ABSTRACT
A cornucopia of literatures has characterized the involvement of a host of functional molecules in liver cancer. Herein, according to online datasets, we found that cytochrome P450 family 2 subfamily C member 8 (CYP2C8) was downregulated in liver cancer, and high CYP2C8 expression was associated with favorable overall survival. Lower levels of CYP2C8 were confirmed in liver cancer cells. CYP2C8 overexpression efficiently attenuated liver cancer cell proliferation and promoted apoptosis. We then discovered that miR-382-3p directly targeted CYP2C8 to inhibit its expression in liver cancer cells based on bioinformatic prediction and experimental confirmation. Moreover, a cytoplasmic long noncoding RNA (lncRNA), growth arrest-specific 5 (GAS5), sponged and down-regulated miR-382-3p, thus positively modulating CYP2C8 expression. Rescue assays indicated that GAS5 overexpression gave rise to decreased proliferation and increased apoptosis of liver cancer cells, while CYP2C8 knockdown counteracted GAS5-mediated anti-carcinogenic effects. In summary, our work offered a solid experimental foundation for understanding the functional role of CYP2C8 and the mechanism of GAS5/miR-382-3p/CYP2C8 axis in cell proliferation and apoptosis of liver cancer.
KEYWORDS: CYP2C8, miR-382-3p, GAS5, liver cancer
Introduction
Liver cancer (LC) is acknowledged as one of the six most common tumors worldwide with rising incidence and dismal prognosis.1,2 The predominant histological type of LC is hepatocellular carcinoma, a major challenge for public health.3 Medical treatments against LC in the past decades have been developed and improved. Nevertheless, effective therapy against its postsurgical recurrence and metastasis remains scanty, giving rise to dreadful consequences for hepatocellular carcinoma patients.4 Depressingly, until now, molecular mechanisms related to hepatocellular carcinoma have not been completely identified. Hence, solid biomarkers to predict its recurrence and survival are urgently required.
Long noncoding RNAs (lncRNAs) are a type of transcripts with vital functions in pathophysiological processes, which have more than 200 nucleotides in length and possess essentially limited protein-coding potentials.5,6 The emerging recognition that lncRNAs mediate tumor initiation, aggravation, recurrence and patient survival has also been widely substantiated in various cancers.7,8 For example, elevated expression of lncRNA AK023391 facilitates tumorigenesis and invasion in gastric cancer through PI3K/Akt signaling pathway activation.9 LncRNA miR503 HG is a prognostic indicator of hepatocellular carcinoma and inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway.10 LncRNA MIR31 HG, which targets HIF1A and P21, promotes cell-cycle progression in head and neck cancer to facilitate its cell proliferation and tumorigenesis.11 In the case of LC, a host of lncRNAs, such as CCAT1, HULC and HOXD-AS1, have been testified to be fundamentally involved in the biological processes of LC and consequently affect its pathogenesis.12–14 Nevertheless, more critical roles of lncRNAs in LC need to be further investigated.
In this study, using available online datasets, we found that CYP2C8 was lowly expressed in LC tissues, and high CYP2C8 expression was strongly associated with LC patient survival. Then, the mechanism of CYP2C8 downregulation in LC was analyzed. Consequently, the regulatory network of lncRNA GAS5/miR-382-3p/CYP2C8 was uncovered, casting new light on the investigation of carcinogenesis and the development of identifying survival biomarkers for LC therapy.
Materials and methods
Cell lines and culture
LC cell lines for our study comprised SK-HEP-1, Hep3B, Huh-7 and HepG2. LC cells and normal liver cell line THLE-3 as the control were all acquired from the Shanghai Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China). Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA), as suggested, was applied for the cultivation of all cell lines in an incubator with 5% CO2 at 37°C, with the addition of 10% fetal bovine serum (Gibco).
Cell transfection
The sequences of GAS5 and CYP2C8 were, respectively, cloned into pcDNA3.1 vectors (Invitrogen, Carlsbad, CA, USA) to construct overexpression plasmids pcDNA3.1/GAS5 and pcDNA3.1/CYP2C8. For CYP2C8 silencing, short hairpin RNAs (shRNAs) targeting CYP2C8 (sh-CYP2C8#1 and sh-CYP2C8#2; GenePharma, Shanghai, China) or negative control (sh-NC; GenePharma) were utilized. MicroRNA (miRNA) mimics and inhibitors (RiboBio, Guangzhou, China) were applied for augmenting and suppressing miRNA expression in LC cells, with NC-mimics and NC-inhibitors as their negative controls. Lipofectamine 2000 (Invitrogen) was adopted as the transfection reagent.
Western blot analysis
Proteins extracted from cells were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. After being blocked for 2 h in 5% skim milk, membranes were cultivated at 4°C overnight with the primary antibodies (Abcam, Cambridge, MA, USA) against GAPDH and CYP2C8, followed by treatment with secondary antibody (Abcam) for 2 h. Enhanced chemiluminescence (ECL) system (Sigma-Aldrich, St. Louis, MO, USA) was applied for protein band visualization.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cultured cells underwent total RNA extraction with TRIzol reagent (Invitrogen). Isolated RNAs were reversely transcribed into complementary DNAs (cDNAs) using M-MLV Reverse Transcriptase (Invitrogen). The expression of specific genes was detected with a standard SYBR-Green method under ABI7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). 2−ΔΔCq method was adopted for calculating relative expression. GAPDH served as the internal control. All reactions were performed in triplicate. The sequences of primers (GenePharma) were listed as follows: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ (forward) and 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ (reverse) for GAPDH; 5ʹ-GGATGCAGTGTGGCTCTGGATA-3ʹ (forward) and 5ʹ-TGTGTGCCAATGGCTTGAGTTAG-3ʹ (reverse) for GAS5; 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAGTGTT-3ʹ (reverse transcription), 5ʹ-ACACTCCAGCTGGGAATCATTCACGGACA-3ʹ (forward) and 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ (reverse) for miR-382-3p; 5ʹ-GTCCACCTTGAGGCCACTAC-3ʹ (forward) and 5ʹ-GCGGAGTGAGTTGATGCATTT-3ʹ (reverse) for CYP2C8; 5ʹ-CCATGATCACGAAGGTGGTTT-3ʹ (forward) and 5ʹ-ATGCAGTCGAGTTTCCCACAT-3ʹ (reverse) for U1.
EdU proliferation assay
According to the user’s manual of the Click-iT®EdU Imaging Kit (Invitrogen), 5-ethynyl-2′-deoxyuridine (EdU) solution was added into 24-well plates with cultured LC cells. After 48 h of incubation at 37°C, LC cells were washed twice with phosphate-buffered saline (PBS), treated with glycine and penetrant (0.5% Triton X-100 in PBS) fixation, and then stained for 30 min with Apollo dyeing reaction solution without light exposure. Penetrant was added, and Hoechst 33342 solution or 4′,6-diamidino-2-phenylindole (DAPI) was utilized for staining cells for 30 min. After incubation, a fluorescence microscope (WuMo, Shanghai, China) photographed the cells. The percentage of EdU-positive cells was determined.
Cell counting kit-8 (CCK-8) assay
LC cell viability was evaluated through CCK-8 assay according to the manufacturer’s specifications from Beyotime (Shanghai, China). After 0, 24, 48, 72 or 96 h of incubation in 96-well plates with 3 × 103 cells distributed per well, cells were treated with CCK-8 reagent for 1 h. TECAN Infinite M200 Multimode microplate reader (Tecan, Männedorf, Switzerland) was eventually utilized to examine the absorbance at a wavelength of 450 nm.
TUNEL apoptosis assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay was adopted for LC cell apoptosis evaluation. One Step TUNEL Apoptosis Assay Kit (Beyotime) was applied. As described above, LC cells were collected into 48-well plates. After attachment, TUNEL detecting solution (50 μl/well) was supplemented into each well. Cells were incubated without light exposure for 1 h at 37°C. After DAPI staining, cells were observed by WMF-CX40 fluorescence microscope (WuMo). The cells labeled by TUNEL reagent were considered as apoptotic cells.
Flow cytometry analysis
After cells were cultured in 6-well plates for 48 h, Annexin V Apoptosis Detection Kit (eBiosciences, Waltham, MA, USA) was applied to perform flow cytometry analysis for evaluating cell apoptosis. The cells were rinsed by PBS (Solarbio, Beijing, China), followed by fixation with 70% ice-cooled ethanol (Solarbio). In the end, BD FACS AriaII Flow Cytometer (BD Biosciences, San Jose, CA, USA) was applied to analyze the rate of apoptotic cells.
Luciferase reporter assay
The sequence of GAS5 containing wild-type miR-382-3p binding site or its mutant was cloned into pmirGLO plasmids (Promega, Madison, WI, USA). The consequent reporter plasmids were referred to as GAS5-WT or GAS5-MUT. SNHG1-WT and NORAD-WT reporters were designed similarly. The sequence of CYP2C8 3′-untranslated region (3′-UTR) with miR-382-3p binding site or mutated site was inserted into pmirGLO plasmids likewise, and the reporters were termed as CYP2C8-WT or CYP2C8-MUT. The constructed luciferase reporters were co-transfected with miR-382-3p mimics or NC-mimics into LC cells as demanded using Lipofectamine™ 2000 (Invitrogen). After 48 h of transfection, Dual-Luciferase Reporter Assay System (Promega) was adopted to detect the relative luciferase activity (firefly luciferase activity/Renilla luciferase activity for normalization) in each group.
RNA immunoprecipitation (RIP) assay
RIP assay was conducted using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). According to the manufacturer’s manual, cell lysates were cultivated with magnetic beads coated by anti-Ago2 antibody (Abcam) or anti-IgG antibody as the control (Abcam) in RIP buffer. qRT-qPCR was used for detecting the enrichment of specific RNAs.
RNA pull-down assay
Biotin-labeled miRNA probes (Bio-miR-NC and Bio-miR-382-3p; GenePharma) were cultivated with Dynabeads M-280 Streptavidin (Invitrogen) as suggested by the directions. Then, cell lysates were incubated with the probe-coated beads. The enriched CYP2C8 messenger RNA (mRNA) was measured by qRT-qPCR analysis.
Fluorescence in situ hybridization (FISH) assay
Cells were cultivated in hybridization buffer (RiboBio) with the addition of GAS5 FISH probes (RiboBio). After DAPI staining, cells were observed using a microscope (WuMo) to show the subcellular distribution of GAS5.
Cytoplasmic and nuclear RNA isolation
With the utilization of the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada), cytoplasmic and nuclear RNAs were separated as instructed by the user’s manual. qRT-PCR examination of the extracted RNAs was conducted to probe the expressions of U1, GAPDH and GAS5. U1 and GAPDH were, respectively, regarded as the nuclear and cytoplasmic controls.
Statistical analysis
Each assay in this study was repeated at least three times. Statistical significance was estimated through comparing mean value ± standard deviation (SD) utilizing Student’s t-test or one-way analysis of variance and was presented by p < .05 (*), p < .01 (**), p < .001 (***). All statistical analyses were carried out by SPSS 18.0 software package (SPSS, Chicago, IL, USA).
Results
CYP2C8 was anti-proliferative and indicative of favorable survival in liver cancer
Previously, it was reported that the overall survival and the recurrence-free survival of hepatocellular carcinoma patients worsened as CYP2C8 in tumor tissues was downregulated.15 In the present study, CYP2C8 expression in tissues from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) datasets was accessible from the GEPIA database (http://gepia.cancer-pku.cn/). The result showed that CYP2C8 was notably downregulated in tumor tissues of liver hepatocellular carcinoma (LIHC) compared with normal ones (Figure 1a). Additionally, a higher level of CYP2C8 indicated better overall survival of LIHC patients (Figure 1b). Subsequently, we detected the expression of CYP2C8 in cell lines using qRT-PCR and western blot. The result showed that the levels of CYP2C8 mRNA and protein were reduced in LC cells compared with normal cells (Figure 1c and d). Among these LC cells, HepG2 and Huh-7 cells exhibited lower levels of CYP2C8 mRNA. Hence, we constructed CYP2C8 overexpression plasmids and transfected them into HepG2 and Huh-7 cells. The efficiency of overexpression was confirmed by qRT-PCR (Figure 1e). EdU assay illustrated that the proliferation of LC cells was attenuated in response to CYP2C8 overexpression (figure 1f). Besides, as validated by CCK-8 assay, LC cells with CYP2C8 overexpression exhibited a restrained trend of viability (Figure 1g). TUNEL assay for cell apoptosis detection showed that elevated CYP2C8 expression led to promoted LC cell apoptosis (Figure 1h). Finally, flow cytometry analysis indicated that cell apoptosis was enhanced by CYP2C8 overexpression (Figure 1i). These data above suggested the anti-carcinogenic function of CYP2C8 in LC.
Figure 1.
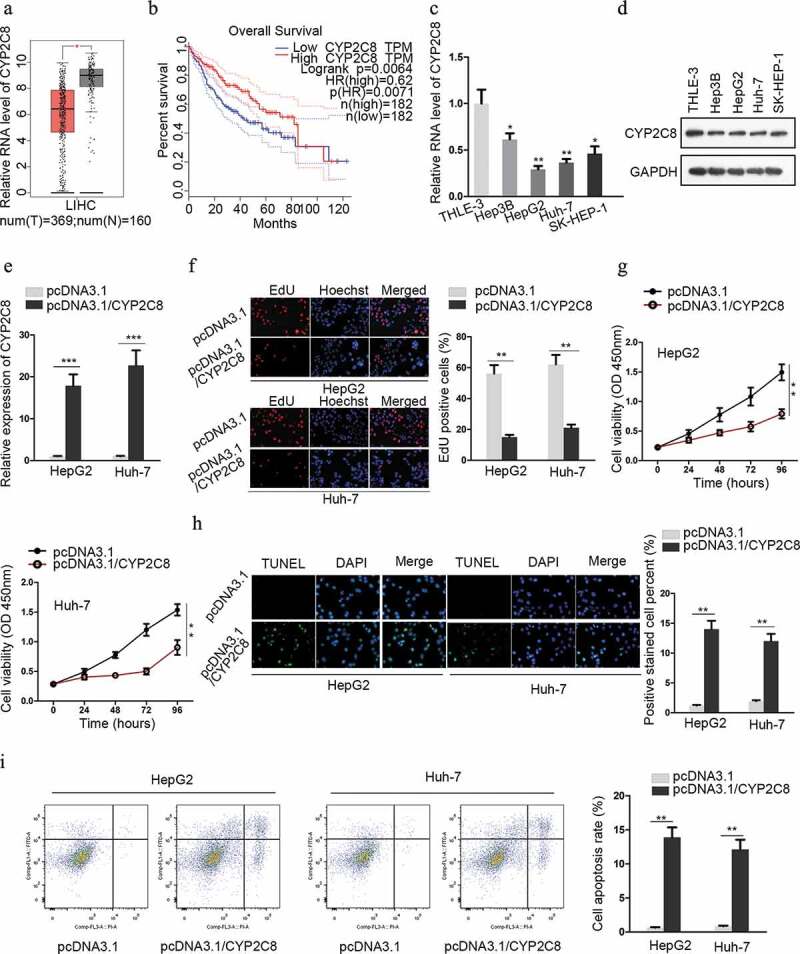
CYP2C8 was anti-proliferative and indicative of favorable survival in liver cancer. (a) The expression of CYP2C8 in LIHC tumor dataset (red) and normal dataset (gray) was plotted. (b) Kaplan-Meier survival analysis illustrated the relationship between CYP2C8 expression and overall survival from LIHC dataset. (c and d) LC cells from four cell lines and normal cells underwent qRT-PCR analysis to detect CYP2C8 mRNA (c) and protein (d) levels. (e) HepG2 and Huh-7 cells transfected with pcDNA3.1 or pcDNA3.1/CYP2C8 underwent qRT-PCR analysis to detect CYP2C8 mRNA level. (f) The proliferation of HepG2 and Huh-7 cells transfected with pcDNA3.1 or pcDNA3.1/CYP2C8 was tested by EdU assay. (g) CCK-8 assay monitored the reduced viability of HepG2 and Huh-7 cells after CYP2C8 overexpression. Optical density (OD) at 450 nm was regarded as the measurement of cell viability. (h) TUNEL assay evaluated the enhanced apoptosis of HepG2 and Huh-7 cells after the transfection of CYP2C8 overexpression plasmids. (i) Flow cytometry analysis examined the influence of CYP2C8 overexpression on LC cell apoptosis. p < .05 (*), p < .01 (**), p < .001 (***)
CYP2C8 was directly targeted by miR-382-3p
Next, we investigated the mechanism of CYP2C8 downregulation in LC. The online bioinformatics database miRDB (http://mirdb.org/) was adopted for predicting miRNAs which could possibly target CYP2C8 to suppress its expression. Among the 17 miRNAs predicted by miRDB, the top three miRNAs (miR-4797-5p, miR-382-3p and miR-3665), whose target scores with CYP2C8 were over 90, were selected as possible candidates. A series of qRT-PCR assay unveiled that after the successful transfection of miR-4797-5p, miR-382-3p or miR-3665 mimics or inhibitors into LC cells (Fig. S1A), CYP2C8 mRNA level was significantly reduced or increased only in response to miR-382-3p up-regulation or down-regulation (Figure 2a). Besides, miR-382-3p mimics or inhibitors also reduced or increased CYP2C8 protein level in LC cells (Figure 2b). Next, the molecular interaction between miR-382-3p and CYP2C8 was evaluated. As presented in the result of RIP assay, CYP2C8 and miR-382-3p were enriched in anti-Ago2 groups, indicating their existence in RNA-induced silencing complexes (RISCs) (Figure 2c). According to bioinformatic prediction, the binding sequences between miR-382-3p and CYP2C8 3ʹ-UTR were shown, along with the CYP2C8 3ʹ-UTR sequence with the mutated binding site (Figure 2d). Through luciferase reporter assay, we discovered that the luciferase activity of CYP2C8-WT reporter was significantly suppressed upon the transfection of miR-382-3p mimics into LC cells (Figure 2e). Furthermore, RNA pull-down determined the binding capacity between bio-miR-382-3p and CYP2C8 mRNA (Figure 2f). Altogether, miR-382-3p inhibited CYP2C8 expression in LC cells by directly targeting the 3ʹ-UTR of CYP2C8 mRNA.
Figure 2.
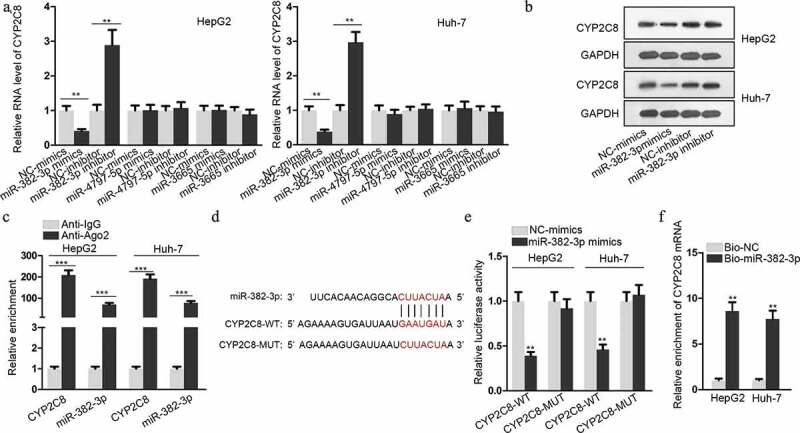
CYP2C8 was directly targeted by miR-382-3p. (a) According to the prediction of miRDB, top three miRNAs (miR-4797-5p, miR-382-3p and miR-3665) with higher target score with CYP2C8 were selected. The alteration of CYP2C8 mRNA levels in HepG2 and Huh-7 cells were evaluated in response to the transfection of mimics or inhibitors of these miRNAs. (b) CYP2C8 protein levels in LC cells were measured in response to miR-382-3p overexpression or inhibition by western blot. (c) RIP assay revealed that CYP2C8 mRNA and miR-382-3p could be enriched with Ago2 protein. (d) The sequences of wild-type CYP2C8 3ʹ-UTR, mutated CYP2C8 3ʹ-UTR and miR-382-3p were shown. The binding sites were highlighted in red. (e) Luciferase reporter assay showed that miR-382-3p overexpression inhibited the luciferase activity of CYP2C8-WT reporters in HepG2 and Huh-7 cells. (f) RNA pull-down demonstrated that CYP2C8 mRNA could be enriched by bio-miR-382-3p probes. p < .01 (**), p < .001 (***)
GAS5 bound to miR-382-3p to positively regulate CYP2C8 expression
LncRNAs, as one participant of competing endogenous RNA (ceRNA) network, can compete with mRNAs for miRNA interaction through sharing miRNA response elements (MREs), thus modulating the available levels of miRNAs and their downstream genes to influence their functions in tumors.16,17 In the present study, ENCORI database (http://starbase.sysu.edu.cn/) was employed to predict the lncRNAs which could sponge miR-382-3p. Among the candidates, the top three lncRNAs (SNHG1, GAS5 and NORAD) with more supported Argonaute cross-linking and immunoprecipitation followed by sequencing (Ago CLIP-sep) experiments were selected. Next, the luciferase activity of SNHG1-WT, GAS5-WT or NORAD-WT reporters was, respectively, monitored in response to the transfection of miR-382-3p mimics or NC-mimics into LC cells. The result illustrated that only the luciferase activity of GAS5-WT reporter was significantly inhibited in response to miR-382-3p mimics (Figure 3a). Therefore, GAS5 was chosen for further assays. To verify that GAS5 could act as a ceRNA to sponge miR-382-3p, the subcellular location of GAS5 was evaluated in LC cells. The result of FISH assay suggested that GAS5 was mainly located in cytoplasm (Figure 3b). The result of nuclear-cytoplasmic RNA isolation assay showed the same tendency (Figure 3c). Furthermore, an online database lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) also indicated that GAS5 was predominantly located in the cytoplasm (Figure 3d). These data clearly testified the post-transcriptional regulatory capacity of GAS5. To determine the molecular interactions, RIP assay confirmed the enrichment of GAS5, miR-382-3p and CYP2C8 mRNA in anti-Ago2 groups (Figure 3e). The binding site between GAS5 and miR-382-3p is presented in Figure 3f, together with the mutated miR-382-3p binding site. Luciferase reporter assay also demonstrated the decline of GAS5-WT luciferase activity upon the transfection of miR-382-3p mimics, while no change of luciferase activity was observed when the potential miR-382-3p targeting site was mutated (Figure 3g). Besides, as shown by qRT-PCR assay detection, GAS5 overexpression led to a decreased level of miR-382-3p in LC cells (Figure 3h). Also, through qRT-PCR and western blot, we also discovered that GAS5 overexpression resulted in upregulated levels of CYP2C8 mRNA and protein (Figure 3i). The above data revealed that GAS5 upregulated CYP2C8 via sponging miR-382-3p.
Figure 3.
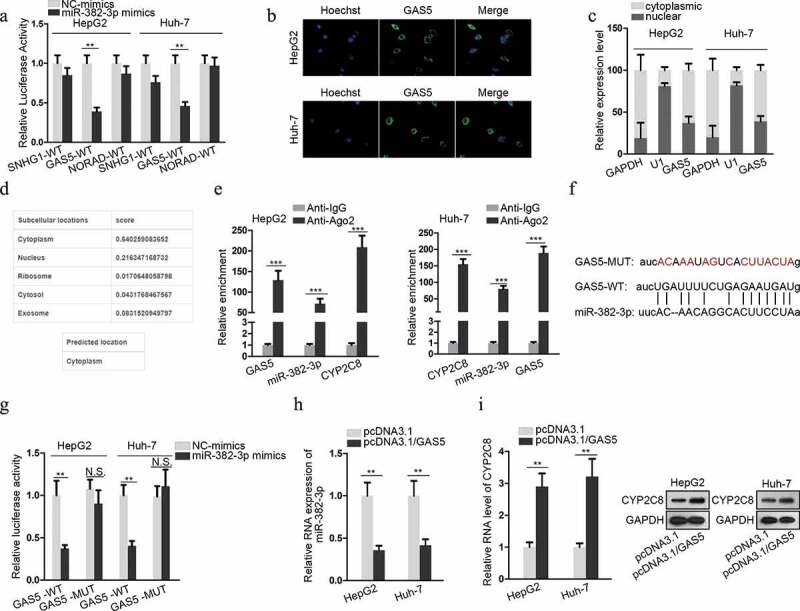
GAS5 bound to miR-382-3p to positively regulate CYP2C8 expression. (a) Top three lncRNAs (SNHG1, GAS5 and NORAD), which could possibly associate with miR-382-3p, were presented using ENCORI. Luciferase reporter assay evaluated the the luciferase activity of SNHG1-WT, GAS5-WT and NORAD-WT reporters after the transfection of NC-mimics or miR-382-3p mimics into HepG2 and Huh-7 cells. (b) FISH assay revealed GAS5 subcellular location in HepG2 and Huh-7 cells. (c) Cytoplasmic and nuclear RNA isolation uncovered the distribution of GAS5 in the cytoplasm or nuclei of HepG2 and Huh-7 cells. (d) lncLocator predicted a high possibility of the cytoplasmic abundance of GAS5. (e) RIP assay illustrated the enrichment of GAS5, miR-382-3p and CYP2C8 with Ago2 protein. (f) Complementary sequences between miR-382-3p and GAS5, together with the mutant site of GAS5 were shown. (g) Luciferase reporter assay showed the decline of GAS5-WT reporter luciferase activity due to the transfection of miR-382-3p mimics. (h) The impact of GAS5 overexpression on miR-382-3p level in LC cells was assessed by qRT-PCR. (i) qRT-PCR and western blot illustrated the effects of altered GAS5 expression on CYP2C8 mRNA and protein levels in LC cells. p < .01 (**), p < .001 (***), N.S.: not significant
CYP2C8 silencing counteracted the impact of GAS5 on LC cell proliferation and apoptosis
Given the above-verified mechanism, we intended to testify the impact of GAS5/miR-382-3p/CYP2C8 in LC cells. First of all, the expression of CYP2C8 was successfully interfered by the transfection of sh-CYP2C8#1/2, and the level of GAS5 was effectively promoted by the transfection of pcDNA3.1/GAS5 into HepG2 cells (Figure 4a). As demonstrated by CCK-8 assay, HepG2 cell viability was reduced due to the upregulation of GAS5, whereas cell viability was promoted via CYP2C8 silencing (Figure 4b). Besides, the result of EdU assay showed that GAS5 overexpression reduced HepG2 cell proliferation, while CYP2C8 silencing reversed the inhibitory trend (Figure 4c). TUNEL assay presented that HepG2 cell apoptosis was promoted in response to GAS5 overexpression, but the promotional effect was later restored by CYP2C8 silencing (Figure 4d). In the end, it was revealed by flow cytometry analysis that the facilitated cell apoptosis in HepG2 cells transfected with pcDNA3.1/GAS5 could be restored by CYP2C8 depletion (Figure 4e). All these results confirmed the regulatory function of GAS5/miR-382-3p/CYP2C8 on LC cell proliferation and apoptosis.
Figure 4.
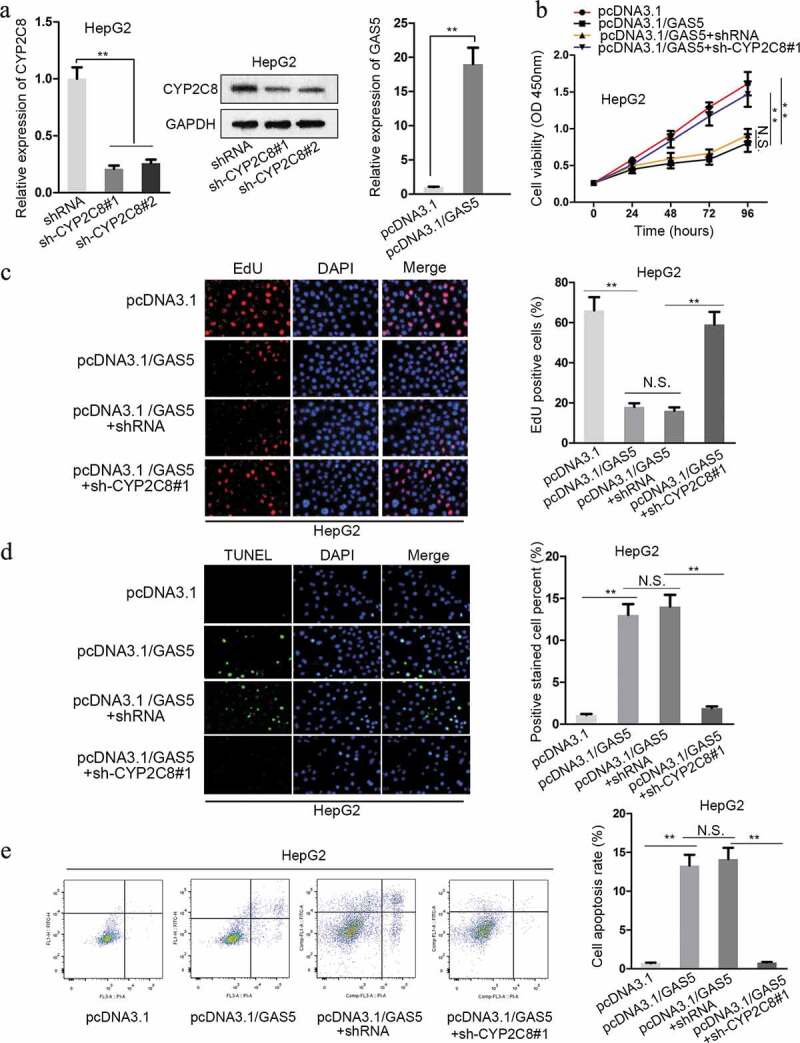
CYP2C8 elevation counteracted the impact of GAS5 on LC cell proliferation and apoptosis. (a) The knockdown efficiency of CYP2C8 and the overexpression efficiency of GAS5 in HepG2 cells were measured by qRT-PCR and western blot. (b) CCK-8 assay indicated that CYP2C8 knockdown reversed the inhibitory effect of GAS5 overexpression on HepG2 cell viability. (c) EdU assay showed that CYP2C8 silencing facilitated HepG2 cell proliferation which had been suppressed by GAS5 overexpression. (d) TUNEL assay presented that CYP2C8 knockdown abolished the enhanced HepG2 cell apoptosis altered by GAS5. (e) Flow cytometry analysis evaluated the cell apoptotic level of the transfected cells. p < .01 (**), N.S.: not significant
Discussion
Herein, CYP2C8 was testified to be highly expressed in LC tissues and cells. A higher level of CYP2C8 indicated favorable overall survival of LC patients. Functionally, CYP2C8 overexpression is pro-apoptotic and anti-proliferative for LC cells. Mechanistically, CYP2C8 3ʹ-UTR was targeted by miR-382-3p, and lncRNA GAS5 sponged miR-382-3p to boost CYP2C8 expression. Therefore, we concluded that GAS5 served as a ceRNA for miR-382-3p to suppress the malignant behaviors of LC cells via the augmentation of CYP2C8 expression.
Previously, CYP2C8 was documented to be involved in hepatocellular carcinoma, and its downregulation denoted unsatisfactory overall survival and recurrence-free survival.15,18 To our knowledge, available literature had not given a solid validation on CYP2C8 function in LC, and a well-elucidated mechanism on the dysregulation of CYP2C8 expression was still elusive. Our study corroborated the biological role of CYP2C8 as a critical inhibitor of LC cell proliferation, which was experimentally proven for the first time.
It has been extensively acknowledged that miRNAs regulate various biological processes, including tumorigenesis, via targeting the 3ʹ-UTR of specific mRNAs to suppress their expression. In the case of LC, for example, SF3B4 targeted by miR-133b stimulates cell proliferation and metastasis in hepatocellular carcinoma.19 MiR-3928 v facilitates hepatocellular carcinoma malignancy by suppressing VDAC3.20 Previously, miR-382-3p was reported to be implicated in ovarian cancer and breast cancer.21,22 In this study, we first discovered that CYP2C8 was a downstream target gene of miR-382-3p, and CYP2C8 expression was suppressed by miR-382-3p in LC cells.
As elucidated before, the interaction between miRNAs and mRNAs can be countervailed by lncRNAs, which serves as ceRNAs to sequester miRNAs, thereby regulating tumorigenesis and cancer progression. For example, lncRNA MIR31HG ameliorates hepatocellular carcinoma malignancy via sponging miR-575 to facilitate ST7L expression.23 LINC01133 hinders gastric cancer progression through sponging miR-106a-3p to promote APC level.24 The role of lncRNA GAS5 as a tumor suppressor has been demonstrated in various tumors, such as in pancreatic cancer,25 esophageal squamous cell carcinoma26 and colorectal cancer.27 Moreover, in the case of liver diseases, GAS5 retards liver fibrogenesis through a ceRNA network of GAS5/miR-222/p27 and suppresses hepatocellular carcinoma cell migration and invasion via miR-21.28,29 Our work first discovered that GAS5 overexpression inhibited LC cell proliferation, and contributed to high LC cell apoptotic levels. Furthermore, GAS5 was mainly located in the cytoplasm of LC cells and was proven to be able to sponge miR-382-3p. Besides, the silencing of CYP2C8, which was targeted by miR-382-3p, counteracted the decreased LC cell proliferation and the increased apoptosis induced by GAS5 overexpression.
Above all, the present study demonstrated a novel ceRNA axis of GAS5/miR-382-3p/CYP2C8 in LC. The anti-carcinogenic role of CYP2C8 may offer a solid experimental basis for LC treatment.
Supplementary Material
Acknowledgments
The help of all laboratory members for this study is appreciated.
Funding Statement
The authors have no funding to report.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A.. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 5.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17:61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Zhang J, Hou L, Wang G, Liu H, Zhang R, Chen X, Zhu J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Liang L, Dong Q, Huan L, He J, Li B, Yang C, Jin H, Wei L, Yu C, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-kappaB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi Q, Lian M, He S, Ma H, Fang J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018;17:162. doi: 10.1186/s12943-018-0916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, Li X, Zheng Q, Pu H, Gui X, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren X, Ji Y, Jiang X, Qi X. Downregulation of CYP2A6 and CYP2C8 in tumor tissues is linked to worse overall survival and recurrence-free survival from hepatocellular carcinoma. Biomed Res Int. 2018;2018:5859415. doi: 10.1155/2018/5859415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S, Wang L, Yang Q, Liu H, Meng Q, Jiang L, Wang S, Jiang W. Systematical analysis of lncRNA–mRNA competing endogenous RNA network in breast cancer subtypes. Breast Cancer Res Treat. 2018;169:267–275. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Liao X, Yang C, Huang K, Yu T, Yu L, Han C, Zhu G, Zeng X, Liu Z, et al. Identification of prognostic biomarkers for patients with hepatocellular carcinoma after hepatectomy. Oncol Rep. 2019;41:1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Li W, Pang Y, Zhou Z, Liu S, Cheng K, Qin Q, Jia Y, Liu S. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68. doi: 10.1016/j.ebiom.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Song G, Yao L, Liu Y, Liu M, Li S, Tang H. miR-3928v is induced by HBx via NF-kappaB/EGR1 and contributes to hepatocellular carcinoma malignancy by down-regulating VDAC3. J Exp Clin Cancer Res. 2018;37:14. doi: 10.1186/s13046-018-0681-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Liu Y, Wang Y, Fu X, Lu Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 2018;109:2188–2198. doi: 10.1111/cas.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu L, Li Z, Zhu J, Wang P, Fan G, Dai Y, Zheng Z, Liu Y. Serum expression levels of microRNA-382-3p, −598-3p, −1246 and −184 in breast cancer patients. Oncol Lett. 2016;12:269–274. doi: 10.3892/ol.2016.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan S, Tang Z, Chen K, Liu Y, Yu G, Chen Q, Dang H, Chen F, Ling J, Zhu L, et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37:214. doi: 10.1186/s13046-018-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, Zhang F, Shang M, Mao A. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Sun J, Zhao H, Li H. Long non-coding RNA (lncRNA) growth arrest specific 5 (GAS5) suppresses esophageal squamous cell carcinoma cell proliferation and migration by inactivating phosphatidylinositol 3-kinase (PI3K)/AKT/Mammalian target of rapamycin (mTOR) signaling pathway. Med Sci Monit. 2018;24:7689–7696. doi: 10.12659/MSM.910867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng K, Zhao Z, Wang G, Wang J, Zhu W. lncRNA GAS5 inhibits colorectal cancer cell proliferation via the miR1825p/FOXO3a axis. Oncol Rep. 2018;40:2371–2380. [DOI] [PubMed] [Google Scholar]
- 28.Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, Guo C, Liu Z, Fan X. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem. 2015;290:28286–28298. doi: 10.1074/jbc.M115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


