Abstract
Purified dimeric human Hsp90α was used to investigate whether Hsp90 affects the FMN–heme interdomain electron transfer (IET), an essential step in nitric oxide synthase (NOS). The IET rate for rat neuronal NOS (nNOS) displayed a noticeable increase upon adding Hsp90 in a dose-saturable manner, and the effect was abolished by a single charge-neutralization mutation at conserved Hsp90 K585. The kinetic results with added Ficoll 70 further suggest that Hsp90 influences the IET through association with nNOS. The Hsp90 effect is presumably via narrowing the available conformational space for the FMN domain motions.
Keywords: Nitric oxide synthase, Heat shock protein 90, Electron transfer, Kinetics, Docking
Introduction
Nitric oxide (NO) is produced by one of the three isoforms of NO synthases (NOSs), neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS) [1]. These tissue-specific NOSs perform vital physiological functions, and dysfunctional NOSs are involved in numerous fatal diseases that currently lack effective treatment, including stroke and Alzheimer’s disease [1]. Each subunit of the homo-dimeric NOS protein contains a C-terminal reductase domain and an N-terminal oxygenase domain, which are connected by a calmodulin (CaM) binding linker. The oxygenase domain binds heme, l-arginine (l-Arg) substrate, and tetrahydrobiopterin (H4B). The reductase domain comprises NADPH/FAD- and FMN-binding subdomains. All NOS isoforms utilize l-Arg and O2 as substrates, and the electron source is NADPH.
The regulation of NO biosynthesis by NOS in vivo is largely dependent on interactions with other proteins in cells, such as CaM and heat shock proteins [2,3]. CaM is the primary modulator of NOS. In nNOS and eNOS, CaM-binding to NOS is Ca2+-dependent, which requires an influx of calcium into the cells, while iNOS binds to CaM at basal Ca2+ level. CaM binding to the linker between the reductase and oxygenase domains is required for release of the FMN domain from the NADPH/FAD domain, and the subsequent large-scale shuttling motion of the FMN domain to transport the NADPH-derived electron to the catalytic heme domain [4,5]. Beside its binding to the canonical linker, CaM docks to the NOS heme domain [6,7], which is mediated by formation of salt bridges [8]. Facilitating the interdomain FMN/heme alignment by CaM is essential for the functions of all the three NOS isoforms [9].
While Ca2+/CaM is the primary means of activating NOS, CaM alone is not sufficient to regulate the NOS function in vivo, in both time and place, in response to a wide variety of stimuli [10]. Hsp90 is another potent regulator of NOS, and the importance of Hsp90 in modulating NO production of all the three NOS isoforms has been well established both in vivo [2,11] and in vitro [12–14]. Sessa and co-workers have presented elegant studies of molecular aspects of eNOS regulation by Hsp90 [2,15,16]. Early steady state kinetic studies proposed that Hsp90 might facilitate electron transfer across the NOS domains [15,16]. More recently, a domain mapping study showed that the middle domain of Hsp90 and four glutamate residues on the bovine eNOS heme domain are involved in the NOS-Hsp90 interface [17]. The interprotein docking interface emerges as an increasingly attractive target for design of small modulators in general. The “interfacial inhibitor” concept becomes mainstream in drug discovery, and many of such new drugs have been FDA-approved for other targets [18]. It is thus interesting and timely to further elucidate molecular mechanism of NOS regulation by Hsp90.
Despite extensive efforts, the structure-function relationship for the Hsp90-NOS complex remains enigmatic. For example, it is unclear which Hsp90 residue(s) may be involved in interacting with the heme(NOS) domain. The mechanistic understanding has been hampered by mainly three factors: (a) lack of access to recombinant Hsp90 protein (the previous studies were mostly done on commercially available Hsp90 proteins isolated from tissues [19], and thus could not scrutinize mutants); (b) insufficient application of rapid kinetics and spectroscopic tools (the previous studies are mostly limited to steady state kinetic assays [12] due to lack of the detailed methods at that time); and (c) lack of Hsp90-NOS docking model because of inadequate structural information on functional states of Hsp90 and NOS, which resulted in rather random exploration of the Hsp90-NOS interface [17,20]. To understand molecular mechanism of any enzyme, besides the steady state activity assays, one must study discrete catalytic step as a function of change at specific sites.
In this work, with recombinant Hsp90 proteins and new Hsp90-NOS docking models at hand, we have overcome these obstacles to better understand whether (and if so, how) Hsp90 modulates discrete step in NOS function. We chose to investigate the FMN–heme interdomain electron transfer (IET) step. This IET is essential in the delivery of electrons required for O2 activation in the heme domain and the subsequent NO synthesis by NOS [6]. The rates of catalytic reactions at the NOS heme site are generally comparable to that of the IET [21,22]. The FMN–heme IET thus plays a substantial role in the NOS function, which justifies our study of the effect of Hsp90 on the IET kinetics. Although the eNOS-Hsp90 interactions have been extensively studied by Sessa and coworkers [2,15,16,20], little information is known about whether Hsp90 influences nNOS electron transfer. Herein we have directly measured the FMN–heme IET kinetics of rat nNOS with added wild type (wt) or mutant Hsp90 protein. Our results indicate that Hsp90 protein enhances the FMN–heme IET through specific interaction with the NOS heme domain.
Materials and Methods
Materials.
2’,5’-ADP Sepharose 4B resin and Mono Q 10/100 ionic exchange column were purchased from GE Healthcare. β-Nicotinamide adenine dinucleotide phosphate disodium salt was purchased from Biosynth International Inc. Native PAGE was product of Invitrogen.
Rat nNOS Purification.
Recombinant rat nNOSμ was expressed and purified as previously described [23], with a few modifications. The rat nNOSμ plasmid on a vector of pCWori was transformed into E. coli BL21 (DE3) cells. Protein expression was induced at OD600 = 0.8 − 1.0 by addition of final concentrations of 0.5 mM IPTG, 500 μM δ-aminolevulinic acid and 3 μM riboflavin. The harvested cell lysate was applied on a 2’,5’-ADP Sepharose affinity column and eluted with a buffer containing 500 mM NaCl and 10 mM β-nicotinamide adenine dinucleotide phosphate disodium. NADP disodium salt was used instead of NADPH because NADPH may lead to production of superoxide free radical and subsequent consumption of redox-sensitive cofactors during purification. The pooled fractions were exchanged into a pH 7.4 storage buffer (50 mM Tris, 250 mM NaCl, 10 % glycerol, 0.1 mM DTT, 0.1 mM EDTA, 10 μM H4B) using a 100-kDa molecular weight cut-off spin concentrator. The purity of isolated nNOS protein was examined by SDS-PAGE.
Human Hsp90α Purification.
The expression and purification of human Hsp90α were performed using previous procedure [24], with some modifications. The human Hsp90α plasmid on a vector of pET15b containing a N-terminal His-tag is a generous gift from Dr. Daniel Gewirth. Harvested cells were lysed using pulsed sonication on ice to avoid protein aggregation. The protein fraction eluted from a 5-mL HisTrap HP column were exchanged into a pH 8.0 buffer (20 mM Tris, 500 mM KCl and 6 mM 2-mercaptoethanol) and were slowly diluted with 20 mM Tris pH 8.0 buffer to reach a final concentration of 20 mM KCl. The manipulation must be very slow during dilution of the Hsp90 protein. The sample was then filtered and loaded on a Mono Q 10/100 ionic exchange column. The oligomeric states of the eluted fractions from the column were determined by native PAGE, and the dimeric fractions, the native state of Hsp90α, were pooled accordingly (see Figure S1 in Supporting Information). The protein was exchanged and concentrated into a pH 7.5 storage buffer (20 mM Tris, 150 mM KCl, 6 mM 2-mercaptoethanol, 10 % glycerol).
Construction and preparation of D552 rat nNOSμ and K585 Hsp90 mutants.
The plasmids of rat nNOSμ and human Hsp90 mutants were constructed by site-directed mutagenesis using Agilent QuikChange II XL Site-Directed Mutagenesis Kit. The primers for introducing the mutations are listed in Table S1 in Supporting Information. The mutations were confirmed by DNA sequencing at Genewiz LLC (New Jersey, USA). The recombinant Hsp90 and nNOS mutant proteins were then overexpressed and purified as described above.
The FMN–heme IET kinetics measurement by laser flash photolysis.
The kinetics experiments were performed on an Edinburgh LP920 laser flash photolysis spectrometer, in combination with a Q-switched Continuum Surelite I-10 Nd:YAG laser and a Continuum Surelite OPO [25]. [CaM] is 2.3-fold of [nNOS] in all the IET experiments because two CaM molecules binds to one dimeric NOS molecule. Human Hsp90α protein was also added when necessary. The laser flash photolysis experiments were conducted at least twice on separate dates, and the averages of the observed IET rates are listed in tables.
Docking of human Hsp90 and rat nNOS holoprotein.
ZDOCK [26] was used to dock the structures of human Hsp90α and rat nNOS holoprotein. Partial open or closed conformation of human Hsp90α [27] was used as one of the input files. The rat nNOS holoprotein model structure was obtained by fitting crystal structures of the individual nNOS domains’ (pdb 1OM4, 1TLL) and CaM-bound nNOS peptide (pdb 2O60) into the electron microscopy density map [28] using UCSF Chimera [29]. Rat nNOS E539, E543, and D552 residues on the heme domain were included as required contacts because the equivalent residues in bovine eNOS heme domain are involved in the Hsp90-NOS association [17]. Human Hsp90 K585 was also selected as the Hsp90-heme(NOS) contact. No further refinement of the docked structure was attempted. The solvent accessible surface area was obtained for the ZDOCK complex using BIOVIA Discovery Studio 2020.
Modeling of the ionic pairing region at the Hsp90-nNOS interface.
The models of the nNOS 552 and Hsp90 585 pairing regions of the mutant proteins were constructed by starting with the obtained docking complex of the wild type rat nNOS and human Hsp90α proteins. The residues were replaced by corresponding mutated amino acids, using the Rotamers tool in UCSF Chimera version 1.14 [29]. The Dunbrack Rotamer library was used [30] and the rotamer was selected based on probability.
Results and Discussions
We first determined whether Hsp90 influences the FMN–heme IET for rat nNOS by probing the kinetics directly [25]. Milligram pure Hsp90 protein is required for such IET kinetics measurements. The native form of Hsp90α protein is dimeric, while Hsp90 tends to aggregate easily during purification [31]. We optimized the purification protocol to reduce the aggregation and isolate the dimeric form. To assess the protein size and purity, each fraction of eluted Hsp90 from the Mono Q column was analyzed by native PAGE. As shown in Figure 1, Hsp90 dimers (lane “frac32”) can be isolated from those oligomers with larger molecular mass (lane “frac35”); see also the chromatogram in Figure S1. The fractions of Hsp90 dimers (lane labelled “final”) were pooled. According to the native PAGE image, the purity of the dimeric Hsp90 is > 85 %, with an average yield of 5 mg of isolated enzyme per liter of cell cultures. Notably, the purity of commercial Hsp90 proteins are often only ~ 70 %, and their oligomeric state is not defined in the product data sheet. Therefore, the previous studies using the commercial Hsp90 proteins at that time may not faithfully reproduce its native dimeric state.
Figure 1.
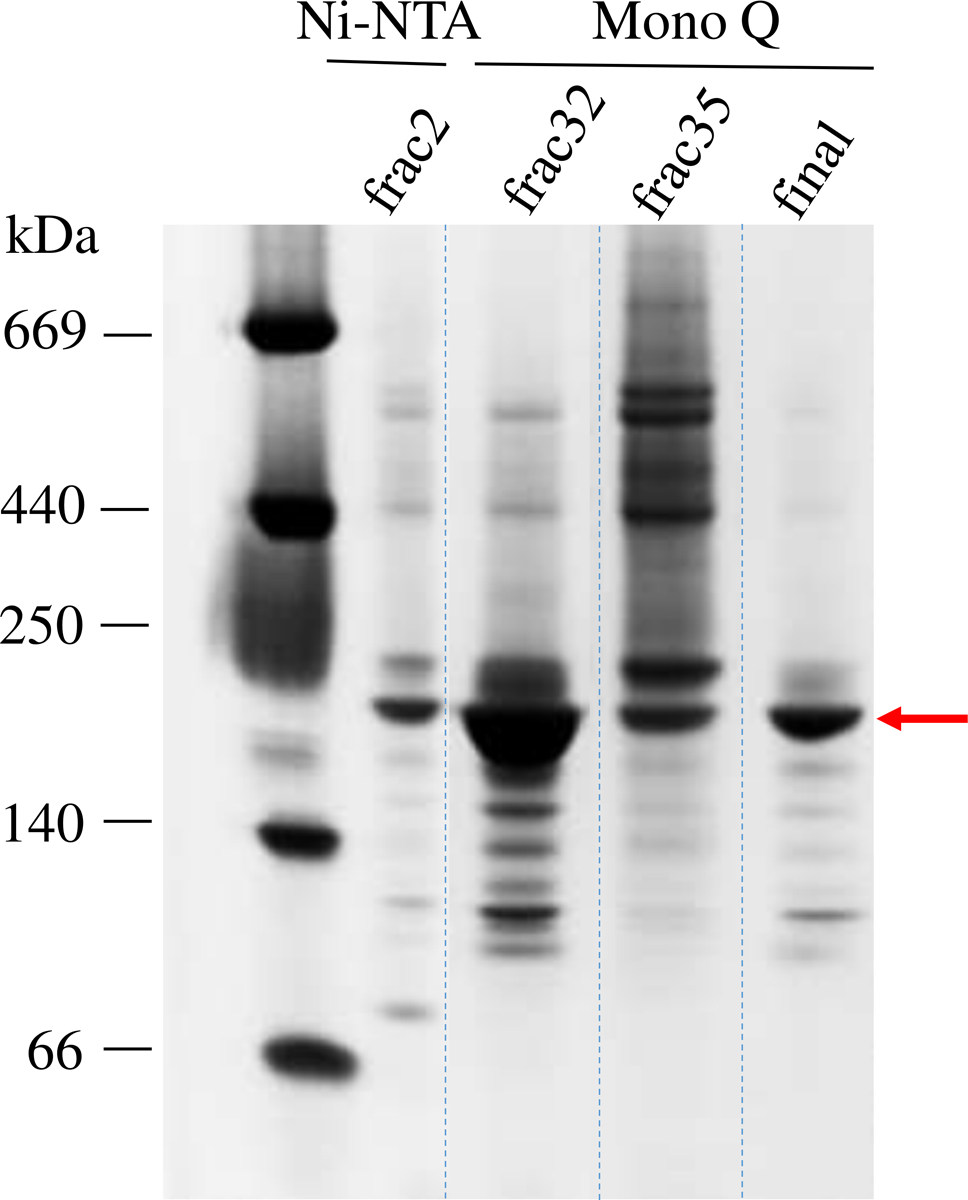
Native PAGE analysis of the fractions eluted from the Ni2+-NTA column and the subsequent Mono Q column. Fraction 32 that showed a majority of dimeric Hsp90 was collected. The last “final” lane shows the size and purity of the isolated Hsp90 protein (marked by an arrow) that was then used for the IET kinetics experiments.
We measured the FMN–heme IET rate in CaM-bound rat nNOS in the presence of the purified dimeric human Hsp90α. CaM was present in all the measurements because it is an essential factor to activate the FMN–heme IET [25]. A typical IET trace at 460 nm in the laser flash photolysis experiments is shown in Figure S2, and the observed IET rates are listed in Table 1. A final concentration of 200 mM NaCl was present in the buffer for the IET measurements since higher ionic strength resulted in dissociation of Hsp90 from NOS [2]. Hsp90 increases the IET rate to a small but noticeable extent (Figure 3A): the observed FMN–heme IET rate of wt nNOS in the presence of 4-fold concentration of wt Hsp90 is 37 ± 2 s−1, which is 37% larger than that of wt nNOS only sample (27 ± 1 s−1); the rate constant for the CaM-bound wt nNOS only sample is the standard of comparison in Figure 3B. Moreover, the enhancement effect is dose-saturable (Figure 3A): the IET rate in the presence of ≥ 4-fold Hsp90 (37 ± 2 s−1) was not further increased; these results indicate that Hsp90 enhances the FMN–heme IET through specific binding to nNOS. The relatively high concentration ratio for saturating the enhancement effect suggests that the Hsp90-NOS binding affinity is modest, which is consistent with the reported value of 200–500 nM [32].
Table 1.
The FMN–heme IET rates of CaM-bound wt rat NOS with or without added wt human Hsp90α
| [nNOS] (μM) | [Hsp90] (μM) | [Hsp90]:[nNOS] | kIET (s−1) |
|---|---|---|---|
| 12 | 0 | 0 | 27 ± 1 |
| 12 | 12 | 1 | 32 ± 1 |
| 12 | 36 | 3 | 35 ± 1 |
| 12 | 48 | 4 | 37 ± 2 |
| 12 | 60 | 5 | 37 ± 2 |
Figure 3.
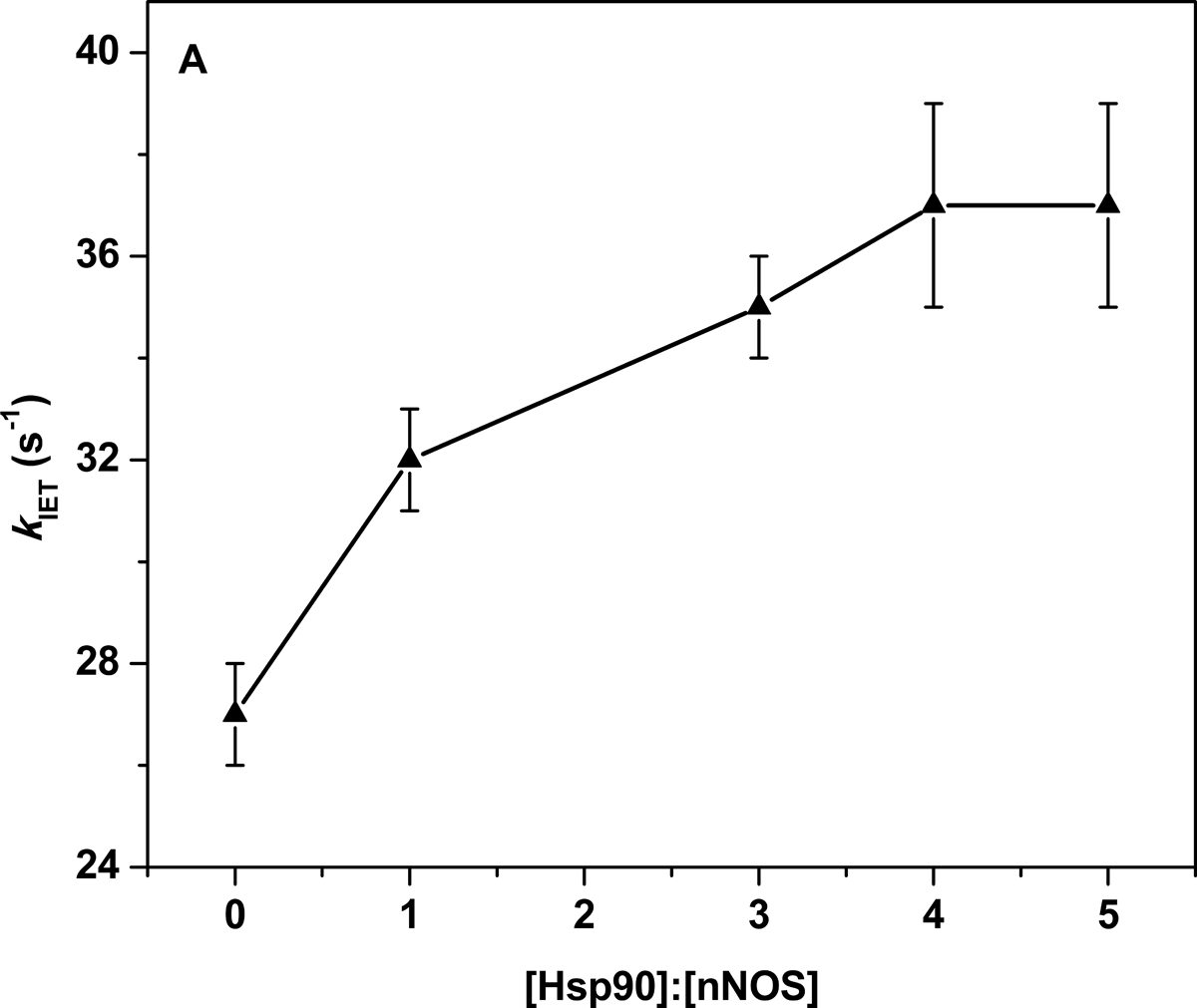
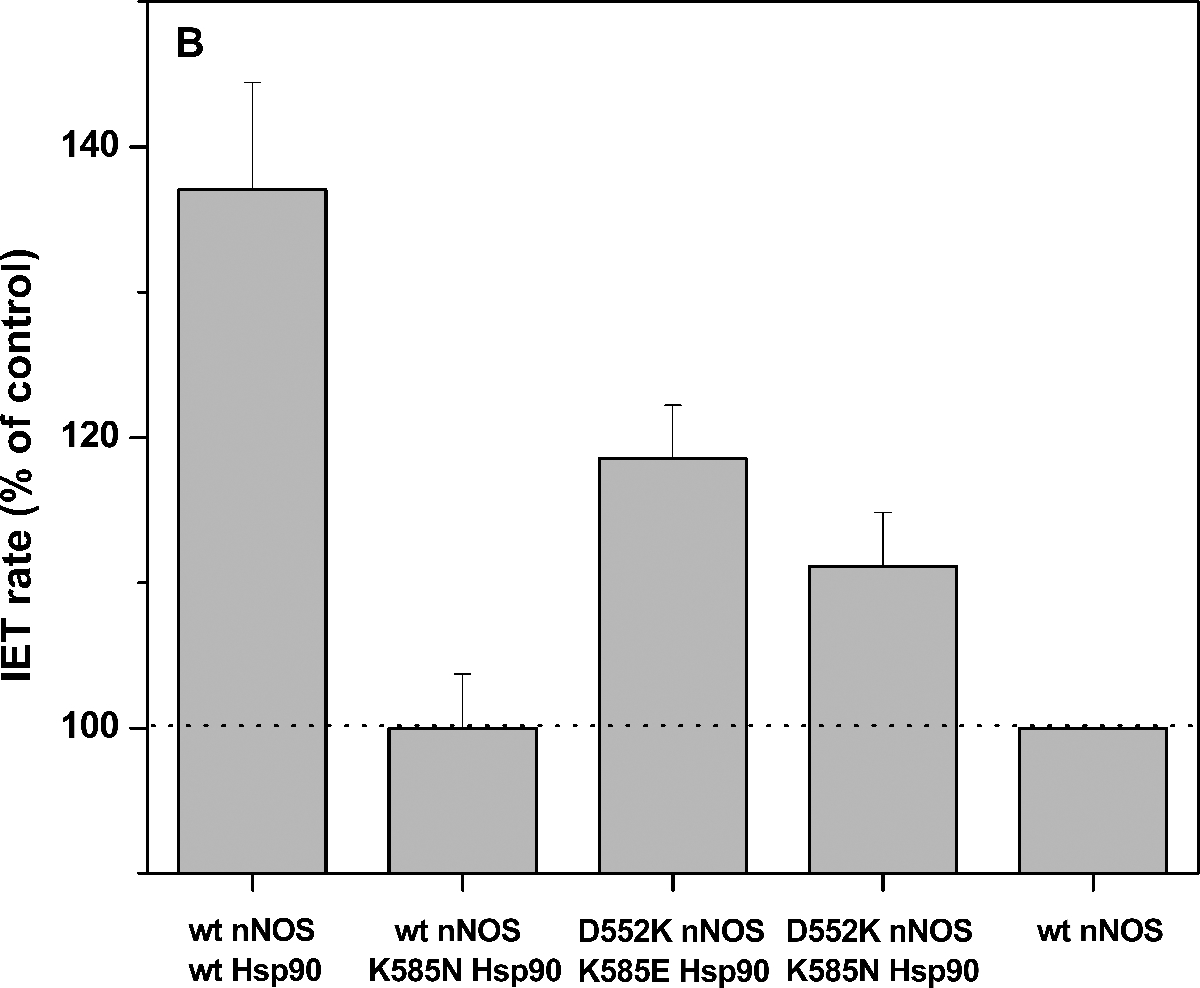
(A) Plot of the FMN–heme IET rates of wt rat nNOS protein with added wt human Hsp90α protein. The enhancement effect saturates at [Hsp90]:[nNOS] ≥ 4:1. (B) Comparison of the FMN–heme IET rates (% of control) of wt or mutant nNOS with added wt or mutant Hsp90α protein ([Hsp90]:[nNOS] = 4:1); dotted line indicates the control, i.e., the CaM-bound wt nNOS only sample. Bars indicate means ± S.D.
We then explored how Hsp90 may enhance the FMN–heme IET in nNOS. To guide rational study of the Hsp90-nNOS interface, we managed to obtain the Hsp90-nNOS docked complex models using ZDOCK [26]. Hsp90 functions through cycling between an open state and an active closed state [33]. On account of the newly available computed conformations [27], either the closed or partially open conformation of human Hsp90α was used as one of the input files for ZDOCK. The crystal structure for the NOS holoenzyme remains elusive due to its large size and flexibility. We generated a model structure of full length rat nNOS by fitting the individual nNOS domains into the recently reported negative-stain EM map [28]. Only a few satisfactory docking models were obtained for rat nNOS holoprotein in complex with Hsp90, where the known NOS heme domain residues and the identified K585(Hsp90) residue (through this work) are implemented at the interprotein interface. Table S2 lists the distances between K585(Hsp90) and the nNOS heme domain residues in the top satisfactory ZDOCK models for the closed Hsp90 conformation. The residues E539, E543 and D552 of rat nNOS are equivalent to those identified for bovine eNOS [17]. Note that the distance should be within 12 Å so that an ionic bond can readily form between the residues.
There are only five ZDOCK models that position the K585(Hsp90) residue at suitable ionic-bonding distance from the nNOS triad (E539, E543 and D552), among which four complexes have a K585(Hsp90)⋯D552(NOS) distance of 2 − 5 Å and one complex has a K585(Hsp90)⋯E543(NOS) distance of 9.1 Å (Table S2). The K585(Hsp90) residue may thus participate in interacting with the triad in the heme(NOS) domain. Figure 2 illustrates a docking model of rat NOS holoprotein docked with Hsp90 in its (active) closed conformation, in which the COO−(D552)⋯NH3+(K585) distance is 4.5 Å. The corresponding residues involved at the docking interface of Hsp90-NOS are shown in the bottom of Figure 2. It is important to note that these residues are conserved in the Hsp90 and NOS isoforms (Figure S3). These conserved interface residues likely assure effective interaction of Hsp90 chaperone with the NOS isoforms.
Figure 2.
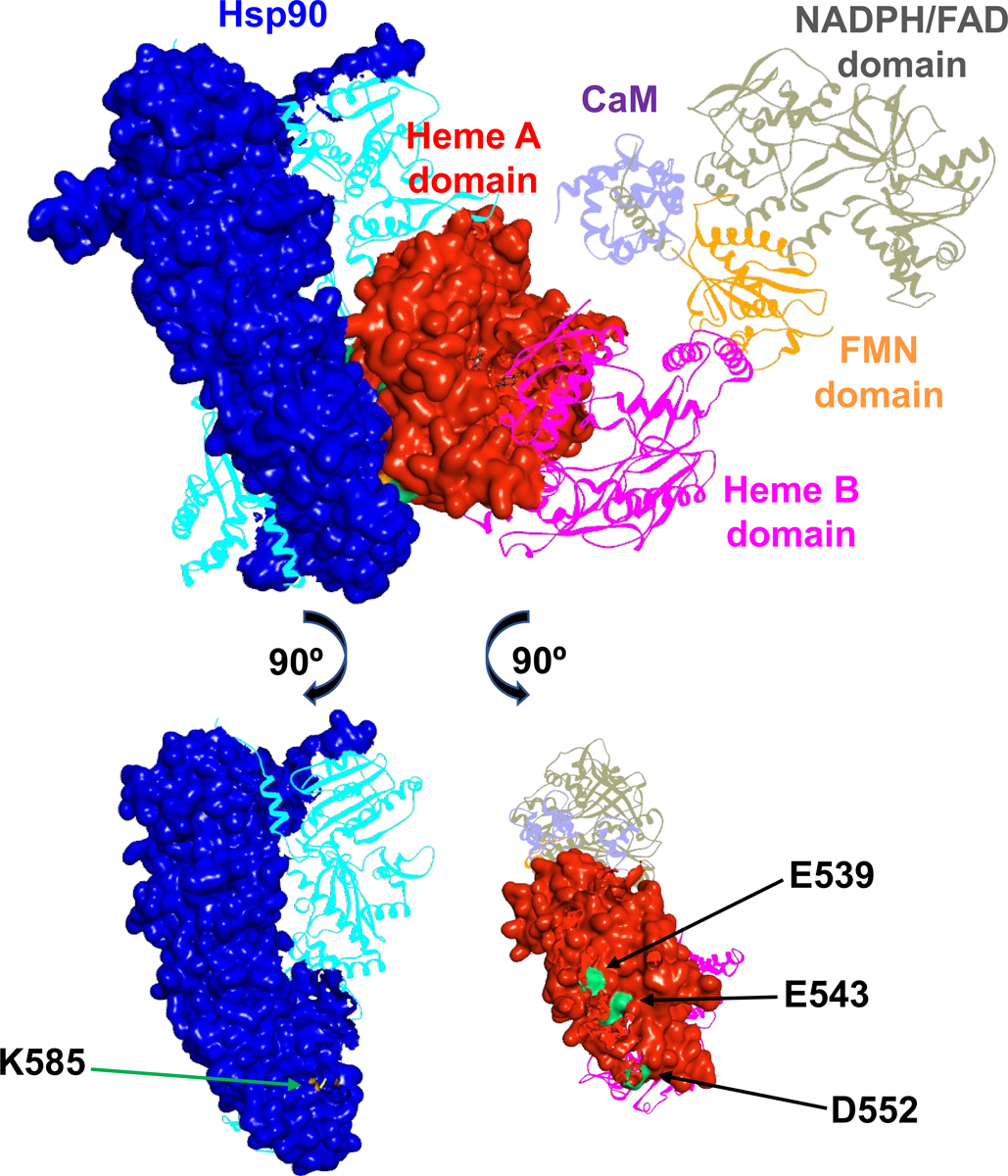
A docking model of rat nNOS holoprotein in complex with human Hsp90α in a closed conformation. The subunits in the dimeric Hsp90 protein are shown in blue and cyan, and the domains of nNOS bound with CaM (purple) are colored differently. Molecular surfaces of the interacting Hsp90 subunit and nNOS heme A domain are depicted, while the other modules are shown in ribbons. The FMN domain departs from the NADPH/FAD domain and is on its way to dock to the heme A domain. Only one reductase domain of nNOS is shown for clarity, along with the dimeric heme-containing oxygenase domain. The bottom panels illustrate the Hsp90 K585 residue (colored in orange) and the rat nNOS heme domain E539, E543 and D552 residues (colored in green) at the predicted Hsp90-nNOS interface.
To assess whether the Hsp90 K585 residue is involved in the docking interface via electrostatic interactions, we measured the IET kinetics of wt nNOS with added K585N Hsp90, a charge-neutralization mutant. The observed IET rate was decreased back to 27 ± 1 s−1 (see Table 2 and Figure 3B), which is identical to that of control sample, i.e., wt nNOS sample in the absence of Hsp90. This single charge-neutralization mutation basically abolished the enhancement effect on the FMN–heme IET by the wt Hsp90. In other words, the Hsp90 K585 residue is indeed involved in the Hsp90-nNOS interactions. We are aware that a classic way to address the essentiality of ion pairs is to use 1 M salt, while 200 mM NaCl was present in the experiments. However, the Hsp90-NOS complex is sensitive to ionic strength and disassembles when washed with 0.5 M NaCl [2].
Table 2.
The FMN–heme IET rates of rat nNOS with added 4-fold final concentration of human Hsp90α protein
| Protein complex | kIET (s−1) |
|---|---|
| wt nNOS + wt Hsp90 | 37 ± 2 |
| wt nNOS + K585N Hsp90 | 27 ± 1 |
| D552K nNOS + K585E Hsp90 | 32 ± 1 |
| D552K nNOS + K585N Hsp90 | 30 ± 1 |
We recognize that the enhancement effect of IET by wt Hsp90 is ~ 37%, which is not that substantial compared to activation of nNOS by CaM (where no IET was observed in the absence of CaM) [34]. On the other hand, the IET rate values for the “wt Hsp90 + wt nNOS” and “wt nNOS only” samples are indeed statistically different: unpaired t test gives a two-tailed P value of 0.0241; by conventional criteria, this difference is considered to be statistically significant. Furthermore, the observed IET rates were obtained on different Hsp90 protein batches. It is also of note that the observed IET rate for the CaM-bound nNOSμ sample (the control in the present work) is in reasonably good agreement with that obtained for a different preparation of nNOSμ protein [23]. Therefore, the difference (~ 37%) is unlikely due to chance. Also note that the small but noticeable increase in the IET rate upon Hsp90 complexing is functionally relevant because similar extent of stimulation in nNOS activity by Hsp90 was reported [35]. The rates of chemical reactions at the NOS heme active site are generally comparable with that of the IET [21,22], and the FMN–heme IET thus plays a substantial role in determining the overall NO production rate. Hence, Hsp90 appears to play an auxiliary but meaningful role in enhancing the essential FMN–heme IET step in nNOS. Moreover, the extent of the IET rates variation by Hsp90 is comparable to those of residues at the FMN-heme docking interface [36].
The distinct difference in the magnitude of effects by CaM and Hsp90 likely stems from their mechanistical roles in the NOS conformational changes. Mounting evidence demonstrated that the IET is gated by conformational change [9], and that the FMN domain explores the conformational space to dock productively onto the heme domain [37,38]. In the absence of CaM, nNOS is predominantly in the electron-accepting input state, with the FMN domain locked to the NADPH/FAD domain [39]. CaM binding releases the FMN domain and enables it to swing between docking positions at the NADPH/FAD and heme domains (input and output states, respectively) through free/open states [40]. Moreover, once CaM docks to the heme domain, the conformational space available to the FMN domain decreases by nearly an order of magnitude compared to the situation when CaM is undocked, because the tether between the bound CaM and FMN domain is much shorter than the full heme-FMN domain tether [37]. This should dramatically increase the probability of formation of IET-competent FMN-heme docking complex. As such, CaM acts as the master switch of the NOS-spanning electron transport. On the other hand, based on our docking complex model (Figure 2), Hsp90 does not seem to have an apparent structural role in the shuttling motions of the FMN domain because it docks onto the heme domain at a site that is apparently apart from where the FMN domain docks (Figure S4). The Hsp90-heme(NOS) docking interface we modelled (Figure 2) shields at a surface area on the heme domain of ~ 677 Å2 (water excluded surface, 1.4 Å probe radius). This value is comparable to that of burial CaM-heme(NOS) surface area (403–568 Å2) [6]. The bound Hsp90 protein occupies a portion of the solvent accessible surface on the heme domain and thus further constrains/limits the free conformational space available for the FMN domain to explore beyond what CaM binding causes by itself. The added constraints caused by bound Hsp90 may further facilitate the docking of the FMN domain at the heme domain. The proposed role of Hsp90 in facilitating the NOS conformational dynamics is supported by the reported shifts in the NOS conformations’ populations upon Hsp90 binding [32]. It is also of note that the separated FMN domain and Hsp90 docking sites on the heme domain (Figure S4) would permit both the FMN domain and Hsp90 to interact with the heme domain spatially and temporally. Perhaps, the interprotein Hsp90-heme(NOS) and intraprotein FMN-heme dockings are relatively independent or synergistic in the context of modulating the FMN–heme IET. These plausible pathways merit investigation.
Importantly, the putative role of Hsp90 in the NOS conformational dynamics is different from the macromolecular crowding effect, although both presumably narrow the available conformational space for the FMN domain motions to enhance the IET. Hsp90 specifically binds to the NOS heme domain, as indicated in this work, whereas the crowding effect is via non-specific interaction [38]. Inert macromolecular solutes can be used to modulate the protein-protein associations in volume-occupied crowding conditions [41]. The experimental observation should fundamentally differ: the enhancement effect by Hsp90 is saturable (Figure 3A), while the macromolecular effect is non-saturable [38]. Indeed, the IET rate of nNOS in the presence of wt Hsp90 (1:4) and 7% Ficoll 70, a commonly used crowder, is 41 ± 2 s−1 (Table 3). Ficoll 70 does not further enhance IET, within statistical error, in comparison to that of NOS with added Hsp90 (37 ± 2 s−1), i.e., the enhancement effect by Hsp90 is saturable and reaches a plateau at this concentration ratio (4:1). Therefore, the observed Hsp90 effect on the IET is primarily via its specific binding to nNOS, not the macromolecular crowding effect. We are aware that the Ficoll 70 concentration (7%) is not high enough to reach the more physiologically relevant fractional volume occupancy. Unfortunately, the nNOS-Hsp90 sample was not stable and became cloudy upon adding more Ficoll 70. We cannot exclude the possibility that macromolecular crowding might have an additive enhancement effect on top of the Hsp90 binding in cellular environments.
Table 3.
The FMN–heme IET rate of wt nNOS in the presence of Hsp90α and Ficoll 70.
| [nNOS] (μM) | [Hsp90] (μM) | [Hsp90]:[nNOS] | Ficoll 70 (%) | kIET (s−1) |
|---|---|---|---|---|
| 12 | 48 | 4 | 7 | 41 ± 2 |
We next started to identify the partnering residues at the Hps90-nNOS interface. Our docking models imply a higher probability of forming the K585(Hsp90)/D552(nNOS) pair (see Table S2). The “Hsp90 K585E + nNOS D552K” pair was used to potentially restore the cross-protein charge interaction between these two sites (Figure 4). Such polarity reversal approach has been used in study of charge interactions in the interdomain interfaces in rat nNOS [42] and human iNOS [8]. The IET rate for this sample (32 ± 1 s−1) exhibits modest recovery toward the wt level (Figure 3B), whereas the effect is still in the expected increasing direction. Additionally, the FMN–heme IET rate for the “Hsp90 K585N + nNOS D552K” sample is 30 ± 1 s−1, also displaying a trend of increase toward the wt level (Figure 3B); although a positively charged lysine residue is introduced at the 552 site in the nNOS D552K mutant, in silico mutation still permits the nNOS K552 and Hsp90 N585 side chains to fit comfortably in the interface area (Figure 4). The observable increased IET rates for the mutants at the nNOS 552 and Hsp90 585 residues imply the existence of pairing interactions between these two sites.
Figure 4.
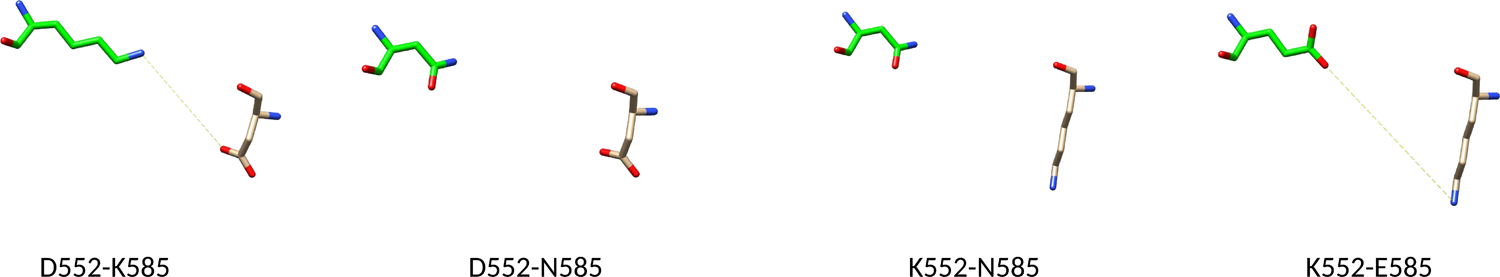
Comparison of the interaction between the rat nNOS 552 and human Hsp90α 585 residues. In the wild type proteins, D552(nNOS) and K585(Hsp90) forms an ionic bond shown as a dotted yellow line.
The much smaller increase in % of control for these two mutant samples (Figure 3B) is presumably due to partial restoration of the ionic bond in the complex because there are two other Glu residues in the triad area of nNOS heme domain (Figure 2). What might be specific in the Hsp90-heme(NOS) interactions is complementary electrostatic surfaces, but not specific ion pairing. Previous work showed that the Hsp90-eNOS association is enabled by electrostatic interactions involving several glutamate residues, which form a cluster of positive charges on the NOS heme domain surface, although each residue does not do much by itself [17].
Hsp90 protein generally cycles among its various conformations including open and closed states [33]. It remains an open and intriguing question whether Hsp90 changes its conformation after it binds nNOS, and whether the D552(nNOS)···K585(Hsp90) ionic bond is maintained during this process. In addition to the docking model for the closed conformation of Hsp90 (Figure 2), we obtained a satisfactory docking model for rat nNOS in complex with a partial open conformation of human Hsp90 (Figure S5), in which the COO−(D552)⋯NH3+(K585) distance is 6.5 Å (Table S3). Therefore, the ionic bond between these two residues may well preserve, at least in the docking complexes of nNOS bound with Hsp90 protein in both closed and partially open conformations. In other words, the interprotein ionic bond would allow some degree of conformational change that is inherent to the Hsp90 protein.
In summary, our data indicated that Hsp90 binding to nNOS enhances the FMN–heme IET rate through specific interactions. The newly identified interface residue (Hsp90 K585) led to selection of several satisfactory docking models, which provides hints on how Hsp90 might facilitate the IET by narrowing the conformational space for the FMN domain to explore. These docking models will not only inspire rational experimental examination of other residues at the dynamic interprotein interface, but also provide initial structures for future molecular dynamics simulations. The proposed role of Hsp90 in modulating the conformational dynamics may be applicable to other interprotein systems.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (GM081811).
Abbreviations
- NO
nitric oxide
- NOS
NO synthase
- nNOS
neuronal NOS
- eNOS
endothelial NOS
- iNOS
inducible NOS
- CaM
calmodulin
- Hsp90
heat shock protein 90
- IET
interdomain electron transfer
- kIET
rate constant for the FMN – heme IET
- wt
wild type
Footnotes
Supporting Information
FPLC chromatograph of Hsp90 purification on a Mono Q 10/100 column; typical kinetic traces in laser flash photolysis experiments; sequence alignment of NOS and Hsp90 proteins; docking model of rat nNOS holoprotein in complex with human Hsp90α in the partial open conformation; primers for constructing human Hsp90α and rat nNOS mutants; distances between interface residues in the top ZDOCK models.
Reference
- [1].Förstermann U and Sessa WC (2012). Nitric oxide synthases: Regulation and function. Eur Heart J 33, 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A and Sessa WC (1998). Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature 392, 821–824. [DOI] [PubMed] [Google Scholar]
- [3].Garcia V and Sessa WC (2019). Endothelial NOS: Perspective and recent developments. 176, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Astashkin AV and Feng C (2015). Solving kinetic equations for the laser flash photolysis experiment on nitric oxide synthases: Effect of conformational dynamics on the interdomain electron transfer. J. Phys. Chem. A 119, 11066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Feng C (2012). Mechanism of nitric oxide synthase regulation: Electron transfer and interdomain interactions. Coordination chemistry reviews 256, 393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Smith BC, Underbakke ES, Kulp DW, Schief WR and Marletta MA (2013). Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci U S A 110, E3577–E3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Astashkin AV, Chen L, Zhou X, Li H, Poulos TL, Liu KJ, Guillemette JG and Feng C (2014). Pulsed electron paramagnetic resonance study of domain docking in neuronal nitric oxide synthase: The calmodulin and output state perspective. J. Phys. Chem. A 118, 6864–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li J, Zheng H, Wang W, Miao Y, Sheng Y and Feng C (2018). Role of an isoform-specific residue at the calmodulin-heme (NO synthase) interface in the FMN - heme electron transfer. FEBS Lett. 592, 2425–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li J, Zheng H and Feng C (2018). Deciphering mechanism of conformationally controlled electron transfer in nitric oxide synthases. Frontiers in bioscience (Landmark edition) 23, 1803–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fulton DJR (2016) Chapter two - Transcriptional and posttranslational regulation of eNOS in the endothelium In Advances in Pharmacology (Raouf AK, ed.êds), pp. 29–64. Academic Press; [DOI] [PubMed] [Google Scholar]
- [11].Vladic N et al. (2011). Decreased tetrahydrobiopterin and disrupted association of Hsp90 with eNOS by hyperglycemia impair myocardial ischemic preconditioning. American Journal of Physiology-Heart and Circulatory Physiology 301, H2130–H2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Song Y, Cardounel AJ, Zweier JL and Xia Y (2002). Inhibition of superoxide generation from neuronal nitric oxide synthase by heat shock protein 90: implications in NOS regulation. Biochemistry 41, 10616–22. [DOI] [PubMed] [Google Scholar]
- [13].Yoshida M and Xia Y (2003). Heat shock protein 90 as an endogenous protein enhancer of inducible nitric-oxide synthase. J. Biol. Chem 278, 36953–8. [DOI] [PubMed] [Google Scholar]
- [14].Chen W, Xiao H, Rizzo AN, Zhang W, Mai Y and Ye M (2014). Endothelial nitric oxide synthase dimerization is regulated by heat shock protein 90 rather than by phosphorylation. PLoS One 9, e105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pritchard KA Jr., Ackerman AW, Gross ER, Stepp DW, Shi Y, Fontana JT, Baker JE and Sessa WC (2001). Heat shock protein 90 mediates the balance of nitric oxide and superoxide anion from endothelial nitric-oxide synthase. J. Biol. Chem 276, 17621–4. [DOI] [PubMed] [Google Scholar]
- [16].Gratton J-P, Fontana J, O’Connor DS, García-Cardeña G, McCabe TJ and Sessa WC (2000). Reconstitution of an endothelial nitric-oxide synthase (eNOS), Hsp90, and caveolin-1 complex in vitro: Evidence that Hsp90 facilitates calmodulin stimulated displacement of eNOS from caveolin-1. J. Biol. Chem 275, 22268–22272. [DOI] [PubMed] [Google Scholar]
- [17].Xu H, Shi Y, Wang J, Jones D, Weilrauch D, Ying R, Wakim B and Pritchard KA (2007). A heat shock protein 90 binding domain in endothelial nitric-oxide synthase influences enzyme function. J. Biol. Chem 282, 37567–37574. [DOI] [PubMed] [Google Scholar]
- [18].Pommier Y and Marchand C (2012). Interfacial inhibitors: Targeting macromolecular complexes. Nature Reviews Drug Discovery 11, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen Y, Jiang B, Zhuang Y, Peng H and Chen W (2017). Differential effects of heat shock protein 90 and serine 1179 phosphorylation on endothelial nitric oxide synthase activity and on its cofactors. PLoS One 12, e0179978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fontana J, Fulton D, Chen Y, Fairchild TA, McCabe TJ, Fujita N, Tsuruo T and Sessa WC (2002). Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ Res 90, 866–873. [DOI] [PubMed] [Google Scholar]
- [21].Chen K and Popel AS (2006). Theoretical analysis of biochemical pathways of nitric oxide release from vascular endothelial cells. Free Radical Biol. Med 41, 668–680. [DOI] [PubMed] [Google Scholar]
- [22].Astashkin AV, Li J, Zheng H, Miao Y and Feng C (2018). A docked state conformational dynamics model to explain the ionic strength dependence of FMN – heme electron transfer in nitric oxide synthase. J. Inorg. Biochem 184, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Panda SP, Li W, Venkatakrishnan P, Chen L, Astashkin AV, Masters BS, Feng C and Roman LJ (2013). Differential calmodulin-modulatory and electron transfer properties of neuronal nitric oxide synthase mu compared to the alpha variant. FEBS Lett. 587, 3973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kirschke E, Goswami D, Southworth D, Griffin PR and Agard DA (2014). Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Feng C, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC and Ghosh DK (2007). Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase. J. Am. Chem. Soc 129, 5621–9. [DOI] [PubMed] [Google Scholar]
- [26].Chen R, Li L and Weng Z (2003). ZDOCK: An initial-stage protein-docking algorithm. Proteins: Structure, Function, and Genetics 52, 80–87. [DOI] [PubMed] [Google Scholar]
- [27].Penkler DL, Atilgan C and Tastan Bishop Ö (2018). Allosteric modulation of human Hsp90α conformational dynamics. J. Chem. Inf. Model 58, 383–404. [DOI] [PubMed] [Google Scholar]
- [28].Yokom AL, Morishima Y, Lau M, Su M, Glukhova A, Osawa Y and Southworth DR (2014). Architecture of the nitric oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J. Biol. Chem 289, 16855–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- [30].Shapovalov Maxim V. and Dunbrack Roland L. (2011). A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 19, 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Moullintraffort L et al. (2010). Biochemical and biophysical characterization of the Mg2+-induced 90-kDa heat shock protein oligomers. J. Biol. Chem 285, 15100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dipak Kumar Ghosh CC, McMury Jonathan L and Salerno John C. (2012). HSP90 Interaction with eNOS and nNOS The FASEB Journal. 26, 756. [Google Scholar]
- 33.[] Hessling M, Richter Kand Buchner J (2009). Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nature Structural &Amp; Molecular Biology 16, 287. [DOI] [PubMed] [Google Scholar]
- 34.[] Feng CJ, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC and Ghosh DK (2007). Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase. J. Am. Chem. Soc 129, 5621–5629. [DOI] [PubMed] [Google Scholar]
- [35].Song Y, Cardounel AJ, Zweier JL and Xia Y (2002). Inhibition of superoxide generation from neuronal nitric oxide synthase by heat shock protein 90: Implications in NOS regulation. Biochemistry 41, 10616–10622. [DOI] [PubMed] [Google Scholar]
- [36].Tejero J, Hannibal L, Mustovich A and Stuehr DJ (2010). Surface charges and regulation of FMN to heme electron transfer in nitric-oxide synthase. J. Biol. Chem 285, 27232–27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Astashkin AV, Li J, Zheng H and Feng C (2019). Positional distributions of the tethered modules in nitric oxide synthase: Monte Carlo calculations and pulsed EPR measurements. The Journal of Physical Chemistry A 123, 7075–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li J, Zheng H and Feng C (2019). Effect of macromolecular crowding on the FMN–heme intraprotein electron transfer in inducible NO synthase. Biochemistry 58, 3087–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Garcin ED et al. (2004). Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem 279, 37918–37927. [DOI] [PubMed] [Google Scholar]
- [40].Campbell MG, Smith BC, Potter CS, Carragher B and Marletta MA (2014). Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci U S A 111, E3614–E3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jarvis TC, Ring DM, Daube SS and von Hippel PH (1990). “Macromolecular crowding”: thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem 265, 15160–7. [PubMed] [Google Scholar]
- [42].Haque MM, Tejero J, Bayachou M, Kenney CT and Stuehr DJ (2018). A cross-domain charge interaction governs the activity of NO synthase. J. Biol. Chem 293, 4545–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


