To the Editor: Detection of replication-competent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the most reliable indicator of contagiousness.1 Although the duration of live-virus shedding is well-characterized in immunocompetent patients with coronavirus disease 19 (Covid-19), little is known about how long immunocompromised patients are contagious. Consequently, the Centers for Disease Control and Prevention (CDC) guidelines on transmission-based precautions for immunocompromised patients are based on limited data.2
In the current study, we used cell cultures to detect viable virus in serially collected respiratory samples (nasopharyngeal and sputum samples) obtained from 20 immunocompromised patients who had Covid-19 (Figure 1A). These patients included 18 recipients of hematopoietic stem-cell transplants or chimeric antigen receptor (CAR) T-cell therapy and 2 patients with lymphoma. Covid-19 was diagnosed between March 10 and April 20, 2020, with the use of a modified CDC nucleic acid amplification test. Live virus was isolated in Vero cells, and genetic variants were identified by whole-genome sequencing of nasopharyngeal and cultured specimens (see the Supplemental Methods section of the Supplementary Appendix, available with the full text of this letter at NEJM.org). The patients’ demographic characteristics, medical history, and clinical course of Covid-19 were abstracted from medical records (Table S1 in the Supplementary Appendix).
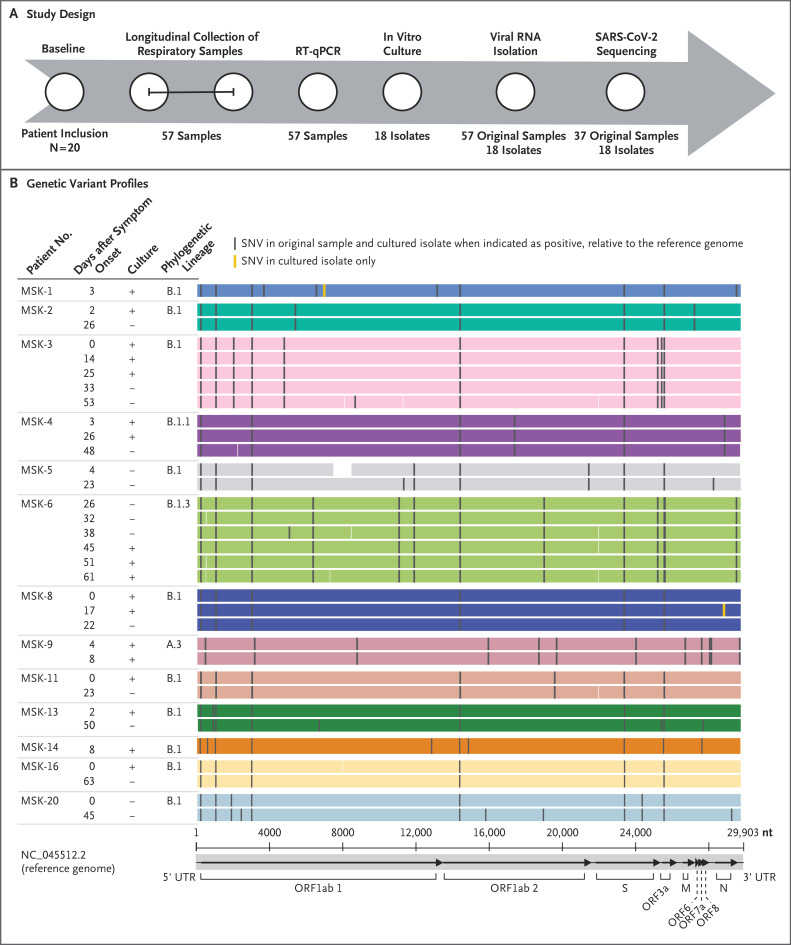
Figure 1. Study Design and Genetic Variant Profiles of Sequenced SARS-CoV-2.
Panel A shows the patient enrollment, respiratory sample collection (all nasopharyngeal samples except for one sputum sample), and testing design. The 𝙸 bar indicates serial collection of samples. RT-qPCR denotes quantitative reverse-transcriptase polymerase chain reaction. Panel B shows the genetic variant profiles of sequenced SARS-CoV-2 relative to the Wuhan-Hu-1 reference genome in 13 patients who had at least one cultured isolate (plus sign), more than one longitudinal sample sequenced, or both. The genomes shown were derived from the original respiratory samples. The mean (±SD) read depth was 2481±1558 reads per base. Assembled genomic regions are indicated by shaded areas colored according to patient, and gaps in coverage are indicated by white areas. Complete genome sequences from the 4 patients who did not have a corresponding culture or follow-up samples are not included. The replicase domains (ORF1ab 1 and 2, ORF3a, ORF6, ORF7a, and ORF8) and the S, M, and N regions of the reference genome are shown on the x axis. UTR denotes untranslated region.
Of the 20 patients, 15 were receiving active treatment or chemotherapy. Eleven had severe Covid-19. A total of 78 samples were collected from the 20 patients; 57 samples were obtained in the time periods shown in Figure S1. Viral RNA was detected for up to 78 days after the onset of symptoms (interquartile range, 24 to 64 days). Viable virus was detected in 10 of 14 nasopharyngeal samples (71%) that were available from the first day of laboratory testing. Follow-up samples obtained from 5 patients (Patients MSK-3, MSK-4, MSK-6, MSK-8, and MSK-9) grew virus in culture for 8, 17, 25, 26, and 61 days after the onset of symptoms (Figure 1). The 3 patients with viable virus for more than 20 days had received allogeneic hematopoietic stem-cell transplants (2 patients) or CAR T-cell therapy (1 patient) within the previous 6 months and remained seronegative for antibodies to viral nucleoprotein; 2 of these patients had severe Covid-19 and received investigational treatments.
Whole-genome sequencing detected viral reads in all the samples and yielded more than 95% complete SARS-CoV-2 genomes for 37 of 57 nasopharyngeal samples obtained from 17 patients and all 18 cultured specimens (accession numbers, EPI_ISL_583426 to EPI_ISL_583480 [55 complete genomes]). Serial sample genomes were obtained for 11 patients, up to day 63 after the onset of symptoms. Each patient was infected by a distinct virus, and there were no major changes in the consensus sequences of the original serial specimens or cultured isolates (Figure 1B); these findings were consistent with persistent infection.
Patients with profound immunosuppression after undergoing hematopoietic stem-cell transplantation or receiving cellular therapies may shed viable SARS-CoV-2 for at least 2 months. The current guidelines for Covid-19 isolation precautions may need to be revised for immunocompromised patients.
Supplementary Appendix
Disclosure Forms
The content of this letter is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This letter was published on December 1, 2020, at NEJM.org.
Footnotes
Supported by an award (P01 CA23766) and a National Institutes of Health (NIH)–National Cancer Institute Cancer Center Support Grant (P30 CA008748) from the NIH, a grant (to Drs. Hohl, Babady, and Kamboj) from the Jack and Dorothy Byrne Foundation, a contract from the National Institute of Allergy and Infectious Diseases (HHSN272201400008C) awarded to the Center for Research on Influenza Pathogenesis –(a Center of Excellence for Influenza Research and Surveillance), philanthropic donations from the JPB Foundation, a research grant (2020-215611 [5384]) from the Open Philanthropy Project, philanthropic donations (to Dr. García-Sastre) from Mount Sinai Philanthropy, awards (S10OD018522 and S10OD026880) from the NIH Office of Research Infrastructure Programs, and a Robin Chemers Neustein Postdoctoral Fellowship Award.(to Dr. Gonzalez-Reiche).
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 2020;382:2081-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Discontinuation of transmission-based precautions and disposition of patients with COVID-19 in healthcare settings (interim guidance). August 10, 2020. (https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.