Abstract
Background: Effective surveillance of COVID-19 highlights the importance of rapid, valid, and standardized information to crisis monitoring and prompts clinical interventions. Minimal basic data set (MBDS) is a set of metrics to be collated in a standard approach to allow aggregated use of data for clinical purposes and research. Data standardization enables accurate comparability of collected data, and accordingly, enhanced generalization of findings. The aim of this study is to establish a core set of data to characterize COVID-19 to consolidate clinical practice.
Methods: A 3-step sequential approach was used in this study: (1) an elementary list of data were collected from the existing information systems and data sets; (2) a systematic literature review was conducted to extract evidence supporting the development of MBDS; and (3) a 2-round Delphi survey was done for reaching consensus on data elements to include in COVID-19 MBDS and for its robust validation.
Results: In total, 643 studies were identified, of which 38 met the inclusion criteria, where a total of 149 items were identified in the data sources. The data elements were classified by 3 experts and validated via a 2-round Delphi procedure. Finally, 125 data elements were confirmed as the MBDS.
Conclusion: The development of COVID-19 MBDS could provide a basis for meaningful evaluations, reporting, and benchmarking COVID-19 disease across regions and countries. It could also provide scientific collaboration for care providers in the field, which may lead to improved quality of documentation, clinical care, and research outcomes.
Keywords: Minimum basic data set, Minimum data set, COVID-19
↑ What is “already known” in this topic:
There is no established national core data set for the COVID- 19 in Iran, which has led to a lack of standardization and variable assessment criteria being used across the country. This hinders the quality and monitoring of COVID-19.
→ What this article adds:
Using a structured approach, we developed a minimal dataset to underpin COVID-19 documentation and practice. It is anticipated that the MBDS will facilitate a more consistent approach to COVID-19 practice. This dataset can also be used by other researchers to apply statistical analyses and machine learning algorithms to compare the characteristics of the pandemic among different countries and to identify characteristics that could bring new insights about the pandemic and how to fight it.
Introduction
In December 2019, a series of cases of pneumonia with mysterious etiology was first identified in Wuhan, China. On January 7, 2020, the novel Coronavirus (COVID-19), previously known as Severe Acute Respiratory Syndrome Coronavirus2 (SARS-CoV-2 or 2019-nCoV) was identified as the causal organism (1-3).COVID-19 is classified as a type of RNA virus, belonging to the family of coronaviruses, which primarily leads to a respiratory system infection and is extensively transmitted among humans and mammals, causing numerous conditions that range from the ‘‘common’’ influenza to death (4, 5). COVID-19 seems to be extremely communicable. The World Health Organization (WHO) has recently confirmed the COVID-19 a public health emergency (6). The WHO is warning countries to expand their efforts to contain the disease and safeguard health care environments and notes that a solution calls for a worldwide ’aggressive preparedness’ (7).
Early, systematic, and active emergency management practices are key points in epidemic prevention and control. The effective surveillance of this emerging outbreak heavily relies on regulatory management and coordinated interventions, which include comprehensive and directed surveillance, antimicrobial stewardship program, education and training, research and epidemiological studies, and policymaking, etc. These interventions highlight the importance of rapid, valid, and reliable information sharing across hospitals and public health authorities for monitoring crisis and early warning. In this situation, high-quality datasets are the prerequisite of necessary analysis for public health, which is inherently a data-intensive domain (8-12). In Iran, most organizations have developed different processes and infrastructure for management and subsequent data collection of COVID-19 patients (13-17). Although current efforts to report COVID-19 are a good start, the absence of information management viewpoint regarding which data elements are critical to be recorded leads to significant inconsistent, unreliable, redundant, or duplicate reports. Thus, this precludes data integration, which limits the share of data across multiple health information systems (18, 19).
Further, standardized clinical documentation is an essential factor for electronic health records (EHRs) and for supporting secondary use of data gathered in the context of clinical daily workflows for other purposes than patient care, eg, for clinical research, quality management, epidemiologic studies, patient outcomes, and interoperability initiatives. MBDS is a data collection tool that aims to identify the common components of data sets as one of the first and most basic steps in foundation and implementation of numerous information systems through minimizing duplication of effort and improving data quality (20-24). COVID-19 monitoring depends on clinical data and reports from widely scattered public and hospital information systems as data input (eg, Hospital information systems (HIS), Iranian Electronic Health Record (so-called SEPAS), Iranian Integrated Health System (known as SIB), and other clinical information systems). Accordingly, as we are in the primary step of this emergency, the need to establish a supportive, standardized, accurate, and updated dataset is of paramount importance. Adopting such dataset is an important step in promoting data (capture) and data exchange with regard to COVID-19. Thus, we conducted a systematic literature review combined with a Delphi survey to establish a minimal dataset that would be regarded as a standardized method of reporting COVID-19 disease, and thus it is expected to improve the quality of clinical and research outcomes.
Methods
Design
In this study, a 3-step sequential approach was used. First, an elementary list of data was collected from the existing information systems and datasets. Next, a search strategy was developed to identify data items for establishing COVID-19 MBDS from an evidence-based perspective. These sources were continuously reviewed until data saturation (maximum data set). Finally, the data included from the review were analyzed using a 2-round Delphi survey to achieve consensus on optimal data set (minimum data set).
Data collection
The initial data elements were extracted from the medical records of patients with COVID-19, reports from Corona National headquarters, and other clinical and public health organizations affiliated to the Iranian Ministry of Health as well as official dataset provided by international organizations, such as the World Health Organization (WHO) reports, European Centre for Disease Prevention and Control dataset (ECDC), Chinese Center for Disease Control and Prevention (Chinese CDC). In addition to mapping available evidence supporting the development of minimal dataset, a systematic review was also conducted to identify probable data elements for inclusion in COVID-19 MBDS. To that end, PubMed, Scopus, Web of Science, and Google Scholar databases were reviewed by the following search terms (designed using English MeSH keywords and Emtree terms): “COVID-19”, “Novel coronavirus 2019”, “2019 nCoV”, “clinical characteristics”, “clinical features” and “clinical findings”. In Table 1, the systematic search strategy is proposed based on Boolean search operators, keywords, and search fields (advance search interface).
Table 1. Search strategy details .
| Databases | Search details |
| PubMed | (((((((((((((("covid-19 "[Title]) OR "novel coronavirus"[Title]) OR "2019 nCoV"[Title]) AND clinical characteristics) [Title/ Abstract]) OR clinical features [Title/ Abstract]) (English [lang]), limited to 2019-2020. |
| Scopus | (TITLE (covid-19) OR TITLE (novel AND coronavirus) OR TITLE (2019 nCoV) AND TITLE-ABS-KEY (clinical AND features) OR TITLE-ABS-KEY (clinical AND characteristics)) AND (LIMIT-TO (PUBSTAGE, "final")) AND (LIMIT-TO ( PUBYEAR, 2020) OR LIMIT-TO ( PUBYEAR, 2019)) AND (LIMIT-TO (DOCTYPE, "ar")) AND (LIMIT-TO (LANGUAGE, "English")). |
| Web of Science | TITLE: (COVID-19) OR TITLE: (novel coronavirus) OR TITLE: (2019 nCoV) AND TOPIC: (clinical findings) OR TOPIC: (clinical characteristics) Refined by: DOCUMENT TYPES: (ARTICLE) AND PUBLICATION YEARS: (2019-2020)) |
| Scholar | allintitle: (COVID-19 OR "novel coronavirus") AND ("clinical features" OR "clinical characteristics") AND English [lang], limited to 2019- 2020. |
Data material
Two authors independently performed electronic literature searches for study identification and screening. The results of the initial search strategy were first screened based on the title and abstract. The full-texts of relevant articles were examined for inclusion and exclusion criteria. This research included all full-text articles extracted from reliable sources in English between December 2019 and April 2020. Short articles, letters to the editor, accepted papers in conferences, thesis, and reports extracted from blogs were not included in this study. The main criterion for selection of research articles was the relevancy of their content with the research title. Due to the large number of available research articles, several criteria were considered for selecting articles and introducing clinical core data elements to report COVID-19. Hence, full articles with at least 2 of the following data classes related to the main objectives of reporting of COVID-19 were selected: (1) clinical, (2) laboratory, (3) radiology, and (4) epidemiological features. Finally, probable data elements to be included in COVID-19 MBDS were introduced in a checklist.
Questionnaire development
A questionnaire was developed using the data elements of the checklist and included 5 columns: “very important”, “important” “neutral”, “slightly important”, and “very slightly important” for each data item. To add necessary data elements by experts, a blank row was provided at the end of the questionnaire. The content validity of the questionnaire was assessed by an expert panel, including 2 infectious specialists and 3 health information management (HIM) experts. To add necessary data elements by experts, a blank row was provided at the end of the questionnaire. Test-retest (at an 8-day interval) was done to determine the reliability of the questionnaires, based on experts’ answers, including 2 health information management (HIM) and 2 medical informatics experts. Finally, the collected data were analyzed using SPSS 16, with the questionnaire showing a Cronbach's alpha of 0.86.
Delphi phase
The data elements were validated using 2 rounds of the Delphi survey by a group of multidisciplinary medical experts (Table 2). The experts participating in the study were asked to score the tabulated list of data elements in terms of their importance using a 5-point Likert scale (ranging from 1:“very slightly important” to 5:“highly important”. The level of agreement was considered to be a criterion for the acceptance of the data elements. Thus, after initial ranking, data elements with ≤50% agreement were excluded in the first round, those with 50%-75% agreement entered the second round, and data elements with ≥75% agreement were included in the primary round.
Table 2. Demographic characteristics of Delphi participants .
| Variables | Frequency | Percentage |
| Specialty | ||
| Infectious disease | 6 | 31.58 |
| Internal medicine | 5 | 26.31 |
| Radiologist | 4 | 21.05 |
| Epidemiologist | 4 | 21.05 |
| Gender | ||
| Female | 6 | 31.58 |
| Male | 13 | 68.42 |
| Age (years) | ||
| 30–40 | 6 | 31.58 |
| 40–50 | 7 | 36.84 |
| 50–60 | 5 | 26.31 |
| >60 | 1 | 5.27 |
| Work experience (years) | ||
| <10 | 7 | 36.84 |
| 10–20 | 8 | 42.10 |
| 20–30 | 3 | 15.78 |
| >30 | 1 | 5.27 |
| Total | 19 | 100 |
Results
A total of 643 articles were obtained from the literature review. After removal of duplicate articles and applying the exclusion criteria, 38 articles were included in the analysis (Fig. 1).
Fig. 1.
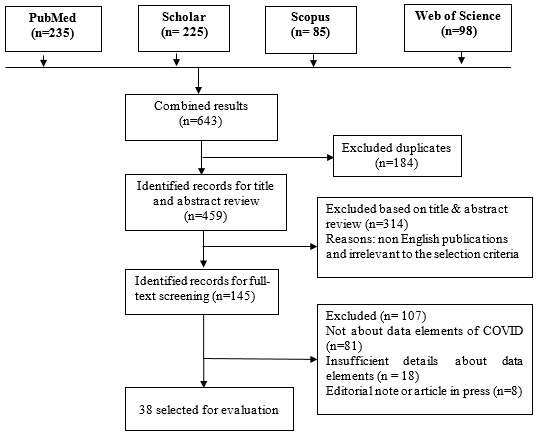
Flow diagram showing publication selection process
The demographic data of the study participants are presented in Table 2. The potential participants consisted of 25 medical specialists involved in COVID-19 care, treatment, and research domains. However, 6 specialists did not participate in the study. Thus, 19 experts contributed.
Overall, 3 data categories, 19 data classes and 149 data items were extracted from the related comprehensive literature review (maximum dataset). These data categories were epidemiological, clinical, and paraclinical. Epidemiological data contained 4 categories, including basic information, exposure history, transmission mode, and susceptible populations. The clinical data category consisted of clinical manifestations, coexisting conditions, treatment and supportive care, physical examinations, complications, time intervals, disease severity, disease status, and outcome data classes. Finally, the paraclinical category was divided into 2 laboratory and radiology indicators. The definitive numbers of data elements for epidemiological, clinical and paraclinical classes were 25, 73, and 51 respectively (Table 3).
Table 3. COVID-19 minimum basic data set .
| Literature review results |
Specialists perspectives ✓ : Accepted in first round × : Rejected in first round : Refer to second round |
||||
| First round | Second round | ||||
| Data classes / items | Frequency |
Mean (percentage) |
Initial decision |
Mean (percentage) |
Final decision |
| A. Epidemiological data | |||||
| Basic information | |||||
| Age (2-4, 25-44) | 23 | 4.26 (85.2) | ✓ | Accept | |
| Sex (2-4, 25, 27-29, 31-44) | 21 | 3.90 (78) | ✓ | Accept | |
| Occupation (40, 42) | 2 | 2.1 (42) | × | Refuse | |
| Nationality / race (40, 42) | 2 | 3.37 (67.36) | 3.32 (66.31) | Refuse | |
| Exposure information | |||||
| Exposure history (33, 35, 36, 41, 42, 45) | 6 | 4.52 (90.52) | ✓ | Accept | |
| Uncertainty (32, 36, 39, 46) | 4 | 4.32 (86.31) | ✓ | Accept | |
| Living in epidemic area (2, 4, 32-34, 46) | 6 | 4.21 (84.21) | ✓ | Accept | |
| Recent travelling (2, 4, 32, 34, 37, 42, 44) | 7 | 4.26 (85.20) | ✓ | Accept | |
| Contact with suspicious person (2, 4, 32-34, 46) | 6 | 4.36 (87.36) | ✓ | Accept | |
| Transmission mode | |||||
| Person-person (2, 33-35, 42, 46-48) | 7 | 3.9 (77.90) | ✓ | Accept | |
| Nosocomial (34, 35, 42, 46, 48) | 5 | 4.26 (85.2) | ✓ | Accept | |
| Inhalation (aerosols) (35, 42, 48) | 3 | 3.32 (66.32) | 2.79 (55.79) | Refuse | |
| Food / water born (35, 42, 48) | 3 | 3.37 (67.36) | 1.87 (37.40) | Refuse | |
| Contaminated surfaces (33, 35, 42, 47) | 4 | 4.16 (83.15) | ✓ | Accept | |
| Sporadic occurrence of zoonotic (35, 42) | 2 | 1.07 (21.4) | × | Refuse | |
| Other (35, 42) | 2 | 3.95 (78.95) | × | Accept | |
| Susceptible population | 2 | 3.95 (78.95) | ✓ | Accept | |
| Elderly people (32, 36, 39, 46) | 4 | 4.47 (89.48) | ✓ | Accept | |
| Current pregnancy (32, 36, 40, 45) | 4 | 4.53 (90.53) | ✓ | Accept | |
| Poor immune function (32, 40, 45) | 3 | 3.84 (76.84) | ✓ | Accept | |
| A. Epide miological data | |||||
| Susceptible population | 2 | 3.95 (78.95) | ✓ | Accept | |
| Chronic co-morbidities (32, 36, 46) | 3 | 3.79 (75.8) | ✓ | Accept | |
| Long-term use of immunosuppressive (32, 40) | 2 | 4.26 (85.2) | ✓ | Accept | |
| Surgery history (32, 40) | 2 | 2.79 (55.79) | 3.32 (66.31) | Refuse | |
| Active smoker (32, 39, 40) | 3 | 4.37 (87.37) | ✓ | Accept | |
| Other (32, 40) | 2 | 3.95 (78.95) | ✓ | Accept | |
| B. Clinical data | |||||
| Clinical manifestations | |||||
| Fever (2-4, 25-27, 29-37, 39-45, 47-54) | 29 | 4.53 (90.53) | ✓ | Accept | |
| Dry cough (2-4, 25, 26, 29-37, 39-43, 45, 47-54) | 27 | 4.37 (87.37) | ✓ | Accept | |
| Sputum / expectoration (2-4, 26, 29, 31, 34, 39, 40, 43, 44, 48-53) | 16 | 3.58 (71.57) | 3.98 (79.6) | Accept | |
| Dyspnea (2-4, 25-27, 29-37, 39, 40, 42-45, 47-54) | 27 | 4.26 (85.2) | ✓ | Accept | |
| Myalgia or fatigue (2-4, 25-27, 29, 31-34, 36, 39, 41-44, 48-54) | 22 | 4.05 (81) | ✓ | Accept | |
| Headache (2-4, 25, 26, 29, 30, 32-36, 39, 42-44, 48-52) | 20 | 2.47 (49.47) | × | Refuse | |
| Sore throat (2-4, 25, 26, 29, 31, 32, 34-36, 39, 40, 42-44, 48-53) | 21 | 3.84 (76.84) | ✓ | Accept | |
| Dizziness (2, 3, 26, 37, 40, 43, 44, 48, 50) | 9 | 2.42 (48.42) | × | Refuse | |
| Rhinorrhea (2, 3, 26, 32, 40, 43, 44, 48, 49, 52) | 10 | 3.53 (70.53) | 3.95 (79) | Accept | |
| Chest pain (2, 3, 25, 26, 32, 34, 35, 40, 42-44, 48, 49, 54) | 14 | 2.47 (49.47) | × | Refuse | |
| Pharyngeal congestion (30, 34, 36, 43, 44, 48, 52, 53) | 8 | 3.78 (75.79) | ✓ | Accept | |
| Chill (2-4, 34, 44, 48) | 6 | 3.35 (67) | 3.65 (73) | Refuse | |
| Abdominal pain (29, 31, 32, 34, 39, 42, 43, 48, 54) | 9 | 2.37 (47.37) | × | Refuse | |
| Diarrhea (2-4, 25-27, 29-33, 36, 37, 39, 40, 43, 44, 48-52, 54) | 23 | 2.15 (43.16) | × | Refuse | |
| Anorexia (2-4, 26, 32, 36, 42, 44, 48-50, 52) | 12 | 2.73 (54.73) | 2.87 (57.4) | Refuse | |
| Vomiting and nausea (4, 29-33, 35, 39, 42-44, 48, 52, 53) | 14 | 3.15 (63) | 3.48 (69.60) | Refuse | |
| More than one sign or symptom (26, 34, 36, 48) | 4 | 4.58 (91.59) | ✓ | Accept | |
| No sign or symptom (asymptomatic) (42, 47, 48, 52) | 4 | 4.16 (83.16) | ✓ | Accept | |
| Co-existing conditions | |||||
| Hypertension (3, 25, 26, 28, 29, 31, 32, 34, 36, 38-40, 42-44, 48-50, 54, 55) | 20 | 3.84 (76.84) | ✓ | Accept | |
| Cardiovascular (3, 25-29, 31, 32, 34, 36, 38-40, 42, 43, 48-50, 54, 55) | 20 | 4.32 (86.32) | ✓ | Accept | |
| Cerebrovascular (3, 25, 26, 40, 43, 49, 50, 54, 55) | 9 | 4.05 (81.05) | ✓ | Accept | |
| Diabetes (3, 25-29, 32, 34, 36, 38, 43, 44, 49, 50, 53-55) | 17 | 3.84 (76.84) | ✓ | Accept | |
| Gastrointestinal disease (26, 36, 50, 51) | 4 | 3.26 (65.26) | 3.97 (79.40) | Accept | |
| Malignant tumors (3, 25, 26, 28, 31, 34, 38, 43, 49, 50, 54, 55) | 12 | 4.05 (81.05) | ✓ | Accept | |
| Neural system disease (25, 26, 29, 34, 39, 40, 43, 49, 50, 53, 54) | 11 | 3.05 (61.05) | 3.63 (72.63) | Refuse | |
| Pulmonary disease (25, 26, 28, 31, 34, 38, 43, 49, 50, 53, 54) | 12 | 4.84 (96.84) | ✓ | Accept | |
| Chronic liver disease (28, 29, 34, 38, 40, 43, 50, 53-55) | 11 | 4.53 (90.53) | ✓ | Accept | |
| Chronic kidney disease (28, 29, 34, 38-40, 42, 43, 50, 53-55) | 12 | 3.79 (75.8) | ✓ | Accept | |
| HIV / immunodeficiency (26, 39, 43, 50, 53) | 5 | 3.84 (76.84) | ✓ | Accept | |
| Virus - bacterial coinfection (26, 36, 50, 51) | 4 | 3.84 (76.84) | ✓ | Accept | |
| None (28, 39) | 2 | 3.79 (75.8) | ✓ | Accept | |
| Treatment & supportive | |||||
| Mechanical ventilation | |||||
| Non-invasive (NMV) (4, 26, 30, 34, 36, 38, 40-42, 48, 51-53) | 13 | 4.11 (82.10) | ✓ | Accept | |
| Invasive (IMV) (4, 26, 30, 34, 36, 38, 40, 41, 44, 48, 51-53) | 13 | 4.21 (84.21) | ✓ | Accept | |
| Extracorporeal Membrane (ECMO) (4, 26, 30, 34, 38, 40, 51, 52) | 8 | 4 (80) | ✓ | Accept | |
| Both ECMO and IMV (30, 42, 51) | 3 | 3.84 (76.84) | ✓ | Accept | |
| Prescription | |||||
| Antibiotic (4, 26, 34, 36, 38, 40, 41, 44, 48, 49, 52, 53) | 12 | 3.79 (75.8) | ✓ | Accept | |
| Antifungal (4, 26, 34, 36, 42, 44, 48, 49, 52, 53) | 10 | 1.84 (36.84) | × | Refuse | |
| Antiviral (4, 26, 34, 40, 42, 44, 48, 49, 51-53) | 11 | 4.21 (84.21) | ✓ | Accept | |
| Glucocorticoids (4, 26, 34, 36, 38, 44, 49, 51) | 8 | 3.79 (75.8) | ✓ | Accept | |
| Immunoglobulin (4, 26, 34, 38, 44, 49, 52) | 7 | 4.26 (85.2) | ✓ | Accept | |
| B. Clini cal data | |||||
| Physical examination | |||||
| Body temperature (25, 26, 29, 32, 34, 36, 42, 45, 51-53) | 10 | 4.11 (82.11) | ✓ | Accept | |
| Respiratory rate (per minute) (32, 34, 36, 51) | 4 | 3.95 (78.95) | ✓ | Accept | |
| Heart rate (beats/ per minute) (32, 34, 36, 51) | 4 | 3.84 (76.84) | ✓ | Accept | |
| Body Mass Index (BMI) (36, 51) | 2 | 2.42 (48.42) | × | Refuse | |
| Systolic blood pressure [mmHg] (32, 34, 36, 51) | 4 | 3.95 (78.95) | ✓ | Accept | |
| Lung auscultation (sounds) (34, 36, 51) | 3 | 2.47 (49.47) | × | Refuse | |
| Disease Complication | |||||
| ARDS (31, 34, 36, 38, 39, 41, 43, 49, 51-53, 55) | 12 | 4 (80) | ✓ | Accept | |
| Acute heart injury (31, 34, 36, 38, 41, 43, 49, 51, 52) | 9 | 4.26 (85.2) | ✓ | Accept | |
| Liver abnormality (34, 36, 38, 39, 43, 49, 51, 52) | 8 | 3.42 (68.42) | 4.05 (81.05) | Accept | |
| Acute kidney injury (31, 36, 38, 41, 43, 49, 51-53, 55) | 10 | 4.05 (81.05) | ✓ | Accept | |
| Secondary infection (31, 34, 36, 38, 43, 51, 52) | 7 | 3.89 (77.89) | ✓ | Accept | |
| Shock (31, 34, 36, 38, 41, 43, 49, 51-53, 55) | 11 | 4.32 (86.31) | ✓ | Accept | |
| Other (31, 34, 36, 38, 43, 51, 52) | 7 | 3.89 (77.89) | ✓ | Accept | |
| Disease severity | |||||
| Mild (27, 28, 30, 32, 47, 49, 50, 53) | 8 | 3.79 (75.8) | ✓ | Accept | |
| Moderate (27, 28, 30, 32, 47, 49, 50, 53) | 8 | 3.95 (78.95) | ✓ | Accept | |
| Severe (27, 28, 30, 32, 47, 49, 50, 53) | 8 | 4 (80) | ✓ | Accept | |
| Critical (27, 28, 30, 32, 47, 49, 50, 53) | 8 | 3.84 (76.84) | ✓ | Accept | |
| Disease status | |||||
| Active (32, 36, 45, 46) | 4 | 3.89 (77.89) | ✓ | Accept | |
| In active(32, 36, 45, 46) | 4 | 4.05 (81.05) | ✓ | Accept | |
| Recovered (32, 36, 45, 46) | 4 | 4 (80) | ✓ | Accept | |
| Outcome | |||||
| Remained in hospital (4, 31, 34, 36, 38, 41, 47, 51, 53) | 10 | 3.79 (75.8) | ✓ | Accept | |
| Healed / discharged (4, 31, 34, 36, 38, 41, 47, 51, 53) | 9 | 3.84 (76.84) | ✓ | Accept | |
| Referred (32, 39, 46, 52, 55) | 5 | 3.12 (62.40) | 3.63(72.63) | Refuse | |
| Critical condition / ICU (4, 31, 34, 36, 38, 41, 47, 51, 53) | 9 | 4 (80) | ✓ | Accept | |
| Death (4, 31, 34, 36, 38, 47, 51, 53, 55) | 9 | 3.89 (77.89) | ✓ | Accept | |
| Partial recovery / follow up (32, 37, 39, 42, 46, 49, 52, 55) | 8 | 2.22 (44.40) | × | Refuse | |
| Time interval | |||||
| Exposure to symptom onset (3, 35-37, 39, 42) | 6 | 4.05 (81.05) | ✓ | Accept | |
| Illness to start treatment (3, 35, 37, 39, 42) | 5 | 4.19 (83.80) | ✓ | Accept | |
| Median incubation period (29, 46, 48, 49) | 4 | 3.79 (75.8) | ✓ | Accept | |
| Hospitalization date (37, 42) | 2 | 2.37 (47.36) | × | Refuse | |
| Diagnosis date (36, 37, 39, 42) | 4 | 4.21 (84.21) | ✓ | Accept | |
| Hospital day (36, 51) | 2 | 2.31 (46.31) | × | Refuse | |
| Discharge date (3, 37, 42) | 3 | 3.95 (78.95) | ✓ | Accept | |
| First round | Second round | ||||
| Data classes / items | Frequency |
Mean (percentage) |
Initial decision |
Mean (percentage) |
Final decision |
| C. Paraclinical Data | |||||
| Laboratory indicators | |||||
| Blood routine tests | |||||
| T lymphocyte count ×109/L ↑↓ (26, 30, 32, 34, 35, 38, 39, 42, 43, 48, 49, 56) | 12 | 4.26 (85.26) | ✓ | Accept | |
| Platelet count ×109/L ↑↓(26, 30, 32, 34, 35, 37-39, 42, 43, 48, 49, 56) | 13 | 3.26 (65.26) | 3.79 (75.8) | Accept | |
| Hemoglobin level (g/L) ↑↓(26, 30, 32, 34, 38, 39, 42, 43, 48, 49, 56) | 11 | 4.21 (84.21) | ✓ | Accept | |
| D-dimer (ug/ml) ↑↓(26, 29, 30, 32, 34, 39, 49, 56) | 9 | 4.16 (83.16) | ✓ | Accept | |
| Prothrombin times, s ↑↓ (29, 30, 34, 42, 43, 49) | 7 | 3.37 (67.37) | 3.32 (66.31) | Refuse | |
| Blood chemistry | |||||
| ALT (U/L) ↑↓ (26, 29, 30, 32, 37, 39, 42, 43, 48, 49, 56) | 11 | 4.31 (85.31) | ✓ | Accept | |
| AST (U/L) ↑↓ (26, 29, 30, 32, 37, 39, 42, 43, 48, 49, 56) | 12 | 4.42 (88.42) | ✓ | Accept | |
| Blood Urea Nitrogen (mmol/L) ↑↓(26, 37, 42, 43, 48, 56) | 6 | 4.26 (85.2) | ✓ | Accept | |
| Serum creatinine (umol/L) ↑↓ (34, 39, 42, 43, 48) | 5 | 3.84 (76.84) | ✓ | Accept | |
| Lactate dehydrogenase (U/L) ↑↓(26, 29, 30, 32, 34, 39, 42, 49, 56) | 3 | 3.63 (72.63) | 4.26 (85.26) | Accept | |
| Albumin (g/L) ↑↓(32, 34, 42, 49, 56) | 5 | 3.59 (71.58) | 3.70 (74) | Refuse | |
| Globulin (g/L) ↑↓(32, 34, 42, 49, 56) | 5 | 3.42 (68.42) | 3.03 (60.6) | Refuse | |
| Total bilirubin(umol/L) ↑↓ (26, 29, 30, 32, 34, 39, 42, 49, 56) | 9 | 3.53 (70.53) | 3.68 (73.68) | Refuse | |
| Direct bilirubin(umol/L) ↑↓(4, 31, 50) | 3 | 2.42 (48.42) | × | Refuse | |
| Infection-related biomarkers | |||||
| C-reactive protein (mg/L) ↑↓(26, 30, 32, 37, 42, 43, 48, 49, 56) | 9 | 3.95 (78.95) | ✓ | Accept | |
| Procalcitonin (ng/ml)↑↓(26, 29, 30, 32, 34, 39, 42, 43, 48, 49, 56) | 11 | 2.95 (58.95) | 3.11 (62.11) | Refuse | |
| Interleukin 6 (pg/ml) ↑↓ (32, 34, 38, 39, 42, 43, 56) | 8 | 4.63 (92.63) | ✓ | Accept | |
| RT-PCR (3, 4, 26, 28-30, 32, 33, 35-38, 40-42, 44-46, 48, 49, 51-54, 57-59) | 28 | 4.47 (89.47) | ✓ | Accept | |
| Radiology information | |||||
| Radiology procedure | |||||
| Chest X-ray (41, 49, 50) | 3 | 2.63 (52.63) | 2.75 (55) | Refuse | |
| CT scan (2-4, 25, 31, 32, 40-43, 52, 58, 59) | 12 | 4.05 (81.05) | ✓ | Accept | |
| Lung ultrasound (41, 50) | 2 | 3.52 (70.53) | 3.84 (76.84) | Accept | |
| Pattern of the lesion | |||||
| Ground glass opacity (2-4, 25, 29-32, 40-43, 45, 48, 49, 52, 58, 59) | 17 | 4.47 (89.47) | ✓ | Accept | |
| Consolidation (2-4, 25, 32, 41-43, 45, 48, 49, 52, 58, 59) | 14 | 4.42 (88.42) | ✓ | Accept | |
| Both (2, 25, 42, 45, 49, 58) | 6 | 3.95 (78.95) | ✓ | Accept | |
| Patchy shadow (3, 41, 42, 52, 58, 59) | 6 | 3.84 (76.84) | ✓ | Accept | |
| Lymphadenopathy (3, 32, 41, 42, 45, 49, 58, 59) | 8 | 3.90 (77.9) | ✓ | Accept | |
| Pleural effusion (32, 41, 42, 45, 49, 58, 59) | 7 | 4.15 (83.15) | ✓ | Accept | |
| Crazy paving (32, 41, 42, 45, 49, 58, 59) | 7 | 3.95 (78.95) | ✓ | Accept | |
| Bronchiectasis (3, 32, 42, 45, 52, 58, 59) | 7 | 4.16 (83.15) | ✓ | Accept | |
| Interlobular septal thickening (3, 41, 42, 45, 52, 58, 59) | 7 | 4.05 (81.05) | ✓ | Accept | |
| Reticulation (3, 41, 42, 52, 58, 59) | 6 | 3.68 (73.68) | 3.89 (77.9) | Accept | |
| Other (3, 41, 52, 58) | 5 | 3.78 (75.79) | ✓ | ||
| Lesion distribution | Accept | ||||
| Unilateral (4, 25, 26, 29, 31, 32, 38, 42, 43, 45, 48, 52, 58, 59) | 14 | 4.42 (88.42) | ✓ | Accept | |
| Bilateral (4, 25, 26, 29, 31, 32, 38, 42, 43, 45, 48, 52, 58, 59) | 14 | 4.42 (88.42) | ✓ | Accept | |
| Lesion morphology | |||||
| Patchy / nodular (25, 32, 45, 47, 48, 52, 58, 59) | 8 | 4.42 (88.42) | ✓ | Accept | |
| Spherical (25, 32, 45, 47, 48, 52, 58, 59) | 8 | 3.89 (77.9) | ✓ | Accept | |
| Both (25, 32, 45, 58, 59) | 5 | 4.10 (82.10) | ✓ | Accept | |
| No lesion (32, 45, 47, 52, 58, 59) | 6 | 3.68 (73.69) | 3.95 (78.95) | Accept | |
| C. Paraclinical Data | |||||
| Lesion staging | |||||
| Early (30, 32, 45, 52) | 4 | 3.84 (76.84) | ✓ | Accept | |
| Progressing (30, 32, 45, 52) | 4 | 4.11 (82.10) | ✓ | Accept | |
| Severe (30, 32, 45, 52) | 4 | 3.89 (77.9) | ✓ | Accept | |
| Lesion location | |||||
| Peripheral (2, 25, 32, 41, 45, 58, 59) | 7 | 3.84 (76.84) | ✓ | Accept | |
| Central (2, 25, 32, 41, 45, 58) | 6 | 3.79 (75.8) | ✓ | Accept | |
| Both (2, 25, 32, 58, 59) | 5 | 4 (80) | ✓ | Accept | |
| Random / diffuse (2, 32, 45, 58) | 4 | 3.79 (75.8) | ✓ | Accept | |
| Involved lobe | |||||
| Right upper lobe (2, 25, 32, 42, 45, 58, 59) | 7 | 4.32 (86.31) | ✓ | Accept | |
| Right middle lobe (2, 25, 32, 42, 45, 58, 59) | 7 | 4.05 (81.05) | ✓ | Accept | |
| Right lower lobe (2, 25, 32, 42, 45, 58, 59) | 7 | 4.05 (81.05) | ✓ | Accept | |
| Left upper lobe (2, 25, 32, 42, 45, 58, 59) | 7 | 3.95 (79) | ✓ | Accept | |
| Left lower lobe (2, 25, 32, 42, 45, 58, 59) | 7 | 4.21 (84.21) | ✓ | Accept | |
| Peripheral/central (2, 25, 32, 41, 42, 45, 58, 59) | 8 | 3.89 (77.9) | ✓ | Accept | |
Finally, 149 primary data elements were included in the Delphi survey, of which 112 data elements were finalized in the first round and 15 were rejected. A total of 22 data elements progressed to the second round of the Delphi survey. Of them, 9 were approved in round 2. Thus, on completion of the survey, 125 data elements were approved. Accordingly, the final data elements for epidemiological, clinical, and paraclinical categories were 22, 57, and 46, respectively.
Discussion
This study reports the basic required data items originally derived from studying the COVID-19 patients’ medical records, existing official data sets, and through conducting a systematic literature review, and Delphi survey. The aim of this study was to identify a set of parameters believed to be essential and sufficient to assist the uniform reporting of data on COVID-19. Through the designed COVID-19 MBDS, it is possible to meet some of the data requirements regarding care practice, leading to reliable framework on which health care experts can base their documentation. These elements give both the clinicians and researchers high-quality data to support diagnosis and analysis, respectively. The resulting MBDS is therefore more likely to be acceptable and practical in clinical practice and biomedical research. It has also the potential to homogenize data capturing among public and medical information systems, so that clinical data on COVID-19 can be merged and compared. In addition, data exchange and interoperability can be enhanced using a proper and reliable data set (60). Development of a required data set is the most fundamental step for construction of any information system in the health care sector. Determining these data elements based on viewpoints and real requirements of their customers or users can help designers and vendors of information systems to facilitate and accelerate the development of such systems and reduce the possibility of their failure (61). Thus, the MBDS established in this study can be used as a basis for developing different information systems for collection and management of COVID-19 data.
In the context of COVID-19, huge volumes of data are generated every day in clinical and public health domains. In such big data area, what can be collected is not an issue; rather attention should be paid to the depth and statistical power of collected data to confirm or disprove a hypothesis, and answer specific questions (62, 63). The anticipated hypothesis and questions to be addressed by a health information system or clinical registry should determine the data items that are preferred, and resource accessibility should inform the scope of the data collected to respond to the expected queries. Part of the problem can be due to lack of comparable data derived from limited sharing, unstructured reporting, and lack of standardized data capture strategies (64, 65). To resolve this, new advances in data collection instruments improve the fundability, accessibility, interoperability and reusability (FAIR) of data, highlighting the need for uniform data that can be integrated from different fragmented resources (66-69). In this regard, the anticipated benefits of the COVID-19 MBDS for investigators can include accelerating study initiation, facilitating data exchange and accumulation, and good data management to reach FAIR data. The COVID-19 MBDS aims to facilitate FAIR data collection from COVID-19 individuals with the context of care, evaluation, and research to improve the comparability of data, interdisciplinary communication, and collaboration within the field of COVID-19.
For developing this MBDS, we performed an extensive literature review to identify COVID-19 variables from an evidence-based perspective in a multiresearch study. Then, a 2-round Delphi methodology narrowed down opinions until consensus was reached, during which parameters that may have importance for some applications were excluded from consideration.
This study reported the development of the first MDS-COVID-19 based on state-of-the-art evidence as well as consultation with future users (experts and clinicians). This method could contribute to establishing a balance between scientific theoretical knowledge and technical knowledge as well as applied wisdom from clinical practice to inform the data set. The resulting MBDS is therefore more possible to be satisfactory and practical in clinical practice. We identified the variables required to analyze fundamental aspects, such as transmission patterns, severity, clinical phenotype, prognostic factors, the effectiveness of therapeutic plans and complications, survival estimation, as well as incidence and prevalence of disease across the country.
The literature review only incorporated the search published in the first 4 months of COVID-19 disease during the review period. A more systematic review may have identified additional relevant studies. However, given that the literature review is aimed to identify potential items for inclusion in the MBDS (rather than identifying every paper that considered COVID-19 parameters), and we drew on the collective wisdom of experts in the COVID-19 field throughout the consensus process, it seems unlikely that any important aspects of COVID-19 would have remained overlooked.
Conclusion
To conclude, the developed MBDS used structured agreement methods that integrated a literature review and expert opinion to consolidate COVID-19 documentation, research, and practice. Data collection in line with the configuration presented in this MBDS contributes to unified reporting, probably leading to improved quality of patient documentation, augmented continuity of care, and improved health outcomes regarding COVID-19. COVID-19 MBDS is not proposed to be inclusive; it is what the consulted professionals arbitrated to be a manageable, minimal, and essential set that would ideally be provided in all COVID-19-related research studies. This core set can be augmented in each particular project according to the project’s purpose and available resources. Future testing in other health care settings is recommended. In the future, further strategies, including a comprehensive search of the literature, should be considered to enhance this MBDS.
Acknowledgments
This article is extracted from a research project supported by Abadan Faculty of Medical Sciences (IR.ABADANUMS.REC.1399.021). We also thank the Research Deputy of Abadan faculty of Medical Sciences for financially supporting this project. We also would like to thank all experts who participated in this study and played a role in the validation of the data elements.
Conflict of Interests
The authors declare that they have no competing interests.
Cite this article as: Shanbehzadeh M, Kazemi-Arpanahi H. Development of minimal basic data set to report COVID-19. Med J Islam Repub Iran. 2020 (1 Sep);34:111. https://doi.org/10.34171/mjiri.34.111
Footnotes
Conflicts of Interest: None declared
Funding: This article is extracted from a research project supported by Abadan Faculty of Medical Sciences (IR.ABADANUMS.REC.1399.021). We also thank the Research Deputy of Abadan faculty of Medical Sciences for financially supporting this project
References
- 1.Peeri NC, Shrestha N, Rahman MS, Zaki R, Tan Z, Bibi S. et al. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned. Int J Epidemiol. 2020 doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C. et al. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J. et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W, Cao Q, Qin L, Wang X, Cheng Z, Pan A. et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): A multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz MP, Santos E, Cervantes MV, Juárez ML. COVID-19, a worldwide public health emergency. Rev Clin Esp.(English Edition) . 2020 [Google Scholar]
- 6.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX. et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Benna S. Availability of COVID-19 Information from National and International Aesthetic Surgery Society Websites. Aesthetic Plast Surg. 2020:1. doi: 10.1007/s00266-020-01751-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idrovo AJ, Manrique-Hernández EF. Data Quality of Chinese Surveillance of COVID-19: Objective Analysis Based on WHO’s Situation Reports. Asia Pac J Public Health. 2020 doi: 10.1177/1010539520927265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu B, Gutierrez B, Mekaru S, Sewalk K, Goodwin L, Loskill A. et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci Data. 2020;7(1):1–6. doi: 10.1038/s41597-020-0448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vokinger KN, Nittas V, Witt CM, Fabrikant SI, von Wyl V. Digital health and the COVID-19 epidemic: an assessment framework for apps from an epidemiological and legal perspective. Swiss Med Wkly. 2020;150(1920) doi: 10.4414/smw.2020.20282. [DOI] [PubMed] [Google Scholar]
- 11.Gong M, Liu L, Sun X, Yang Y, Wang S, Zhu H. Cloud-Based System for Effective Surveillance and Control of COVID-19: Useful Experiences From Hubei, China. J Med Internet Res. 2020;22(4):e18948. doi: 10.2196/18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grange ES, Neil EJ, Stoffel M, Singh AP, Tseng E, Resco-Summers K. et al. Responding to COVID-19: The UW Medicine Information Technology Services Experience. Appl Clin Inform. 2020;11(02):265–75. doi: 10.1055/s-0040-1709715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javanmard SH, Nasirian M, Ataei B, Vaseghi G, Vaezi A, Changiz T. Isfahan COvid-19 REgistry (I-CORE): Design and methodology. J Res Med Sci. 2020;25(1):32. doi: 10.4103/jrms.JRMS_271_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talebpour M, Hadadi A, Oraii A, Ashraf H. Rationale and Design of a Registry in a Referral and Educational Medical Center in Tehran, Iran: Sina Hospital Covid-19 Registry (SHCo-19R) Adv J Emerg Med. 2020;4(2s) [Google Scholar]
- 15.Mounesan L, Eybpoosh S, Haghdoost A, Moradi G, Mostafavi E. Is reporting many cases of COVID-19 in Iran due to strength or weakness of Iran’s health system? Iran J Microbiol. 2020;12(2):73–6. [PMC free article] [PubMed] [Google Scholar]
- 16.Zareie B, Roshani A, Mansournia MA, Rasouli MA, Moradi G. A model for COVID-19 prediction in Iran based on China parameters. medRxiv. 2020 doi: 10.34172/aim.2020.05. [DOI] [PubMed] [Google Scholar]
- 17.Kazemi-Arpanahi H, Moulaei K, Shanbehzadeh M. Design and development of a web-based registry for Coronavirus (COVID-19) disease. Med J Islam Repub Iran. 2020;34(1):482–90. doi: 10.34171/mjiri.34.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Wang L, Guo S, Xie N, Yao L, Cao Y. et al. The Data set for Patient Information Based Algorithm to Predict Mortality Cause by COVID-19. Data Brief. 2020:105619. doi: 10.1016/j.dib.2020.105619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashofteh A, Bravo JM. A study on the quality of novel coronavirus (COVID-19) official datasets. Stat J IAO. (Preprint):1–11. [Google Scholar]
- 20.Shanbehzadeh M, Ahmadi M. Identification of the necessary data elements to report AIDS: A systematic review. Electron Physician. 2017;9(12):5920. doi: 10.19082/5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazemi-Arpanahi H, Vasheghani-Farahani A, Baradaran A, Mohammadzadeh N, Ghazisaeedi M. Developing a Minimum Data Set (MDS) for Cardiac Electronic Implantable Devices Implantation. Acta Inform Med. 2018;26(3):164–8. doi: 10.5455/aim.2018.26.164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazemi-Arpanahi H, Vasheghani-Farahani A, Baradaran A, Ghazisaeedi M, Mohammadzadeh N, Bostan H. Development of a minimum data set for cardiac electrophysiology study ablation. J Educ Health Promot. 2019:8. doi: 10.4103/jehp.jehp_232_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varghese J, Holz C, Neuhaus P, Bernardi M, Boehm A, Ganser A. et al. editors Key Data Elements in Myeloid Leukemia. MIE. 2016 [PubMed] [Google Scholar]
- 24.Davis J, Morgans A, Burgess S. Information management for aged care provision in Australia: development of an aged care minimum dataset and strategies to improve quality and continuity of care. Health Inf Manag. 2016;45(1):27–35. doi: 10.1177/1833358316639453. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Sun Y, Duong TQ, Nguyen LD, Hanzo L. Risk-Aware Identification of Highly Suspected COVID-19 Cases in Social IoT: A Joint Graph Theory and Reinforcement Learning Approach. IEEE Access. 2020;8:115655–61. doi: 10.1109/ACCESS.2020.3003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Duan Y, Zhang W, Liang J, Xu J, Zhang Y. et al. Epidemiologic and Clinical Characteristics of 26 Cases of COVID-19 Arising from Patient-to-Patient Transmission in Liaocheng, China. Clin Epidemiol. 2020;12:387. doi: 10.2147/CLEP.S249903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Zou R, Zeng L, Kou S, Lan J, Li X. et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm Res. 2020:1–8. doi: 10.1007/s00011-020-01342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surveillances V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly. 2020;2(8):113–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu T, Chen C, Zhu Z, Cui M, Chen C, Dai H. et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP. et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med Infect Dis. 2020:101623. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Wu J, Wu F, Guo D, Chen L, Fang Z. et al. The Clinical and Chest CT Features Associated with Severe and Critical COVID-19 Pneumonia. Invest Radiol. 2020 doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YK, Peng S, Li LQ, Wang Q, Ping W, Zhang N. et al. Clinical and Transmission Characteristics of Covid-19—A Retrospective Study of 25 Cases from a Single Thoracic Surgery Department. Curr Med Sci. 2020:1–6. doi: 10.1007/s11596-020-2176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y. et al. Clinical Features and Treatment of COVID‐19 Patients in Northeast Chongqing. J Med Virol. 2020 doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia J, Hu X, Yang F, Song X, Dong L, Zhang J. et al. Epidemiological characteristics on the clustering nature of COVID-19 in Qingdao City, 2020: a descriptive analysis. Disaster Med Public Health Prep. 2020:1–17. doi: 10.1017/dmp.2020.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia XY, Wu J, Liu HL, Xia H, Jia B, Huang WX. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J Clin Virol. 2020:104360. doi: 10.1016/j.jcv.2020.104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Zhang ZZ, Chen YK, Long QX, Tian WG, Deng HJ. et al. The clinical and immunological features of pediatric COVID-19 patients in China. Genes Dis. 2020 doi: 10.1016/j.gendis.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, Chen Y, Lin R, Han K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Zhou H, Yang R, Xu Y, Feng X, Gong P. Clinical characteristics of 36 non-survivors with COVID-19 in Wuhan, China. medRxiv. 2020 [Google Scholar]
- 40.Tabata S, Imai K, Kawano S, Ikeda M, Kodama T, Miyoshi K. et al. The clinical characteristics of COVID-19: a retrospective analysis of 104 patients from the outbreak on board the Diamond Princess cruise ship in Japan. medRxiv. 2020 doi: 10.1016/S1473-3099(20)30482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyu P, Liu X, Zhang R, Shi L, Gao J. The performance of chest CT in evaluating the clinical severity of COVID-19 pneumonia: identifying critical cases based on CT characteristics. Invest Radiol. 2020 doi: 10.1097/RLI.0000000000000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu H, Xu H, Zhang N, Xu H, Li Z, Chen H. et al. Association between Clinical, Laboratory and CT Characteristics and RT-PCR Results in the Follow-up of COVID-19 patients. medRxiv. 2020 [Google Scholar]
- 43.Pormohammad A, Ghorbani S, Baradaran B, Khatam A, Turner R, Mansournia MA. et al. Clinical Characteristics, Laboratory Findings, Radiographic Signs and Outcomes of 52,251 Patients with Confirmed COVID-19 Infection: A Systematic Review and Meta-Analysis. Microb Pathog. 2020 doi: 10.1016/j.micpath.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, Lin W, Wen J, Chen G. Clinical and pathological characteristics of 2019 novel coronavirus disease (COVID-19): a systematic reviews. medRxiv. 2020 [Google Scholar]
- 45.Lomoro P, Verde F, Zerboni F, Simonetti I, Borghi C, Fachinetti C. et al. COVID-19 pneumonia manifestations atthe admission on chest ultrasound, radiographs, and CT: single-center study and comprehensive radiologic literature review. Eur J Radiol Open. 2020:100231. doi: 10.1016/j.ejro.2020.100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ. et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang TH, Wu JL, Chang LY. Clinical characteristics and diagnostic challenges of pediatric COVID-19: A systematic review and meta-analysis. J Formos Med Assoc. 2020 doi: 10.1016/j.jfma.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020;92(6):568–76. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hassan S, Sheikh FN, Jamal S, Ezeh JK, Akhtar A. Coronavirus (COVID-19): a review of clinical features, diagnosis, and treatment. Cureus. 2020;12(3) doi: 10.7759/cureus.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siordia Jr JA. Epidemiology and Clinical Features of COVID-19: A Review of Current Literature. J Clin Virol. 2020:104357. doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J. et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol. 2020:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu L, Wang B, Yuan T, Chen X, Ao Y, Fitzpatrick T. et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: a systematic review and meta-analysis. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lovato A, de Filippis C. Clinical Presentation of COVID-19: A Systematic Review Focusing on Upper Airway Symptoms. Entechnology. 2020:0145561320920762. doi: 10.1177/0145561320920762. [DOI] [PubMed] [Google Scholar]
- 54.Zhao X, Zhang B, Li P, Ma C, Gu J, Hou P. et al. Incidence, clinical characteristics and prognostic factor of patients with COVID-19: a systematic review and meta-analysis. medRxiv. 2020 [Google Scholar]
- 55.Fang Z, Yi F, Wu K, Lai K, Sun X, Zhong N. et al. Clinical Characteristics of 2019 Coronavirus Pneumonia (COVID-19): An Updated Systematic Review. medRxiv. 2020 [Google Scholar]
- 56.Zheng Y, Xu H, Yang M, Zeng Y, Chen H, Liu R. et al. Epidemiological Characteristics and Clinical Features of 32 Critical and 67 Noncritical Cases of COVID-19 in Chengdu. J Clin Virol. 2020:104366. doi: 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Fang J, Zhu Y, Chen L, Ding F, Zhou R. et al. Clinical characteristics of non-critically ill patients with novel coronavirus infection (COVID-19) in a Fangcang Hospital. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. CT scans of patients with 2019 novel coronavirus (COVID-19) pneumonia. Theranostics. 2020;10(10):4606. doi: 10.7150/thno.45016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Gao ZH, Liu YL, Xu DY, Guan TM, Li ZP. et al. Clinical and CT imaging features of 2019 novel coronavirus disease (COVID-19) J Infect. 2020 doi: 10.1016/j.jinf.2020.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey CJ, Slade SV, Shickle D. A proposed minimum data set for international primary care optometry: a modified Delphi study. Ophthalmic Physiol Opt. 2017;37(4):428–39. doi: 10.1111/opo.12372. [DOI] [PubMed] [Google Scholar]
- 61.Hashemi N, Sheikhtaheri A, Hashemi N, Rawassizadeh R. Electronic Medical Records for Mental Disorders: What Data Elements Should These Systems Contain? Stud Health Technol Inform. 2019;260:25–32. [PubMed] [Google Scholar]
- 62.Shaghaghi A, Bhopal RS, Sheikh A. Approaches to recruiting ‘hard-to-reach’populations into research: a review of the literature. Health Promot Perspect. 2011;1(2):86. doi: 10.5681/hpp.2011.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadler GR, Lee HC, Lim RSH, Fullerton J. Recruitment of hard‐to‐reach population subgroups via adaptations of the snowball sampling strategy. Nurs Health Sci. 2010;12(3):369–74. doi: 10.1111/j.1442-2018.2010.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lapinlampi N, Melin E, Aronica E, Bankstahl JP, Becker A, Bernard C. et al. Common data elements and data management: Remedy to cure underpowered preclinical studies. Epilepsy Res. 2017;129:87–90. doi: 10.1016/j.eplepsyres.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 65.Grinnon ST, Miller K, Marler JR, Lu Y, Stout A, Odenkirchen J. et al. National institute of neurological disorders and stroke outcome set for hip fracture trials. Bone Joint J. 2014;96(8):1016–23. [Google Scholar]
- 66.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016:3. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haywood K, Griffin X, Achten J, Costa M. Developing a core common data element project–approach and methods. Clin Trials J. 2012;9(3):322–9. doi: 10.1177/1740774512438980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lutomski JE, Baars MAE, Schalk BWM, Boter H, Buurman BM, den Elzen WPJ. et al. The development of the Older Persons and Informal Caregivers Survey Minimum DataSet (TOPICS-MDS): a large-scale data sharing initiative. PLoS One. 2013;8(12):e81673–e. doi: 10.1371/journal.pone.0081673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riley WT, Glasgow RE, Etheredge L, Abernethy AP. Rapid, responsive, relevant (R3) research: a call for a rapid learning health research enterprise. Clin Transl Med. 2013;2(1):10. doi: 10.1186/2001-1326-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


