Abstract
Background
Recent studies have demonstrated that microRNAs (miRNAs) in the blood circulation can serve as promising diagnostic markers for cancers. This four-stage study aimed at finding serum miRNAs as potential biomarkers for lung adenocarcinoma (LA) diagnosis.
Methods
The study was carried out between 2016 and 2017. The Exiqon miRNA qPCR panel (3 LA vs. 1 normal control [NC] pooled serum samples) was used for initial screening to acquire miRNA profiles. Thirty-five dysregulated miRNAs were further evaluated in the training (24 LA vs. 24 NCs) and testing stages (110 LA vs. 110 NCs) using quantitative real-time polymerase chain reaction assays.
Results
Four serum miRNAs (miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p) were significantly overexpressed in LA patients compared with NCs. The diagnostic value of the four-miRNA panel was validated by an external cohort (36 LA vs. 36 NCs). The areas under the receiver operating characteristic curve of the four-miRNA panel in the training, testing, and external validation stages were 0.734, 0.803, and 0.894 respectively. Meanwhile, the expression level of miR-221-3p was much higher in LA tumor samples than that in the adjacent normal tissues (19 LA vs. 19 NCs). The expression level of miR-10b-5p was also elevated in the serum-derived exosomes samples (18 LA vs. 18 NCs). The expression of miR-133a-3p, miR-584-5p, and miR-10b-5p was significantly elevated in LA patients with epidermal growth factor receptor mutation compared with NCs.
Conclusion
The study established a four-miRNA signature in serum that could improve the diagnostic capability of LA.
Keywords: Serum microRNA, Lung adenocarcinoma, Diagnostic biomarker, Exosomes, Epidermal growth factor receptor
Introduction
Lung cancer is the most lethal disease worldwide and has two main histological types: non-small-cell lung cancer (NSCLC) and small-cell lung cancer,[1] with NSCLC representing 80% to 85%.[2] If NSCLC is discovered at an early stage, surgical resection can offer a satisfying prognosis, with a 5-year survival rate of 70% to 90% for stage I patients.[3] However, approximately 75% of patients have advanced diseases (stage III/IV) at the time of diagnosis for the lack of early symptoms. Although significant developments have been achieved in the oncological management of advanced-stage lung cancer in recent years, the prognosis is still poor with a 5-year survival rate of less than 15%. Low dose computed tomography screening has been recommended as an effective tool in the early detection of lung cancer. However, the extra radiation exposure, high misdiagnosis rate, and costly inspection fee restricted its wide application.[4] The protein biomarkers such as carcino-embryonic antigen, cytokeratin-19-fragment (CYFRA21-1) have limited sensitivity and specificity, which restricted their further application in NSCLC detection.[5] Thus, it is pivotal to discover novel non-invasive biomarkers with high diagnostic power for NSCLC.
MicroRNAs (miRNAs) are a class of small (21–23 nucleotides) and highly conserved non-coding RNAs. Acting through translational inhibition or degradation of messenger RNA targets, miRNAs are essential regulators of gene expression.[6,7] Accumulating evidence has proved that circulating miRNAs are reliable non-invasive biomarkers for various cancers, including NSCLC.[8–11] However, the results so far are inconsistent. Lung adenocarcinoma (LA) is the most common histological subtype of NSCLC.[12] The molecular profiling characteristics drawn by the Cancer Genome Atlas project revealed the different genomic profiles between LA and lung squamous cell carcinoma which provided the foundation for NSCLC classification.[13] Moreover, NSCLC patients with point driver mutations can greatly benefit from personalized targeted therapy. By now, the most common driver mutation in NSCLC is the mutant activation of the epidermal growth factor receptor (EGFR). More than 70% of NSCLC patients carrying the point mutation in exon 21 (p.L858R) or deletion in exon 19 achieved a favorable response to tyrosine kinase inhibitors of EGFR (EGFR-TKIs) treatment.[14] Several circulating miRNAs have been reported as a potential supplement to genetic testing and could predict the response to EGFR-TKIs, which further emphasized the critical roles of circulating miRNAs in LA.[15,16] In a previous study, we had developed a six-miRNA signature in plasma for the diagnosis of LA.[9] Here, we further explored miRNA expression in serum to identify a specific serum miRNA signature that might be used as a non-invasive biomarker for LA diagnosis.
Methods
Ethical approval
The study was conducted with the approval of the Institutional Review Board and the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (ID: 2016-SRFA-148).
Patient selection and study design
Our study enrolled 170 LA patients and 170 healthy individuals (as normal controls [NCs]) between 2016 and 2017 from the First Affiliated Hospital of Nanjing Medical University and Jiangsu Cancer Hospital. All the LA patients were accurately diagnosed by pathology evidence. Briefly, only tumors with clear morphologic features of adenocarcinoma or supported by specific immunohistochemical stains (ie, thyroid transcription factor-1 positive) were included in this study. Patients who met any of the following exclusion criteria were excluded from the experiment: (i) metastasis of other cancers confirmed by judicious immunohistochemical stains and clinical evaluation; (ii) LA patients who had received clinical treatment (ie, chemotherapy, radiotherapy, surgery, immunotherapy) before the study; (iii) patients without informed consents or considered to be unsuitable for the experiment. The clinical and histopathological baseline data including age, sex, smoking status, histologic classification, tumor location, and tumor nodes metastases (TNM) stage (based on the seventh edition American Joint Committee on Cancer) were collected from each patient retrospectively.
The experimental process was separated into four phases [Figure 1]. In the screening stage, 30 serum samples from LA patients and ten from NCs were randomly selected. Per ten serum samples were combined to get three LA and one NC pooled serum samples. Exiqon miRCURY-Ready-to-Use PCR-Human-panel-I+II-V1.M (Exiqon, Vedbaek, Denmark) was used to initially select the differentially expressed miRNAs in LA according to our previous study.[9] In the next training stage, the dysregulated miRNAs identified in the screening stage were examined by quantitative real-time polymerase chain reaction (qRT-PCR) in the serum samples of 24 LA and 24 NCs. In the following testing stage, the selected miRNAs were confirmed in 110 LA patients and 110 NCs. The identified miRNA panel was validated in an external cohort of 36 LA patients and 36 NCs. The expression levels of these identified miRNAs were also evaluated among tissue (19 LA vs. 19 NCs) and serum exosomes samples (18 LA vs. 18 NCs).
Figure 1.
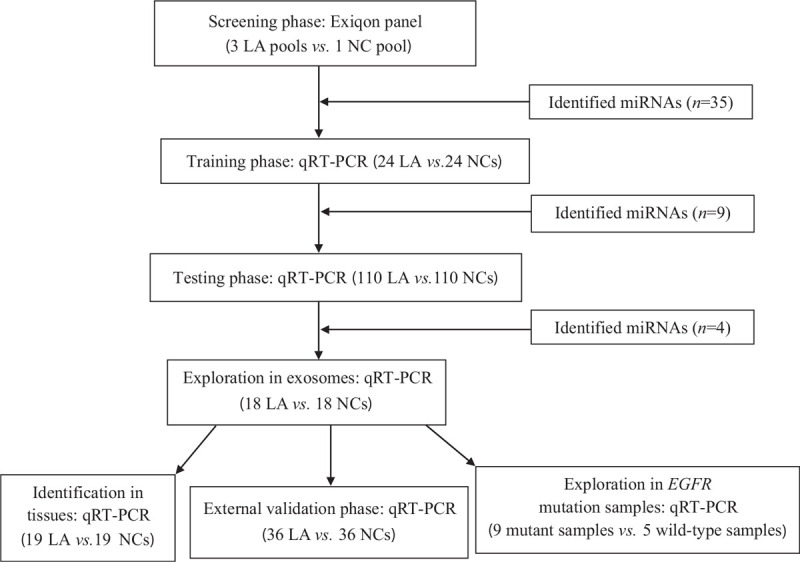
Flow chart of the experiment design. EGFR: Epidermal growth factor receptor; LA: Lung adenocarcinoma; mi-RNA: MicroRNA; NC: Normal control; qRT-PCR: Quantitative real-time polymerase chain reaction.
Sample preparation
Venous blood samples (5 mL) were collected and stored in a serum separator tube. Then, the cell-free serum sample was separated from whole blood within 6 h by centrifugation at 2000 ×g for 10 min, and then kept at –80°C for further usage. Liquid nitrogen was used to store tissue specimens.
Isolation of exosomes
With the usage of ExoQuick™ (System Biosciences, Palo Alto, CA, USA), exosomes were isolated from serum in accordance with the manufacturer's protocol. Exosome granules extracted from 200 μL serum and 50 μL ExoQuick exosome precipitation solutions were dissolved in 200 μL RNase-free water until further usage.
RNA extraction
The mirVana PARIS Kit (Ambion, Austin, TX, USA) was utilized to extract total RNA from 200 μL serum or exosomes according to the manufacturer's protocol. The 5 μL of synthetic Caenorhabditis elegans miR-39 (5 mmol/L; RiboBio, Guangzhou, China) was spiked into each sample normalization. Trizol (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA from tissue specimens. RNA was eluted with 100 μL RNase-free water and stored at –80°C for further processing. The concentration and purity of the RNA samples were quantified by the ultraviolet spectrophotometer.
qRT-PCR
The expression of each miRNA was quantified in triplicate with the specific primers of reverse transcription (RT) and polymerase chain reaction (PCR) (RiboBio). The processes of RT and PCR were carried out as described in our previous study.[17–19] A LightCycler 480 (Roche 480, Penzberg, Germany) real-time thermal cycler was used to amplify and detect miRNA expression with SYBR Green dye. The melting analysis was added to assess our PCR products’ specificity. The 2–ΔΔCt method relative to the combination of cel-miR-39 and endogenous reference gene (miR-103a-3p) was used to calculate the expression of serum and exosomal miRNAs (ΔCt = CtmiRNA − 1/2(Ctcel-miR-39 + CtmiR-103a-3p; Ct: cycle threshold).[20] miRNA expression in tissue samples was determined using the 2-ΔΔCt method relative to RNU6B (U6) and cel-miR-39.
Statistical analysis
We compared the miRNA expression levels between the two groups using the Mann-Whitney U test. Multiple logistic regression analysis was used to construct the miRNA panel. The association between miRNAs and the clinical characteristics was evaluated by the one-way analysis of variance or Chi-square test. Receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were evaluated to estimate the diagnostic performance of the identified miRNA panel. SPSS software (version 23.0, IBM, Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) were used for statistical analyses and graph construction. A two-sided P value < 0.05 was considered to be statistically significant.
Results
Characteristics of patient samples
The study enrolled 170 LA patients and 170 NCs in a whole. Their clinicopathological characteristics in the training, testing, and external validation stage were listed in Table 1. There was no significant difference in the distribution of age, gender, or smoking status between LA patients and NCs in each stage (P > 0.05).
Table 1.
Characteristics of the 170 lung adenocarcinoma patients and 170 normal controls enrolled in the study.
Discovery of candidate miRNAs from pooled samples
To select candidate miRNAs for LA diagnosis, we examined miRNAs profiles from three LA and one NC pooled serum samples with Exiqon miRCURY-Ready-to-Use PCR-Human-panel-I+II-V1.M. This panel can detect the expression of 168 miRNAs which are relatively higher expressed in blood circulation. The miRNAs with a Ct 5 lower than the negative control or at least 1.5-fold altered expressed were included. Furthermore, miRNAs with limited expression levels (Ct-value > 37) were removed from the candidate list. As a result, 35 miRNAs were selected as candidate miRNAs for further examination via qRT-PCR [Supplementary Table 1].
Evaluation of stable internal reference genes
Hemolysis would limit the application of certain miRNAs as disease biomarkers. Therefore, our study evaluated two hemolysis markers (miR-23a, miR-451) in serum samples.[21] In this study, a ΔCt (CtmiR-23a − CtmiR-451) greater than 5 indicated a high risk of hemolysis.[21] Samples with high hemolysis probability would be filtered out. The result turned out that the expression levels of miR-23a and miR-451 were not altered between serum samples of NSCLC and NCs [Supplementary Table 2]. MiR-16-5p[22,23] and miR-103a-3p[24,25] were potential internal reference genes according to previous studies. However, the expression of serum miR-16-5p was unstable between LA patients and NCs [Supplementary Figure 1]. Therefore, miR-103a-3p was selected as the endogenous reference gene in this study. Both two miRNAs were not stable enough to act as internal reference genes in serum exosomes [Supplementary Figure 2].
Confirmation of candidate serum miRNAs by qRT-PCR
Based on the qRT-PCR analysis, the candidate miRNAs chosen in the screening phase were further validated in the training phase (24 LA vs. 24 NCs). Nine miRNAs that demonstrated differential expression were tested in the testing phase (110 LA vs. 110 NCs). In this larger cohort, four (miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p) of the nine miRNAs demonstrated consistent up-regulation (P < 0.05). The expression level of the identified miRNAs was further validated in an independent cohort of 36 LA patients and 36 NCs. All the miRNAs were up-regulated with fold change >2 compared with NCs [Supplementary Table 3]. What's more, when the data of the three cohorts were combined, the four miRNAs showed consistently higher expression in the serum samples of LA patients than NCs [Figure 2].
Figure 2.
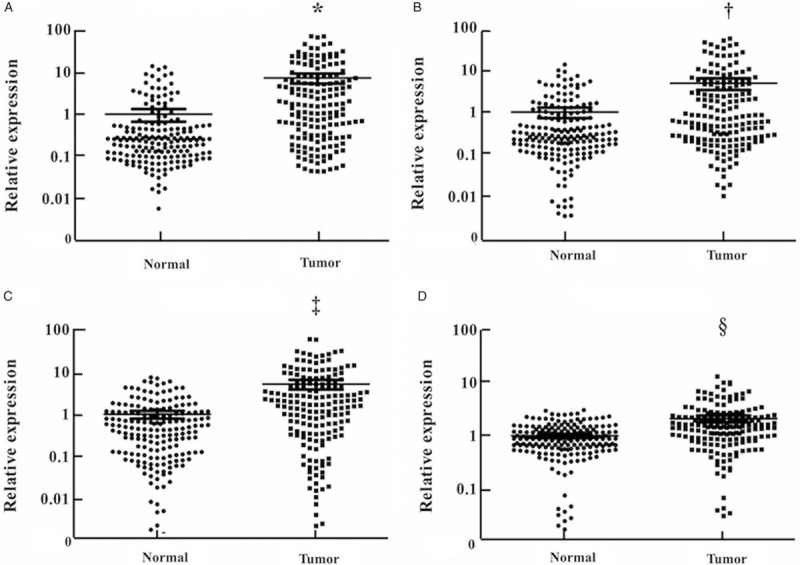
Expression of the four miRNAs in the serum of 170 lung adenocarcinoma (LA) patients and 170 normal controls (NCs). The relative expression levels of miRNAs were compared between LA patients and NCs using the Mann-Whitney U test. Horizontal line: Mean with 95% confidence interval (CI). (A) miR-133a-3p: ∗P < 0.001, Z = –4.839; (B) miR-584-5p: †P < 0.001, Z = –5.230; (C) miR-10b-5p: ‡P < 0.006, Z = –4.723; (D) miR-221-3p: §P < 0.001, Z = –4.924. mi-RNA: MicroRNA.
Diagnostic value of the candidate miRNAs
The diagnostic accuracy of the four candidate miRNAs in serum was measured by ROC curves. When the data of training, testing, and external validation stages were combined, the corresponding AUCs were 0.754, 0.652, 0.713, and 0.668 for miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p, respectively [Supplementary Figure 3]. We then combined the identified serum miRNAs and established a four-miRNA panel to distinguish LA patients from NCs using logistic regression analysis. The predicted probability of PTC detection was figured by the formula: Logit(P) = 0.915 − 0.036 × miR-133a-3p + 0.013 × miR-584-5p − 0.066 × miR-10b-5p – 0.135 × miR-221-3p. The combination of the four-miRNAs showed further improvement in diagnostic capability (AUC = 0.765, 95% confidence interval [CI]: 0.714–0.816, sensitivity: 66.5%, specificity: 76.5%; Figure 3A). The AUCs of the four-miRNA panel in the separated training, testing, and external validation stages were 0.734 (95% CI: 0.572–0.897; sensitivity: 70.8%, specificity: 79.7%; Figure 3B), 0.803 (95% CI: 0.745–0.860; sensitivity: 78.5%, specificity: 68.2%; Figure 3C), and 0.894 (95% CI: 0.882–0.967; sensitivity: 72.2%, specificity: 97.2%; Figure 3D), respectively.
Figure 3.
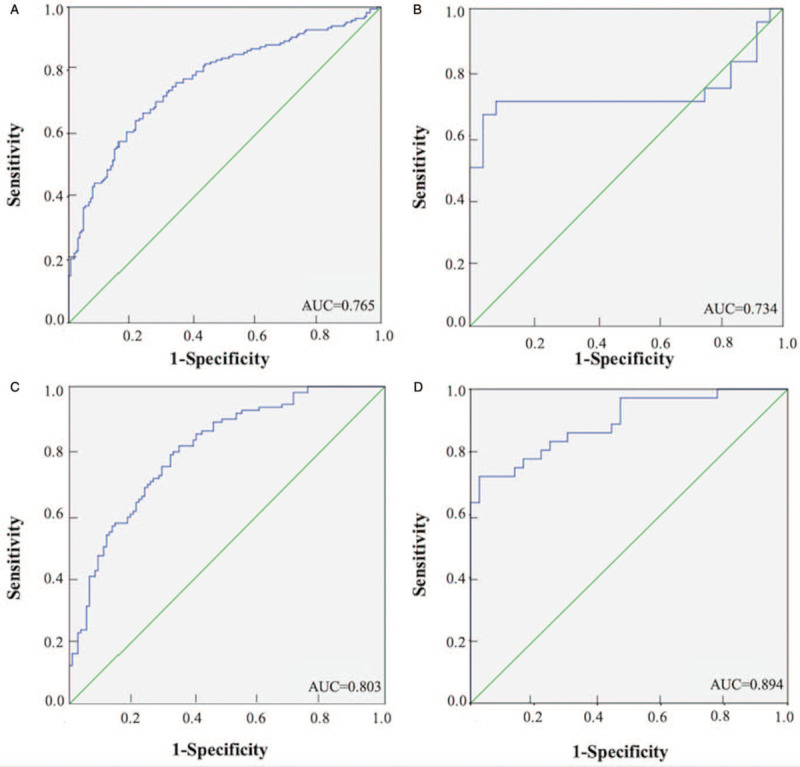
Receiver-operating characteristic (ROC) curves of the four-miRNA panel to discriminate lung adenocarcinoma (LA) patients from normal controls (NCs). (A): the combined three phases (170 LA vs. 170 NCs): AUC = 0.765, 95% CI: 0.714–0.816, P = 0.001; (B): the training phase (24 LA vs. 24 NCs): AUC = 0.734, 95% CI: 0.572–0.897, P = 0.002; (C): the testing phase (110 LA vs. 110 NCs): AUC = 0.803, 95% CI: 0.745–0.860, P < 0.001; (D): the external validation phase (36 LA vs. 36 NCs): AUC = 0.894, 95% CI: 0.882–0.967, P < 0.001.AUC: Areas under the curve; CI: Confidence interval; mi-RNA: MicroRNA.
Sub-group analysis of the diagnostic efficiency of the four-miRNA panel
The expression levels of the identified miRNAs were further compared among LA patients with different clinicopathologic features [Supplementary Table 4]. In addition, we further explored the performance of the established miRNA panel in discriminating LA patients with different clinicopathological features from NCs [Table 2]. The corresponding AUCs for patients at TNM stages I, II, III, and IV were 0.736, 0.762, 0.811, and 0.865, respectively. Moreover, the diagnostic value of the four-miRNA panel was assessed according to different lymph node status. For those patients without lymph node metastasis, the AUC was 0.740, while for those patients with positive lymph node metastasis, the AUC was 0.796. The comparison was also conducted among samples with different features of tumor location, general type, and histologic classification. These results turned out that this serum four-miRNA panel could well discriminate LA patients with different clinicopathologic features from NCs.
Table 2.
Subgroup analysis of the diagnostic value of the four-miRNA panel.
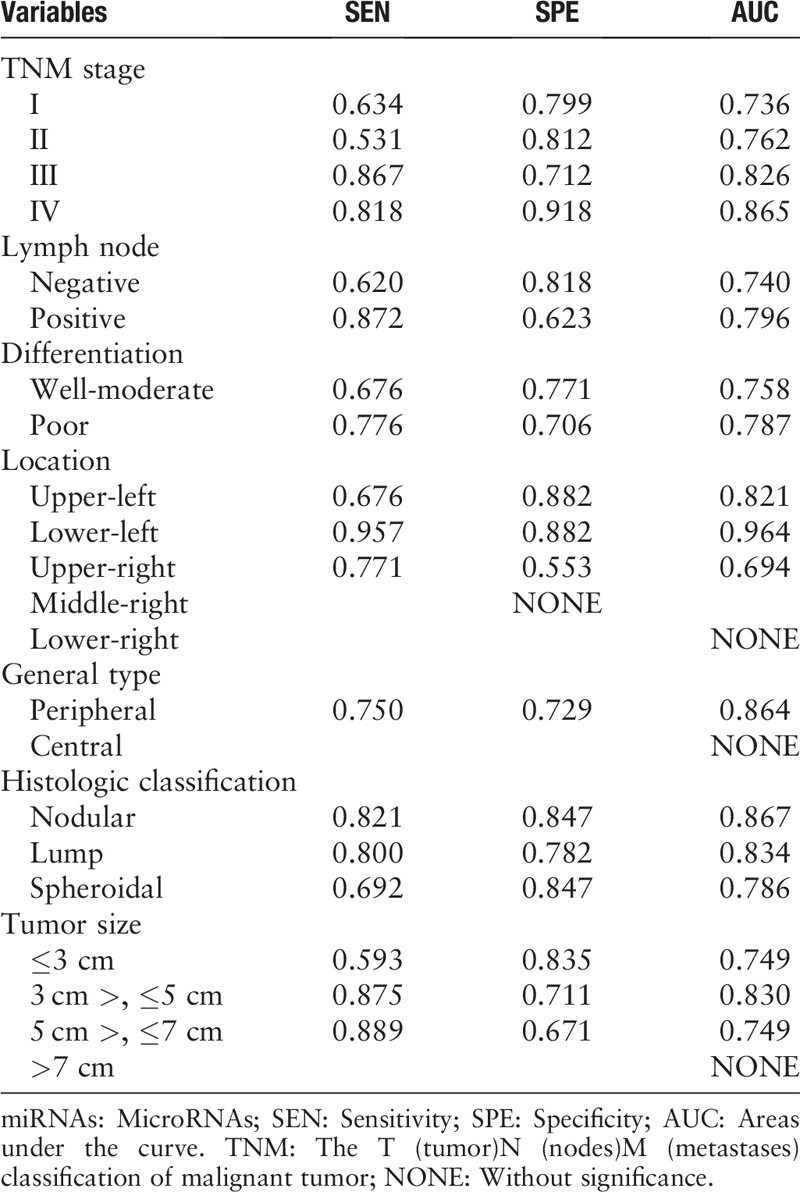
Identification of miRNA expression levels in tissues
To investigate the origin of the four serum miRNAs, we examined the expression of the four miRNAs in 24 paired LA tissue specimens and adjacent normal tissue samples. As shown in Figure 4, only miR-221-3p showed consistent up-regulation in tumor tissues with P < 0.05. No significant expression difference was observed in tissue samples for the other three miRNAs.
Figure 4.
Expression of the four miRNAs in the 24 pairs of tissue samples. The relative expression levels of miRNAs were compared between lung adenocarcinoma tumor tissues and the adjacent normal tissues using a paired non-parametric test. (A): miR-221-3p: ∗P = 0.008, Z = –2.704; (B): miR-133a-3p: †P = 0.179, Z = –1.265; (C): miR-584-5p: ‡P = 0.429, Z = –0.932; (D): miR-10b-5p: §P = 0.407, Z = –1.118. mi-RNA: MicroRNA.
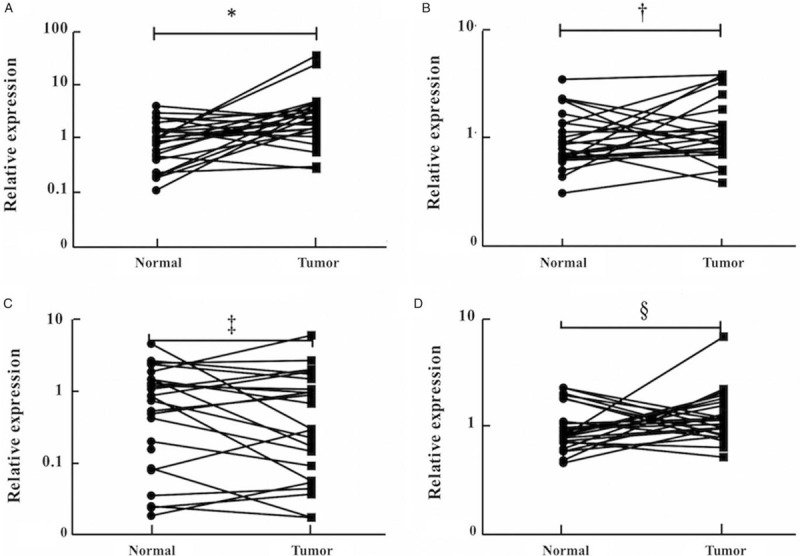
Exploration of miRNA expression in exosomes derived from serum
To assess the potential form of the four miRNAs in peripheral serum, we tested exosomal miRNA expression in the serum samples of 18 LA patients and 18 NCs. Only miR-10b-5p was significantly up-regulated in the serum exosomes of LA patients [Table 3].
Table 3.
Relative expression levels of the four miRNAs in the serum-derived exosomes samples of 18 lung adenocarcinoma patients and 18 normal controls.
Exploration of miRNA expression in LA patients with EGFR mutation
To further decipher the link between miRNA expression and the EGFR mutation status, the serum samples from 14 patients were tested (three LA patients with E19 DelE746-A750 mutation vs. six LA patients with E21 p. L858 mutation vs. five LA patients with EGFR-wild type). As shown in Figure 5, the expression of serum miR-133a-3p, miR-584-5p, and miR-10b-5p was higher in patients with EGFR mutation compared to NCs (P < 0.05). The AUC of the three-miRNA panel to discriminate LA patients with EGFR mutation status from NCs was 0.855 (95% CI: 0.673–1.00; sensitivity: 77.8%, specificity: 92.31%; Supplementary Figure 4). Although without significant difference, the expression levels of miR-133a-3p and miR-10b-5p were also elevated in patients with EGFR mutation, compared to those with the wild type [Supplementary Figure 5]. Moreover, the comparison of the serum exosomal miRNA expression in LA patients with different EGFR mutation status (seven mutant cases vs. four wide-type cases) was conducted, however, no difference of the four-miRNA expression was observed (data not shown).
Figure 5.
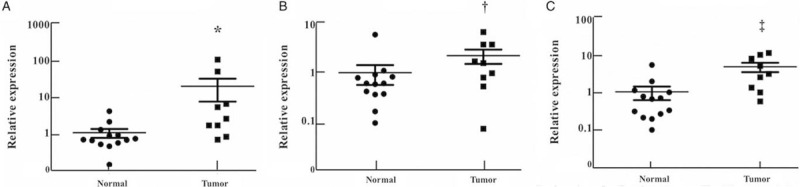
Expression of the three miRNAs in the serum samples of nine lung adenocarcinoma patients with epidermal growth factor receptor (EGFR) mutation (E19 DelE746-A750 mutation or E21 p. L858R mutation). The relative expression levels of the serum miRNAs were compared between patients with EGFR mutation and normal controls using the Mann-Whitney U test. Horizontal line: Mean with SEM. (A): miR-133a-3p: ∗P < 0.05, Z = –2.532; (B): miR-584-5p: †P < 0.001, Z = –3.890; (C): miR-10b-5p: ‡P < 0.05, Z = –2.164. mi-RNA: MicroRNA.
Bioinformatics analysis of miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p
DIANA-TarBase v7.0 was applied to identify the potential target genes of miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p. The DIANA-miRPath v3.0 was then used for KEGG pathway analysis [Supplementary Table 5] and GO category analysis [Supplementary Table 6] to explore the pathways related to the four miRNAs. The results turned out that both miR-10b-5p and miR-221-3p have tight connections with the molecular mechanisms of cancer (ie, the p53 signaling pathway).
Discussion
Accumulating studies have confirmed the potential of tumor-specific miRNAs as new markers for the diagnosis of malignancies, including NSCLC. However, NSCLC comprised a class of subtypes with histological heterogeneity.
The multi-phase study focused on the specific type of NSCLC-LA and tried to discover differentially expressed serum miRNAs for the diagnosis of LA patients. The Exiqon miRNA qPCR panel used in the screening phase was proved to have better linearity than the TaqMan platform. To reduce the false-positive rate, the research carried out three phases of qRT-PCR validation after the miRNA screening phase, which included a total of 170 patients and 170 NCs. Currently, the quantification of miRNA expression did not have a recognized endogenous control. Previous studies have discovered that miR-103a-3p might be a promising internal reference gene.[24,25] Meanwhile, in this study, the expression of serum miR-103a-3p was confirmed to be stable both in LA patients and NCs in the training stage. Hence, miR-103a-3p was selected as the reference miRNA in serum. However, in exosomes, miR-103a-3p was not stable enough to act as a reference gene. To investigate the probability of hemolysis during sampling, the expression of serum miR-23a and miR-451 were assessed. The relatively stable expression of the two miRNAs helped to eliminate the effect of hemolysis. As a result, four significantly up-regulated miRNAs (miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p) showed relatively high accuracy in the detection of LA. However, only the expression of 221-3p was significantly higher in tumor samples than that in paired adjacent normal tissues; conversely, miR-584-5p had lower expression in tumor samples.
All the four miRNAs (miR-133a-3p, miR-584-5p, miR-10b-5p, and miR-221-3p) have been found separately to have certain diagnostic value in NSCLC.[26,27] Furthermore, plasma miR-221-3p and miR-584-5p were confirmed by our previous study as reliable biomarkers for the early diagnosis of LA.[9] This indicates that the expressing characteristics of serum miRNAs and plasma miRNAs are similar to some extent. Among the four miRNAs, miR-10b-5p is the most widely studied. Former studies have provided evidence that overexpression of miR-10b-5p can lead to the development of lung cancer. The high expression of miR-10b-5p could promote the migration and invasion of A549 cells.[28,29] Hung et al[30] reported that the expression of miR-10b was higher in NSCLC tissues compared with normal tissues. Evaluated miR-10b could promote tumorigenesis via targeting the tumor suppressor Fas, FasL, Bax, and caspase. Moreover, up-regulated miR-10b-5p led to a poor prognosis of NSCLC by targeting E-cadherin.[31] An evaluated level of circulating miR-10b-5p was also observed in breast cancer,[32] gastric cancer,[33] and could predict the advanced disease status and liver metastasis in colorectal cancer.[34]
The role of miR-584-5p in lung cancer is still unclear. Our finding suggested that its expression was up-regulated in LA serum samples but not in LA tissues. Previous studies have revealed that miR-584-5p may play distinct roles in different cancer types.[35,36] Considering the theory that circulating miRNAs are released from cells,[37,38] it is possible that circulating miR-584-5p acts as a passenger but not a driver in LA development.
To our knowledge, the role of miR-221-3p in NSCLC is controversial. Heegaard et al[27,40] reported that serum miR-221-3p expression was reduced in early-stage NSCLC patients.[39] But in other studies, the up-regulation of miR-221-3p was observed in NSCLC patients both in circulation and tissue. Our previous study also reported that miR-221-3p could act as a reliable biomarker for LA.[9] Reduction of miR-221 inhibited cell proliferation and induced mitochondrial-mediated apoptosis in the A549 lung cancer cell line by targeting PUMA21042732/ TIMP2.[40]Meanwhile, it could inactivate the tumor suppressor PTEN and TIMP3, induce TRAIL resistance, and enhance cellular migration via activating the AKT pathway and metallopeptidases.[41] However, Yamashita et al[42] found the tumor-suppressive effects (intra-S-phase arrest or apoptosis) of miR-221 in lung cancer cells. The mechanism of miR-221-3p in lung cancer needs to be further explored.
MiR-133a-3p was reported to inhibit cell invasiveness and growth via suppressing IGF-1R, TGFBR1, and EGFR.[43] However, there was no significant difference of miR-133a-3p expression in neither serum nor tumor tissues of NSCLC patients in previous studies.[44] However, our study found that miR-133a-3p was significantly up-regulated in serum samples of LA patients, but its expressing difference was not significant in tumor tissues. Although these results might be contradictory, this phenomenon showed exactly the duplicity of miRNA function in cancer. Previous studies have reported different miRNA profiles in extracellular and intracellular components and suggested the existence of a cellular selection mechanism for the miRNA release.[45] As a result, we speculated that the LA cells may selectively secret or release intracellular miR-133a-3p into the serum. Both our previous studies and other research have discovered the inconsistent miRNA expression between tissue samples and blood samples.[9,46] This phenomenon could possibly be explained by the existence of miRNA-releasing or -capturing mechanisms in the cells and the blood.
Exosomes, secreted by most types of cells including cancer cells, are 40 to 100 nm nano-sized vesicles that protect miRNAs against degradation by ribonuclease.[47] Therefore, this study further investigated the expression of the candidate miRNAs in exosomes to identify its potential form in the extracellular environment and seek the diagnostic and therapeutic value in clinical practice. MiR-10b-5p showed markedly higher expression in serum exosomes of LA patients than the NCs. Liu et al[48] have reported that elevated expression of exosomal miR-10b-5p was independently associated with poor overall survival in NSCLC. The other two up-regulated serum exosomal miRNAs have not been explored in lung cancer and further investigations are urgently needed. Previous studies have reported that circulating miRNAs are not only co-fractionated with exosomes but also co-purified with the Ago2 ribonucleoprotein complex. Thus, we assumed that those miRNAs might bind to proteins in serum rather than exosomes molecules. The mechanisms of this phenomenon need further research.
EGFR is a crucial growth factor receptor and a critical target of lung cancer treatment.[49] As an important member of the receptor tyrosine 53 kinase family, it could participate in NSCLC progression by regulating various biological processes, including proliferation, apoptosis, and differentiation.[50] NSCLC patients with EGFR mutations could benefit from the tyrosine kinase inhibitor (EGFR-TKIs) therapy. Many biopsy materials can be used to detect the EGFR status including tumor tissues acquired from surgery or biopsy and cell-free tumor DNA. Besides, certain serum miRNAs have been reported to correlate with the EGFR signaling pathway, such as miR-21 and miR-10b.[16] miR-10b is highly expressed in several progressive diseases rather than complete remission or stable disease.[16] In this study, we also discovered that miR-10b-5p is up-regulated in EGFR mutation cases compared with NCs. These results indicated that circulating miRNA expression differed between healthy individuals and patients with positive or negative EGFR mutation status. Further study will continue to explore the potential role of miR-10b-5p in monitoring EGFR-TKI treatment effectiveness and its prognostic value for LA patients.
Several limitations should be considered in this study. First, only two types of EGFR mutation (exon 21 mutation and exon 19 deletions) were recorded in our study, other sensitizing mutations such as T790M mutation and ALK rearrangement should also be included in further study to acquire a comprehensive understanding. Second, our study was conducted only in Chinese populations, but different ethnicities might act differently in treatment response. Third, the sample size was relatively small. Further prospective clinical trials containing larger cohorts with diverse populations are warranted. Last but not least, the relationship between tissue miRNA expression and EGFR status was not identified. Future studies focusing on the association between miRNA expression and EGFR status are needed.
In conclusion, our study identified a four-miRNA signature in serum that could distinguish LA patients from healthy controls. Our study may provide a basis for the application of the circulating miRNAs as reliable biomarkers for the LA diagnosis. More efforts were needed to explore the functions of these identified miRNAs in LA.
Funding
The work was supported by grants from the National Natural Science Foundation of China (No. 81672400, and No. 81672788), Natural Science Foundation of Jiangsu Provincial Department of Education (No. 17KJB320007), and CSCO-Hausoh Cancer Research Foundation (No. Y-HS2017-032).
Conflicts of interest
None.
Supplementary Material
Supplementary Material
Footnotes
How to cite this article: Shan X, Zhang L, Zhu DX, Zhou X, Zhang H, Liu QX, Tang JW, Wen W, Wang TS, Zhu W, Liu P. Serum microRNA expression profiling revealing potential diagnostic biomarkers for lung adenocarcinoma. Chin Med J 2020;133:2532–2542. doi: 10.1097/CM9.0000000000001100
Supplemental digital content is available for this article.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chang H, Liu YH, Wang LL, Wang J, Zhao ZH, Qu JF, et al. MiR-182 promotes cell proliferation by suppressing FBXW7 and FBXW11 in non-small cell lung cancer. Am J Transl Res 2018; 10:1131–1142. doi: 10.1186/s12920-019-0648-7. [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Cui X, Li Y, Zhang T, Li S. Upregulated expression of miR-421 is associated with poor prognosis in non-small-cell lung cancer. Cancer Manag Res 2018; 10:2627–2633. doi: 10.2147/CMAR.S167432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sestini S, Boeri M, Marchiano A, Silva M, Calareso G, Galeone C, et al. Lung cancer screening in high-risk subjects: early detection with LDCT and risk stratification using miRNA-based blood test (In Italian). Epidemiol Prev 2016; 40:42–50. doi: 10.19191/EP16.1S1.P042.029. [DOI] [PubMed] [Google Scholar]
- 5.Abtahi S, Malekzadeh M, Nikravan G, Ghaderi A. Measurement of lung cancer tumor markers in a glass wool company workers exposed to respirable synthetic vitreous fiber and dust. Int J Occup Environ Med 2018; 9:23–31. doi: 10.15171/ijoem.2018.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017; 8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu SR, Wu Q, Shi YQ. Recent advances of miRNAs in the development and clinical application of gastric cancer. Chin Med J 2020; 133:1856–1867. doi: 10.1097/CM9.0000000000000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li CY, Zhang WW, Xiang JL, Wang XH, Li J, Wang JL. Identification of microRNAs as novel biomarkers for esophageal squamous cell carcinoma: a study based on The Cancer Genome Atlas (TCGA) and bioinformatics. Chin Med J 2019; 132:2213–2222. doi: 10.1097/CM9.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalela R, Curull V, Enriquez C, Pijuan L, Bellosillo B, Gea J. Lung adenocarcinoma: from molecular basis to genome-guided therapy and immunotherapy. J Thorac Dis 2017; 9:2142–2158. doi: 10.21037/jtd.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toschi L, Rossi S, Finocchiaro G, Santoro A. Non-small cell lung cancer treatment (r)evolution: ten years of advances and more to come. Ecancermedicalscience 2017; 11:787.doi: 10.3332/ecancer.2017.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Ren S, Li X, Wang Y, Garfield D, Zhou S, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014; 83:146–153. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Tang D, Yao R, Wang M, Wang Y, Yao Y, et al. microRNA expression profiles associated with survival, disease progression, and response to gefitinib in completely resected non-small-cell lung cancer with EGFR mutation. Med Oncol 2013; 30:750.doi: 10.1007/s12032-013-0750-1. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Zhou X, Zhu J, Cheng W, Zhu W, Shu Y, et al. MiR-4728-3p could act as a marker of HER2 status. Cancer Biomark 2015; 15:807–814. doi: 10.3233/CBM-150524. [DOI] [PubMed] [Google Scholar]
- 18.Zhao DS, Chen Y, Jiang H, Lu JP, Zhang G, Geng J, et al. Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc Pathol 2013; 22:444–450. doi: 10.1016/j.carpath.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, et al. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett 2014; 588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013; 59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci 2012; 57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 23.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, et al. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer 2013; 109:2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahdipour M, van Tol HT, Stout TA, Roelen BA. Validating reference microRNAs for normalizing qRT-PCR data in bovine oocytes and preimplantation embryos. BMC Dev Biol 2015; 15:25.doi: 10.1186/s12861-015-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One 2013; 8:e83025.doi: 10.1371/journal.pone.0083025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan Y, et al. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer 2017; 114:6–11. doi: 10.1016/j.lungcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res 2014; 15:149.doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Li Y, Liu J, Fan Y, Li X, Dong M, et al. Expression levels of microRNA-145 and microRNA-10b are associated with metastasis in non-small cell lung cancer. Cancer Biol Ther 2016; 17:272–279. doi: 10.1080/15384047.2016.1139242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Li M, Zhang G, Pang Z. MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur J Med Res 2013; 18:41.doi: 10.1186/2047-783X-18-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Sun C, Wang S, He Q, Li D. microRNA miR-10b inhibition reduces cell proliferation and promotes apoptosis in non-small cell lung cancer (NSCLC) cells. Mol Biosyst 2015; 11:2051–2059. doi: 10.1039/c4mb00752b. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Xu L, Yang Z, Lu H, Hu D, Li W, et al. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin Transl Oncol 2015; 17:209–214. doi: 10.1007/s12094-014-1213-7. [DOI] [PubMed] [Google Scholar]
- 32.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015; 5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzaei H, Khataminfar S, Mohammadparast S, Sales SS, Maftouh M, Mohammadi M, et al. Circulating microRNAs as potential diagnostic biomarkers and therapeutic targets in gastric cancer: current status and future perspectives. Curr Med Chem 2016; 23:4135–4150. doi: 10.2174/0929867323666160818093854. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Liu J, Chen Y, Ma C, Li B, Hao T. Up-regulation of mir-10b predicate advanced clinicopathological features and liver metastasis in colorectal cancer. Cancer Med 2016; 5:2932–2941. doi: 10.1002/cam4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Wen W, Zhu J, Huang Z, Zhang L, Zhang H, et al. A six-microRNA signature in plasma was identified as a potential biomarker in diagnosis of esophageal squamous cell carcinoma. Oncotarget 2017; 8:34468–34480. doi: 10.18632/oncotarget.16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaedcke J, Grade M, Camps J, Sokilde R, Kaczkowski B, Schetter AJ, et al. The rectal cancer microRNAome--microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res 2012; 18:4919–4930. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornby RJ, Starkey Lewis P, Dear J, Goldring C, Park BK. MicroRNAs as potential circulating biomarkers of drug-induced liver injury: key current and future issues for translation to humans. Expert Rev Clin Pharmacol 2014; 7:349–362. doi: 10.1586/17512433.2014.904201. [DOI] [PubMed] [Google Scholar]
- 38.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer 2011; 128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 39.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012; 130:1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene 2017; 620:46–53. doi: 10.1016/j.gene.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009; 16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med 2015; 4:551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang LK, Hsiao TH, Hong TM, Chen HY, Kao SH, Wang WL, et al. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS One 2014; 9:e96765.doi: 10.1371/journal.pone.0096765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petriella D, De Summa S, Lacalamita R, Galetta D, Catino A, Logroscino AF, et al. miRNA profiling in serum and tissue samples to assess noninvasive biomarkers for NSCLC clinical outcome. Tumour Biol 2016; 37:5503–5513. doi: 10.1007/s13277-015-4391-1. [DOI] [PubMed] [Google Scholar]
- 45.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010; 5:e13515.doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 2012; 316:196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- 47.Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol 2018; 8:66.doi: 10.3389/fonc.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017; 8:13048–13058. doi: 10.18632/oncotarget.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun W, Yuan X, Tian Y, Wu H, Xu H, Hu G, et al. Non-invasive approaches to monitor EGFR-TKI treatment in non-small-cell lung cancer. J Hematol Oncol 2015; 8:95.doi: 10.1186/s13045-015-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F, Chan LW, Law HK, Cho WC, Tang P, Yu J, et al. Exploring microRNA-mediated alteration of EGFR signaling pathway in non-small cell lung cancer using an mRNA:miRNA regression model supported by target prediction databases. Genomics 2014; 104:504–511. doi: 10.1016/j.ygeno.2014.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


