Abstract
Radial glial cells (RGCs) are progenitors of the cerebral cortex which produce both neurons and glia during development. Given their central role in development, RGC dysfunction can result in diverse neurodevelopmental disorders. RGCs have an elongated bipolar morphology that spans the entire radial width of the cortex and ends in basal endfeet connected to the pia. The basal process and endfeet are important for proper guidance of migrating neurons and are implicated in signaling. However, endfeet must function at a great distance from the cell body. This spatial separation suggests a role for local gene regulation in endfeet. Endfeet contain a local transcriptome enriched for cytoskeletal and signaling factors. These localized mRNAs are actively transported from the cell body and can be locally translated in endfeet. Yet, studies of local gene regulation in RGC endfeet are still in their infancy. Here, we draw comparisons of RGCs with foundational work in anatomically and phylogenetically related cell types, neurons and astrocytes. Our review highlights a striking overlap in the types of RNAs localized, as well as principles of local translation between these three cell types. Thus, studies in neurons, astrocytes and RGCs can mutually inform an understanding of RNA localization across the nervous system.
Keywords: astrocyte, endfeet, local translation, neuron, radial glia, RNA localization
1 |. INTRODUCTION
Humans possess an expanded cerebral cortex relative to other mammals. This feature is thought to contribute to our higher neurological functions. Proper formation of the cortex requires precisely coordinated production and organization of multiple cell types, including neurons and glia. Signaling cues orchestrate the timing of these processes throughout embryonic development. Perturbation of cortical development can result in neurological disorders associated with impairment of cognitive function such as microcephaly, autism spectrum disorder and intellectual disability. Especially important in the production and organization of cells in the developing brain are the radial glial cells (RGCs) which function as progenitors in the ventricular zone (VZ) giving rise to first neurons and then glia.1,2 Neurogenesis begins with symmetric divisions of neuroepithelial cells to expand the progenitor pool (Figure 1). These progenitors are replaced by RGCs which undergo either symmetric or asymmetric divisions in the VZ to produce new RGCs, intermediate progenitors (IPs) and neurons. IPs also directly produce neurons. In higher order species with an expanded neocortex, such as humans and other primates, neurogenesis is driven largely by outer radial glia (oRG or basal radial glia), which are also derived from RGCs.3 Neuron production concludes at the end of embryonic development, at which time RGCs switch their potency and begin to produce glia.
FIGURE 1.
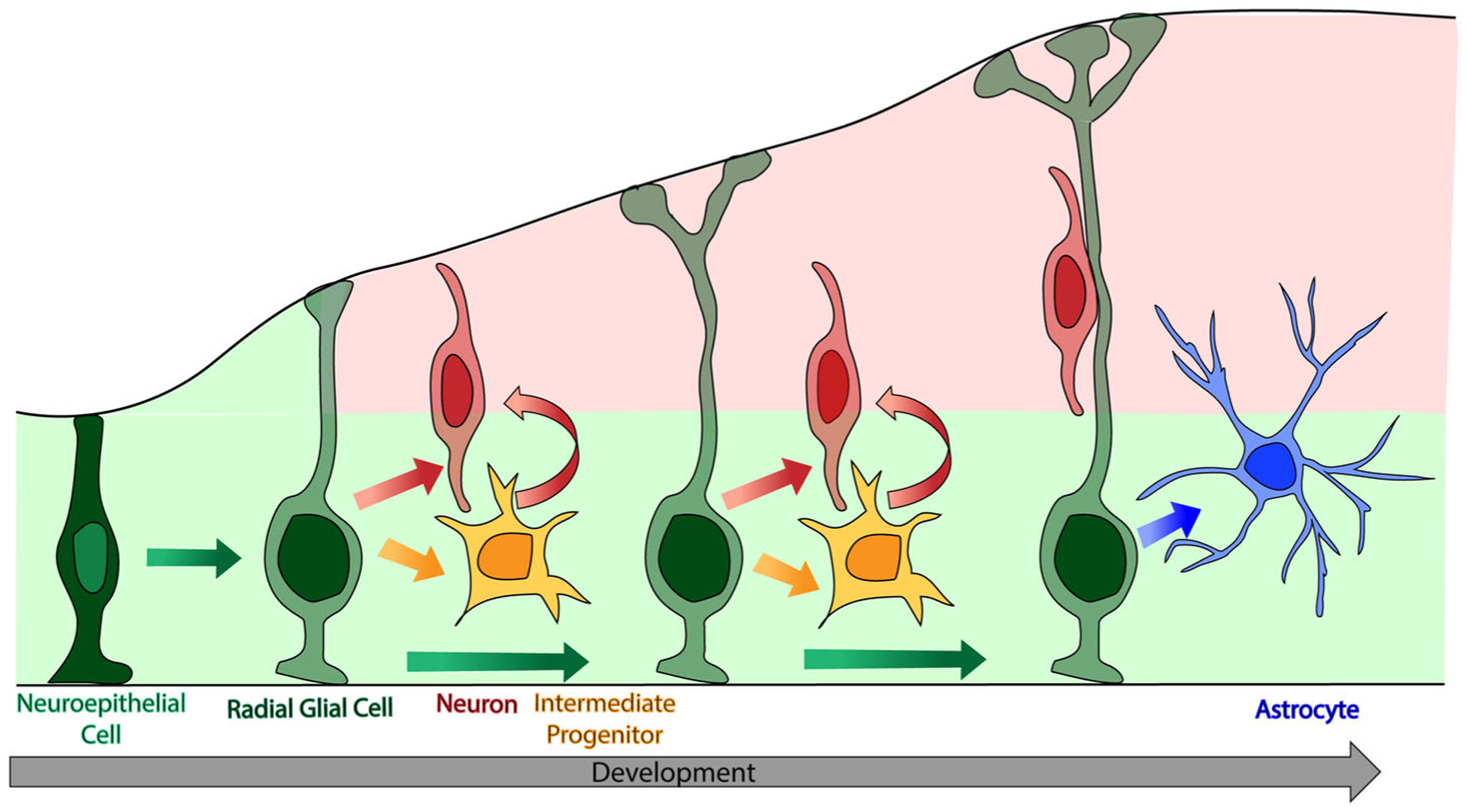
Radial glial cells (RGCs) play a vital role in cortical development. This simplified cartoon reflects neurogenesis in mice, between embryonic day (E) 11.5 and 18.5, followed by gliogenesis. Neuroepithelial cells (light green nucleus) initially expand the precursor pool. RGCs (dark green nucleus) are the main progenitors of the cerebral cortex during development producing both neurons and glia. Early in development RGCs produce neurons (red) either directly or indirectly through their production of intermediate progenitors (yellow). Following the conclusion of neurogenesis, RGCs change potency and produce glia, including astrocytes (blue). As cortical development progresses, RGC morphology matures from having one endfoot to multiple
2 |. RGCs HAVE A COMPLEX, BIPOLAR MORPHOLOGY THAT IS FUNCTIONALLY IMPORTANT
RGCs have a unique morphology which consists of a cell body located close to the ventricle and a long basal process that spans the radial width of the cortex ending in one or more protrusions called basal endfeet. As cortical development progresses, branching of the basal process becomes increasingly complex and RGCs mature from having a single endfoot to multiple endfeet.4 The length of the basal process can range from several hundred micrometers in mice to several centimeters in humans. Importantly, oRGs, the major progenitor in primates, also share similar anatomical features of RGCs, including a basal process and endfeet.5 Lengthening of the basal process goes hand in hand with radial expansion of the cortex over the course of development. This suggests that RGCs (and potentially oRGs) require synthesis of new membrane and cytoskeletal proteins to support cellular growth.
Distant from the ventricle and cell body, the basal endfeet connect to the basement membrane (BM) at the pia.6,7 The endfoot niche is comprised of extracellular matrix proteins including collagens and laminins that make up the BM as well as fibroblasts and blood vessels in the pia.8 The basal processes act as a scaffold which is used by new neurons to migrate from their birthplace in the VZ to their final destination in the cortical plate. The endfeet lining the pia provide a physical barrier between the cortical plate and the BM and ensure neurons do not migrate outside the cortex9,10 (Figures 1 and 2). Genetic models inducing endfoot detachment from the BM demonstrate defects in the continuity of the BM and over-migration of neurons into the pia.9–12 These misplaced neurons form ectopic structures on top of the cortex reminiscent of malformations observed in human cobblestone lissencephaly.
FIGURE 2.
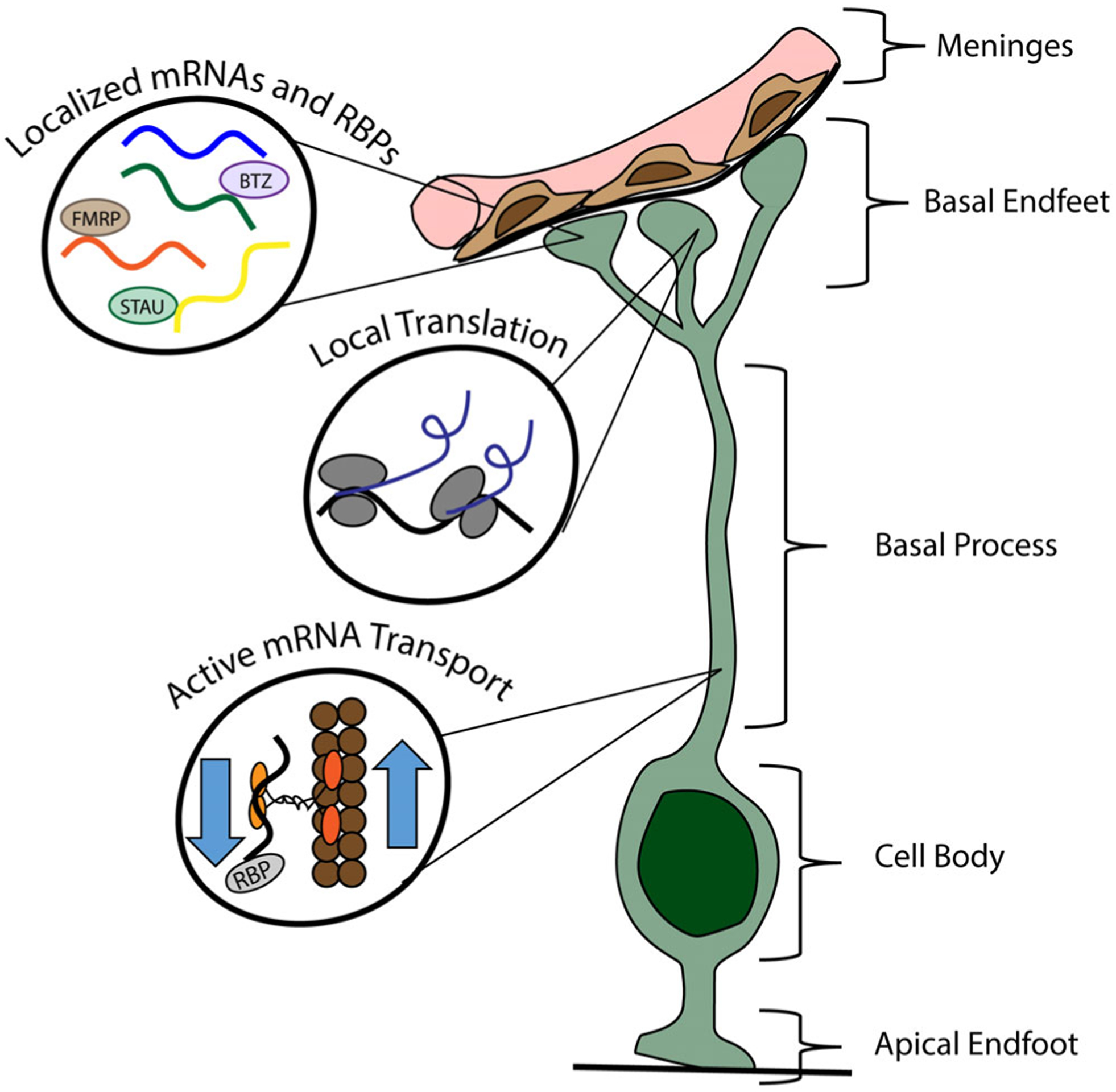
Current knowledge of local gene regulation in radial glial cells (RGCs). Basal endfeet function far from the cell body embedded in a local niche comprised of basement membrane, fibroblasts and blood vessels. Three major discoveries in RGCs have been published: (a) mRNAs are actively transported from the cell body to basal endfeet. (b) Endfeet have a unique local transcriptome. (c) Endfoot localized mRNAs are competent for local translation
Beyond their physical function, additional roles have been putatively attributed to endfeet. By connecting to the BM, endfeet are hypothesized to enable signaling between the RGCs and niche cells that reside in the pia and meninges.13,14 Extrinsic cues from the basal niche, such as retinoic acid, influence RGC behavior.15–18 Thus, it is logical that the endfeet physically embedded in this niche may be responsible for relaying signals. The basal process and endfeet may also contain fate-determining signals. This hypothesis is supported by studies showing that the basal process and endfeet are preferentially inherited by the newborn progenitor following mitosis.19–21 Those daughter cells retaining basal structures are more likely to maintain the stem-like nature of RGCs and undergo proliferative divisions. The importance of endfeet in RGC function and cortical development is clear, but a mechanistic understanding of their role is not. A major step toward filling this gap is expanding our knowledge of local gene regulation in these distal structures.
3 |. LOCAL GENE REGULATION IN RGC ENDFEET
Subcellular local gene regulation is well conserved among many highly polarized cells comprising the central nervous system as it allows for spatial and temporal control. Roles for localized gene regulation in the processes of cell migration and division have also been established in diverse cell types and species, such as fibroblasts and dividing yeast, respectively.22 RGCs are a prime model for studying local gene regulation in the CNS due to the extended distance between the cell body and basal endfeet and the functional need to relay information from the pia to the cell body. Despite this conservation and the strength of RGCs as a model system for RNA localization, the field of local gene regulation in RGCs is still in its infancy. We begin with a discussion of the knowledge in the field thus far.
3.1 |. mRNAs are actively transported from the RGC cell body to basal endfeet
RNA localization to endfeet from the cell body is not limited to passive diffusion. Instead, RNAs can be actively transported in a process regulated by both cis- and trans-factors. Tsunekawa et al were the first to show that cis-elements could drive the localization of reporter mRNAs to endfeet.23 In doing so, they identified the 3′UTR of Cyclin D2 (Ccnd2) as sufficient for endfoot localization, including the minimal cisendfoot localization sequence. Our lab has also shown that the 3′UTR of Kif26a is sufficient to localize its mRNA to endfeet.24 While a handful of cis-localization elements have been determined, it is unclear whether there are consensus motifs for endfoot localization. Additionally, it is unknown if either the nucleotide sequence and/or secondary structure of a transcript mediates its subcellular localization in RGCs.
Trans-factors are also critical for transcript localization. Multiple RNA-binding proteins, including Fragile X mental retardation protein (FMRP), Stau2, APC and Pum, are present in endfeet and thus poised to influence RNA localization in RGCs.24,25 However, to date the only trans-factor shown to localize transcripts to RGC endfeet is FMRP, whose mutation is linked to Fragile X syndrome, a form of autism.26 FMRP is enriched in endfeet where it binds to over 100 RNAs.24 Further investigation using Fmr1 knockout mice revealed that some transcripts normally localized to endfeet, such as Kif26a, fail to localize in the absence of FMRP. In contrast, other transcripts, such as Apc, localize without FMRP. This suggests that beyond FMRP, there are additional trans-factors at play which have transcript specificity. Additionally, beyond RNA-binding proteins, other molecules such as noncoding RNAs may also act as trans-factors for transcript localization to endfeet.
To observe the dynamics of transcript localization, we utilized the MS2 reporter system27 coupled with live imaging of ex vivo brain slices to image mRNA movement in real time.24 Dynamics of mRNA transport were characterized using previously published Ccnd2 reporters23 containing the localization element. Ccnd2 reporter transcripts moved at speeds of about 2 μm/s during developmental stages from E14.5 to E16.5, similar to that seen in other systems, including neurons, and suggesting a microtubule-based mechanism of transport.24 Transcripts were actively transported in both the apical and basal directions and this directionality shifted over the course of development. For example, at E14.5 only about 15% of observed movements were toward the apical direction and the majority were basally directed, whereas at E16.5 this bias was lost. One caveat of this methodology is that the MS2 system relies on overexpression of reporters which could lead to artifacts and results lacking physiological relevance.28 However, an MS2 reporter without the localization element was used as a negative control and did not exhibit directed motility. The mechanism and relevance of directional transport in RGCs is unknown, although it could be linked to differences in progenitor potency and/or cytoskeletal organization.
3.2 |. Characterization of RGC endfoot localized transcripts
Important to our understanding of mRNA localization in RGCs is knowledge of which transcripts are present in endfeet. Nestin, Abba and Ccnd2 were among the first transcripts observed to localize to endfeet through traditional in situ hybridization (ISH) in mouse brain sections.23,29,30 Our lab expanded upon these findings by identifying 115 transcripts that are localized to endfeet.24 This was made possible by localization of the RNA-binding protein, FMRP, to endfeet. EGFP-FMRP was introduced into RGCs by in utero electroporation and endfeet preparations (containing BM, endfeet and older neurons) were collected 24 hours later by mechanical isolation. Importantly, this strategy ensured that EGFP-FMRP expression was restricted to RGCs, and was not in newborn neurons. Using RNA immunoprecipitation of EGFP-FMRP followed by microarray (RIP-Chip), 115 transcripts bound to FMRP specifically in endfeet were isolated and identified. Endfeet were enriched for several classes of transcripts, including those related to cytoskeletal dynamics and signaling. One limitation of these findings is they only identified FMRP-bound transcripts in endfeet. It is likely that there are additional transcripts present in endfeet which are not FMRP-bound. For example, Ccnd2 was not a high-affinity FMRP target despite being endfoot localized.23 Therefore, it is of interest to determine whether the class enrichment observed is representative of the entire endfoot transcriptome or simply the FMRP interactome. To address this, global transcriptome analysis of endfeet is required.
3.3 |. Localized transcripts are locally translated in RGC endfeet
In a 1991 study of brain development in embryonic rats, Astrom and Webster reported the presence of ribosomes and other translational machinery in basal endfeet suggesting their competence for translation.31 Twenty-one years later, Tsunekawa et al performed initial investigations into the competency of endfeet for local translation.23 Their experimental paradigm was designed using an EGFP reporter containing a nuclear localization signal (NLS-EGFP) with the assumption that protein produced in the cell body would be immediately transported to the nucleus while distally translated protein would not. In this way, they attempted to circumvent the possibility that protein trafficked from the cell body to endfeet would be mistaken for local translation. Inclusion of the Ccnd2 3′UTR within an EGFP reporter forced the localization to endfeet. Introduction of the reporter into RGCs in vivo revealed expression of EGFP in endfeet far from the cell body, hinting at the possibility of local translation. However, this study did not physically separate the endfeet from the cell body so the possibility of protein trafficking or diffusing from the cell body and/or basal process remained.
Our lab further advanced this question using ex vivo time-lapse imaging to directly observe active translation in endfeet in real time.24 To achieve this, the photoconvertable reporter Dendra2 with a Ccnd2 endfoot localization sequence was introduced into embryonic mouse brains and then endfeet preparations were generated by mechanical separation. It is important to note that by completely separating the endfeet from the cell body this eliminates the possibility of newly observed protein being translated elsewhere and then trafficked to the endfeet. Dendra2 protein already present in endfeet was irreversibly photoconverted from green to red allowing for the detection of new green protein, which serves as a proxy for de novo protein synthesis. Time-lapse imaging of ex vivo endfoot preparations revealed that active translation does in fact occur but is not uniform across endfeet. Throughout the 45-minute imaging window, some endfeet recovered up to 75% of their initial green fluorescence intensity, while others showed no signs of active translation with a Ccnd2 reporter. Interestingly, even though the majority of endfeet in these preparations contained markers of translation machinery, such as ribosomal RNA and RPS6, both the proportion of translationally active endfeet and the percentage of protein recovery in each endfoot varied across different reporters. This suggests that local translation in endfeet is a highly regulated process. It begs the questions of how this process is controlled, and which transcripts are translated in endfeet.
3.4 |. Putative functions of locally produced gene products in RGCs
Our understanding of the mechanism of local gene regulation in RGC endfeet and the transcripts and proteins involved is steadily increasing; however, the functional implications of RNA localization in endfeet remain unknown. There are two overarching hypotheses for the function of gene products localized to endfeet. First, locally produced proteins function within endfeet to support spatially relevant processes such as morphology and signaling. Second, proteins produced in endfeet may be stored there and then trafficked back to function in the cell body. It is important to note that these need not be mutually exclusive. We can begin to evaluate these hypotheses based upon the functional gene classes enriched in endfeet, as identified through FMRP RIP-Chip and GO analysis. The most highly enriched gene classes, based on these data, were cytoskeletal regulation and cell signaling.24 As genes belonging to these classes may promote RGC morphology and relay intracellular and/or extracellular signals back to the cell body, these data imply a local function. Supporting this notion, we recently discovered a Rho GTPase regulator whose local translation in endfeet is critical for their morphology and further for cortical organization.32 Intriguingly, chromatin regulatory factors and other genes with canonical nuclear functions were also identified as FMRP targets in endfeet. It is possible such factors function in noncanonical roles in the basal endfeet. However, their presence also lends support for a model in which locally translated proteins are trafficked back down the basal process to function in the cell body. In the context of an asymmetrically dividing cell, this would allow RGCs to faithfully segregate “stem cell” components far away, so that they are not inherited by a neuron. Indeed, Tsunekawa et al put forth this hypothesis when they suggested a role for endfoot localized Ccnd2 in regulating cell cycle and cell fate of RGCs during cortical development.23,33 Going forward, it is a high priority to understand in which cellular compartments locally produced proteins act, and how this influences RGCs.
3.5 |. Unique challenges of studying RNA localization in RGCs
The studies discussed above only begin to scratch the surface of understanding the mechanisms and functions of local gene regulation in RGCs. There are technical challenges that must be overcome in order to broaden our understanding of subcellular gene expression in RGC endfeet. First and foremost, RGCs do not readily grow endfeet in cell culture prohibiting use of in vitro assays. This requires all experiments to be performed in vivo, adding technical complexity. Further, endfeet are closely juxtaposed to the BM and meninges; these produce strong autofluorescence which can hinder imaging capabilities in cortical tissue sections. Additionally, no specific endfoot marker has yet been determined requiring most experiments to rely on in utero electroporation to label and image these structures. Furthermore, endfeet in the developing mouse brain range from about 5 to 10 μm in diameter. As a result, individual endfeet contain small amounts of RNA and protein, decreasing the feasibility of biochemical experiments which require abundant material.
As the field works to circumvent these technical challenges, we may look to work done in related cell types for inspiration to better understand potential functions of subcellular gene regulation in RGCs. Toward this, the most relevant cell types are neurons and astrocytes (Figure 3A). Both have long cellular processes in which local gene regulation has been studied in depth. RGCs produce both neurons and astrocytes during cortical development and all three cell types function in the cortex. Below we will discuss the lessons gained from RNA localization studies of neurons and astrocytes. Probing available data, we discuss overlap between transcripts localized in RGC endfeet, astrocyte processes, neuronal axons and synapses. Further, we highlight exciting new discoveries pertaining to the mechanisms and functions of local gene regulation in neurons and astrocytes that may be conserved in RGCs.
FIGURE 3.
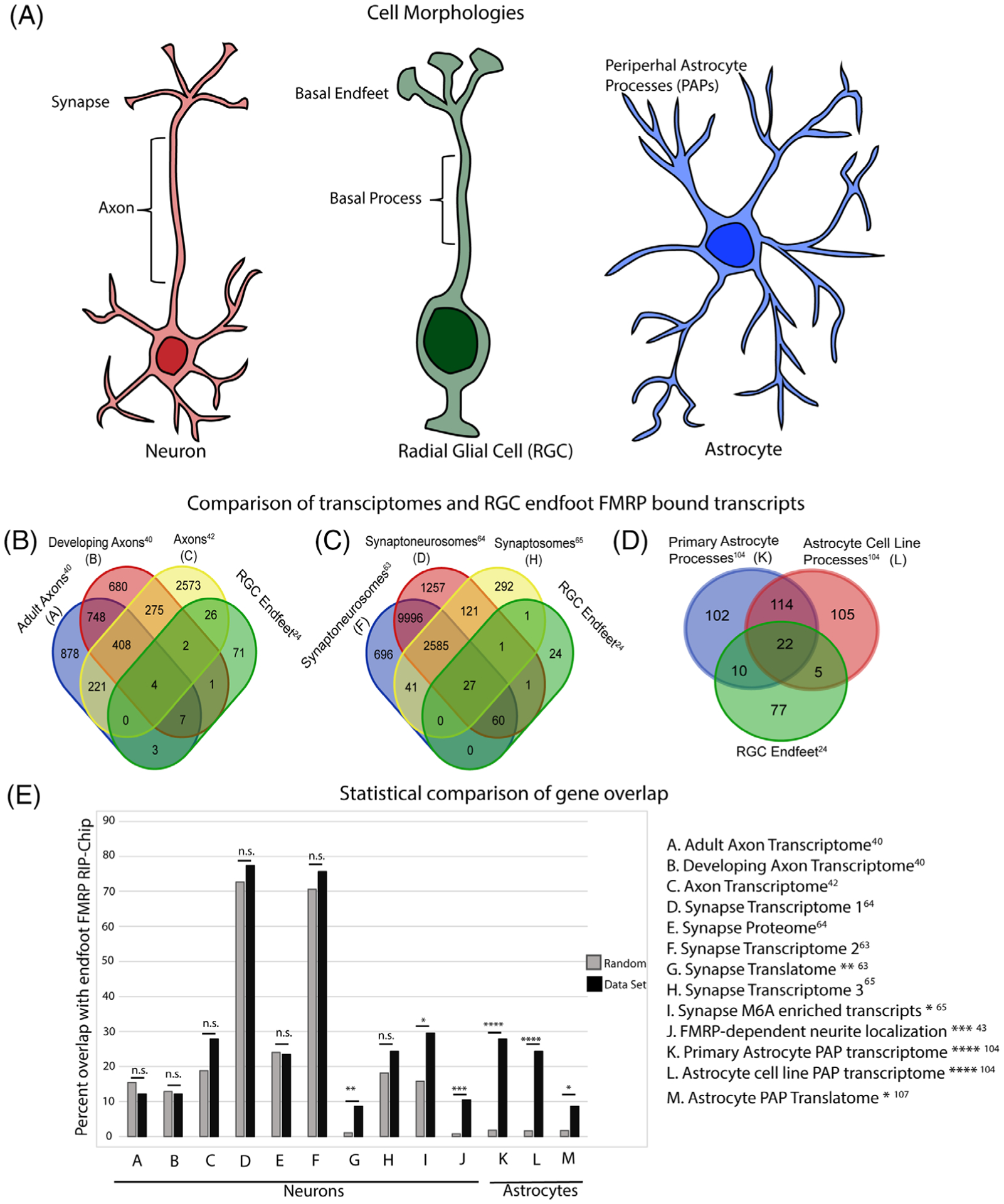
Comparison of radial glial cell (RGC) endfeet, neuronal axons, neuronal synapses and peripheral astrocyte processes. (A) Cartoon representations of neurons, RGCs and astrocytes to depict their unique morphology and emphasize subcellular compartments distant from the cell body. (B-D) Venn diagrams show the comparison of endfoot localized transcripts to mRNAs localized in neuronal axons (B), neuronal synapses (C) and peripheral astrocyte processes (D). (E) Graphical representation of the percent of genes shared between the FMRP RIP-Chip in endfeet and relevant localization datasets compared to a random gene list. Random gene lists were generated through Bootstrap sampling of all the genes present in the FMRP RIP-Chip input. For each comparison, 100 random lists were generated and the mean overlap is represented. P-values were calculated using a two-tailed Fisher’s exact test. *<.05, **<.01, ***<.001, ****<.0001
4 |. RNA LOCALIZATION AND LOCAL TRANSLATION IN NEURONS
Proper positioning and morphology of neurons are vital for brain function as they process and transmit information. This function relies on the highly complex and bipolar morphology of neurons which allows for compartmentalization of signaling.34,35 With a long axon and multiple shorter dendrites extending from the cell body, neurons can receive and initiate signals at abundant locations distant from each other and interact with multiple cells simultaneously. Axons extend from the cell body to relay information at a distance. For example, the axons that make up the corpus callosum stretch from one side of the cortex to the other to connect both hemispheres. To achieve this, during development the axon grows and responds to external cues for successful pathfinding. Axons and dendrites have small highly plastic structures called synapses, which are junctions between two neurons composed of presynaptic and postsynaptic compartments. Synapses transmit chemical and electrical signals between cells allowing for intercellular communication and circuit formation.
The complex morphology and requirement for compartmentalized functions suggests the need for local gene regulation. Indeed, mRNA localization and local translation in both axons and synapses have been studied for decades.36,37 Cumulative studies have produced a wealth of knowledge regarding which transcripts are localized and locally translated, cis- and trans-factors involved in mRNA transport and signals which regulate local gene regulation. These findings have been comprehensively reviewed recently.34,35 Below, we outline key results that have potential to inform our understanding of RGC endfeet, starting first with a discussion of localized transcripts in axons, then local translation in axons, and finally a discussion of the same processes in synapses.
4.1 |. Localized transcripts in neuronal axons
Numerous studies have characterized transcripts localized to axons across broad neuronal types. Here, we focus on a handful of studies that represent diverse neuronal cell states including in vivo, in vitro, injury and development.
mRNA localization and local translation support regeneration of injured axons in Caenorhabditis elegans. In this system, several genes, including Dlk-1 and Cebp1, were identified as regulators of axon regeneration.38 To obtain a more complete picture of localized regeneration factors in mammalian axons, Taylor et al separated the axons from cell bodies of in vitro dorsal root ganglion neurons (DRGs) for analysis by microarray.39 The authors then tested the hypothesis that injured axons are more likely to contain transcripts important for regeneration compared to uninjured axons. Of the mRNAs identified, two transcripts from the uninjured axons and three from the injured axons were also present in RGC endfeet (not shown). These overlaps represent 1.7% and 2.6% of the FMRP-bound endfoot transcripts, respectively, and are not statistically significant over a random comparison.
Gumy et al compared mRNAs localized to axons in developing and adult rat neurons.40 Using a two-chamber system developed by Vogelaar et al to culture DRG in vitro they isolated neuronal axons and then identified mRNAs by microarray.40,41 This comparison revealed enrichment of unique gene classes at each stage. For example, mRNAs involved in cell cycle, transport and cytoskeletal organization were enriched in the developing axons. Meanwhile, transcripts with roles in inflammation and immune response were enriched in adult axons. These observations suggest that developmental stage is a factor influencing which mRNAs are transported to axons.
It is interesting to consider how this finding may be applicable to RGCs. As development progresses, RGC morphology becomes increasingly complex and their potency evolves from producing neurons to astrocytes.1,2,4 Localized mRNAs may contribute to these changes; thus, by characterizing transcriptomes of endfeet at different developmental stages, one can increase understanding of these dynamic and important processes. To investigate whether the composition of RGC endfeet are more like that of developing or adult axons, we compared the developing and adult transcriptomes with our RGC endfoot RIP-Chip data24,40 (Figure 3B). Fourteen transcripts are enriched in both developing axons and RGC endfeet, representing a 12.2% overlap (Figure 3B,E). Comparison of adult axons and RGC endfeet also yielded 14 transcripts (12.2%) in common. Both comparisons are not statistically significant compared to random (Figure 3E), suggesting that this particular axonal transcriptome is not representative of the FMRP endfoot transcriptome. Comparisons with a total endfoot transcriptome may yield a different answer. However, it is notable that in total 11 transcripts were conserved between adult axons, developing axons and RGC endfeet. This observation begs the question of whether there are staple localized transcripts which universally influence the maintenance of highly bipolar and elongated morphologies shared by neurons and RGCs.
Further expanding the list of axonal transcripts, Cajigas et al performed deep sequencing on mRNAs isolated from hippocampal neuropils in vivo and identified 2550 neuronal transcripts.42 This is a significant increase from the approximately 300 transcripts characterized by microarray.39,40 It is important to note that this study reported transcripts present in the neuropil but not necessarily enriched there; 27.8% of transcripts (total = 32) are shared between the neuropils and RGC endfeet, which was not quite significant (P = .16) (Figure 3B,E). However, the overlap between RGCs and neuropil was higher than that of isolated developing and adult neurons. It is possible that the in vivo external neuronal environment is more similar to that of RGC endfeet in vivo, conferring common gene expression changes. Technical differences between microarray and deep sequencing could be responsible for observed differences using both axonal datasets. Finally, a caveat to note, the isolated neuropils contain both axons and dendrites unlike the purer axonal populations of in vitro systems. Therefore, presence of dendritic transcripts is a possible explanation for increased similarity in localized transcripts. Regardless, these comparisons suggest there is localization of conserved mRNAs in both neuronal and RGC processes.
A recent study examined the composition of mRNAs which depend upon FMRP for their localization to neurites, the immature state of an axon or dendrite. Goering et al used CAD cells, a mouse immortalized neuronal cell line, that were either WT or lacked FMRP.43 Using a Boyden assay they isolated neurites and cell bodies, discovering a subset of transcripts which are FMRP-dependent in their localization. The Boyden chamber consists of two isolated compartments, in which culture conditions can be unique, separated by a microporous membrane. Classically used to study invasion potential of cells, it is also valuable to physically segregate cellular structures of neurons and fibroblast protrusions.44,45 Among transcripts identified, 12 are also present in RGC endfeet, representing a statistically significant 10.4% of all FMRP targets in endfeet (Figure 3E). The authors also identified domains in FMRP that mediate this localization, separate from translation. It would be interesting to investigate whether there are subsets of FMRP targets in endfeet that differentially interact with distinct FMRP domains.
4.2 |. Local translation in axons
Local translation in axons has been observed in model systems ranging from Xenopus to mammals. Roles for spatially targeted and rapid translation, including cytoskeletal proteins, have been reported in axon pathfinding and axon regeneration.46–50 In vitro, axons physically severed from their somas are still capable of reacting to external stimuli in a protein synthesis-dependent manner.51 Interestingly, local translation is not required for axon extension even though additional proteins are needed to lengthen the neuronal process.51–53 This foundational work established the presence and function of local translation in axons. However, it also sparked additional questions including which transcripts are translated in axons and how is local translation spatially regulated?
To tackle the first question, Biever et al used a ribosome footprinting approach to produce an unbiased list of mRNAs undergoing active translation in neuronal processes in vivo.54 Similar to Cajigas et al, they isolated hippocampal neuropils containing both axons and dendrites. Of the ribosome-bound transcripts reported, 81 overlapped with those localized to RGC endfeet representing 70.4% of the endfoot FMRP interactome,24,54 13 of which were specifically characterized as neuronal (not shown).54 It is unclear whether the remaining transcripts are from neurons or from other sources of RNA in the neuropils, such as astrocytes.
Adding another layer of complexity, Biever et al assessed whether individual transcripts were translated by monosomes or polysomes. Canonically, polysomes have been considered the main sites of active translation.55–57 However, very few polysomes have been observed in neuronal processes and monosomes are most populous in these compartments. Consistent with their high numbers, monosomes were shown to be important sources of translation in neuronal processes requiring protein production, as well as in synapses.54 Further analysis revealed that certain transcripts show strong preferences for translation by either polysomes or monosomes. Interestingly, for some transcripts this preference is associated with the subcellular location of translation. This suggests a new spatial layer of translational control. The preference of monosomes within neuronal processes (and synapses) is thought to be due to the small spatial constraints. This could also be true in RGC endfeet, which are only 5 to 10 μm in diameter. In support of this idea, assessing the list of monosome-associated transcripts in axons, 20 of these localized to RGC endfeet (not shown). Meanwhile, eight were preferentially translated by polysomes and the remaining 53 showed no preference (not shown).
Vesicular trafficking may enable mRNA transport and spatial regulation of local translation in axons. In rat neurons, most moving RNA granules in the axon co-traffick with lysosomes.58 A molecular tether, Annexin AII, links RNA granules to lysosomes and is required for axonal RNA transport. Other types of vesicles are also capable of mRNA transport in the axon. Cioni et al observed that ribonucleoproteins (RNPs), known to transport mRNAs, associate with late endosomes in the axons of Xenopus retinal ganglion cells.59 These endosomes are capable of interacting with translation machinery and mitochondria to form pockets of translation along the axon. Active translation occurs at these sites, producing survival factors and mitochondrial proteins which may support functional mitochondria throughout the axon. Likewise, the authors suggest that mitochondria may provide the energy needed for local translation. A similar role for the endosome pathway has been described in fungi, suggesting that this mechanism may be evolutionarily conserved.60,61
Indeed, it is possible that endosome and vesicle dependent translation hot spots could occur in mammalian RGCs. Mitochondria have been observed in RGC basal structures, but roles for endosomes in endfeet are unknown.62 Transport of RNPs by endosomes and docking at mitochondria for translation could explain the “stop and go” kinetics of RNA granules as they traffic up the RGC basal process.24 Translation hotspots along the basal process may also support morphological features of RGCs, including branching and filopodia extension. It is tantalizing to speculate that perhaps translationally competent endfeet contain both endosomes and mitochondria.
4.3 |. RNA localization at neuronal synapses
What is the role of localized RNAs in synapses? Early studies of RNA localization in neurons were limited to brain regions with laminar organization of dendrites, such as the hippocampus, to allow for mechanical separation of the cell body and projections by microdissection. Ouwenga et al used biochemical fractionation of membrane-enclosed particles to isolate synaptoneurosomes (both pre- and postsynapses) from cortical neurons.63 Deep sequencing of these fractions elucidated the first local transcriptomes of cortical neuron synapses. It is important to note that while many of the transcripts enriched in the synaptoneurosome fraction were neuronal, glia-specific transcripts were also evident.63 Comparison of the synaptic transcriptome and endfoot localized FMRP interactome reveals 87 transcripts in common between synapses and endfeet, accounting for over 75% of all known endfoot localized transcripts24,63 (Figure 3C,E). This overlap is also evident with two other published synaptic transcriptomes in which 89 and 29 transcripts overlap with RGC endfeet, 77.4% and 24.3%, respectively (Figure 3C,E).64,65 Despite the high number of conserved transcripts, the overlap for these datasets was not statistically significant (Figure 3E).63–65
However, the synaptic transcriptome is not static. Indeed, the synaptic transcriptome can be modulated by circadian cycles, alcohol exposure, aging and in schizophrenia.64,66–68 Interestingly, comparison of synaptoneurosomes to homogenized forebrain revealed that 70% of all synapse localized transcripts were altered at least 2-fold with daily circadian cycles. Ninety-three percent of oscillating transcripts showed this behavior only in synaptoneurosomes.64 The oscillations were found to be a result of posttranscriptional regulation. It is intriguing to consider whether circadian cycle also influences localization of transcripts in RGC endfeet. If so, it could provide insights into transcript regulation in RGCs at both a spatial and temporal level.
Identification of unique features among localized transcripts in synapses may provide generalizable clues to how mRNAs are transported to distal locations and locally translated. Of those transcripts enriched in cortical neuron synapses, common features include longer average transcript and 3′UTR as well as higher GC content.63 Additionally, based upon primary sequence, localized transcripts were predicted to have more stable secondary structures and show alternative splicing. In addition to encoding unique functions in the synapse, RNA processing may also provide a means for directing a transcript to synapses or retaining it in the soma.
Furthermore, some synapse localized transcripts contain m6A modifications. Merkurjev et al defined the synaptic epitranscriptome identifying 2921 transcripts with 4469 m6A sites selectively enriched in the synapse.65 Knockdown of m6A readers alters transcript levels at the synapse and disrupts spine morphology and synaptic transmission suggesting localization is functionally significant.65 However, the mechanism of m6A regulation of localization is unknown. Previously reported roles for m6A in splicing, trafficking, stability and translation are all likely candidates.69 Although analysis of RNA modifications in endfeet has not been reported, 34 of the m6A-enriched transcripts identified at the synapse are localized to RGC endfeet.24 This represents a statistically significant 29.6% overlap, suggesting that these and other transcripts may be methylated in RGCs and that modifications could influence their localization and function in endfeet (Figure 3E).
4.4 |. Local translation at neuronal synapses
Several studies have also characterized local translatomes in synapses. The importance of proper regulation of local translation at synapses is reflected in human disease. Perturbation of synaptic local translation has been linked to autism spectrum disorders, Fragile X syndrome and other intellectual disorders.26,70,71
In addition to characterizing local synaptic transcriptomes, the Ouwenga et al study also identified ribosome-bound transcripts in cortical neuronal synapses.63 They isolated synapses specifically from cortical neurons by coupling sucrose-Percoll subcellular fractionation with a method called translating ribosome affinity purification (TRAP).63,72 Adding TRAP to standard synapse isolation (termed SynapTRAP) allowed for the segregation of ribosome-bound RNAs from neurons and glia, both of which are present in fractionated synaptoneurosomes. Using this technique, 153 ribosome-bound mRNAs were identified, which were enriched for functions related to cell morphology including cytoskeletal, motor and cell junction pathways. Cytoskeletal elements alone accounted for 17.6% of identified transcripts.56 Furthermore, 46 of the 153 transcripts are known FMRP targets. These classes of localized transcripts are reminiscent of those enriched in RGC endfeet. Indeed, 10 ribosome-bound transcripts are also present in RGC endfeet representing a statistically significant 8.7% of the endfoot localized transcripts (Figure 3E).24,63 Interestingly, a comparison of transcriptomes and translatomes from cortical synapses reveals a stark contrast, as there are over 13 000 transcripts present in synapses but only 153 ribosome-bound transcripts enriched there.63 This suggests that only a subset of transcripts in the synapse may be translated at any given time, arguing for translational regulation at the synapse.
Evidence for tight regulation of local translation is further echoed by Noya et al who investigated changes in the proteome of synapses as a result of sleep-wake cycles.64 They discovered fluctuations in the proteome linked to sleep-wake cycles. Intriguingly, these oscillations were segregated by functional protein classes. For example, proteins involved in metabolism and translation were abundant before the resting phase while synaptic signaling proteins accumulated prior to the active phase. Proteomic analysis of RGC endfeet has not been reported; however, 27 of the proteins identified in synapses are present in endfeet at the transcript level (Figure 3E).24,64 Although this overlap does not achieve statistical significance, it suggests that these transcripts are competent to be locally translated far from the cell body and could be locally translated in endfeet.
The question of how local translation is controlled at the synapse remains under investigation. Two possible contributors are FMRP and M6A. FMRP is important for translational control at the synapse.26,73 M6A is another possible regulator present at synapses which controls translation dynamics.74–76 Altogether, these studies of RNA localization and translation in neuronal processes and synapses highlight intriguing overlap of transcripts with RGC endfeet and suggest potential mechanisms that may be at play in RGCs.
5 |. RNA LOCALIZATION AND LOCAL TRANSLATION IN ASTROCYTES
Following the conclusion of neurogenesis, RGCs switch potency and begin producing glia including astrocytes. Astrocytes are a highly populous cell type in the mammalian brain and actively participate in broad brain functions including circadian rhythm, learning and memory, and wakefulness and sleep.77–83 More specifically, astrocytes form a scaffold for migrating neurons, support the blood-brain barrier and interact with and promote neural synapse formation and function.84–86 Human astrocytes are more numerous and complex than their rodent counterparts. A single human astrocyte can contact up to 2 million distinct synapses.87,88
Similar to RGCs, astrocytes have branched processes that stretch far from the cell body to interact with neurons and vasculature. Astrocytes are even more complex than RGCs extending multiple processes each with their own branches claiming spherical territories 40 to 60 μm in diameter in mice.89,90 The main processes, secondary processes and finest processes are referred to as the branches, branchlets and peripheral astrocyte processes (PAPs), respectively. Astrocytes also have protrusions called endfeet which, in this context, are specialized structures that physically contact blood vessels. The PAPs interact with neuronal synapses and show reversible structural remodeling in response to behavior, brain state or synaptic activity. The remodeling is modulated by coordinated polymerization and depolymerization of cytoskeletal elements.91–95 The functional significance of PAPs is illustrated by their plasticity in response to a changing microenvironment and the resulting impact of this plasticity on spine stability and synaptic efficacy and maturation.92,94,96–98
Emerging evidence indicates that PAP morphology and function may be controlled by local gene regulation. The membrane cytoskeletal linker protein, Ezrin, localizes to PAPs and is required for the formation and motility of filopodia.95,96,99–101 Additionally, glial fibrillary acidic protein (GFAP) mRNA is localized to the branch points and distal ends of protrusions and the RNA-binding protein APC localizes to the leading edge of migrating astrocytes.102,103 Below we first discuss localized RNAs and then local translatomes identified in astrocytes.
5.1 |. Transcriptome analysis of astrocyte protrusions
Thomsen et al discovered transcriptomes isolated from astrocyte cell bodies and processes in vitro using a modified Boyden chamber.104 Thomsen and Lade Nielsen adapted this system for astrocytes by decreasing the pore size of the membrane and coating it with ECM components to encourage protrusion growth.105 In a pilot study, several RNAs including Pkp4, Ankrd25, Inpp1 and 18-s rRNA were identified in the cellular processes of both primary astrocytes and the astrocyte cell line C8-S.105 To obtain a more complete characterization of the protrusion transcriptome, single-molecule direct RNA sequencing (DRS) was used to analyze transcripts recovered from the cell body and protrusion fractions.104,106 The top 250 protrusion-enriched transcripts represented a wide variety of functional groups and were present in both primary astrocytes and immortalized astrocyte cell lines.
Interestingly, the most enriched transcript was Nestin, which also localizes to RGC endfeet.29,104 In astrocytes, the Nestin 3′UTR is sufficient to localize the mRNA to protrusions in an FMRP-dependent manner.104 This is similar to roles for 3′UTRs in localization to endfeet.23,24 Comparison of the 250 most enriched transcripts in primary astrocyte protrusions and the 115 FMRP-bound transcripts in RGC endfeet reveal 32 in common (Figure 3D).24 Additionally, 28 genes are conserved between C8-S protrusion-enriched transcripts and RGC endfoot localized transcripts (Figure 3D). The overlap with primary astrocytes and C8-S cells represented 28% and 24% of the FMRP endfoot localized transcripts, respectively (Figure 3E). Both comparisons are statistically significant and 22 genes were shared between all three groups (Figure 3D,E).24
The overall number of transcripts common to RGCs and astrocytes is high but may still underrepresent the actual extent of overlap. Astrocyte transcripts were filtered for those enriched in processes compared to the cell body while the RGC transcripts were present in endfeet, but only enriched relative to the local niche and not cell body.24,104,107 In addition, the RGC endfoot localized transcripts were limited to those bound to FMRP, suggesting there may be more overlap when comparing a global endfoot transcriptome. We have already seen two examples of this with Nestin and Cyclin D2. Both Nestin and Cyclin D2 are localized in astrocytes and found in endfeet but not significantly enriched in the FMRP RIP-Chip analysis.24,104
5.2 |. Local translation in astrocyte protrusions
Do localized transcripts undergo translation in astrocytes? Thomson et al hinted at this possibility as their western blot analyses of protrusions showed the presence of Nestin protein.104 Sakers et al then identified ribosomes in PAPs near synapses in brain slices.107 Furthermore, in this study they observed active translation in PAPs using puromycilation assays in slice culture. Up to 73% of translation measured in astrocytes occurred greater than 9 μm from the nucleus. To expand upon this discovery and identify transcripts likely to be locally translated, the authors performed in-depth characterization of ribosome-bound transcripts in PAPs.107 To do so, they developed and employed the PAP-TRAP method, similar to other TRAP methods, to isolate and sequence ribosome-bound mRNAs in astrocytes in vivo. 224 transcripts were significantly enriched in PAP ribosomes compared to soma. The enriched transcripts were biased for genes involved in glutamate and GABA metabolism and biosynthesis of fatty acids as well as motor and cytoskeletal proteins. This is perhaps unsurprising considering the importance of these classes in PAP function and morphology. However, it does support the idea that local translation is a regulated process that controls spatial production of functionally relevant proteins. There are also 10 transcripts (8.7%) shared between the astrocyte ribosome-bound transcripts and RGC endfoot FMRP interactome,24 which were statistically significant107 (Figure 3E). Interestingly, seven of these transcripts are also ribosome bound in synapses.63 Importantly, classes of locally translated genes are related to established PAP functions. This foreshadows that characterization of endfeet will provide insights into genes and signaling cascades important for endfoot functionality and potentially uncover new roles for endfeet in cortical development.
5.3 |. Comparisons of astrocytes and RGCs: Roles for extrinsic cues
Several lines of evidence suggest that mRNA localization in astrocytes is not entirely preprogramed, but rather is influenced by external cues. Differences in growth conditions between astrocytes in vivo and in vitro are reflected by striking differences between their localized transcriptomes. Furthermore, work by Foster et al shows that treatment of astrocytes with pyridazine derivatives is sufficient to activate the local translation of specific subsets of transcripts.108 This provides evidence that an external stimulus is capable of regulating local gene regulation in astrocyte processes.
How can this inform what happens in RGCs? Transcripts localized to RGC endfeet highly overlap those of primary astrocytes in vitro. In contrast, there was less overlap between RGC endfeet transcripts and those in astrocyte processes in vivo, and no transcripts in common with astrocyte endfeet in vivo.109 This may reflect similar microenvironments between astrocyte in vitro culture and RGC endfeet; astrocytes were cultured on Type I collagen and RGC endfeet directly contact the BM containing multiple ECM components including Type I collagens.
It is possible that external cues control which and when mRNAs are transported to RGC endfeet. Influence of external stimuli on local gene regulation in RGC endfeet is consistent with its location in a unique niche composed of vasculature, fibroblasts, ECM, Cajal-Retzius neurons and interneurons. Signals from this niche are known to influence neurogenesis15–18 and may activate production of proteins necessary to trigger signaling which in turn influences cellular responses.
6 |. SUMMARY
RGCs have a complex and bipolar morphology including basal endfeet that are located hundreds of microns from the cell body. This great distance between the cell body and endfeet raises the question of how genes are locally regulated to support endfoot function. The field of local gene regulation in RGCs is still in its infancy, but several key discoveries have already been made. RNAs are actively transported to endfeet in a manner regulated by both cis- and trans-factors.24 Endfeet contain a local transcriptome of which over 100 transcripts have been identified.23,24 The endfoot transcriptome is enriched for cytoskeletal and signaling elements which may inform endfoot functionality. Additionally, mRNAs can be locally translated in endfeet independent of the cell body.24
Despite this initial foundation of knowledge, many questions remain unanswered regarding which mRNAs are localized, when they are transported, how their fate is controlled and how these processes influence stem cell function. For insight, in this review, we turned to neurons and astrocytes which have been heavily studied in the context of RNA localization and local translation. Our analysis of published datasets reveals extensive overlap between transcripts localized in RGC endfeet, neuronal axons and synapses, and especially with astrocyte protrusions. This cellular conservation suggests common transcriptomes within subcellular compartments and perhaps common mechanisms. Future studies will be invaluable toward understanding functions of RGC endfeet and how it may play roles in brain development and disease. Further, as we delve deeper into studies of endfeet this can also generate fundamental new insights into basic mechanisms of RNA localization and local translation in neurons and astrocytes.
ACKNOWLEDGMENTS
This work was supported by funding from NIH R01NS083897, NIH R21MH119813 and NIH R01NS110388 (D.L.S.); NSF-GRFP (B.R.D.). The authors apologize to those authors whose work could not be included due to space.
Funding information
National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: R01NS083897, R01NS110388, R21MH119813; National Science Foundation
Footnotes
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12769.
REFERENCES
- 1.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. [DOI] [PubMed] [Google Scholar]
- 2.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127(24):5253–5263. [DOI] [PubMed] [Google Scholar]
- 3.Silver D, Rakic P, Grove EA, et al. Evolution and ontogenetic development of cortical structures In: Singer W, Sejnowski TJ, Rakic P, eds. The Neocortex. Vol 27 Cambridge, MA: MIT Press; 2019;61–93. [Google Scholar]
- 4.Lu X, Duan M, Song L, et al. Morphological changes of radial glial cells during mouse embryonic development. Brain Res. 2015;1599:57–66. [DOI] [PubMed] [Google Scholar]
- 5.Miller DJ, Bhaduri A, Sestan N, Kriegstein A. Shared and derived features of cellular diversity in the human cerebral cortex. Curr Opin Neurobiol. 2019;56:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadisseux JF, Kadhim HJ, van den Bosch de Aguilar P, Caviness VS, Evrard P. Neuron migration within the radial glial fiber system of the developing murine cerebrum: an electron microscopic autoradio-graphic analysis. Brain Res Dev Brain Res. 1990;52(1–2):39–56. [DOI] [PubMed] [Google Scholar]
- 7.Schmechel DE, Rakic P. A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat Embryol (Berl). 1979;156(2):115–152. [DOI] [PubMed] [Google Scholar]
- 8.Bjornsson CS, Apostolopoulou M, Tian Y, Temple S. It takes a village: constructing the neurogenic niche. Dev Cell. 2015;32(4):435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Jin Z, Koirala S, et al. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28(22):5817–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myshrall TD, Moore SA, Ostendorf AP, et al. Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. J Neuropathol Exp Neurol. 2012;71(12):1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal sub-types. Development. 2006;133(16):3245–3254. [DOI] [PubMed] [Google Scholar]
- 12.Graus-Porta D, Blaess S, Senften M, et al. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31(3):367–379. [DOI] [PubMed] [Google Scholar]
- 13.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43(5):647–661. [DOI] [PubMed] [Google Scholar]
- 14.Rash BG, Ackman JB, Rakic P. Bidirectional radial Ca(2+) activity regulates neurogenesis and migration during early cortical column formation. Sci Adv. 2016;2(2):e1501733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegenthaler JA, Ashique AM, Zarbalis K, et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell. 2009;139(3):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seuntjens E, Nityanandam A, Miquelajauregui A, et al. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12(11):1373–1380. [DOI] [PubMed] [Google Scholar]
- 17.Griveau A, Borello U, Causeret F, et al. A novel role for Dbx1-derived Cajal-Retzius cells in early regionalization of the cerebral cortical neuroepithelium. PLoS Biol. 2010;8(7):e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartfuss E, Forster E, Bock HH, et al. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development. 2003;130(19):4597–4609. [DOI] [PubMed] [Google Scholar]
- 19.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31(10):3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosodo Y, Toida K, Dubreuil V, et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008;27(23):3151–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosodo Y, Huttner WB. Basal process and cell divisions of neural progenitors in the developing brain. Dev Growth Differ. 2009;51(3):251–261. [DOI] [PubMed] [Google Scholar]
- 22.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31(8):1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilaz LJ, Lennox AL, Rouanet JP, Silver DL. Dynamic mRNA transport and local translation in radial glial progenitors of the developing brain. Curr Biol. 2016;26(24):3383–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31(4):1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16(2):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia JF, Parker R. MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA. 2015;21(8):1393–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res. 1995;84(1):109–129. [DOI] [PubMed] [Google Scholar]
- 30.Saarikangas J, Hakanen J, Mattila PK, Grumet M, Salminen M, Lappalainen P. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J Cell Sci. 2008;121(Pt 9):1444–1454. [DOI] [PubMed] [Google Scholar]
- 31.Astrom KE, Webster HD. The early development of the neopallial wall and area choroidea in fetal rats. A light and electron microscopic study. Adv Anat Embryol Cell Biol. 1991;123:1–76. [PubMed] [Google Scholar]
- 32.Pilaz L-J, Joshi K, Liu J, et al. Subcellular mRNA localization and local translation of Arhgap11a in radial glial cells regulates cortical development. bioRxiv. 2020. 10.1101/2020.07.30.229724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsunekawa Y, Kikkawa T, Osumi N. Asymmetric inheritance of Cyclin D2 maintains proliferative neural stem/progenitor cells: a critical event in brain development and evolution. Dev Growth Differ. 2014;56(5):349–357. [DOI] [PubMed] [Google Scholar]
- 34.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80(3):648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glock C, Heumuller M, Schuman EM. mRNA transport & local translation in neurons. Curr Opin Neurobiol. 2017;45:169–177. [DOI] [PubMed] [Google Scholar]
- 36.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21(4):741–751. [DOI] [PubMed] [Google Scholar]
- 37.Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci. 1996;16(4):1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138(5):1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gumy LF, Yeo GS, Tung YC, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogelaar CF, Gervasi NM, Gumy LF, et al. Axonal mRNAs: character-isation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42(2):102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74(3):453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goering R, Hudish LI, Guzman BB, et al. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. Elife. 2020;9:e52621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smit M, Leng J, Klemke RL. Assay for neurite outgrowth quantification. Biotechniques. 2003;35(2):254–256. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Ding SJ, Wang W, et al. Methods for pseudopodia purification and proteomic analysis. Sci STKE. 2007;2007(400):pl4. [DOI] [PubMed] [Google Scholar]
- 46.Zheng JQ, Kelly TK, Chang B, et al. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21(23):9291–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma P, Chierzi S, Codd AM, et al. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25(2):331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J Neurotrauma. 2006;23(3–4):295–308. [DOI] [PubMed] [Google Scholar]
- 49.Willis DE, Twiss JL. The evolving roles of axonally synthesized proteins in regeneration. Curr Opin Neurobiol. 2006;16(1):111–118. [DOI] [PubMed] [Google Scholar]
- 50.Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110(2):223–235. [DOI] [PubMed] [Google Scholar]
- 51.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32(6):1013–1026. [DOI] [PubMed] [Google Scholar]
- 52.Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blackmore M, Letourneau PC. Protein synthesis in distal axons is not required for axon growth in the embryonic spinal cord. Dev Neurobiol. 2007;67(7):976–986. [DOI] [PubMed] [Google Scholar]
- 54.Biever A, Glock C, Tushev G, et al. Monosomes actively translate synaptic mRNAs in neuronal processes. Science. 2020;367(6477):eaay4991. [DOI] [PubMed] [Google Scholar]
- 55.Warner JR, Knopf PM. The discovery of polyribosomes. Trends Biochem Sci. 2002;27(7):376–380. [DOI] [PubMed] [Google Scholar]
- 56.Warner JR, Knopf PM, Rich A. A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci U S A. 1963;49:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warner JR, Rich A. The number of soluble RNA molecules on reticulocyte polyribosomes. Proc Natl Acad Sci U S A. 1964;51:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao YC, Fernandopulle MS, Wang G, et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179(1):147–164.e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cioni JM, Lin JQ, Holtermann AV, et al. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176(1–2):56–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumann S, Konig J, Koepke J, Feldbrugge M. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higuchi Y, Ashwin P, Roger Y, Steinberg G. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204(3):343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rash BG, Micali N, Huttner AJ, Morozov YM, Horvath TL, Rakic P. Metabolic regulation and glucose sensitivity of cortical radial glial cells. Proc Natl Acad Sci U S A. 2018;115(40):10142–10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouwenga R, Lake AM, O’Brien D, Mogha A, Dani A, Dougherty JD. Transcriptomic analysis of ribosome-bound mRNA in cortical neurites in vivo. J Neurosci. 2017;37(36):8688–8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noya SB, Colameo D, Bruning F, et al. The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science. 2019;366(6462):eaav2642. [DOI] [PubMed] [Google Scholar]
- 65.Merkurjev D, Hong WT, Iida K, et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21(7):1004–1014. [DOI] [PubMed] [Google Scholar]
- 66.Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA. The synaptoneurosome transcriptome: a model for profiling the emolecular effects of alcohol. Pharmacogenomics J. 2015;15(2):177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen BJ, Ueberham U, Mills JD, et al. RNA sequencing reveals pronounced changes in the noncoding transcriptome of aging synaptosomes. Neurobiol Aging. 2017;56:67–77. [DOI] [PubMed] [Google Scholar]
- 68.Hall LS, Medway CW, Pain O, et al. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet. 2020;29(1):159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Zhao JC. Update: mechanisms underlying N(6)-methyladenosine modification of eukaryotic mRNA. Trends Genet. 2016;32(12):763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kelleher RJ 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–406. [DOI] [PubMed] [Google Scholar]
- 71.Santini E, Huynh TN, MacAskill AF, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doyle JP, Dougherty JD, Heiman M, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer KD, Patil DP, Zhou J, et al. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi J, Ieong KW, Demirci H, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol. 2016;23(2):110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herculano-Houzel S The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 2014;62(9):1377–1391. [DOI] [PubMed] [Google Scholar]
- 78.Brancaccio M, Edwards MD, Patton AP, et al. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science. 2019; 363(6423):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diniz DG, de Oliveira MA, de Lima CM, et al. Age, environment, object recognition and morphological diversity of GFAP-immunolabeled astrocytes. Behav Brain Funct. 2016;12(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dere E, De Souza-Silva MA, Frisch C, et al. Connexin30-deficient mice show increased emotionality and decreased rearing activity in the open-field along with neurochemical changes. Eur J Neurosci. 2003;18(3):629–638. [DOI] [PubMed] [Google Scholar]
- 81.Alberini CM, Cruz E, Descalzi G, Bessieres B, Gao V. Astrocyte glycogen and lactate: new insights into learning and memory mechanisms. Glia. 2018;66(6):1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papouin T, Dunphy JM, Tolman M, Dineley KT, Haydon PG. Septal cholinergic neuromodulation tunes the astrocyte-dependent gating of hippocampal NMDA receptors to wakefulness. Neuron. 2017;94(4):840–854.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poskanzer KE, Yuste R. Astrocytes regulate cortical state switching in vivo. Proc Natl Acad Sci U S A. 2016;113(19):E2675–E2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen NJ, Barres BA. Neuroscience: Glia—more than just brain glue. Nature. 2009;457(7230):675–677. [DOI] [PubMed] [Google Scholar]
- 85.Morest DK, Silver J. Precursors of neurons, neuroglia, and ependymal cells in the CNS: what are they? Where are they from? How do they get where they are going? Glia. 2003;43(1):6–18. [DOI] [PubMed] [Google Scholar]
- 86.Allen NJ, Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96(3):697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oberheim NA, Takano T, Han X, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29(10):3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han X, Chen M, Wang F, et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 2013;12(3):342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci. 2002;22(1):183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ogata K, Kosaka T. Structural and quantitative analysis of astrocytes in the mouse hippocampus. Neuroscience. 2002;113(1):221–233. [DOI] [PubMed] [Google Scholar]
- 91.Theodosis DT. Oxytocin-secreting neurons: a physiological model of morphological neuronal and glial plasticity in the adult hypothalamus. Front Neuroendocrinol. 2002;23(1):101–135. [DOI] [PubMed] [Google Scholar]
- 92.Bernardinelli Y, Randall J, Janett E, et al. Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr Biol. 2014;24(15):1679–1688. [DOI] [PubMed] [Google Scholar]
- 93.Ding F, O’Donnell J, Xu Q, Kang N, Goldman N, Nedergaard M. Changes in the composition of brain interstitial ions control the sleep-wake cycle. Science. 2016;352(6285):550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perez-Alvarez A, Navarrete M, Covelo A, Martin ED, Araque A. Structural and functional plasticity of astrocyte processes and dendritic spine interactions. J Neurosci. 2014;34(38):12738–12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lavialle M, Aumann G, Anlauf E, Prols F, Arpin M, Derouiche A. Structural plasticity of perisynaptic astrocyte processes involves ezrin and metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2011;108(31):12915–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nishida H, Okabe S. Direct astrocytic contacts regulate local maturation of dendritic spines. J Neurosci. 2007;27(2):331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oliet SH, Piet R, Poulain DA, Theodosis DT. Glial modulation of synaptic transmission: insights from the supraoptic nucleus of the hypothalamus. Glia. 2004;47(3):258–267. [DOI] [PubMed] [Google Scholar]
- 98.Oliet SH, Piet R, Poulain DA. Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science. 2001;292(5518):923–926. [DOI] [PubMed] [Google Scholar]
- 99.Haber M, Zhou L, Murai KK. Cooperative astrocyte and dendritic spine dynamics at hippocampal excitatory synapses. J Neurosci. 2006;26(35):8881–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Derouiche A, Frotscher M. Peripheral astrocyte processes: monitoring by selective immunostaining for the actin-binding ERM proteins. Glia. 2001;36(3):330–341. [DOI] [PubMed] [Google Scholar]
- 101.Kim HS, Bae CD, Park J. Glutamate receptor-mediated phosphorylation of ezrin/radixin/moesin proteins is implicated in filopodial protrusion of primary cultured hippocampal neuronal cells. J Neurochem. 2010;113(6):1565–1576. [DOI] [PubMed] [Google Scholar]
- 102.Landry CF, Watson JB, Kashima T, Campagnoni AT. Cellular influences on RNA sorting in neurons and glia: an in situ hybridization histochemical study. Brain Res Mol Brain Res. 1994;27(1):1–11. [DOI] [PubMed] [Google Scholar]
- 103.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421(6924):753–756. [DOI] [PubMed] [Google Scholar]
- 104.Thomsen R, Pallesen J, Daugaard TF, Borglum AD, Nielsen AL. Genome wide assessment of mRNA in astrocyte protrusions by direct RNA sequencing reveals mRNA localization for the intermediate filament protein nestin. Glia. 2013;61(11):1922–1937. [DOI] [PubMed] [Google Scholar]
- 105.Thomsen R, Lade Nielsen A. A Boyden chamber-based method for characterization of astrocyte protrusion localized RNA and protein. Glia. 2011;59(11):1782–1792. [DOI] [PubMed] [Google Scholar]
- 106.Ozsolak F, Platt AR, Jones DR, et al. Direct RNA sequencing. Nature. 2009;461(7265):814–818. [DOI] [PubMed] [Google Scholar]
- 107.Sakers K, Lake AM, Khazanchi R, et al. Astrocytes locally translate transcripts in their peripheral processes. Proc Natl Acad Sci U S A. 2017;114(19):E3830–E3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foster JB, Zhao F, Wang X, et al. Pyridazine-derivatives enhance structural and functional plasticity of tripartite synapse via activation of local translation in astrocytic processes. Neuroscience. 2018;388:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boulay AC, Saubamea B, Adam N, et al. Translation in astrocyte distal processes sets molecular heterogeneity at the gliovascular interface. Cell Discov. 2017;3:17005. [DOI] [PMC free article] [PubMed] [Google Scholar]


