The Hedgehog pathway can be activated by cilia-associated sterol and oxysterol lipids that bind to Smoothened. Findakly et al. identify two enzymes near the ciliary base that activate the Hedgehog pathway, shedding light on how the lipid composition of cilia is established and regulated.
Abstract
Vertebrate Hedgehog signals are transduced through the primary cilium, a specialized lipid microdomain that is required for Smoothened activation. Cilia-associated sterol and oxysterol lipids bind to Smoothened to activate the Hedgehog pathway, but how ciliary lipids are regulated is incompletely understood. Here we identified DHCR7, an enzyme that produces cholesterol, activates the Hedgehog pathway, and localizes near the ciliary base. We found that Hedgehog stimulation negatively regulates DHCR7 activity and removes DHCR7 from the ciliary microenvironment, suggesting that DHCR7 primes cilia for Hedgehog pathway activation. In contrast, we found that Hedgehog stimulation positively regulates the oxysterol synthase CYP7A1, which accumulates near the ciliary base and produces oxysterols that promote Hedgehog signaling in response to pathway activation. Our results reveal that enzymes involved in lipid biosynthesis in the ciliary microenvironment promote Hedgehog signaling, shedding light on how ciliary lipids are established and regulated to transduce Hedgehog signals.
Introduction
The Hedgehog pathway directs gene expression programs that are essential for development and adult tissue homeostasis, but misactivation of the Hedgehog pathway can cause congenital disorders and cancer (Briscoe and Thérond, 2013). Smith-Lemli-Opitz syndrome (SLOS) is an autosomal recessive condition caused by inactivating mutations in the gene encoding 3β-hydroxysterol-Δ7-reductase (DHCR7), a rate-limiting enzyme that catalyzes the last step of cholesterol biosynthesis and regulates Hedgehog signal transduction (Jira et al., 2003). SLOS patients have multiple congenital malformations, such as craniofacial abnormalities, holoprosencephaly, syndactyly, and polydactyly. Many of these phenotypic manifestations occur in tissues where Hedgehog signaling is important for embryonic patterning and are also observed in patients who were exposed to teratogenic cholesterol biosynthesis inhibitors in utero or who have Hedgehog ligand mutations (Cooper et al., 1998, 2003). These data suggest that sterol biosynthesis is important for Hedgehog signaling in humans, but DHCR7’s role in Hedgehog signal transduction is controversial (Blassberg et al., 2016; Koide et al., 2006; Kinnebrew et al., 2019). Broadly, sterol lipids are required for vertebrate Hedgehog signaling, and both sterols and oxysterols can bind to Smoothened to activate the downstream pathway (Xiao et al., 2017; Raleigh et al., 2018; Byrne et al., 2016; Cooper et al., 2003; Corcoran and Scott, 2006; Luchetti et al., 2016; Dwyer et al., 2007; Myers et al., 2017, 2013; Nachtergaele et al., 2012, 2013; Nedelcu et al., 2013; Huang et al., 2018; Qi et al., 2019; Deshpande et al., 2019). Nevertheless, biochemical mechanisms underlying lipid biosynthesis–based regulation of the Hedgehog pathway and accumulation of Smoothened-activating lipids in primary cilia are incompletely understood (Kong et al., 2019).
The primary cilium is a microtubule-based cellular projection that is required for Hedgehog morphogens to pattern tissues during development (Huangfu et al., 2003). Ciliary proteins and lipids are different from those of other cellular membranes, and the molecular structure of primary cilia is critical for signal transduction (Garcia et al., 2018). Phosphoinositide lipids confer distinct molecular identities to subcellular compartments, and inositol polyphosphate-5-phosphatase E (INPP5E) localizes to cilia (Balla, 2013; Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Moreover, loss of INPP5E attenuates Hedgehog signaling and can inhibit the growth of Hedgehog-associated medulloblastomas, the most common malignant brain tumors in children (Conduit et al., 2017; Ostrom et al., 2017). We have shown that cilia are also enriched in oxysterol lipids that activate Smoothened and that hydroxysteroid 11-β-dehydrogenase 2 (HSD11β2), an enzyme participating in the production of Smoothened-activating oxysterols, is dramatically enriched in Hedgehog-associated medulloblastomas (Raleigh et al., 2018). Both genetic and pharmacologic inhibition of HSD11β2 attenuate Hedgehog signaling and Hedgehog-associated medulloblastomas in mice (Raleigh et al., 2018), but, like most enzymes involved in sterol biosynthesis, HSD11β2 localizes to the endoplasmic reticulum (Odermatt et al., 2001). Thus, how Smoothened-activating sterols and oxysterols accumulate in the ciliary microenvironment remains unknown, and proteomic studies have not identified enzymes involved in sterol or oxysterol biosynthesis in primary cilia (Mick et al., 2015; Kohli et al., 2017; Sigg et al., 2017).
We hypothesized that enzymes at the ciliary base may produce sterol or oxysterol lipids to establish the unique lipid composition of cilia and regulate Hedgehog signal transduction. To test this, we performed confocal and superresolution microscopy to discover that DHCR7 localizes near the ciliary base. Our results further suggest that DHCR7 may function through Smoothened to promote the Hedgehog pathway, but that Smoothened accumulation in cilia in response to pathway activation does not require DHCR7. Using mass spectrometry–based sterolomics and microscopy, we show that Hedgehog stimulation negatively regulates DHCR7 activity and removes DHCR7 from the ciliary microenvironment. In contrast, confocal and superresolution microscopy, CRISPR interference (CRISPRi), and mass spectrometry–based sterolomics reveal that Hedgehog stimulation positively regulates cytochrome P450 family 7 subfamily A member 1 oxysterol synthase (CYP7A1), an enzyme that has overlapping functions with HSD11β2 (Russell, 2003). In contrast to our findings in HSD11β2, our results demonstrate that CYP7A1 accumulates near the ciliary base and promotes Hedgehog signaling in response to Hedgehog pathway activation but does not regulate Smoothened accumulation in primary cilia. In sum, our results identify enzymes involved in lipid biosynthesis near the ciliary base that promote Hedgehog signaling, shedding light on how the lipid composition of cilia is established and regulated to transduce Hedgehog signals.
Results and discussion
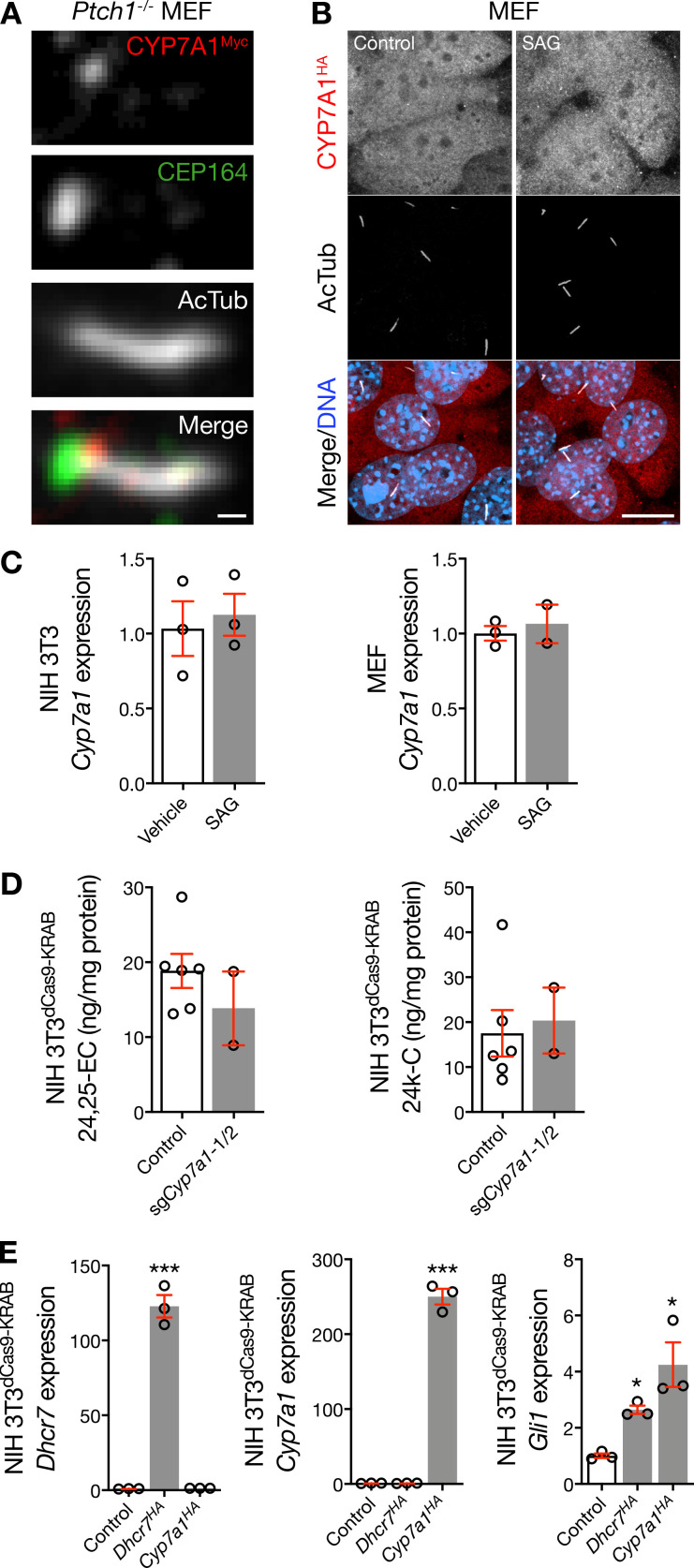
DHCR7 activates the Hedgehog pathway by producing cholesterol, which binds and activates Smoothened (Jira et al., 2003; Blassberg et al., 2016; Kinnebrew et al., 2019). We performed immunofluorescence confocal microscopy for DHCR7 in ciliated NIH 3T3 cells and found that endogenous DHCR7 predominantly localized near the ciliary base (Fig. 1 A and Fig. S1 A). Superresolution microscopy, which has provided new insights into the relationships between ciliary structure and function (Yang et al., 2018; Shi et al., 2017), confirmed that DHCR7 localized near the ciliary base in NIH 3T3 cells and ciliated mouse embryonic fibroblasts (MEFs; Fig. 1 B and Fig. S1 B). In further support of these findings, overexpressed exogenous DHCR7 with a Myc tag could be found at the ciliary base in MEFs (Fig. S1 C). However, exogenous DHCR7 with an HA tag that was overexpressed in MEFs using a different transfection reagent localized to the cytoplasm and endoplasmic reticulum (Fig. S1 D), which was also the case for the majority of overexpressed DHCR7 with a Myc tag. These data are consistent with previous reports (Koczok et al., 2019), but contrast with our results for endogenous DHCR7, underscoring the importance of localization studies that focus on endogenous gene products rather than on overexpression of exogenous constructs that may be influenced by expression conditions or epitope tags.
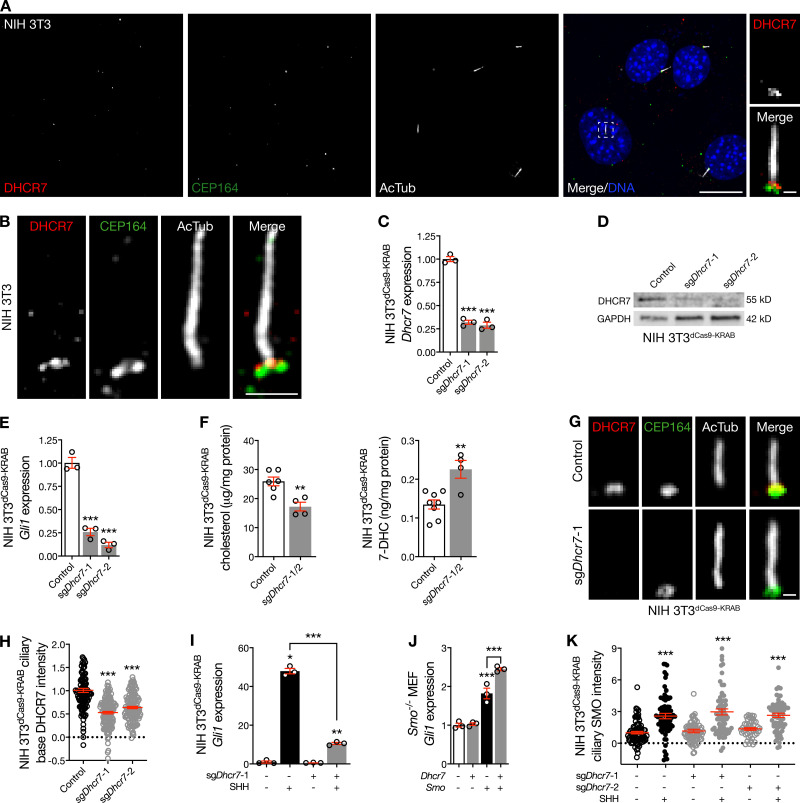
Figure 1.
DHCR7 localizes near the ciliary base and activates the Hedgehog pathway. (A) Immunofluorescence confocal microscopy for DHCR7, the ciliary protein acetylated tubulin (AcTub), and the centriole protein CEP164 demonstrates that DHCR7 localizes near the ciliary base in NIH 3T3 cells. DNA is marked with Hoechst 33342. Main scale bar, 10 µm. Inset scale bar, 1 µm. (B) Superresolution microscopy validates DHCR7 localization near the ciliary base, at the level of CEP164, in NIH 3T3 cells. Scale bar, 1 µm. (C) qRT-PCR assessment of Dhcr7 expression in NIH 3T3 cells expressing the CRISPRi components dCas9-KRAB (NIH 3T3dCas9-KRAB) after transduction of sgRNAs targeting Dhcr7 (sgDhcr7) demonstrates Dhcr7 suppression compared with transduction control (ANOVA). (D) Immunoblot assessment of NIH 3T3dCas9-KRAB lysates after transduction with sgDhcr7 validates DHCR7 suppression compared with transduction control. (E) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells after transduction of sgDhcr7 compared with control demonstrates that Dhcr7 suppression inhibits the Hedgehog transcriptional program (ANOVA). (F) Mass spectrometry–based sterolomics demonstrate reduced expression of cholesterol and increased expression of the 7-DHC in NIH 3T3dCas9-KRAB cells transduced with sgDhcr7 compared with control (Student’s t test). (G and H) Quantitative immunofluorescence confocal microscopy for DHCR7 after transduction of sgDhcr7 in NIH 3T3dCas9-KRAB cells confirms that DHCR7 localizes near the ciliary base and Dhcr7 suppression reduces DHCR7 intensity near the ciliary base (ANOVA). Scale bar, 1 µm. (I) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells transduced with sgDhcr7 reveals Dhcr7 that suppression attenuates the Hedgehog transcriptional program in response to SHH (ANOVA). (J) qRT-PCR assessment of Gli1 expression in Smo−/− MEFs transfected with Dhcr7, Smo, or both demonstrates that Smo is required for Dhcr7 to activate the Hedgehog transcriptional program (ANOVA). (K) Quantitative immunofluorescence confocal microscopy for Smoothened in NIH 3T3dCas9-KRAB cells after sgDhcr7 transduction demonstrates that Dhcr7 suppression fails to block Smoothened accumulation in cilia in response to SHH compared with either control cells treated with vehicle or cells expressing sgDhcr7 treated with vehicle (ANOVA). *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates.
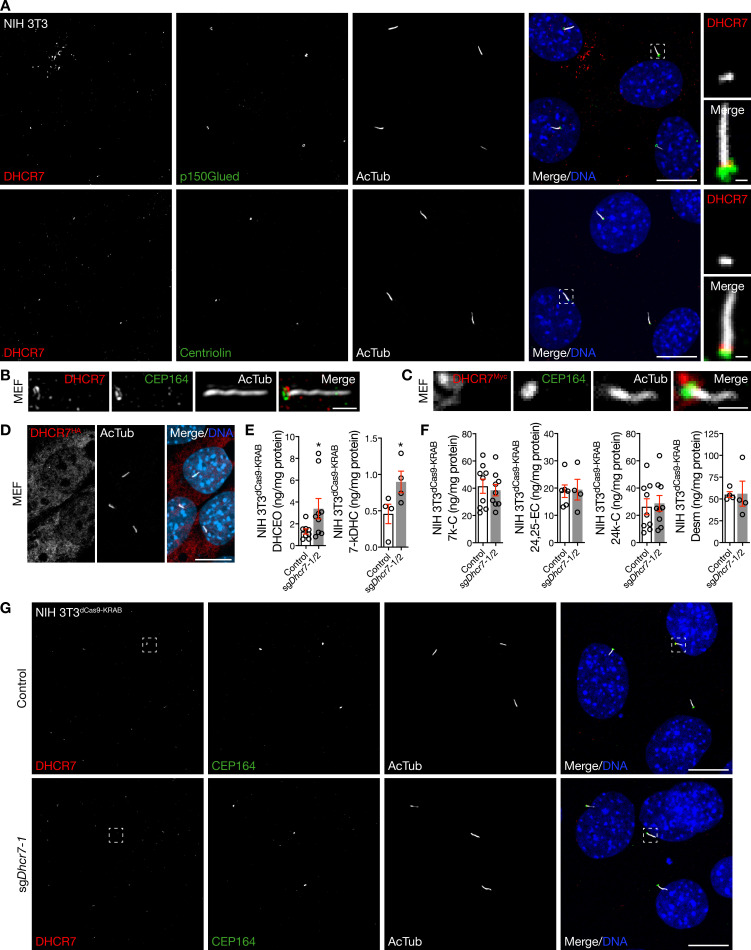
Figure S1.
DHCR7 localizes near the ciliary base. (A) Immunofluorescence confocal microscopy for DHCR7, the ciliary protein acetylated tubulin (AcTub), and the centriole proteins p150Glued or centriolin, demonstrates that DHCR7 localizes near the ciliary base in NIH 3T3 cells. DNA is marked with Hoechst 33342. Main scale bar, 10 µm. Inset scale bar, 1 µm. (B) Superresolution microscopy validates DHCR7 localization near the ciliary base (marked by AcTub) at the level of CEP164 in MEFs. Scale bar, 1 µm. (C) Immunofluorescence confocal microscopy of MEFs transfected with Dhcr7Myc in pCMV6-Entry using Lipofectamine demonstrates that exogenous DHCR7 can localize near the ciliary base, as marked by the centriole protein CEP164. Scale bar, 1 µm. (D) Immunofluorescence confocal microscopy of MEFs transfected with Dhcr7HA in pCDNA3 using FuGENE demonstrates that exogenous DHCR7 can localize away from the ciliary base. DNA is marked with Hoechst 33342. Scale bar, 10 µm. (E and F) Mass spectrometry–based sterolomics demonstrate increased expression of the 7-DHC oxidation by-products DHCEO and 7-keto-7-dehydroxycholesterol (7-kDHC), but no change in expression of Smoothened-activating oxysterols produced by other enzymes, in NIH 3T3dCas9-KRAB cells transduced with sgDhcr7 compared with transduction control (Student’s t test). (G) Immunofluorescence confocal microscopy for DHCR7 after transduction of sgDhcr7 in NIH 3T3dCas9-KRAB cells confirms DHCR7 localization near the ciliary base. DNA is marked with Hoechst 33342. Boxes correspond to Fig. 1 G. Note that speckled immunofluorescent staining away from the ciliary microenvironment does not attenuate with sgDhcr7 transduction, suggestive of nonspecific staining. Scale bar, 10 µm. *, P ≤ 0.05. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates. 24,25-EC, 24,25-epoxycholesterol; 24k-C, 24-keto-cholesterol; Desm, desmosterol.
With these contrasting data in mind, we sought to assess the specificity of DHCR7 antibodies for studying endogenous DHCR7. To do so, we generated NIH 3T3 cells stably expressing the CRISPRi components dCas9-KRAB (NIH 3T3dCas9-KRAB), a system that relies on single-guide RNAs (sgRNAs) to sterically and epigenetically inhibit gene transcription (Gilbert et al., 2013). We transduced NIH 3T3dCas9-KRAB cells with lentiviral particles harboring sgRNAs targeting Dhcr7 (sgDhcr7) and validated Dhcr7 suppression in monoclonal cell lines using quantitative RT-PCR (qRT-PCR) and immunoblots (Fig. 1, C and D). qRT-PCR for the Hedgehog target gene glioma-associated oncogene 1 (Gli1) showed that Dhcr7 suppression inhibited basal Hedgehog signaling compared with control (Fig. 1 E), suggesting that DHCR7 contributes to Hedgehog signal transduction even without pathway activation. Single-molecule imaging studies have identified Smoothened in cilia in the absence of pathway activation and have revealed that Smoothened accumulation in cilia is associated with changing diffusion coefficients suggestive of interactions with proteins or lipids in the ciliary microenvironment (Weiss et al., 2019; Milenkovic et al., 2015). Although the absence of transient activating interactions with the products of DHCR7 activity in cilia may account for the decrease in Gli1 expression we observed after Dhcr7 suppression (Fig. 1 E), it is also possible that DHCR7 may contribute to Hedgehog signal transduction downstream of Smoothened or through noncanonical pathways. Nevertheless, mass spectrometry–based sterolomics demonstrated that Dhcr7 suppression decreased cellular levels of cholesterol, the product of DHCR7 activity that activates Smoothened, and increased levels of 7-dehydroxycholesterol (7-DHC), the substrate of DHCR7 activity (Fig. 1 F). Consistently, the levels of nonenzymatic oxidation products of 7-DHC (i.e., oxysterols such as 3β,5α-dihydroxycholest-7-en-6-one [DHCEO] and 7-keto-7-dehydroxycholesterol [7-kDHC]) were also increased with Dhcr7 suppression compared with control (Fig. S1 E; Xu et al., 2011a,b; 2013). Notably, Dhcr7 suppression did not change the cellular levels of other Smoothened-activating sterol or oxysterol lipids, such as 24,25-epoxycholesterol, 24-keto-cholesterol, or desmosterol (Fig. S1 F). Moreover, immunofluorescence confocal microcopy revealed that Dhcr7 suppression attenuated DHCR7 labeling near the ciliary base in NIH 3T3dCas9-KRAB cells (Fig. 1, G and H; and Fig. S1 G), validating the specificity of our endogenous localization studies.
DHCR7 promotes Hedgehog pathway activity (Blassberg et al., 2016; Kinnebrew et al., 2019), and we found that suppression of Dhcr7 in NIH 3T3dCas9-KRAB cells inhibited activation of the Hedgehog transcriptional program after treatment with recombinant Sonic Hedgehog (SHH; Fig. 1 I). Smoothened is necessary for HSD11β2 to promote the Hedgehog pathway (Raleigh et al., 2018). To shed light on the mechanistic relationship between DHCR7 and Smoothened, we transfected ciliated Smo−/− MEFs with Dhcr7, Smoothened (Smo), or both, and we assessed Hedgehog pathway activity using qRT-PCR for Gli1. Exogenous Smoothened accumulates in cilia even in the absence of pathway stimulation (Raleigh et al., 2018), and, consistently, transfection of Smo into Smo−/− MEFs mildly activated the Hedgehog transcriptional program (Fig. 1 J). Although transfection of Dhcr7 did not activate the Hedgehog transcriptional program in the absence of Smoothened, we found that Gli1 expression after cotransfection of Dhcr7 and Smo was greater than Gli1 expression after transfection of Smo alone (Fig. 1 J). These data suggest that DHCR7 may contribute to Smoothened activation upon Hedgehog pathway stimulation, and they further suggest that DHCR7 does not activate Hedgehog signaling downstream of Smoothened or through noncanonical pathways. To determine if we could localize the impact of DHCR7 on Smoothened activation or Smoothened accumulation in primary cilia, we performed quantitative immunofluorescence confocal microscopy for Smoothened in NIH 3T3dCas9-KRAB cells treated with SHH or vehicle control. Interestingly, we found that Dhcr7 suppression failed to alter Smoothened accumulation in primary cilia with or without Hedgehog pathway stimulation with SHH (Fig. 1 K). These data indicate that DHCR7 contributes to Hedgehog pathway activation but does not regulate Smoothened accumulation in primary cilia.
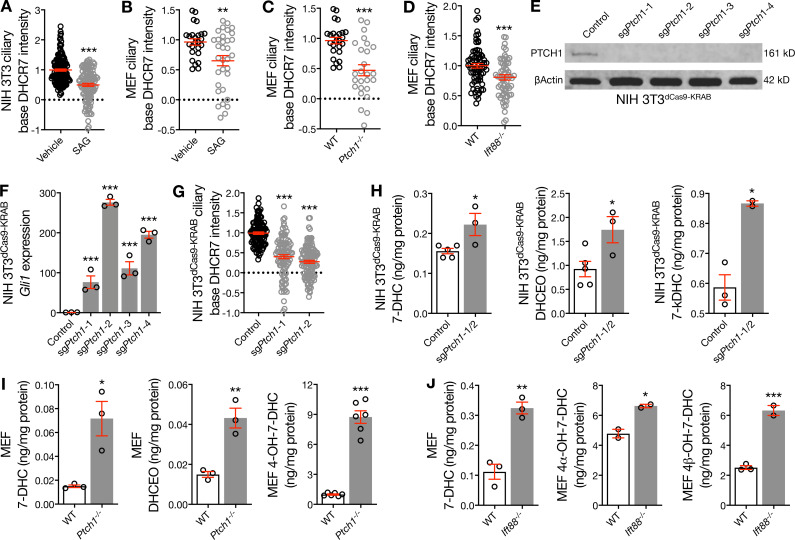
Hedgehog ligand interaction with Patched1 (PTCH1) accumulates Smoothened in cilia and activates the Hedgehog transcriptional program (Corbit et al., 2005). To determine if Hedgehog stimulation alters DHCR7 localization, we treated ciliated cells with Smoothened agonist (SAG) and performed quantitative immunofluorescence confocal microscopy for DHCR7. Surprisingly, pharmacologic stimulation of the Hedgehog pathway removed DHCR7 from the ciliary microenvironment in NIH 3T3 cells and MEFs (Fig. 2, A and B). Ptch1−/− MEFs display constitutive Hedgehog signaling due to Smoothened de-repression, and quantitative immunofluorescence confocal microscopy in wild-type and Ptch1−/− MEFs demonstrated that genetic activation of the Hedgehog pathway also attenuated DHCR7 localization near the ciliary base (Fig. 2 C). Ift88−/− MEFs, which lack cilia and cannot transduce Hedgehog signals, also had less DHCR7 near the centriole than wild-type MEFs (Fig. 2 D). In sum, these data suggest that DHCR7 accumulates near the ciliary base in the absence of Hedgehog signals but is removed upon Hedgehog pathway activation or loss of cilia. To further test this hypothesis, we suppressed Ptch1 in NIH 3T3dCas9-KRAB cells by transducing lentiviruses harboring sgRNAs targeting Ptch1 (sgPtch1). We validated monoclonal cell lines using immunoblots (Fig. 2 E) and performed qRT-PCR to validate that Ptch1 suppression in NIH 3T3dCas9-KRAB cells activated the Hedgehog transcriptional program (Fig. 2 F). Quantitative immunofluorescence confocal microscopy further demonstrated that Ptch1 suppression in NIH 3T3dCas9-KRAB cells removed DHCR7 from the ciliary microenvironment (Fig. 2 G). Importantly, neither pharmacologic nor genetic activation of the Hedgehog pathway altered the overall expression of Dhcr7 transcript or protein (Fig. S2, A–G). Nevertheless, Hedgehog pathway activation in NIH 3T3dCas9-KRAB cells after transduction of sgPtch1 was associated with increased cellular levels of 7-DHC and 7-DHC oxysterols (Fig. 2 H), suggesting that removal of DHCR7 from the ciliary microenvironment was associated with decreased DHCR7 activity. In support of this hypothesis, cellular levels of 7-DHC and 7-DHC oxysterols were also increased in Ptch1−/− MEFs (Fig. 2 I) and Ift88−/− MEFs (Fig. 2 J) compared with wild-type MEFs. Moreover, cellular levels of cholesterol were not decreased in NIH 3T3dCas9-KRAB cells after transduction of sgPtch1 or in Ptch1−/− or Ift88−/− MEFs compared with respective their controls (Fig. S2 H). In sum, these data suggest that increases in the activity of DHCR24, oxidosqualene cyclase, or other enzymes participating in the cholesterol shunt pathway may compensate for changes in DHCR7 activity in some contexts (Russell, 2003).
Figure 2.
Hedgehog pathway activation removes DHCR7 from the ciliary microenvironment and attenuates DHCR7 activity. (A–C) Quantitative immunofluorescence confocal microscopy for DHCR7 in NIH 3T3 cells, WT MEFs, and Ptch1−/− MEFs shows that pharmacologic stimulation and genetic de-repression of the Hedgehog pathway remove DHCR7 from the ciliary microenvironment (Student’s t test). Dotted lines denote average background intensity. (D) Quantitative immunofluorescence confocal microscopy for DHCR7 in WT and Ift88−/− MEFs shows that genetic deletion of cilia removes DHCR7 from near the centriole (Student’s t test). (E) Immunoblot assessment of NIH 3T3dCas9-KRAB lysates after transduction with sgPtch1 validates PTCH1 suppression compared with transduction control. (F) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells demonstrates activation of the Hedgehog transcriptional program after transduction of sgPtch1 compared with control (ANOVA). (G) Quantitative immunofluorescence confocal microscopy for DHCR7 in NIH 3T3dCas9-KRAB cells shows that Ptch1 suppression leading to Hedgehog pathway activation removes DHCR7 from the ciliary microenvironment (ANOVA). (H) Mass spectrometry–based sterolomics of NIH 3T3dCas9-KRAB cells show that transduction of sgPtch1 inhibits the activity of DHCR7 compared with transduction control (Student’s t test). (I) Mass spectrometry–based sterolomics of WT and Ptch1−/− MEFs show that genetic de-repression of the Hedgehog pathway inhibits DHCR7 activity (Student’s t test). (J) Mass spectrometry–based sterolomics of WT and Ift88−/− MEFs show that genetic deletion of cilia inhibits DHCR7 activity (Student’s t test). *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates. 7-kDHC, 7-keto-7-dehydroxycholesterol; 4-OH-7-DHC, 4-hydroxy-7-dehydroxycholesterol.
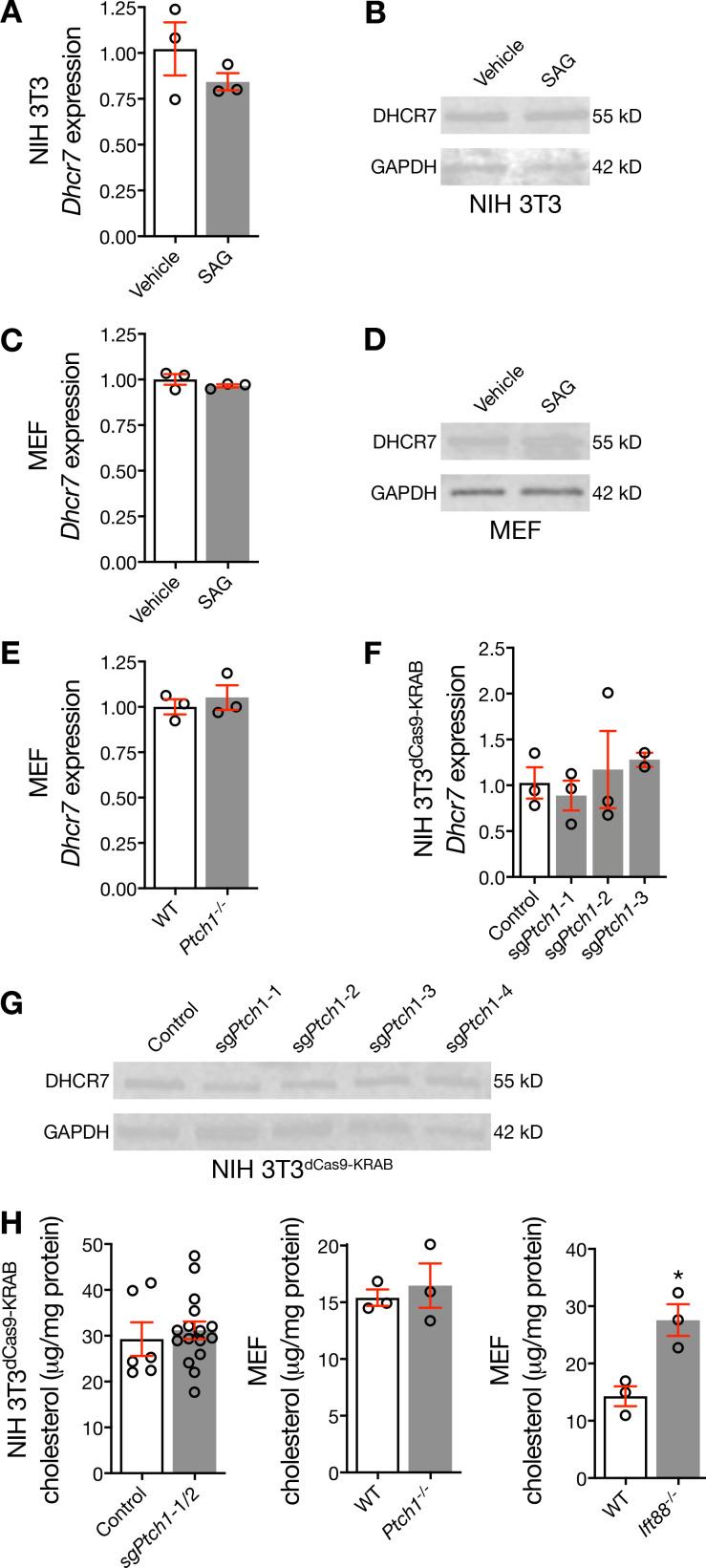
Figure S2.
Hedgehog pathway activation does not influence DHCR7 expression. (A and B) qRT-PCR and immunoblot assessment of NIH 3T3 cells demonstrates that pharmacologic activation of the Hedgehog pathway fails to alter expression of Dhcr7 or DHCR7. (C and D) qRT-PCR and immunoblot assessment of MEFs demonstrates that pharmacologic activation of the Hedgehog pathway fails to alter expression of Dhcr7 or DHCR7. (E) qRT-PCR assessment of WT and Ptch1−/− MEFs demonstrates that genetic de-repression of the Hedgehog pathway fails to alter expression of Dhcr7. (F and G) qRT-PCR and immunoblot assessment of NIH 3T3dCas9-KRAB cells transduced with sgPtch1 validates that genetic de-repression of the Hedgehog pathway fails to alter expression of Dhcr7 or DHCR7. (H) Mass spectrometry–based sterolomics of NIH 3T3dCas9-KRAB cells and MEFs show that genetic inhibition of Ptch1 does not alter levels of cellular cholesterol, but genetic deletion of Ift88 increases levels of cellular cholesterol (Student’s t test). *, P ≤ 0.05. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates.
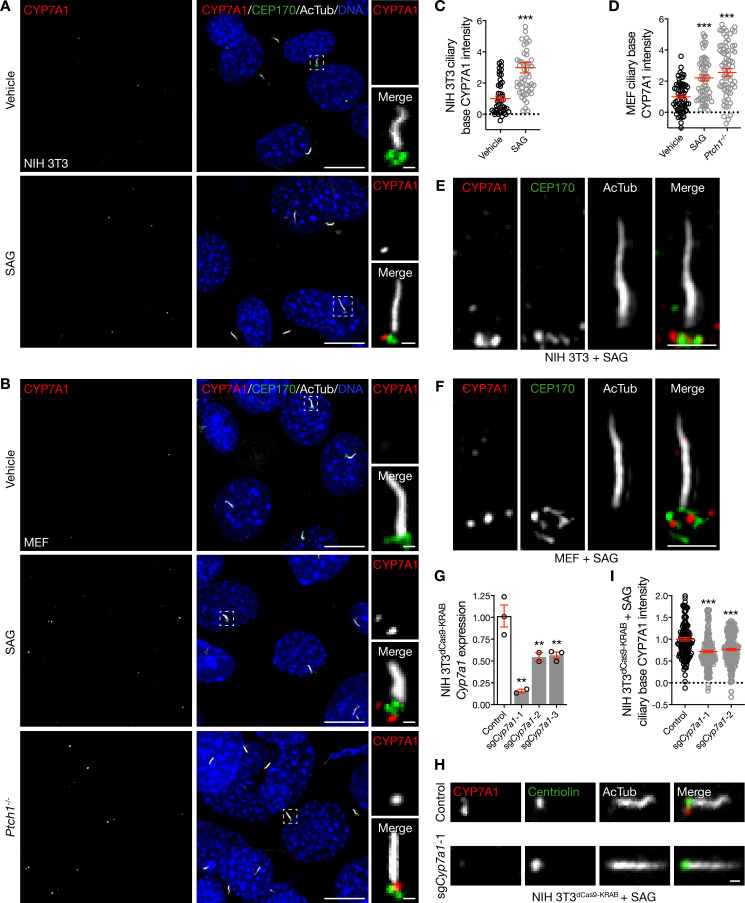
Our results demonstrate that DHCR7 localizes near the ciliary base and promotes Hedgehog signal transduction but is negatively regulated by pathway activation. These data perhaps explain why conflicting studies have variably defined DHCR7 as both a positive and a negative regulator of the Hedgehog pathway (Koide et al., 2006; Blassberg et al., 2016; Kinnebrew et al., 2019). Alternatively, it is possible that aspects of DHCR7 activity on a shorter time scale than we were able to resolve in fibroblasts (as has been observed for INPP5E) may account for the different functions ascribed to DHCR7 with respect to Hedgehog signal transduction. Nevertheless, our data raise the possibility that DHCR7 contributes to cholesterol enrichment in primary cilia (Raleigh et al., 2018). Like ciliary cholesterol, ciliary oxysterol regulation is incompletely understood. HSD11β2 produces 7-keto-cholesterol (7k-C), which is converted into oxysterols that bind and activate Smoothened by CYP27A1 (Dwyer et al., 2007; Raleigh et al., 2018; Myers et al., 2017). Because HSD11β2 does not localize to primary cilia (Odermatt et al., 2001), we hypothesized that CYP7A1, which also generates 7k-C, may activate the Hedgehog pathway from the ciliary microenvironment. To test this hypothesis, we performed immunofluorescence confocal microscopy of unstimulated NIH 3T3 cells and MEFs, but we did not detect CYP7A1 near the ciliary base (Fig. 3, A and B). However, activation of the Hedgehog pathway with SAG accumulated CYP7A1 near the ciliary base in both cell lines (Fig. 3, A–D). Moreover, superresolution microscopy confirmed that CYP7A1 accumulated near the ciliary base after pharmacologic stimulation of the Hedgehog pathway (Fig. 3, E and F).
Figure 3.
Hedgehog pathway activation accumulates CYP7A1 near the ciliary base. (A–D) Quantitative immunofluorescence confocal microscopy for CYP7A1 in relation to the ciliary protein acetylated tubulin (AcTub) and the centriole protein CEP170 demonstrates that CYP7A1 localizes near the ciliary base in NIH 3T3 cells and MEFs after pharmacologic stimulation or genetic de-repression of the Hedgehog pathway. DNA is marked with Hoechst 33342. Main scale bar, 10 µm. Inset scale bar, 1 µm. Student’s t test in C; ANOVA in D. Dotted lines denote average background intensity. (E and F) Superresolution microscopy validates CYP7A1 localization near the ciliary base at the level of CEP170 in NIH 3T3 cells and MEFs. Scale bars, 1 µm. (G) qRT-PCR assessment of Cyp7a1 expression in NIH 3T3dCas9-KRAB cells after transduction of sgCyp7a1 demonstrates Cyp7a1 suppression compared with transduction control (ANOVA). (H and I) Quantitative immunofluorescence confocal microscopy for CYP7A1 after transduction of sgCyp7a1 in NIH 3T3dCas9-KRAB cells and pharmacologic activation of the Hedgehog pathway confirms that CYP7A1 localizes near the ciliary base (ANOVA). Scale bar, 1 µm. **, P ≤ 0.01; and ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates.
Similar to DHCR7, exogenous CYP7A1 with a Myc tag could be found near the ciliary base in Ptch1−/− MEFs (Fig. S3 A), but exogenous CYP7A1 with an HA tag that was overexpressed using a different transfection reagent primarily localized to the cytoplasm and endoplasmic reticulum in wild-type MEFs, even after activation of the Hedgehog pathway with SAG (Fig. S3 B). Thus, to assess the specificity of CYP7A1 antibodies for studying endogenous CYP7A1, we transduced NIH 3T3dCas9-KRAB cells with sgCyp7a1 and validated suppression of Cyp7a1 in monoclonal cell lines using qRT-PCR (Fig. 3 G). After Hedgehog pathway stimulation with SAG, we performed quantitative immunofluorescence confocal microscopy to discover that Cyp7a1 suppression attenuated labeling near the ciliary base compared with control (Fig. 3, H and I). Importantly, pharmacologic activation of the Hedgehog pathway did not alter the expression of Cyp7a1 (Fig. S3 C), suggesting that changes in CYP7A1 accumulation in the ciliary microenvironment were the product of coordinated signaling events. In sum, these data demonstrate that CYP7A1 accumulates near the ciliary base upon Hedgehog pathway activation.
Figure S3.
CYP7A1 localizes near the ciliary base. (A) Immunofluorescence confocal microscopy of Ptch1−/− MEFs transfected with Cyp7a1Myc in pCMV6-Entry using Lipofectamine demonstrates that exogenous CYP7A1 can localize near the ciliary base, as marked by the centriole protein CEP164, after genetic Hedgehog pathway activation. Scale bar, 1 µm. (B) Immunofluorescence confocal microscopy of MEFs treated with vehicle control or SAG, and transfected with Cyp7a1HA in pCDNA3 using FuGENE, demonstrates that exogenous CYP7a1 can localize away from the ciliary microenvironment. DNA is marked with Hoechst 33342. Scale bar, 10 µm. (C) qRT-PCR assessment of NIH 3T3 cells and MEFs reveals that pharmacologic activation of the Hedgehog pathway fails to alter expression of Cyp7a1. (D) Mass spectrometry–based sterolomics of NIH 3T3dCas9-KRAB cells show that transduction of sgCyp7a1 does not alter cellular levels of Smoothened-activating oxysterols that are produced by other enzymes. (E) qRT-PCR assessment of NIH 3T3 cells demonstrates that overexpression of Dhcr7HA or Cyp7a1HA activates the Hedgehog transcriptional program (ANOVA). *, P ≤ 0.05; ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates. 24,25-EC, 24,25-epoxycholesterol; 24k-C, 24-keto-cholesterol.
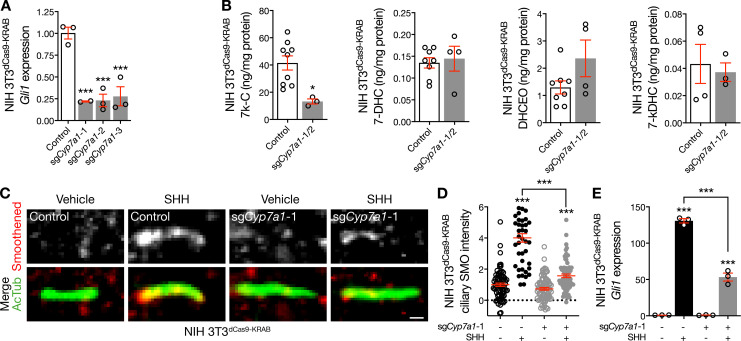
To determine if CYP7A1 participates in Hedgehog signal transduction, we assessed Gli1 expression using qRT-PCR in monoclonal NIH 3T3dCas9-KRAB cells transduced with sgCyp7a1. Cyp7a1 suppression in unstimulated NIH 3T3dCas9-KRAB cells inhibited the Hedgehog transcriptional program (Fig. 4 A), likely resulting from a lack of oxysterols diffusing to the ciliary microenvironment from CYP7A1 activity elsewhere in the cell (Russell, 2003). Furthermore, Cyp7a1 suppression reduced cellular levels of 7k-C but did not influence levels of 7-DHC, 7-DHC oxysterols, or other Smoothened-activating oxysterols (Fig. 4 B and Fig. S3 D). Consistent with suppression of Dhcr7 (Fig. 1 K), quantitative immunofluorescence confocal microscopy revealed that Cyp7a1 suppression failed to block Smoothened accumulation in NIH 3T3dCas9-KRAB cilia in response to Hedgehog pathway activation with SHH (Fig. 4, C and D). However, in contrast to suppression of Dhcr7, Cyp7a1 suppression attenuated Smoothened accumulation relative to SHH treatment of control cells. Cyp7a1 suppression in NIH 3T3dCas9-KRAB cells also attenuated the Hedgehog transcriptional program in response to SHH (Fig. 4 E). Moreover, overexpression of either Cyp7a1 or Dhcr7 activated the Hedgehog transcriptional program even without pathway activation (Fig. S3 E), suggesting that enzymatic activity in the ciliary microenvironment may promote transient interactions in cilia that activate Hedgehog signaling (Weiss et al., 2019; Milenkovic et al., 2015). In sum, these data demonstrate CYP7A1 near the ciliary base promotes Hedgehog signaling.
Figure 4.
CYP7A1 activates the Hedgehog pathway. (A) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells after transduction of sgCyp7a1 compared with control demonstrates inhibition of the Hedgehog transcriptional program after Cyp7a1 suppression (ANOVA). (B) Mass spectrometry–based sterolomics reveal reduced expression of 7k-C and no change in 7-DHC or 7-DHC oxidation by-products in NIH 3T3dCas9-KRAB cells transduced with sgCyp7a1 compared with control (Student’s t test). (C and D) Quantitative immunofluorescence confocal microscopy for Smoothened in relation to the ciliary protein acetylated tubulin (AcTub) in NIH 3T3dCas9-KRAB cells after transduction of sgCyp7a1 demonstrates that Cyp7a1 suppression fails to block Smoothened accumulation in cilia in response to SHH compared with either control cells treated with vehicle or cells expressing sgCyp7a1 treated with vehicle (ANOVA, Student’s t test). Scale bar, 1 µm. Dotted lines denote average background intensity. (E) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells after transduction of sgCyp7a1 reveals that Cyp7a1 repression attenuates the Hedgehog transcriptional program in response to SHH (ANOVA). *, P ≤ 0.05; ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates.
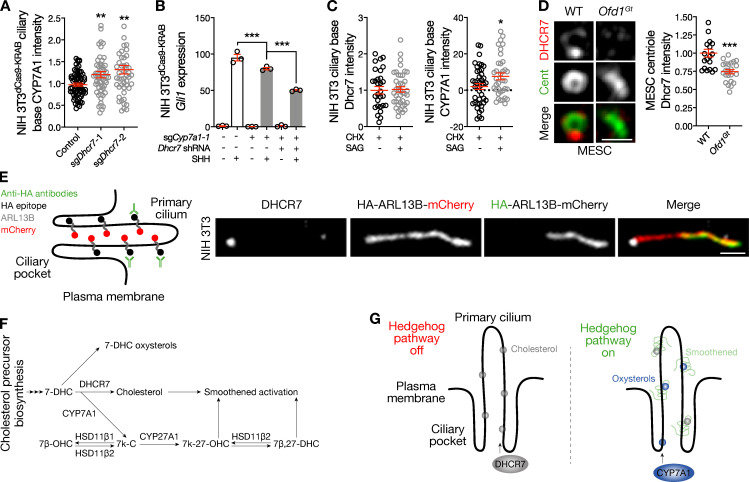
To investigate the functional relationship between Cyp7a1 and Dhcr7, we performed quantitative immunofluorescence confocal microscopy for CYP7A1 in NIH 3T3dCas9-KRAB cells transduced with sgDhcr7, and we found that Dhcr7 suppression, which activated the Hedgehog pathway (Fig. 1 E), promoted accumulation of CYP7A1 near the ciliary base (Fig. 5 A). These data raise the possibility that CYP7A1 may compensate for decreased DHCR7 activity upon Hedgehog pathway activation by maintaining Smoothened-activating lipid levels in the ciliary microenvironment. In support of this hypothesis, we found that concurrent suppression of Cyp7a1 and Dhcr7 in NIH 3T3dCas9-KRAB cells inhibited SHH activation of the Hedgehog transcriptional program more than Cyp7a1 suppression alone (Fig. 5 B).
Figure 5.
DHCR7 and CYP7A1 cooperate to activate the Hedgehog pathway near the ciliary base. (A) Quantitative immunofluorescence confocal microscopy for CYP7A1 in NIH 3T3dCas9-KRAB cells shows that Dhcr7 suppression leading to Hedgehog pathway activation accumulates CYP7A1 near the ciliary base (ANOVA). (B) qRT-PCR assessment of Gli1 expression in NIH 3T3dCas9-KRAB cells transduced with sgCyp7a1 and shRNAs targeting Dhcr7 reveals that concurrent suppression of Cyp7a1 and Dhcr7 attenuates the Hedgehog transcriptional program greater than suppression of either enzyme alone in response to SHH (ANOVA). (C) Quantitative immunofluorescence confocal microscopy for DHCR7 and CYP7A1 in NIH 3T3 cells treated the translation inhibitor CHX cells shows that protein synthesis is required to remove DHCR7 from the ciliary base, but is not required to accumulate CYP7A1 at the ciliary base, in response to Hedgehog pathway activation with SAG (Student’s t test). (D) Quantitative immunofluorescence confocal microscopy for DHCR7 in WT and Ofd1Gt mouse embryonic stem cells (MESCs) reveals disruption of centriole structure, as marked by centriolin (Cent), reduces DHCR7 localization near the ciliary base. Scale bar, 1 µm (Student’s t test). (E) Superresolution microscopy and the IN/OUT assay using NIH 3T3 cells stably expressing ARL13B with extracellular HA and intracellular mCherry tags validates DHCR7 localization near the ciliary base. Scale bar, 1 µm. (F) Network of Smoothened-activating sterol and oxysterol biosynthesis constructed from the data in this paper and published literature (Raleigh et al., 2018). (G) Model of lipid enrichment in the ciliary microenvironment and regulation of lipids in response to Hedgehog pathway activity. *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Error bars represent SEM. The sample size of each experiment is represented by the number of independent data points on each graph. Each experiment is representative of at least three independent biological replicates.
The Hedgehog pathway regulates development through gene expression programs and signaling events that are independent of gene expression (Briscoe and Thérond, 2013). Moreover, some functions of the Hedgehog pathway, such as cell cycle progression, involve expression of indirect target genes requiring translation of intermediate transcription factors (Raleigh et al., 2017). Thus, to shed light on the biochemical mechanisms regulating DHCR7 and CYP7A1 accumulation near the ciliary base, we inhibited translation in NIH 3T3 cells with cycloheximide (CHX) and performed quantitative immunofluorescence confocal microscopy after pathway stimulation with SAG. We found that CHX blocked DHCR7 removal from the ciliary microenvironment, but it did not block CYP7A1 accumulation near the ciliary base in response to pathway activation (Fig. 5 C). These data suggest that independent mechanisms downstream of Hedgehog pathway activation regulate the subcellular localization of these enzymes.
To shed light on how centriole structure at the ciliary base contributes to sterol synthase accumulation in the ciliary microenvironment, we performed confocal microscopy of mouse embryonic stem cells with a gene trap mutation in the Ofd1 locus (Ofd1Gt) that disrupts centriole length and distal structure (Singla et al., 2010). We found that neither wild-type nor Ofd1Gt mouse embryonic stem cells expressed CYP7A1 and that DHCR7 accumulation near the centriole in Ofd1Gt cells was reduced compared with wild-type mouse embryonic stem cells (Fig. 5 D). Thus, like ciliary structure (Fig. 2 D), centriole structure appears to be important for sterol synthase localization near the ciliary base.
DHCR7 is a nine-pass transmembrane protein (Jira et al., 2003) and is likely embedded in a lipid bilayer at the ciliary base. Thus, to determine if DHCR7 localized to the ciliary pocket, we used the IN/OUT assay, which relies on NIH 3T3 cells stably expressing a transmembrane ADP-ribosylation factor–like protein 13B (ARL13B) construct constitutively localizing to cilia and tagged with an extracellular HA epitope and an intracellular mCherry fluorophore (Kukic et al., 2016). Staining for the HA epitope in the absence of cell permeabilization labels the ciliary body projecting from the cell surface. In contrast, mCherry fluorescence in this system projects both above and below the reflection of the plasma membrane that forms the ciliary pocket (Fig. 5 E). When subsequently permeabilized and stained for DHCR7, the IN/OUT assay validated our confocal and superresolution microscopy results showing that DHCR7 localized near the ciliary base (Fig. 5 E) and not along the length of the ciliary pocket. These data suggest that DHCR7 may localize to endosomes near the ciliary base or perhaps at the nadir of the ciliary pocket, either of which may also be true for CYP7A1, a predicted single-pass transmembrane protein.
In conclusion, our results place DHCR7 and CYP7A1 in the network of sterol and oxysterol synthases that activate the Hedgehog pathway (Fig. 5 F). These data are consistent with a model of lipid enrichment and regulation in cilia where DHCR7 produces cholesterol in the absence of Hedgehog signals to prime the ciliary microenvironment for Hedgehog pathway activation and CYP7A1 contributes oxysterols to cilia to modulate Hedgehog signaling when the pathway is on (Fig. 5 G). According to this model, DHCR7 may provide an initial pool of cholesterol near the ciliary base to activate Smoothened, and, once activated, Hedgehog signal transduction may be refined by exchanging DHCR7 for CYP7A1 (and cholesterol for oxysterols) in the ciliary microenvironment. Consistently, we have shown that primary cilia are enriched in cholesterol even in the absence of Hedgehog pathway activation (Raleigh et al., 2018). It remains to be established why DHCR7 is removed from the ciliary microenvironment upon pathway activation, but it is notable that nonenzymatic oxidation products of 7-DHC, such as DHCEO, antagonize Smoothened (Sever et al., 2016). Thus, it is possible that accumulation of DHCEO near the ciliary base (in the absence of DHCR7) opposes the activity of CYP7A1 and serves as a negative feedback mechanism to restrain the intensity of Hedgehog signaling after the pathway is activated. Indeed, current paradigms of Hedgehog signaling and lipid biosynthesis are reminiscent of negative feedback loops observed elsewhere in the Hedgehog pathway, such as induction of the pathway inhibitor Ptch1 in response to activation of the Hedgehog transcriptional program.
The contributions that DHCR7 and CYP71 make to the Hedgehog pathway suggest that both cholesterol and oxysterols can function as endogenous Smoothened agonists. Dissecting the potentially redundant functions of these lipids may be challenging due to tissue- or organism-specific expression of enzymes participating in the production of Smoothened-activating lipids. Indeed, human SLOS patients display multiple congenital malformations, but developmental phenotypes in mice lacking Dhcr7 are subtle, and mice lacking Cyp7a1 do not have overt Hedgehog phenotypes (Pullinger et al., 2002). Furthermore, we found that combined suppression of Dhcr7 and Cyp7a1 fails to completely inhibit Hedgehog signal transduction (Fig. 5 B), suggesting that other mechanisms or other enzymes must contribute to Hedgehog pathway activation.
The list of enzymes participating in the production of Smoothened-activating lipids is growing (Kinnebrew et al., 2019), but our ability to understand mechanistic relationships between Hedgehog signaling and ciliary lipids depends on the emergence of new technologies to isolate primary cilia for mass spectrometry–based sterolomics. Indeed, an important limitation of our study is that our sterolomics were performed on whole cells. The genetic perturbations necessary to repeat these experiments in model systems where primary cilia can be isolated for biochemistry, such as Stongylocentrotus purpuratus, Salpingoeca rosetta, Nematostella vectensis, Chlamydomonas reinhardtii, or Sus scrofa cells, are currently not possible. Thus, significant technical advances are necessary to improve our understanding of Hedgehog signaling and ciliary lipids. Chief among these is likely the improvement of cellular probes for localizing cellular cholesterol (Courtney et al., 2018); these probes are not as refined as reagents for studying phosphoinositides in primary cilia (Balla, 2013; Chávez et al., 2015; Garcia-Gonzalo et al., 2015).
Materials and methods
Cell culture, molecular biology, and overexpression
NIH 3T3 (CRL-1658; American Type Culture Collection), 293T (CRL-3216; American Type Culture Collection), and MEF cells were cultured in DMEM (10313021; Life Technologies) supplemented with 10% FBS (Life Technologies) and GlutaMAX (Life Technologies). Mouse embryonic stem cells were cultured on 0.1% gelatin in KnockOut DMEM (10929-018; Life Technologies) supplemented with 15% FBS, nonessential amino acids (Life Technologies), GlutaMAX, 2-mercaptoethanol (BP176-100; Thermo Fisher Scientific), the mitogen-activated protein kinase kinase inhibitor PD0325901 (S1036; Selleck Chemicals), and the glycogen synthase kinase 3 inhibitor CHIR99021 (S2924; Selleck Chemicals).
For ciliation and Hedgehog signaling assays, NIH 3T3 and MEF cultures were transitioned to OptiMEM (31985062; Thermo Fisher Scientific) and treated with recombinant Sonic Hedgehog 1 µg/ml (1845; R&D Systems), Smoothened agonist 100 nM (566660; Calbiochem), or vehicle control, with or without CHX 25 µg/ml (C1988; Sigma-Aldrich) for 24 h. Of note, this dose, concentration, and duration of CHX treatment are effective for blocking expression of indirect Hedgehog target genes in NIH 3T3 cells (Raleigh et al., 2017).
For overexpression experiments, Myc-tagged Dhcr7 (RC200480) and Cyp7a1 (RC211019L1) in pCMV6-Entry were purchased from OriGene and transfected using Lipofectamine (A12621; Thermo Fisher Scientific) according to the manufacturer’s instructions. HA-tagged Dhcr7 and Cyp7a1 coding DNA sequences were synthesized by Integrated DNA Technologies, cloned into pcDNA3 (Addgene), and transfected using FuGENE HD reagent (E2311; Promega) according to the manufacturer’s instructions. For all constructs, cells were ciliated 48 h after transfection as described above.
CRISPR targeting and cloning
To suppress gene expression, CRISPRi was used to target dCas9-KRAB to the promoter regions of target genes to sterically repress transcription initiation. Protospacer sequences were chosen from a previously generated genome-wide library (Horlbeck et al., 2016) and were cloned into either pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro (71236; Addgene) or pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-GFP (71237; Addgene) lentiviral plasmids following a BsmBI (R0580; New England Biolabs) restriction digest. Notably, Dhcr7 was found to have two transcriptional start sites (TSSs) >1 kb apart. Thus, this gene was targeted for knockdown by transduction of two protospacer sequences to target both TSSs (i.e., Dhcr7-A1 + Dhcr7-A2). Cyp7a1, Dhcr7-A, and Ptch1 protospacers were cloned into the pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro, and Dhcr7-B protospacers were cloned into pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-GFP.
The specific TSSs targeted are Cyp7a1 (chromosome 4, strand −, primary TSS 5′-6275629, primary TSS 3′-6275634, secondary TSS 5′-6275629, secondary TSS 3′-6275634), Dhcr7-A (chromosome 7, strand +, primary TSS 5′-143830204, primary TSS 3′-143830249, secondary TSS 5′-143830204, secondary TSS 3′-143830249), Dhcr7-B (chromosome 7, strand +, primary TSS 5′-143823057, primary TSS 3′-143823113, secondary TSS 5′-143823057, secondary TSS 3′-143823113), and Ptch1 (chromosome 13, strand −, primary TSS 5′-63573400, primary TSS 3′-63573430, secondary TSS 5′-63573400, secondary TSS 3′-63573430).
The specific protospacer sequences for each gene of interest, and their genomic position relative to TSSs, are Cyp7a1-1 (5′-GAAGCAGGAAAACGTCCCAA-3′, TSS+37), Cyp7a1-2 (5′-GCAAGACGGTCCAGAAACTG-3′, TSS−7), Cyp7a1-3 (5′-GTAAGCACAGATTCTCCCCT-3′, TSS+19), Dhcr7-1 (A: 5′-GCCTGAGTGCGGAATCCAGG-3′, TSS+44; B: 5′-GTCCAGGACGCTCTGACCTT-3′, TSS+59), Dhcr7-2 (A: 5′-GTCACCTGAGTGCGGAATCC-3′, TSS+47; B: 5′-CTCTGACCTTGGGCAACAG-3′, TSS+50), Ptch1-1 (5′-GGCACTCCGCCGAAAGCCTG-3′, TSS+23), Ptch1-2 (5′-GGGCCGCCGCAGGCTTTCGG-3′, TSS+31), and Ptch1-3 (5′-GACGGAAAGTGTAAAAACCC-3′, TSS+60).
Lentivirus production and transduction
pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro or pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-GFP was cotransfected into 293T cells with lentiviral packaging plasmids psPAX2 and pMD2.G (12260 and 12259; Addgene). Briefly, 3.5 × 106 293T cells were seeded in poly-D-lysine–coated (P6407; Sigma-Aldrich) 10-cm plates. For each 10-cm dish, the following DNA was diluted into 600 µl of OptiMEM: 4.5 µg of construct, 6 µg of psPAX2, and 1.5 µg of pMD2.G. Separately, 36 µl of TransIT-Lenti transfection reagent (6603; Mirus Bio) was diluted into 600 µl of OptiMEM, vortexed, and incubated for 5 min. The DNA and Mirus Bio solutions were combined and allowed to incubate at room temperature for 20 min. Meanwhile, the cell culture media on 293T cells was replaced with 9 ml of prewarmed DMEM supplemented with 1% FBS and GlutaMAX. The transfection mixture was added dropwise onto cells. Viral particles were harvested 48 h following transfection by passing the cell culture medium through a 0.45-µm polyvinylidene difluoride syringe filter. For transduction, viral supernatant was added directly onto cells in complete growth media supplemented with polybrene (TR-1003-G; EMD Millipore) to a final concentration of 10 µg/ml.
For CRISPR targeting, lentiviral particles for three sgRNAs targeting either Cyp7a1 or Ptch1 and six sgRNAs targeting Dhcr7 were produced, and cells were transduced. Cells were then selected with 2.5 µg/ml of puromycin and/or sorted for GFP fluorescence. Because Dhcr7 had two putative promoters targeted, cells were sequentially transduced with “A” protospacers cloned into pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro, followed by “B” protospacers cloned into pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-GFP, resulting in three cell lines expressing two protospacers each. For each sgRNA, single cells were seeded into 96-well plates, and the resulting clones were tested for successful knockdown of the targeted gene by immunofluorescence, immunoblotting, and/or qRT-PCR before experiments were carried out.
For shRNAs targeting Dhcr7, we used 5′-GCCATGACCACTTTGGGTG-3′ (Dharmacon SMARTvector lentiviral shRNA) cloned into pSMART mCMV/TurboGFP (V3SM11241-230979455; GE Healthcare Life Sciences). Lentiviral particles were produced, and cells were transduced following the protocol described above.
Immunoblotting
Immunoblot analysis was used to detect the expression of proteins. Proteins were separated on precast SDS-polyacrylamide 4–20% gels (456–2093; Bio-Rad Laboratories), transferred to nitrocellulose membrane, blocked in Odyssey blocking buffer (927-50000; LI-COR Biosciences) for 30 min at room temperature, and labeled with PTCH1 (ab53715, rabbit; Abcam), DHCR7 (ab103296, rabbit; Abcam), CYP7A1 (ab65596, rabbit; Abcam), β-actin (20536-1-AP, rabbit; Proteintech), or GAPDH (MA5-15738, mouse; Invitrogen) primary antibodies at 4°C overnight. Secondary antibodies of donkey antimouse IgG polyclonal antibody (925-32212; LI-COR Biosciences) and goat antirabbit IgG polyclonal antibody (925-68071; LI-COR Biosciences) were used for 1 h at room temperature. Proteins of interest were detected using a LI-COR Odyssey imaging system (model 2800).
Immunofluorescence
Immunofluorescence for NIH 3T3 and MEF cilia was performed on glass coverslips that were coated with gelatin 1% for MEFs. Cells were fixed in 4% PFA, blocked in 2.5% BSA (Sigma-Aldrich) and 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 30 min at room temperature, and they were labeled with Smoothened (20787-1-AP, mouse; Proteintech), DHCR7 (PA5-48204, rabbit; Invitrogen), CYP7A1 (ab65596, rabbit; Abcam), p150 Glued (610473, mouse; BD Biosciences), centrosomal protein 164 (Cep164; sc-240226, goat; Santa Cruz Biotechnology), Cep170 (72-413-1, mouse; Invitrogen), centriolin (sc-365521, mouse; Santa Cruz Biotechnology), or acetylated tubulin (see below) antibodies at room temperature for 3 h. Cells were subsequently labeled with donkey antimouse linked with Alexa Fluor 488 (A21202; Invitrogen), donkey antirabbit linked with Alexa Fluor 568 (A10042; Invitrogen), or donkey antigoat linked with Alexa Fluor 647 (A32849; Invitrogen) secondary antibodies; Hoechst 33342 to mark DNA (H3570; Life Technologies); and acetylated tubulin Alexa Fluor 647 conjugate (sc-23950, mouse; Santa Cruz Biotechnology) to mark cilia, all for 1 h at room temperature. Cells were mounted in ProLong Diamond Antifade Mountant (Thermo Fisher Scientific).
The IN/OUT assay was performed in NIH 3T3 cells stably expressing HA-CD8-Arl13b-mCherry with a modified protocol (Kukic et al., 2016). In brief, cells were grown to confluency on coverslips, fixed with 4% PFA in PBS, and stained for HA (ab9110, rabbit; Abcam) in the absence of detergent followed by a second fixation and permeability for DHCR7 staining as described above in the presence of detergent. Of note, the IN/OUT assay was attempted for CYP7A1 but was not successful due to antibody incompatibility with the sequential fixation/permeabilization protocol necessitated by this assay.
Quantitation of sterols and oxysterols
Sterol and oxysterol quantitation was performed by ultrahigh-performance liquid chromatography–tandem mass spectrometry using stable isotope–labeled internal standards (d7-cholesterol, d7-7-dehydrocholesterol, 13C3-desmosterol, and 13C3-lanosterol for sterols and d7-7-ketocholesterol for oxysterols) as described previously (Herron et al., 2018; Raleigh et al., 2018). Each sterol or oxysterol level was normalized to the corresponding protein weight of the sample. In some instances, multiple monoclonal gene edited cell lines were combined for sterolomics, as indicated in the figure legends.
Microscope image acquisition and analysis
Fluorescence microscopy was performed on a Zeiss LSM 800 confocal laser-scanning microscope with a Plan Apo, 63×, NA 1.4, oil-immersion objective using a Zeiss Axiocam 503 mono camera. Fluorochromes are indicated alongside secondary antibodies. Images were acquired at room temperature using Zen software and were processed and quantified from five distinct regions per coverslip using ImageJ. Fluorescence intensity was quantified by subtracting background intensity immediately adjacent to the region of interest and normalizing the resulting intensity to the average intensity within the control condition for each experiment. In rare instances, background intensity immediately adjacent to the region of interest was higher than intensity in the region of interest, especially for conditions where DHCR7 or CYP7A1 was absent from the ciliary base. In these cells, image quantification yielded negative normalized intensities, which are interpreted as within the margin of error for fluorescence intensity quantification. For presentation, image intensity was adjusted to suppress background staining from nonspecific interactions that were identified for each experiment using secondary only controls. For superresolution microscopy, including the IN/OUT assay, 3D structured illumination data were collected on a DeltaVision OMX SR microscope equipped with a Plan Apo N, 1.42 NA, oil-immersion 60× objective using a Zeiss Axiocam 503 mono camera. Images were acquired at room temperature, with fluorochromes indicated alongside secondary antibodies, and were processed using ImageJ.
qRT-PCR
RNA was isolated from cells using the RNeasy Mini Kit and a QIAcube (QIAGEN), and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories) and a ProFlex thermocycler (Thermo Fisher Scientific). Target genes were amplified using PowerUp SYBR Green Master Mix and a QuantStudio 6 thermocycler (Thermo Fisher Scientific). For each experiment, expression of target genes was compared with Gapdh expression using the 2−ΔΔCt method with normalization to the control condition. Normalized gene expression changes are shown in each figure. All qRT-PCR primers span exon–intron–exon boundaries and were located in the middle of target genes, far downstream of sgRNA binding sites. The specific qRT-PCR primary sequences and their genomic position relative to the TSS(s) of each gene of interest are Gapdh (sense, 5′-TGCCCCCATGTTTGTGATG-3′, TSS+3274; antisense, 5′-TGTGGTCATGAGCCCTTCC-3′, TSS+3552), Ptch1 (sense, 5′-CTCTGGAGCAGATTTCCAAGG-3′, TSS+8049; antisense, 5′-TGCCGCAGTTCTTTTGAATG-3′, TSS+9692), Dhcr7 (sense, 5′-GCCAAGACACCACCTGTGACAG-3′, TSS+7718; antisense, 5′-TGGACGCCTCCCACATAACC-3′, TSS+10277), Cyp7a1 (sense, 5′-GAAGCAATGAAAGCAGCCTC-3′, TSS+4395; antisense, 5′-GTAAATGGCATTCCCTCCAG-3′, TSS+4462), and Gli1 (sense, 5′-GGTGCTGCCTATAGCCAGTGTCCTC-3′, TSS+10623; antisense, 5′-GTGCCAATCCGGTGGAGTCAGACCC-3′, TSS+10701).
Statistical analysis
All experiments were performed with at least three biological replicates. Histograms and scatterplots show mean ± SEM. Two-tailed Student’s unpaired t tests, one-way ANOVA, and two-way ANOVA were used to compare groups, as indicated in the figure legends, with statistical significance defined as *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. The sample size of each experiment is represented by number of independent data points on each graph.
Online supplemental material
Fig. S1 demonstrates DHCR7 localization near the ciliary base. Fig. S2 demonstrates that Hedgehog pathway activation does not influence DHCR7 expression. Fig. S3 demonstrates that CYP7A1 localizes near the ciliary base.
Supplementary Material
Acknowledgments
This study was supported by the UCSF Physician Scientist Scholar Program and by National Institutes of Health grants K08 CA212279-01 (to D.R. Raleigh) and R01 HD092659 (to L. Xu).
The authors declare no competing financial interests.
Author contributions: S. Findakly: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, supervision, validation, visualization, writing – original draft, and writing – review and editing. V. Daggubati: formal analysis, investigation, methodology, resources, supervision, and writing – review and editing. G. Garcia: formal analysis, investigation, methodology, resources, supervision, and writing – review and editing. S. LaStella and A. Choudhury: investigation and writing – review and editing. C. Tran, A. Li, and P. Tong: data curation, formal analysis, and methodology. J.Q. Garcia and N. Puri: investigation and writing – review and editing. J.F. Reiter: conceptualization, funding acquisition, project administration, supervision, and writing – review and editing. L. Xu: data curation, formal analysis, funding acquisition, investigation, methodology, resources, supervision, and writing – review and editing. D.R. Raleigh: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing – original draft, and writing – review and editing.
References
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg R., Macrae J.I., Briscoe J., and Jacob J.. 2016. Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum. Mol. Genet. 25:693–705. 10.1093/hmg/ddv507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., and Thérond P.P.. 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14:416–429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Byrne E.F.X., Sircar R., Miller P.S., Hedger G., Luchetti G., Nachtergaele S., Tully M.D., Mydock-McGrane L., Covey D.F., Rambo R.P., et al. 2016. Structural basis of Smoothened regulation by its extracellular domains. Nature. 535:517–522. 10.1038/nature18934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez M., Ena S., Van Sande J., de Kerchove d’Exaerde A., Schurmans S., and Schiffmann S.N.. 2015. Modulation of ciliary phosphoinositide content regulates trafficking and Sonic Hedgehog signaling output. Dev. Cell. 34:338–350. 10.1016/j.devcel.2015.06.016 [DOI] [PubMed] [Google Scholar]
- Conduit S.E., Ramaswamy V., Remke M., Watkins D.N., Wainwright B.J., Taylor M.D., Mitchell C.A., and Dyson J.M.. 2017. A compartmentalized phosphoinositide signaling axis at cilia is regulated by INPP5E to maintain cilia and promote Sonic Hedgehog medulloblastoma. Oncogene. 36:5969–5984. 10.1038/onc.2017.208 [DOI] [PubMed] [Google Scholar]
- Cooper M.K., Porter J.A., Young K.E., and Beachy P.A.. 1998. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 280:1603–1607. 10.1126/science.280.5369.1603 [DOI] [PubMed] [Google Scholar]
- Cooper M.K., Wassif C.A., Krakowiak P.A., Taipale J., Gong R., Kelley R.I., Porter F.D., and Beachy P.A.. 2003. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 33:508–513. 10.1038/ng1134 [DOI] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y.R., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Corcoran R.B., and Scott M.P.. 2006. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl. Acad. Sci. USA. 103:8408–8413. 10.1073/pnas.0602852103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney K.C., Fung K.Y., Maxfield F.R., Fairn G.D., and Zha X.. 2018. Comment on ‘Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol’. eLife. 7:e38493. 10.7554/eLife.38493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande I., Liang J., Hedeen D., Roberts K.J., Zhang Y., Ha B., Latorraca N.R., Faust B., Dror R.O., Beachy P.A., et al. 2019. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 571:284–288. 10.1038/s41586-019-1355-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer J.R., Sever N., Carlson M., Nelson S.F., Beachy P.A., and Parhami F.. 2007. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem. 282:8959–8968. 10.1074/jbc.M611741200 [DOI] [PubMed] [Google Scholar]
- Garcia G. III, Raleigh D.R., and Reiter J.F.. 2018. How the ciliary membrane is organized inside-out to communicate outside-in. Curr. Biol. 28:R421–R434. 10.1016/j.cub.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Phua S.C., Roberson E.C., Garcia G. III, Abedin M., Schurmans S., Inoue T., and Reiter J.F.. 2015. Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev. Cell. 34:400–409. 10.1016/j.devcel.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 154:442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron J., Hines K.M., and Xu L.. 2018. Assessment of altered cholesterol homeostasis by xenobiotics using ultra-high performance liquid chromatography-tandem mass spectrometry. Curr. Protoc. Toxicol. 78:e65 10.1002/cptx.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., et al. 2016. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5:e19760. 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Zheng S., Wierbowski B.M., Kim Y., Nedelcu D., Aravena L., Liu J., Kruse A.C., and Salic A.. 2018. Structural basis of smoothened activation in Hedgehog signaling. Cell. 174:312–324.e16. 10.1016/j.cell.2018.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., and Anderson K.V.. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Jira P.E., Waterham H.R., Wanders R.J.A., Smeitink J.A.M., Sengers R.C.A., and Wevers R.A.. 2003. Smith-Lemli-Opitz syndrome and the DHCR7 gene. Ann. Hum. Genet. 67:269–280. 10.1046/j.1469-1809.2003.00034.x [DOI] [PubMed] [Google Scholar]
- Kinnebrew M., Iverson E.J., Patel B.B., Pusapati G.V., Kong J.H., Johnson K.A., Luchetti G., Eckert K.M., McDonald J.G., Covey D.F., et al. 2019. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. eLife. 8:e50051. 10.7554/eLife.50051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczok K., Gurumurthy C.B., Balogh I., Korade Z., and Mirnics K.. 2019. Subcellular localization of sterol biosynthesis enzymes. J. Mol. Histol. 50:63–73. 10.1007/s10735-018-9807-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli P., Höhne M., Jüngst C., Bertsch S., Ebert L.K., Schauss A.C., Benzing T., Rinschen M.M., and Schermer B.. 2017. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 18:1521–1535. 10.15252/embr.201643846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide T., Hayata T., and Cho K.W.Y.. 2006. Negative regulation of Hedgehog signaling by the cholesterogenic enzyme 7-dehydrocholesterol reductase. Development. 133:2395–2405. 10.1242/dev.02393 [DOI] [PubMed] [Google Scholar]
- Kong J.H., Siebold C., and Rohatgi R.. 2019. Biochemical mechanisms of vertebrate hedgehog signaling. Development. 146:dev166892 10.1242/dev.166892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukic I., Rivera-Molina F., and Toomre D.. 2016. The IN/OUT assay: a new tool to study ciliogenesis. Cilia. 5:23 10.1186/s13630-016-0044-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchetti G., Sircar R., Kong J.H., Nachtergaele S., Sagner A., Byrne E.F., Covey D.F., Siebold C., and Rohatgi R.. 2016. Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife. 5:e20304. 10.7554/eLife.20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., and Nachury M.V.. 2015. Proteomics of primary cilia by proximity labeling. Dev. Cell. 35:497–512. 10.1016/j.devcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L., Weiss L.E., Yoon J., Roth T.L., Su Y.S., Sahl S.J., Scott M.P., and Moerner W.E.. 2015. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc. Natl. Acad. Sci. USA. 112:8320–8325. 10.1073/pnas.1510094112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers B.R., Sever N., Chong Y.C., Kim J., Belani J.D., Rychnovsky S., Bazan J.F., and Beachy P.A.. 2013. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev. Cell. 26:346–357. 10.1016/j.devcel.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers B.R., Neahring L., Zhang Y., Roberts K.J., and Beachy P.A.. 2017. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. USA. 114:E11141–E11150. 10.1073/pnas.1717891115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., Mydock L.K., Krishnan K., Rammohan J., Schlesinger P.H., Covey D.F., and Rohatgi R.. 2012. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 8:211–220. 10.1038/nchembio.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., Whalen D.M., Mydock L.K., Zhao Z., Malinauskas T., Krishnan K., Ingham P.W., Covey D.F., Siebold C., and Rohatgi R.. 2013. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife. 2:e01340 10.7554/eLife.01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu D., Liu J., Xu Y., Jao C., and Salic A.. 2013. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol. 9:557–564. 10.1038/nchembio.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt A., Arnold P., and Frey F.J.. 2001. The intracellular localization of the mineralocorticoid receptor is regulated by 11beta-hydroxysteroid dehydrogenase type 2. J. Biol. Chem. 276:28484–28492. 10.1074/jbc.M100374200 [DOI] [PubMed] [Google Scholar]
- Ostrom Q.T., Gittleman H., Liao P., Vecchione-Koval T., Wolinsky Y., Kruchko C., and Barnholtz-Sloan J.S.. 2017. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-oncol. 19(suppl 5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullinger C.R., Eng C., Salen G., Shefer S., Batta A.K., Erickson S.K., Verhagen A., Rivera C.R., Mulvihill S.J., Malloy M.J., et al. 2002. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J. Clin. Invest. 110:109–117. 10.1172/JCI0215387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Liu H., Thompson B., McDonald J., Zhang C., and Li X.. 2019. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature. 571:279–283. 10.1038/s41586-019-1286-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh D.R., Sever N., Choksi P.K., Sigg M.A., Hines K.M., Thompson B.M., Elnatan D., Jaishankar P., Bisignano P., Garcia-Gonzalo F.R., et al. 2018. Cilia-Associated Oxysterols Activate Smoothened. Molecular Cell. 72:316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh David R., Choksi Pervinder K., Krup Alexis Leigh, Mayer Wasima, Santos Nicole, and Reiter Jeremy F.. 2017. Hedgehog signaling drives medulloblastoma growth via CDK6. Journal of Clinical Investigation. 128(1):120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72:137–174. 10.1146/annurev.biochem.72.121801.161712 [DOI] [PubMed] [Google Scholar]
- Sever N., Mann R.K., Xu L., Snell W.J., Hernandez-Lara C.I., Porter N.A., and Beachy P.A.. 2016. Endogenous B-ring oxysterols inhibit the Hedgehog component Smoothened in a manner distinct from cyclopamine or side-chain oxysterols. Proc. Natl. Acad. Sci. USA. 113:5904–5909. 10.1073/pnas.1604984113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Garcia G. III, Van De Weghe J.C., McGorty R., Pazour G.J., Doherty D., Huang B., and Reiter J.F.. 2017. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 19:1178–1188. 10.1038/ncb3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg M.A., Menchen T., Lee C., Johnson J., Jungnickel M.K., Choksi S.P., Garcia G. III, Busengdal H., Dougherty G.W., Pennekamp P., et al. 2017. Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev. Cell. 43:744–762.e11. 10.1016/j.devcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V., Romaguera-Ros M., Garcia-Verdugo J.-M., and Reiter J.F.. 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 18:410–424. 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L.E., Milenkovic L., Yoon J., Stearns T., and Moerner W.E.. 2019. Motional dynamics of single Patched1 molecules in cilia are controlled by Hedgehog and cholesterol. Proc. Natl. Acad. Sci. USA. 116:5550–5557. 10.1073/pnas.1816747116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Tang J.-J., Peng C., Wang Y., Fu L., Qiu Z.-P., Xiong Y., Yang L.-F., Cui H.-W., He X.-L., et al. 2017. Cholesterol modification of smoothened is required for Hedgehog signaling. Mol. Cell. 66:154–162.e10. 10.1016/j.molcel.2017.02.015 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu W., Sheflin L.G., Fliesler S.J., and Porter N.A.. 2011a Novel oxysterols observed in tissues and fluids of AY9944-treated rats: a model for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52:1810–1820. 10.1194/jlr.M018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Korade Z., Rosado D.A. Jr., Liu W., Lamberson C.R., and Porter N.A.. 2011b An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52:1222–1233. 10.1194/jlr.M014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Korade Z., Rosado D.A. Jr., Mirnics K., and Porter N.A.. 2013. Metabolism of oxysterols derived from nonenzymatic oxidation of 7-dehydrocholesterol in cells. J. Lipid Res. 54:1135–1143. 10.1194/jlr.M035733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Chong W.M., Wang W.-J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.-D., Weng R.R., Huang C.-E., et al. 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.