Abstract
Transfer RNAs (tRNAs) are non-coding RNAs essential for protein synthesis. tRNAs are heavily decorated with a variety of post-transcriptional modifications (tRNA modifications). Recent methodological advances provide new tools for rapid profiling of tRNA modifications and have led to discoveries of novel modifications and their regulation. Here, we provide an overview of the techniques for investigating tRNA modifications and of the expanding knowledge of their chemistry and regulation.
Keywords: tRNA modification, RNA mass spectrometry, High-throughput sequencing
Introduction
Transfer RNAs (tRNAs) are essential adaptor molecules that bridge amino acid sequences to the genetic codes of mRNAs. tRNAs are heavily decorated with a variety of post-transcriptional modifications (tRNA modifications) [1,2]. These modifications are synthesized by dedicated site-specific enzymes (tRNA modifying enzymes). The structures of tRNA modifications are diverse; to date, more than 100 species of modifications have been reported [2] (Figure 1A). The types and sites of modification differ among tRNA species. Some modifications, e.g., T at 54 and Ψ at 55, are ubiquitously introduced into all tRNA species whereas other modifications are incorporated only into a subset of tRNAs. Specific positions in tRNAs are frequently modified. For example, the first letter of the anticodon (position 34) and the nucleotide 3’ adjacent to the anticodon (position 37) are frequently converted to diverse modified nucleosides in many tRNA species (Figure 1B). These modifications alter the strength and specificity of codon recognition, and aminoacylation specificity [3]. Modifications of nucleosides outside of the anticodon loop generally increase tRNA stability [4]. Both of these effects alter translation efficiency at each individual codon which in turn controls the speed of translation, protein folding, and protein expression level [5,6].
Figure 1. Structures and sites of tRNA modifications.
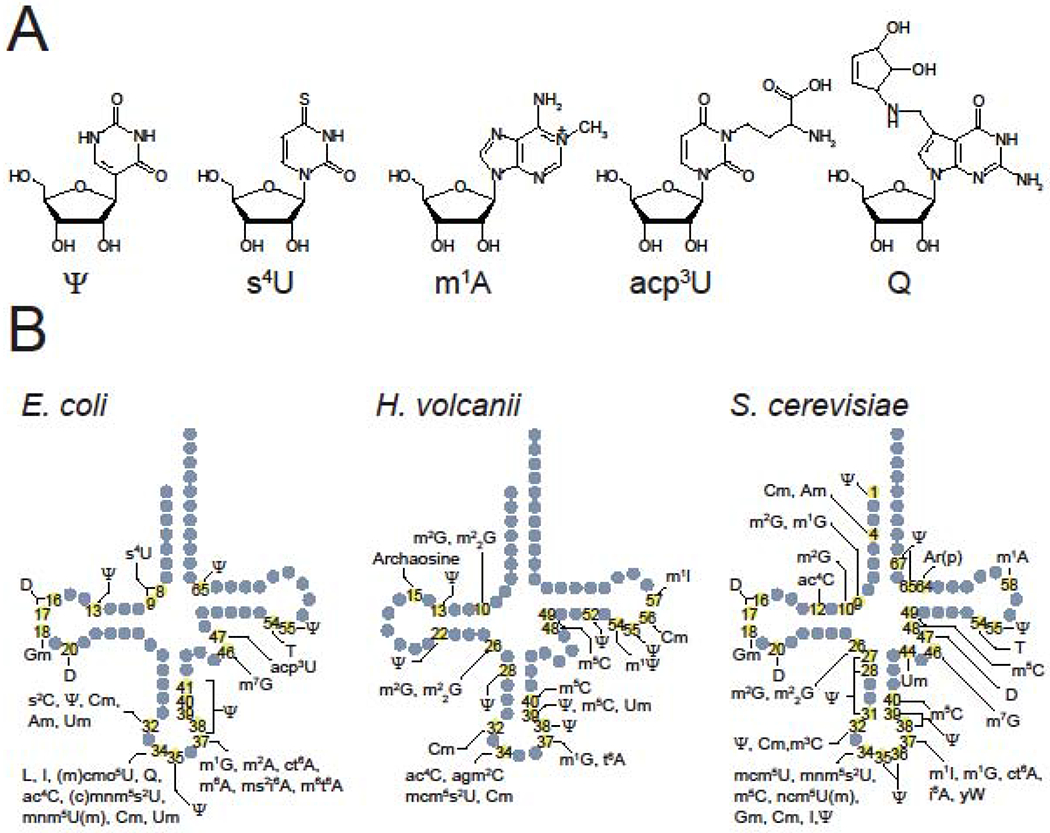
(A) Chemical structures of tRNA modifications: pseudouridine (Ψ), 4-thiouridine (s4U), 1-methyladeno (m1A), 3-(3-amino-3-carboxypropyl)uridine (acp3U), and queuosine (Q).
(B) Modification sites and types in E. coli, H. volcanii, and S. cerevisiae tRNAs. The positions and types of modifications present in one or more tRNA species are indicated on the schematic secondary structure of tRNAs. The information is based on [1,54].
Protein translation systems, including tRNA modifications, have not been well characterized in non-model organisms. Moreover, there have been few studies of regulation of tRNA modifications. A major impediment to comprehensive studies of tRNA modification and its regulation is the technical hurdle to rapidly chart modifications in dozens of tRNA species at once.
Recent methodological advances offer tools for rapid profiling of tRNA modifications in microorganisms. These tools open up new ways to reveal novel chemical modifications and to probe the links between tRNA modification and cellular conditions. Here, we provide an overview of current techniques for profiling tRNA modifications and the application of such techniques to identify new modifications and to study the regulation of modification.
Methods for identifying chemical structures and sites of tRNA modifications
RNA mass spectrometry
Charting tRNA modifications entails determination of both the chemical identities and the sites of modifications. Each modified nucleoside has unique physical properties, which enable its separation from other nucleosides by chromatography. A nucleoside pool is prepared by digesting tRNAs with non-specific nucleases and a phosphatase. Modified and canonical nucleosides can be separated by different retardation factors (Rf) in thin-layer chromatography (TLC) [7] or different retention times in liquid chromatography (LC) [2]. Catalogs of previously determined Rf values and retention times of known modification are used as references to identify nucleosides that have chromatographic properties matching to the references [7]. In liquid chromatography mass spectrometric analysis (LC-MS), retention times and mass values of nucleosides are also used to distinguish one modification from other modifications and canonical nucleosides [8] (Figure 2). A nucleoside with unknown Rf or retention time or mass values can represent a new modification. It is noteworthy that the detected signals can be derived from molecules other than modified nucleosides, such as contaminants in the sample or the instrument. Some sample preparation processes also can convert a modification into an artificial adduct [9]. Careful characterization of detected signals are required to verify the presence of a new modification [10].
Figure 2. Mass spectrometry-based profiling tRNA modifications.
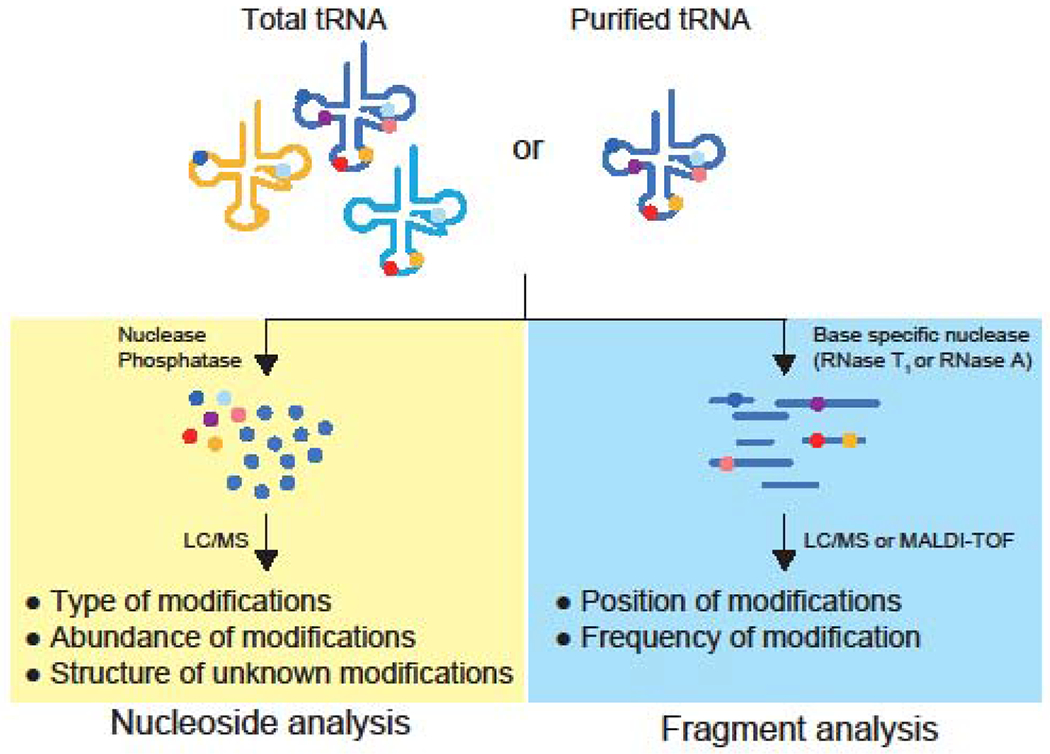
Schematics of nucleoside analysis (left) and fragment analysis (right) are shown.
To elucidate the chemical structure of an unknown modification, chemical formula can be predicted based on an accurate mass value determined by high-resolution mass spectrometric analysis (Figure 2). There are several methods to predict the chemical structure of modifications. Tandem mass spectrometry (MS/MS) provides nucleoside fragmentation patterns that are useful for determining the nucleoside structure [11–13]. Once the structure of a new modification is proposed, a standard can be chemically synthesized to verify that an unknown nucleoside has the same mass value, retention time, and MS/MS fragmentation as those of the standard. Furthermore, in-depth structural analyses, such as NMR and X-ray crystallography, are required to robustly validate proposed structures of unknown modifications[14,15].
Metabolic labeling experiments are useful to investigate the origin of a chemical moiety in a modification [11,16]. Using this approach, the incorporation of stable isotope labelled atoms from metabolites is detected as a mass shift of the target nucleoside. MS/MS analysis of labeled nucleosides yields the positions of labeled atoms in the nucleosides. Ex vivo deutrium exchange experiments are also useful for estimating the number of hydrogens exchangeable with solvent [11].
LC-MS analysis of the total tRNA fraction prepared from an organism reveals what modifications are present. Although LC-MS is not intrinsically quantitative, with appropiate internal standards (Box 1), this analysis also yields quantification of each modification, enabling the study of the link between the amount of tRNA modification and a cellular condition. The abundance of a modification is influenced by the modification frequency on each tRNA molecule (i.e., the ratio of modified tRNA to total tRNA of that species) and the abundance of the tRNA species containing that modification. However, studies of pooled nucleosides do not allow assignment of modifications to specific positions or specific tRNA species.
Box 1. MS-based quantification of tRNA modifications.
A mass spectrometer detects ions generated by an ionization source. Several confounding factors can complicate quantification of modifications using LC-MS. First, ionization efficiency largely depends on the chemical nature of analytes; thus, signal intensities among different modifications are intrinsically not comparable. Second, the signal intensity of an analyte can be suppressed by other compounds eluted at the same retention time due to the capacity of the detector. Third, the condition of a mass spectrometer, including the status of the ion source or the detector, can vary between runs, altering the ion detection efficiency.
Several calibration methods are used to eliminate these confounding factors. When a synthetic standard is available, the abundance of a modification can be determined with an external calibration curve generated by serial dilutions of the synthetic standard. However, signal suppression effects by other compounds can differ between the external standard and experimental samples; normalization with an internal reference can cancel such signal suppression. The ratio of the signal of a target modification to that of an internal reference canonical nucleoside (i.e., A, U, G, C) or to a presumably consistent modification can be used for monitoring the differences in the amount of the target modification in different samples [46].
Stable isotope-labeled internal standards (SIL-IS) are ideal for absolute quantification. Given that a SIL-IS has the same chemical properties as the compound of interest, it has the same ionization efficiency and retention time, circumventing the confounding factors described above. A hurdle for SIL-IS based calibration is the lack of availability of SIL-IS for a variety of modifications. The Helm and Kellner group have prepared mixtures of SIL-IS from bacteria and yeast cells grown with stable isotope-labeled metabolites [52,53] . The absolute amount of each labeled modification is calculated with an external calibration curve of synthetic chemical standards. These mixtures of SIL-IS enable the simultaneous absolute quantification of many modifications.
RNA Mass spectrometry can also be used to identify modification sites through fragment analysis (Figure 2). A mixture of tRNA fragments produced by digestion with base-specific ribonucleases are analyzed by LC-MS or matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) [17,18]. Observation of a fragment whose mass deviates from the predicted value indicates the presence of one or more modifications. Furthermore, MS/MS analysis enables assignment of a modification to a specific nucleotide in the tRNA fragment [19]. The shift in mass value enables prediction of the type of modification, such as methylation or acetylation, but not the exact chemical structure of the modified nucleoside. These two complementary methods, nucleoside analysis and fragment analysis, are generally used together for profiling tRNA modifications.
The presence of dozens of tRNA species in an organism complicates fragment analyses of total tRNA fractions because fragments from different tRNAs may share the same mass value. Purification of individual tRNAs is often conducted to accurately map modifications on specific tRNAs. Hybridization based purification is commonly used for isolation of individual tRNAs [20,21], whereas 2D polyacrylamide gel electrophoresis is used to isolate multiple tRNAs at once [22]. The nucleoside and fragment analyses of purified tRNAs yield the chemical identity and positions of modifications in a specific tRNA [23]. Recent improvements in LC separation, MS sensitivity, and computational techniques have heightened the power of fragment analysis for direct profiling of tRNA modification in total tRNA preparations, without the need to purify individual tRNAs [24–27].
Sequencing-based methods
High-throughput RNA sequencing is an alternative way to locate modification sites on RNAs. In tRNA sequencing (tRNA-seq), tRNAs are converted to complementary DNA by reverse transcription (RT). Some modifications, especially modifications in the Watson-Crick base pairing interface, perturb reverse transcription, resulting in incorporation of a wrong base (misincorporation) or termination [28]. Such reverse transcription derived signatures (RT-signatures) in the sequencing data enables the prediction of modified sites (Figure 3). In Escherichia coli, tRNA-seq produces RT-signatures that can be mapped to half of the known modifications [13]. The other half of modified nucleosides do not generate RT-signatures likely because the structural changes in the modifications do not significantly disrupt Watson-Crick base pairing.
Figure 3. Sequencing-based method to predict sites of modification on tRNAs.
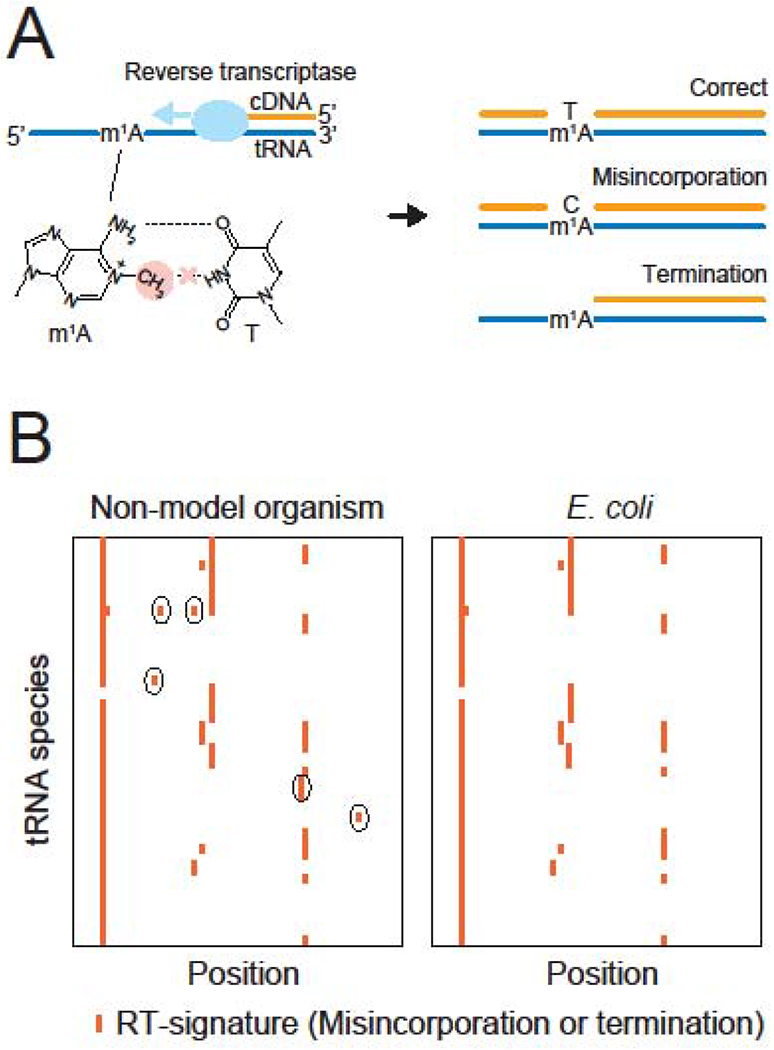
(A) Modifications located in the Watson-Crick base pairing interface inhibit the incorporation of correct basesduring reverse transcription. For example, steric hindrance between the methyl-modification in m1A (red circle) and thymine (T) prevents the canonical A-T Watson-Crick base pairing (dashed lines). This disruption generates RT-signatures, including incorporation of a wrong base (misincorporation) and termination of RT.
(B) Schematic heatmaps represent frequencies of misincorporation or termination of RT in non-model organism (left) and E. coli (right). Each column and row represents a position and a species of tRNA, respectively. Detected RT-signatures are depicted as red lines. The RT-signatures observed in non-model organism but absent in model organism (E. coli) are circled. These RT-signatures represent potential organism-specific modifications.
Several ex-vivo chemical treatments have been described for making specific silent modifications detectable, broadening the scope of identifiable modifications [29,30]. Some tRNA methylations interfere with reverse transcription, resulting in RT-signatures. Two groups developed tRNA-sequencing techniques coupled with ex-vivo demethylation (DM-tRNA-seq and ARM-seq). In these techniques, promiscuous demethylases are used to eliminate RT-signatures derived from methylated nucleosides, allowing rapid assignment of methyl-modifications [31,32]. The demethylation procedure also increases sequencing coverage of the 5’ end region of tRNA through elimination of premature RT termination caused by methyl-modifications [31,32]. Although these additional chemical or enzymatic treatment provide hints to the types of modification, RT-signatures are not usually in themselves sufficient to determine the structures of modifications. A recent study takes advantage of machine learning technology, attempting to predict modification species by the RT signatures [33].
Other methods
There are several tRNA modification detection methods that are based on gel electrophoresis supplemented with chemicals that interact with specific modifications. These chemicals retard the mobility of specific modified tRNA molecules. For example, borate in APB gels and mercury in APM gels cause gel shifts for tRNAs with queuosine [34] and thiolated modifications [35], respectively. Such electrophoretic methods are combined with northern blotting analysis to quantitatively measure the modified fraction in a specific tRNA species.
RNA modifications that have an impact on Watson-Crick base pairing also suppress hybridization of complementary DNA probes to the modified region. A recent study took advantage of such inhibitory effects of RNA modifications on in situ hybridization to detect modifications in the cell. Microscopic analysis enables the detection of modifications at the single cell level [36].
Applications of tRNA modification profiling
Identification of new tRNA modifications
While conventional MS techniques are invaluable for discovery of new tRNA modifications [25,37,38], advanced MS techniques have also proved useful for characterizing modifications, even in well-studied organisms. For example, a triple quadrupole mass spectrometer (QqQ) was used to analyze E. coli tRNA modifications [10,39]. Unlike traditional mass spec, QqQ has higher sensitivity because of its ability to enrich for ions showing specific fragmentation patterns during MS/MS. A multiple reaction monitoring (MRM) method using QqQ enables highly sensitive and quantitative analyses of known tRNA modifications [40]. To search for new modified nucleosides, distinguishing signals of putative modifications from other signals derived from contaminants is critical. Neutral loss scan (NLS) enables specific detection of putative nucleosides by screening for ions that lose a ribose moiety (132 Da) during MS/MS fragmentation [10]. NLS analysis revealed several new modifications in E. coli [10,39]. One of the modifications is generated by a non-enzymatic methylation of a thiolated nucleoside with alkylation reagents. Addition of an alkylation reagent to cultures drastically increased the level of this modification, which is rare under normal condition. These findings revealed the presence of a novel modification even in an extensively studied organism (like E. coli) and that new modifications can exist only in specific conditions.
tRNA modifications have only been well characterized in a few organisms in the three domains of life, including E. coli in bacteria, Saccharomyces cerevisiae in eukaryotes, and Haloferax volcanii in archaea [1] (Figure 1B). The tRNA modification profiles of most non-model organisms are largely uncharacterized. These organisms can have known as well as previously uncharacterized modifications. A non-model organism bearing homologs of genes encoding tRNA modifying enzymes that are well-characterized in model organisms can be predicted to have the same modifications. However, some modifying enzymes are known to have different substrate specificity among their homologs [41]; thus, the modification sites or tRNA substrate species may differ from a model organism even if the modifying enzymes are quite similar.
In the past few years, LC-MS analysis of total tRNA fractions has been carried out in a variety of non-model organisms [40,42–44]. These studies, which used both MRM and NLS analyses, identified sets of known modifications as well as potential new modifications [42]. In addition, a conventional fragment analysis of individually isolated tRNAs enabled the discovery of a new but as yet uncharacterized modification in Staphylococcus aureus [22]. Further investigation of non-model organisms will likely lead to many more discoveries of new modifications.
Sequencing based analyses have also been applied to non-model organisms to profile known modifications and explore new organism-specific modifications. In several bacterial species, DM-tRNA-seq identified RT-signatures that are not present in E. coli [45]. However, as mentioned above, the determination of the chemical structures of modifications is not feasible using sequencing-based methods alone.
Recently, our group combined tRNA sequencing with RNA mass spectrometric analysis to rapidly survey the tRNA modifications in Vibrio cholerae, an organism whose complement of tRNA modifications were largely uncharacterized [13]. tRNA-seq enables rapid prediction of modification sites based on RT-signatures. A comparison of the maps of RT-signatures in E. coli and V. cholerae pointed to sites of potential V. cholerae specific modifications. Through in-depth mass spectrometric analyses, including nucleoside and fragment analyses with purified tRNAs, a new modification -acacp3U (acetylated acp3U)- and a new base conversion process -C-to-Ψ editing-were discovered in V. cholerae tRNAs. Given that V. cholerae and E. coli are phylogenetically close, these observations suggests that many new modifications await discovery. Moreover, this discovery suggests that comparative tRNA-seq, i.e., comparisons of the RT signatures of well-characterized organisms, like E. coli, to those found in non-model organisms will enable rapid screening for organism specific modifications.
Uncovering the links between tRNA modification profiles and cellular conditions
tRNA modification profiles are associated with intra- and extracellular conditions. Some chemical purturbations are known to alter the abundance and frequency of specific tRNA modifications. When a modification modulates tRNA decoding ability, the frequency of the modification can affect protein synthesis. Total tRNA analyses using LC-MS revealed changes in the abundance of tRNA modification in several organisms in different growth conditions. For example, in S. cerevisiae, the level of thio-modifications are controlled by the availability of intracellular methionine and cysteine [46]. In Mycobacterium bovisBCG, the first letter of the anticodon in a threonyl-tRNA is converted to four different modified nucleosides. The frequencies of these modifications drastically changed in aerobic and hypoxic conditions [40]. Plasmodium falciparum, a causative agent of malaria, showed changes in the abundance of many modifications during the intra-erythrocytic stage of its developmental cycle [43]. The synthesis of t6A, one of the modifications in human mitochodria critical for respiratory activity, is sensitive to CO2 availability [47]. The latter three studies also conducted fragment analysis with purified tRNAs and validated the changes in the frequencies of modifications; furthermore, the changes in frequencies of tRNA modifications were linked to alterations in protein synthesis [40,43,46,47].
High-throughput sequencing is also well-suited for tracking the frequency of many modifications at a time. The diverse and complex microbial communities that constitute the microbiota consist of thousands of species of bacteria, making studies of their tRNA modifications impossible without high-throughput methods. tRNA-seq was used for determining the sites and frequency of modifications in tRNAs from the murine gut microbiota [45]. This clever application of tRNA-seq showed that the frequency of two modifications, s4U and nHA, is linked to the host diet, and can partly account for changes in translation in the microbiota [45].
The mechanisms that control modifications can also vary among organisms. The level of acp3U modification is altered between log and stationary phase in E. coli and V. cholerae but in opposite directions. How the frequency of modification in each bacteria depends on the growth phases remains to be investigated [13]. tRNA-seq has also been used to detect the depletion of multiple tRNA modifications caused by a pathogenic mutation in human mitochondrial tRNA [48].
Electrophoresis-based methods are also useful for tracking changes in specific modifications. Using the APB-gel, which can separate tRNAs with and without queuosine, the frequency of this anticodon modification was found to vary between different developmental stages in the fly [49] and in mice fed different diets [50]. These changes in modifications were linked to the modulation of global translation. These examples suggest that diverse mechanisms dynamically control tRNA modification profiles depending on environmental conditions and cellular states in order to optimize translation. Elucidating these mechanisms is an exciting area for future research.
Perspectives
Recent technological advances, especially enhanced LC-MS methods and emerging tRNA-seq techniques, enable the rapid surveillance of tRNA modification profiles. Since our knowledge of the full complement of tRNA modifications is restricted to very few model organisms, many diverse chemical modifications await discovery. New sequencing technologies, such as direct RNA sequencing with PacBio and nanopore technologies have promise for accelerating discovery of novel modifications, though many challenges must be overcome.
These advances will facilitate investigation of the mechanisms that govern tRNA modifications, a topic that has received relatively little attention to date, as well as studies of the functional roles of known and new tRNA modifications. We speculate that new modifications will include cellular metabolites in their structures and that such modifications will function as conduits, linking specific metabolic pathways to control of protein synthesis. Ribosome profiling will enable systematic evaluation of the impact of tRNA modifications on translation at both the gene and codon level. As illustrated by a global analysis in yeast [5], coupling ribosome profiling-based analyses of translation with modification deficient conditions will enable systematic investigation of the functions of tRNA modifications on translation.
Discovery of new chemical structures of tRNA modification will also lead to discovery of new tRNA modifying enzymes and pathways. Coupling knowledge of the presence or absence of specific tRNA modifications in a range of organisms with comparative genomics is a fruitful approach for identifying candidate tRNA modification enzymes[13,16,41]. We speculate that as the number of available modification profiles in different organisims increases, systematic comparative genomic approaches will become increasingly feasible to identify the genes responsible for their biogenesis. Ultimately, understanding the mechanisms of such enzymes will enable their re-engineering, potentially yielding new reagents for modifying nucleic acids for a variety of applications including new base editors, a precise technology for genome editting that was derived from a tRNA modifying enzyme[51].
Highlights.
Diverse chemical modifications of tRNAs (tRNA modifications) modulate protein synthesis
Recent approaches based on high-throughput tRNA sequencing and RNA mass spectrometry enable rapid identification of the structures and sites of tRNA modifications
Novel modifications have been discovered in a variety of organisms.
tRNA modification frequencies are regulated by environmental conditions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
None.
References
- 1.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J: tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009, 37:D159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. : MODOMICS: a database of RNA modification pathways−-2013 update. Nucleic Acids Res 2013, 41:D262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjork GR, Hagervall TG: Transfer RNA Modification: Presence, Synthesis, and Function. EcoSal Plus 2014, 6. [DOI] [PubMed] [Google Scholar]
- 4.Lorenz C, Lunse CE, Morl M: tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ: Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol Cell 2017, 68:978–992 e974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nedialkova DD, Leidel SA: Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosjean H, Droogmans L, Roovers M, Keith G: Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol 2007, 425:55–101. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y: Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol 2007, 425:211–229. [DOI] [PubMed] [Google Scholar]
- 9.Miyauchi K, Kimura S, Suzuki T: A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 2013, 9:105–111. [DOI] [PubMed] [Google Scholar]
- 10.Dal Magro C, Keller P, Kotter A, Werner S, Duarte V, Marchand V, Ignarski M, Freiwald A, Muller RU, Dieterich C, et al. : A Vastly Increased Chemical Variety of RNA Modifications Containing a Thioacetal Structure. Angew Chem Int Ed Engl 2018, 57:7893–7897. [DOI] [PubMed] [Google Scholar]; Highly sensitive MS analysis of E. coli tRNAs discovered dozens of putative nucleosides with unknown mass values that do not correspond to known modified nucleosides. The structure of one of these nucleosides was determined.
- 11.Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T, Suzuki T: Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat Chem Biol 2010, 6:277–282. [DOI] [PubMed] [Google Scholar]
- 12.Jora M, Burns AP, Ross RL, Lobue PA, Zhao R, Palumbo CM, Beal PA, Addepalli B, Limbach PA: Differentiating Positional Isomers of Nucleoside Modifications by Higher-Energy Collisional Dissociation Mass Spectrometry (HCD MS). J Am Soc Mass Spectrom 2018, 29:1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura S, Dedon PC, Waldor MK: Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat Chem Biol 2020, in press 10.1038/s41589-020-0558-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper combines comparative tRNA-seq with RNA mass spectrometry for profiling the landscape of tRNA modifications in Vibrio cholerae. This approach led to the discovery of a new modification and should be widely applicable.
- 14.Matuszewski M, Wojciechowski J, Miyauchi K, Gdaniec Z, Wolf WM, Suzuki T, Sochacka E: A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res 2017, 45:2137–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T: A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem 1988, 263:9261–9267. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi T, Miyauchi K, Sakaguchi Y, Yamashita S, Soma A, Tomita K, Suzuki T: Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat Chem Biol 2018, 14:1010–1020. [DOI] [PubMed] [Google Scholar]
- 17.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA: A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res 1993, 21:4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douthwaite S, Kirpekar F: Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol 2007, 425:3–20. [DOI] [PubMed] [Google Scholar]
- 19.Ross R, Cao X, Yu N, Limbach PA: Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry. Methods 2016, 107:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsurui H, Kumazawa Y, Sanokawa R, Watanabe Y, Kuroda T, Wada A, Watanabe K, Shirai T: Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal Biochem 1994, 221:166–172. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Suzuki T: Chaplet column chromatography: isolation of a large set of individual RNAs in a single step. Methods Enzymol 2007, 425:231–239. [DOI] [PubMed] [Google Scholar]
- 22.Antoine L, Wolff P, Westhof E, Romby P, Marzi S: Mapping post-transcriptional modifications in Staphylococcus aureus tRNAs by nanoLC/MSMS. Biochimie 2019, 164:60–69. [DOI] [PubMed] [Google Scholar]; In this paper, fragment analysis of purified Staphylococcus aureus tRNAs determined the sites of a majority of known tRNA modifications and suggested the presence of a novel modified nucleoside.
- 23.Suzuki T, Suzuki T: A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 2014, 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu N, Jora M, Solivio B, Thakur P, Acevedo-Rocha CG, Randau L, de Crecy-Lagard V, Addepalli B, Limbach PA: tRNA Modification Profiles and Codon-Decoding Strategies in Methanocaldococcus jannaschii. J Bacteriol 2019, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai Y, Miyauchi K, Kimura S, Suzuki T: Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res 2016, 44:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulines MJ, Wetzel C, Limbach PA: Using spectral matching to interpret LC-MS/MS data during RNA modification mapping. J Mass Spectrom 2019, 54:906–914. [DOI] [PubMed] [Google Scholar]
- 27.Wein S, Andrews B, Sachsenberg T, Santos-Rosa H, Kohlbacher O, Kouzarides T, Garcia BA, Weisser H: A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat Commun 2020, 11:926. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes NucleicAcidSearchEngine (NASE), a free open source software that enables assignment of known RNA modifications to reference RNA sequences in fragment analysis. The software allowed for identification of some sites of modifications in human tRNAs, without purification of any specific tRNAs.
- 28.Kellner S, Burhenne J, Helm M: Detection of RNA modifications. RNA Biol 2010, 7:237–247. [DOI] [PubMed] [Google Scholar]
- 29.Helm M, Motorin Y: Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 2017, 18:275–291. [DOI] [PubMed] [Google Scholar]
- 30.Motorin Y, Helm M: Methods for RNA Modification Mapping Using Deep Sequencing: Established and New Emerging Technologies. Genes (Basel) 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T: Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 2015, 12:835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM: ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 2015, 12:879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner S, Schmidt L, Marchand V, Kemmer T, Falschlunger C, Sednev MV, Bec G, Ennifar E, Hobartner C, Micura R, et al. : Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igloi GL, Kossel H: Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res 1985, 13:6881–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igloi GL: Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 1988, 27:3842–3849. [DOI] [PubMed] [Google Scholar]
- 36.Ranasinghe RT, Challand MR, Ganzinger KA, Lewis BW, Softley C, Schmied WH, Horrocks MH, Shivji N, Chin JW, Spencer J, et al. : Detecting RNA base methylations in single cells by in situ hybridization. NatCommun 2018, 9:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagao A, Ohara M, Miyauchi K, Yokobori SI, Yamagishi A, Watanabe K, Suzuki T: Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria. Nat Struct Mol Biol 2017, 24:778–782. [DOI] [PubMed] [Google Scholar]
- 38.Kang BI, Miyauchi K, Matuszewski M, DAlmeida GS, Rubio MAT, Alfonzo JD, Inoue K, Sakaguchi Y, Suzuki T, Sochacka E, et al. : Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res 2017, 45:2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reichle VF, Petrov DP, Weber V, Jung K, Kellner S: NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli. Nat Commun 2019, 10:5600. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, highly sensitive MS analysis uncovered non-enzymatic tRNA alkylation on a thiolated nucleoside in E. coli, revealing the presence of a new modification, in a well-studied organism. Alkylation is promoted by the addition of alkylating reagents and removed from tRNAs by AlkB family demethylase.
- 40.Chionh YH, McBee M, Babu IR, Hia F, Lin W, Zhao W, Cao J, Dziergowska A, Malkiewicz A, Begley TJ, et al. : tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat Commun 2016, 7:13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takakura M, Ishiguro K, Akichika S, Miyauchi K, Suzuki T: Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat Commun 2019, 10:5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grobe S, Doberenz S, Ferreira K, Krueger J, Bronstrup M, Kaever V, Haussler S: Identification and Quantification of (t)RNA Modifications in Pseudomonas aeruginosa by Liquid Chromatography-Tandem Mass Spectrometry. Chembiochem 2019, 20:1430–1437. [DOI] [PubMed] [Google Scholar]
- 43.Ng CS, Sinha A, Aniweh Y, Nah Q, Babu IR, Gu C, Chionh YH, Dedon PC, Preiser PR: tRNA epitranscriptomics and biased codon are linked to proteome expression in Plasmodium falciparum. Mol Syst Biol 2018, 14:e8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Delft P, Akay A, Huber SM, Bueschl C, Rudolph KLM, Di Domenico T, Schuhmacher R, Miska EA, Balasubramanian S: The Profile and Dynamics of RNA Modifications in Animals. Chembiochem 2017, 18:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz MH, Wang H, Pan JN, Clark WC, Cui S, Eckwahl MJ, Pan DW, Parisien M, Owens SM, Cheng BL, et al. : Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat Commun 2018, 9:5353. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper used tRNA-seq to analyze the sites and frequency of tRNA modifications in bacteria grown in vitro and in the murine gut microbiota, revealing a link between host dietary composition and tRNA modification frequency in the microbiota.
- 46.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP: Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 2013, 154:416–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin H, Miyauchi K, Harada T, Okita R, Takeshita E, Komaki H, Fujioka K, Yagasaki H, Goto YI, Yanaka K, et al. : CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat Commun 2018, 9:1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richter U, Evans ME, Clark WC, Marttinen P, Shoubridge EA, Suomalainen A, Wredenberg A, Wedell A, Pan T, Battersby BJ: RNA modification landscape of the human mitochondrial tRNA(Lys) regulates protein synthesis. Nat Commun 2018, 9:3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA: A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 2014, 12:e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Muller M, Ehrenhofer-Murray AE, Dittmar G, Grone HJ, Lyko F: Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that queuosine modification of tRNAs facilitates translation speed at individual codons to prevent protein misfolding in human cell lines and a mouse model.
- 51.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR: Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borland K, Diesend J, Ito-Kureha T, Heissmeyer V, Hammann C, Buck AH, Michalakis S, Kellner S: Production and Application of Stable Isotope-Labeled Internal Standards for RNA Modification Analysis. Genes (Basel) 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M: Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res 2014, 42:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosjean H, Gaspin C, Marck C, Decatur WA, de Crecy-Lagard V: RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics 2008, 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]


