Abstract
Defects in innate immunity affect many different physiologic systems and several studies of patients with primary immunodeficiency disorders demonstrated the importance of innate immune system components in disease prevention or colonization of bacterial pathogens. To assess the role of the innate immune system on nasal colonization with Staphylococcus aureus, innate immune responses in pediatric S. aureus nasal persistent carriers (n=14) and non-carriers (n=15) were profiled by analyzing co-clustered gene sets (modules). We stimulated previously frozen peripheral blood mononuclear cells (PBMCs) from these subjects with i) a panel of TLR ligands, ii) live S. aureus (either a mixture of strains or stimulation with respective carriage isolates), or iii) heat-killed S. aureus. We found no difference in responses between carriers and non-carriers when PBMCs were stimulated with a panel of TLR ligands. However, PBMC gene expression profiles differed between persistent and non-S. aureus carriers following stimulation with either live or dead S. aureus. These observations suggest that individuals susceptible to persistent carriage with S. aureus may possess differences in their live/dead bacteria recognition pathway and that innate pathway signaling is different between persistent and non-carriers of S. aureus.
Keywords: Staphylococcus aureus, targeted assay, gene expression, Vita-PAMPs, RNAseq, nasal carriage
1. Introduction
The spectrum of diseases resulting from S. aureus infections includes serious skin infections, endocarditis, arthritis, osteomyelitis, and sepsis as a consequence of its ability to colonize a variety of different tissues and its ability to circumvent various immune surveillance systems [1, 2]. Approximately 20–50% of adults and children nasal S. aureus carriers (persistent or intermittent carriers vs. non-carriers) based on the presence or absence of S. aureus in nasal cultures collected over time [3, 4]. Nasal sampling over a defined time period allows classification of S. aureus carrier phenotypes as either persistent carriers (those testing positive ≥75% of the time), intermittent carriers (individuals <75% of the time) and non-carriers (negative for S. aureus over the sampling period) [5]. Although a genome-wide association study conducted by our group suggests that each carriage phenotype is in part shaped by host genetic profiles [6], characterization of antibody responses and S. aureus re-colonization studies suggest that the intermittent and non-carrier phenotypes are more similar to each other compared to persistent carriers [7, 8].
A combination of environmental, genetic, and immune factors play roles in defining the respective S. aureus carriage phenotypes and candidate gene studies have linked specific genes with persistent S. aureus nasal carriage [9–19]. For example, polymorphisms in immune-related genes (IL-4, mannose binding lectin [complement pathway], C1 inhibitor [complement pathway], complement factor H, C-reactive protein, vitamin D receptor polymorphisms, β-defensin production, and Toll-like receptor 2) have been shown to contribute to persistent S. aureus nasal carriage [9–18]. It is important to keep in mind that different innate and acquired immune effector functions play different role protection against colonization versus infection [18].
Despite these insights into immune components that promote persistent S. aureus carriage, no effective human vaccine that prevents colonization or infections has been developed [18, 20]. This failure can be partially explained in part by our lack of understanding regarding the nature of the immune response(s) required to prevent S. aureus infections/colonization. Due to the clear differences in anti-S. aureus immune responses observed between persistent and intermittent/non-carriers [7, 8] and based on previous work by our group showing innate immune transcriptional profiles that reflect disease manifestations of pediatric patients infected with S. aureus, we examined differences in innate immune responses between persistent and non-carriers of S. aureus using high-throughput methods for transcript profiling [21, 22]. Our goal was to provide a better understanding of which innate immune gene modules are associated with these distinct carriage profiles in a pediatric cohort.
The successful assessment of innate immune responsiveness to respective pathogens could result in the development of novel vaccines for patients presenting with recurrent and/or severe infections. We have previously reported on the design of modules consisting of sets of coordinately expressed transcripts [23]. The modular framework has been successfully implemented in a wide range of studies, covering infections and autoimmune diseases [22, 24, 25]. Recently, we reported on the creation of a set of modules designed to further investigate gene profiles associated with innate immune activation signals [26]. The resulting 66 modules are divided into 7 clusters based on their response specificities; some are specific for bacterial stimuli while others are specific for stimuli via a TIR (Toll-IL-1 receptor) domain.
Here we use the dimension-reduction concept we have developed for modular microarray analysis and use it for gene selection for a focused qPCR-based assay in order to develop a time- and cost-efficient tool that can be used to assess potential susceptibilities in individual patients or groups. We used this targeted tool kit for the assessment of innate immune response profiles of peripheral blood mononuclear cells (PBMCs) isolated from pediatric S. aureus persistent and non-carriers stimulated with either i) a panel of TLR ligands, ii) live S. aureus, or iii) heat-killed S. aureus. This analysis demonstrates that children who are persistently colonized with S. aureus displayed unique responses to live vs. dead bacteria compared to responses observed by non-carriers of S. aureus suggesting potential differences in Vita-PAMP (pathogen associated molecular patterns) signaling pathways between these two carriage phenotypes. This suggests that recognition of Vita-PAMPs via innate immune pattern recognition receptors present on host cells between persistent and non-carriers of S. aureus may differ and may be a contributing factor to S. aureus colonization.
2. Materials and methods
2.1. Study population and sample collection
Pediatric patients from the Northwest Assistance Ministries Clinic, Houston, TX, were eligible for participation in the current study if they were participants of a prior parent study (unpublished results) investigating pediatric S. aureus nasal carriage (n=438). All children enrolled into the parent study were enrolled during their wellness visits; there were no apparent health or clinical difference between children determined to be persistent carriers of S. aureus compared to non-carriers. In the parent study, we established nasal carriage phenotypes (persistent, intermittent, or non-carriers) based on the identification of S. aureus on 2 consecutive nasal swabs collected 12–17 days apart [6, 27, 28]. Participants in the parent study testing positive for S. aureus on both swabs were classified as persistent carriers. Non-carriers were S. aureus negative at both time points and intermittent carriers were positive for one of the two initial swabs collected. No intermittent carriers are included in the present study. Using univariate statistics, we compared demographic characteristics between the two patient groups, using the chi-squared test for categorical variables (Fisher’s exact p-value if the comparison included a frequency ≤5) or ANOVA for continuous variables, using Stata 14 (Table 1) (College Station, TX).
Table 1.
Characteristics of the Study Population (n=30).
| Non-Carriers | Persistent Carriers | ||
|---|---|---|---|
| 15 (50%) | 15 (50%) | p-value | |
| <6 | 6 (40%) | 7 (47%) | 1.000 |
| ≥6 | 9 (60%) | 8 (53%) | |
| Sex | |||
| Female | 7 (47%) | 5 (33%) | 0.710* |
| Male | 8 (53%) | 10 (67%) | |
| Race/Ethnicity | |||
| Caucasian, non-Hispanic | 0 (0%) | 2 (13%) | 0.636 * |
| Latino/Hispanic | 12 (80%) | 9 (60%) | |
| African American | 2 (13%) | 3 (20%) | |
| Other | 1 (7%) | 1 (7%) | |
| History of Culture-Positive Staphylococcal Infection | |||
| No 15 (100%) | 10 (67%) | 0.042 *‡ | |
| Yes | 0 (0%) | 5§ (33%) | |
| Number of Nasal Swabs Completed | |||
| 3 | 10 (67%) | 13 (87%) | 0.390 |
| 4 | 5 (33%) | 2 (13%) | |
Fisher’s exact p-value.
Statistically significant at p<0.05.
Four patients had MRSA infections; 3 of those patients were MRSA persistent carriers.
As participants were enrolled and consented into the present study, we took this opportunity to confirm their nasal carriage status established in the parent study by collecting and analyzing a confirmatory third swab. Enrollment was concluded after 15 confirmed persistent and non-carriers of S. aureus were identified. Third swabs were collected between 6–931 days after the second swab (median 207 days) (Table 2) and processed for the detection of S. aureus as described previously [6, 27, 28]. After carriage status was confirmed, participants were asked to return to the clinic for collection of the PBMCs. S. aureus isolates from 12 of the 15 persistent carriers were available for analysis in the current study, and they were used individually (denoted “Carrier Strain”) or combined (denoted “Carrier Mix”). In all, a persistent carrier could have up to 6 S. aureus positive cultures from the 3 swabs collected at different time points (Table 2).
Table 2.
Strain characteristics of S. aureus strains isolated from pediatric persistent carriers.
| Pediatric Persistent Carrier* | Antibiotic Resistance of Carriage Isolate(s) | Ridom spa Type 1st Nasal Swab† | Ridom spa Type 2nd Nasal Swab | Ridom spa Type 3rd Nasal Swab |
|---|---|---|---|---|
| P1 | MSSA‡ | t1077/ t1077 | t1077/ t1077 | t1077/ t1077 |
| P2 | MSSA | New32/New32** | New32/New32 | NEG¥/t008 |
| P3 | MSSA | NEG/t9964 | NEG/t9964 | t9964/t9964 |
| P4 | MSSA | NEG/t13031 | t13031/t13031 | t13031/t13031 |
| P5 | MSSA | t338/t338 | t338/t338 | t338/t338 |
| P6 | Both MSSA | t002/t002 | t620/NEG | t620/t620 |
| PC7 | MSSA | t164/t164 | t164/t164 | t164/t164 |
| PC8 | MSSA | t164/t164 | t164/t164 | t164/t164 |
| PC9 | MSSA | t165/t165 | t165/t165 | t165/t165 |
| PC10 | MRSA§ and MSSA, respectively | t008/t008 | t008/t008 | NEG/t1978 |
| PC11 | MRSA | t1978/t1978 | t1978/t1978 | t1978/t1978 |
| PC12 | MRSA | t008/t008 | t008/t008 | t008/NEG |
Persistent carriage is defined by the identification of S. aureus at each sampling time point.
At each sampling time point swabs were used to inoculate MSA plates. The swab tips were cultured in TSB for 48 h and then used to inoculate a second MSA plates. Therefore, for each collection time point, an individual could have two S. aureus colonies available for analysis. S. aureus spa type in bold designates the spa type identified at the time of the last nasal sampling and also the isolate used to stimulate respective persistent carriers and in the generation of the Carrier Mix.
MSSA, methicillin susceptible S. aureus
This provisional new spa type with the following repeats: r07-r23-r12-r21-r12-r17-r20-r17-r12-r12-r24
NEG, S. aureus not detected.
MRSA, methicillin resistant S. aureus
Peripheral blood was drawn into ACD Vacutainer® tubes from participants enrolled in the study using standard phlebotomy techniques. Specimen tubes were labeled and transported at 4°C to the University of Texas Health Science Center at Houston School of Public Health for PBMC isolation.
2.2. Ethics of experimentation
This study was approved by the Institutional Review Boards of the University of Texas Health Science Center and Benaroya Research Institute at Virginia Mason. Informed written consent was obtained from each participant’s parent or guardian. Child assent was obtained from all participants 7–11 years of age and adolescent consent from all participants ages 12–17 years.
2.3. PBMC stimulation
Blood was collected at the Northwest Assistance Ministries Clinic (Houston, TX) and PBMCs were isolated from whole blood within 4 h of collection using Ficoll Paque plus (GE Healthcare) and frozen down in 7% DMSO in fetal bovine serum and cooled at −1°C/min in a freezing container (Nalgene, ThermoFisher Scientific). Frozen PBMCs were stored at −80°C before shipping on dry ice to Benaroya Research Institute for further processing and stimulation as described above.
PBMCs were stimulated for 6 h with (1) TNF (20 ng/ml, R&D Systems), (2) heat-killed S. aureus (107 cells/ml, InvivoGen), (3) S. aureus (107 cells/ml) isolated from the subject’s own nasal swab (“Carrier Strain”) (i.e., only carriers were stimulated using their own carriage isolate), (4) a mixture (107 cells/ml) of all S. aureus isolates obtained from carrier subjects (“Carrier Mix”), (5) IL-1β (2 ng/ml, R&D Systems), (6) PAM3CSK4 (10 ng/ml, InvivoGen), (7) CpG (1.75 μM, InvivoGen), or (8) un-stimulated, for the same time period. Due to the limited cell numbers that could be obtained from each pediatric donor, each treatment and control group/participant were carried out as singletons. After the incubation period, cells were washed with PBS and lysed using RLT buffer (Qiagen) supplemented with 1% β-mercaptoethanol (Sigma). Cell lysates were stored at −80°C until RNA extraction. One of the donor’s PBMCs were lost viability during processing leaving 14 persistent and 15 non-carriers of S. aureus for further analysis.
2.4. RNA extraction and the targeted assay (TA)
RNA was extracted from PBMCs using the RNeasy Mini Kit (Qiagen) and from blood tempus mix tubes using MagMAX™−96 Total RNA Isolation Kit (Applied Biosystems, ThermoFisher Scientific). RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
After conversion of RNA to cDNA (50 ng total RNA from PBMCs and 100 ng total RNA from WB) (no globin reduction was performed) targeted genes were amplified and analyzed with Fast Gene Expression Analysis using EVAGreen® on the BioMark™ HD System (Fluidigm, San Francisco, CA).
Raw CT values were exported from Fluidigm Real-Time PCR Analysis (version 3.1.3) and ΔCT and ΔΔCT values were calculated using R (version 3.4.1). Genes or samples that had a fail call rate of 50% or more were excluded from downstream analyses (all samples and genes passed this criterion in the experiments included in this study).
Transcripts differentially regulated upon stimulation were defined based on a minimum 2-fold change (up- or down-regulation) compared to respective unstimulated samples. Differentially-expressed genes were tested using the Student’s t-test and the Fligner-Killeen test was used to test variance. Differences between genes between a commercial S. aureus strain, the unique carrier strain, and the mix of all unique carrier strains were tested using paired t-tests and the paired variance test. To determine if a gene was expressed or not in a module for the TA, fold change cutoffs were applied (log2 FC > ±2). Differences in module activity in the TA was tested using a mixed linear model (the lme function from nlme package in R) with age, gender, and carrier history added as fixed effects and family as random effect. Heat maps were generated using R (version 3.4.1).
2.5. RNA sequence processing
RNAseq was performed on a representative subset of the samples from the respective S. aureus carriage phenotype groups. Donors were randomly selected from each group based on how well they represented the average response of the group in the TA, aiming for age and gender matched samples. A total of 6 donors, 3 carriers (2 females and 1 male age 4, 5, and 10 years, respectively) and 3 non-carriers (2 females, 1 male, ages 2, 4, and 11 years) were treated as described above (together with their untreated matched samples) and analyzed. Samples were subjected to cell deconvolution using xCell to estimate the cell composition in each RNAseq sample. No detectable differences were observed (data not shown) [29].
Sequencing libraries were constructed from total RNA using TruSeq RNA Sample Preparation Kits v2 (Illumina, San Diego, CA). Libraries were clustered onto a flowcell using a cBOT amplification system with a HiSeq SR v3 Cluster Kit (Illumina). Single-read sequencing was carried out on an Illumina HiScanSQ sequencer using a HiSeq SBS v3 Kit to generate 100-base single-end reads with a target of approximately 10 million reads per sample. The raw reads were processed in Galaxy using TopHat (with bowtie) for alignment (GRCh37 as reference genome), BAM-to-SAM, Picard Alignment Summary Metrics, and Picard RNAseq Metrics. Genes were quantified using htseq-count. Raw counts were used for downstream analysis and Fragments Per Kilobase Million (FPKM) values were used for plotting. FPKMs s for genes were obtained using edgeR [30, 31].
2.6. RNAseq analysis
Raw counts were used for downstream analysis and FPKM values were used for plotting. To identify differentially-expressed genes, a general linear model using negative binomial distribution with no random effects was applied using edgeR [30–32]. Genes having a false discovery rate (FDR) <0.05 were considered differentially expressed.
Defining altered responses to live bacteria was done in one of two ways: (1) by applying the edgeR model with an FDR <0.05 as cutoff or (2) by calculating the difference between the log2FC values of live bacteria from non-carriers and carriers and applying standard deviation (SD) cutoffs. Normal variation was assumed to be ±2 SD of the mean. All analyses and calculations were performed using R (version 3.4.1). Functional analysis and network enrichments of the differentially expressed genes and the genes identified as showing altered responses between live and dead bacteria were done using Pathway Studio® (version 12.0.1.9, Elsevier).
2.7. Analysis of module-level data
The modules are designed based on uniform expression of genes within respective modules. Therefore, module activity was calculated as the difference between percent up- and down-regulated genes within each module. To obtain module results for the RNAseq data we first converted the Illumina microarray probe ID for each gene included in the modules to Ensembl gene IDs. Some probes shared Ensembl IDs and were dealt with as follows: i) multiple probes/Ensembl Gene IDs in the same module were kept as one input and ii) multiple probes/Ensembl Gene ID in different modules can appear in multiple, different modules. This was done to avoid picking the “best” fit. To determine if a gene was expressed or not in a module for the TA, fold change cutoffs were applied (log2 FC > ±2) and for the RNAseq data, the DEG cutoff FDR<0.05 was used.
2.8. Statistics
To compare demographic characteristics between the two patient groups univariate statistics were used, chi-squared test for categorical variables (Fisher’s exact p-value if the comparison included a frequency ≤5) or ANOVA for continuous variables using Stata 14.
Differentially-expressed genes from the TA experiment were tested using the Student’s t-test and paired t-tests when suitable. Differences in module activity in the TA was tested using Student’s t-test and paired t-tests when suitable. As the groups were balanced regarding age and gender, these factors were not included as factors in the tests. All p-values were False Discovery Rate (FDR) adjusted and all analyses were done in R (version 3.4.1).
To identify differentially-expressed genes from the RNAseq experiment, a general linear model using negative binomial distribution with no random effects was applied using edgeR [30–32]. Genes having a FDR <0.05 were considered differentially expressed.
2.9. Data availability
The data described in this publication have been deposited in NCBI’s Gene Expression Omnibus [33] and are accessible through GEO Series accession number GSE153122 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153122).
3. Results
3.1. Pediatric S. aureus carriage study population
Five persistent carriers had prior medical histories of culture-positive Staphylococcal infections compared to non-carriers that lacked any infection history (p=0.042) (Table 1). No participant had an S. aureus infection at the time of enrollment. Participants enrolled in the present study were at the clinic for either a viral upper respiratory infection, athletes’ foot, a wound needing treatment, eczema, abdominal pain, or were at the clinic for their well child checkup. MRSA (methicillin resistant S. aureus) persistent carriers were more likely to have developed an S. aureus infection than MSSA (methicillin susceptible S. aureus) persistent carriers (75% vs. 9%, respectively, p=0.033). Most persistent carriers harbored MSSA (n=12; 80%) and 3 (20%) were MRSA persistent carriers. S. aureus isolates recovered from 12 persistent carriers were available for our stimulation assay (Table 2). PBMCs harvested from these 12 carriers were stimulated with their respective, live carriage isolate. In addition, the 12 isolates were then combined to form the ‘Carrier Mix’ for the stimulation of PBMCs (Table 2).
3.2. PBMC responses to different stimuli
We used the Targeted Assay (TA, details on development and testing can be found in the Supplementary Results) to assess gene expression profiles associated with innate immunity by stimulating PBMCs harvested from pediatric persistent nasal carriers or non-carriers of S. aureus with various stimuli.
Although the responses observed to the various stimuli differed, responses to respective stimuli were similar between groups (Fig. 1 and Fig. S5). Differences in responses observed between persistent carriers and non-carriers were observed for M7.5 following PAM3 stimulation (p=0.04, Fig. S5), M2.5 following stimulation with the Carrier Mix (p=0.01, Fig. 1), and M2.13 following CpG stimulation (p=0.004, Fig. S5). Some modules showed large donor-to-donor variation (e.g., M1.6 showed donor to donor variation for all stimulations) while other module responses were relatively stable across donors (e.g., M5.4 and M8.2 in the Carrier Mix panel). M8.2 is down-regulated across all donors after Carrier Mix stimulation but up-regulated after PAM3 stimulation (Fig. 1). The other stimulation profiles showed donor to donor variation. For example, M5.4 is up-regulated in all donors after Carrier Mix stimulation, but only in a few donors following stimulation with PAM3 or other stimuli, and responses in M1.2 varied between donors following stimulation with either Carrier Mix (Fig. 1), TNF, or CpG, whereas M1.2 responses are up-regulated following stimulation with either PAM3, IL-1β, or HKSA (Fig. S5).
Fig. 1.
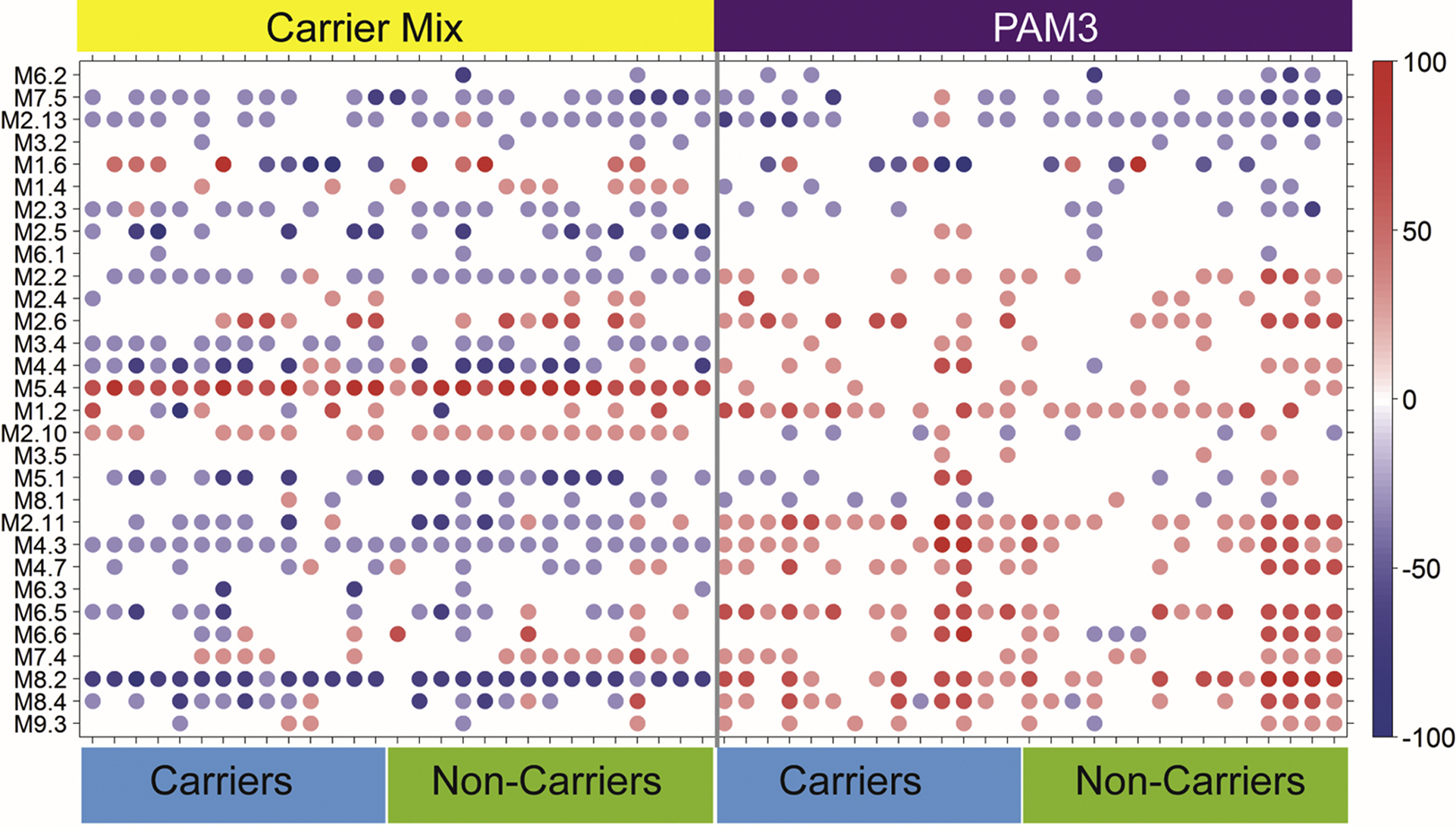
Responses to a 6 h stimulation with either a mix of all clinical carrier strains (Carrier Mix) or PAM3 are similar between S. aureus carrier status groups. The module plot represents the modular activity of each donor for the 30 TA modules. Spot intensity indicates changes in transcript abundance from baseline. No clustering was applied to either samples or modules. Intensity for each module represents the percent of module probes that are up (red) or down (blue) regulated in respective samples. The modular responses are different between the mix of all clinical carrier strains and PAM3, but no differences were found between carriers and non-carriers. Some modules show larger donor-to-donor variations (e.g., M1.6) while others are relatively stable across donors (e.g., M5.4 and M8.2 in the Carrier Mix panel).
The responses to the Carrier Strain and the Carrier Mix were, although not statistically significant, slightly different both at a gene level (Fig. 2A) and the module level (Fig. 2B). Some modules showed large donor-to-donor variation (e.g., M1.6, and M1.2) while others were relatively stable across donors (e.g., M5.4, M4.3, and M8.2) pointing to the relative importance of these modules following stimulation with bacterial antigens. When looking at the module responses in an individual stimulated with the Carrier Mix compared to the response elicited following stimulation with their respective carrier strain, small and subtle differences were observed (Fig. 2B). Although not statistically significant, these differences still may contribute to the understanding persistent carriage of specific S. aureus isolates.
Fig. 2.
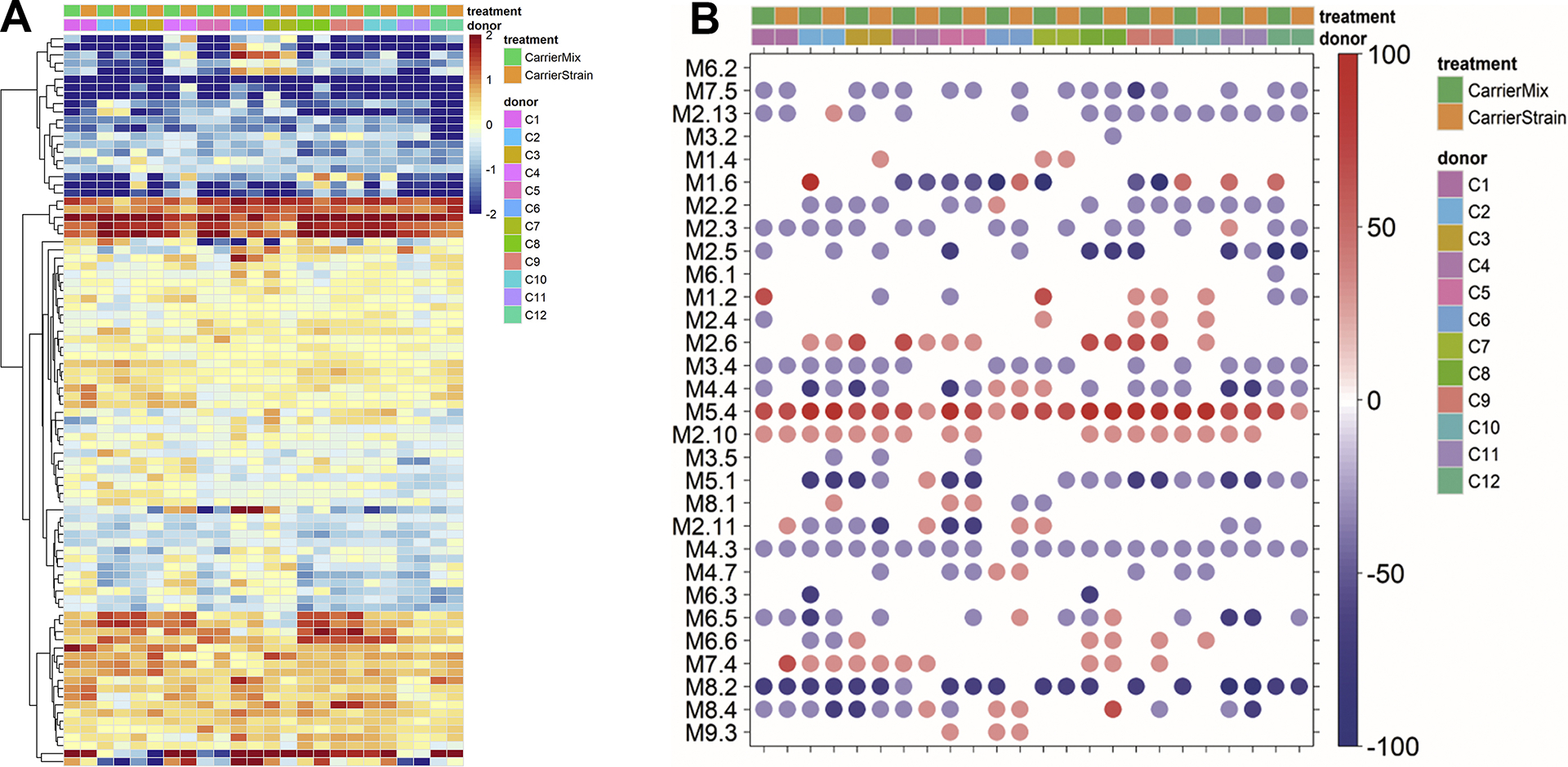
Responses by PBMCs isolated by persistent carriers to donor specific clinical carrier strains or the mix of all clinical carrier strains following stimulation for 6 h. (A) Heat map comparing gene expression profiles after stimulation with either the donor specific clinical carrier strain or a mix of all clinical carrier strains. Heat maps represent a hierarchical clustering of transcripts responding to each stimulus (compared to non-stimulated self). Changes versus the non-stimulated condition are represented by a color scale: red = up regulated; blue = down regulated; yellow = no change. Some donors show more variability than others. (B) The module plot represents the modular activity of each donor for the 30 TA modules. Spot intensity indicates changes in transcript abundance from baseline. No clustering was applied to either samples or modules. Intensity for each module represents the percent of module probes that are up (red) or down (blue) regulated in a sample. Some modules show larger donor-to-donor variation (e.g., M1.6) while others are relatively stable across donors (e.g., M5.4 and M8.2) suggesting the relative importance of these modules in bacterial responses.
3.3. PBMC responses to live or dead bacteria
Clear differences were observed in responses elicited following stimulation with heat-killed S. aureus compared to stimulation with live carriage isolates (Fig. 3 and Table 3). Modules showing statistically significant differences in gene expression profiles following stimulation with either heat-killed or live bacteria were annotated as “Type I IFN” (M1.2, M2.2, M2.6, M5.4), “Inflammation” (M4.3, M8.4, M2.11), “Apoptosis” (M4.4), “Transcriptional Regulation” (M7.4, M5.1, M2.10, M1.4), “Phagocytosis” (M2.5, M8.2), or none or poorly defined annotations (M2.3, M2.13, M3.4, M4.7, M6.5, M7.5). Only 9 of 30 modules were not different following stimulation with either live or heat-killed bacteria and 16 modules showed a consistent pattern in both carriers and non-carriers. In addition, seven modules (M1.2, M1.4, M1.6, M2.6, M2.10, M4.7, and M5.1) showed small differences to stimulation with live bacteria between carriers and non-carriers (Fig. 3). Especially interesting are the small differences observed in the expression profiles observed for modules M1.4 and M1.6 (the two modules from the poly [I:C] specific cluster) that were up-regulated in non-carriers stimulated with live bacteria, but down-regulated after heat-killed bacteria stimulation. The expression profiles for these same modules were unchanged when PBMCs harvested from persistent carriers were stimulated with either live or dead bacteria (Fig. 3). Recently, induction of IFNβ via a TIR-domain-containing adaptor protein (TRIF)-dependent pathway [34] following stimulation with live pathogenic bacteria was identified as a potential mechanism for distinguishing between stimulation with either live or dead organisms or between pathogenic or commensal bacteria [34]. This suggests that individuals susceptible to persistent carriage with S. aureus may possess differences in their live/dead bacteria recognition pathway.
Fig. 3.
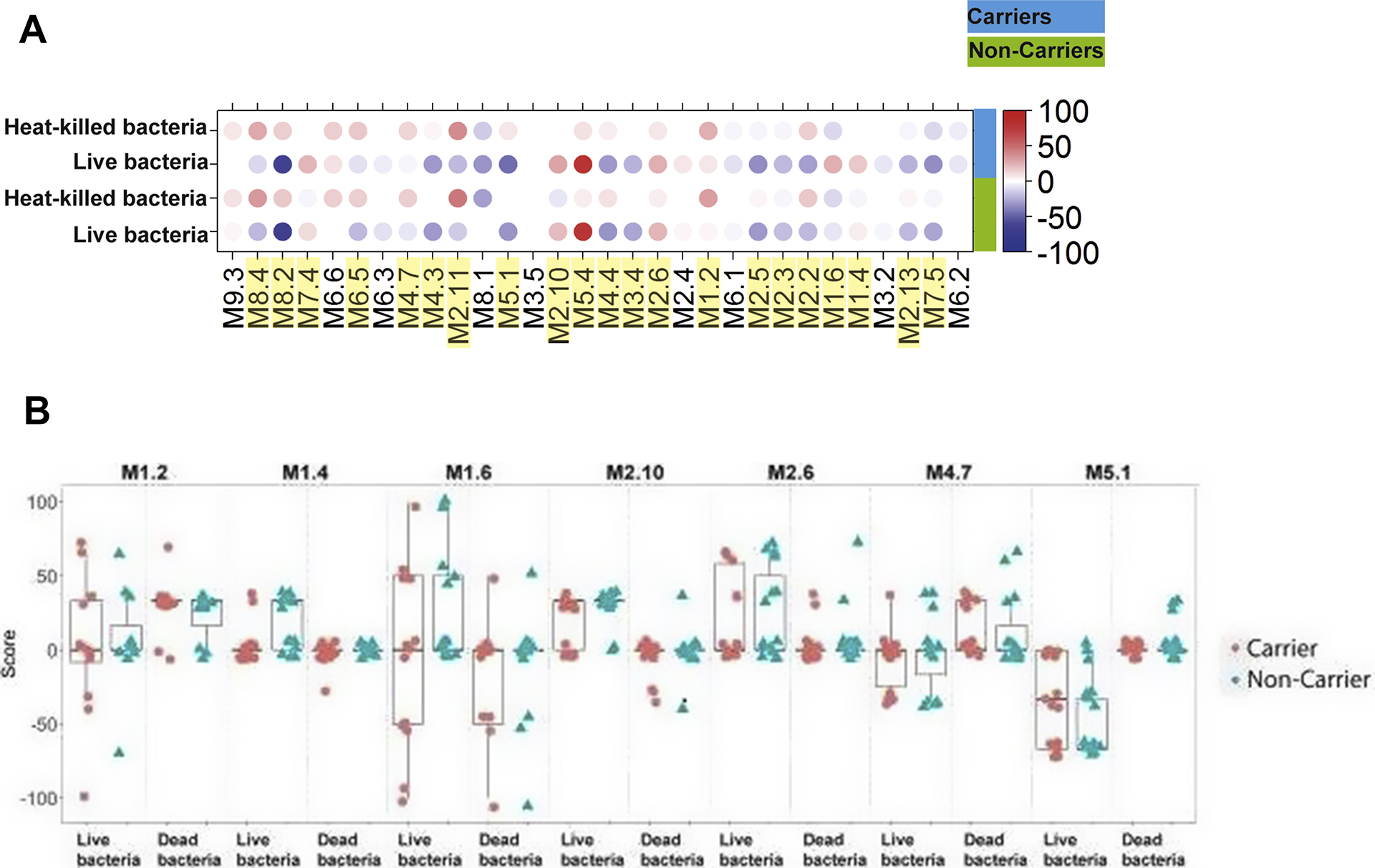
Responses to live verses dead bacteria. (A) The module plot represents the mean modular activity of each donor group for the 30 TA modules. Spot intensity indicates changes in transcript abundance from baseline. No clustering was applied to either samples or modules. Intensity for each module represents the percent of module probes that are up (red) or down (blue) regulated in a sample. Yellow highlights denote modules where the module score is statistically significantly different between live and dead bacteria in at least one of carriers and non-carriers, or both groups combined (B) The individual module score for the 7 modules where Carriers showed a different pattern in module scores than seen in Non-carriers.
Table 3.
Differences in module scores between heat-killed and live bacteria.
| Module | Non-carriers | Persistent Carriers | Combined |
|---|---|---|---|
| P-value | P-value | P-value | |
| M9.3 | 0.2821 | 0.2803 | 0.1175 |
| M8.4 | 0.0028 | 0.0002 | 6.81E-07 |
| M8.2 | 1.59E-08 | 4.99E-09 | 5.24E-18 |
| M7.4 | 0.0012 | 0.0344 | 4.92E-05 |
| M6.6 | 0.4344 | 0.1985 | 0.1368 |
| M6.5 | 0.0184 | 0.0022 | 0.0001 |
| M6.3 | 0.1828 | 0.2088 | 0.0741 |
| M4.7 | 0.0742 | 0.0170 | 0.0026 |
| M4.3 | 6.95E-11 | 4.35E-07 | 1.89E-20 |
| M2.11 | 4.01E-05 | 5.38E-06 | 2.53E-10 |
| M8.1 | 0.0740 | 0.5400 | 0.7963 |
| M5.1 | 4.85E-06 | 0.0040 | 2.16E-08 |
| M3.5 | NA | NA | NA |
| M2.10 | 7.50E-06 | 0.0002 | 9.22E-10 |
| M5.4 | 1.72E-10 | 2.07E-08 | 1.93E-19 |
| M4.4 | 0.0046 | 0.0024 | 1.27E-05 |
| M3.4 | 0.0001 | 0.0002 | 1.06E-08 |
| M2.6 | 0.0833 | 0.0622 | 0.0073 |
| M2.4 | 0.1041 | 0.6070 | 0.1175 |
| M1.2 | 0.1078 | 0.1289 | 0.0204 |
| M6.1 | 0.1815 | 0.3792 | 0.1129 |
| M2.5 | 0.0041 | 0.0146 | 0.0001 |
| M2.3 | 0.0111 | 0.0240 | 0.0003 |
| M2.2 | 3.65E-08 | 0.0002 | 4.75E-12 |
| M1.6 | 0.0255 | 0.8528 | 0.1175 |
| M1.4 | 0.0015 | 0.2088 | 0.0006 |
| M3.2 | 0.0625 | NA | 0.0598 |
| M2.13 | 0.0027 | 0.0011 | 3.00E-06 |
| M7.5 | 0.0028 | 0.0034 | 1.63E-05 |
| M6.2 | 0.7374 | NA | 0.7645 |
FDR corrected P-values for each module. p-values in bold are <0.05. NA indicates that the tested values were identical so the t-test did not return a significant p-value.
To obtain a better understanding of the different responses observed following stimulation with live versus dead bacteria between persistent carriers and non-carriers, we performed RNAseq on unstimulated cells, cells stimulated with heat-killed bacteria, and cells stimulated with live bacteria from a subset of the donors representing the average response of the respective groups (Fig. S6). A principal component analysis (PCA) shows that samples stimulated with live bacteria are different from those stimulated with dead bacteria or left unstimulated (Fig. 4A). Furthermore, differential expression of genes (Table 4 and Table S4) and modules (Fig. 4B) is greater following stimulation with live bacteria compared to stimulation with heat-killed bacteria. Furthermore, non-carriers and persistent carriers differ in their module responses and differences between persistent carriers and non-carriers detected by the targeted assay are more pronounced when using RNAseq (Fig. 4C).
Fig. 4.
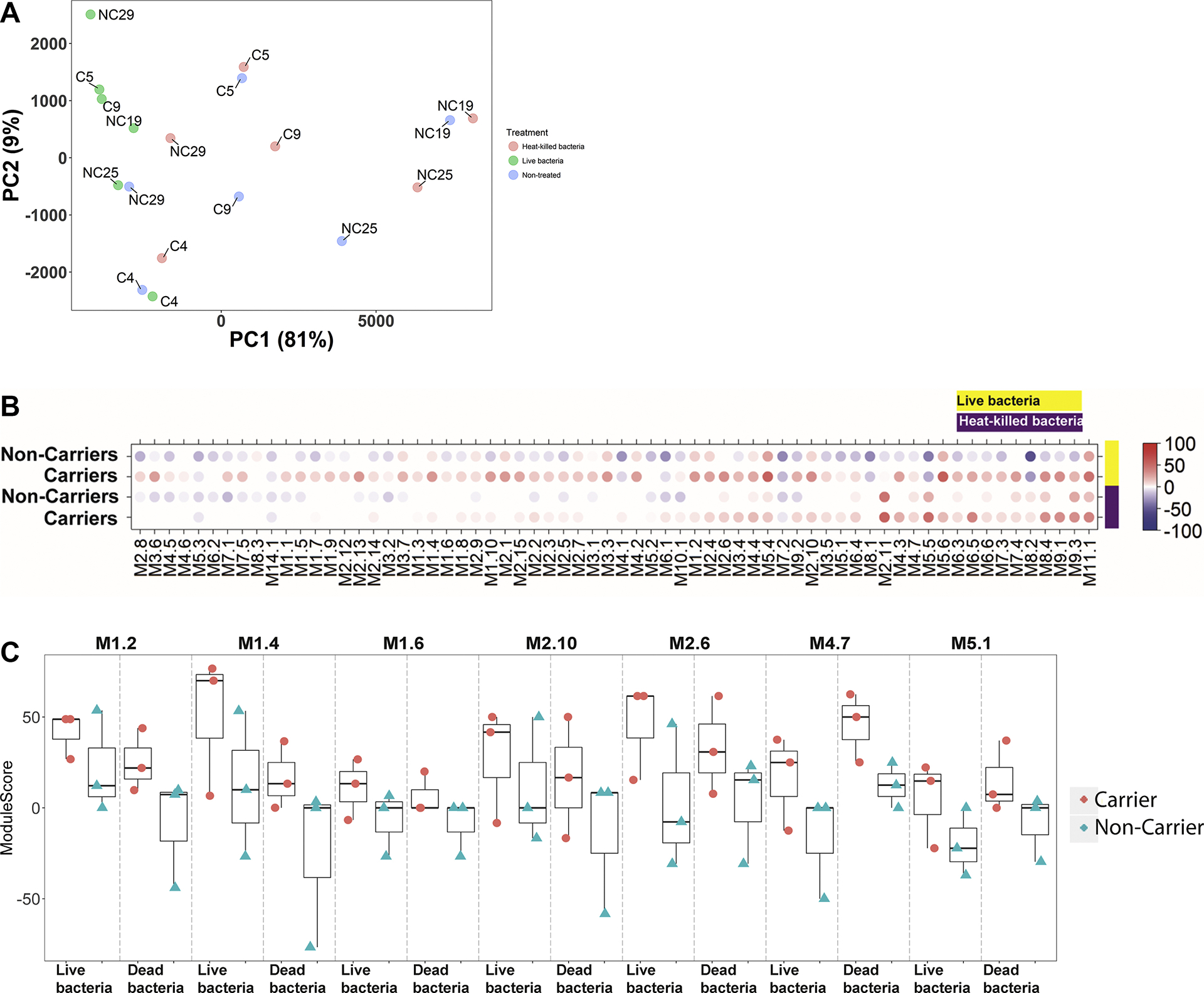
Principle Component Analysis plot and Module plot. (A) Principal Component Analysis (PCA) plot showing variability in the data. Treatments are indicated by different colors: blue = NS [not stimulated], red = Heat-killed bacteria, green = Live bacteria. (B) Mean modular activity of each donor group for the 30 TA modules. Spot intensity indicates changes in transcript abundance from baseline. No clustering was applied to either samples or modules. Intensity for each module represents the percent of module probes that are up (red) or down (blue) regulated in a sample. (C) The individual module scores for the modules identified in Fig.3A–B.
Table 4.
Differentially expressed genes from RNAseq analysis.
| Donor group | Comparison | FDR<0.05 | Top 5 Biological functions from Pathway Studio |
|---|---|---|---|
| Persistent Carrier | Live bacteria vs. NS | 5171 | Microtubule Cytoskeleton Centriole Duplication and Separation P2RXs -> Synaptic Transmission Kynurenine Metabolites in Neurotoxicity and Neuroprotection SWI/SNF BRG1/BAF in Chromatin Remodeling |
| Persistent Carrier | HKSA vs. NS | 101 | mRNA Transcription and Processing Epinephrine/Norepinephrine SWI/SNF BRG1/PBAF in Chromatin Remodeling Mast-Cell Activation without Degranulation through CYSLTR1/CYSLTR2 Signaling Histone Methylation |
| Non-carrier | Live bacteria vs. NS | 6535 | Golgi to Endosome Transport SRCAP in Chromatin Remodeling Actin Cytoskeleton PTGER2/3 -> Inflammation-Related Expression Targets Synaptic Potentiation by PKMZ in LTP Maintenance - Basal State |
| Non-carrier | HKSA vs. NS | 20 | Telomere Maintenance Single-Strand Nucleotide Excision Tight Junction Assembly (JAMs) Neutrophil Recruitment and Priming SIRT2 Signaling in Aging |
We were next interested in identifying genes differentially activated in carriers and non-carriers. Because we hypothesized that differences would be minor due to our small data set, we tried two different approaches aimed at identifying potential differences: (1) statistical testing using a general linear model using negative binomial distribution with no random effects (edgeR) and (2) measuring the difference between the log2FC of persistent and non-carriers of S. aureus and applying standard deviation cutoffs to identify groups of genes with a difference greater than normal variation. This could be due to a difference in responses to live bacteria in the different donor groups. When applying these approaches to our data, 20 genes were identified as different between carriers and non-carriers when using the statistical model (Table 5) and a total of 367 (only including genes passing FDR for either or both groups) genes showed an altered profile for persistent carriers compared to non-carriers when applying a 2 ± SD difference (Fig. 5 and Table S5). The overlap between the two different gene lists were rather low, only 5 genes were found with both methods (C5AR1, SIGLEC9, MIR210HG, SLC22A15, GK-AS1), but the remaining genes were all close to the SD cutoff used for the log2FC differences (Table S5).
Table 5.
Differentially expressed genes for live vs. dead S. aureus between persistent and non-carriers of S. aureus.
| Gene | EnsemblGeneID | Log2FC | FDR |
|---|---|---|---|
| C5AR1 | ENSG00000197405 | −2.3 | 0.007 |
| CPEB4 | ENSG00000113742 | −0.9 | 0.007 |
| C1orf122 | ENSG00000197982 | −1.2 | 0.007 |
| SIGLEC9 | ENSG00000129450 | −2.3 | 0.016 |
| MIR210HG | ENSG00000247095 | 2.8 | 0.016 |
| EMILIN2 | ENSG00000132205 | −1.4 | 0.016 |
| APOBR | ENSG00000184730 | −1.1 | 0.016 |
| SLC22A15 | ENSG00000163393 | −2.7 | 0.024 |
| MT1XP1 | ENSG00000233929 | −2.0 | 0.024 |
| GK-AS1 | ENSG00000243055 | −4.2 | 0.024 |
| PLA2G4C | ENSG00000105499 | −1.9 | 0.024 |
| RAB31 | ENSG00000168461 | −1.0 | 0.024 |
| RP1–232L22_B.1 | ENSG00000178146 | −1.2 | 0.024 |
| FTH1P20 | ENSG00000226564 | −0.91 | 0.024 |
| ARNTL2 | ENSG00000029153 | −1.78 | 0.026 |
| MFSD1 | ENSG00000118855 | −0.77 | 0.026 |
| CTSZ | ENSG00000101160 | −0.94 | 0.029 |
| FTH1 | ENSG00000167996 | −0.95 | 0.029 |
| CD63 | ENSG00000135404 | −0.88 | 0.043 |
| IGF2R | ENSG00000197081 | −0.76 | 0.046 |
Fig. 5.
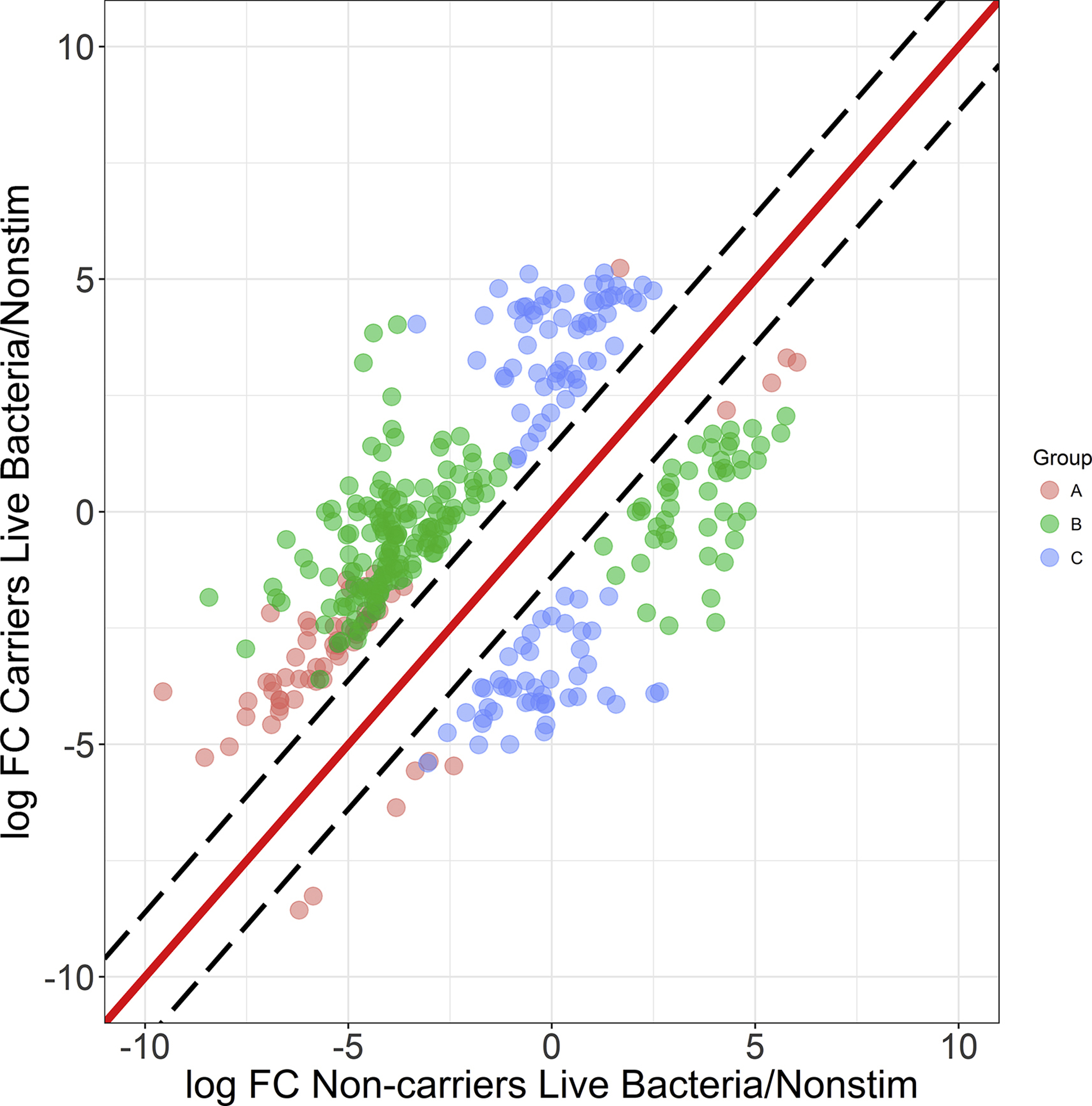
Defining the variation between carrier and non-carrier gene signatures after co-culture with live bacteria. Black dashed lines indicate the 2 ± SD interval (assumed to be normal variation), grey dashed lines indicate 3 ± SD. All points indicate genes outside the normal variation and with an FDR-corrected p-value <0.05 in at least one donor group (live bacteria vs. non-stimulated) (n = 367). Three groups of genes can be identified based on the cutoffs they pass: Both: 2 ± SD and FDR<0.05 for both non-carriers and carriers (red points, n = 60), Non-carrier: 2 ± SD and FDR<0.05 for non-carriers but not persistent carriers (blue points, n= 198), Carriers: 2*SD and FDR<0.05 for persistent carriers but not non-carriers (green points, n = 109).
4. Discussion
Interrogation of the human genome can be carried out by conducting analyses that define either its architecture or measure its output. Genome-wide association studies represent an example of the former, designed to identify genes or pathways that are determined by heredity and do not change over time that may be associated with various immune-related or chronic conditions or infectious diseases [35–40]. By contrast, genetic approaches that measure transcript abundance e.g. the TA, are largely affected by environmental stimuli such as infection/colonization status.
The modular transcriptional repertoire analysis pioneered by our laboratories was used as a basis for the development of the TA and used to identify differences in the transcriptome between pediatric individuals determined to be either persistent nasal carriers or nasal non-carriers of S. aureus [21, 23, 26, 41, 42]. The module concept is based on co-expression of genes that may or may not be related and individual genes represented in respective modules may not necessarily be directly involved with the function of the modules. This assay is meant to be analyzed and interpreted at the modular level where individual genes comprising each module contribute to a profile that can be used to understand potential defects in innate immune responsiveness, not as a tool to pinpoint the exact gene responsible for respective phenotypes.
The modules used in the present study were specifically developed for analyses of stimulated whole blood [26], but the underlying framework is the same as what has been previously described [41]. More importantly, the TA described in the present report allowed for frozen PBMCs to be analyzed, producing results indistinguishable from similarly treated fresh whole blood.
Gene expression profiles that differed between persistent carriers and non-carriers were observed in M2.13 (cytoskeleton, cell cycle), M2.5 (phagocytosis, phagosomes, respiratory burst), and M7.5 (IL-8, differentiation) following stimulation with CpG, Carrier Mix, and PAM3, respectively. When PBMCs isolated from persistent carriers were stimulated with either their respective live carrier strains or the Carrier Mix, differences in expression profiles, although not statistically significant, differed both at the gene and module levels suggesting that host responses to this organism could be impacted by the stimulating strain/strains. This indicated that an intimate association exists between persistent carriers and their respective persistent carriage isolates.
Differences in gene expression profiles were also observed between persistent carriers and non-carriers following stimulation with either heat-killed or live bacteria. A change in gene expression profiles of non-carriers was observed in modules associated with type I interferons, inflammation, apoptosis, transcriptional regulation, or phagocytosis (M1.4, M1.6) when stimulated with live bacteria but not with dead bacteria. Surprisingly, there was little change in the expression profile observed for persistent carriers when stimulated with either live or dead bacteria. Additional RNAseq analyses were then conducted to uncover additional differences in responses to live/dead bacteria between persistent and non-carriers by comparing unstimulated cells, cells stimulated with heat-killed bacteria, and with live bacteria on a subset of participants. This analysis also uncovered differences in gene expression profiles between persistent carriers and non-carriers. Specifically, unique process networks for non-carriers were related to IL-1 and IL-6 signaling, apoptosis, inflammasome related regulators, and inflammation compared to IL-5 signaling, apoptosis, and basophil related processes associated with persistent carriers. This suggested that persistent carriers may have alterations in their ability to respond to Vita-PAMPs (pathogen associated molecular patterns) that may have contributed to persistent colonization with S. aureus [34, 43].
Vita-PAMPs are closely related to PAMPs in that they activate the innate immune system following recognition by the host’s pattern recognition receptors (PRRs), e.g. Toll-like receptors (TLRs). Vita-PAMPs such as bacterial mRNA, pyrophosphates, quorum sensing molecules, bacterial second messengers, and isopyrenoids, however, serve as microbial signatures of viability, resulting in heightened innate immune activation following recognition by cognate PRRs [34, 43, 44]. For example, phagocytosis of live pathogens expressing both PAMPs and Vita-PAMPs, but not dead pathogens expressing PAMPs alone resulted in the activation of a TRIF-dependent signaling pathway that triggered inflammasome activation and subsequent caspase-1 mediated production of INFβ [34]. Activation of the inflammasome eventually results in a highly inflammatory form of cell death (pyroptosis) that is beneficial to pathogen containment/clearance since the release of endocytosed organisms results in more efficient clearance by neutrophils [45].
TLRs and other PRRs are the ‘guardians at the gate’ that define whether or not an immune response will be initiated by activated dendritic cells, and depending on the nature of the stimulus these receptors will alter the type and quality of the adaptive response that develops. Ten TLRs have been described in humans and each TLR homodimer or heterodimer combination (e.g., TLR1:TLR2 and TLR2:TLR6) recognize unique PAMPs. TLR2 is particularly important in the recognition of S. aureus lipopeptides/lipoteichoic acid and individuals with mutations to adaptor molecules such as MAL (MyD88-adaptor-like), required to initiate activations signals delivered via TLR2, are highly predisposed to S. aureus infections [17, 18]. All TLRs described to date utilize the MyD88/MAL signaling adaptor molecules with the exception of TLR4 that can also bind to the TRIM/TRAM adaptor molecule complex at the membrane of endosomal compartments [46]; however, data suggested that TRIM/TRAM can also serve as an adaptor protein in TLR2:TLR6 signaling [47]. Although TLR4 has long been recognized as the receptor for the Gram negative PAMP lipopolysaccharide (LPS) as well as a receptor for host-derived danger associated molecular patterns (DAMPs), it has also been shown to play a role in immune responses to Gram positive bacteria that generate lipoproteins and lipoteichoic acids typically recognized by TLR2 [46, 48]. For example, TRAM-deficient macrophages infected with either herpes simplex virus or S. aureus displayed impaired type I interferon responses suggesting a role for TRAM in TLR2-dependent responses. Further complicating the TLR2 (Gram positive)/TLR4 (Gram negative) dichotomy are data demonstrating that almost half of mice deficient in TLR4 died from an S. aureus infection suggesting that both TLR2 and TLR4 are important to abscess containment in addition to coordination of the T-cell response and mediating wound healing [18, 49]. In addition, TLR4 knockdown modulated the inflammatory response to S. aureus by down-regulating TNFα concentrations suggesting that both TLR2 and TLR4 contributed to the anti-S. aureus response [50, 51].
Our results suggested that although there are no or only few statistically significant differences between responses observed between persistent carriers and non-carriers following stimulation with either TLR ligands or bacteria, our data suggest that there may be potential alterations in the previously described Vita-PAMP pathway among S. aureus persistent carriers. These alterations appear to be localized to the TRIF/TRAM-related pathway suggested to be part of the recognition of live versus dead bacteria [52]. The implications of these potential differences in TRIF/TRAM-related pathway activation between persistent carriers and non-carriers indicated that although the persistent carriers included in this study are fully capable of recognizing and responding to TLR ligands and dead bacteria, their responses to live bacteria are different and may be one determinant resulting in persistent colonization with this organism.
That the persistent carrier phenotype may be the result of altered signaling via the TRIF/TRAM-related pathway may seem contradictory to data that clearly demonstrates persistent carriers make a more robust antibody response to S. aureus virulence factors compared to intermittent and non-carriers [7, 53, 54]. But, how can a potential ‘defect’ or polymorphism(s) in innate immune responses result in a more robust/focused acquired response? One explanation is that the altered innate response observed in persistent carriers prevents S. aureus clearance leading to persistent colonization that over time results in the activation of acquired immune responses that culminate in the production of neutralizing antibodies to an array of staphylococcal adhesins and virulence factors [21, 51, 53, 55]. By contrast, TRIF/TRAM signaling in intermittent and non-carriers may more effectively prevent or reduce the level of colonization thereby sequestering S. aureus antigens from acquired immune responses. The possibility that persistent carriers develop a better focused acquired immune response may help explain why persistent carriers although more likely to become infected with their colonizing strain, are also at a reduced risk of death from bacteremia resulting from S. aureus infection [7, 53–56].
Supplementary Material
Acknowledgments:
This project was funded by the National Institutes of Health grant AI085014 and the Kleberg Foundation to E.L. Brown
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors declare no conflicts of interest.
References
- [1].Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal innate immune evasion. Trends Microbiol 2005;13:596–601. [DOI] [PubMed] [Google Scholar]
- [2].de Jong NWM, van Kessel KPM, van Strijp JAG. Immune Evasion by Staphylococcus aureus. Microbiol Spectr 2019;7. [DOI] [PubMed] [Google Scholar]
- [3].Sakr A, Bregeon F, Mege JL, Rolain JM, Blin O. Staphylococcus aureus Nasal Colonization: An Update on Mechanisms, Epidemiology, Risk Factors, and Subsequent Infections. Front Microbiol 2018;9:2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 1997;10:505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nouwen JL, Ott A, Kluytmans-Vandenbergh MF, Boelens HA, Hofman A, van Belkum A, et al. Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a “culture rule”. Clin Infect Dis 2004;39:806–11. [DOI] [PubMed] [Google Scholar]
- [6].Brown EL, Below JE, Fischer RS, Essigmann HT, Hu H, Huff C, et al. Genome-Wide Association Study of Staphylococcus aureus Carriage in a Community-Based Sample of Mexican-Americans in Starr County, Texas. PLoS One 2015;10:e0142130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, et al. Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis 2009;199:1820–6. [DOI] [PubMed] [Google Scholar]
- [8].Nouwen J, Boelens H, van Belkum A, Verbrugh H. Human factor in Staphylococcus aureus nasal carriage. Infect Immun 2004;72:6685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ruimy R, Angebault C, Djossou F, Dupont C, Epelboin L, Jarraud S, et al. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis 2010;202:924–34. [DOI] [PubMed] [Google Scholar]
- [10].Emonts M, de Jongh CE, Houwing-Duistermaat JJ, van Leeuwen WB, de Groot R, Verbrugh HA, et al. Association between nasal carriage of Staphylococcus aureus and the human complement cascade activator serine protease C1 inhibitor (C1INH) valine vs. methionine polymorphism at amino acid position 480. FEMS Immunol Med Microbiol 2007;50:330–2. [DOI] [PubMed] [Google Scholar]
- [11].Emonts M, Uitterlinden AG, Nouwen JL, Kardys I, Maat MP, Melles DC, et al. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J Infect Dis 2008;197:1244–53. [DOI] [PubMed] [Google Scholar]
- [12].van Belkum A, Emonts M, Wertheim H, de Jongh C, Nouwen J, Bartels H, et al. The role of human innate immune factors in nasal colonization by Staphylococcus aureus. Microbes Infect 2007;9:1471–7. [DOI] [PubMed] [Google Scholar]
- [13].van den Akker EL, Nouwen JL, Melles DC, van Rossum EF, Koper JW, Uitterlinden AG, et al. Staphylococcus aureus nasal carriage is associated with glucocorticoid receptor gene polymorphisms. J Infect Dis 2006;194:814–8. [DOI] [PubMed] [Google Scholar]
- [14].Sollid JU, Furberg AS, Hanssen AM, Johannessen M. Staphylococcus aureus: determinants of human carriage. Infect Genet Evol 2014;21:531–41. [DOI] [PubMed] [Google Scholar]
- [15].Nurjadi D, Herrmann E, Hinderberger I, Zanger P. Impaired beta-defensin expression in human skin links DEFB1 promoter polymorphisms with persistent Staphylococcus aureus nasal carriage. J Infect Dis 2013;207:666–74. [DOI] [PubMed] [Google Scholar]
- [16].Panierakis C, Goulielmos G, Mamoulakis D, Maraki S, Papavasiliou E, Galanakis E. Staphylococcus aureus nasal carriage might be associated with vitamin D receptor polymorphisms in type 1 diabetes. Int J Infect Dis 2009;13:e437–43. [DOI] [PubMed] [Google Scholar]
- [17].Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 2011;11:505–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Miller LS, Fowler VG, Shukla SK, Rose WE, Proctor RA. Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol Rev 2020;44:123–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].de Vor L, Rooijakkers SHM, van Strijp JAG. Staphylococci evade the innate immune response by disarming neutrophils and forming biofilms. FEBS Lett 2020. [DOI] [PubMed] [Google Scholar]
- [20].Projan SJ, Nesin M, Dunman PM. Staphylococcal vaccines and immunotherapy: to dream the impossible dream? Curr Opin Pharmacol 2006;6:473–9. [DOI] [PubMed] [Google Scholar]
- [21].Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007;109:2066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Banchereau R, Jordan-Villegas A, Ardura M, Mejias A, Baldwin N, Xu H, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One 2012;7:e34390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 2008;29:150–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010;466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, et al. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol 2009;10:R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alsina L, Israelsson E, Altman MC, Dang KK, Ghandil P, Israel L, et al. A narrow repertoire of transcriptional modules responsive to pyogenic bacteria is impaired in patients carrying loss-of-function mutations in MYD88 or IRAK4. Nat Immunol 2014;15:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leung NS, Padgett P, Robinson DA, Brown EL. Prevalence and behavioural risk factors of Staphylococcus aureus nasal colonization in community-based injection drug users. Epidemiol Infect 2014;1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Reid MJA, Fischer RSB, Mannathoko N, Muthoga C, McHugh E, Essigmann H, et al. Prevalence of Staphylococcus aureus Nasal Carriage in Human Immunodeficiency Virus-Infected and Uninfected Children in Botswana: Prevalence and Risk Factors. Am J Trop Med Hyg 2017;96:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 2012;8:e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol 2012;12:215–25. [DOI] [PubMed] [Google Scholar]
- [35].Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol 2009;27:363–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet 2009;10:43–55. [DOI] [PubMed] [Google Scholar]
- [37].Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007;317:944–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009;41:1105–9. [DOI] [PubMed] [Google Scholar]
- [39].Ye Z, Vasco DA, Carter TC, Brilliant MH, Schrodi SJ, Shukla SK. Genome wide association study of SNP-, gene-, and pathway-based approaches to identify genes influencing susceptibility to Staphylococcus aureus infections. Front Genet 2014;5:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nelson CL, Pelak K, Podgoreanu MV, Ahn SH, Scott WK, Allen AS, et al. A genome-wide association study of variants associated with acquisition of Staphylococcus aureus bacteremia in a healthcare setting. BMC Infect Dis 2014;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol 2014;14:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol 2010;28:535–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Blander JM, Barbet G. Exploiting vita-PAMPs in vaccines. Curr Opin Pharmacol 2018;41:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mourao-Sa D, Roy S, Blander JM. Vita-PAMPs: signatures of microbial viability. Adv Exp Med Biol 2013;785:1–8. [DOI] [PubMed] [Google Scholar]
- [45].Kitur K, Wachtel S, Brown A, Wickersham M, Paulino F, Penaloza HF, et al. Necroptosis Promotes Staphylococcus aureus Clearance by Inhibiting Excessive Inflammatory Signaling. Cell Rep 2016;16:2219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stack J, Doyle SL, Connolly DJ, Reinert LS, O’Keeffe KM, McLoughlin RM, et al. TRAM is required for TLR2 endosomal signaling to type I IFN induction. J Immunol 2014;193:6090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sacre SM, Lundberg AM, Andreakos E, Taylor C, Feldmann M, Foxwell BM. Selective use of TRAM in lipopolysaccharide (LPS) and lipoteichoic acid (LTA) induced NF-kappaB activation and cytokine production in primary human cells: TRAM is an adaptor for LPS and LTA signaling. J Immunol 2007;178:2148–54. [DOI] [PubMed] [Google Scholar]
- [48].Molteni M, Gemma S, Rossetti C. The Role of Toll-Like Receptor 4 in Infectious and Noninfectious Inflammation. Mediators Inflamm 2016;2016:6978936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stenzel W, Soltek S, Sanchez-Ruiz M, Akira S, Miletic H, Schluter D, et al. Both TLR2 and TLR4 are required for the effective immune response in Staphylococcus aureus-induced experimental murine brain abscess. Am J Pathol 2008;172:132–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chantratita N, Tandhavanant S, Seal S, Wikraiphat C, Wongsuvan G, Ariyaprasert P, et al. TLR4 genetic variation is associated with inflammatory responses in Gram-positive sepsis. Clin Microbiol Infect 2017;23:47 e1–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Liu B, Fu Y, Feng S, Zhang X, Liu Z, Cao Y, et al. Involvement of RP105 and toll-like receptors in the activation of mouse peritoneal macrophages by Staphylococcus aureus. Scand J Immunol 2013;78:8–16. [DOI] [PubMed] [Google Scholar]
- [52].Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 2011;474:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Holtfreter S, Roschack K, Eichler P, Eske K, Holtfreter B, Kohler C, et al. Staphylococcus aureus carriers neutralize superantigens by antibodies specific for their colonizing strain: a potential explanation for their improved prognosis in severe sepsis. J Infect Dis 2006;193:1275–8. [DOI] [PubMed] [Google Scholar]
- [54].Verkaik NJ, de Vogel CP, Boelens HA, Grumann D, Hoogenboezem T, Vink C, et al. Anti-staphylococcal humoral immune response in persistent nasal carriers and noncarriers of Staphylococcus aureus. J Infect Dis 2009;199:625–32. [DOI] [PubMed] [Google Scholar]
- [55].Kolata J, Bode LG, Holtfreter S, Steil L, Kusch H, Holtfreter B, et al. Distinctive patterns in the human antibody response to Staphylococcus aureus bacteremia in carriers and non-carriers. Proteomics 2011;11:3914–27. [DOI] [PubMed] [Google Scholar]
- [56].Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004;364:703–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data described in this publication have been deposited in NCBI’s Gene Expression Omnibus [33] and are accessible through GEO Series accession number GSE153122 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153122).


