCG-3′, 5′-TTGTAGAATTAAACGATAAAATATAATATTTCTTCACATAGTTTAGATATCAT-3′, and 5′-CGATGAATTCGAGCTCG-3′, 5′-GACAATTCAGTAAACGCCGTG-3′, 5′-CTACGTTTGCATTGGGTGGA-3′; pLB157 integration for CDC5 overexpression: 5′-GCGCTATACGTGCATATGTTC-3′ and 5′-GGAACGGGTATTCAATTGCTTATC-3′.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 8 software. Statistical significance was calculated at the 95% confidence level.
Online supplemental material
Fig. S1 shows the localization of GTP-Cdc42 in asynchronously dividing cells. Fig. S2 shows GTP-Cdc42 localization and cell size in cells treated with nocodazole or latrunculin. Fig. S3 shows the timing of cell cycle events and the orientation of the anaphase spindle in cells released from a prolonged metaphase arrest. Fig. S4 shows the budding pattern in cells released from a prolonged metaphase arrest. Fig. S5 shows the localization of GTP-Cdc42 in cdc15-2 mutant cells.
Acknowledgments
We thank Erfi Bi (University of Pennsylvania, Perelman School of Medicine, Philadelphia, PA), Peter Matthias (ETH Zurich, Institute of Biochemistry, Zurich, Switzerland), Mark Hall (Purdue University, Lafayette, IN), Yanchang Wang (Florida State University, Tallahassee, FL), and Christopher Puccia (Indiana University, Bloomington, IN) for strains and plasmids. We are grateful to the Light Microscopy and Imaging Center at Indiana University, especially Jim Powers, for assistance with microscopy.
This work was supported by National Institutes of Health grant GM105755 (to S. Lacefield).
The authors declare no competing financial interests.
Author contributions: G.M. Gihana and S. Lacefield made yeast strains; conceived, designed, and performed experiments; and wrote the manuscript. A.A. Cross-Najafi made yeast strains and performed experiments. S. Lacefield secured funding.
References
- Adams A.E., and Pringle J.R.. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934–945. 10.1083/jcb.98.3.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A.E., Johnson D.I., Longnecker R.M., Sloat B.F., and Pringle J.R.. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131–142. 10.1083/jcb.111.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins B.D., Yoshida S., Saito K., Wu C.F., Lew D.J., and Pellman D.. 2013. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 202:231–240. 10.1083/jcb.201301090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., and Deshaies R.J.. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 305:516–519. 10.1126/science.1099402 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin R., and Amon A.. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31. 10.1016/S0092-8674(00)00007-6 [DOI] [PubMed] [Google Scholar]
- Beach D.L., Thibodeaux J., Maddox P., Yeh E., and Bloom K.. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497–1506. 10.1016/S0960-9822(00)00837-X [DOI] [PubMed] [Google Scholar]
- Bi E., and Park H.O.. 2012. Cell polarization and cytokinesis in budding yeast. Genetics. 191:347–387. 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchetot C., Chagnon M., Dubé N., Hallé M., and Tremblay M.L.. 2005. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 35:44–53. 10.1016/j.ymeth.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Breeden L.L. 1997. Alpha-factor synchronization of budding yeast. Methods Enzymol. 283:332–341. 10.1016/S0076-6879(97)83027-3 [DOI] [PubMed] [Google Scholar]
- Carminati J.L., and Stearns T.. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641. 10.1083/jcb.138.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou J.G., Balasubramanian M.K., and Lew D.J.. 2017. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 33:77–101. 10.1146/annurev-cellbio-100616-060856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko Z.N., and Blagosklonny M.V.. 2008. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 7:3355–3361. 10.4161/cc.7.21.6919 [DOI] [PubMed] [Google Scholar]
- Di Talia S., Wang H., Skotheim J.M., Rosebrock A.P., Futcher B., and Cross F.R.. 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 7:e1000221 10.1371/journal.pbio.1000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D.G., Miller K.G., and Botstein D.. 1988. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107:2551–2561. 10.1083/jcb.107.6.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissler C.L., Mazón G., Powers B.L., Savinov S.N., Symington L.S., and Hall M.C.. 2014. The Cdk/cDc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Mol. Cell. 54:80–93. 10.1016/j.molcel.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein A.E., and Drubin D.G.. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19:287–332. 10.1146/annurev.cellbio.19.111401.093127 [DOI] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu L.A., Vissers S., Jauniaux J.C., van Vliet-Reedijk J.C., Planta R.J., and Gibbons I.R.. 1993. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA. 90:11172–11176. 10.1073/pnas.90.23.11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkaš V., Kovarík J., Kosinová A., and Bauer S.. 1974. Autoradiographic study of mannan incorporation into the growing cell walls of Saccharomyces cerevisiae. J. Bacteriol. 117:265–269. 10.1128/JB.117.1.265-269.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F., Colomina N., Palmisano A., Garí E., Gallego C., Csikász-Nagy A., and Aldea M.. 2012. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat. Commun. 3(1012):1–11. 10.1038/ncomms2015 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Walker P.A., Johnston L.H., and Sedgwick S.G.. 2003. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278:14591–14594. 10.1074/jbc.C300059200 [DOI] [PubMed] [Google Scholar]
- Gulli M.P., Jaquenoud M., Shimada Y., Niederhäuser G., Wiget P., and Peter M.. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 6:1155–1167. 10.1016/S1097-2765(00)00113-1 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., and Unger M.W.. 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75:422–435. 10.1083/jcb.75.2.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., Stinus S., Bellows E., Berry L.K., Wood H., and Thorpe P.H.. 2020. Asymmetric Transcription Factor Partitioning During Yeast Cell Division Requires the FACT Chromatin Remodeler and Cell Cycle Progression. Genetics. 216:701–716. 10.1534/genetics.120.303439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., and Schiebel E.. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862. 10.1093/emboj/cdf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.S., and Lew D.J.. 2012. Morphogenesis and the cell cycle. Genetics. 190:51–77. 10.1534/genetics.111.128314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., and Elledge S.J.. 2002. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle. 1:351–355. 10.4161/cc.1.5.154 [DOI] [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., and Elledge S.J.. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 107:655–665. 10.1016/S0092-8674(01)00580-3 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Tinker-Kulberg R.L., and Morgan D.O.. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:2803–2817. 10.1091/mbc.9.10.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.I. 1999. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54–105. 10.1128/MMBR.63.1.54-105.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G.C., Pringle J.R., and Hartwell L.H.. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98. 10.1016/0014-4827(77)90154-9 [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V., and Adams A.E.. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922–933. 10.1083/jcb.98.3.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus M., Pelli-Gulli M.P., van Drogen F., Springer S., Jaquenoud M., and Peter M.. 2007. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 26:4501–4513. 10.1038/sj.emboj.7601873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J., Meyer A., Snyder M.P., and Barral Y.. 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16:1627–1639. 10.1101/gad.222602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laabs T.L., Markwardt D.D., Slattery M.G., Newcomb L.L., Stillman D.J., and Heideman W.. 2003. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 100:10275–10280. 10.1073/pnas.1833999100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Frenz L.M., Wells N.J., Johnson A.L., and Johnston L.H.. 2001. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11:784–788. 10.1016/S0960-9822(01)00228-7 [DOI] [PubMed] [Google Scholar]
- Leitao R.M., and Kellogg D.R.. 2017. The duration of mitosis and daughter cell size are modulated by nutrients in budding yeast. J. Cell Biol. 216:3463–3470. 10.1083/jcb.201609114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., and Reed S.I.. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305–1320. 10.1083/jcb.120.6.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y., Yeh E., Hays T., and Bloom K.. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 90:10096–10100. 10.1073/pnas.90.21.10096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., and Wang Y.. 2007. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol. Cell. Biol. 27:5067–5078. 10.1128/MCB.00095-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Marquitz A.R., Harrison J.C., Bose I., Zyla T.R., McMillan J.N., and Lew D.J.. 2002. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 21:4012–4025. 10.1093/emboj/cdf416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanka E., Alexander J., Yeh B.J., Charoenpong P., Lowery D.M., Yaffe M., and Weiss E.L.. 2008. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 6:e203 10.1371/journal.pbio.0060203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R., Neutzner A., and Seufert W.. 2001. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr. Biol. 11:345–350. 10.1016/S0960-9822(01)00095-1 [DOI] [PubMed] [Google Scholar]
- Miller R.K., and Rose M.D.. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377–390. 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Matheos D., and Rose M.D.. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963–975. 10.1083/jcb.144.5.963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Stuchell-Brereton M.D., and Cooper J.A.. 2009. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil. Cytoskeleton. 66:546–555. 10.1002/cm.20364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurohr G.E., Terry R.L., Lengefeld J., Bonney M., Brittingham G.P., Moretto F., Miettinen T.P., Vaites L.P., Soares L.M., Paulo J.A., et al. 2019. Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence. Cell. 176:1083–1097.e18. 10.1016/j.cell.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Leda M., Hanna J., Savage N.S., Bi E., and Goryachev A.B.. 2013. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell. 26:148–161. 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye M.A., Noll B., and Hausser A.. 2019. Spatiotemporal Control of Intracellular Membrane Trafficking by Rho GTPases. Cells. 8:8 10.3390/cells8121478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R.E., Sullivan D.S., Huffaker T., and Koshland D.. 1992. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 119:583–593. 10.1083/jcb.119.3.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell E.J., and Stillman D.J.. 2008. Getting a transcription factor to only one nucleus following mitosis. PLoS Biol. 6:e229 10.1371/journal.pbio.0060229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Höfken T., Grindlay J., Manson C., and Schiebel E.. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10. 10.1016/S1097-2765(05)00017-1 [DOI] [PubMed] [Google Scholar]
- Pereira G., Manson C., Grindlay J., and Schiebel E.. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367–379. 10.1083/jcb.200112085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers B.L., Hall H., Charbonneau H., and Hall M.C.. 2017. A Substrate Trapping Method for Identification of Direct Cdc14 Phosphatase Targets. Methods Mol. Biol. 1505:119–132. 10.1007/978-1-4939-6502-1_10 [DOI] [PubMed] [Google Scholar]
- Pringle J.R., Bi E., Harkins H.A., Zahner J.E., De Virgilio C., Chant J., Corrado K., and Fares H.. 1995. Establishment of cell polarity in yeast. Cold Spring Harb. Symp. Quant. Biol. 60:729–744. 10.1101/SQB.1995.060.01.079 [DOI] [PubMed] [Google Scholar]
- Pruyne D., and Bretscher A.. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365–375. [DOI] [PubMed] [Google Scholar]
- Rodal A.A., Kozubowski L., Goode B.L., Drubin D.G., and Hartwig J.H.. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell. 16:372–384. 10.1091/mbc.e04-08-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarfone I., and Piatti S.. 2015. Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1. Small GTPases. 6:196–201. 10.1080/21541248.2015.1109023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., Bardin A.J., and Amon A.. 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12:2098–2110. 10.1016/S0960-9822(02)01388-X [DOI] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Shevchenko A., Charbonneau H., and Deshaies R.J.. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 97:233–244. 10.1016/S0092-8674(00)80733-3 [DOI] [PubMed] [Google Scholar]
- Smith G.R., Givan S.A., Cullen P., and Sprague G.F. Jr. 2002. GTPase-activating proteins for Cdc42. Eukaryot. Cell. 1:469–480. 10.1128/EC.1.3.469-480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead J.L., Sullivan M., Lowery D.M., Cohen M.S., Zhang C., Randle D.H., Taunton J., Yaffe M.B., Morgan D.O., and Shokat K.M.. 2007. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem. Biol. 14:1261–1272. 10.1016/j.chembiol.2007.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J.C., Figeys D., and Andrews B.J.. 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26:4487–4500. 10.1038/sj.emboj.7601847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Shochet N.R., Kashman Y., and Groweiss A.. 1983. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 219:493–495. 10.1126/science.6681676 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., and Amon A.. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232. 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., and Amon A.. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220. 10.1016/S0092-8674(02)00618-9 [DOI] [PubMed] [Google Scholar]
- Taylor G.S., Liu Y., Baskerville C., and Charbonneau H.. 1997. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J. Biol. Chem. 272:24054–24063. 10.1074/jbc.272.38.24054 [DOI] [PubMed] [Google Scholar]
- Theesfeld C.L., Irazoqui J.E., Bloom K., and Lew D.J.. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019–1032. 10.1083/jcb.146.5.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld C.L., Zyla T.R., Bardes E.G., and Lew D.J.. 2003. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell. 14:3280–3291. 10.1091/mbc.e03-03-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz J.S., and Lampen J.O.. 1972. Wall replication in saccharomyces species: use of fluorescein-conjugated concanavalin A to reveal the site of mannan insertion. J. Gen. Microbiol. 72:243–247. 10.1099/00221287-72-2-243 [DOI] [PubMed] [Google Scholar]
- Traverso E.E., Baskerville C., Liu Y., Shou W., James P., Deshaies R.J., and Charbonneau H.. 2001. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 276:21924–21931. 10.1074/jbc.M011689200 [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J., and Pelham H.R.. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13:1636–1640. 10.1016/j.cub.2003.09.001 [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., and Amon A.. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718. 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., and Amon A.. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 398:818–823. 10.1038/19775 [DOI] [PubMed] [Google Scholar]
- Visintin R., Stegmeier F., and Amon A.. 2003. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell. 14:4486–4498. 10.1091/mbc.e03-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheals A.E. 1982. Size control models of Saccharomyces cerevisiae cell proliferation. Mol. Cell. Biol. 2:361–368. 10.1128/MCB.2.4.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods B., and Lew D.J.. 2019. Polarity establishment by Cdc42: Key roles for positive feedback and differential mobility. Small GTPases. 10:130–137. 10.1080/21541248.2016.1275370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T.C., and Bretscher A.. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015. 10.1038/35023024 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Asakawa K., and Toh-e A.. 2002. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12:944–950. 10.1016/S0960-9822(02)00870-9 [DOI] [PubMed] [Google Scholar]
- Young M.E., Cooper J.A., and Bridgman P.C.. 2004. Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166:629–635. 10.1083/jcb.200404159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Hart M.J., Shinjo K., Evans T., Bender A., and Cerione R.A.. 1993. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J. Biol. Chem. 268:24629–24634. [PubMed] [Google Scholar]
- Zheng Y., Cerione R., and Bender A.. 1994. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 269:2369–2372. [PubMed] [Google Scholar]