Abstract
IDH1/2 hotspot mutations occur in glioma, cholangiocarcinoma, chondrosarcoma, sinonasal carcinoma and T-cell lymphoma and have diagnostic, prognostic and/or therapeutic value. Availability of immunohistochemistry (IHC) protocols to specific IDH2 mutations detection is limited. A targeted exome sequencing assay MSK-IMPACT cohort comprising >38,000 cancer cases was explored for the presence of IDH1/2 mutations in solid malignancies and select T-cell lymphomas. Seventy-four formalin-fixed paraffin-embedded IDH1/2 mutated (n=62) and wild-type (n=12) samples were used for testing and optimization of anti-IDH2 monoclonal antibodies (mAbs) 14H7, 3C11, MMab1 targeting R172K, R172G and R172M mutant proteins, respectively. IDH1/2 mutations were common in glioma (26.8% and 1.6%), intrahepatic cholangiocarcinoma (23.1% and 5.7%) chondrosarcoma (19.4% and 10.7%), sinonasal undifferentiated/large-cell neuroendocrine carcinoma (0 and 84.2%), angioimmunoblastic T-cell lymphoma (0 and 22%) and peripheral T-cell lymphoma (0 and 5.1%). In other cancers, IDH2 mutations were rare. IDH2 R172 variants included R172K (39%), R172S (29%), R172W (12%), R172G (10%), R172M (5%) and R172T (4%). 14H7, 3C11 and MMab1 detected all IDH2 R172K, R172G and R172M, respectively and produced a crisp, granular cytoplasmic staining pattern. 3C11 was also positive in 5 of 6 IDH1 R132G mutants showing a homogeneous, smooth cytoplasmic staining. All 3 mAbs were negative in other IDH1/2 mutant or wild-type cases. IHC employing mAbs 14H7, 3C11, MMab1 can facilitate molecular diagnosis as a reliable, fast and inexpensive alternative for specific IDH2 variants detection. Given the distinct distribution of IDH2 R172 mutations in cancers, these mAbs could serve also as useful pathologic diagnostic markers.
Keywords: IDH2 immunohistochemistry, 14H7, 3C11, MMab1, glioma, intrahepatic cholangiocarcinoma, chondrosarcoma, sinonasal undifferentiated carcinoma, angioimmunoblastic T-cell lymphoma
INTRODUCTION
Hotspot mutations in IDH1 and IDH2 have been detected in various human malignancies. Gliomas (1) acute myeloid leukemia (AML) (2), chondrosarcoma (3) and intrahepatic cholangiocarcinoma (IHCC) (4) predominantly harbor IDH1 hotspot variants, while other cancers, such as sinonasal undifferentiated/large cell neuroendocrine carcinoma (SNUC/LCNEC) (5, 6), angioimmunoblastic T-cell lymphoma (AITL) (7) and solid papillary breast carcinoma with reverse polarity (SPCRP) are characterized by highly recurrent IDH2 R172 mutations (8). In gliomas, detection of IDH1/2 mutations is essential for prognosis and classification (9). In chondrosarcomas, these mutations were associated with longer disease-free survival (DFS) (10). Similarly, IDH2 mutated sinonasal carcinomas tended to show longer DFS and relatively lower propensity for lung metastasis in comparison to their IDH1/2 wild-type counterparts (11). A favorable outcome was also associated with in IDH1/2 mutations in IHCC (12). In addition to the prognostic and diagnostic value, IDH1/2 mutations in solid tumors including gliomas, chondrosarcomas and IHCC have been recently explored as therapeutic targets for IDH-inhibitor therapy, which further emphasizes the importance of specific variants detection (13, 14). Given their prognostic (9, 11, 15), therapeutic (16) and diagnostic (8, 9, 17) significance, an accurate detection of IDH1/2 mutated proteins by immunohistochemistry (IHC) may represent a simple and rapid alternative to more expensive and time-consuming molecular assays. Recently, we have demonstrated that IHC is a reliable tool for the detection of IDH2 R172S/T mutated proteins (17). In the present study, we explored the frequency of IDH2 R172 mutations in a large clinical sequencing cohort and the clinical usability of monoclonal antibodies (mAb) to identify IDH2 R172M, R172G and R172K mutant proteins.
METHODS
1. Cases
A targeted exome sequencing assay MSK-IMPACT cohort comprising >38,000 clinical cancer cases profiled as previously described (18, 19) was explored for the presence of IDH1/2 mutations in solid malignancies and select T-cell lymphomas. An addition of previously published 15 sinonasal cancer research samples was also included in the study (11). Formalin-fixed paraffin-embedded (FFPE) material on 74 malignant tumors with the established IDH1/2 mutation status by MSK-IMPACT was retrieved from archives of the Department of Pathology at Memorial Sloan Kettering Cancer Center including select cases of glioma (n=31), carcinoma (n=24), lymphoma/leukemia (n=10), sarcoma (n=7) and melanoma (n=2; Table S1).
2. Antibodies
Commercial availability of serological reagents to various IDH2 mutations was investigated by on-line searches. Monoclonal antibodies to the following mutations were identified: IDH2 R172K: clone 14H7 (Neweast Biosciences, #26163; mouse), IDH2 R172G: clone 3C11 (Neweast Biosciences, Malvern, PA; #26231; mouse), IDH2 R172M: clone MMab1 (MBL, Woburn, MA; D337; rat) (20), IDH2 R172W: clone 13B7 (Neweast Biosciences, #26164; mouse) and clone WMab-1 (MBL, 338; rat). All antibodies underwent testing and optimization by initial titration on a panel of ten normal tissues and at least two lesions with molecularly proven presence of corresponding IDH2 R172 mutation. All assays were performed on Leica Bond-3 (Leica, Buffalo Grove, IL) automated stainer platform. The standard protocol comprised of 30 minutes primary incubation time and a 30 minutes heat-based antigen retrieval step. Low pH (Leica ER1) and high pH (Leica ER2) retrieval buffers were tested. A polymeric secondary (Refine, Leica) was used for detection of the primary. Since two of the reagents were rat derived primaries, a rabbit-anti-rat antibody (1:400; rabbit-anti-rat (H&L), #AI-4000; Vector Labs, Burlingame, CA) was employed as a link antibody for the polymer detection step.
RESULTS
1. Frequency and the spectrum of IDH1/2 variants in cancer
IDH1 mutations affecting exclusively codon R132 (n=749) and IDH2 hotspot mutations (n=125), including 117 (93.6%) codon R172 and 8 (6.4%) codon R140 variants were detected in various malignancies of epithelial and mesenchymal lineage (Table S2). IDH1 hotspot mutations were highly recurrent in glioma (26.8%), intrahepatic cholangiocarcinoma (IHCC, 23.1%) and chondrosarcoma (22.2%) and less common in extrahepatic/perihepatic cholangiocarcinoma (5%, Table 1). IDH1 R132 variants were also detected in small intestine adenocarcinoma (3%), melanoma (2.5%), mostly cutaneous, and in adenoid cystic carcinoma (2.2%). In other cancers, including colorectal, non-small cell lung, prostatic, pancreatic, breast, ovarian, thyroid, germ cell and bladder cancer R132 variants occurred in up to 0.5% cases or were absent. R132H was the most prevalent (62%) largely due to its highest prevalence in gliomas (89%). R132C was the most frequent variant in IHCC (71%) and chondrosarcomas (44%, Figure 1).
Table 1.
Frequency of IDH1 R132 and IDH2 R172 mutations in cancer.
| Tumor type/category | All cases (n=38,163) | IDH1/2 mutations (%) | IDH1 mutations (%) | IDH2 mutations (%) |
|---|---|---|---|---|
| Glioma | 1839 | 523 (28.4) | 493 (26.8) | 30 (1.6) |
| Intrahepatic cholangiocarcinoma | 472 | 136 (28.8) | 109 (23.1) | 27 (5.7) |
| Cholangiocarcinoma, NOS* | 297 | 20 (6.4) | 16 (5.3) | 4 (1.3) |
| Chondrosarcoma | 81 | 28 (34.6) | 18 (22.2) | 10 (12.3) |
| SNUC/LCNEC | 19 | 16 (84.2) | 0 | 16 (84.2)10 |
| Angioimmunoblastic T-cell lymphoma | 59 | 13 (22) | 0 | 13 (22) |
| Peripheral T-cell lymphoma | 99 | 5 (5.1) | 0 | 5 (5.1) |
| Small intestine carcinoma | 132 | 4 (3) | 4 (3) | 0 |
| Melanoma | 1506 | 39 (2.6) | 37 (2.5) | 2 (0.1) |
| Adenoid cystic carcinoma | 178 | 4 (2.2) | 4 (2.2) | 0 |
| Colorectal and appendiceal carcinoma | 4435 | 25 (0.6) | 20 (0.5) | 5 (0.1) |
| Non-small cell lung carcinoma | 6091 | 22 (0.4) | 19 (0.3) | 3 (<0.1) |
| Prostate adenocarcinoma | 2574 | 11 (0.4) | 11 (0.4) | 0 |
| Endometrial carcinoma | 1882 | 6 (0.3) | 6 (0.3) | 0 |
| Pancreatic cancer | 2459 | 4 (0.2) | 4 (0.2) | 0 |
| Breast cancer ** | 5422 | 4 (<0.1) | 4 (<0.1) | 0 |
| Ovarian cancer | 1729 | 1 (<0.1) | 1 (<0.1) | 0 |
| Thyroid carcinoma | 711 | 1 (0.1) | 1 (0.1) | 0 |
| Germ cell tumor | 623 | 1 (0.2) | 1 (0.2) | 0 |
| Bladder cancer | 1578 | 1 (<0.1) | 1 (<0.1) | 0 |
| Soft tissue sarcoma | 1734 | 0 | 0 | 0 |
| Esophagogastric carcinoma | 1319 | 0 | 0 | 0 |
| Renal cell carcinoma | 894 | 0 | 0 | 0 |
| Head and neck carcinoma *** | 674 | 0 | 0 | 0 |
| Uterine sarcoma | 306 | 0 | 0 | 0 |
| Small cell lung carcinoma | 385 | 0 | 0 | 0 |
| Cervical carcinoma | 279 | 0 | 0 | 0 |
| Gastrointestinal neuroendocrine carcinoma | 174 | 0 | 0 | 0 |
| Ampullary carcinoma | 110 | 0 | 0 | 0 |
| Anal carcinoma | 102 | 0 | 0 | 0 |
Including any cholangiocarcinoma except intrahepatic cholangiocarcinoma
No cases of solid papillary carcinoma with reverse polarity are tested.
Including salivary gland tumors except adenoid cystic carcinoma and excluding sinonasal IDH2 mutated carcinoma.
Figure 1.
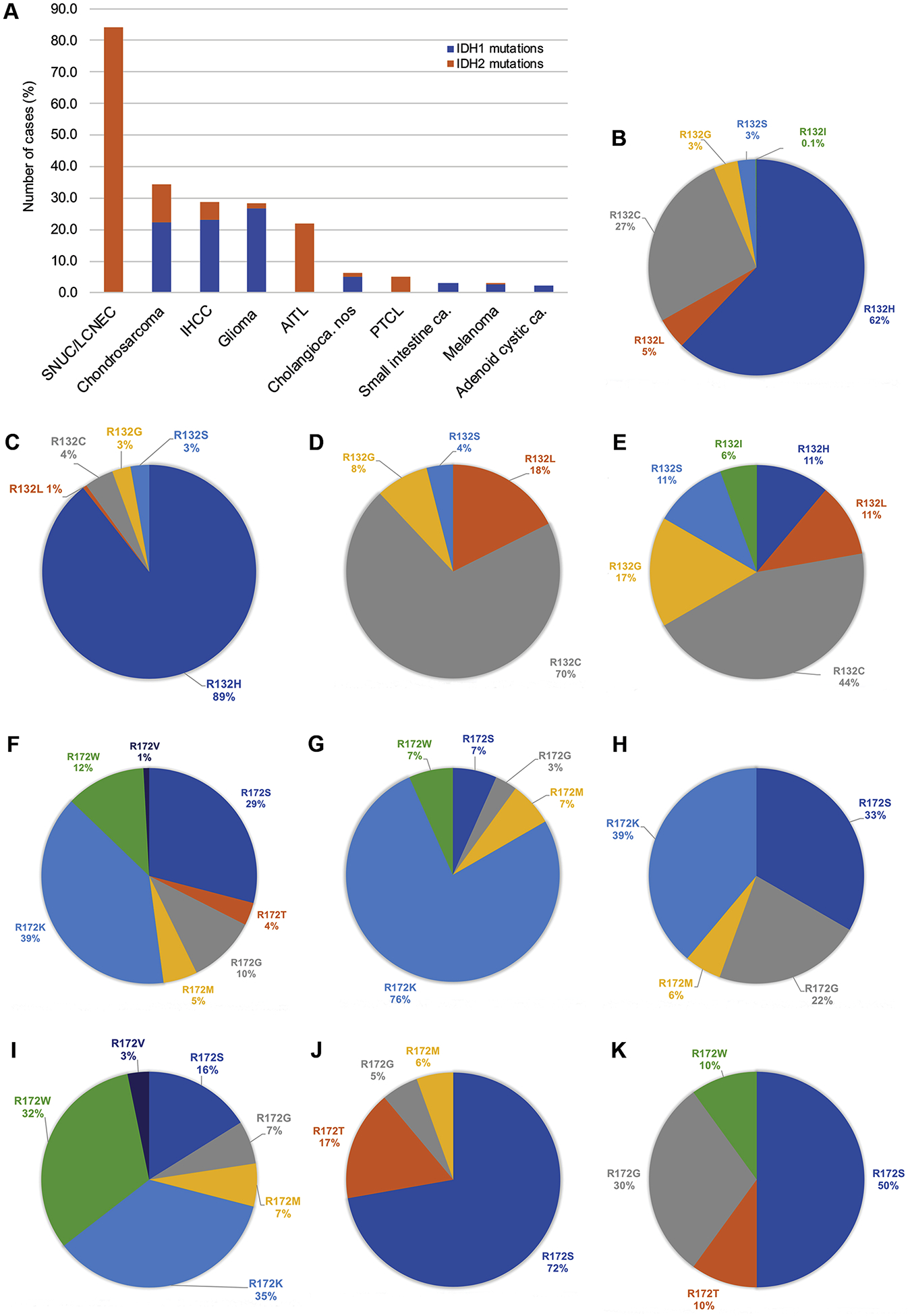
Frequency of IDH1 R132 and IDH2 R172 mutations in various malignancies (A). All IDH1 R132 mutated tumors (B), glioma (C), cholangiocarcinoma (D) and chondrosarcoma (E). IDH2 R172 mutations in all tumor types (F), glioma (G), AITL/PTCL (H), cholangiocarcinoma (I), sinonasal carcinoma (including 16 SNUC/LCNEC and 2 poorly differentiated carcinomas with glandular differentiation [ref. 10], J), and chondrosarcoma (K). Abbreviations: IHCC, intrahepatic cholangiocarcinoma; PTCL, peripheral T-cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; SNUC/LCNEC, sinonasal undifferentiated/large cell neuroendocrine carcinoma; nos, not otherwise specified; ca, carcinoma.
Exclusively IDH2 variants were present in the majority of SNUC/LCNEC (84.2%) (11), in a significant proportion of AITL (22%) and less often in peripheral T-cell lymphoma (PTCL, 5.1%, Table 1). The overall most common variant was R172K (39%), also predominated in glioma (76%), AITL/PTCL (39%) and IHCC (35%). R172S (29%) was the most frequent mutation in sinonasal carcinomas (72%) and chondrosarcoma (50%). R172G was common in chondrosarcoma (30%) and AITL/PTCL (22%) and occurred less often in all other cancers. R172W was common in IHCC (32%) but was not found in AITL/PTCL or sinonasal carcinomas. R172M was detected in 6–7% in all tumors but chondrosarcoma and R172T was present only in sinonasal carcinomas (17%) and chondrosarcoma (10%; Figure 1). IDH2 R172 mutations were exceedingly rare in other cancers including 2 cutaneous melanoma, 5 colorectal and 3 lung adenocarcinomas (Table 1). A full list of variants is provided in Table S2.
2. Immunohistochemical protocol
The conditions were successfully optimized for anti-IDH2 R172K, R172G and R172M antibodies (Table 2A), which all displayed best immunoreactivity at rather high dilutions and employing high pH (ER2) retrieval buffer. These antibodies gave a good and consistent staining in the tested tumors carrying the respective mutations. Normal tissues and tumors without the corresponding mutation remained negative. Testing of either anti-IDH2 R172W antibody, clone 13B7 and clone WMab-1 generated staining unrelated to the presence of IDH2 172W and consequently were not pursued (data not shown).
Table 2A.
Optimized working conditions for anti-IDH2-mutation reactive monoclonal antibodies.
| Mutation | Clone | Species | Provider (order #) | Final working concentration (dilution) |
|---|---|---|---|---|
| IDH2 R172K | 14H7 | Mouse | Neweast Biosciences (26163) | 0.5ug/ml (1:2K) |
| IDH2 R172G | 3C11 | Mouse | Neweast Biosciences (26231) | 0.1ug/ml (1:10K) |
| IDH2 R172M | MMab1 | Rat | MBL (D337) | 0.5ug/ml (1:2K) |
3. Immunohistochemical results
Test cases of the present study consisted 32 IDH1 R132 mutants, 30 IDH2 R172 mutants, and 12 IDH1/2 wild-type tumors. Anti-IDH2 R172K mAb 14H7 was positive in all 13 tumors carrying the R172K mutation; 14H7 remained negative in IDH1/2 wild-type cases and any case harboring any other IDH1/2 mutation (n=50, Table 2B). The staining intensity ranged from strong and diffuse to focal and weak, but the cytoplasmic pattern was consistently crisp and granular consistent with mitochondrial localization of IDH2 mutated proteins (Figure 2). Anti-IDH2 R172G clone 3C11 was positive in all six R172G mutated tumors and was diffuse and strong in all cases except in areas of suboptimal tissue quality due to crush artifact (Figure 3). Given the similarity in IDH1 R132 and IDH2 R172 protein sequences, 3C11 was also positive in 5 of 6 IDH1 R132G mutated cases yet producing a rather smooth, homogeneous cytoplasmic staining consistent with cytosolic distribution of IDH1 mutated protein and was notably distinct from the characteristic granular pattern seen in IDH2 R172G mutated cases (Figure 4). The staining in IDH1 R132G mutant cases, a moderate smooth, homogeneous cytoplasmic staining pattern was similar to that observed in IDH1 R132S mutants stained by anti-IDH2 R172S clone 11C8B1 (17). 3C11 remained negative in all other tested tumors (n=49, Table 2B). MMab1 showed consistent immunoreactivity in all 3 IDH2 R172M mutated cases and was immunonegative in all other tumors (n=55) including IDH1/2 wild-type cases (Table 2B). MMab1 generated a consistently strong, granular cytoplasmic staining similar to that seen in with 3C11 mAb in IDH2 R172G mutated cases (Figure 5).
Table 2B.
Immunohistochemistry of IDH1/2 mutated malignancies using monoclonal anti-IDH2 R172 antibodies.
| Gene | Variant | 14H7 (IDH2R172K) | 3C11 (IDH2R172G) | MMab1 (IDH2R172M) |
|---|---|---|---|---|
| IDH2 | R172K (n=13) | 13/13 | 0/13 | 0/13 |
| R172G (n=6) | 0/5 | 6/6 | 0/5 | |
| R172M (n=3) | 0/3 | 0/2 | 3/3 | |
| R172S (n=7) | 0/7 | 0/6 | 0/6 | |
| R172T (n=1) | 0/1 | 0/1 | 0/1 | |
| IDH1 | R132G (n=6) | 0/5 | 5/6 | 0/5 |
| R132S (n=6) | 0/4 | 0/4 | 0/4 | |
| R132H (n=8) | 0/7 | 0/7 | 0/8 | |
| R132L (n=6) | 0/4 | 0/4 | 0/5 | |
| R132C (n=6) | 0/6 | 0/6 | 0/6 | |
| IDH1/IDH2 | wild-type (n=12) | 0/8 | 0/12 | 0/9 |
Figure 2.
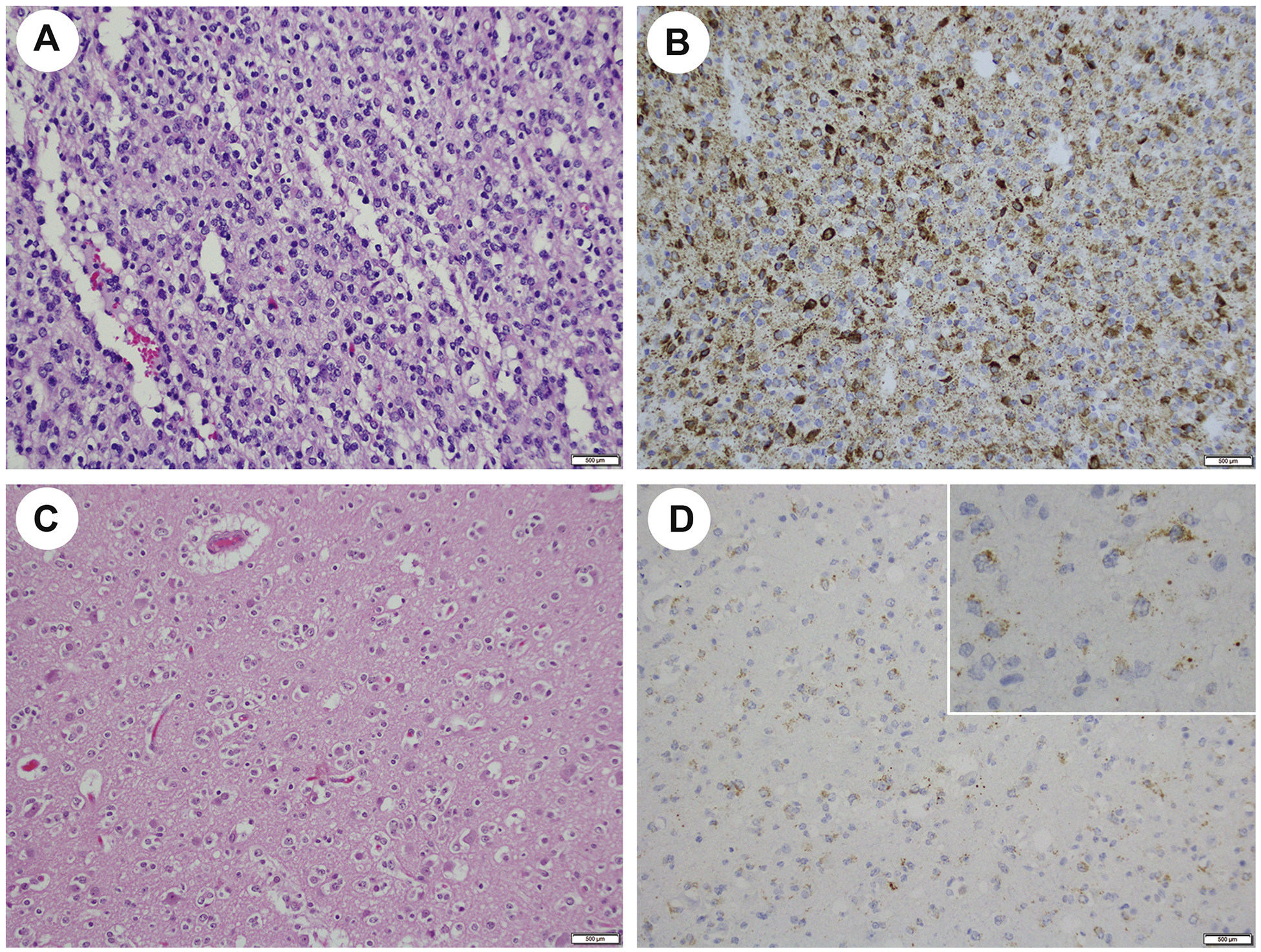
14H7 mAb shows a range in staining intensity in IDH2 R172K mutated gliomas. Anaplastic oligodendroglioma (H&E, A) stains strongly for 14H7 (B) and in another case (H&E, C), anti-IDH2 R172K is rather weak (D) but a distinct granular pattern indicative of mitochondrial localization of the mutant IDH2 protein is apparent at high magnification (inset, D).
Figure 3.

IDH2 R172G detection by 3C11. IDH2 R172G mutated chondrosarcoma (H&E, A) shows patchy and strong cytoplasmic granular staining by 3C11 (B, inset, 400X). AITL (H&E, C), shows a similar staining pattern by 3C11 (D). In SNUC (H&E, E) the quality of staining is absent in areas of crush artifact (F, inset, 400X, red arrow). Abbreviations: AITL, angioimmunoblastic T-cell lymphoma; SNUC, sinonasal undifferentiated carcinoma.
Figure 4.

IDH2 protein sequence in the proximity of codon R172 is remarkably similar to IDH1 protein sequence around codon R132. IDH2 aminoacids 168–172 with R172G mutated protein (black frame, left) and IDH1 aminoacids 128–136 with R132G mutated protein (black frame, right, A). A strong, diffuse, granular staining by 3C11 is seen in IDH2 R172G mutated IHCC (B) and homogeneous, smooth cytoplasmic staining is seen in IDH1 R132G mutated IHCC (C). Anaplastic oligodendroglioma with IDH2 R172G mutation (D). Anaplastic oligodendroglioma with IDH1 R132G mutation shows a weak cytoplasmic staining best appreciated at high power (inset, E). Abbreviations: IHCC, intrahepatic cholangiocarcinoma.
Figure 5.

IDH2 R172M detection in oligodendroglioma (H&E, A) using MMab1 IHC (B). IDH2 R172M mutated AITL (H&E, C) stained by MMab1 (D, inset, 400X). Abbreviations: AITL, angioimmunoblastic T-cell lymphoma;
DISCUSSION
In this study, we have provided the overall frequency of IDH1/2 hotspot variants in a large sequencing cohort of predominantly solid cancers with an emphasis on the type and distribution of IDH2 R172 mutations. We have used a selection of IDH1/2 mutated and wild-type tumors to establish and clinically validate three IHC protocols targeting IDH2 R172M, R172G and R172K mutant proteins, respectively and have demonstrated their usability as reliable alternatives for specific IDH2 variants detection.
Our sequencing data showed that the frequencies of IDH1/2 mutations in glioma, chondrosarcoma, SNUC/LCNEC, IHCC and AITL are similar to those previously reported in literature (1, 3, 4, 7, 11). Interestingly, IDH2 mutations showed predilection for certain tumor types, such as SNUC/LCNEC and AITL/PTCL, and the overall distribution of R172 variants was distinct among different tumor types. R172K was the most common in gliomas, AITL/PTCL, and cholangiocarcinoma, but was not found in SNUC/LCNEC. Instead, R172S was the most common variant in sinonasal carcinomas and chondrosarcoma. Outside of the main tumor types known to harbor recurrent IDH1/2 mutations, in our study IDH1 mutations rarely occurred in other cancers such as adenoid cystic carcinoma, melanoma and adenocarcinoma of small intestine, colon and lung, while IDH2 variants remained extremely uncommon and limited only to rare examples of melanoma, and colorectal and lung adenocarcinomas.
IHC detection of the mutant IDH1/2 proteins may represent a quick and convenient alternative to often complex, time-consuming and more expensive molecular testing (21). However, availability of IDH1/2 antibodies and clinically validated IHC protocols is primarily determined by the extent of their usability in clinical settings. Given the higher frequency of IDH1 variants and their occurrence in relatively common cancers, such as glioma, anti-IDH1 IHC, namely anti-IDH1 R132H represents a prototype of IDH IHC that is widely used surgical pathology practice (9). In contrast, less common IDH2 mutations have not been the focus of in situ protein expression analyses and IHC protocols for their detection have been mostly limited to methods using multispecific antibodies for simultaneous targeting of multiple IDH1 R132 and IDH2 R172 variants (22–24). While multispecific IDH1/2 antibodies might certainly be a very practical solution to facilitate the diagnosis of glioma for example, these antibodies do not inform on the particular IDH1 or IDH2 variant. Distinguishing IDH1/2 variants is important if treatment with a specific IDH-inhibitor is considered (13, 14). Moreover, the differences in response to selective IDH2-inhibitors was found to depend on the IDH2 mutation variant, further emphasizing the importance of an exact variant detection (25).
Reports or clinically validated methods for specific targeting of individual IDH2 R172 variants are limited and include our previously reported 11C8B1 IHC for IDH2 R172S and R172T variants detection (17) and IDH2 R172K IHC detection in AITL (21). One possible challenge in the development and clinical validation of specific IDH2 IHC protocols could be related to their relative rarity and lacking positive controls. Here, we took advantage of our sizable sequencing cohort comprised of different tumor types with established molecular profiles and a full spectrum of IDH1/2 variants. This allowed for a robust testing and cross-validation of different IDH2 mAbs to determine their sensitivity and specificity and to examine the cross-reactivity between different molecular subgroups. All three IHC protocols proved to be sensitive and mutation-specific rendering a distinct granular cytoplasmic staining pattern consistent with mitochondrial localization of IDH2 mutated proteins and suggesting that mAbs 14H7, 3C11 and MMab1 can serve as reliable markers for detection of IDH2 R172K, R172G and R172M variants, respectively. A smooth, homogeneous, cytoplasmic staining by 3C11 is in concordance with its non-mitochondrial localization and can indicate the presence of IDH1 R132G mutation. In addition to the anti-IDH2 R172S IHC by 11C8B1, the three IHC protocols presented here can identify up to 87% of IDH2 R172 hotspot variants overall. Most notably, in tumors harboring only IDH2 hotspot mutations, AITL/PTCL and SNUC/LCNEC, 93% and 100% R172 variants, respectively could be identified. From a practical perspective, a distinct distribution of R172 variants among different tumor types can help prioritize a specific IDH2 mAb and enable a cost-effective IHC performed in a sequential manner. Based on our findings, immunostains presented here can be useful in pathologic diagnosis; given the infrequency of IDH2 R172 mutations outside of several major tumor groups, an addition of anti-IDH2 IHC to the diagnostic work-up of metastasis of unknown primary.
In summary, here we examined the type and frequency of IDH1/2 hotspot mutations with an emphasis on IDH2 R172 variants in, to our knowledge the largest sequencing cohort comprising the most human solid malignancies and select T-cell lymphomas. We have established 3 anti-IDH2 IHC protocols for detection of R172K, R172G and R172M proteins and propose their use as a reliable method for the respective IDH2 mutations detection. In addition to providing a molecular diagnosis, the present mutation-specific IHC can potentially be useful diagnostic markers in the work-up of metastasis/unknown primary. Finally, in view of the clinical significance of IDH1/2 mutations in cancer and the feasibility of these mutation specific anti-IDH2 IHC, it is reasonable to expect their applicability in surgical pathology practice in the near future.
Supplementary Material
Disclosure Statement:
No competing financial interests exist for all contributory authors.
Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcucci G, Maharry K, Wu Y-Z et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amary MF, Bacsi K, Maggiani F et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–43. [DOI] [PubMed] [Google Scholar]
- 4.Borger DR, Tanabe KK, Fan KC et al. Frequent mutation of isocitrate dehydrogenase (IDH) 1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dogan S, Chute DJ, Xu B, et al. Frequent IDH2 R172 mutations in undifferentiated and poorly-differentiated sinonasal carcinomas. J Pathol. 2017;242:400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jo VY, Chau NG, Hornick JL, Krane JF, Sholl LM. Recurrent IDH2 R172X mutations in sinonasal undifferentiated carcinoma. Mod Pathol. 2017;30(5):650–9. [DOI] [PubMed] [Google Scholar]
- 7.Cairns RA, Iqbal J, Lemonnier F et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang S, Weigelt B, Wen HC et al. IDH2 Mutations Define a Unique Subtype of Breast Cancer with Altered Nuclear Polarity. Cancer Res. 2016;76:7118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann C, Hentschel B, Wick W et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–18. [DOI] [PubMed] [Google Scholar]
- 10.Zhu GG, Nafa K, Agaram N et al. Genomic Profiling Identifies Association of IDH1/IDH2 Mutation with Longer Relapse-Free and Metastasis-Free Survival in High-Grade Chondrosarcoma. Clin Cancer Res. 2020;26:419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogan S, Vasudevaraja V, Xu B et al. DNA methylation-based classification of sinonasal undifferentiated carcinoma. Mod Pathol. 2019;32:1447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma B, Meng H, Tian Y et al. Distinct clinical and prognostic implication of IDH1/2 mutation and other most frequent mutations in large duct and small duct subtypes of intrahepatic cholangiocarcinoma. BMC Cancer. 2020;20:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan B, Mellinghoff IK, Wen PY et al. Clinical pharmacokinetics and pharmacodynamics of ivosidenib, an oral, targeted inhibitor of mutant IDH1, in patients with advanced solid tumors. Invest New Drugs. 2019:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konteatis Z, Artin E, Nicolay B et al. Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma. ACS Med Chem Lett. 2020;11:101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiNardo CD, Ravandi F, Agresta S et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90:732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNardo CD, Stein AS, Stein EM et al. Mutant IDH (mIDH) inhibitors, ivosidenib or enasidenib, with azacitidine (AZA) in patients with acute myeloid leukemia (AML). ASCO: 2018. J Clin Oncol. 36, no.15_suppl 7042–7042. [Google Scholar]
- 17.Dogan S, Frosina D, Fayad M et al. The role of a monoclonal antibody 11C8B1 as a diagnostic marker of IDH2-mutated sinonasal undifferentiated carcinoma. Mod Pathol. 2019;32:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zehir A, Benayed R, Shah RH et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko MK, Morita S, Tsujimoto Y et al. Establishment of novel monoclonal antibodies KMab-1 and MMab-1 specific for IDH2 mutations. Biochem Biophys Res Commun. 2013;432:40–5. [DOI] [PubMed] [Google Scholar]
- 21.Dupuy A, Lemonnier F, Fataccioli V et al. Multiple ways to detect IDH2 mutations in angioimmunoblastic T-cell lymphoma from immunohistochemistry to next-generation sequencing. J Mol Diagn. 2018;20:677–85. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko MK, Ogasawara S, Kato Y. Establishment of a multi-specific monoclonal antibody MsMab-1 recognizing both IDH1 and IDH2 mutations. Tohoku J Exp Med. 2013;230:103–9. [DOI] [PubMed] [Google Scholar]
- 23.Ogasawara S, Kaneko MK, Tsujimoto Y, Liu X, Kato Y. Multi-specific monoclonal antibody MsMab-2 recognizes IDH1-R132L and IDH2-R172M mutations. Monoclon Antib Immunodiagn Immunother. 2013;32:377–81. [DOI] [PubMed] [Google Scholar]
- 24.Kato Y. Specific monoclonal antibodies against IDH1/2 mutations as diagnostic tools for gliomas. Brain Tumor Pathol. 2015;32:3–11. [DOI] [PubMed] [Google Scholar]
- 25.Gao M, Zhu H, Fu L et al. Pharmacological characterization of TQ05310, a potent inhibitor of isocitrate dehydrogenase 2 R140Q and R172K mutants. Cancer Sci. 2019;110:3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


