Abstract
The mammalian Cytochrome P450 (Cyp) gene superfamily encodes enzymes involved in numerous metabolic pathways and are frequently expressed in the liver. Despite the remarkably high sequence similarity of Cyp2a4 and Cyp2a5 genes and their surrounding genomic regions, they exhibit differences in expression in the adult mouse liver. For example, Cyp2a4 is highly female-biased whereas Cyp2a5 is only moderately female-biased and Cyp2a4, but not Cyp2a5, is activated in liver cancer. We hypothesized that the limited sequence differences may help us identify the basis for this differential expression. An antisense expressed sequence tag had been uniquely annotated to the Cyp2a4 gene which led us to investigate this transcript as a possible regulator of this gene. We characterized the full-length antisense transcript and also discovered a similar transcript in the Cyp2a5 gene. These transcripts are nuclear long noncoding RNAs that are expressed similarly to their sense mRNA counterparts. This includes the sex-biased and liver tumor differences seen between the Cyp2a4 and Cyp2a5 genes, but we also find that these two genes and their antisense transcripts are expressed within different zones of the liver structure. Interestingly, while the differences in sex-biased expression of the mRNAs are established 1-2 months after birth, the antisense transcripts exhibit these expression differences earlier, at 3-4 weeks after birth. By analyzing published genomic data, we have identified candidate transcription factor binding sites that could account for differences in Cyp2a4/Cyp2a5 expression. Taken together, these studies characterize the first antisense RNAs within the Cyp supergene family and identify potential transcriptional and post-transcriptional mechanisms governing different Cyp2a4 and Cyp2a5 expression patterns in mouse liver.
Keywords: natural antisense RNA, lncRNA, cytochrome p450, gene duplication, sex-biased expression
1. Introduction
The mammalian Cytochrome P450 (Cyp) gene superfamily encodes numerous enzymes involved in a wide range of processes in adult liver, including metabolism of pharmaceuticals, foreign chemical and pollutants; cholesterol, sterol and bile acid biosynthesis; and steroid synthesis (Nelson et al., 2004). Humans and mice contain 58 and 105 Cyp genes, respectively (Hrycay and Bandiera, 2009). While many Cyps are expressed in a variety of tissues, the majority of mouse Cyps are highly and/or inducibly expressed in the liver, the major detoxification organ (Renaud et al., 2011). Thus, understanding differential Cyp gene regulation can provide valuable information towards understanding molecular pathways that affect chemical toxicity risks and that predispose individuals to disease, including cancer.
In mammals, the liver must perform a number of new metabolic functions at birth that coincide with new sources of nutrition. Levels of many Cyp mRNAs increase dramatically during postnatal liver development and different Cyp genes exhibit distinct developmental expression patterns, governed by a variety of factors including different transcription factors and hormones (Cui et al., 2012). In the liver, some, but not all, mouse and human Cyp genes are also known to exhibit sex-biased expression (Waxman and Holloway, 2009) and to be zonally regulated (Braeuning et al., 2006; Loeppen et al., 2005). In mice, sex-biased gene expression differences are established during the first few months after birth (Conforto and Waxman, 2012) whereas zonal expression initiates shortly before birth (Burke et al., 2018).
The mouse Cyp2a4 and Cyp2a5 genes are highly similar at the DNA sequence level; the proteins encoded by these two genes differ by only 11 amino acids (Lindberg and Negishi, 1989). Interestingly, while Cyp2a4 and Cyp2a5 both catalyze hydroxylation reactions, their substrate specificity differs significantly; Cyp2a4 catalyzes testosterone hydroxylation (testosterone 15α-hydroxylase) whereas Cyp2a5 catalyzes coumarin hydroxylation (coumarin 7-hydroxylase). Cyp2a5 is the mouse ortholog of human CYP2A6, based on their shared catalytic hydroxylation of coumarin and a variety of procarcinogens (Hrycay and Bandiera, 2009). However, humans do not have a Cyp2a4 ortholog. In fact, Cyp2a4 is only found in domestic mouse strains; a wild mouse strain, Mus spretus, only contains Cyp2a5, indicating that Cyp2a4 arose via duplication of Cyp2a5 in Mus musculus domesticus within the last 3 million years (Aida et al., 1994).
Despite their high sequence similarity, these two genes do not have the same expression patterns. Both are expressed in the liver, kidney, and olfactory mucosa, but at varying levels (Squires and Negishi, 1988; Su and Ding, 2004; Su et al., 1996). However, while both are female biased in the liver (Squires and Negishi, 1988), Cyp2a4 is strongly female-biased (female/male ratio of ~1000) while Cyp2a5 is only modestly female-biased (female/male ratio of ~3) (Creasy et al., 2016). Female mice therefore express both Cyp2a4 and Cyp2a5 while male mice express Cyp2a5 nearly exclusively. Liver Cyp2a4 expression is increased in male mice that contain a hepatocyte-specific deletion of the Zinc fingers and homeobox domain (Zhx2) gene (Zhx2ΔHep) whereas Cyp2a5 expression is more modestly increased in male Zhx2ΔHep mice. Finally, in a model of diethylnitrosamine (DEN)-induced liver tumors in male mice, Cyp2a4 expression was much higher in the tumors compared to normal liver, whereas Cyp2a5 expression was not altered in these same samples (Creasy et al., 2016).
Given the high sequence homology but dramatic expression pattern differences, especially the sex-biased expression, we sought to understand how the mouse Cyp2a4 and Cyp2a5 genes are differentially regulated. Because an antisense expressed sequence tag (EST) had been annotated to overlap the promoter region of the Cyp2a4 gene, we hypothesized that antisense transcripts may contribute to differential regulation of Cyp2a4 and Cyp2a5. We fully characterized this Cyp2a4 antisense transcript and its multiple alternatively spliced variants and discovered a similar spliced Cyp2a5 antisense transcript. We then characterized the expression patterns of the Cyp2a4 and Cyp2a5 mRNAs and their corresponding antisense RNAs in male and female developing and adult liver and in liver tumors. In general, we observed that the mRNA and antisense RNAs are expressed with similar sex-biased expression patterns. Interestingly, during post-natal development, the antisense RNAs show statistically significant difference between males and females at earlier timepoints than do the mRNAs. The antisense RNAs are primarily located in the nucleus, which has implications for potential regulatory roles of these transcripts. We compiled available DNase I hypersensitivity (DHS) and chromatin modification data for the Cyp2a4 and Cyp2a5 gene regions and identify differences in predicted transcription factor binding sites within the DHS sites that may contribute to distinct aspects of Cyp2a4 and Cyp2a5 expression. Taken together, these studies characterize the first antisense long non-coding RNAs (lncRNAs) within the Cyp supergene family and identify potential transcriptional and post-transcriptional mechanisms governing different Cyp2a4 and Cyp2a5 expression patterns in mouse liver.
2. Materials and methods
2.1. Mice
All mice were housed in the University of Kentucky Division of Laboratory Animal Research facility according to Institutional Animal Care and Use Committee approved protocols. All mice had ad libitum access to food and water and were maintained on a 14/10 light/dark cycle. Six hours prior to sacrifice, all mice were restricted to water only and all mice were sacrificed between 12–4 pm to limit circadian rhythm variances in gene expression.
2.2. Euthanasia and Mouse Tissue Collection
C57BL/6 male and female mice were euthanized by CO2 asphyxiation at 8-weeks of age. Livers were dissected and immediately snap frozen in a 100% ethanol and dry ice bath and stored at −80°C until RNA isolation. For developmental timepoint experiments, female C3H/HeJ mice were bred with C57BL/6J mice and female mice were monitored for vaginal plugs to estimate the time of fertilization. Pregnant females were euthanized by CO2 asphyxiation at 17.5 days post-conception and embryos were removed. Neonatal and perinatal pups were euthanized by decapitation, and pups older than 14 days were euthanized by CO2 asphyxiation.
2.3. RNA Isolation and Analysis
Liver pieces were weighed and homogenized using tube pestle and RNAzol®RT (1mL/100mg tissue; Molecular Research Center, Inc) following the product protocol for total RNA isolation. RNA concentrations were measured using NanoDrop One (Thermo Scientific).
For reverse transcriptase – quantitative polymerase chain reaction (RT-qPCR) assays, cDNA was synthesized from 1.0 μg of liver tissue RNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). qPCR reactions were prepared with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and amplified in a Bio-Rad CFX96 real-time PCR system. All RT-qPCR runs were done in duplicate and included non-template controls. qPCR Ct values were normalized to Arginine Rich Splicing Factor 4 (Sfrs4) and reported as normalized expression of the indicated gene using the ΔCt method unless otherwise noted.
Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA; Supplemental Table 1). Because the Cyp2a4 and Cyp2a5 genes were highly homologous, primers were designed to place nucleotide differences at the 3’ ends or to span introns when possible. The Cyp2a4- and Cyp2a5-specific qPCR primers were located to the exon 3-to-exon 4 splice junction and within the region of exon 4 where sequence differences between these genes exist. The melting curves for each reaction were examined for single peaks above 80°C to ensure primer specificity. In addition, amplicons from some RT-qPCR reactions were analyzed by polyacrylamide or agarose gel electrophoresis. Based on this type of analysis (see Supplemental Figure 1), RT-qPCR reactions to quantitate Cyp2a4as were carried out with a 70°C annealing temperature to ensure specific amplification, whereas Cyp2a5as RT-qPCR reactions were carried out with a 60°C annealing temperature. The GeneRuler 100 bp plus ladder (Thermo Scientific) was used as the size marker on the gels.
For standard RT-PCR reactions, cDNA was prepared as described above and was amplified using Q5 High Fidelity Polymerase (New England Biolabs) and the gene-specific primers identified in the figures.
2.4. Rapid Amplification of cDNA Ends (RACE)
5’ and 3’ RACE was performed using the 5’/3’RACE Kit 2nd Generation (Roche) following the product protocol and using gene-specific primers (Supplemental Table 1). For 5’ RACE, cDNA was synthesized from 8-week old C57BL/6 female liver total RNA using kit components and Cyp2a4as (primer e) or Cyp2a5as (primer c) specific primers. The cDNA was purified using the Wizard®SV Gel and PCR Clean-Up System (Promega) and poly A-tailing was completed using kit components. Primary PCR amplification of the dA-tailed cDNA used the oligo-dT-Anchor Primer and Cyp2a4as-specific primer (primer g) or Cyp2a5as-specific primer (primer f) and Q5 High Fidelity Polymerase (New England Biolabs). The secondary PCR amplification used 1.0 μL of 1:20 diluted primary PCR reaction, PCR Anchor Primer and Cyp2a4as-specific primer (primer h) or Cyp2a5as specific primer (primer i) and Q5 High Fidelity Polymerase.
For 3’ RACE, cDNA was synthesized from the same female liver total RNA using kit components and the oligo-dT-Anchor Primer. Purified cDNA was amplified using the PCR Anchor Primer and the Cyp2a4as-specific primer (primer k) or Cyp2a5as-specific primer (primer d). For the secondary PCRs, 1.0 μL of the 1:10 diluted primary reaction, PCR Anchor Primer and the Cyp2a4as-specific primer (primer j) or Cyp2a5as-specific primer (primer l) were used.
2.6. Cloning and Sequencing
Amplicons were isolated from agarose gel using the Wizard®SV Gel and PCR Clean-Up System (Promega), ligated into pGEM®-T Easy Vector and transformed into DH5α E.coli. Colonies were screened for inserts by colony PCR and positive colonies were grown for miniprep plasmid isolation (Thermo Scientific GeneJET Plasmid Miniprep Kit) and sequencing (ACGT Inc., Wheeling, IL). Sequences obtained for full-length antisense transcripts were submitted to GenBank, accession numbers MT019821 – MT019825.
2.7. Nuclear and Cytoplasmic RNA Isolation and analysis
Livers were removed from 6–7 week old C57BL/6 male (2) and female (4) mice and immediately prepared for separation of nuclear and cytoplasmic fractions over two sequential sucrose cushions (Schibler et al., 1983). RNA was isolated from the pellet of purified nuclei in 800 μL of TRIzol® reagent following the product protocol (Life Technologies). Cytoplasmic RNA was isolated from 200 μL of the cytoplasmic fraction and 1 ml of TRIzol reagent. One μg of RNA from each fraction was made into cDNA as described above and the abundance of various RNAs were quantitated by qPCR. A relative cytoplasmic to nuclear RNA abundance ratio for each transcript in a single animal was calculated and then this ratio was compared to the relative cytoplasmic to nuclear ratio for ribosomal protein L30 mRNA in the same animal. These relative ratios were averaged among the six animals. This provides a relative enrichment ratio for the antisense transcripts, when compared with known cytoplasmic (mRNAs) and nuclear (unspliced mRNAs and U6 snRNA) RNAs.
2.8. Liver Regeneration and liver tumors
Mouse liver regeneration was induced by a single intraperitoneal (i.p.) injection of carbon tetrachloride (CCl4) as described (Creasy et al., 2016). Eight-month old C3H male mice were administered either mineral oil (MO) or 0.05 mL 10 % CCl4 diluted in MO. After 3 days, a time when most pericentral hepatocytes have been eliminated, animals were euthanized by CO2 asphyxiation, livers were removed and RNA was isolated for RT-qPCR analysis as described previously. For liver tumors, 14-day old C57BL/6 male mice were injected i.p. with diethylnitrosamine (DEN; diluted in PBS) or PBS alone as described (Creasy et al., 2016). After 36 weeks, mice were euthanized by CO2 asphyxiation and livers removed from age-matched controls (PBS-treated; no tumors) and DEN-treated mice; tumor and non-tumor tissue were dissected from DEN-treated mice. RNA was isolated for RT-qPCR analysis as previously described.
2.9. Statistics
All values within a group were averaged and plotted as mean with standard deviation. p-values were calculated as detailed in each figure legend. Data was graphed and statistical tests were performed in GraphPad PRISM.
3. Results
3.1. The sequences surrounding the Cyp2a4 and Cyp2a5 genes are highly similar
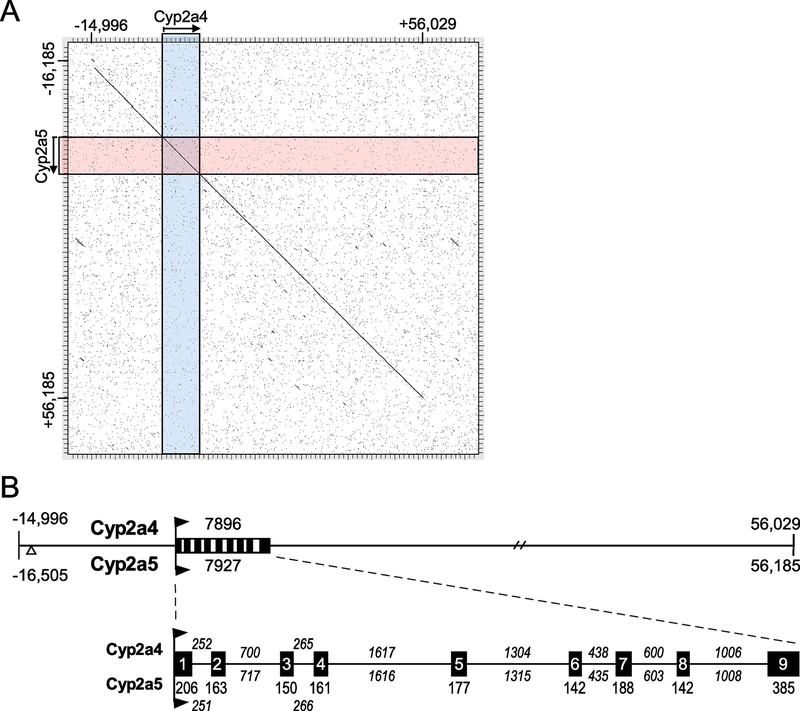
Cyp2a4 arose from a duplication of Cyp2a5 in domesticated mice within the last 3 million years (Aida et al., 1994). While the two enzymes encoded by these genes have acquired different substrate specificity, their DNA sequence remains highly similar. To delimit the extent of the DNA sequence conservation, the genomic sequence regions surrounding these two genes were aligned. This found that a region of strong homology extends roughly 15 kb upstream and 48 kb downstream of these two genes (Figure 1). This ~71 kb region is 96% identical, but the homology completely disappears outside this region. Across the region of the genes, the Cyp2a4 (7,8967 bp) and Cyp2a5 (7,9276 bp) exons are identical in size and their sequence identity ranges from 96% to 99%. The introns are similar, but not identical, in size and their sequence identity ranges from 94% to 98%. There are 31 nucleotide differences between the two mRNAs, 23 in the coding region, 7 in the 3’ untranslated region (UTR) and 1 in the 5’ UTR. Overall, the high sequence similarity over 71 kb of DNA surrounding the Cyp2a4 and Cyp2a5 genes is consistent with a recent duplication event.
Figure 1. The sequences of the mouse Cyp2a4 and Cyp2a5 genes are highly similar over 71 kb.
A. Matrix alignment of the C57BL/6 genomic sequences upstream and downstream of the Cyp2a4 and Cyp2a5 genes using DNA Strider 3.0 (Marck, 1988). A ~71 kb region of 96% identity exists; homology abruptly declines upstream and downstream of this region. B. The duplicated Cyp2a4 and Cyp2a5 regions are shown in more detail. The regions upstream (negative numbers) and downstream are relative to the transcription start site (designated +1) and the sizes of the two genes are shown. The Cyp2a5 upstream region contains a 1312 bp insertion of unrelated sequence, shown as an open triangle. The exon (black boxes) and intron (lines between boxes) sizes for the Cyp2a4 and Cyp2a5 genes are shown below. All exons are the same size while the intron sizes, as designated by numbers above and below lines, show minor variations. Individual exons and introns vary from 94% to 99% identical.
3.2. A potential antisense transcript from the Cyp2a4 gene
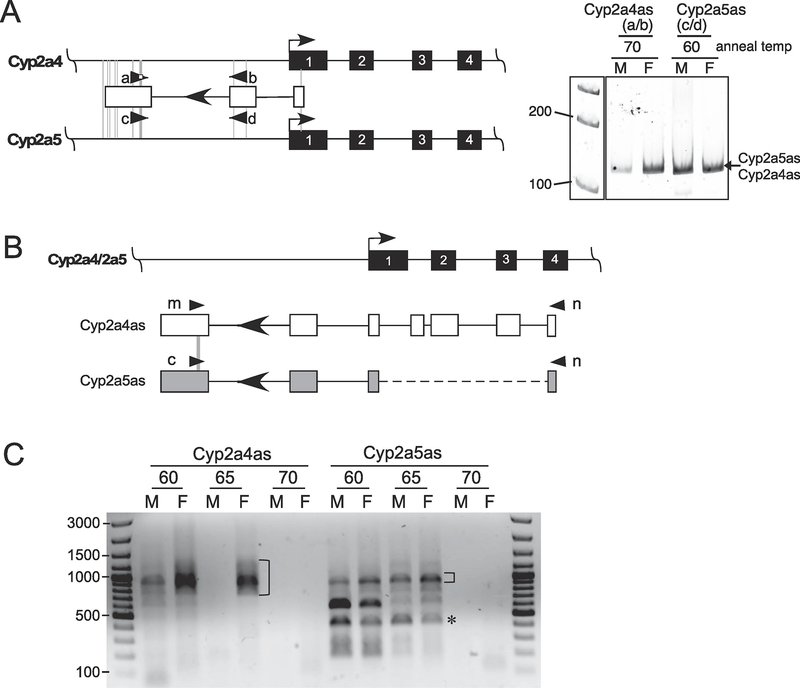
An EST, Gm42375, is annotated in genome databases as an antisense RNA that overlaps with the 5’ end of the Cyp2a4 gene. No such EST is annotated within the Cyp2a5 gene. This led us to consider whether this antisense transcript may contribute to the differential expression patterns of Cyp2a4 and Cyp2a5. To test this, PCR primers were designed for Gm42375 and a possible orthologous antisense transcript in the Cyp2a5 gene and RT-PCR was performed using liver RNA from male and female mice (Figure 2A). Because the sequences of these two genes are so similar, we designed primers that took advantage of a four-nucleotide gap to ensure they specifically amplified transcripts from Cyp2a4 (Figure 2A, primers a and b) or from Cyp2a5 (Figure 2A, primers c and d), should they exist. Initial RT-PCR reactions using Cyp2a4 primers contained multiple bands, suggesting there may be some non-specific amplicons. By titrating the annealing temperature, and sequencing the PCR products, we were able to identify specific RT-PCR products from the antisense strands of both the Cyp2a4 and Cyp2a5 genes using annealing temperatures of 70°C and 60°C, respectively (Figure 2A, right and Supplemental Figure 1, A and B). The antisense RT-PCR product from Cyp2a4, which we called Cyp2a4as, is most prominent in the female sample whereas the antisense RT-PCR product from Cyp2a5, called Cyp2a5as, was amplified to a similar extent in male and female liver RNA. By sequencing, these two products were found to be similarly spliced and the Cyp2a4 product matched the original Gm42375 EST.
Figure 2. Characterizing transcripts antisense to the Cyp2a4 and Cyp2a5 genes.
A. The 5’ ends of the Cyp2a4 and Cyp2a5 genes are shown; exons are black boxes and arrows denote the transcriptional direction. The EST that was annotated antisense to the Cyp2a4 gene is shown as open boxes, lines are the introns and the arrowhead within the intron denotes the transcriptional direction. Vertical thin gray lines designate the nucleotide differences between the two genes in this region; primers to distinguish potential antisense transcripts within Cyp2a4 and Cyp2a5 are diagrammed as arrowheads. cDNA from a male (M) or female (F) liver was amplified with primers a/b and c/d for Cyp2a4 and Cyp2a5, respectively, and products were separated on a 6% polyacrylamide gel, as shown to the right. The PCR annealing temperatures are shown, as established in Supplemental Figure 1. B. Primers specific for Cyp2a4as (m) and Cyp2a5as (c) and a common primer (n) were used to amplify potential fulllength antisense transcripts. The transcript structure based on 5’ RACE results for Cyp2a4as (open boxes) and Cyp2a5as (gray boxes) is shown. Nucleotide differences between Cyp2a4as and Cyp2a5as in the region of the primers are marked by gray lines. C. cDNA was synthesized from male (M) and female (F) nuclear RNA using random hexamers. PCR reactions from different annealing temperatures (60, 65, 70°C) were resolved on a 1% agarose gel; an overexposed view of the gel is shown. Amplicons isolated, cloned and sequenced are marked for Cyp2a4as (bracket) and Cyp2a5as (bracket and *); the bracketed bands were gene-specific while the * amplicon was nonspecific. The size marker is shown on either side of the gel.
3.3. Cyp2a4as and Cyp2a5as transcripts overlap with the Cyp2a4 and Cyp2a5 gene regions
To further characterize the Cyp2a4as and Cyp2a5as transcripts, we first performed 5’ and 3’ RACE using C57BL/6 female liver RNA (Supplemental Figure 1, C and D). The 5’ RACE sequence of several clones ended within exon 4 of the Cyp2a4 gene (see Figure 2B) and, using an online program to predict core promoter elements (Sloutskin et al., 2015), an initiator and bridge sequences were located close to the 5’ end identified by 5’ RACE, consistent with this region being the transcription start site of the antisense RNA. The 3’ RACE amplicons from both Cyp2a4as and Cyp2a5as were found to have the same 3’ end as the initial EST sequence, about 2770 nt upstream from the transcription start site of the overlapping Cyp2a4 and Cyp2a5 genes. A cleavage-polyadenylation signal, AAUAAA, was conserved between Cyp2a4as and Cyp2a5as at an appropriate distance from the 3’ end, suggesting that both transcripts use the same signal for 3’ end processing.
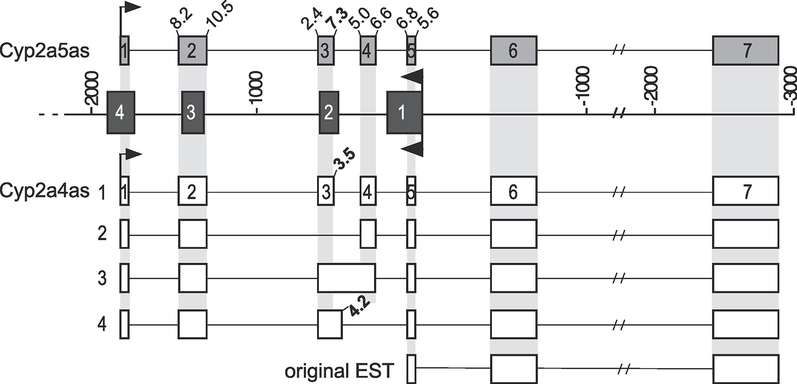
To characterize the full length Cyp2a4as and Cyp2a5as transcripts, we used gene-specific 3’ primers that incorporate the 4 nt gap in the last exon of the transcripts (Figure 2B, primers m and c for Cyp2a4as and Cyp2a5as, respectively) and a 5’ primer located at the 5’ end of the antisense RNAs that could not distinguish Cyp2a4- from Cyp2a5-derived transcripts (Figure 2B, primer n). RT-PCR was carried out using male and female nuclear RNA fractions to enrich for the antisense transcripts (see below). By titrating the annealing temperature, we could ignore some non-specific amplicons that were eliminated at the higher temperatures; the multiple bands that were cloned and sequenced are shown (Figure 2C). This identified four Cyp2a4as spliced isoforms from the broad amplified band in female mice and one Cyp2a5as spliced isoform that is present in both male and female mice (summarized in Figure 3). The Cyp2a4as isoforms all varied within the region that overlaps Cyp2a4 intron 1 and exon 2, while the other Cyp2a4as exons were constant. The single Cyp2a5as transcript we identified was very similar to Cyp2a4as isoform 1 (Figure 3). Interestingly, the Cyp2a5as exon 3 5’ splice junction sequence contains a G at the +1 position whereas Cyp2a4as has an A at this position, making it incapable of being recognized as a functional splice junction. Instead, Cyp2a4as, isoform 1, is spliced using a 5’ splice junction 7 nt further downstream. Based on the maximum entropy score for predicting splice junction strengths (Yeo and Burge, 2004), the 5’ splice junction in Cyp2a5as is stronger (MaxEnt score of 7.3) than the one used in Cyp2a4as, isoform 1 (MaxEnt score of 3.5). This likely accounts for the Cyp2a4as alternative splicing that occurs in this region. Thus, in addition to isoform 1, we found isoforms that skipped exon 3 (isoform 2), that retained intron 3 (isoform 3) and that used an alternative exon 3 5’ splice site but skipped exon 4 (isoform 4). These results are consistent with the exon definition model of splice site choice (Berget, 1995).
Figure 3. Summary of the Cyp2a4as and Cyp2a5as transcripts.
The orientation of the gene and antisense transcripts have been flipped relative to previous figures but the numbering along the gene and the fill styles remain the same. The mRNA genes are shown in the center by black boxes with the transcriptional direction to the left, as noted by the arrows. The antisense transcripts identified by cloning and sequencing multiple RT-PCR products are shown with the transcriptional direction to the right, as noted by the arrows. The single Cyp2a5as transcript is shown above and the four Cyp2a4as transcripts are shown below the genes, with the original Cyp2a4as EST shown at the bottom. The exons of Cyp2a5as and Cyp2a4as isoform 1 are numbered. The light gray highlighted bars denote common exons and splice junctions among the transcripts. The numbers linked to the splice junctions in the region of alternative splicing are the predicted 5’ and 3’ splice junction strengths for the Cyp2a4as and Cyp2a5as transcripts, as predicted by MaxEntScan (Yeo and Burge, 2004). Those that differ between Cyp2a5 and Cyp2a4 are shown in bold. The single nucleotide difference at the 5’ splice junction of antisense exon 3 is the likely driver of the alternative splicing seen in Cyp2a4as.
Cyp2a4as transcript lengths ranged between 1098 nt and 1333 nt; the Cyp2a5as transcript was 1235 nt. We used the Coding Potential Assessment Tool to predict the open reading frames (ORF) in each of these antisense transcripts and to assess the probability that they code for proteins (Wang et al., 2013). The ORFs identified were all relatively short, with non-optimal hexamer use, and thus all antisense transcripts were judged by this assessment to be lncRNAs.
3.4. Cyp2a4 and Cyp2a5 mRNA and their antisense RNA levels are higher in female liver
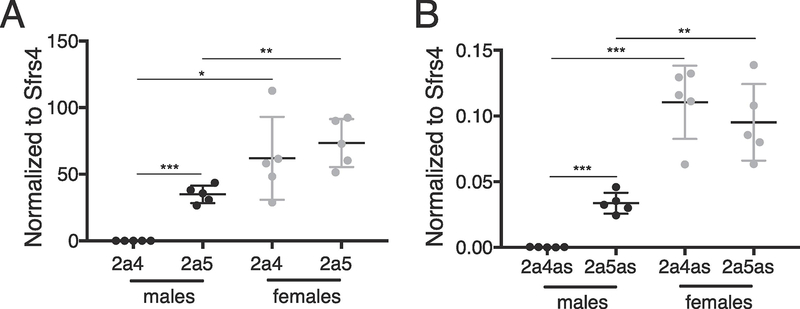
To compare the expression patterns of the Cyp2a4 and Cyp2a5 mRNAs with their corresponding antisense RNAs, we quantitated their RNA levels in liver RNA from 8-week old adult C57BL/6 male and female mice by RT-qPCR (Figure 4). As seen previously (Creasy et al., 2016), Cyp2a4 is much more highly female-biased (>1000-fold) than Cyp2a5 ( ~ 2–3-fold). The corresponding antisense RNAs also show a female bias, with Cyp2a4as being highly female biased ( ~ 500-fold) and Cyp2a5as having a modest female bias ( ~3-fold). Similar results were obtained using adult liver from C3H/HeJ mice (not shown). In total adult liver RNA from both sexes, Cyp2a4as and Cyp2a5as RNA levels range from about 100- to 800-fold lower than their sense mRNA counterparts. This data also shows that in males, Cyp2a5 is expressed much higher than Cyp2a4, whereas both mRNAs are present in roughly equivalent levels in females. The expression patterns of Cyp2a5as and Cyp2a4as are similar to their corresponding mRNAs in males and females.
Figure 4. Sex-biased Cyp2a4as and Cyp2a5as expression mimics those of their overlapping mRNA counterparts.
A. Liver Cyp2a4 and Cyp2a5 mRNA abundance was measured by RT-qPCR and normalized to Serine and Arginine Rich Splicing Factor 4 (Sfrs4) from male (black, n=5) and female (gray, n=5) C57BL/6 mice. B. Cyp2a4as and Cyp2a5as transcripts levels were measured by RT-qPCR, normalized to the control Sfrs4 mRNA in the same samples as in A. Statistics: Student’s unpaired t-test. *=p<.05, **=p<.01, ***p<.001.
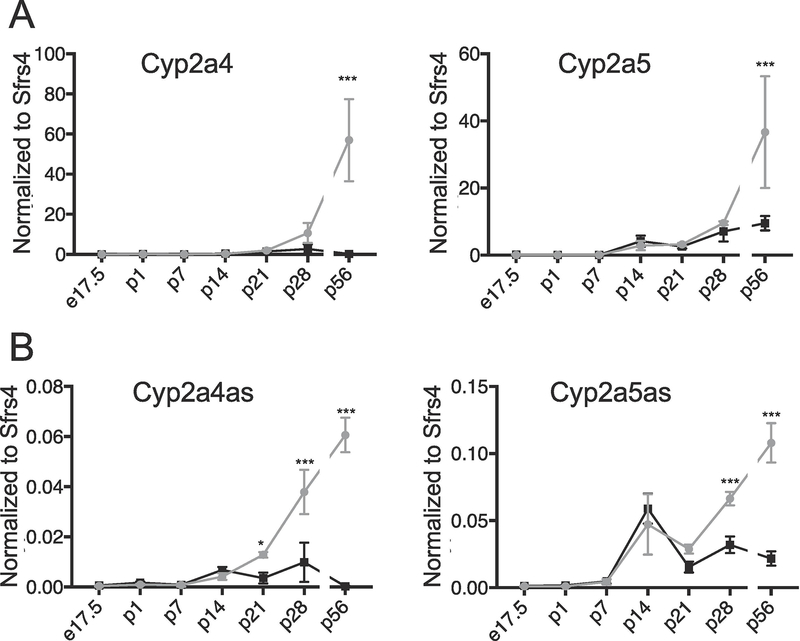
3.5. Expression of Cyp2a4 and Cyp2a5 and their antisense RNAs increases after birth
Many Cyps and other genes involved in drug metabolism show sex differences beginning at sexual maturity (Conforto and Waxman, 2012; Hart et al., 2009). We previously showed Cyp2a4 mRNA levels gradually increased during male and female mouse liver development until postnatal day 28 (p28). However, by p56, male gene expression had decreased, while female expression increased dramatically (Creasy et al., 2016). To compare developmental expression patterns of Cyp2a4as and Cyp2a5as RNAs with their corresponding mRNAs, we measured their levels in female and male liver, beginning at embryonic day 17.5, one day after birth (p1), weekly to day 28 (p28) and again at 8 weeks (p56) in mature adult mice (Figure 5). We found that Cyp2a5 mRNA levels increase similarly in male and female mouse liver until p28; expression is then markedly increased from p28 to p56 specifically in females whereas expression remains constant in males (Figure 5A). Thus, the adult levels of Cyp2a4 and Cyp2a5 mRNA (Figure 4A) are not established until after p28, which is approximately when puberty begins in mice. Interestingly, measuring Cyp2a4as and Cyp2a5as expression at these same developmental timepoints gave slightly different results than their corresponding mRNAs. Cyp2a4as expression in female mice begins to increase and becomes statistically different between males and females at p21 (Figure 5B, left panel). Cyp2a4as expression continues to increase from p21 to p56 in females. Cyp2a5as expression in females becomes statistically different from expression in males by p28 and increases further by p56 (Figure 5B, right panel). Thus, sex-biased expression for both Cyp2a4as and Cyp2a5as occurs earlier in development, preceding their corresponding mRNAs by at least a week.
Figure 5. Cyp2a4 and Cyp2a5 mRNA levels, along with Cyp2a4as and Cyp2a5as, increase during mouse postnatal liver development.
A. Cyp2a4 and Cyp2a5 mRNA levels and B. Cyp2a4as and Cyp2a5as transcripts levels were measured by RT-qPCR, normalized to the control Sfrs4 mRNA, in liver samples from B6C3F1 male (black squares) and female (gray circles) mice at embryonic day 17.5 (M and F, n=3) and postnatal day 1 (p1; M and F n=3), p7 (M and F n=3), p14 (M n=6, F n=3), p21 (M n=5, F n=3), p28 (M n=5, F n=3) and p56 (M and F n=4). The gap in the lines indicates a longer time period between timepoints p28 and p56. Statistics: Twoway ANOVA was used to compare females to males across developmental timepoints. *=p<.05, ***p<.001.
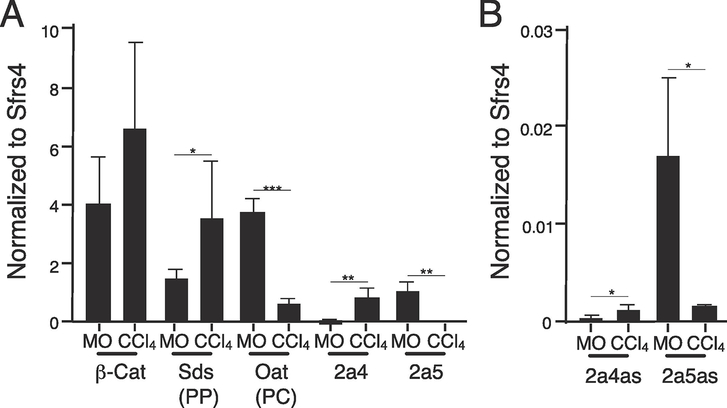
3.6. Cyp2a4 and Cyp2a5 and their antisense RNAs display different zonal expression patterns
Many liver-expressed genes, including a number of the Cyp genes, are expressed in different hepatocyte subpopulations, depending on their location relative to the central vein (pericentral or PC) or portal triad (periportal or PP) within the three-dimensional liver structure, a phenomenon termed zonal expression (Oinonen and Lindros, 1998; Spear et al., 2006). A single-cell analysis of gene expression within the liver of male mice identified Cyp2a5 mRNA as having a PC expression pattern whereas Cyp2a4 had a PP pattern (Halpern et al., 2017). To examine zonal gene expression patterns of Cyp2a4, Cyp2a5 and their antisense transcripts, we used a well-characterized experimental model of liver regeneration as a proxy to measure zonal expression (Weber et al., 2003). A single i.p. injection of adult male mice with the hepatotoxin CCl4, dissolved in MO, causes acute necrosis of PC hepatocytes after three days, compared to injection with MO alone. Thus, at this time point, the CCl4-treated liver contains mainly hepatocytes that have a PP gene expression pattern whereas the MO-treated liver contains both PC and PP hepatocytes. We isolated RNA from these livers and analyzed the expression of genes with known zonal expression patterns to validate this zonal proxy model. As expected, β-catinen (β-cat) mRNA levels, which do not exhibit zonal expression, do not change with CCl4 treatment (Figure 6A). Serine dehydratase (Sds) mRNA levels were significantly higher in CCl4-treated mice, as expected based on its known PP expression pattern (Braeuning et al., 2006). Levels of Ornithine aminotransferase (Oat) mRNA, a known PC-expressed gene, were lower in CCl4-treated mice, as expected (Kuo et al., 1991). Consistent with the single-cell RNA-Seq data, Cyp2a5 mRNA levels were lower after CCl4 treatment, indicating a PC expression pattern and Cyp2a4 mRNA levels were higher after CCl4 treatment, suggesting a PP expression pattern. While this experiment used only male mice and Cyp2a4 is not well-expressed, we detected expression changes consistent with a zonal pattern nonetheless. We also measured expression of the antisense transcripts in these same samples. Both antisense transcripts exhibit expression patterns that mirror their sense counterparts; Cyp2a4as is PP enriched, whereas Cyp2a5as is PC enriched (Figure 6B). These data demonstrate that Cyp2a4 and Cyp2a5 have contrasting responses to CCl4 treatment, suggesting they are expressed in different liver zones and the antisense transcripts are expressed similarly to their corresponding mRNAs.
Figure 6. Cyp2a4 and Cyp2a5 mRNA, along with Cyp2a4as and Cyp2a5as, display different zonal expression patterns.
Liver RNA was prepared three days after adult male C3H mice were injected i.p. with either mineral oil (MO, n=5) or carbon tetrachloride (CCl4, n=5) and analyzed by RT-qPCR, using Sfrs4 for normalization. A. Expression of β-catenin (not zonally expressed), Sds (a known PP gene), Oat (a known PC gene), Cyp2a4, Cyp2a5 are shown. Cyp2a4 mRNA is periportally enriched whereas Cyp2a5 mRNA is pericentrally enriched. B. Cyp2a4as and Cyp2a5as exhibit similar changes in expression as their sense counterparts after CCl4 treatment, indicating the same zonal expression. Statistics: Student’s unpaired t-test. *=p<.05, **=p<.01, ***p<.001.
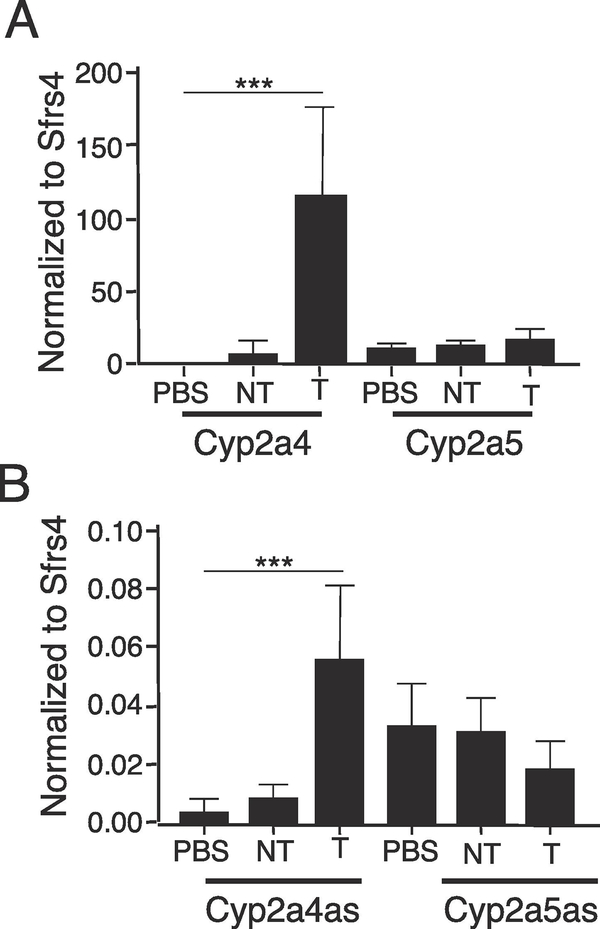
3.7. Cyp2a4as, but not Cyp2a5as, is activated in liver tumors
We previously showed that Cyp2a4 was activated in DEN-induced liver tumors, whereas Cyp2a5 expression remained unchanged, which led us to analyze Cyp2a4as and Cyp2a5as expression in tumors (Figure 7). Consistent with previous studies, Cyp2a4 mRNA levels increased dramatically in tumor samples compared with non-tumor liver regions and control livers whereas Cyp2a5 levels did not change. The Cyp2a4as levels also exhibited increased expression in tumors compared to non-tumor samples, whereas there was no significant difference in Cyp2a5as levels in the different liver samples.
Figure 7. Expression of Cyp2a4as, but not Cyp2a5as, increased in DEN-induced liver tumors.
RNA was prepared from livers of control mice (PBS; n=8) and from non-tumorous tissue (NT) and tumors (T) of DEN-treated mice (n=9) and analyzed by RT-qPCR, using Sfrs4 for normalization. A, B. Expression of Cyp2a4 and Cyp2a4as, but not Cyp2a5 and Cyp2a5as, increase in tumors compared to non-tumor tissue. Statistics: One-way ANOVA was used to compare genes between tumor and non-tumor tissue, ***p<.001.
3.8. Cyp2a4as and Cyp2a5as are nuclear-enriched transcripts
Functional studies of natural antisense RNAs and other lncRNAs have shown that they can repress or activate gene expression through multiple different mechanisms (Cech and Steitz, 2014; Faghihi and Wahlestedt, 2009; Kung et al., 2013; Pelechano and Steinmetz, 2013; Rinn and Chang, 2012). For example, lncRNAs localized to the cytoplasm can impact target mRNA stability or translational efficiency and, when in the nucleus, can regulate target gene expression through epigenetic modifications, transcriptional interference or by directing alternative RNA splicing. To determine the subcellular localization of these antisense transcripts, liver nuclear and cytoplasmic RNA was isolated and analyzed by RT-qPCR for known cytoplasmically-enriched spliced mRNAs (encoding ribosomal protein L30, Cyp2a4, Cyp2a5) and known nuclear-enriched RNAs, including unspliced pre-mRNA (L30-U) and U6 small nuclear RNA (Table 1). The relative cytoplasmic to nuclear RNA abundance for each transcript was calculated as described in section 2.7. By comparing the relative cytoplasmic to nuclear ratios of the Cyp2a4as and Cyp2a5as transcripts to the control cytoplasmic and nuclear RNAs, both antisense RNAs are clearly enriched in the nucleus. Thus, if the antisense transcripts contribute to Cyp2a4 and Cyp2a5 mRNA regulation, their location would require that they act through a nuclear-restricted mechanism. In addition, this analysis shows that the antisense RNAs are only about 20–30-fold less abundant than their corresponding mRNAs in the nucleus whereas, when measured in total RNA, they are 100–800-fold less abundant.
Table 1.
Cyp2a4as and Cyp2a5as are nuclear-enriched transcripts.
| Transcript | relative cytoplasmic/nuclear localization | known localization |
|---|---|---|
| L30 mRNA | 1.0 | cytoplasmic |
| Cyp2a4 mRNA | 0.47 ± 0.01 | cytoplasmic |
| Cyp2a5 mRNA | 0.46 ± 0.12 | cytoplasmic |
| L30 pre-mRNA | 0.0027 ± 0.001 | nuclear |
| U6 snRNA | 0.0056 ± 0.006 | nuclear |
| Cyp2a4as | 0.014 ± 0.008 | nuclear |
| Cyp2a5as | 0.014 ± 0.005 | nuclear |
RNA was isolated from cytoplasmic and nuclear fractions of adult C57BL/6 male (n=2) and female (n=4) livers and analyzed by RT-qPCR. Spliced mRNAs are known to be cytoplasmicallyenriched [ribosomal protein L30 (L30), Cyp2a4, and Cyp2a5] whereas unspliced L30 (L30-U) mRNA and U6 small nuclear RNA (U6) are known to be nuclear-enriched RNAs. The relative abundance of the Cyp2a4as and Cyp2a5as transcripts was compared to these known transcripts to determine their location within liver cells. The cytoplasmic to nuclear RNA ratio (cyto/nuc) relative to L30 mRNA was calculated in two steps. The cytoplasmic to nuclear RNA abundance for each transcript in a single animal was calculated and then this ratio was compared to the cytoplasmic to nuclear ratio for L30 mRNA in the same animal. The average and standard deviation of the six animals are shown. The Cyp2a4as and Cyp2a5as cyto/nuc ratios more closely resembled the nuclear RNAs, rather than the cytoplasmic mRNAs, indicating Cyp2a4as and Cyp2a5as transcripts are enriched in the nuclear fraction.
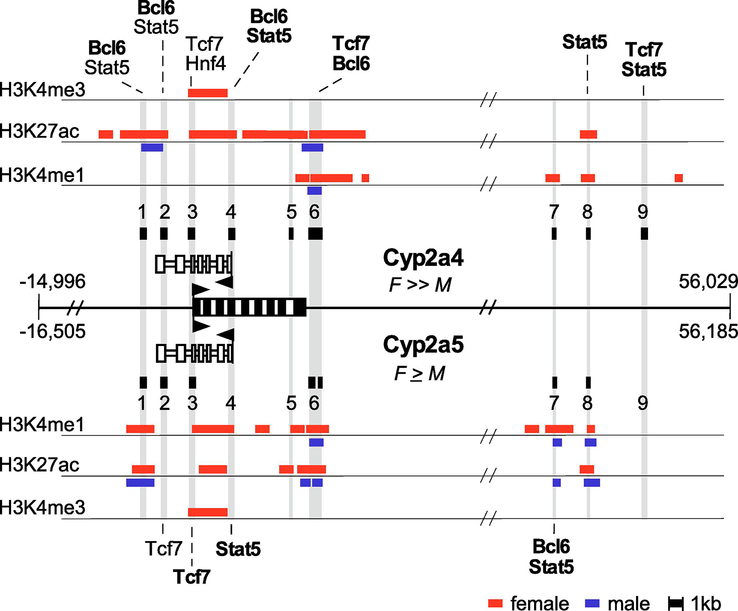
3.9. Potential transcription factor binding differences between the Cyp2a4 and Cyp2a5 genes
Since we have documented differences in developmental, sex-biased and zonal expression of Cyp2a4 and Cyp2a5 and their corresponding antisense RNAs, we considered whether predicted transcription factor binding site differences between these two genes may provide clues to transcriptional mechanisms that contribute to their differential regulation. To begin, we localized DHS sites within the 71 kb genomic region of homology between Cyp2a4 and Cyp2a5 (Figure 8) from published genome-wide DHS data from male and female mouse liver (Ling et al., 2010). DNase hypersensitivity identifies accessible chromatin regions that are often closely associated with transcriptional regulatory regions. This analysis identified six high-stringency DHS sites for Cyp2a4 (sites 1 – 4, 6 and 9) and three additional standard-stringency DHS sites (sites 5, 7 and 8); all of these DHS sites were categorized as female-enriched. In Cyp2a5, only site 1 was identified as a high-stringency DHS site, the others were standard-stringency sites and the regions corresponding to Cyp2a4 sites 4, 5 and 9 were not identified as DHS sites at all. Cyp2a5 DHS sites 1, 2 and 3 were female-enriched while DHS sites 7 and 8 were sex-independent. DHS site 6 in Cyp2a5 was divided into two distinct sites; one was female-biased and the other was sex-independent. The locations of several of the DHS sites correlate with relevant features of the genes; site 3 is over the Cyp2a4 and Cyp2a5 promoters and site 4 is over the antisense promoter region. In a global analysis of six specific chromatin modifications in male and female livers, multiple combinations of activating and repressive marks were identified over different classes of sex-biased genes (Sugathan and Waxman, 2013). The data from male and female liver chromatin modifications over the Cyp2a4 and Cyp2a5 genomic regions were extracted and incorporated into Figure 8. The activating chromatin marks associated with enhancers (H3K4me1 and H3K27ac) were found in both Cyp2a4 and Cyp2a5, with the signals in female livers being more prevalent than in male livers, consistent with their differential expression. All the DHS sites, except for DHS site 9, were associated with one or both of these sites in these data sets. The H3K4me3 mark, often found associated with active promoters, was found in female livers spanning both the mRNA and antisense transcript promoter regions, consistent with Cyp2a4 and Cyp2a5 both being expressed in females. Although data were available for other chromatin marks (H3K36me3, H3K9me3 and H3K27me3), none of these were associated with the Cyp2a4 or Cyp2a5 genes.
Figure 8. DNase I hypersensitivity (DHS) regions, chromatin modifications and predicted differential transcription factor binding sites within the mouse Cyp2a4 and Cyp2a5 genes.
The nearly identical 71 kb genomic regions of the Cyp2a4 and Cyp2a5 genes (see Figure 1) is shown in the middle; exons are black boxes, the arrows show the transcriptional direction and the relative female-to-male bias for each gene is indicated. The antisense transcripts associated with each gene are shown as open boxes above and below the gene diagram with the arrows showing transcriptional direction. The DHS sites from Supplemental Table S5A (Ling et al., 2010) within this genomic region are shown as black boxes above the Cyp2a4 gene, numbered 1 through 9. Six of the nine corresponding regions in the Cyp2a5 gene also had DHS sites and these are marked with solid black boxes that are connected to the sites in Cyp2a4 by light grey bars. The chromatin modifications (H3K4me3, H3K27ac, H3K4me1) from Supplemental Table S3 (Sugathan and Waxman, 2013) over these genomic regions in male (blue bars) and female (red bars) liver samples are shown and aligned with the DHS data. The overall figure is not to scale, but the sizes of the DHS regions and the extent of the chromatin modifications is drawn approximately to scale, as noted below the figure. Potential binding sites for transcription factors associated with sex-biased (Stat5a/b, Bcl6, Hnf4) and zonal (Tcf7, Hnf4) regulation were identified within the DHS sites using JASPAR. Transcription factors that are predicted to bind uniquely in one gene are in bold. Those factors predicted to be present in both genes, but that have higher relative scores in one compared to the other are shown associated with the gene having the higher score. If a transcription factor is shown associated with the same DHS in both genes, this means that each gene has these sites predicted at different locations with the DHS region.
The homologous sequences of Cyp2a4 and Cyp2a5 from the nine Cyp2a4 DHS sites were submitted to JASPAR (http://jaspar.genereg.net) to identify potential transcription factor binding sites (Khan et al., 2018; Sandelin et al., 2004). We scanned for transcription factors that are known to contribute to sex-biased (Stat5a/b, Bcl6, Hnf4, Cux2) and zonal (Tcf7, Hnf4) transcriptional regulation (Clinkenbeard et al., 2012; Clodfelter et al., 2007; Conforto et al., 2012; Gougelet et al., 2014; Stanulovic et al., 2007; Sugathan and Waxman, 2013; Udy et al., 1997; Wiwi et al., 2004; Zhang et al., 2012). High confidence sites for each of these were identified in multiple regions. However, we focused on those factors that were predicted to uniquely bind, or have a higher relative binding score, in one gene compared to the other (Figure 8). Of particular interest was the higher number of predicted Bcl6 and Stat5 sites in Cyp2a4, since these factors repress female-biased genes in male mice. There were multiple predicted sites for Cux2, a female-specific activator of female-specific genes, within DHS sites 1 and 2, but they were not different between Cyp2a4 and Cyp2a5 and therefore are not shown in the figure. In addition, all the DHS sites also contained predicted binding sites for several liver-enriched transcription factors (Cebpa, Hnf1, Foxa; not included in the figure). This analysis provides a short list of potential transcription factors that may contribute to the different sex-biased and zonal Cyp2a4 and Cyp2a5 expression patterns.
4. Discussion
Despite the 96% sequence identity in the ~71 kb genomic regions containing the Cyp2a4 and Cyp2a5 genes, these two genes exhibit different expression patterns in the adult mouse liver, including different sex-bias, tumor and zonal expression. The limited sequence differences between Cyp2a4 and Cyp2a5 could potentially provide insight into the mechanisms governing their differential expression, which may be due to transcriptional or post-transcriptional mechanisms or a combination of these. Because there are seven sequence differences between Cyp2a4 and Cyp2a5 mRNAs within their 208 nt 3’ UTRs (the mRNAs are 98.4% identical over their coding region but 96.6% identical over the 3’UTR) and 3’UTRs often impact mRNA stability (Matoulkova et al., 2012), we considered whether there may be different miRNA target sequences or sequence targets for RNA-binding proteins that could contribute to differential stability of the mRNAs. Using multiple miRNA target prediction programs, we found several predicted miRNA binding sites differences between the two mRNAs. However, none of the miRNAs predicted to bind these sites are known to be expressed in liver or to be sex-biased miRNAs (Hao and Waxman, 2018), suggesting that differential miRNA binding could not account for different Cyp2a4 and Cyp2a5 mRNA levels. In addition, none of the sequence elements known to be bound by RNA-binding proteins that either stabilize or destabilize mRNA are found in the Cyp2a4 or Cyp2a5 3’UTRs.
A single EST had been uniquely annotated to the Cyp2a4 gene, which raised the possibility this could contribute to the difference in gene expression patterns between Cyp2a4 and Cyp2a5. However, while we validated this partial EST as an antisense RNA (Cyp2a4as), we also discovered a similar antisense transcript for the Cyp2a5 gene (Cyp2a5as) and characterized the structure and expression of these antisense transcripts. To our knowledge, these are the first antisense transcripts characterized within the Cyp supergene family. The antisense RNAs are nuclear-enriched, spliced, polyadenylated, likely non-coding and, as with most natural antisense RNAs, are present at lower levels than their associated mRNA (Faghihi and Wahlestedt, 2009; Pelechano and Steinmetz, 2013). Both Cyp2a4as and Cyp2a5as initiate within the fourth exon and end at the same location, roughly 2770 bp upstream of their respective genes, and thus overlap with the gene promoters. A single nt difference between the two primary transcripts that occurs at a splice junction, results in four alternatively spliced isoforms of Cyp2a4as. However, it is not likely that there are functional differences among them. We did find it curious that several of the antisense RNA exons closely overlapped with exons of the mRNAs. This is especially true for the mRNA exons 2 and 3 that closely overlap with antisense exons 3 and 2 respectively (see Figure 3). This could reflect the differential nucleosome localization and modification that is known to exist over exons compared to introns (Carrillo Oesterreich et al., 2011; de Almeida and Carmo-Fonseca, 2012; Naftelberg et al., 2015). The transcripts being made from opposite DNA strands would likely be similarly affected by the chromatin structure and this may help guide the RNA splicing machinery to suitable splice junction sequences within the primary transcripts.
Because Cyp2a4as and Cyp2a5as are nuclear RNAs and generally are expressed in the same patterns as their sense mRNA counterparts, this limits the types of mechanisms involved if they were to contribute to mRNA regulation. The one situation where we detect some differences between the mRNA and antisense RNA patterns is during postnatal liver development, where the female-biased expression of the antisense transcripts is established at least a week or more before the mRNA sex-biased patterns. This earlier expression could facilitate activation of, and potentially contribute to, the sex-biased patterns of Cyp2a4 and Cyp2a5 in adult mouse liver. For example, one model that is consistent with our observations is that transcription from the antisense promoters could open the chromatin in the region of the mRNA promoters, making these regions more accessible to transcription factors. This transcription activity, or the antisense transcripts themselves, could recruit chromatin modifiers associated with active transcription (Cech and Steitz, 2014; Faghihi and Wahlestedt, 2009; Kornienko et al., 2013; Pelechano and Steinmetz, 2013; Rinn and Chang, 2012). While published data are available for DHS sites and chromatin marks present in adult (8 week) male and female liver (Figure 8), data for these chromatin marks during postnatal liver development, when the antisense RNAs are differentially expressed, for example on days p14, p21 and p28, would be needed to specifically address this hypothesis. Such studies could identify the regions that are differentially modified and occupied by transcription factors, specifically in the region of antisense transcription, in male in female mouse liver and identify mechanisms contributing to Cyp2a4 and Cyp2a5 differential regulation.
Because Cyp2a4as and Cyp2a5as are expressed similarly to Cyp2a4 and Cyp2a5 mRNAs, it is likely that the same transcription factors are governing expression of the mRNAs and their antisense transcripts during liver development and disease and that transcriptional regulation is a major determinant of the different expression patterns of these two genes. By analyzing available genomic data, we identified multiple candidate transcription factor binding site differences between these two genes within DHS sites (Ling et al., 2010). Since Stat5b, the major Stat5 isoform in the liver, is an activator of male-biased genes and a repressor of female-biased genes in male mice (Clodfelter et al., 2007; Udy et al., 1997), we might expect more unique Stat5 sites in the highly female-biased Cyp2a4 gene. In fact, this is the case, as DHS sites 1, 2, 4, 8 and 9 within the Cyp2a4 gene have either unique or stronger Stat5 sites, whereas the Cyp2a5 gene has unique Stat5 sites within DHS sites 4 and 7. Bcl6, a male-biased repressor of female-biased genes, often competes with Stat5 for binding DNA (Sugathan and Waxman, 2013). Consistent with the more extreme female-biased expression of Cyp2a4, we found unique Bcl6 sites within the Cyp2a4 high-stringency DHS sites 1, 2, 4, and 6. In contrast, the weakly female-biased Cyp2a5 gene has one predicted unique Bcl6 site, located within DHS site 7. Interestingly, the Cyp2a4 DHS site 4, which is most closely associated with the Cyp2a4as promoter region, has predicted overlapping Stat5 and Bcl6 sites that would be predicted to enforce a strict female-biased Cyp2a4as expression, whereas these sites are not uniquely present within the Cyp2a4 promoter region covered by DHS site 3. Hnf4, which is associated with repression of female genes in males (Wiwi et al., 2004) is the only sex-biased regulator we examined that is predicted to have a differential binding between the genes within the mRNA promoter region. ChIP-seq datasets are available for many of these transcription factors and, in general, their signals overlap well with the DHS sites (Conforto et al., 2015; Zhang et al., 2012). However, because the sites we’ve highlighted here are only those that are different between Cyp2a4 and Cyp2a5 and high-scoring binding sites that are identical between the two genes exist throughout this region, we did not analyze this data further. How the multiple general, liver-enriched and sex-biased transcription factors interact to direct the different expression patterns of these genes is likely more complex than the presence or absence of a single binding site. However, our work identifies DNA regions that likely contain relevant regulatory information that can be used for future studies.
Only a few factors have been identified to contribute to zonal regulation in the liver; β-catenin, through interactions with Tcf7, is known to drive pericentral gene expression, and Hnf4a represses pericentral genes in periportal regions, but much less is known about contributors to periportal gene expression (Benhamouche et al., 2006). Cyp2a5 and Cyp2a4 have pericentral and periportal patterns of expression, respectively, as do their antisense RNAs. This would predict that there may be more Tcf7 and Hnf4 sites in Cyp2a5 than in Cyp2a4. In fact, this was not the case as both genes had unique or stronger Tcf7 sites located within different DHS sites and an Hnf4 site was predicted only in DHS3 of Cyp2a4. However, because Tcf7 and Hnf4a are not likely the only regulators of zonal expression in the liver, a better understanding of this regulatory mechanism is needed to elucidate differential zonal expression of Cyp2a4 and Cyp2a5.
Numerous studies have analyzed the expression of Cyp2a5 and, to a lesser extent, Cyp2a4, in response to liver toxins (Abu-Bakar et al., 2013; Camus-Randon et al., 1996; Poca et al., 2017). Several luciferase-based transfection analyses have identified multiple clusters of regulatory elements within the first 3 kb upstream of the Cyp2a5 gene (Abu-Bakar et al., 2013; Ulvila et al., 2004). These include a “stress-responding cluster” in the −2524 to −2377 region that corresponds to DHS site 2 (Figure 8) and the proximal promoter region from −271 to +10 which corresponds to DHS site 3 (Figure 8). Both Cyp2a4 and Cyp2a5 are also regulated in a circadian pattern (Lavery et al., 1999). However, since many of these earlier RNA studies used Northern blots with probes that could not distinguish between Cyp2a4 and Cyp2a5, their induction by various chemicals may need to be revisited using methods to distinguish these highly related mRNAs. Future studies will also be needed to determine whether Cyp2a4as and Cyp2a5as are responsive to various toxins or show circadian changes in expression. It will also be of interest to determine whether antisense RNAs are encoded within other Cyp genes.
Supplementary Material
A 71kb genomic region surrounding the mouse Cyp2a4 and Cypa5 genes is 96% identical. The highly homologous Cyp2a4 and Cypa5 genes are differentially expressed in liver. Spliced nuclear antisense long noncoding RNAs overlap with the Cyp2a4 and Cypa5 genes. Limited sequences differences between genes target potential regulatory regions.
Acknowledgements
We would like to thank Kate Townsend Creasy, Guofong (Shirley) Qiu and Jieyun Jiang for providing samples and advice for this work. Statistical support was provided by the Applied Statistics Lab in the Department of Statistics that is partially funded by NIH P20GM103436. This work was supported by the National Institutes of Health grant number NIDDK-DK074816.
Abbreviations List
- Cyp
Cytochrome P450
- Zhx2
Zinc fingers and homeobox domain
- DEN
diethylnitrosamine
- EST
expressed sequence tag
- DHS
DNase I hypersensitivity
- lncRNA
long non-coding RNA
- RT-qPCR
reverse transcriptase – quantitative polymerase chain reaction
- Sfrs4
Arginine Rich Splicing Factor 4
- RACE
rapid amplification of cDNA ends
- CCl4
carbon tetrachloride
- MO
mineral oil
- i.p.
intraperitoneal
- UTR
untranslated region
- p
postnatal day
- PP
periportal
- PC
pericentral
- β-cat,
β-catinen
- Sds
Serine dehydratase
- Oat
Ornithine aminotransferase
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Bakar A, Hakkola J, Juvonen R, Rahnasto-Rilla M, Raunio H and Lang MA, 2013. Function and regulation of the Cyp2a5/CYP2A6 genes in response to toxic insults in the liver. Curr Drug Metab. 14, 137–50. [PubMed] [Google Scholar]
- Aida K, Moore R and Negishi M, 1994. Lack of the steroid 15 alpha-hydroxylase gene (Cyp2a-4) in wild mouse strain Mus spretus: rapid evolution of the P450 gene superfamily. Genomics. 19, 564–6. [DOI] [PubMed] [Google Scholar]
- Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, Vasseur-Cognet M, Kuo CJ, Kahn A, Perret C and Colnot S, 2006. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 10, 759–70. [DOI] [PubMed] [Google Scholar]
- Berget S, 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A and Schwarz M, 2006. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 273, 5051–61. [DOI] [PubMed] [Google Scholar]
- Burke ZD, Reed KR, Yeh SW, Meniel V, Sansom OJ, Clarke AR and Tosh D, 2018. Spatiotemporal regulation of liver development by the Wnt/beta-catenin pathway. Sci Rep. 8, 2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus-Randon AM, Raffalli F, Bereziat JC, McGregor D, Konstandi M and Lang MA, 1996. Liver injury and expression of cytochromes P450: evidence that regulation of CYP2A5 is different from that of other major xenobiotic metabolizing CYP enzymes. Toxicol Appl Pharmacol. 138, 140–8. [DOI] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Bieberstein N and Neugebauer KM, 2011. Pause locally, splice globally. Trends Cell Biol. 21, 328–35. [DOI] [PubMed] [Google Scholar]
- Cech TR and Steitz JA, 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 157, 77–94. [DOI] [PubMed] [Google Scholar]
- Clinkenbeard EL, Butler JE and Spear BT, 2012. Pericentral activity of alpha-fetoprotein enhancer 3 and glutamine synthetase upstream enhancer in the adult liver are regulated by beta-catenin in mice. Hepatology. 56, 1892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodfelter KH, Miles GD, Wauthier V, Holloway MG, Zhang X, Hodor P, Ray WJ and Waxman DJ, 2007. Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis. Physiol Genomics. 31, 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Steinhardt G.F.t. and Waxman DJ, 2015. Cross Talk Between GH-Regulated Transcription Factors HNF6 and CUX2 in Adult Mouse Liver. Mol Endocrinol. 29, 1286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL and Waxman DJ, 2012. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol Sex Differ. 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Zhang Y, Sherman J and Waxman DJ, 2012. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol Cell Biol. 32, 4611–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy KT, Jiang J, Ren H, Peterson ML and Spear BT, 2016. Zinc Fingers and Homeoboxes 2 (Zhx2) Regulates Sexually Dimorphic Cyp Gene Expression in the Adult Mouse Liver. Gene Expr. 17, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Renaud HJ and Klaassen CD, 2012. Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos. 40, 1226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF and Carmo-Fonseca M, 2012. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem Sci. 37, 248–53. [DOI] [PubMed] [Google Scholar]
- Faghihi MA and Wahlestedt C, 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 10, 637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougelet A, Torre C, Veber P, Sartor C, Bachelot L, Denechaud PD, Godard C, Moldes M, Burnol AF, Dubuquoy C, Terris B, Guillonneau F, Ye T, Schwarz M, Braeuning A, Perret C and Colnot S, 2014. T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 59, 2344–57. [DOI] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I and Itzkovitz S, 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 542, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao P and Waxman DJ, 2018. Functional Roles of Sex-Biased, Growth Hormone-Regulated MicroRNAs miR-1948 and miR-802 in Young Adult Mouse Liver. Endocrinology. 159, 1377–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD and Zhong XB, 2009. Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab Dispos. 37, 116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycay EG and Bandiera SM, 2009. Expression, function and regulation of mouse cytochrome P450 enzymes: comparison with human P450 enzymes. Curr Drug Metab. 10, 1151–83. [DOI] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F and Mathelier A, 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP and Pauler FM, 2013. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D and Lee JT, 2013. Long noncoding RNAs: past, present, and future. Genetics. 193, 651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo FC, Hwu WL, Valle D and Darnell JE Jr., 1991. Colocalization in pericentral hepatocytes in adult mice and similarity in developmental expression pattern of ornithine aminotransferase and glutamine synthetase mRNA. Proc Natl Acad Sci U S A. 88, 9468–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U and Bonfils C, 1999. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 19, 6488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg RL and Negishi M, 1989. Alteration of mouse cytochrome P450coh substrate specificity by mutation of a single amino-acid residue. Nature. 339, 632–4. [DOI] [PubMed] [Google Scholar]
- Ling G, Sugathan A, Mazor T, Fraenkel E and Waxman DJ, 2010. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol. 30, 5531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeppen S, Koehle C, Buchmann A and Schwarz M, 2005. A beta-catenin-dependent pathway regulates expression of cytochrome P450 isoforms in mouse liver tumors. Carcinogenesis. 26, 239–48. [DOI] [PubMed] [Google Scholar]
- Marck C, 1988. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16, 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B and Hrstka R, 2012. The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 9, 563–76. [DOI] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G and Kornblihtt AR, 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 84, 165–98. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM and Nebert DW, 2004. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 14, 1–18. [DOI] [PubMed] [Google Scholar]
- Oinonen T and Lindros KO, 1998. Zonation of hepatic cytochrome P-450 expression and regulation. Biochem J. 329 ( Pt 1), 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V and Steinmetz LM, 2013. Gene regulation by antisense transcription. Nat Rev Genet. 14, 880–93. [DOI] [PubMed] [Google Scholar]
- Poca KS, Parente TE, Chagas LF, Leal BS, Leal HS, Paumgartten FJ and De-Oliveira AC, 2017. Interstrain differences in the expression and activity of Cyp2a5 in the mouse liver. BMC Res Notes. 10, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M and Klaassen CD, 2011. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci. 124, 261–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL and Chang HY, 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 81, 145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW and Lenhard B, 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Hagenbuchle O, Wellauer PK and Pittet AC, 1983. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 33, 501–8. [DOI] [PubMed] [Google Scholar]
- Sloutskin A, Danino YM, Orenstein Y, Zehavi Y, Doniger T, Shamir R and Juven-Gershon T, 2015. ElemeNT: a computational tool for detecting core promoter elements. Transcription. 6, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear BT, Jin L, Ramasamy S and Dobierzewska A, 2006. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell. Mol. Life Sci. 63, 2922–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires EJ and Negishi M, 1988. Reciprocal regulation of sex-dependent expression of testosterone 15 alpha-hydroxylase (P-450(15 alpha)) in liver and kidney of male mice by androgen. Evidence for a single gene. J Biol Chem. 263, 4166–71. [PubMed] [Google Scholar]
- Stanulovic VS, Kyrmizi I, Kruithof-de Julio M, Hoogenkamp M, Vermeulen JL, Ruijter JM, Talianidis I, Hakvoort TB and Lamers WH, 2007. Hepatic HNF4alpha deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology. 45, 433–44. [DOI] [PubMed] [Google Scholar]
- Su T and Ding X, 2004. Regulation of the cytochrome P450 2A genes. Toxicol Appl Pharmacol. 199, 285–94. [DOI] [PubMed] [Google Scholar]
- Su T, Sheng JJ, Lipinskas TW and Ding X, 1996. Expression of CYP2A genes in rodent and human nasal mucosa. Drug Metab Dispos. 24, 884–90. [PubMed] [Google Scholar]
- Sugathan A and Waxman DJ, 2013. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol Cell Biol. 33, 3594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ and Davey HW, 1997. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 94, 7239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J, Arpiainen S, Pelkonen O, Aida K, Sueyoshi T, Negishi M and Hakkola J, 2004. Regulation of Cyp2a5 transcription in mouse primary hepatocytes: roles of hepatocyte nuclear factor 4 and nuclear factor I. Biochem J. 381, 887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP and Li W, 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ and Holloway MG, 2009. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 76, 215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber LW, Boll M and Stampfl A, 2003. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 33, 105–36. [DOI] [PubMed] [Google Scholar]
- Wiwi CA, Gupte M and Waxman DJ, 2004. Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4alpha-deficient mice. Mol Endocrinol. 18, 1975–87. [DOI] [PubMed] [Google Scholar]
- Yeo G and Burge CB, 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 11, 377–94. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Laz EV and Waxman DJ, 2012. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol. 32, 880–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.