Abstract
Background:
The inbred mouse strain C57BL/6 is widely used in both models of addiction and immunological disease. However, there are pronounced phenotypic differences in ethanol consumption and innate immune response between B6 substrains. The focus of this study was to examine the effects of substrain on innate immune response and neuroimmune-induced escalation of voluntary ethanol consumption. The main goal was to identify if substrain differences in immune response can account for differences in ethanol behavior.
Methods:
We compared acute innate immune response to a viral dsRNA mimic, polyinosinic:polycytidylic acid (poly(I:C)), in brain using qRT-PCR in both C57BL/6N and C57BL/6J mice. Next, we used a neuroimmune model of escalation using poly(I:C) to compare drinking behavior between substrains. Finally, we compared brain neuroimmune response to both ethanol and repeated poly(I:C) in both strains as a way to account for differences in ethanol behavior.
Results:
We found that C57BL/6 strains have differing immune response and drinking behaviors. C57BL/6N mice have a shorter but more robust inflammatory response to acute poly(I:C). In contrast, C57BL/6J mice have a smaller but longer lasting acute immune response to poly(I:C). In our neuroimmune-induced escalation model, C57BL/6J mice but not C57BL/6N mice escalate ethanol intake after poly(I:C). Finally, only C57BL/6J mice show enhanced proinflammatory transcript abundance after poly(I:C) and ethanol, suggesting that longer lasting immune responses are critical to neuroimmune drinking phenotypes.
Conclusions:
Altogether, this work has elucidated additional influences that substrain has on both innate immune response and drinking phenotypes. Our observations highlight the importance of considering and reporting the source and background used for production of transgenic and knockout mice. This data provides further evidence that genetic background must be carefully considered when investigating the role of neuroimmune signaling in ethanol abuse.
Keywords: neuroimmune, TLR3, alcohol, C57BL/6J, C57BL/6N, poly I:C
Introduction
The genetic background of a mutant or transgenic mouse plays an important role in its phenotype. Over the past 68 years, there has been substantial genetic drift between two substrains of the C57BL/6 strain, C57BL/6J (B6J) and C57BL/6N (B6N). Comparison of the genome between the B6N and B6J substrains revealed substantial overlap with 40 single nucleotide repeats (SNPs) and 30 indels (Simon et al., 2013, Vanden Berghe et al., 2015, Mekada et al., 2009). This difference is particularly interesting in the addiction field because different substrains have different behavioral responses to drugs. Specifically, there are substantial phenotypic differences among the C57BL/6 strains in response to drugs, like ethanol (Mulligan et al., 2008, Blum et al., 1982) and cocaine (Kumar et al., 2013). Moreover, commercially available mice are often on a mixed background with potential genetic contributions from both substrains. Therefore, it is crucial to understand how substrains influence both addiction and immunological studies.
B6J mice are widely used to study ethanol phenotypes because they have high ethanol preference, consume more ethanol, and have fewer withdrawal-induced seizures than other inbred strains (McClearn GE, 1959, Belknap et al., 1993). The B6J phenotype of high ethanol preference and consumption has remained stable over time, likely because ethanol phenotypes are complex polygenic traits (Wahlsten et al., 2006). B6J differ from B6N for several ethanol-related phenotypes, including consumption (Blum et al., 1982), sensitivity to fetal ethanol syndrome (Green et al., 2007), ethanol deprivation effect (Khisti et al., 2006) and brain dopamine release after exposure to ethanol (Ramachandra et al., 2007). There are also immune function differences between B6N and B6J substrains. Differences in immune response include a more robust proinflammatory response in B6N mice but increased interleukin-mediated activation B6J mice (Simon et al., 2013). B6N mice also show enhanced clearance of bacterial infections and neutrophil recruitment compared with B6J mice (Simon et al., 2013, Ulland et al., 2016).
The genetic differences between these substrains can explain some of the phenotypic differences in drug and immune response. B6N mice are homozygous for Cyfip2M1N, this mutation is not found in B6J substrain and the mutations result in lower acute response to cocaine (Kumar et al., 2013). For ethanol consumption, there is not a single regulatory element that accounts for the gene expression differences observed between substrains (Mulligan et al., 2008). However, several genes divergent between the substrains relate to ethanol phenotypes including H2afz, Psen1, D14Ertd449e, Wdfy1 and Clu (Mulligan et al., 2008). The difference in immune function can be explained by a missense mutation in NLRP12. NLRP12 was initially identified as a member of the nucleotide-binding domain and leucine-rich repeat containing receptor (NLR) family reported to attenuate inflammatory responses (Schroder and Tschopp, 2010). The missense mutation in NLRP12 in B6J mice results in diminished NLRP12 function such as blunted neutrophil recruitment after inflammatory stimulus (Ulland et al., 2016). Therefore, to understand differences in behavioral phenotypes, the influence of substrain on both immune function and response to drugs must be taken into account.
These behavioral and immune differences also extend to mice from mixed N/J backgrounds. Before CRISPR, many commercially available knockout mice were made from B6N blastocysts and then bred with B6J mice, resulting in confounding contributions from B6N and B6J backgrounds. Because we were interested in the role of the neuroimmune system in regulation of ethanol consumption, we originally wanted to use Tlr3 mutant mice to test if TLR3 is necessary for ethanol consumption and neuroimmune-induced escalations in ethanol intake. However, the wild-type Tlr3(+/+) mice did not replicate drinking behavior or immune responses found in B6J mice (Warden unpublished observations). We hypothesized this was due to the mixed N/J background, suggesting that mutant mice may phenocopy certain behaviors from each substrain that was crossed. Therefore, we chose to test the hypothesis that B6N and B6J mice have different responses to viral activation and that this difference in immune response can regulate drinking behavior.
Here, we compared male B6N and B6J mice for response to TLR3 agonist polyinosinic:polycytidylic acid (poly(I:C)) and escalation of ethanol consumption, a known model of neuroimmune-related drinking behaviors (Randall et al., 2019, Warden et al., 2019a). B6N mice had a more robust but shorter inflammatory response to poly(I:C). Only B6J mice had lasting immune response after poly(I:C), with upregulation of inflammatory mediators lasting beyond 48 h post-injection. Hierarchical clustering supported that B6J neuroimmune response was unique. Next, we tested ethanol consumption differences between substrains using our poly(I:C) model of neuroimmune-induced escalation. B6N mice did not escalate ethanol consumption after poly(I:C). Only B6J mice escalated ethanol consumption and had a lasting immune response after chronic poly(I:C) and ethanol, suggesting that prolonged changes in inflammatory response are critical to how neuroimmune activation escalates ethanol consumption. Together, this suggests substrain influences both immune function and drinking behavior and should be considered when designing and interpreting studies.
Materials & Methods
Mice
Male C57BL/6J (B6J) and C57BL/6N (B6N) mice were purchased from Jackson Laboratories, Bar Harbor, ME at 8 weeks of age. Drinking experiments began when the mice were between 8–10 weeks old. Mice were weighed every 4 days during drinking experiments. All experiments were approved by The University of Texas at Austin Institutional Animal Care and Use Committee.
Drug Preparation and Administration
Poly(I:C) HMW obtained from Invivogen (San Diego, CA) was prepared previously described (Warden et al., 2019a). Briefly, 50 mg of poly(I:C) HMW was diluted in sterile physiological water (NaCl 0.9%) to a concentration of 1 mg/mL, heated for 10 minutes at 65–70 C then cooled for 1 h at room temperature (RT) to ensure proper annealing of the dsRNA.
For the qRT-PCR timecourse of poly(I:C), we injected poly(I:C) (5 mg/kg) intraperitoneally (i.p.) and then euthanized mice at 3 and 24 h post-injection before qRT-PCR analysis. The 5 mg/kg dose has been shown to have a peak immune response between 3–8 h after injection, minimizes sickness response and prevents any adverse effect on drinking behavior (Qin and Crews, 2012, Randall et al., 2019, Warden et al., 2019a).
For ethanol drinking studies we used the same protocol previously published by our lab (Warden et al., 2019a). Briefly, poly(I:C) (5 mg/kg, i.p.) was administered every 4th day during no alcohol access periods, 24–36 h after peak poly(I:C) immune response. For the control injection, 0.9% sterile saline (volume matched) was administered. Single use, sterile needles (27.5 gauge) were used. All injections we made between 8 am and 9 am to male animals 8 −16 weeks of age.
Two-bottle choice every-other-day procedure
Mice were given every-other-day (EOD) access to ethanol (15% v/v) as previously described (Warden et al., 2019a). Ethanol consumption was calculated as g/kg body weight per 24 h period. Total fluid intake was calculated as g/kg body weight per 24 h period. Poly(I:C) was injected every 4th day during the EOD procedure.
qRT-PCR
For RNA isolations, brains were harvested and the prefrontal cortex dissected before being snap frozen in liquid nitrogen. Total RNA was extracted using the MagMAX-96 Total RNA Isolation Kit (Life Technologies, Grand Island, NY) and quantified using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Grand Island, NY). RNA quality was additionally assessed using the Agilent TapeStation (Agilent Technologies, Santa Clara, CA). We used a predetermined quality metric of RIN > 8 as a cutoff for inclusion in data analysis. Relative quantification of mRNA levels was determined using BIORAD software as previously described (Warden et al., 2019a, Osterndorff-Kahanek et al., 2013). The endogenous control for qRT-PCR experiments was Gusb.
Statistical analysis
Unless noted, data are reported as mean ± SEM values. 2-way ANOVAs, one-way ANOVAs, and Student’s t-tests were done using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). All drinking experiments were analyzed by repeated measures 2-way ANOVA and Bonferroni post-hoc tests, escalation data was analyzed by one-way ANOVA and Tukey’s post-hoc tests. qRT-PCR data were analyzed by one-way ANOVA and/or t-test. Substrain comparison and heatmap generation was performed by using the R software program hierarchical clustering function.
RESULTS
B6N have a shorter immune response compared with B6J mice
First, we chose to investigate mRNA expression changes in the medial prefrontal cortex because this region is the only one to show differences in TLR expression after EOD drinking and is the region with the strongest inflammatory response to acute poly(I:C) (McCarthy et al., 2018; Warden unpublished observations). In C57BL/6J (B6J) mice, acute poly(I:C) increases transcript abundance of inflammatory mediators which return to saline levels 48 h post-injection (Warden et al., 2019a). Because C57BL/6N (B6N) mice have more rapid bacterial clearance, we hypothesized that B6N mice would have an abbreviated immune response to poly(I:C) (Ulland et al., 2016). First, with no injections, we determined if there were any differences in transcript abundance of TLR3/TRIF-dependent pathway components between B6N and B6J mice (Figure 1A). Without injections, B6N mice have higher transcript abundance of TLR3/TRIF-dependent pathway components (Ticam1: t (19) = 3.39, P = 0.004; Ikkε: t (19) = 5.99, P = 0.0001; Irf3: t (19) = 7.54, P < 0.0001; Tbk1: t (19) = 6.86, P < 0.0001) and MyD88-depedent pathway components (Myd88: t (19) = 4.51, P < 0.0001; Ikkβ: t (19) = 3.21, P = 0.006) compared with B6J mice (Figure 1B).
Figure 1:

B6N mice have higher levels of innate immune transcripts compared with B6J mice. (A) Experiment schematic, To assess transcript abundance between substrains, B6N and B6J mice were euthanized, the mPFC freshly dissected, RNA extracted and qRT-PCR analysis was performed. (B) Levels for each transcript are shown as log2 fold-change values, normalized to Gusb (n=6/group). Data represented as mean + SEM (analyzed by Welch’s t-test, **** P <0.0001, *** P < 0.0002, ** P < 0.0021, * P < 0.05).
Next, we sought to determine how substrain influences the intensity and timecourse of poly(I:C) response in brain using a separate cohort of mice. To test this hypothesis we injected B6N mice with either saline or poly(I:C) (5 mg/kg) then euthanized animals at 3 and 24 h post-injection. We then measured prefrontal cortical (mPFC) transcript abundance of genes previously implicated in poly(I:C) response and compared them with our previously published data from B6J mice treated on the same poly(I:C) timecourse (Figure 2A) (Warden et al., 2019a). Poly(I:C) produced distinct changes in transcript abundance and duration between B6 substrains. Three hours after poly(I:C), B6N mice had similar transcript abundance of TLR3 pathway components and inflammatory factors compared with B6J mice (Figure 2B, Figure S1). By 24 h post-injection, inflammatory mediator transcript abundance returned to saline levels for B6N mice (Figure 2, Figure S1). However, B6J mice at 24 h post-injection still had elevated transcript abundance of both TLR pathways (Tlr3: t (19) = 6.48, P = 0.001; Tlr2: t (19) = 7.79, P = 0.003; Myd88: t (19) = 7.18, P = 0.004) and inflammatory mediators (Il1β: t (19) = 5.83, P = 0.01; Ccl5: t (19) = 4.23, P = 0.0017) (Figure 2, Figure S1). Interestingly, the saline injection itself changed the expression of many TLR components between substrains (Figure S2) and compared with the no-injection group (Figure 1). We hypothesize the stress of restraint and i.p. injection may alter the expression of TLR pathways due to the link between stress and immune response (Bollinger et al., 2017, Malki et al., 2015, Sathyanesan et al., 2017). Taken together, the shorter inflammatory response to poly(I:C) in B6N mice suggests that the longer lasting changes in gene expression that occur in B6J mice may be crucial to how poly(I:C) increases ethanol intake.
Figure 2:

B6N mice have a shorter poly(I:C) immune response. (A) Experiment schematic: To assess differences in poly(I:C) immune response timecourse between substrains, B6N and B6J mice were either injected with saline or poly(I:C) (5mg/kg) euthanized 3 h or 24 h post-injection, the mPFC freshly dissected, RNA extracted and qRT-PCR analysis was performed. (B) Heatmap showing transcript abundance for each group: saline, 3 h post-injection poly(I:C), 24 h post-injection poly(I:C). Levels for each transcript are shown as log2 fold-change values, normalized to Gusb (n=6/group). For the heatmap, yellow represents high transcript abundance, purple is low transcript abundance. Heirarchical clustering, one-way ANOVA, and Tukey’s post-hoc tests were used to analyze the data (**** P < 0.0001, *** P < 0.0002, ** P < 0.0021, * P < 0.05).
B6N mice do not escalate ethanol intake with repeated poly(I:C) treatment
Without longer lasting immune responses, we hypothesized that B6N mice would not escalate ethanol intake with repeated poly(I:C) injections. To test this hypothesis, we injected poly(I:C) (5 mg/kg) or saline every four days during EOD for 32 days into both B6N and B6J mice (Figure 3A). As shown in Figure 3B–D, poly(I:C) escalated ethanol consumption (Ftreatment (1,16) = 4.52, P = 0.002) and preference (Ftreatment (1,16) = 17.42, P = 0.0007) in B6J mice. Total fluid intake was unaffected by poly(I:C) in B6J mice (Ftreatment (1,16) = 1.41, P = 0.24). In contrast, in B6N mice, poly(I:C) did not escalate ethanol consumption (Ftreatment (1,16) = 1.78, P = 0.20) nor preference (Ftreatment (1,16) = 2.11, P = 0.17) (Figure 3E–F). Poly(I:C) did not alter total fluid intake in B6N mice (Ftreatment (1,16) = 0.01, P = 0.91) (Figure 3G). Finally, we compared escalation phenotypes by comparing drinking behavior at the end of the experiment (day 32) to the beginning (day 4). Only B6J mice treated with poly(I:C) escalate ethanol consumption (Figure 3H, F (3,34) = 24.04, P < 0.001; there was no significant difference between B6N Saline, B6J Saline and B6N poly(I:C)-treated mice (Ps > 0.05)). Similarly, only B6J mice treated with poly(I:C) escalate ethanol preference (Figure 3I, F (3,34) = 12.63, P < 0.001; there was no significant difference between B6N Saline, B6J Saline and B6N poly(I:C)-treated mice (Ps > 0.05)). There was no significant difference in total fluid intake between substrains or treatments (Figure 3H, F (3,34) = 2.214, P = 0.11)). We noted increased variability in the effect of poly(I:C) on B6N mice ethanol consumption across time therefore we compared variance statistical measures and the effect of poly(I:C) on B6N and B6J mice directly (Figure S2). Poly(I:C) reduced bodyweight in B6N mice similar to B6J mice (t (18) = 0.48, P = 0.86) (Figure S3), suggesting substrain does not influence this phenotype. Taken together this suggests that the effect of TLR3 activation on drinking behavior is substrain specific.
Figure 3:
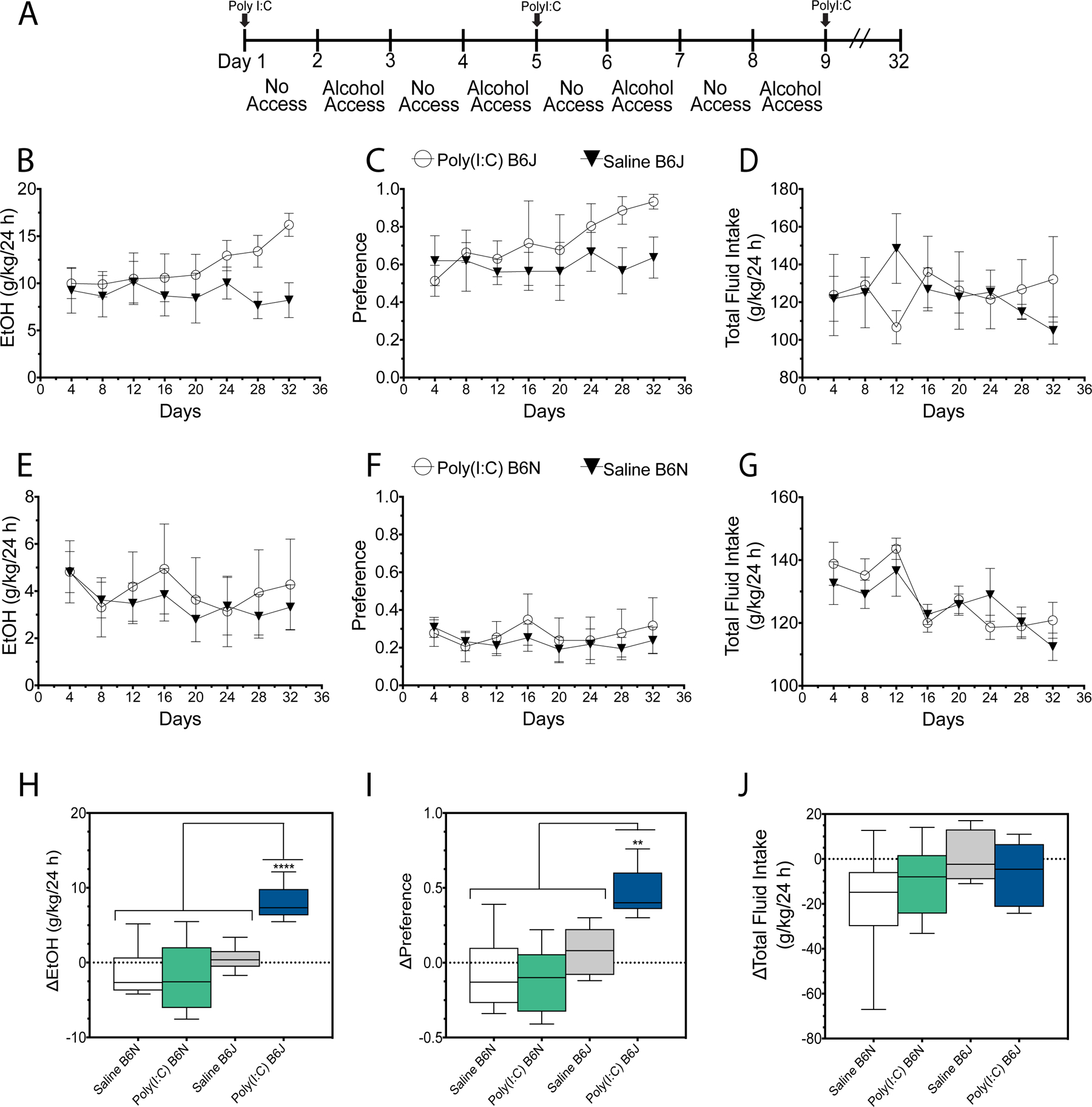
Poly(I:C) does not change drinking in B6N mice. (A) Experiment schematic: poly(I:C) or saline was injected every 4th day while male B6J and B6N mice underwent EOD drinking. The procedure lasted for 32 days (i.e. 8 injections). (B,E) Ethanol intake (g/kg/24 h) for B6J and B6N mice, respectively; (C,F) preference for EtOH for B6J and B6N mice, respectively; (D,G) Total fluid intake (g/kg/24 h) for B6J and B6N mice, respectively. (H) Escalation of ethanol intake (g/kg/24 h) for B6J and B6N mice, respectively; (I) Escalation of preference for EtOH for B6J and B6N mice, respectively; (J) Escalation of total fluid intake (g/kg/24 h) for B6J and B6N mice, respectively. Drinking data represented as mean + SEM analyzed by two-way ANOVA with Bonferroni post-hoc tests. Escalation data represented as mean + SEM analyzed by one-way ANOVA with Tukey post-hoc tests (**** P < 0.0001, *** P < 0.0002, ** P < 0.0021, * P < 0.05).
Poly(I:C) and ethanol produce larger immune responses in B6J mice
Because B6N mice did not escalate ethanol intake after poly(I:C), we hypothesized B6N mice would have few changes in innate immune receptors and inflammatory mediators after chronic ethanol consumption and poly(I:C) treatment. To test this hypothesis we euthanized the animals that had undergone 32 days of EOD drinking with poly(I:C) injections every 4 days (8 injections total). We dissected the mPFC 24 h after last ethanol exposure and measured transcript abundance of innate immune components using qRT-PCR. As shown in Figure 4A, after poly(I:C) and ethanol, there were no changes in transcript abundance of TLR3/TRIF-dependent components or MyD88-dependent components in B6N mice. There were also no changes in inflammatory mediators except for an increase in transcript abundance of Ccl5, (t (16) = 5.25, P = 0.001) (Figure 4A). In contrast, B6J mice had significant increases in transcript abundance of TLR3/TRIF-dependent pathway components and inflammatory genes as well as significant decreases in MyD88-dependent pathway genes (Figure 4B–C) (Warden et al., 2019a). Taken together, in contrast to B6J mice, B6N did not escalate ethanol intake after poly(I:C) and there were almost no accompanying changes in inflammatory response after poly(I:C) and ethanol, suggesting that the prolonged changes in immune function observed in B6J mice are necessary for neuroimmune-induced escalations in ethanol consumption.
Figure 4:

Chronic poly(I:C) and ethanol increases inflammatory response in B6J but not B6N mice. (A) B6N mice treated with poly(I:C) have no significant changes in transcript abundance for innate immune components compared with B6N saline mice. (B) B6J mice treated with poly(I:C) have many significant increases in transcript abundance for innate immune components compared with B6J saline mice. (C) Heatmap showing transcript abundance normalized to saline control for each substrain. Levels for each transcript are shown as log2 fold-change values, normalized to Gusb (n=10/group). Yellow represents high transcript abundance, purple is low transcript abundance. Heirarchical clustering, Welch’s t-test and one-way ANOVA were used to analyze the data (**** P < 0.0001, *** P < 0.0002, ** P < 0.0021, * P < 0.05).
Discussion
Here we provide evidence that genetic differences between B6J and B6N mice influence immune and ethanol consumption phenotypes. B6N mice consume less ethanol compared with B6J mice and have a shorter but more robust response to viral infection. Additionally, B6N mice did not escalate ethanol intake after neuroimmune activation with poly(I:C) or have long lasting gene expression changes after chronic poly(I:C) injections and EOD drinking. This neuroimmune behavioral and immune phenotype is only found in B6J mice. We hypothesize these differences in immune response timecourse between substrains underlies the difference neuroimmune behavioral phenotypes.
Similar to previous reports we found that B6N mice consume less ethanol compared with B6J mice in an EOD procedure (Mulligan et al., 2008, Blum et al., 1982). Additionally, we demonstrate B6N mice do not escalate ethanol intake or preference after poly(I:C), whereas, B6J mice escalate ethanol intake after poly(I:C). This supports our initial hypothesis that the reason TLR3(+/+) wild-type did not phenocopy B6J mice after poly(I:C) was because of contributions from the B6N background. This suggests that substrain influences are different for consumption and immune-induced escalation.
Consistent with the literature about the peripheral immune system, we found B6N have a stronger proinflammatory response compared with B6J mice in brain (Simon et al., 2013). After acute poly(I:C), B6N mice show higher transcript levels of proinflammatory mediators in the medial prefrontal cortex. However, B6N mice have a much shorter immune activation timecourse compared with B6J animals, with all inflammatory response back to saline levels within 24 h post-injection. This suggests a much more rapid and robust inflammatory response in B6N animals. Similarly, B6N mice clear bacterial infections such as Listeria, much faster than their B6J counterparts (Simon et al., 2013). B6J mice have longer lasting immune responses to infection or contact hypersensitivity, suggesting that although the immune response is less robust, the duration of immune response prevents clearance of infection leading to longer-lasting changes in immune function (Simon et al., 2013). Only B6J mice had significant increases in transcript levels of inflammatory mediators and innate immune receptors 24 h post-injection. Hierarchical clustering further demonstrated that the prolonged inflammatory response after TLR3 activation is a facet of the B6J substrain alone. We hypothesized that these prolonged changes in inflammatory response were necessary for how repeated TLR3 activation increases ethanol intake and this is supported by our finding that B6N mice have no changes in innate immune components after chronic poly(I:C) and EOD drinking. In contrast, B6J mice had broad changes in immune transcript abundance after poly(I:C) and ethanol. This suggests that attenuated but longer lasting immune responses are critical for neuroimmune-induced escalations in ethanol intake. One caveat of this study is that only the mPFC was investigated for transcriptional changes after poly(I:C) and ethanol. Due to observed the brain regional differences in response to ethanol and poly(I:C) (McCarthy et al., 2018, Randall et al., 2019, Warden et al., 2019a) a future direction of this work will be to investigate differences in poly(I:C) effect between substrains in different brain regions important for addiction.
Interestingly, in both substrains, poly(I:C) increases transcript abundance of both TLR3/TRIF-dependent and MyD88-dependent components, suggesting a potential role for both branches of TLR signaling for acute responses to poly(I:C). It has been well established that the MyD88-dependent pathway is important for regulation of ethanol consumption in backcrossed B6J mice (Mayfield et al., 2016). However, these foundational knockout studies have not been studied in B6N mice, most likely because B6N consume significantly less ethanol than their B6J counterparts (Blum et al., 1982, Mulligan et al., 2008). In B6J mice, both the acute the poly(I:C) response and the poly(I:C) effect on drinking behavior is, at least partially, dependent on the TLR3/dsRNA complex (Warden et al., 2019a). Additionally, the effect of poly(I:C) on drinking is not dependent on Myd88 in B6J backcrossed mice (Warden et al., 2019a). Future studies will determine whether the acute poly(I:C) response in B6N mice is dependent on the TLR3/dsRNA complex or MyD88-dependent signaling. It has been shown that TLR4-expressing cells with B6N functional Nlrp12 are more sensitive to LPS stimulation and produce a significantly greater inflammatory response than cells with the B6J missense mutations in Nlrp12, suggesting MyD88-dependent signaling may be critical for immune responses in B6N mice (Ulland et al., 2016). Therefore, we postulate that only B6J mice escalate ethanol consumption after poly(I:C) because of the missense mutation in NLRP12 in B6J mice resulting in longer lasting immune responses (Simon et al., 2013, Ulland et al., 2016).
An interesting future direction is to determine the specific genetic differences between these substrain that leads to this difference in both immune response and neuroimmune-induced escalations in ethanol intake. A candidate gene that may mediate both phenotypes is NLRP12. B6J carry a missense variant in NLRP12 (Simon et al., 2013). NLRP12 is associated with inflammatory disorders in humans (Jeru et al., 2008). NLRP12 knockout mice have fewer inflammatory changes after contact hypersensitivity and reduced neutrophil recruitment after bacterial infection, recapitulating B6J immune phenotypes (Arthur et al., 2010, Ulland et al., 2016). Ineffective neutrophil recruitment leads to slower recovery and longer periods of inflammation after specific bacterial infections (Ulland et al., 2016). Therefore, future studies will need to determine if NLRP12 knockout mice phenocopy B6J inflammatory response to TLR3 activation and escalation of ethanol consumption.
A caveat of this work is that all studies were done in B6N and B6J male mice. Female B6J mice also consume more ethanol compared with female B6N mice (Mulligan et al., 2008). Similarly, female B6N mice clear bacterial infections faster than their female B6J female counterparts (Ulland et al., 2016), suggesting that there is not a sex-specific effect between substrains for baseline drinking or immune response. However, poly(I:C) does have a sex-specific, only male B6J mice escalate ethanol intake after TLR3 activation (Warden et al., 2019a, Warden et al., 2019b). This is due to differences in immune response timecourse between sexes in B6J mice, with longer lasting changes occurring in female B6J mice (Warden et al., 2019a, Warden et al., 2019b). Therefore, the role of substrain in neuroimmune-induced escalation in ethanol consumption could only be tested in males. We hypothesize that since female B6N mice have more robust immune response and faster clearance of bacterial infections compared with female B6J mice, that female B6N would also have a shorter, more robust immune response to poly(I:C).
One of the underlying principles of the neuroimmune hypothesis of addiction is that lasting changes in gene expression must occur to alter drinking behavior (Mayfield and Harris, 2017). Our study demonstrates that substrain influences the intensity and duration of immune response and these differences in immune response can directly influence drinking phenotypes. However, a critical question is, which inbred substrain most closely follows human immune response? Many drugs and human pathogens are species-specific and there are many differences in innate immune molecules between species, including the lack of a functional TLR10 in mice and the expression of TLR11, TLR12, and TLR13 in mice that are not present in humans (Zschaler et al., 2014). Stimulation with poly(I:C) induces activation of NF-kB, production of cytokines like IL-6 and a strong anti-viral response via IFNβ in many different types of human cell lines such as corneal epithelial cells, bronchial epithelial cells, human metastatic pharynx carcinoma cell line, and even prostate cell lines (Kumar et al., 2006, Matijevic and Pavelic, 2012, Dauletbaev et al., 2015, Palchetti et al., 2015). Both this strong anti-inflammatory response and antiviral response are mimicked in B6J and B6N mice. Therefore, the question of timecourse and duration of inflammatory response becomes key in determining which substrain is a better model for human inflammatory responses to poly(I:C). In human corneal epithelial cells, IFNβ, IFNα and IP-10 were elevated up to 8 h after poly(I:C) challenge (Kumar et al., 2006). In human PMBCs treated with poly(I:C) and then profiled 3 h and 24 h post-stimulation, there were two distinct patterns of activation (Mian et al., 2013). One cluster of genes were upregulated at 3 h post-poly(I:C) and another cluster had higher expression at 24 h. Genes that remained upregulated at 24 h included many genes also upregulated in B6J mice 24 h post-stimulation, CCL2, STAT1, MYD88, IL-6, IFNγ, and TNFα (Mian et al., 2013). Similarly, the primary and secondary responses between B6J and human PMBCs were similar: cytokines/chemokines were transient and effectors of primary response genes were changed for a longer period of time to control response to infection (Mian et al., 2013, Warden et al., 2019a). Therefore, B6J mice with their prolonged response to TLR3 activation may be a better model for human TLR3 responses.
Supplementary Material
Acknowledgements and Disclosures:
This work was supported by the National Institutes of Health/National Institute of Alcohol Abuse and Alcoholism [U01 AA020926, R01 AA006399, F31 AA025499, R01 AA012404]. The authors report no biomedical financial interests or potential conflicts of interest.
References
- ARTHUR JC, LICH JD, YE Z, ALLEN IC, GRIS D, WILSON JE, SCHNEIDER M, RONEY KE, O’CONNOR BP, MOORE CB, MORRISON A, SUTTERWALA FS, BERTIN J, KOLLER BH, LIU Z & TING JP 2010. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol, 185, 4515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELKNAP JK, CRABBE JC & YOUNG ER 1993. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl), 112, 503–10. [DOI] [PubMed] [Google Scholar]
- BLUM K, BRIGGS AH, DELALLO L, ELSTON SF & OCHOA R 1982. Whole brain methionine-enkephalin of ethanol-avoiding and ethanol-preferring c57BL mice. Experientia, 38, 1469–70. [DOI] [PubMed] [Google Scholar]
- BOLLINGER JL, COLLINS KE, PATEL R & WELLMAN CL 2017. Behavioral stress alters corticolimbic microglia in a sex- and brain region-specific manner. PLoS One, 12, e0187631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAULETBAEV N, CAMMISANO M, HERSCOVITCH K & LANDS LC 2015. Stimulation of the RIG-I/MAVS Pathway by Polyinosinic:Polycytidylic Acid Upregulates IFN-beta in Airway Epithelial Cells with Minimal Costimulation of IL-8. J Immunol, 195, 2829–41. [DOI] [PubMed] [Google Scholar]
- GREEN ML, SINGH AV, ZHANG Y, NEMETH KA, SULIK KK & KNUDSEN TB 2007. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn, 236, 613–31. [DOI] [PubMed] [Google Scholar]
- JERU I, DUQUESNOY P, FERNANDES-ALNEMRI T, COCHET E, YU JW, LACKMY-PORT-LIS M, GRIMPREL E, LANDMAN-PARKER J, HENTGEN V, MARLIN S, MCELREAVEY K, SARKISIAN T, GRATEAU G, ALNEMRI ES & AMSELEM S 2008. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A, 105, 1614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHISTI RT, WOLSTENHOLME J, SHELTON KL & MILES MF 2006. Characterization of the ethanol-deprivation effect in substrains of C57BL/6 mice. Alcohol, 40, 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUMAR A, ZHANG J & YU FS 2006. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology, 117, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUMAR V, KIM K, JOSEPH C, KOURRICH S, YOO SH, HUANG HC, VITATERNA MH, DE VILLENA FP, CHURCHILL G, BONCI A & TAKAHASHI JS 2013. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science, 342, 1508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALKI K, MINEUR YS, TOSTO MG, CAMPBELL J, KARIA P, JUMABHOY I, SLUYTER F, CRUSIO WE & SCHALKWYK LC 2015. Pervasive and opposing effects of Unpredictable Chronic Mild Stress (UCMS) on hippocampal gene expression in BALB/cJ and C57BL/6J mouse strains. BMC Genomics, 16, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATIJEVIC T & PAVELIC J 2012. Poly(I:C) treatment influences the expression of calreticulin and profilin-1 in a human HNSCC cell line: a proteomic study. Tumour Biol, 33, 1201–8. [DOI] [PubMed] [Google Scholar]
- MAYFIELD J, ARENDS MA, HARRIS RA & BLEDNOV YA 2016. Genes and Alcohol Consumption: Studies with Mutant Mice. Int Rev Neurobiol, 126, 293–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAYFIELD J & HARRIS RA 2017. The Neuroimmune Basis of Excessive Alcohol Consumption. Neuropsychopharmacology, 42, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCARTHY GR, WARDEN A, BRIDGES C, BLEDNOV Y, MAYFIELD R & HARRIS R 2018. Chronic ethanol consumption: Role of TLR3/TRIF-dependent signaling. Addiction Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCLEARN GE RD 1959. Differences in alcohol preference among inbred strains of mice. Q J Stud Alcohol, 20, 691–695. [Google Scholar]
- MEKADA K, ABE K, MURAKAMI A, NAKAMURA S, NAKATA H, MORIWAKI K, OBATA Y & YOSHIKI A 2009. Genetic differences among C57BL/6 substrains. Exp Anim, 58, 141–9. [DOI] [PubMed] [Google Scholar]
- MIAN MF, AHMED AN, RAD M, BABAIAN A, BOWDISH D & ASHKAR AA 2013. Length of dsRNA (poly I:C) drives distinct innate immune responses, depending on the cell type. J Leukoc Biol, 94, 1025–36. [DOI] [PubMed] [Google Scholar]
- MULLIGAN MK, PONOMAREV I, BOEHM SL 2ND, OWEN JA, LEVIN PS, BERMAN AE, BLEDNOV YA, CRABBE JC, WILLIAMS RW, MILES MF & BERGESON SE 2008. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav, 7, 677–89. [DOI] [PubMed] [Google Scholar]
- OSTERNDORFF-KAHANEK E, PONOMAREV I, BLEDNOV YA & HARRIS RA 2013. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One, 8, e59870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALCHETTI S, STARACE D, DE CESARIS P, FILIPPINI A, ZIPARO E & RICCIOLI A 2015. Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J Biol Chem, 290, 5470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIN L & CREWS FT 2012. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. J Neuroinflammation, 9, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMACHANDRA V, PHUC S, FRANCO AC & GONZALES RA 2007. Ethanol preference is inversely correlated with ethanol-induced dopamine release in 2 substrains of C57BL/6 mice. Alcohol Clin Exp Res, 31, 1669–76. [DOI] [PubMed] [Google Scholar]
- RANDALL PA, VETRENO RP, MAKHIJANI VH, CREWS FT & BESHEER J 2019. The Toll-Like Receptor 3 Agonist Poly(I:C) Induces Rapid and Lasting Changes in Gene Expression Related to Glutamatergic Function and Increases Ethanol Self-Administration in Rats. Alcohol Clin Exp Res, 43, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATHYANESAN M, HAIAR JM, WATT MJ & NEWTON SS 2017. Restraint stress differentially regulates inflammation and glutamate receptor gene expression in the hippocampus of C57BL/6 and BALB/c mice. Stress, 20, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRODER K & TSCHOPP J 2010. The inflammasomes. Cell, 140, 821–32. [DOI] [PubMed] [Google Scholar]
- SIMON MM, GREENAWAY S, WHITE JK, FUCHS H, GAILUS-DURNER V, WELLS S, SORG T, WONG K, BEDU E, CARTWRIGHT EJ, DACQUIN R, DJEBALI S, ESTABEL J, GRAW J, INGHAM NJ, JACKSON IJ, LENGELING A, MANDILLO S, MARVEL J, MEZIANE H, PREITNER F, PUK O, ROUX M, ADAMS DJ, ATKINS S, AYADI A, BECKER L, BLAKE A, BROOKER D, CATER H, CHAMPY MF, COMBE R, DANECEK P, DI FENZA A, GATES H, GERDIN AK, GOLINI E, HANCOCK JM, HANS W, HÖLTER SM, HOUGH T, JURDIC P, KEANE TM, MORGAN H, MÜLLER W, NEFF F, NICHOLSON G, PASCHE B, ROBERSON LA, ROZMAN J, SANDERSON M, SANTOS L, SELLOUM M, SHANNON C, SOUTHWELL A, TOCCHINI-VALENTINI GP, VANCOLLIE VE, WESTERBERG H, WURST W, ZI M, YALCIN B, RAMIREZ-SOLIS R, STEEL KP, MALLON AM, DE ANGELIS MH, HERAULT Y & BROWN SD 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol, 14, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ULLAND TK, JAIN N, HORNICK EE, ELLIOTT EI, CLAY GM, SADLER JJ, MILLS KA, JANOWSKI AM, VOLK AP, WANG K, LEGGE KL, GAKHAR L, BOURDI M, FERGUSON PJ, WILSON ME, CASSEL SL & SUTTERWALA FS 2016. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat Commun, 7, 13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDEN BERGHE T, HULPIAU P, MARTENS L, VANDENBROUCKE RE, VAN WONTERGHEM E, PERRY SW, BRUGGEMAN I, DIVERT T, CHOI SM, VUYLSTEKE M, SHESTOPALOV VI, LIBERT C & VANDENABEELE P 2015. Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice. Immunity, 43, 200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAHLSTEN D, BACHMANOV A, FINN DA & CRABBE JC 2006. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc Natl Acad Sci U S A, 103, 16364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARDEN AS, AZZAM M, DACOSTA A, MASON S, BLEDNOV YA, MESSING RO, MAYFIELD RD & HARRIS RA 2019a. Toll-like receptor 3 activation increases voluntary alcohol intake in C57BL/6J male mice. Brain Behav Immun, 77, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARDEN AS, AZZAM M, DACOSTA A, MASON S, BLEDNOV YA, MESSING RO, MAYFIELD RD & HARRIS RA 2019b. Toll-like receptor 3 dynamics in female C57BL/6J mice: Regulation of alcohol intake. Brain Behav Immun, 77, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZSCHALER J, SCHLORKE D & ARNHOLD J 2014. Differences in innate immune response between man and mouse. Crit Rev Immunol, 34, 433–54. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


