Abstract
Symmetric echo-planar spectroscopic imaging (EPSI) supports higher spectral bandwidth and improves signal-to-noise efficiency compared to flyback EPSI with the same readout bandwidth, but suffers from artifacts that are associated with non-uniform temporal sampling in k-t space. Our goal is to eliminate these artifacts and enhance observation of hyperpolarized [1-13C] pyruvate and its metabolites using symmetric EPSI. We used symmetric EPSI to efficiently acquire radially encoded spectroscopic imaging projections with a spectral under-sampling scheme that was optimized for HP pyruvate and its metabolites. A simple approach called selective correction of off-resonance effects (SCORE) was developed and applied to eliminate spectral artifacts. Simulations were used to assess the relative SNR performance of this technique, and a phantom study was carried out at 3T to evaluate this method and compare it with alternative strategies. SCORE correction eliminated spectral artifacts due to chemical shift and non-uniform sampling in time. It is also compatible with established methods to eliminate artifacts caused by eddy currents. SCORE corrected symmetric EPSI supported maximal EPSI spectral bandwidth and improved SNR efficiency. Symmetric EPSI with SCORE correction offers a straightforward, efficient, and effective framework for assessment of hyperpolarized [1-13C] pyruvate and its metabolites.
Keywords: hyperpolarized pyruvate, EPSI, k-t space, SNR, chemical shift, spectroscopic imaging
Graphical Abstract
Introduction
Magnetic resonance spectroscopic imaging of hyperpolarized (HP) [1-13C]-pyruvate and its downstream metabolites [1, 2] offers remarkable spectral, spatial, and temporal resolution for non-invasive assessment of metabolism in vivo [2–10]. The conversion of HP pyruvate to lactate has been established as a potential biomarker to characterize tumor metabolism [3, 4, 11]. However, HP signal pools are short-lived and non-renewable, requiring robust and efficient methods for imaging and characterizing the spatiotemporal evolution of HP [1-13C]-pyruvate and its metabolites in vivo.
Echo-planar spectroscopic imaging (EPSI) [12] is one of the most widely used among methods for imaging HP pyruvate and its metabolites [13–19]. This approach relies on oscillating gradients along the readout direction (kr) to generate a series of echoes that evolve over time and which can be transformed to reveal spatial and spectral information. For spectral encoding, a symmetric EPSI [20, 21] with bi-directional readout (+kr, −kr), or alternatively the fly-back EPSI [22–24], with a monopolar readout (+kr, for example) and a rapid rewind can be used. While the latter is robust to phase errors and simple to reconstruct, symmetric EPSI has a higher SNR efficiency [22–24], is less demanding of imaging gradients, and supports higher spectral bandwidth (sBW) for a given readout gradient amplitude.
It is challenging for EPSI acquisitions to accommodate the broad sBW required for [1-13C]-pyruvate (170.6 ppm) and its metabolites, which include lactate (183.2 ppm), pyruvate hydrate (179 ppm), alanine (176.5 ppm) and bicarbonate (160.9 ppm), spanning a full sBW of more than 22 ppm. Coverage of this relatively high sBW with EPSI requires low echo spacing, which can lead to low spatial resolution, or low SNR due to high readout bandwidth. Spectral under-sampling can reduce this problem, but care must be taken to ensure that aliased peaks do not interfere with the quantification of signal evolution [13].
For symmetric EPSI, the non-equidistant, zig-zag sampling pattern in k-t space and the resulting non-uniform temporal sampling can lead to N/2 spectral “ghost” artifacts that appear ± sBW/2 from the ‘parent’ peak location [12], which could obfuscate quantification of metabolites within a voxel. These artifacts can be exacerbated by chemical shift offsets in the readout direction that alternate according to the direction of readout gradients. Several methods have been developed to remove these artifacts [20, 25–28]. One method is to separately reconstruct the even and odd echoes of the sampled data then combine them, at the cost of halving the spectral bandwidth [20]. Interlaced Fourier transform-based reconstruction can eliminate ghost artifacts by removing the temporal inconsistency between echoes, but this may lead to amplified noise [25–27] and these technique are not effective for managing signals that alias from outside of the sBW [28]. In addition, those methods were originally developed for [1H]-spectroscopy, which aimed to resolve many metabolites over a narrower bandwidth compared to the spectrum of [1-13C]-pyruvate. Fortunately, the sparse and broad spectrum of [1-13C] can be undersampled (see Figure 1), but new strategies are needed to manage the artifacts that are associated with symmetric EPSI acquisitions of these aliased signals.
Figure 1.
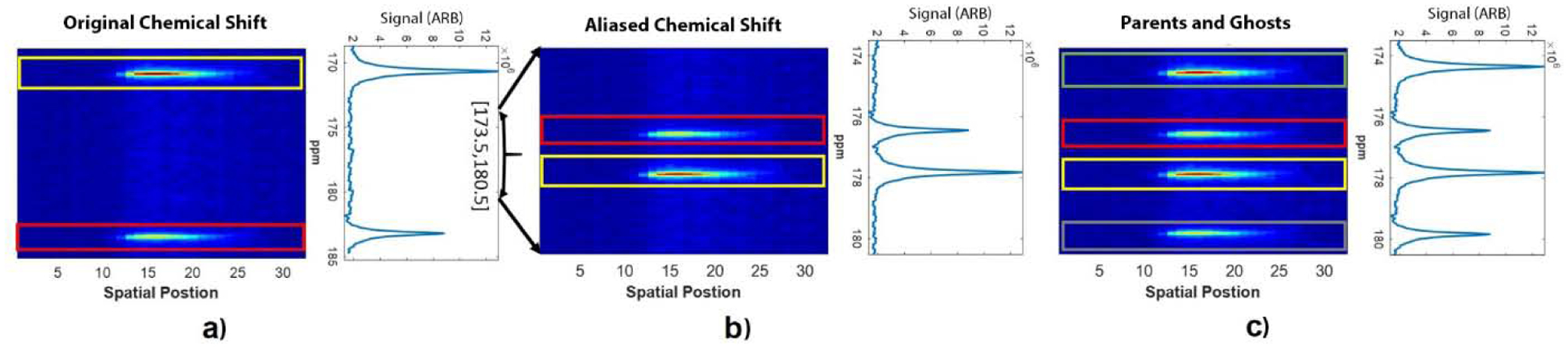
Illustration of the spectral aliasing and ghost artifacts associated with symmetric EPSI sampling. (a) A full spectrum bandwidth (13 ppm) shows peak positions of pyruvate at 170.6 ppm (yellow rectangle) and lactate at 183.2 ppm (red rectangle). (b) These peaks alias to different positions when the spectrum is spectrally under-sampled (7.1 ppm). (c) Non-uniform sampling in time (due to the zig-zag trajectory in symmetric EPSI) leads to spectral ghosts that are offset by half of the spectral bandwidth (pyruvate is denoted by yellow and green boxes, lactate is denoted by the red and grey boxes).
In this work, we describe a new framework to maximize SNR and reduce artifacts for under-sampled symmetric EPSI of HP [1-13C] pyruvate and its downstream metabolites. The first step was to optimize the aliasing pattern of the relatively sparse spectrum of HP [1-13C] pyruvate and its metabolites over an under-sampled sBW. Second, we developed a new reconstruction approach, Selective Correction of Off-Resonance Effects (SCORE), that corrected the imaging artifacts associated with the non-uniform temporal sampling pattern. The SCORE technique eliminates spectral ghosts and refocuses aliased signals in a frequency selective manner, and must be repeatedly applied to all metabolites of interest within the spectrum. We demonstrate this technique in silico and in vitro to confirm its performance relative to alternative strategies.
Theory
Chemical Shift Artifacts and N/2 ghosts
Symmetric EPSI sampling in k-t space follows a zig-zag trajectory, with nonequidistant samples in time [12], as illustrated in Figure 2a, b. The signal expected from a given metabolite in k-t space, Scs(k,t), can be written as:
| (1) |
where r signifies position in the spatial domain, δcs is the difference between the center frequency of the scanner and the chemical shift frequency of a given metabolite (in Hz), Mcs(r) is the magnitude of transverse magnetization (immediately after excitation) of the spin species having chemical shift = δcs, indicates the position along readout in k-t space, γ is the gyromagnetic ratio, Gr is the magnitude of the readout gradient (T/m), and T2* is the transverse relaxation constant for that metabolite. We assume here for simplicity that T2* is uniform across the FOV for a given metabolite.
Figure 2.
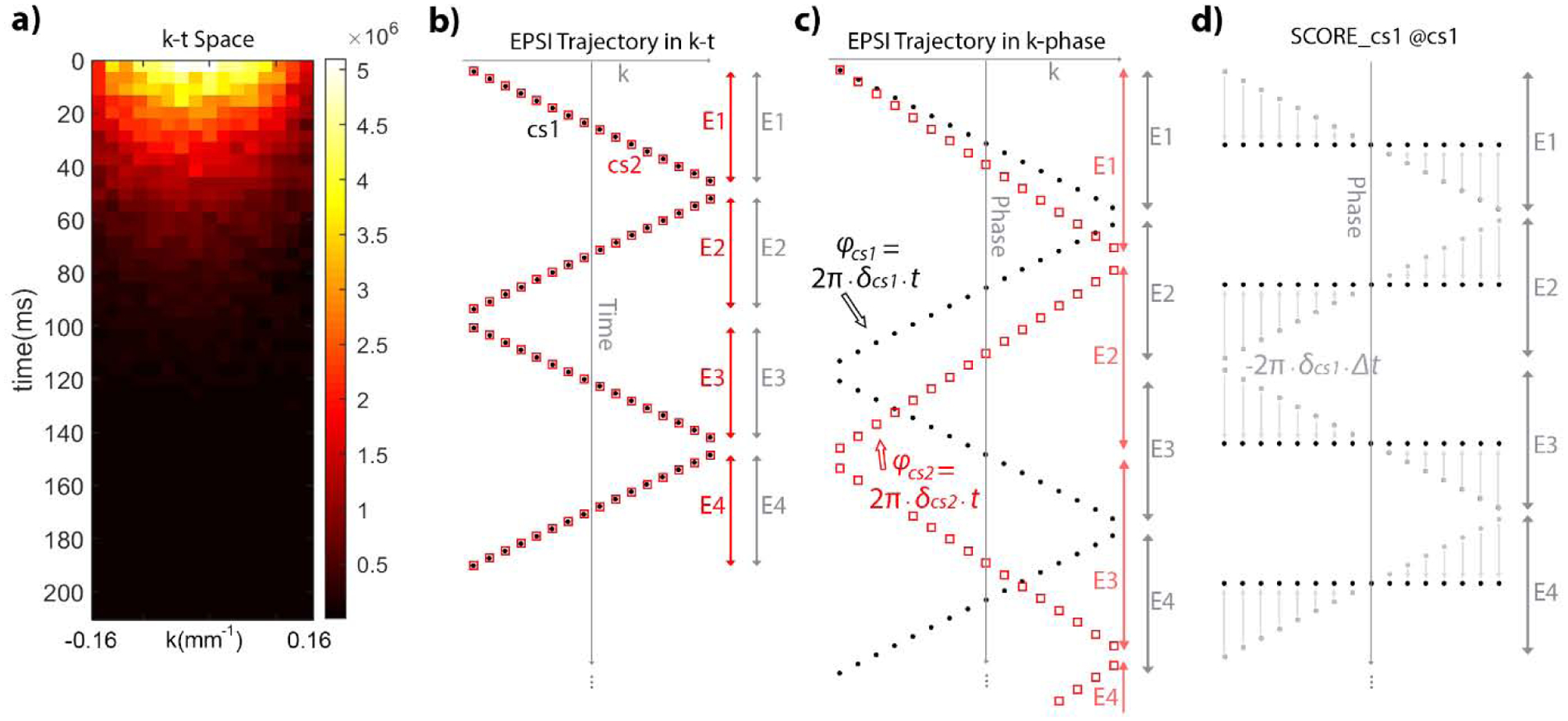
k-t sampling trajectory and phase progression for off-resonance magnetization. (a) The k-t space signal of pyruvate phantom acquired after one excitation. (b) The logical layout of the sampled data in k-t space, which is in a zig-zag pattern. Black dots and red squares represent two metabolites with distinct chemical shifts indicated as δcs1 and δcs2. (c) Phase accumulates differently for each of these metabolites as the product of their chemical shift offset and the ti, time at which the ith sample is acquired. (d) Selective correction of off-resonance effects (SCORE) applied to the metabolite at δcs1 (gray dots) reverses phase progression within each echo (to black dots) to eliminate non-uniform sampling for that metabolite.
Spectral artifacts can arise from several sources when analyzing symmetric EPSI data. First, although the sampling interval between all even echoes is constant and equal to the sampling interval between all odd echoes, the sampling interval between even and odd echoes is not uniform, and depends on position along k-space. Direct Fourier Transform of this data will lead to spectral artifacts [26–28] that appear at ± sBW/2 from the expected peak location. The linewidth of these spectral peaks is not affected by the non-uniform sampling pattern. Note that peaks that alias from outside of the sBW will appear at δ′cs = δcs ± m · sBW, while m = 0 for metabolites that lie within the sBW. Second, metabolites that are not on resonance (δcs ≠ 0) will have a slight spatial offset from their true position due to the chemical shift effect [12, 21, 26, 28], and the direction of that offset varies with gradient readout direction between even and odd readout lines. Without correction, this would lead to artifacts at edges of the signal source at the expected spectral location and at ± sBW/2. It is important to note that on-resonance metabolites will not suffer from either of these effects. With δcs = 0, the spatial chemical shift offset vanishes, the chemical-shift dependent phase term in equation (1) becomes constant, and the non-equidistant phase accumulation between echoes also vanishes. This concept is fundamental to SCORE.
Imperfections in the EPSI gradient waveform can also lead to a mis-alignment of k-space sampling locations between even and odd echoes, which also lead to spectral artifacts at ± sBW/2. SCORE does not correct for this effect, which has been addressed elsewhere [29–32], but SCORE can be combined with these methods as needed.
Selective Correction of Off Resonance Effects (SCORE)
SCORE is a simple and straightforward method to correct for off-resonance effects that lead to blurring in the readout direction and spectral artifacts at ± sBW/2. This is accomplished for one metabolite at a time by multiplying the observed signal by the conjugate of the chemical-shift induced phase modulation of that metabolite:
| (2) |
Here, t indicates the time at which each sample in k-t space was acquired, relative to a fixed reference, t0, which could be chosen as the time at the center of the excitation pulse or the time at which the first sample in k-t space was acquired. In effect, this operation digitally demodulates k-t space so that the target metabolite appears to be on-resonance; the chemical shift offset along readout vanishes, and the non-uniform phases between echoes along t-axis also vanishes. Following standard Fourier transform from k-t space to r-f space, signal will be circularly shifted along the spectral dimension, and signal from the metabolite that would otherwise have been split between f = δ′cs and f = δ′cs ± sBW/2 will appear at f = 0 with no spectral artifacts at f = ± sBW/2. This process can be repeated for each of the chemical species or metabolites of interest, in turn, either to extract the relevant metabolite-specific information for further processing, or to synthesize a complete and corrected r-f space.
Alternatively, SCORE can be applied in a manner that cancels off-resonance effects within each echo, but preserves phase progression between echoes:
| (3) |
Here, (t − TEn) indicates the difference in time between each sample point, within an echo, and the center of that echo. This operation eliminates the chemical shift offset along the readout direction for each echo, and restores a rectilinear sampling pattern for that metabolite (compare Fig 2c vs. Fig 2d). After Fourier transform, the center frequency of r-f space reflects the center frequency of the scanner, and signal from the target metabolite will appear at f = δ′cs with no spectral artifacts at f = δ′cs ± sBW/2. In the following sections, we demonstrate this approach to implementing SCORE which preserves the fixed reference center frequency.
It is important to note that SCORE corrections of symmetric EPSI data for one metabolite will correct artifacts only for that metabolite, while artifacts for other metabolites will be altered according to their distance from the correction frequency; artifacts for some metabolites may be reduced, while others may become more exaggerated. Only data from the corrected metabolite should be retained for further processing. The spectral and spatial artifacts that result from to zig-zag sampling of off-resonance metabolites and SCORE correction are illustrated in Figure 3. The offset frequency in Figures 3h,i refers to the spectral distance between the SCORE correction frequency and the frequency of any other metabolites observed. In uncorrected data, the offset frequency represents the spectral distance from the scanner center frequency to any metabolites observed.
Figure 3.
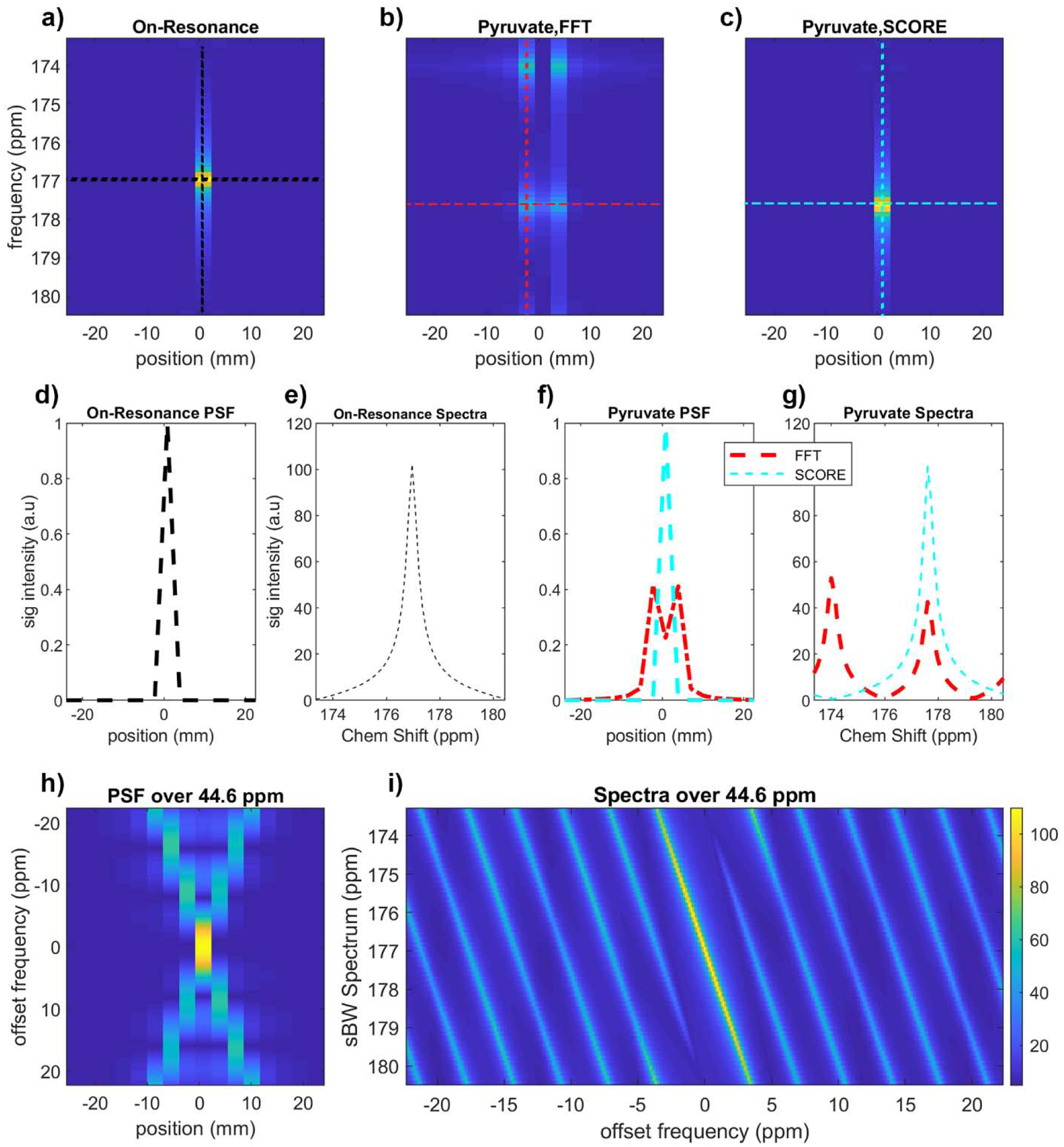
Simulated spectra and point spread function (PSF) using a point source for symmetric EPSI sampling of off-resonance metabolites. (a) Signal from ideal on-resonance metabolites are not subject to spectral or spatial artifacts. (b) The same signal offset by −6.3 ppm to simulate pyruvate appears at an aliased frequency (dcs’), with an N/2 spectral ghost and blurring along the spatial dimension. (c) SCORE correction for this frequency offset eliminates these artifacts. (d,f) The spatial profile for on-resonance, off-resonance, and SCORE corrected data. (e,g) Spectral profiles for on-resonance, off-resonance, and SCORE corrected data (FWHM = 0.60 ppm, 0.59 ppm, FWHM = 0.60 ppm). (h) The spatial PSF of off-resonance metabolites. (i) N/2 spectral ghosts as a function of offset frequency. The frequency range in (h) and (i) encompass the maximum frequency difference between all corrected and uncorrected metabolites, from bicarbonate (160.9 ppm) to lactate (183.2 ppm).
Methods
EPSI Spectral Bandwidth Optimization
The relatively sparse spectrum of pyruvate and its downstream metabolites can be spectrally under-sampled in a controlled manner to preserve the fidelity of peaks associated with pyruvate and lactate, the primary metabolites of interest for assessing tumor metabolism. In simulation, we measured the minimum distance between peaks associated with pyruvate, lactate, pyruvate hydrate, alanine, and bicarbonate, and their N/2 spectral ghosts, as they would appear in correct or aliased locations when sampled using spectral bandwidths ranging from 0.1 ppm to 25 ppm. A spectral bandwidth of 7.08 ppm resulted in both pyruvate and lactate peaks being aliased, but with nearly uniform separation between pyruvate and lactate (parent or ghost) peaks, and this sBW ensured at least 0.5 ppm separation between pyruvate/lactate and all other peaks (Supplementary Figure S1. Spectral separation of 0.5ppm ensures that the full-width at half-max (FWHM) of peaks with T2*=20ms will not overlap. This is a conservative estimate; we expect longer T2* and narrower linewidths at 3T, in practice [33].
Simulation
A Monte Carlo simulation was performed to evaluate the signal-to-noise ratio (SNR) performance of SCORE and reconstruction of symmetric EPSI. Three numerical phantoms were synthesized with different 1D profiles: first, a delta function at the center was used to evaluate PSF and spectra from various reconstruction approaches; second, a Gaussian function with a root-mean-square width of 2.5 mm, which approximates our phantom measurements; and third, a larger numerical phantom (30mm) to evaluate fidelity of SCORE in the presence of eddy currents. The numerical phantom was set on-resonance, off-resonance at δcs= −208 Hz (−6.3ppm, half the spectral distance between pyruvate and lactate at 3T) to simulate Pyruvate, 208 Hz (6.3ppm) to simulate lactate, or −528 Hz (−15.7 ppm) to simulate bicarbonate, and . Signal was numerically sampled using Eq. [1] assuming a symmetric EPSI waveform, a readout bandwidth of 4098 Hz, 16 samples per echo, and an echo train length (ETL) of 56. Complex Gaussian noise was added to synthetic k-t space at the level required for a peak SNR=100 in SCORE-corrected data.
Synthetic noisy k-t space data were processed using several methods. First, even and odd echoes were processed separately to eliminate the non-uniform sampling pattern at cost of half of the spectral bandwidth. Then, r-f space from even and odd echoes were combined using a sum-of-squares approach [34]. The effects of SCORE were then assessed by Fourier transform of uncorrected and SCORE-corrected k-t space. Each of these methods was repeated 1000 times, with fresh noise added to the static signal model at each iteration. The mean and standard deviation of all points in r-f space was recorded, and SNR was calculated as the ratio of the mean to the standard deviation over 1000 repetitions for all points in r-f space[35].
To examine the fidelity of SCORE under imperfect gradient, we simulated symmetric eddy currents artifacts by introducing an echo-shift of one k-space step along the readout direction to each echo, and asymmetric eddy currents by adding a phase delay of 20° between echoes. A data-driven eddy current correction for symmetric EPSI [30] was applied first, then SCORE corrected
Data Acquisition
Imaging data were acquired with a 2D radial EPSI sequence as implemented on a 3T MR scanner (Discovery MR750; GE Healthcare, Chicago, IL, USA) using the GE MNS Research Package. A dual tuned 13C/1H volume resonator with a 35-mm inner diameter (RAPID Biomedical GmbH, Rimpar, Germany) was used to acquire signal from a 1mL syringe phantom with a 4.7-mm inner diameter containing heavily Gd-doped 8M 13C-Urea in water. The symmetric EPSI sequence was designed to cover 48mm over 16 points along the readout direction, with a readout bandwidth of 4098 Hz and sBW =7.08ppm over an ETL of 56. The sequence utilized a 1ms self-refocusing 90° excitation pulse, with TR/TE = 5000/4.9 ms; the long TR was set to ensure complete T1 relaxation. Image was acquired from a single slice with a 10mm slice thickness. Proton images were acquired with a matching in-plane FOV using a 2D gradient echo sequence (TR/TE=8.2/3.1 ms, matrix size = 256×256, in-plane field-of-view = 48 mm, slice thickness = 5 mm), providing the underlay images depicting the phantom position in figure 6.
For assessment of SNR, 96 k-t space projections were acquired without incrementing the readout projection angle. For imaging, 96 k-t space projections were acquired at approximately golden angle increments of 111°.
The center frequency of the scanner was shifted by the appropriate amounts to impart off-resonance effects to the 13C-urea signal that would mimic those of [1-13C]-pyruvate and its metabolites [17, 36], if the center frequency of the scanner were set halfway between resonance frequencies for HP pyruvate and lactate. The scanner was set 6.3ppm above and below the 13C-urea resonance (163.1ppm), for example, to mimic the signals that would be observed at 170.6ppm (pyruvate) and 183.2ppm (lactate) with the scanner centered in between, at 176.9ppm. This data was acquired at multiple frequency offsets and for multiple phantom positions (Supplementary Table S1). Then, the acquired k-t space data were either analyzed separately to assess one spatial and frequency offset at a time, or combined to mimic data from a multispectral acquisition. A representative k-t space signal of the thermal phantom from one projection is shown in Figure 2a.
Reconstruction
Each 16×56 k-t space projection was apodized using 20Hz line broadening, then processed by multiple approaches including standard FT to characterize artifacts due to the zig-zag sampling pattern, the use of even- or odd-only data, their sum-of-square combination, and SCORE. The SCORE method was applied using the chemical shift offset for each metabolite to correct for spectral displacement and ghosting at aliased peak locations. After FT, the spatial distribution of a given metabolite was derived from SCORE-corrected projections by integrating over the full-width at half maximum at f = δ′cs for the associated spectral peak. Images were reconstructed from the resulting integration of SCORE-corrected radial projections using an inverse Radon transform.
SNR Measurement
The SNR of each reconstruction method was measured from acquisitions wherein the projection angle was held constant across 96 repetitions. All SNR comparisons based on data from the 3T MRI scanner were derived from the same raw data that was processed in different ways as described above. As before, signal and noise were defined as the mean and standard deviation of signal intensity for each point in x-f space over the 96 repetitions, respectively.
Results
Simulations under ideal gradient condition indicate that SCORE correction permits use of the full spectral bandwidth from symmetric EPSI acquisitions with optimal SNR performance (Figure 4). Transform of only even or odd echoes also avoids spectral artifacts, but the reduced efficiency of the down-sampled acquisitions leads to an expected reduction in SNR [22]. Simulation under a large frequency offset (bicarbonate) and a low SNR=5 also demonstrate the SCORE correction of spatial and spectral artifacts(Supplementary Figure S2). Simulation under imperfect gradient condition shows that SCORE only corrects for the N/2 ghosting caused from chemical shift, while ghosting from other sources remain. However, the ghosting could be eliminated independently [30] to reduce eddy currents artifacts (Supplementary Figure S3). SCORE is compatible with those corrections methods to reduce ghosting and distorted PSF from multiple sources.
Figure 4.
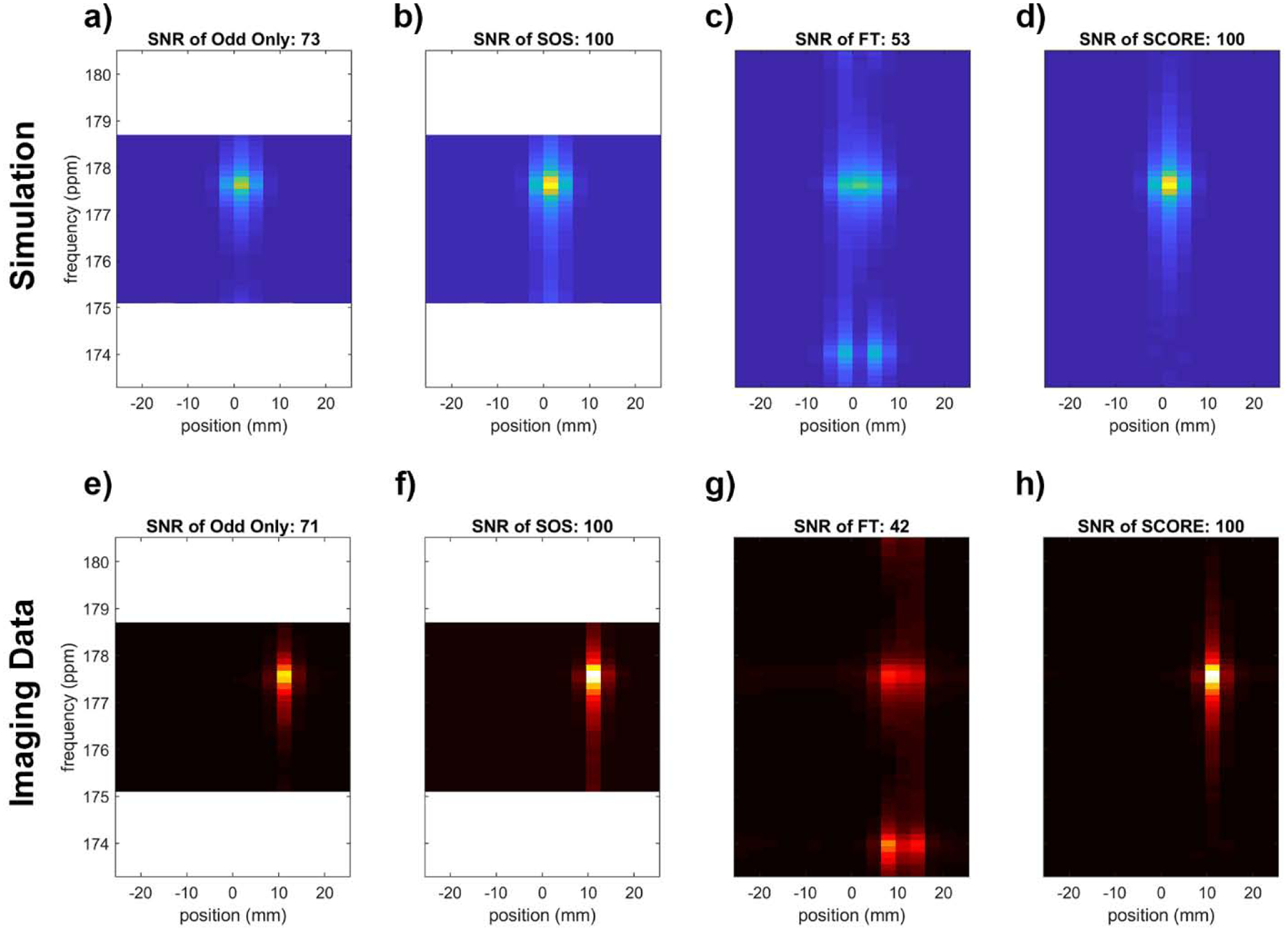
Comparison of methods to correct artifacts associated with symmetric EPSI. Data from (a-d) simulation that approximate phantom measurements and (e-h) actual phantom measurements were processed using (a,e) only odd echoes, (b,f) the sum-of-squares (SOS) combination of data reconstructed from only even and only odd lines of k-t space, (c,g) naïve Fourier transform (FT) to demonstrate artifacts in uncorrected data, and (d,h) FT following SCORE corrections. The FWHM of (a-d) is 0.58ppm, 0.60 ppm, 0.62ppm, 0.60ppm, and the FWHM of (e-h) is 0.63ppm, 0.62 ppm, 0.60ppm, 0.61ppm, respectively. SNR for the spectral peaks was normalized to a maximum value of 100 for each group (a-d, e-h) and each group is displayed with the same color scale. The aliased peak for pyruvate appears at 177.6 ppm, and the N/2 spectral artifact from uncorrected data appears at 174 ppm. SCORE corrections eliminate spectral artifacts and maintain high SNR associated with combination of separately processed even/odd data, without sacrifice of spectral bandwidth.
In the usual case when multiple metabolites are present, SCORE corrections must be applied to k-t space for each individual metabolite. This operation will correct blurring and spectral artifacts for the target metabolite, but will not correct and may even exacerbate artifacts associated with other metabolites in the spectrum. Careful planning of the EPSI sBW ensures that artifacts associated with other peaks will not interfere with quantification of the target metabolite. Figure 5 illustrates the propagation of artifacts when processing phantom data containing multiple metabolites. Here, separate acquisitions of k-t space with the scanner frequency offset to mimic the chemical shifts for pyruvate and lactate were added together with a 2:1 weighting for pyruvate:lactate. Without SCORE correction, signal from both metabolites are blurred along readout, with obvious artifacts for each peak at ± sBW/2. SCORE corrections for pyruvate eliminate artifacts for the pyruvate signal but worsen those for lactate. Artifacts associate with the lactate signal do not interfere with FWHM integration of the pyruvate signal (Fig 5b, dashed box). Likewise, SCORE corrections for lactate eliminate artifacts for lactate but worsen those for pyruvate. The 2:1 SNR ratio of pyruvate:lactate is maintained by the FWHM measurement from rf-space using SCORE correction at 1.97 ± 0.05, while the ratio from Odd only, SOS, and naïve reconstruction are 1.99 ± 0.06, 1.97 ± 0.04, and 2.03 ± 0.10, respectively.
Figure 5.
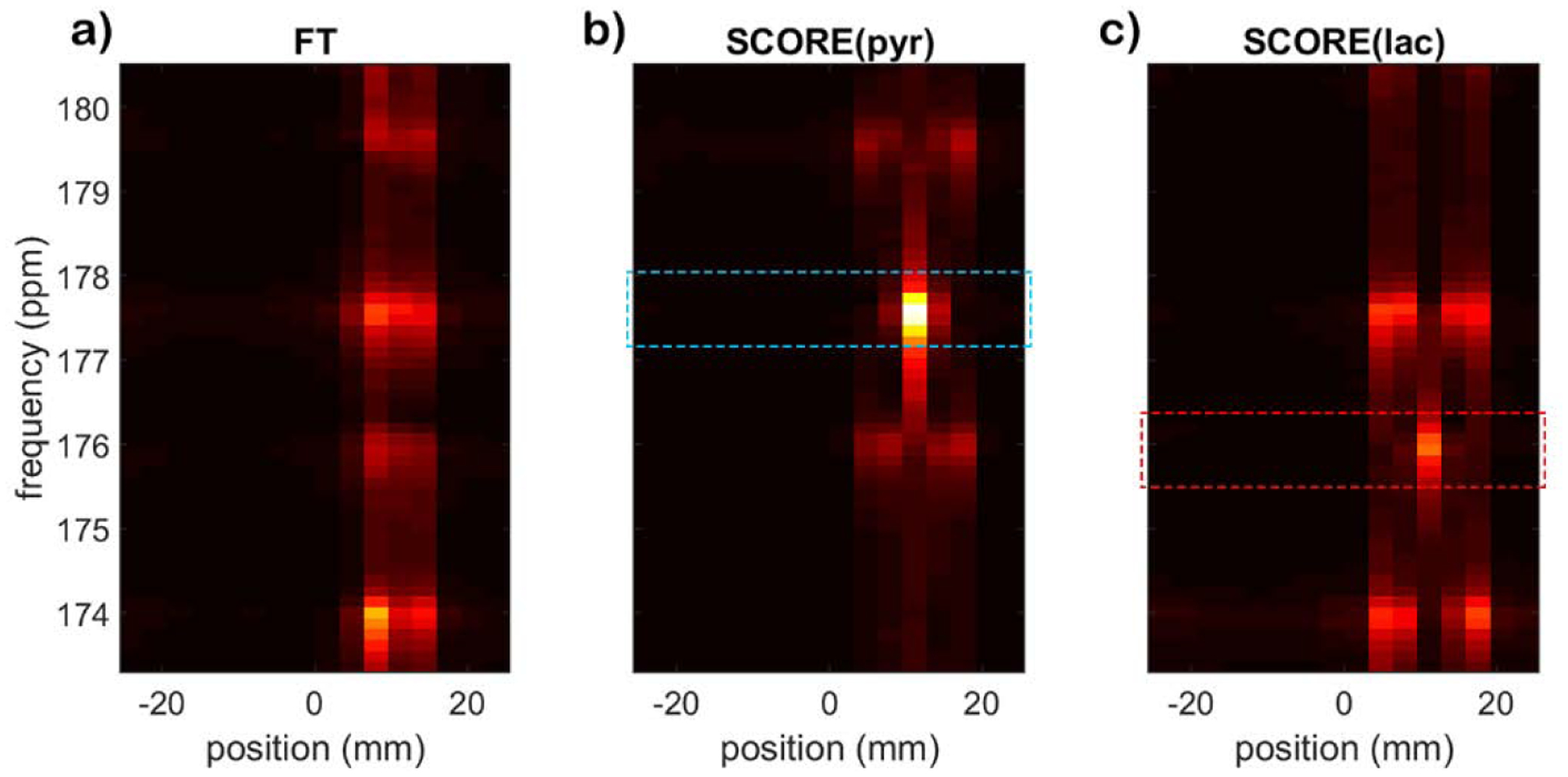
Reconstruction of multiple metabolites at the same spatial location. The spectral signals for pyruvate and lactate alias to 177.6 ppm and 175.9 ppm, respectively. (a) Naïve Fourier transform of symmetric EPSI data (FT) shows spectral artifacts for pyruvate and lactate at 174 ppm and 179.5 ppm, respectively. (b) SCORE correction for pyruvate eliminates the ghost peak for pyruvate but does not correct artifacts associated with the lactate signal. (c) Likewise, SCORE correction for lactate eliminates artifacts for lactate but not for pyruvate. The dash boxes indicate FWHM for signal integration.
After SCORE correction and transform, r-f space projections can be used to form metabolite-specific maps. Good agreement between metabolite maps for pyruvate and lactate and reference proton images of the phantom in multiple locations (Figure 6) indicates that SCORE-corrected symmetric EPSI with radial spatial encoding can be used to generate high quality spectroscopic images of HP [1-13C]-pyruvate and its metabolites.
Figure 6.
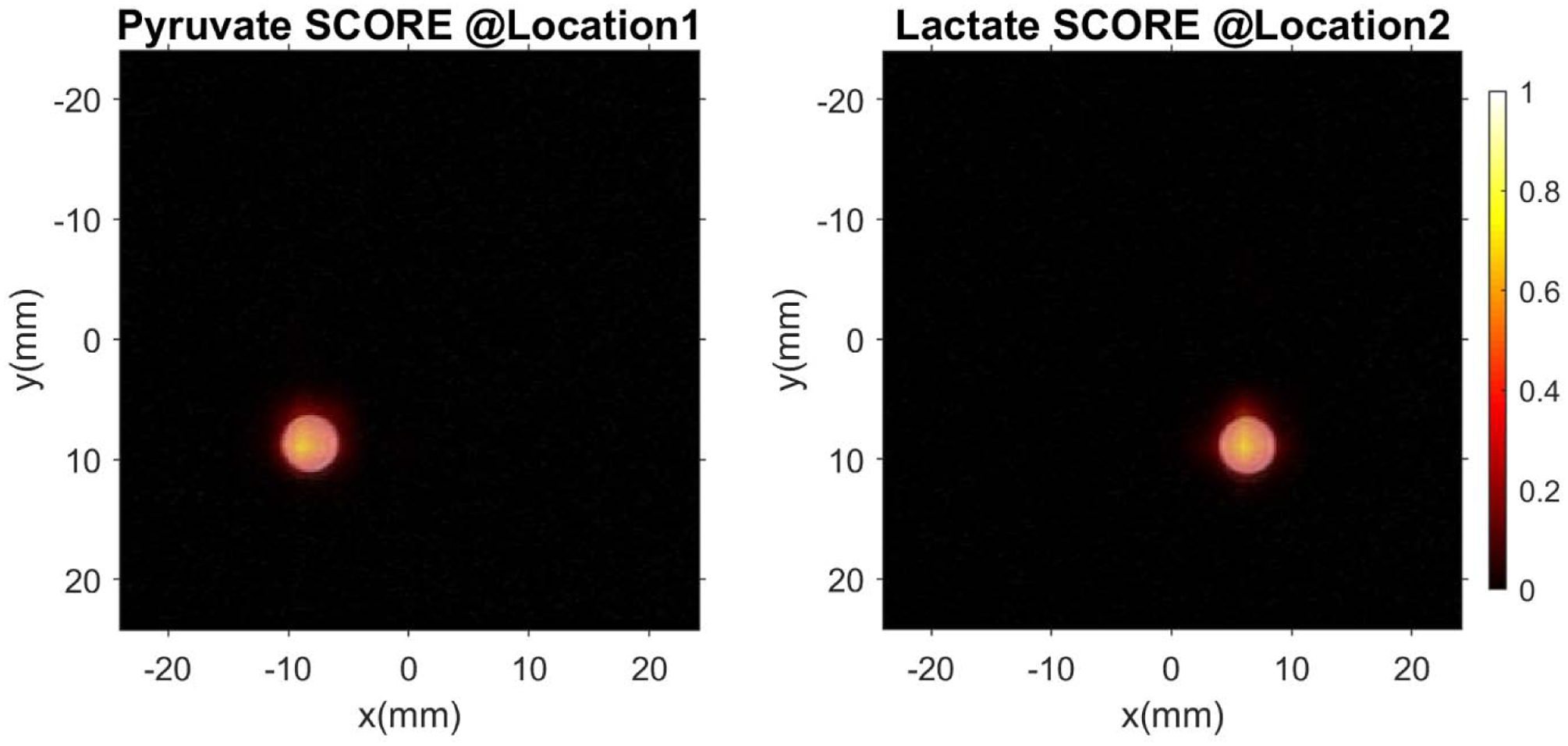
2D radial spatial overlay of phantom data representing [1-13C]-pyruvate and [1-13C]-lactate over reference proton images. These images demonstrate that SCORE permits artifact-free reconstruction of symmetric EPSI data with good spatial fidelity (0.1875 mm in-plane resolution for 1H, 3 mm for 13C).
Discussion
Selective Correction of Off-Resonance Effects (SCORE) permits correction of artifacts associated with symmetric EPSI. Further, SCORE can be applied to spectral peaks that alias from outside of the Nyquist spectral bandwidth of the acquisition. This is particularly interesting for application to spectroscopic imaging of hyperpolarized imaging agents that have a relatively broad but sparse spectrum. SCORE corrections can be applied to minimize spatial chemical shift and N/2 spectral ghosts as long as no parent peaks overlap with other parent peaks or ghost peaks.
The primary goal of this work is to eliminate artifacts in the reconstruction of EPSI acquisitions due to the non-uniform temporal sampling. The zig-zag data pattern acquired with EPSI in k-t space is known to cause spectral artifacts [12, 26, 28]. The simple SCORE correction extends the phenomenon by which non-equidistant symmetric EPSI sampling patterns vanish for metabolites that are on-resonance. A linear metabolite-dependent 1st-order phase correction referenced to the center point of each echo eliminates phase progression within that echo and reformats the zig-zag k-t space sampling (Fig. 2c) to a Cartesian layout (Fig. 2d). After correction, SCORE permits a SNR and metabolites ratio quantification as nearly identical to those from SOS reconstruction, because both methods successfully combined signal from even and odd echoes. The main advantage of SCORE was its ability to maintained the full spectral bandwidth (sBW) with no loss of SNR.
Careful selection of a reduced EPSI sBW enables acquisition of spectra of multiple metabolites with aliased but not overlapped peaks. SCORE is designed exclusively for symmetric EPSI with reduced sBW where aliased signals are expected. Park et al [37] recently used symmetric EPSI to acquire HP MRI data from patients with brain cancer, using a full sBW to cover a chemical shift range encompassing pyruvate and lactate. SCORE correction was not necessary because they did not use a spectrally undersampled acquisition for pyruvate and lactate, while demodulation of the bicarbonate signal may have been accomplished using SCORE. Mayer et al [38] described corrections similar to SCORE which was used to correct blurring caused by off-resonance effects in their spiral CSI acquisition. Here we show that this approach will also correct spectral N/2 artifacts and spatial blurring when applied to spectrally undersampled, symmetric EPSI. It is important to recognize that SCORE correction for one metabolite does not eliminate artifacts for other metabolites, and that a fully corrected spectrum requires separate correction of raw data for each of the metabolites of interest. Repeated SCORE corrections support accurate quantification of relative levels of pyruvate and lactate, which is critical for deriving reproducible metabolic imaging biomarkers for cancer and many other diseases and injuries.
In this work, we integrated the FWHM area under the peak at each spatial location to calculate the relative signal from different peaks and/or reconstruction methods, and then summed that signal along the readout direction. This approach is acceptable when the underlying linewidth is consistent across peaks and datasets, as was the case for our simulations and phantom measurements from our small phantom near isocenter. Other methods for quantifying signal would be needed when linewidth varies between peaks, or between projections. Care must be taken during selection of analysis methods since the coarse temporal sampling resolution of undersampled EPSI leads to a spectral shape with longer tails compared to an ideal Lorentzian lineshape.
It is also important to note that SCORE only eliminates artifacts resulting from non-uniform sampling in symmetric EPSI, and not those that arise from eddy currents. We did not observe any artifacts due to eddy currents in data acquired from the small thermal phantom placed near isocenter. However, gradient delays due to eddy currents also cause spatial and spectral artifacts. SCORE doesn’t correct eddy-current induced artifacts, but it is compatible with established eddy current correction methods. We have demonstrated a combination of eddy current and SCORE correction in numerical simulation, but further confirmation in vivo is still needed.
This approach may not be successful in the presence of excessively broad lines such as may be observed in the presence of poor shimming or other sources of inhomogeneity. Overlap between a target metabolite and another strong signal (or its N/2 spectral ghost) would lead to signal contamination that can’t be eliminated by SCORE. It is important to inspect data to ensure that linewidths are narrow enough to avoid cross-contamination of signals. Modest overlap between spectral peaks could be resolved by fitting those peaks to the appropriate lineshape, but excessive overlap may require prospective utilization of higher sBW. In this work, we selected an aliasing scheme that maximizes separation between the strongest signals (pyruvate and lactate) and their N/2 spectral ghosts, and ensures at least 0.5 ppm separation between pyruvate/lactate and other peaks of potential interest. If necessary, a higher sBW could be selected (such as 10.75ppm as illustrated in Supplementary Figure S1c) to account for pyruvate-hydrate, bicarbonate and alanine peaks.
The SNR of spectral peaks are improved by using symmetric EPSI with SCORE correction. Symmetric EPSI permits a 10.6% more efficient readout [22, 23] with a higher sBW per maximum readout gradient amplitude, compared to flyback EPSI. Careful selection of the sampling bandwidth to avoid overlap of aliased peaks and their N/2 artifacts allowed use of a lower acquisition bandwidth, with corresponding reduction in thermal noise. SCORE correction eliminated blurring along the readout direction and the N/2 spectral artifact for each metabolite, which ensured that signals were properly attributed to the correct spectral peak and spatial location. Successful SCORE corrections require accurate knowledge of the spectral offset between the scanner center frequency and metabolites of interest.
In this work, only pyruvate and lactate spectra were analyzed, but SCORE could be used to reconstruct many more metabolites via a repeated process. Given the spectral resolution and sBW, each line of data in x-f space corresponds to a unique chemical shift. Therefore, any non-overlapping metabolites of interest could be analyzed sequentially using the individual chemical shift offset (δcs) for correction and the (potentially aliased) spectral location (δ′cs) in SCORE-corrected x-f space.
This work was tailored for imaging HP [1-13C] pyruvate and its metabolites, although SCORE would be useful for correcting symmetric EPSI applied to any set of spectra that are well-separated and sparse, and where spectral undersampling can be employed to reduce the readout bandwidth, readout gradient amplitude, and observed noise. These assumptions should be valid for many applications involving X-nucleus MRI and hyperpolarized imaging agents.
Conclusion
A new framework, including data acquisition and image reconstruction, was developed for imaging hyperpolarized [1-13C] pyruvate and its metabolites. Selective Correction of Off-Resonance Effects (SCORE) was used to minimize artifacts in undersampled symmetric EPSI data. The spectral acquisition parameters were set to improve SNR by reducing the readout bandwidth and carefully under-sampling the spectrum of [1-13C] pyruvate and its metabolites. In simulation and phantom measurements, the simple SCORE correction method improved SNR while eliminating spectral artifacts due to non-uniform temporal sampling and spatial artifacts due to chemical shift offsets.
Supplementary Material
Highlight:
Symmetric EPSI maximizes SNR efficiency and spectral bandwidth
Under-sampled spectrum of 7.1ppm reduces the need for high readout bandwidth
Controlled aliasing of HP [1–13C]-pyruvate and its metabolites ensures no overlap
New correction method eliminates N/2 ghost from non-equidistant samples
Acknowledgements
This work was supported, in part, by the National Institutes of Health (P30CA016672, R01CA211150) and GE Healthcare. The content is solely the responsibility of the authors and does not necessarily represent the views of its sponsors.
The authors would also like thank to Dr. Anna Romanowska-Pawliczek for help in preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K, Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR, Proc Natl Acad Sci USA, 100 (2003) 10158–10163, DOI: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH, Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis, Cancer Res, 66 (2006) 10855–10860, DOI: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- [3].Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM, Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy, Nat Med, 13 (2007) 1382–1387, DOI: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- [4].Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ, Vigneron DB, Kurhanewicz J, Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading, Cancer Res, 68 (2008) 8607–8615, DOI: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, Reed G, Carvajal L, Small EJ, Munster P, Weinberg VK, Ardenkjaer-Larsen JH, Chen AP, Hurd RE, Odegardstuen LI, Robb FJ, Tropp J, Murray JA, Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate, Sci Transl Med, 5 (2013) 198ra108, DOI: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cunningham CH, Lau JY, Chen AP, Geraghty BJ, Perks WJ, Roifman I, Wright GA, Connelly KA, Hyperpolarized 13C Metabolic MRI of the Human Heart: Initial Experience, Circ Res, 119 (2016) 1177–1182, DOI: 10.1161/CIRCRESAHA.116.309769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Aggarwal R, Vigneron DB, Kurhanewicz J, Hyperpolarized 1-[(13)C]-Pyruvate Magnetic Resonance Imaging Detects an Early Metabolic Response to Androgen Ablation Therapy in Prostate Cancer, Eur Urol, 72 (2017) 1028–1029, DOI: 10.1016/j.eururo.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, DeAngelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, Sosa RE, Guo YW, Chen AP, Tropp J, Robb F, Keshari KR, Metabolic Imaging of the Human Brain with Hyperpolarized 13C Pyruvate Demonstrates 13C Lactate Production in Brain Tumor Patients, Cancer Res, (2018), DOI: 10.1158/0008-5472.CAN-18-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Grist JT, McLean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, Patterson I, Slough R, Gill AB, Chhabra A, Eichenberger R, Laurent MC, Comment A, Gillard JH, Coles AJ, Tyler DJ, Wilkinson I, Basu B, Lomas DJ, Graves MJ, Brindle KM, Gallagher FA, Quantifying normal human brain metabolism using hyperpolarized [1-(13)C]pyruvate and magnetic resonance imaging, Neuroimage, 189 (2019) 171–179, DOI: 10.1016/j.neuroimage.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, Bankson JA, Brindle K, Cunningham CH, Gallagher FA, Keshari KR, Kjaer A, Laustsen C, Mankoff DA, Merritt ME, Nelson SJ, Pauly JM, Lee P, Ronen S, Tyler DJ, Rajan SS, Spielman DM, Wald L, Zhang X, Malloy CR, Rizi R, Hyperpolarized (13)C MRI: Path to Clinical Translation in Oncology, Neoplasia, 21 (2019) 1–16, DOI: 10.1016/j.neo.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Harris T, Eliyahu G, Frydman L, Degani H, Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells, Proc Natl Acad Sci USA, 106 (2009) 18131–18136, DOI: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mansfield P, Spatial mapping of the chemical shift in NMR, Magn Reson Med, 1 (1984) 370–386, DOI: 10.1002/mrm.1910010308. [DOI] [PubMed] [Google Scholar]
- [13].Yen YF, Kohler SJ, Chen AP, Tropp J, Bok R, Wolber J, Albers MJ, Gram KA, Zierhut ML, Park I, Zhang V, Hu S, Nelson SJ, Vigneron DB, Kurhanewicz J, Dirven HA, Hurd RE, Imaging considerations for in vivo 13C metabolic mapping using hyperpolarized 13C-pyruvate, Magn Reson Med, 62 (2009) 1–10, DOI: 10.1002/mrm.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Larson PEZ, Hu S, Lustig M, Kerr AB, Nelson SJ, Kurhanewicz J, Pauly JM, Vigneron DB, Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies, Magn Reson Med, 65 (2011) 610–619, DOI: 10.1002/mrm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ramirez MS, Lee J, Walker CM, Sandulache VC, Hennel F, Lai SY, Bankson JA, Radial spectroscopic MRI of hyperpolarized [1-(13) C] pyruvate at 7 tesla, Magn Reson Med, 72 (2014) 986–995, DOI: 10.1002/mrm.25004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Durst M, Koellisch U, Frank A, Rancan G, Gringeri CV, Karas V, Wiesinger F, Menzel MI, Schwaiger M, Haase A, Schulte RF, Comparison of acquisition schemes for hyperpolarised (1)(3)C imaging, NMR Biomed, 28 (2015) 715–725, DOI: 10.1002/nbm.3301. [DOI] [PubMed] [Google Scholar]
- [17].Chen HY, Larson PEZ, Gordon JW, Bok RA, Ferrone M, van Criekinge M, Carvajal L, Cao P, Pauly JM, Kerr AB, Park I, Slater JB, Nelson SJ, Munster PN, Aggarwal R, Kurhanewicz J, Vigneron DB, Technique development of 3D dynamic CS-EPSI for hyperpolarized (13) C pyruvate MR molecular imaging of human prostate cancer, Magn Reson Med, (2018), DOI: 10.1002/mrm.27179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gordon JW, Larson PEZ, Pulse Sequences for Hyperpolarized MRS, eMagRes, (2016) 1229–1246, DOI: 10.1002/9780470034590.emrstm1451. [DOI] [Google Scholar]
- [19].Topping GJ, Hundshammer C, Nagel L, Grashei M, Aigner M, Skinner JG, Schulte RF, Schilling F, Acquisition strategies for spatially resolved magnetic resonance detection of hyperpolarized nuclei, MAGMA, 33 (2020) 221–256, DOI: 10.1007/s10334-019-00807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D, High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding, Magn Reson Med, 33 (1995) 34–40, DOI: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- [21].Sarkar S, Heberlein K, Metzger GJ, Zhang X, Hu X, Applications of high-resolution echoplanar spectroscopic imaging for structural imaging, J Magn Reson Imaging, 10 (1999) 1–7, DOI: . [DOI] [PubMed] [Google Scholar]
- [22].Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, Kelley DA, Pauly JM, Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging, Magn Reson Med, 54 (2005) 1286–1289, DOI: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- [23].Chen AP, Cunningham CH, Ozturk-Isik E, Xu D, Hurd RE, Kelley DA, Pauly JM, Kurhanewicz J, Nelson SJ, Vigneron DB, High-speed 3T MR spectroscopic imaging of prostate with flyback echoplanar encoding, J Magn Reson Imaging, 25 (2007) 1288–1292, DOI: 10.1002/jmri.20916. [DOI] [PubMed] [Google Scholar]
- [24].Zierhut ML, Ozturk-Isik E, Chen AP, Park I, Vigneron DB, Nelson SJ, (1)H spectroscopic imaging of human brain at 3 Tesla: comparison of fast three-dimensional magnetic resonance spectroscopic imaging techniques, J Magn Reson Imaging, 30 (2009) 473–480, DOI: 10.1002/jmri.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sekihara K, Kohno H, New reconstruction technique for echo-planar imaging to allow combined use of odd and even numbered echoes, Magn Reson Med, 5 (1987) 485–491, DOI: 10.1002/mrm.1910050512. [DOI] [PubMed] [Google Scholar]
- [26].Metzger G, Hu X, Application of interlaced Fourier transform to echo-planar spectroscopic imaging, J Magn Reson, 125 (1997) 166–170, DOI: 10.1006/jmre.1997.1114. [DOI] [PubMed] [Google Scholar]
- [27].Bruder H, Fischer H, Reinfelder HE, Schmitt F, Image reconstruction for echo planar imaging with nonequidistant k-space sampling, Magn Reson Med, 23 (1992) 311–323, DOI: 10.1002/mrm.1910230211. [DOI] [PubMed] [Google Scholar]
- [28].Hanson LG, Schaumburg K, Paulson OB, Reconstruction strategy for echo planar spectroscopy and its application to partially undersampled imaging, Magn Reson Med, 44 (2000) 412–417, DOI: . [DOI] [PubMed] [Google Scholar]
- [29].Klose U, In vivo proton spectroscopy in presence of eddy currents, Magn Reson Med, 14 (1990) 26–30, DOI: 10.1002/mrm.1910140104. [DOI] [PubMed] [Google Scholar]
- [30].Du W, Du YP, Fan X, Zamora MA, Karczmar GS, Reduction of spectral ghost artifacts in high-resolution echo-planar spectroscopic imaging of water and fat resonances, Magn Reson Med, 49 (2003) 1113–1120, DOI: 10.1002/mrm.10485. [DOI] [PubMed] [Google Scholar]
- [31].Noll DC, Fessler JA, Sutton BP, Conjugate phase MRI reconstruction with spatially variant sample density correction, IEEE Trans Med Imaging, 24 (2005) 325–336, DOI: 10.1109/tmi.2004.842452. [DOI] [PubMed] [Google Scholar]
- [32].Coello E, Noeske R, Burns BL, Gordon JW, Jakary A, Menze B, Haase A, Larson PEZ, Li Y, Schulte RF, High-resolution echo-planar spectroscopic imaging at ultra-high field, NMR Biomed, 31 (2018) e3950, DOI: 10.1002/nbm.3950. [DOI] [PubMed] [Google Scholar]
- [33].Joe E, Lee H, Lee J, Yang S, Choi YS, Wang E, Song HT, Kim DH, An indirect method for in vivo T2 mapping of [1-(13) C] pyruvate using hyperpolarized (13) C CSI, NMR Biomed, 30 (2017), DOI: 10.1002/nbm.3690. [DOI] [PubMed] [Google Scholar]
- [34].Wright SM, Wald LL, Theory and application of array coils in MR spectroscopy, NMR Biomed, 10 (1997) 394–410, DOI: . [DOI] [PubMed] [Google Scholar]
- [35].Sun CY, Walker CM, Michel KA, Venkatesan AM, Lai SY, Bankson JA, Influence of parameter accuracy on pharmacokinetic analysis of hyperpolarized pyruvate, Magn Reson Med, 79 (2018) 3239–3248, DOI: 10.1002/mrm.26992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shang H, Sukumar S, von Morze C, Bok RA, Marco-Rius I, Kerr A, Reed GD, Milshteyn E, Ohliger MA, Kurhanewicz J, Larson PEZ, Pauly JM, Vigneron DB, Spectrally selective three-dimensional dynamic balanced steady-state free precession for hyperpolarized C-13 metabolic imaging with spectrally selective radiofrequency pulses, Magn Reson Med, 78 (2017) 963–975, DOI: 10.1002/mrm.26480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Park I, Larson PEZ, Gordon JW, Carvajal L, Chen HY, Bok R, Van Criekinge M, Ferrone M, Slater JB, Xu D, Kurhanewicz J, Vigneron DB, Chang S, Nelson SJ, Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies, Magn Reson Med, 80 (2018) 864–873, DOI: 10.1002/mrm.27077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM, Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging, Magn Reson Med, 56 (2006) 932–937, DOI: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


