Abstract
The HELLS (helicase, lymphoid specific, also known as lymphoid-specific helicase) protein is related to the SNF2 (sucrose non-fermentable 2) family of chromatin remodeling ATPases. It is required for efficient DNA methylation in mammals, particularly at heterochromatin-located repetitive sequences. In this study, we investigated the interaction of HELLS with chromatin and used an ATPase-deficient HELLS variant to address the role of ATP hydrolysis in this process. Chromatin fractionation experiments demonstrated that, in the absence of the ATPase activity, HELLS is retained at the nuclear matrix compartment, defined in part by lamin B1. Microscopy studies revealed a stronger association of the ATPase-deficient mutant with heterochromatin. These results were further supported by fluorescence recovery after photobleaching measurements, which showed that, at heterochromatic sites, wild-type HELLS is very dynamic, with a recovery half-time of 0.8 s and a mobile protein fraction of 61%. In contrast, the ATPase-deficient mutant displayed 4.5-s recovery half-time and a reduction in the mobile fraction to 30%. We also present evidence suggesting that, in addition to the ATPase activity, a functional H3K9me3 signaling pathway contributes to an efficient release of HELLS from pericentromeric chromatin. Overall, our results show that a functional ATPase activity is not required for the recruitment of HELLS to heterochromatin, but it is important for the release of the enzyme from these sites.
Keywords: chromatin remodeling, ATP hydrolysis, protein dynamics, fluorescence recovery after photobleaching, heterochromatin
Introduction
The genetic material of eukaryotic cells is organized into chromatin, which consists of genomic DNA, associated proteins and RNA molecules. The fundamental unit of this complex polymer is the nucleosome, consisting of 147 bp of DNA wrapped around two copies of each core histone H2A, H2B, H3 and H4. Nucleosomal building blocks are connected by about 20–50 bp of linker DNA [1]. Based on the extent of compaction, chromatin can be broadly divided into euchromatin and heterochromatin. Euchromatin has an open conformation, is gene rich and is favorable to active gene transcription. By contrast, heterochromatin is highly compacted and depleted of active genes [2]. Heterochromatin can be further subdivided into two main classes. While facultative heterochromatin is developmentally regulated and assembled when needed to permanently silence genes, constitute heterochromatin remains condensed throughout the organism’s lifespan and occurs at gene-poor areas [3]. A paradigm for the latter is the tightly condensed heterochromatin present at pericentromeres. Due to its high content of repeats and transposable elements, repression of these sequences is essential for the maintenance of genome integrity [4]. Gene silencing is achieved through repressive epigenetic modifications, the most prominent being the methylation of CpG dinucleotides by DNA methyltransferase enzymes (DNMTs) and tri-methylation of the H3 tail at lysine 9 (H3K9me3) by Suv39H (suppressor of variegation 3–9 homolog) enzymes [5,6]. A functional cross-talk between these two silencing pathways reinforces the stability of heterochromatic subdomains and thereby protects genome integrity [7].
Chromatin packaging is an important part of the regulatory network, as it creates inherent barriers for biological processes that utilize DNA as a template. Consequently, the chromatin structure must be dynamic to enable transcription, DNA replication and DNA repair [8]. The active mobilization of chromatin is performed by ATP-dependent chromatin remodeling enzymes [9–11]. These specialized motor proteins utilize the energy generated from ATP hydrolysis to reorganize, reposition, modify or evict nucleosomes from DNA. To date, four subfamilies of ATP-dependent chromatin remodeling enzymes have been identified: SWI/SNF (switch/sucrose non-fermentable), INO80 (inositol-requiring mutant 80), ISWI (imitation switch) and CHD (chromodomain helicase DNA binding) [9–11]. Each family is defined by a characteristic ATPase domain that is related to the DEAD/H (Asp-Glu-Ala-Asp box helicase) superfamily of DNA helicases and a set of unique domains that mediate binding to nucleosomes and individual complex subunits [8]. Specific histone tail modifications have been shown to recruit remodeling complexes, demonstrating a functional cooperation between these two epigenetic mechanisms [12,13].
HELLS [helicase, lymphoid specific, also known as lymphoid-specific helicase] is a putative chromatin remodeler belonging to the SWI/SNF subfamily that plays a central role at repetitive pericentromeric heterochromatin [14]. This SNF2-related protein was originally identified in T cell precursors, though its expression is ubiquitous and especially high in proliferating tissues [15,16]. The postnatal lethality of HELLS knockout (KO) in mice points to an important role of HELLS during mammalian development [17–19]. This is remarkable, since not all SNF2 homologs play an essential role, for example, neither targeted deletion of Brm nor Rad54 is lethal in mice [20,21]. Apart from multiple developmental defects, such as reduced embryonal growth, hematopoietic deficiencies and death of germ cells, HELLS KO mice also show a severely altered epigenetic landscape [18,22–24]. One of the most striking changes is a 50–70% reduction of global DNA methylation levels, particularly at pericentromeric repetitive sequences, including minor and major satellite repeats. This methylation loss is associated with a reactivation of endogenous retroviral elements, leading to abnormal mitosis with centrosome amplification and multipolar spindle formation [24–27]. A similar reduction in DNA methylation was observed in mutants of DDM1 (deficient in DNA methylation 1), a homolog of HELLS in Arabidopsis thaliana, indicating a conserved function of these proteins [28,29]. In addition to strong DNA hypomethylation, HELLS KO cells also displayed altered levels of H3K4me3, H3K27me3 and H2AK116 ubiquitylation at specific loci [27,30–32].
The importance of HELLS as an epigenetic player is further highlighted by previous studies that demonstrated an interaction of HELLS with the DNA methyltransferases DNMT3A and DNMT3B, as well as an indirect interaction via DNMT3B with DNMT1 and histone deacetylases HDAC1 and HDAC2 [33,34]. However, the precise molecular pathway through which HELLS influences the epigenetic landscape is not understood. With respect to DNA methylation, it was proposed that HELLS may promote the access of DNA methyltransferases to highly compacted chromatin [35,36]. This hypothesis was recently supported by a study showing that DDM1 is specifically required for DNA methylation at heterochromatic sequences that are rich in the linker histone H1 [37], suggesting that DDM1 is important for regions with tightly packaged nucleosomes. However, while nucleosomal remodeling activity has been reported for DDM1, an ATP-dependent chromatin remodeling by HELLS has not been demonstrated in vitro [37,38]. Therefore, it remains unclear, if and how a putative chromatin remodeling function of HELLS, together with its associated ATPase activity, contributes to this wide range of biological effects. According to its classification, HELLS contains the signature motifs of SNF2 enzymes, an ATP binding pocket encompassing Walker A and B motifs [39]. One conserved residue within this domain is lysine 237 (K237). The substitution of the corresponding residue with arginine has been shown to disable the ATPase activity of different SWI/SNF proteins [40–43]. Similar results were obtained for the HELLS K237Q variant [38].
In this study, we used the ATPase-deficient HELLS K237Q mutant to investigate whether ATP hydrolysis plays a role in the interaction between HELLS and chromatin. By employing a combination of biochemical fractionation and microscopy-based techniques, we show that the ATPase activity is not essential for the recruitment of HELLS to chromatin. However, this activity is important for the release of HELLS from highly compacted heterochromatin. We also present evidence suggesting that the release of HELLS from these sites requires a functional H3K9me3 pathway.
Results
Wild-type and ATPase-deficient HELLS show distinct nuclear extraction patterns
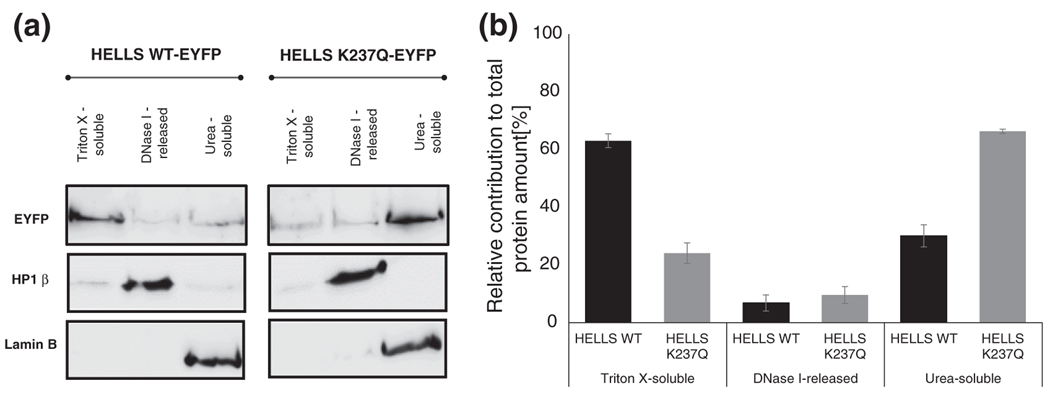
Members of the SNF2 family disrupt histone–DNA interactions and perform chromatin remodeling reactions in part via nucleosome sliding and by altering the accessibility of nucleosomal DNA [9]. In agreement with its classification as a SNF2 family member, HELLS has previously been shown to exclusively localize in the nuclear compartment [44]. We tested the hypothesis that the ATPase activity of HELLS plays a role in the recruitment of the protein to nuclear components. To this end, we have biochemically fractionated NIH 3T3 cells that had been transiently transfected with an YFP-tagged fusion of either wild-type HELLS or its K237Q variant, which has lost the ability to hydrolyze ATP [38]. In the first fractionation step, soluble proteins were extracted by incubating the cells with a high concentration of the detergent Triton X. This treatment leads to the release of many DNA-binding factors, such as MeCP2 and PCNA, in addition to cytoplasmic proteins [44,45]. Next, the Triton-X-resistant fraction was further eluted by digestion with the nuclease DNase I and high-salt washes. Under these conditions, tightly chromatin bound proteins, such as heterochromatic protein HP1β, are separated from the nuclear matrix. Finally, the nuclear matrix, containing scaffolding components, such as lamin B1 and matrix-associated proteins, like BRG1 and SWI/SNF complexes, was solubilized in urea [45]. The fractionation procedure has been established using NIH 3T3 cells transiently transfected with EYFP-NLS. The efficient extraction of this soluble protein predominantly in the TritonX fraction validated the method (Fig. S1a). As shown in Fig. 1a, we were also able to recapitulate the fractionation pattern of HP1β and lamin B1 [44,45]. The distribution of these two markers indicated that there was no cross-contamination between the different fractions. Interestingly, we detected striking fractionation differences between the wild-type HELLS protein and its ATPase-deficient K237Q mutant, particularly in the Triton X and urea-soluble fractions. Densitometric quantification of the Western blots revealed that 60% of the total wild-type HELLS protein was Triton X extractable, while only 25% of mutant HELLS was present in this fraction (Fig. 1b). By contrast, these proportions were inversed in the urea-soluble fraction, where 66% of the mutant HELLS but only 30% of wild type were detected (Fig. 1b). The differences in chromatin fractionation between the wild type and the mutant HELLS were highly reproducible. They were not due to different transfection yields or expression levels of the wild type and the mutant, as the overall signal was comparable between the wild type and the mutant (Fig. 1a). They were also not caused by variations in the fractionation procedure (Fig. S1b). Taken together, these data suggest that the ATPase activity of HELLS is involved in the release of the protein at the nuclear matrix compartment that is associated with lamin B1.
Fig. 1.
The wild-type and ATPase-deficient HELLS show distinct nuclear extraction patterns. (a) NIH 3T3 were biochemically fractionated 72 h post-transient transfection with EYFP-tagged fusions of either wild-type HELLS (HELLS WT) or the ATPase-deficient protein (HELLS K237Q). The cells were extracted stepwise with buffers that contain high concentrations of Triton X, DNase I and urea, as described in Materials and Methods. Equal volume protein extracts were prepared and analyzed by Western blotting using the indicated antibodies. A representative image of three biological repeats is shown. The samples for the wild-type and mutant HELLS were run on the same gel and Western blot; therefore, the overall signal can be taken as a control for similar expression levels of both variants. (b) Densitometric quantification of Western blot results shown in (a). For each protein variant, the signal obtained for the three fractions was summed up to 100% and the percentage of the protein variant in each fractionation was calculated accordingly. Error bars represent standard error of the mean for three independent biological replicates.
Wild-type and ATPase-deficient HELLS show distinct localization patterns in paraformaldehyde-fixed cells
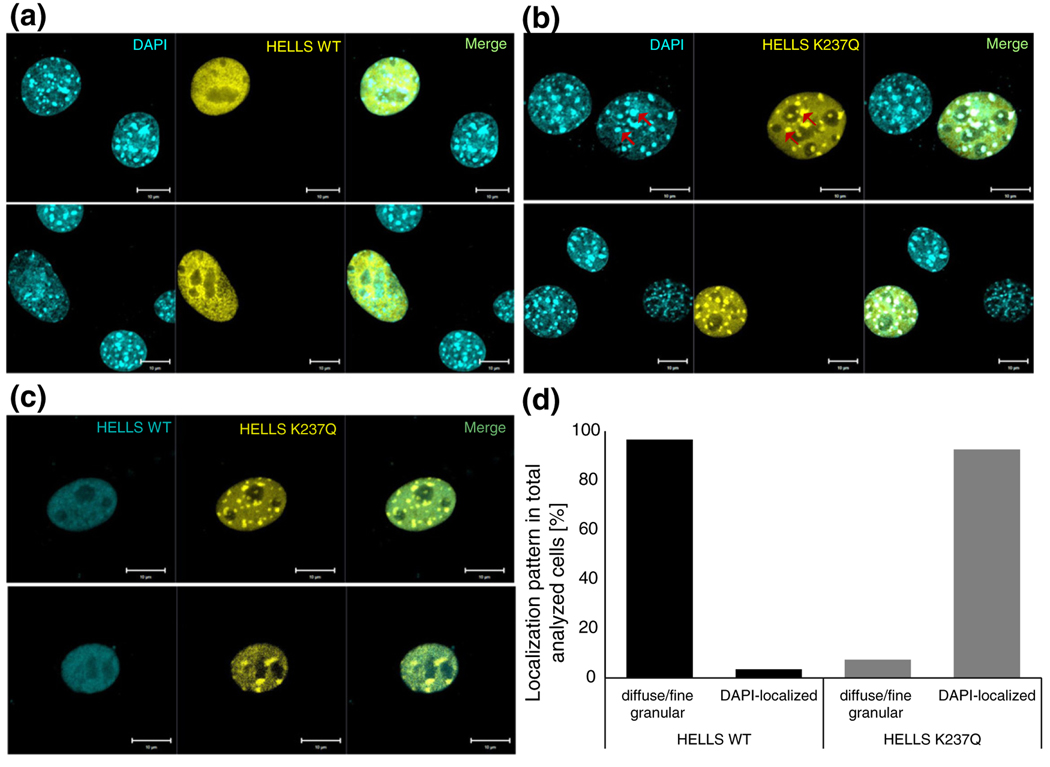
Prompted by the biochemical fractionation results that indicated significant differences between the wild-type and the mutant HELLS protein in their ability to associate with nuclear compartments, we next aimed to resolve these changes using microscopy-based techniques. As revealed by confocal microscopy analysis on fixed cells, both proteins were found exclusively in the nucleus; however, the wild type and the K237Q mutant showed distinct distribution patterns. While wild-type HELLS exhibited mostly a diffuse/fine granular pattern (Fig. 2a), the ATPase-deficient variant displayed a strong spotty localization at 4′,6-diamidino-2-phenylindole (DAPI)-dense regions (Fig. 2b). In murine cells, DAPI spots consist of pericentromeric heterochromatin rich in repetitive sequences [44,46]. The difference in localization was also evident after co-transfection of both wild-type and mutant HELLS (Fig. 2c). After quantification, we found that HELLS K237Q co-localized with DAPI-dense spots in 92% of the analyzed cells. By contrast, wild-type HELLS showed this pattern in only 4% of transfected cells, while the rest showed a diffuse/fine granular pattern (Fig. 2d). The relatively weak association of wild-type HELLS with heterochromatin was some-how unexpected, since previous analysis found a stronger accumulation of the HELLS to DAPI-dense spots [44,46]. Therefore, we applied different fixation protocols. Following the protocol of [44] that used 2% paraformaldehyde instead of 4% as in our previous experiment (Fig. 2), we detected an increased number of cells displaying spotty localization for the wild-type HELLS (Fig. S2a). Even under these conditions, the difference in localization between the wild type and the mutant HELLS was still clearly discernible, with the ATPase-deficient HELLS showing spots of stronger intensity (Fig. S2). Taken together, these results indicate that the ATPase deficiency affects the interaction of HELLS with chromatin, particularly at heterochromatic regions, and the fixation protocol influences the overall recovery of heterochromatically bound HELLS.
Fig. 2.
The wild-type and ATPase-deficient HELLS show distinct localization patterns in paraformaldehyde-fixed cells. (a) Representative confocal laser scanning microscopy images of DAPI-stained NIH 3T3 cells post-transient transfection with EYFP-fused wild-type HELLS. (b) Representative confocal laser scanning microscopy images of DAPI-stained NIH 3T3 cells post-transient transfection with EYFP-fused mutant HELLS. The red arrows indicate illustrative areas of co-localization between the protein and the DAPI-dense foci. (c) Representative confocal laser scanning microscopy images of NIH 3T3 cells post-transient co-transfection with ECFP-fused wild-type HELLS and EYFP-fused mutant HELLS. Scale bars correspond to 10 μm. (d) Quantification of localization patterns observed in wild-type or mutant HELLS transfected cells as illustratively shown in (a) and (b). We analyzed 29 and 27 cells for the wild-type and mutant HELLS transfected cells, respectively.
ATPase-deficient HELLS shows increased localization at pericentromeric heterochromatin in live cells
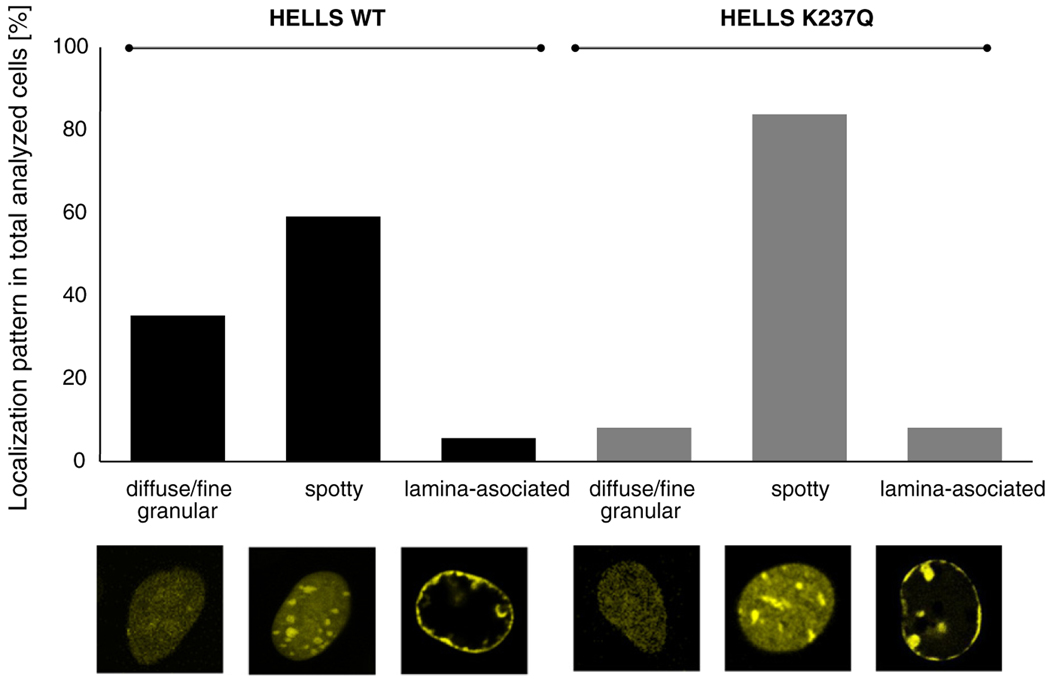
To circumvent the variability introduced by paraformaldehyde fixation, we decided to employ live cell imaging and compare the localization of wild-type and mutant HELLS in transiently transfected NIH 3T3 cells. As presented in Fig. 3, the localization of wild-type and mutant HELLS followed three distinct patterns: diffuse/fine granular, spotty and lamina associated. The distribution within these categories was very different for the two protein variants. While the majority of cells transfected with the HELLS K237Q mutant showed a clear spotty pattern (84% of the population), for the wild-type transfected cells, this type of localization was reduced to 59%. This difference is statistically highly significant because, assuming a Gaussian distribution, the large fraction of K237Q expressing cells showing a spotty localization has a probability of only 2.04 × 10−8 to occur by chance. DRAQ5 DNA staining confirmed that the observed spots represent pericentromeric heterochromatin foci (Fig. S3). In addition, we identified a novel (small) subpopulation of cells, where HELLS localized to the nuclear lamina. This localization was present in both wild-type and mutant HELLS transfected cells, with 6% and 8% of cells affected, respectively (Fig. 3). Collectively, live cell imaging revealed a different distribution in localization patterns between the wild-type HELLS and its K237Q mutant, with the ATPase-deficient variant showing increased localization to pericentromeric heterochromatin. This is in agreement with the results from fixed cells and biochemical fractionation as described above.
Fig. 3.
The ATPase-deficient HELLS shows increased localization at pericentromeric heterochromatin in live cells. Quantification of localization patterns observed in wild-type or mutant HELLS transfected cells. We analyzed 142 and 111 cells for the wild-type and mutant HELLS transfected cells, respectively. Representative examples of protein localization types are shown below the graph.
ATPase-deficient HELLS shows altered dynamics at pericentromeric heterochromatin repeats
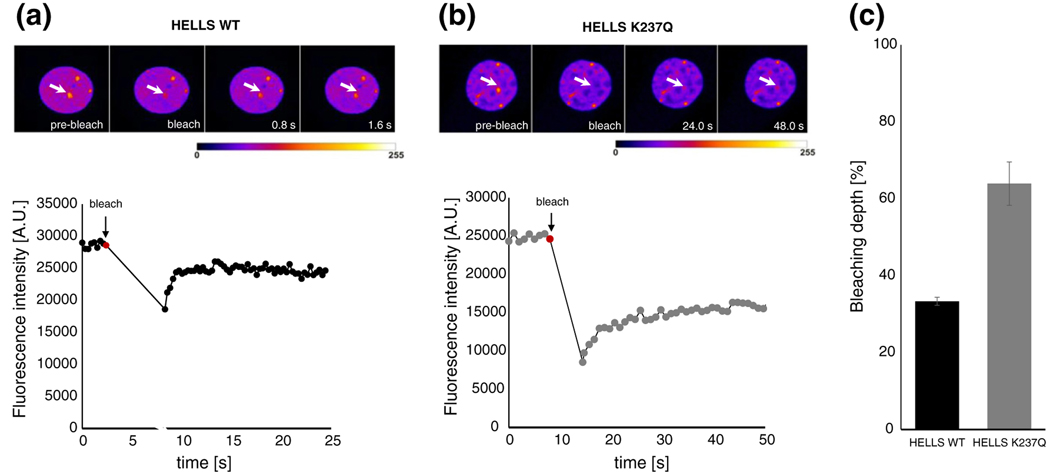
Based on the localization differences between the wild type and the mutant HELLS, we hypothesized that the mutation in the ATPase domain might influence the dynamics of HELLS association with chromatin. An altered interaction of HELLS with chromatin, for example, may then lead to a more efficient fixation at pericentromeric heterochromatin. To assay the chromatin association dynamics, we conducted fluorescence recovery after photobleaching (FRAP) measurements. To this end, we bleached an area with pronounced HELLS accumulation corresponding to one of the heterochromatic spots. Then, we measured the efficiency of bleaching and followed the recovery of fluorescence. Representative images are shown in Fig. 4 and Fig. S4 and in Movies S1 and S2. Upon bleaching of the wild-type HELLS, we consistently observed that the fluorescence signal was only reduced by about 34% (Fig. 4a and c). This was accompanied by a general decrease in the fluorescence intensity of the whole nucleus (compare the changes that occur upon bleaching in the fluorescence of the nucleoplasm in Fig. 4a and Fig. S4a or Movie S1). By contrast, under identical experimental settings, HELLS K237Q mutant could be bleached by 64% on average (Fig. 4b and c and Fig. S4b or Movie S2). In this case, the overall fluorescence in the regions neighboring the bleached area was hardly reduced. It is expected that a more dynamic protein exchanges faster with the unbleached nucleoplasm, resulting in a lower bleaching depth at the bleached site and a global decrease in the overall nuclear fluorescence. By contrast, a more tightly chromatin associated protein, with a low exchange rate, displays a high bleaching depth and an unaffected overall nuclear fluorescence. Hence, our results consistently hint toward a decrease in the mobility of the HELLS K237Q mutant compared to the wild-type protein.
Fig. 4.
The wild-type and ATPase-deficient HELLS show different bleaching behaviors at pericentromeric heterochromatin. Representative time frames of exemplary FRAP series (pseudocolored) for the wild-type (a) and mutant HELLS (b). The arrows denote the circular area that was bleached and where the fluorescence intensity was monitored over time. Images were taken at the indicated time points after the bleach pulse. In the lower half of the graph, the fluorescence intensity of the measured area was plotted as a function of time. Every point of the curve represents data stemming from single images recorded with a period of 0.3 and 1.0 s for wild-type and mutant transfected cells, respectively. The bleaching start point was correspondingly annotated (red point). (c) The average bleaching depth observed for wild-type and mutant HELLS transfected cells. Error bars represent standard error of the mean, based on measurements performed on 9 cells for wild-type HELLS and on 11 cells for the mutant protein.
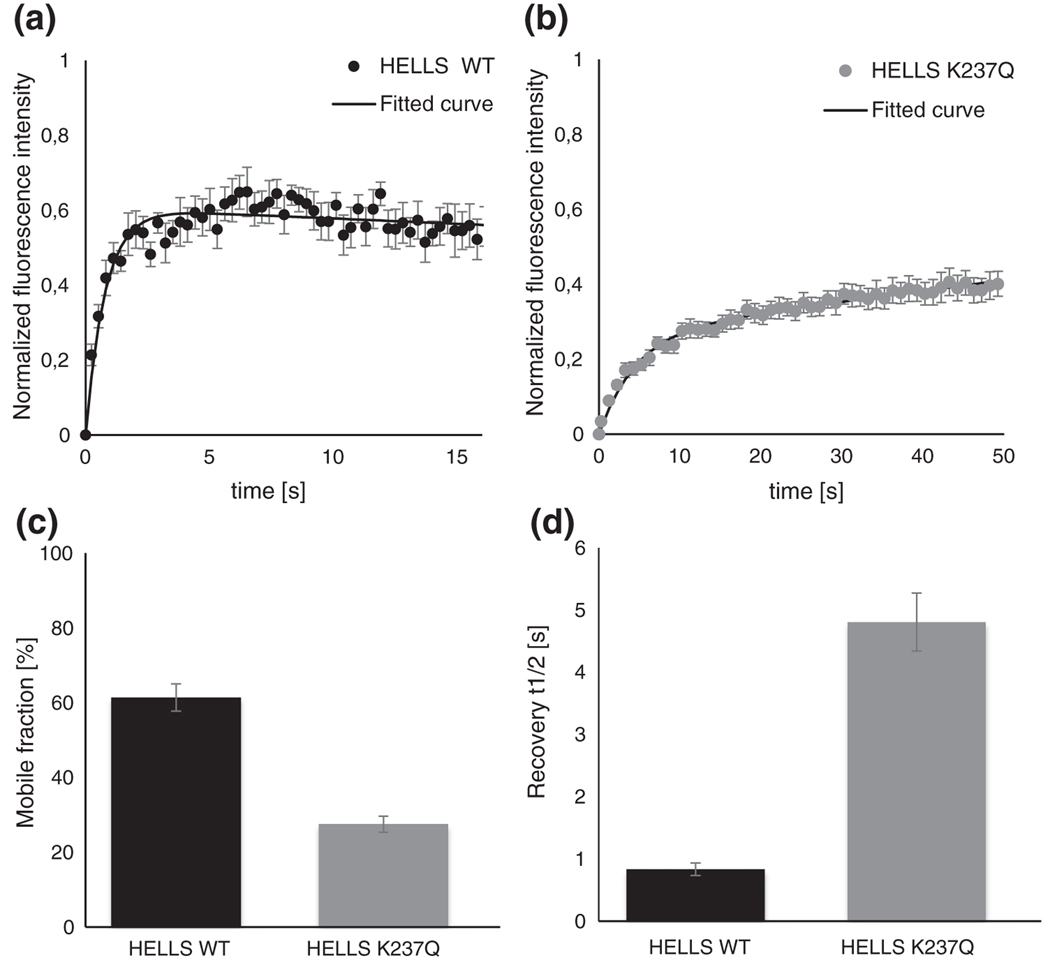
Next, we determined the half-fluorescence recovery half-time (t1/2) and the mobile fraction percentage for both wild-type and mutant HELLS. As shown in Fig. 5a and b, the fluorescence recovery curves for the wild-type HELLS reached a plateau faster compared to the curves generated with the HELLS K237Q mutant. Additionally, the final steady state of the fluorescence was higher for the wild-type HELLS, further emphasizing differences in the dynamic behavior of the two proteins. The analysis of the FRAP recovery curves using a two-component exponential fit model revealed a 2-fold increase in the mobile fraction for the wild-type HELLS compared to its ATPase-deficient mutant (61% versus 28%) (Fig. 5c). The recovery time required to reach half of the fluorescence also showed striking differences between the two protein variants. While the wild-type HELLS displayed a t1/2 of 0.8 s, the HELLS K237Q mutant showed an over 5-fold increase in t1/2 to 4.5 s (Fig. 5d). Taken together, these results demonstrate that the inability of HELLS to hydrolyze ATP leads to a trapping of the protein at pericentromeric heterochromatin repeats, indicating that ATP hydrolysis is necessary for HELLS exchange at these sites.
Fig. 5.
The wild-type and ATPase-deficient HELLS show different dynamics at pericentromeric heterochromatin. Mean FRAP recovery curves for EYFP-tagged wild-type (a) and mutant HELLS (b). The intensity of the bleached area was normalized to 0 for the first image recorded after bleaching. Every point of the curve represents data stemming from single images recorded with a period of 0.3 and 1.0 s for wild-type and mutant transfected cells, respectively. Individually fit curves were used to derive the percentage of mobile protein fraction (c) and the half-recovery time (d). Error bars represent standard error of the mean based on measurements performed on 9 cells for wild-type HELLS and on 11 cells for the mutant protein.
Changes in HELLS localization follow disruption of the H3K9me3 pathway
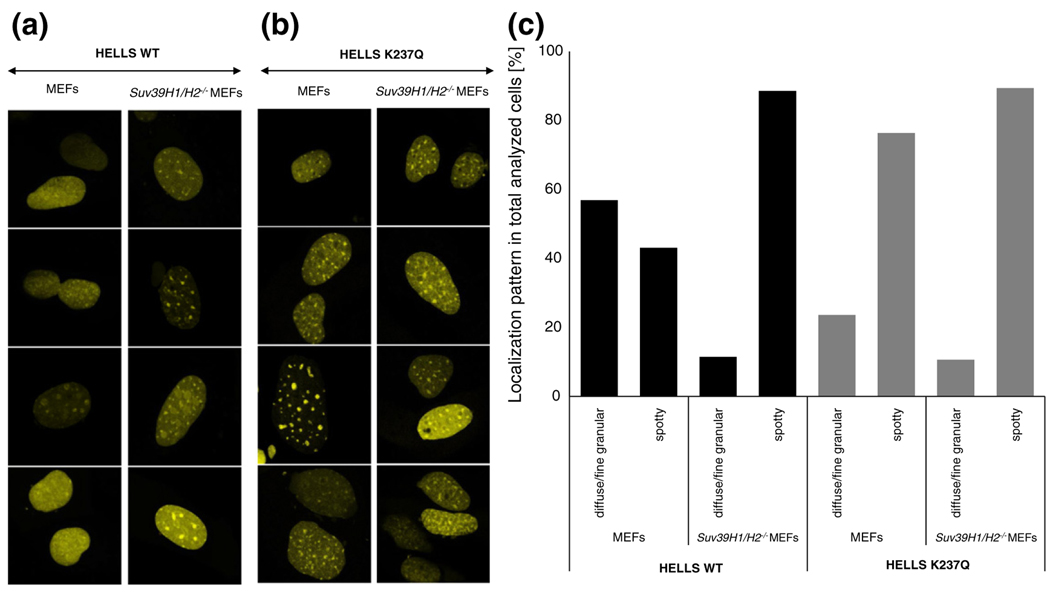
Taking into account the important role that HELLS plays in the epigenetic control of pericentromeric chromatin repeats, we next asked whether the localization or binding dynamics of HELLS is also regulated by specific chromatin modifications. Since pericentromeric heterochromatin repeats are enriched in tri-methylated H3K9 [7,47], we examined the localization of HELLS in Suv39H1/H2 double-KO mouse embryonic fibroblasts (MEFs). Notably, although these cells lack pericentromeric H3K9me3, they still contain DAPI-dense foci [48]. We observed that HELLS was still able to localize to pericentromeric satellite repeats in these cells, indicating that H3K9me3 pathway is not required for the recruitment of HELLS to these sites (Fig. 6a and b). Intriguingly, live cell imaging revealed a strong increase in the wild-type HELLS localization at pericentromeric chromatin in the Suv39H1/H2 double-KO MEFs in comparison to the wild-type MEFs (Fig. 6a). We observed that, in 43% of all counted wild-type cells, HELLS showed a spotty localization (Fig. 6c) similar to what we previously observed in NIH 3T3 cells. By contrast, this percentage increased to 89% for the Suv39H1/H2 double-KO cells (Fig. 6a and c). We did not observe major localization differences between the two chromatin environments for the ATPase-deficient HELLS (Fig. 6b and c). These results suggest that, while HELLS is recruited to its target sites independently of the H3K9me3, its release from pericentromeric heterochromatin requires an intact H3K9me3 pathway comprising the H3K9me3 mark but perhaps also associated factors and downstream modifications.
Fig. 6.
Changes in HELLS localization follow disruption of the H3K9me3 pathway. Representative confocal laser scanning microscopy images of live wild-type and Suv39H1/H2−/− MEFs post-transient transfection with either wild-type HELLS (a) or HELLS K237Q (b). (c) Quantification of localization patterns observed in wild-type or mutant HELLS transfected cells as representatively shown in (a) and (b). We analyzed 50–60 cells per cell type for every construct.
Discussion
Based on the high level of sequence homology of HELLS to other SNF2 family members that were experimentally shown to perform chromatin remodeling, it was proposed that HELLS may similarly alter chromatin structure in vivo [18]. Members of the SNF2 family use the energy derived from ATP hydrolysis to disrupt histone–DNA interactions and slide nucleosomes, thereby making nucleosomal DNA accessible to transcription factors [11]. Even though HELLS has been discovered almost two decades ago [49], any direct evidence for a chromatin remodeling ability of this protein is still missing. In this study, we investigated the interaction of HELLS with chromatin and used the ATPase-deficient HELLS variant K237Q to address the role of ATP hydrolysis in this process.
Localization of the wild-type HELLS differs between live and paraformaldehyde-fixed cells
We investigated the subnuclear localization of HELLS in live and fixed cells and observed accumulation of the protein at DAPI-dense pericentromeric heterochromatic foci, which contain sequences rich in repetitive and retrotransposable elements. The role of HELLS as an epigenetic silencer of these sequences, largely via collaboration with de novo DNA methyltransferases, is well documented [14,33,50–52], and accumulation of HELLS at these sites has been previously reported [44]. Surprisingly, we observed a discrepancy in the localization of HELLS in fixed cells compared to live cells. While 59% of HELLS was recruited to the heterochromatic foci in live cells, this percentage was decreased to only 4% after paraformaldehyde fixation. Notably, similar localization differences of the heterochromatin-bound MeCP2 proteins between live and fixed cells imaging have been documented by Schmiedeberg et al. [53]. Using a series of mutants of the MeCP2 protein, the authors reported that proteins residing on chromatin for less than 5 s could not be efficiently captured by paraformaldehyde cross-link chemistry. Therefore, our results suggest that HELLS is highly mobile and not tightly associated with pericentromeric chromatin. Indeed, our FRAP measurements confirmed that HELLS exchanges rapidly between the bound state and the unbound state at pericentromeric heterochromatin with a residence time of 0.8 s. It should be noted that, due to the 300-ms time resolution imposed by our experimental setup, the recovery time of the wild-type HELLS may be overestimated, and the value of 0.8 s should be taken as an upper limit. Quantitative measurements of the mobility parameters for chromatin remodeling proteins are still sparse in literature. However, the value obtained for HELLS here is in the range of what has been previously reported for other chromatin remodelers, for example, for the human ISWI family members, SNF2H/SNF2L and Acf1 [54].
ATPase-deficient HELLS shows altered dynamics at pericentromeric heterochromatin
To assess the significance of ATP hydrolysis in the interaction of HELLS with chromatin, we investigated the localization and dynamics of the ATPase-deficient HELLS variant K237Q. Similarly to the wild-type HELLS, we observed an accumulation of the mutant protein at DAPI-dense pericentromeric heterochromatic foci, suggesting that the ATPase activity is not necessary for the recruitment of HELLS to chromatin. Interestingly, in contrast to the wild-type HELLS, the ATPase-deficient mutant did not show significant localization differences between fixed and live cells. The fact that the two protein variants show different degrees of sensitivity to paraformaldehyde fixation suggests that the absence of a functional ATPase domain impairs the dynamics of HELLS at pericentromeric heterochromatin foci. Indeed, in FRAP assays, the HELLS K237Q mutant displayed a 5-fold lower recovery time than its wild-type counterpart. This was accompanied by a concomitant 2-fold decrease in the fraction of mobile protein. The increased residence time translates into a higher cross-linking efficiency of the ATPase-deficient HELLS compared to the wild-type protein and, consequently, leads to the discrepancies observed in the localization of the two proteins in fixed cells. Furthermore, a decreased exchange rate culminates in a higher accumulation of the ATPase-deficient HELLS at pericentromeric heterochromatin sites in live cells by comparison to the wild-type protein (84% versus 59%).
The notion that HELLS becomes more tightly associated with chromatin in the absence of ATPase activity was also supported by cellular fractionation experiments. While the wild-type protein was readily extractable with detergent, the majority of the ATPase-deficient mutant remained associated with the nuclear matrix, which includes lamin B1. The results obtained for the wild-type HELLS are in agreement with previous reports, where HELLS was predominantly found in Triton-X-extractable fraction in confluent NIH 3T3 cultures [44]. Together with cellular studies, these data indicate that its high mobility and intrinsic loose chromatin binding make wild-type HELLS more accessible to detergent, thus resulting in the higher protein extractability. In contrast, due to its impaired exchange rate at compacted chromatin, the ATPase-deficient mutant is shielded from both detergent extraction and DNase I activity. Interestingly, in the microscopy studies, we were able to directly visualize a small subpopulation of cells where both HELLS variants are associated with the nuclear periphery. The lamin B1 protein belongs to the nuclear matrix and is located close to the inner nuclear membrane [55]. Recently, genome-wide studies identified genomic sequences interacting with lamin B1, termed the lamina-associated domains [56]. These are large (0.1–10 Mb) silent domains, enriched in repetitive sequences and containing few genes [57]. Noteworthy, HELLS is important for the establishment of DNA methylation over large genomic regions that overlap to a high extent with lamina-associated domains [52]. Indeed, major satellite repeats and other types of repetitive elements are among the sequences with the greatest loss in DNA methylation upon HELLS deletion [52]. Taking this into account, our results indicate that, in the absence of a functional ATPase domain, the release of HELLS from these highly compacted genomic regions is severely impaired. A role for the ATP-dependent release of remodelers from chromatin was also documented for Rad54, another SWI2/SNF2 protein family member [58]. The ATPase-deficient Rad54 protein displayed a 2-fold lower recovery time and a 10% increase in the fraction of immobile protein when compared to its wild-type counterpart [58].
A functional H3K9me3 pathway contributes to the release of HELLS from pericentromeric heterochromatin
Chromatin structure may play a role in the recruitment of HELLS to pericentromeric regions as well. For example, prolonged treatment with histone deacetylase inhibitors disrupts higher-order heterochromatin organization, and this is accompanied by dissociation of HELLS from pericentromeric heterochromatin [44]. However, the specific chromatin signal leading to HELLS localization has not been identified. Since pericentromeric chromatin is highly enriched in the H3K9me3 repressive modification [7,47], we tested whether the H3K9me3 pathway could be involved in the recruitment of HELLS. We found that HELLS localized to pericentromeric regions in the absence of H3K9me3, demonstrating that this histone modification and its associated chromatin environment is not involved in the recruitment of HELLS. However, we observed a 2-fold increase in the wild-type HELLS localization at pericentromeric heterochromatin in Suv39H1/H2 double-KO MEFs compared to wild-type MEFs, indicating that release of HELLS from chromatin is impaired in the absence of H3K9me3. By contrast, the ATPase-deficient HELLS mutant showed strong association with pericentromeric heterochromatin with or without H3K9me3 modification. Importantly, our results are supported by a recent publication showing, with the use of mass spectrometry, that HELLS is enriched at pericentromeric heterochromatin in Suv39H1/H2 double-KO mouse embryonic stem cells [59]. Together, our data suggest that H3K9me3-associated chromatin environment is not critical for the initial recruitment of HELLS to pericentromeric satellite repeats, but it may play a role in the release of HELLS from these sites. Further studies are needed to uncover whether the H3K9me3 mark directly, any component associated with it (like HP1 for example) or any of its downstream modifications (like for example H4K20me3) is responsible for this effect.
Conclusions
Collectively, the data presented in this study show for the first time the importance of a functional ATPase activity and an intact H3K9me3 pathway for the interaction between HELLS and chromatin. We find that ATP hydrolysis is not required for the recruitment of HELLS to chromatin but is an important factor for HELLS release from compacted loci. While ATP hydrolysis might play a role in the release of HELLS from other sites as well, we anticipate that the enzymatic function of the protein is particularly important at highly condensed loci. The fast exchange of HELLS at these repetitive sequences might enhance the local recruitment of other epigenetic enzymes, such as HDACs and DNMTs, and subsequently locally stabilize silencing complexes. This hypothesis was very recently supported by a study showing that the remolding activity of HELLS is in part required for de novo methylation of repeats and for promoting stable association of DNMT3b with these sequences [60]. The existence of an ATP dependent cycle of HELLS binding and release from heterochromatin may point toward an ATP-dependent remodeling activity of this enzyme. Since our experiments were performed under overexpression conditions, they need to be interpreted with caution. Further studies will be required to refine the importance of ATP hydrolysis and dynamics in the function of endogenous HELLS.
Materials and Methods
Tissue culture and transfections
NIH 3T3 cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium and high glucose (Sigma), supplemented with 10% heat-inactivated calf serum and 2 mM L-glutamine (Sigma), at 37 °C in a saturated humidity atmosphere containing 5% CO2. Transient transfections with Fugene HD (Promega) were performed on cells showing 60–70% confluence, according to the manufacturer’s recommendations. Transiently transfected cells were cultured for 24–72 h and HELLS expression was examined by fluorescence microscopy or Western blot analysis. Wild-type and Suv39H1/H2 double-KO MEFs (a kind gift of Dr. T. Jenuwein) were grown at 37 °C in a humidified atmosphere, 5% CO2 using Dulbecco’s modified Eagle’s medium and high glucose supplemented with 10% heat-inactivated calf serum, 1 × non-essential amino acids (Gibco), 1 × sodium pyruvate (Sigma), 0.1 mM β-mercaptoethanol (Gibco) and 2 mM L-glutamine. Transient transfections with jetPRIME (peqlab) were performed on cells showing 50–60% confluence, according to the manufacturer’s recommendations. Transiently transfected cells were cultured for 24–48 h before fluorescence microscopy.
Vector construction
The sequence encoding mouse HELLS was subcloned from pGeneLSHF [44] as an N-terminal fusion into pEYFP-C1 and pECFP-C1 vectors (Clontech). The K237Q mutant was created using a megaprimer mutagenesis protocol [61]. The introduction of the mutation and the integrity of the remaining sequence were verified by sequencing.
Western blot analysis and protein fractionation
Cell extraction with Triton X-100 (Triton X), chromatin fractionation and nuclear matrix isolation were performed according to a protocol described previously [44,45,62]. Briefly, 72 h post-transfection, 1.5 × 106 cells were subjected to two washes with PBS (phosphate-buffered saline) containing Ca2+ and Mg2 +, followed by incubation on ice for 3 min with 98 μL cytoskeleton buffer [CSK; 10 mM Pipes (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and 1 mM ethylene glycol bis(β-aminoethyl ether) N,N′-tetraacetic acid] supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol and 0.5% (vol/vol) Triton X. The Triton-X-resistant nuclear skeleton was separated by centrifugation at 5000g for 3 min at 4 °C. To release chromatin associated proteins, we digested the pellet for 15 min at 37 °C with 20 μL RNase-free DNase I (1 U/μL) in 58 μL CSK buffer with NaCl adjusted to 50 mM. To elute the digested chromatin, we added 20 μL ammonium sulfate directly from a stock of 2 M to a final concentration of 0.25 M. Following a 5-min incubation at room temperature, we isolated the chromatin fraction by centrifugation at 5000g for 3 min at 4 °C. The remaining nuclear matrix pellet was then solubilized in 50 μL of 8 M urea and equilibrated with 50 μL of SDS-PAGE running buffer. Protease inhibitors were present throughout the fractionation procedure (protease inhibitory cocktail, 50×; Promega).
Equal volumes of protein extracts were then separated on a 15% SDS-PAGE by electrophoresis and transferred onto a nitrocellulose membrane. The samples for the wild-type and mutant HELLS were run on the same gel. For detection, the following primary antibodies were used: EYFP (Clontech, 632592), Lamin B (Santa Cruz, sc-6217) and HP1β (Active Motif, 39979). Detection of horseradish-peroxidase-coupled antibodies was performed by enhanced chemiluminescence with Western Lightning Plus ECL reagent (PerkinElmer) according to the manufacturer’s instructions. The resulting signal was visualized on the FUSION Solo (peqlab) system and quantified with the FUSION-CAPT Advance Solo 4 software. The values obtained for the HELLS signal from Triton-X-soluble, DNase-I-released and urea-soluble fractions were summed up to a total of 100%, and the contribution of each fraction to the total amount was calculated accordingly. The similar fractionation and loading of the wild type and K237Q HELLS transfected cells has been further verified by the quantification of the signals obtained for the reference markers HP1β and Lamin B between the two cell populations.
Preparation and imaging of fixed cells
Cells were seeded on glass coverslips to 60–70% confluence. Unless stated otherwise, 24 h post-transfection, the cells were fixed with 4% paraformaldehyde solution prepared in PBS with Ca2+ and Mg2+, for 10 min at room temperature. For 2% paraformaldehyde solution fixation, the original 4% stock was diluted in PBS shortly before fixation. Prior to DAPI staining, the cells were permeabilized for 5 min at 4 °C with 0.5% Triton X solution. The Mowiol-mounted coverslips were then imaged on a LSM 710 Zeiss confocal microscope. Very strongly overexpressing cells (~5% of the population) were excluded from statistical analysis.
Live cell imaging and FRAP
Cells were seeded on 35-mm Fluorodish cell culture dishes (World Precision Instruments) to 60–70% confluence. At 24–48 h after transfection, the growth media was replaced with imaging media containing no phenol red. Live cell imaging was performed on a LSM 710 Zeiss confocal microscope equipped with a Plan-Apochromat 63 ×/1.40 Oil DIC M27 objective and an XL-LSM 710 S1 incubation chamber for temperature and CO2 control. ECFP imaging was performed with an attenuated 405-nm diode laser, to minimize cytotoxicity. For nuclear staining, DRAQ5 (Thermo Scientific) was added to 5 μM and allowed to incubate for 10 min at 37 °C before confocal imaging.
For FRAP experiments, 256 × 256, 16-bit images were recorded with a pixel size of 0.1 μm. The workflow included the acquisition of 9 pre-bleach images, of 5 bleaching cycles with 100% laser intensity and of up to 100 post-bleach images. To prevent unwanted fluorophore bleaching by imaging, we tuned the 25.0-mW, 514-nm laser line to 4% during acquisition. For bleaching, a circular region of interest with a diameter of 1 μm was chosen. This was sufficient to cover the pericentromeric heterochromatin structures without major extensions in the nucleoplasm. To accommodate for differences in protein mobility, for wild-type HELLS, we recorded images at an interval of 300 ms. We note that this limit was set by the 242-ms time interval needed for image acquisition. For the HELLS K237Q variant, the image recording time was increased to 1 s.
For analysis, the mean pixel intensity values for the bleached region of interest and the whole cell were extracted as a time series with the MeasureStack macro from ImageJ. Only time points showing no movement of the cellular structures were included in the analysis. Fluorescence values were normalized for fluctuations in laser intensity, by dividing against the average mean fluorescence intensity of the 9 pre-bleached images. Compensation for bleaching by imaging was incorporated by scaling the fluorescence mean intensity values within the bleached area with the ratio of fluorescence decrease observed for the whole cell, during post-bleaching acquisition. The normalized time series were next fitted with the Solver module in Microsoft Excel to a double-component exponential curve by non-linear least squares minimization. We analyzed 9 cells for the wild type and 11 cells for the mutant protein. Average values and standard deviations of the half-recovery time t1/2 and mobile fraction percentage were determined by individual fitting of each FRAP curve.
Supplementary Material
Acknowledgements
C.L. and R.Z.J. have been financed by fellowships from the Carl Zeiss foundation. K.M. has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- FRAP
fluorescence recovery after photobleaching
- KO
knockout
- MEF
mouse embryonic fibroblast
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2015.03.014.
References
- [1].van Steensel B Chromatin: constructing the big picture. EMBO J 2011;30:1885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature 2007;447:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Craig JM. Heterochromatin—many flavours, common themes. BioEssays 2005;27:17–28. [DOI] [PubMed] [Google Scholar]
- [4].Almouzni G, Probst AV. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus 2011;2:332–8. [DOI] [PubMed] [Google Scholar]
- [5].Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012;13:343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci 2014;39:310–8. [DOI] [PubMed] [Google Scholar]
- [7].Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003;13:1192–200. [DOI] [PubMed] [Google Scholar]
- [8].Swygert SG, Peterson CL. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta 2014;1839:728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem 2002;71:247–73. [DOI] [PubMed] [Google Scholar]
- [10].Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 2003;25: 1192–200. [DOI] [PubMed] [Google Scholar]
- [11].Hopfner KP, Gerhold CB, Lakomek K, Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr Opin Struct Biol 2012;22:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Duan MR, Smerdon MJ. Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin). J Biol Chem 2014;289: 8353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006;442: 86–90. [DOI] [PubMed] [Google Scholar]
- [14].Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res 2004;32:5019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Geiman TM, Durum SK, Muegge K. Characterization of gene expression, genomic structure, and chromosomal localization of Hells (Lsh). Genomics 1998;54:477–83. [DOI] [PubMed] [Google Scholar]
- [16].Raabe EH, Abdurrahman L, Behbehani G, Arceci RJ. An SNF2 factor involved in mammalian development and cellular proliferation. Dev Dyn 2001;221:92–105. [DOI] [PubMed] [Google Scholar]
- [17].Lee DW, Zhang K, Ning ZQ, Raabe EH, Tintner S, Wieland R, et al. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res 2000; 60:3612–22. [PubMed] [Google Scholar]
- [18].Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta 2001; 1526:211–20. [DOI] [PubMed] [Google Scholar]
- [19].Briones V, Muegge K. The ghosts in the machine: DNA methylation and the mystery of differentiation. Biochim Biophys Acta 2012;1819:757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, et al. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 1997;89:195–204. [DOI] [PubMed] [Google Scholar]
- [21].Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J 1998;17: 6979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Geiman TM, Muegge K. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci USA 2000;97:4772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev 2004;18:1035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fan T, Yan Q, Huang J, Austin S, Cho E, Ferris D, et al. Lshdeficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res 2003;63: 4677–83. [PubMed] [Google Scholar]
- [25].Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 2001;15:2940–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res 2011; 21:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci USA 2011;108:5626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science 1993;260:1926–8. [DOI] [PubMed] [Google Scholar]
- [29].Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 2001; 411:212–4. [DOI] [PubMed] [Google Scholar]
- [30].Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA 2007;104:14366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tao Y, Xi S, Briones V, Muegge K. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS ONE 2010;5:e9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dunican DS, Cruickshanks HA, Suzuki M, Semple CA, Davey T, Arceci RJ, et al. Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol 2013;14:R146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, et al. Lsh is involved in de novo methylation of DNA. EMBO J 2006;25:335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol 2008;28: 215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Meehan RR, Pennings S, Stancheva I. Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes Dev 2001;15:3231–6. [DOI] [PubMed] [Google Scholar]
- [36].Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem 2003;278:823–8. [DOI] [PubMed] [Google Scholar]
- [37].Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013;153:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci 2012; 125:5524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 2006;34:2887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res 1996;24:3685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 2000;5:355–65. [DOI] [PubMed] [Google Scholar]
- [42].Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 2000;406:541–4. [DOI] [PubMed] [Google Scholar]
- [43].Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell 2011;42:285–96. [DOI] [PubMed] [Google Scholar]
- [44].Yan Q, Cho E, Lockett S, Muegge K. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol Cell Biol 2003;23:8416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol 1997;137: 263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J 2003;22:5154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 1999;18:1923–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 2002;30:329–34. [DOI] [PubMed] [Google Scholar]
- [49].Jarvis CD, Geiman T, Vila-Storm MP, Osipovich O, Akella U, Candeias S, et al. A novel putative helicase produced in early murine lymphocytes. Gene 1996;169:203–7. [DOI] [PubMed] [Google Scholar]
- [50].Muegge K Lsh, a guardian of heterochromatin at repeat elements. Biochem Cell Biol 2005;83:548–54. [DOI] [PubMed] [Google Scholar]
- [51].De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol 2006;8:1448–54. [DOI] [PubMed] [Google Scholar]
- [52].Yu W, McIntosh C, Lister R, Zhu I, Han Y, Ren J, et al. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res 2014;24: 1613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS ONE 2009;4:e4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Erdel F, Schubert T, Marth C, Langst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci USA 2010;107:19873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Luperchio TR, Wong XR, Reddy KL. Genome regulation at the peripheral zone: lamina associated domains in development and disease. Curr Opin Genet Dev 2014;25:50–61. [DOI] [PubMed] [Google Scholar]
- [56].Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat Protoc 2007;2:1467–78. [DOI] [PubMed] [Google Scholar]
- [57].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008;453:948–51. [DOI] [PubMed] [Google Scholar]
- [58].Agarwal S, van Cappellen WA, Guenole A, Eppink B, Linsen SE, Meijering E, et al. ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol 2011; 192:735–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Saksouk N, Barth Teresa K, Ziegler-Birling C, Olova N, Nowak A, Rey E, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 2014;56:580–94. [DOI] [PubMed] [Google Scholar]
- [60].Ren J, Briones V, Barbour S, Yu W, Han Y, Terashima M, et al. The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences. Nucleic Acids Res 2015;43:1444–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jeltsch A, Lanio T. Site-directed mutagenesis by polymerase chain reaction. Methods Mol Biol 2002;182:85–94. [DOI] [PubMed] [Google Scholar]
- [62].He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol 1990;110:569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.