Abstract
Ovarian cancer ranks 7th among the most common cancer types affecting women worldwide. A number of studies have confirmed that multiple long non-coding RNAs participate in the occurrence and progression of ovarian cancer. Small nucleolar RNA host gene 9 (SNHG9) serves a role in the progression of glioblastoma and pancreatic cancer. However, the specific biological function of SNHG9 in ovarian cancer has not yet been fully investigated. The present study aimed to determine the biological role and potential molecular mechanism underlying the influence of SNHG9 in ovarian cancer. SNHG9 expression in ovarian cancer cell lines and tissues were measured via reverse transcription-quantitative PCR analysis, and cell proliferation was detected via Cell Counting Kit-8 and colony formation assays. Flow cytometry was performed to assess cell cycle progression, and Transwell and wound healing assays were performed to assess cell invasion and migration abilities. Bioinformatics software was utilized to determine the target genes of SNHG9, which were subsequently verified via dual-luciferase reporter and RNA immunoprecipitation assays. The results demonstrated that SNHG9 expression was remarkably lower in ovarian cancer cell lines and tissues compared with the negative controls. Cell function assays demonstrated that decreased SNHG9 expression notably induced the migration, colony formation, proliferation and invasiveness of ovarian cancer cells. Furthermore, the inhibitory effect of SNHG9 on the migration, colony formation, proliferation and invasion of ovarian cancer cells was partially reversed by miR-214-5p upregulation. Thus, taken together, the current results suggest that SNHG9 may serve as a tumor suppressor gene in ovarian cancer by regulating the miR-214-5p/cryptochrome circadian regulator 2 axis.
Keywords: ovarian cancer, SNHG9, microRNA-214-5p, cell proliferation, cell invasion, cell cycle
Introduction
Ovarian cancer ranks 7th among the most common cancer types globally, and is the 8th leading cause of cancer-associated mortality in women, as reported in 2013 (1), with a 5-year overall survival rate <45% (2). Ovarian cancer frequently occurs during perimenopause and has the highest mortality rate among different types of gynecological cancer, due to unclear manifestations and lack of effective diagnosis in the early stages (3). The following long non-coding RNAs (lncRNAs), Human Satellite II (4), Antisense Non-coding RNA in the INK4 Locus (5) and Colon Cancer Associated Transcript 1 (6,7) have been demonstrated to participate in the occurrence and progression of ovarian cancer, which suggests their role as biomarkers for early cancer diagnosis. However, this knowledge does not decrease the threat of ovarian cancer to female health. Thus, the molecular mechanisms underlying ovarian cancer progression need to be further investigated to determine novel effective therapeutic targets and prolong the survival of patients with ovarian cancer.
lncRNAs containing >200 nucleotides are a type of novel transcript that cannot code proteins (8,9). lncRNAs have been reported to influence crucial cell functions, including histone modification, chromatin rearrangement, regulation of gene expression and alternative splicing gene modification (10–12). They also participate in dosage compensation (13), genomic imprinting (14), cell differentiation, organogenesis and tumor formation (15–17). A number of lncRNAs influence the onset and progression of ovarian cancer and may serve as therapeutic targets and markers for its diagnosis and prognosis (18–20). Small nucleolar RNA host gene 9 (SNHG9) is located on chromosome 16p13.3, and while its involvement in the progression of ovarian cancer remains unclear, it is a novel prognostic marker for pancreatic cancer (21).
Yu et al (22) reported that CRY2 suppresses proliferation and migration in osteosarcoma cells. Fang et al (23) demonstrated that CRY2 degradation influences chemoresistance in colorectal cancer. Furthermore, Tokunaga et al (24) suggested that CRY2 may serve a role in the progression of ovarian cancer. Hence, CRY2 was selected for further analyses in the present study.
The present study aimed to determine the biological role and potential molecular mechanism of SNHG9 in ovarian cancer. SNHG9 expression levels in ovarian cancer cell lines and tissues were analyzed by performing in vitro cell function assays.
Materials and methods
Ethical compliance
The present study was approved by the Institutional Ethics Committee of The Second People's Hospital of Taizhou City (Taizhou, China) and written informed consent was obtained from all patients prior to the study start.
Human tissue specimens
A total of 25 pairs of ovarian cancer and paracancerous normal tissue samples (mean age, 53 years; age range, 41–69 years) were collected from patients undergoing surgery at The Second People's Hospital of Taizhou City (Taizhou, China). Patients treated with radiotherapy and chemotherapy prior to surgery were excluded from the study. Fresh tissues were collected during surgery, frozen in liquid nitrogen within 5 min following resection, and stored at −80°C until further experimentation. The clinicopathological characteristics of patients with ovarian cancer are presented in Table I.
Table I.
Association between SNHG9 expression and clinicopathological characteristics of patients with ovarian cancer.
| Characteristic | Low expression, n=13 | High expression, n=12 | P-value |
|---|---|---|---|
| Age, years | |||
| <50 | 6 | 4 | |
| ≥50 | 7 | 8 | 0.6882 |
| Histological subtype | |||
| Serous | 10 | 8 | |
| Others | 3 | 4 | 0.6728 |
| FIGO stage | |||
| I–II | 1 | 6 | |
| III–IV | 12 | 6 | 0.0302a |
| Residual tumor diameter, cm | |||
| <1 | 6 | 7 | |
| ≥1 | 7 | 5 | 0.6951 |
| Lymph node metastasis | |||
| Absent | 10 | 4 | |
| Present | 3 | 8 | 0.0472a |
P<0.05. Data were analyzed using χ2 or Fisher's exact tests. The groups were stratified by median expression level. FIGO, International Federation of Gynaecology and Obstetrics; SNHG9, small nucleolar RNA host gene 9.
Cell lines and cell culture
A total of three ovarian cancer cell lines (SKOV3, OVCAR-3 and A2780) and a normal ovarian cell line (IOSE-80) were purchased from Shanghai Institute for Biological Science and cultured in DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific Inc.) at 37°C with 5% CO2.
Reverse transcription-quantitative (RT-q)PCR
microRNAs (miRNAs/miR) were harvested from the cultured cells and tumor tissues using a mirVana™ miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). miR-214-5p expression levels were measured using the SYBR Primescript miRNA RT-qPCR kit and SYBR1 Green I (Toyobo Life Science), according to manufacturer's protocol. Total RNA was extracted from the ovarian cancer cells and tissues using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA using a RT kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocols. The following primer sequences (Takara Biotechnology Co., Ltd.) were used for qPCR: SNHG9 forward, 5′-GACTGCAGACCCCTAACCTT-3′ and reverse, 5′-ACCCGCATGCAGTGAGTTA-3′; cryptochrome circadian regulator 2 (CRY2) forward, 5′-TCCCAAGGCTGTTCAAGGAAT-3′ and reverse, 5′-TGCATCCCGTTCTTTCCCAAA-3′; GAPDH forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′; miR-214-5p forward, 5′-ACACTCCAGCTGGGCGTGTCGTTCACATCT-3′ and reverse, 5′-CUACAAAGGGAAGCGACAGGCA-3′ and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The following thermocycling conditions were used: Initial denaturation step (95°C for 20 sec), followed by 40 cycles of denaturation (95°C for 3 sec) and annealing (60°C for 30 sec). miR-214-5p and SNHG9 mRNA expression levels were measured using the 2−ΔΔCq method (25) and normalized to the internal reference genes U6 and GAPDH, respectively.
Cell transfection
Short hairpin (sh)RNAs synthesized against SNHG9 (sh-SNHG9) and matched negative control sequences (sh-NC) were subcloned into the pGPU6/GFP/Neo vector (Shanghai GenePharma Co., Ltd.). Subsequently, SKOV3 cells were transfected with 100 pmol sh-SNHG9 or sh-NC using Lipofectamine® 2000 (Thermo Fisher Scientific Inc.), according to the manufacturer's protocol. Stable cell clones were selected using 0.5 mg/ml of G418 (Sigma-Aldrich; Merck KGaA). Thereafter, the cells were transfected with 0.1 nmol miR-214-5p mimics, negative control (NC) mimics (miR-NC), miR-214-5p inhibitors (anti-miR-214-5p) or anti-miR-NC (Guangzhou RiboBio Co., Ltd.) using Lipofectamine® 2000, according to the manufacturer's protocol. The sequences of miR-214-5p mimics are as follows: Sense, 5′-UGCCUGUCUACACUUGCUGUGC-3′ and antisense, 5′-ACAGCAAGUGUAGACAGGCAUU-3′. The sequences of miR-214-5p inhibitor are as follows: Sense, 5′-GCACAGCAAGUGUAGACAGGCA-3′. Subsequent experimentation was performed 48 h post-transfection.
Cytotoxicity assay
A total of 5×103 of the transfected SKOV3 cells were seeded into a 96-well plate and incubated with CCK-8 reagent (Beyotime Institute of Biotechnology) for 2 h at 37°C in the dark, at 24, 48, 72 and 96 h post-transfection, according to the manufacturer's protocol. Cell proliferation was analyzed as a wavelength of 450 nm using a Benchmark Plus™ Microplate spectrometer (Bio-Rad Laboratories, Inc.). All experiments were performed in triplicate.
Colony formation assay
SKOV3 cells that demonstrated a stable decrease in SNHG9 expression were seeded into a 6-well plate at a density of 1×103 cells/well and incubated at 37°C with 5% CO2 for 2 weeks. Cells were fixed with 96% ethanol for 10 min and subsequently stained with 1% crystal violet for 5 min at room temperature (both from Sigma-Aldrich; Merck KGaA). Cells were counted and the visible colonies were imaged using an inverted light microscope (magnification, ×10; Olympus Corporation).
Cell cycle distribution analysis
The transfected SKOV3 cells were fixed with 70% ethanol (Beyotime Institute of Biotechnology) overnight at 4°C and rinsed twice with PBS, prior to incubation with 0.1 mg/ml of propidium iodide and 1 mg/ml of RNase A (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. Cell cycle distribution was assessed using a FACScan flow cytometer (BD Biosciences) and cell percentages in the G0/G1, S and G2/M phases were determined using FlowJo software (version 7.6.1; Tree Star, Inc.).
Wound healing assay
The transfected SKOV3 cells were seeded into a 6-well plate and incubated with DMEM at 37°C with 5% CO2 until they reached ~100% confluence. Cells were rinsed three times with PBS and serum-starved for 24 h. Subsequently, the cell monolayer was scratched using a 10 µl pipette tip and the migratory cells were imaged at 0 and 24 h using an inverted light microscope (magnification, ×10; Olympus Corporation). AxioVision software (version 4.7; Carl Zeiss AG) was used to assess wound healing.
Transwell and Matrigel assays
Transwell chambers (Corning Inc.) coated with Matrigel at 37°C for 30 min (BD Biosciences) were applied to assess cell invasion. A total of 2.0×104 of the transfected SKOV3 cells were plated in the upper chambers of Transwell plates in DMEM without FBS, while the lower Transwell chamber was incubated in DMEM supplemented with 10% FBS as a chemo-attractant. Following incubation for 48 h at 37°C, the non-invasive cells in the upper chamber were removed using cotton swabs, and the invasive cells in the lower chamber were fixed in 4% paraformaldehyde for 20 min and subsequently stained with 1% crystal violet for 15 min at room temperature. Stained cells were counted in five randomly-selected fields using an inverted light microscope (magnification, ×20; Olympus Corporation).
Subcellular fractionation location
The location of SNHG9 in SKOV3 cells was determined using the PARIS kit (Thermo Fisher Scientific, Inc.), whereby the nuclear and cytoplasmic fractions were isolated, according to the manufacturer's protocol. SNHG9 expression levels were quantified via RT-qPCR analysis (as above), and normalized to the internal reference genes GAPDH (cytoplasmic control) and U6 (nuclear control), respectively.
Dual-luciferase reporter assay
Putative miRNA binding sites on SNHG9 and CRY2 sequences were speculated using miRcode (http://www.mircode.org) and confirmed via the dual-luciferase assay. Briefly, SNHG9 and CRY2 3′-untranslated region (UTR) sequences with the predicted wild-type (WT) or mutant (MUT) miR-214-5p binding sites (Shanghai GenePharma Co., Ltd.) were subcloned into the following firefly luciferase reporter psiCHECK-2 vectors; SNHG9-WT, SNHG9-MUT, CRY2-3′-UTR-WT and CRY2-3′-UTR-MUT (Promega Corporation). miR-214-5p mimics or miR-NC mimics, SNHG9-WT or SNHG9-MUT and CRY2-3′-UTR-WT or CRY2-3′-UTR-MUT reporter plasmids were co-transfected in SKOV3 cells using Lipofectamine® 2000 (Beyotime Institute of Biotechnology). Following incubation at 37°C for 48 h, cells were collected and firefly and Renilla luciferase activities were detected using a Dual-luciferase reporter assay system (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using the EZ-Magna RIP kit (EMD Millipore), according to the manufacturer's protocol. SKOV3 cells were lysed using RIP lysis buffer, prior to incubation with magnetic beads conjugated with anti-Ago2 antibody (1:30; cat. no. ab186733; Abcam) or anti-IgG antibody (1:150; cat. no. 12-370; Merck Millipore), which was used as a control. RNAs were purified using the TRIzol™ Plus RNA Purification kit (cat. no. 12183555, Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and detected via RT-qPCR analysis.
Statistical analysis
Statistical analysis was performed using SPSS software (version 18.0; SPSS, Inc.). Data are presented as the mean ± standard deviation. The paired Student's t-test was performed to detect the differential expression of SNHG9, miR-214-5p, and CRY2 between ovarian cancer tissues and paracancerous normal tissues. The difference was evaluated by unpaired Student's t-test (other 2-group comparisons) or one-way ANOVA followed by the post hoc Bonferroni's correction (multigroup comparisons) as appropriate. The target genes of miR-214-5p were screened using TargetScan 7.2 (http://www.targetscan.org) and the miRDB database (http://mirdb.org). Patients were divided into low and high SNHG9 expression groups based on the median expression in ovarian cancer tumor tissues (cut-off=0.055), and Kaplan-Meier survival curves were generated, and overall survival rates were analyzed via the log-rank test. The association between SNHG9, miR-214-5p and CRY2 expression were validated by Pearson's correlation analysis. Association between SNHG9 expression and clinicopathological characteristics of patients with ovarian cancer were analyzed using χ2 or Fisher's exact tests. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate.
Results
SNHG9 is downregulated in ovarian cancer cell lines and tissues
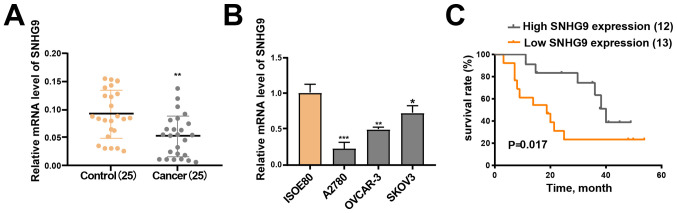
RT-qPCR analysis demonstrated that SNHG9 expression was significantly downregulated in the 25 ovarian cancer tissue samples compared with the matched paracancerous normal tissue samples (P<0.01; Fig. 1A). Furthermore, low SNHG9 expression was observed across all three ovarian cancer cell lines (SKOV3, OVCAR-3 and A2780) compared with IOSE-80 cells (all P<0.05; Fig. 1B), of which SKOV3 demonstrated the highest SNHG9 expression. Thus, this cell line was chosen for subsequent experimentation.
Figure 1.
SNHG9 expression decreases in ovarian cancer cell lines and tissues. (A) Reverse transcription-quantitative PCR was performed to measure SNHG9 expression in ovarian cancer tissues and paracancerous normal tissues (n=25). (B) SNHG9 expression in ovarian cancer and normal ovarian cell lines. (C) Kaplan-Meier survival curve and the log-rank test were used to assess the association between SNHG9 expression and the overall survival of patients with ovarian cancer. The groups were stratified by median expression level. All experiments were performed in triplicate and data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001 vs. control group or ISOE80 group. SNHG9, small nucleolar RNA host gene 9.
SNHG9 expression is associated with clinicopathological characteristics and good prognosis in ovarian cancer
Analysis between SNHG9 expression and clinicopathological characteristics revealed that SNHG9 expression was significantly associated with FIGO stage (26) (P=0.0302) and lymph node metastasis (P=0.0472). However, no significant associations were observed between SNHG9 expression and other clinicopathological characteristics (Table I).
Patients were divided into low and high SNHG9 expression groups based on mean median expression level in ovarian cancer tumor tissues (cut-off=0.055), in order to determine the association between SNHG9 expression and the outcomes of ovarian cancer. The Kaplan-Meier curve demonstrated that patients in the high SNHG9 expression group had higher survival rates compared with patients in the low SNHG9 expression group (P=0.017; Fig. 1C). The current results suggest that SNHG9 may serve as a tumor suppressor gene in ovarian cancer.
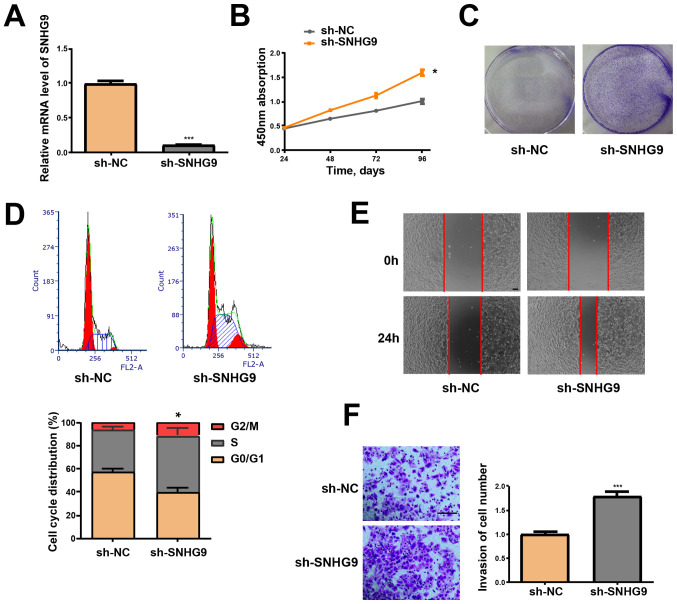
SNHG9-knockdown induces migration, proliferation and invasiveness of ovarian cancer cells
SNHG9-knockdown, via transfection of SKOV3 cells with sh-SNHG9, was performed to determine the effect of SNHG9 in tumor formation of ovarian cancer. RT-qPCR analysis demonstrated that SNHG9 expression was significantly decreased in the sh-SNHG9 group compared with the sh-NC group (P<0.001; Fig. 2A). Furthermore, the CCK-8 assay revealed that low SNHG9 expression induced the viability of SKOV3 cells (P<0.05; Fig. 2B). Cells in the sh-SNHG9 group demonstrated a stronger capacity to form colonies compared with the sh-NC group (Fig. 2C). Flow cytometry analysis demonstrated that low SNHG9 expression decreased the percentage of SKOV3 cells in the G1 phase but increased the number of cells in the S phase (P<0.05, Fig. 2D). The wound healing assay results revealed that SNHG9-knockdown significantly enhanced the migrative ability of SKOV3 cells (Fig. 2E). Results from the Transwell and Matrigel assays verified that low SNHG9 expression significantly induces the invasive ability of SKOV3 cells (P<0.001; Fig. 2F).
Figure 2.
Low SNHG9 expression induces the cell cycle, migration, proliferation and invasiveness of ovarian cancer cells. (A) SNHG9 levels in SKOV3 cells transfected with sh-SNHG9 or sh-NC were detected via reverse transcription-quantitative PCR. (B) Effect of low SNHG9 expression on the proliferation of SKOV3 cells was determined via Cell Counting Kit-8 assay. (C) Colony formation in SKOV3 cells transfected with sh-SNHG9 or sh-NC. (D) Cell cycle progression of SKOV3 cells treated with sh-SNHG9 or sh-NC was assessed via flow cytometry. (E) Wound healing assay was performed to determine the effect of low SNHG9 expression on the migrative ability of SKOV3 cells (scale bar, 100 µm). (F) Matrigel assays were performed to determine the effect of low SNHG9 expression on the invasive ability of SKOV3 cells (scale bar, 100 µm). All experiments were performed in triplicate and data are presented as the mean ± standard deviation. *P<0.05; ***P<0.001 vs. sh-NC group. SNHG9, small nucleolar RNA host gene 9; sh, short hairpin; NC, negative control.
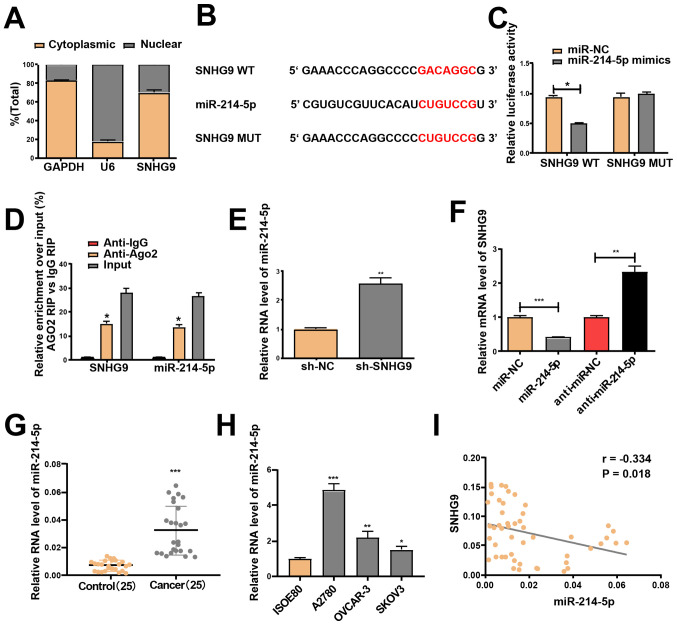
miR-214-5p is a direct target of SNHG9 in ovarian cancer cells
lncRNAs function in accordance to their subcellular distribution (27). Thus, RT-qPCR was performed to determine SNHG9 expression in the nucleus and cytoplasm of SKOV3 cells. SNHG9 was demonstrated to be primarily expressed in the cytoplasm of SKOV3 cells (Fig. 3A). This indicates that SNHG9 may participate in the development of ovarian cancer via post-transcriptional regulation, and thus was assumed to serve as a competing endogenous RNA in the progression of ovarian cancer. This result was confirmed using miRcode software, which identified the underlying miRNA target of SNHG9. As the SNHG9 transcript contains a putative miR-214-5p binding site, miR-214-5p was hypothesized to be a potential binding target of the gene (Fig. 3B).
Figure 3.
miR-214-5p is a target of SNHG9. (A) Reverse transcription-quantitative PCR was performed to evaluate SNHG9 expression in the nucleus and cytoplasm of SKOV3 cells, with U6 as the nuclear control and GAPDH as the cytoplasmic control. (B) Mutant and binding sites between miR-214-5p and SNHG9. (C) Following co-transfection with SNHG9-WT or SNHG9-MUT reporter plasmids, and miR-214-5p/miR-NC mimics, SKOV3 cells were subjected to a dual-luciferase reporter assay. (D) Anti-Ago2 RIP assay with miR-214-5p mimics demonstrated that miR-214-5p and SNHG9 accumulated in Ago2 precipitates compared with the control (IgG) in SKOV3 cells. (E) miR-214-5p expression in SKOV3 cells subjected to transfection with sh-SNHG9 or sh-NC. (F) SNHG9 expression in SKOV3 cells treated with miR-214-5p/miR-NC or anti-miR-214-5p/anti-miR-NC. (G) miR-214-5p expression in ovarian cancer tissues and paracancerous normal tissues (n=25). (H) miR-214-5p expression in ovarian cancer and normal ovarian cell lines. (I) Pearson's correlation analysis demonstrated a negative correlation between the mRNA expression levels of SNHG9 and miR-214-5p in ovarian cancer tissues. All experiments were performed in triplicate and data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001 vs. anti-igG, sh-NC, control group or ISOE80 group. miR, microRNA; SNHG9, small nucleolar RNA host gene 9; WT, wild type; MUT, mutant; NC, negative control; RIP, RNA immunoprecipitation; sh, short hairpin.
A Dual-luciferase reporter assay was performed to determine whether miR-214-5p is a downstream target of SNHG9. Transfection with miR-214-5p mimics did not change the luciferase activity of the SNHG9 -MUT but significantly decreased the activity of the SNHG9 WT in SKOV3 cells (P<0.05, Fig. 3C). miR-214-5p mimics were used for the anti-Ago2 RIP assay, and the results revealed that SNHG9 and miR-214-5p were enriched in the immunoprecipitation pulled down by the anti-Ago2 antibody in SKOV3 cells (Fig. 3D). Furthermore, low SNHG9 levels prominently elevated miR-214-5p expression in SKOV3 cells (Fig. 3E), and transfection with the miR-214-5p mimics significantly downregulated SNHG9 expression, which was reversed by transfection with miR-214-5p inhibitors in SKOV3 cells (Fig. 3F). Significantly increased miR-214-5p expression in ovarian cancer cell lines and tissues, relative to normal samples, was confirmed via RT-qPCR analysis (Fig. 3G and H). Pearson's correlation analysis indicated a negative correlation between the expression levels of miR-214-5p and SNHG9 (r=−0.334; P<0.05; Fig. 3I).
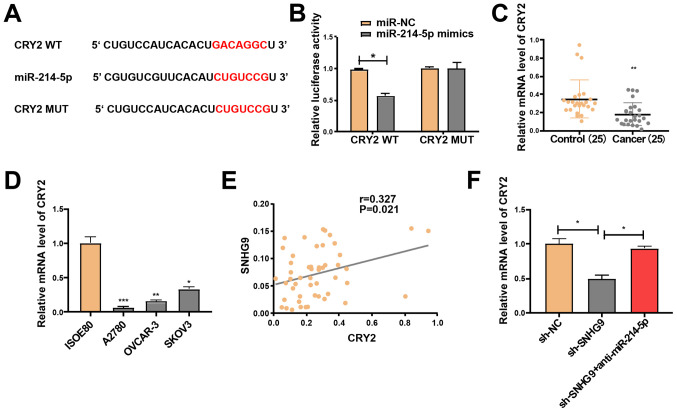
SNHG9 regulates CRY2 expression via miR-214-5p
The target genes of miR-214-5p were screened using TargetScan software and the miRDB database, in order to investigate the potential role of miR-214-5p in the progression of ovarian cancer. Following intersection of the target genes (score, >85) from both databases, a total of 11 genes were selected, including CRY2, which is closely associated with cancer progression (23,28). Following their construction, the luciferase plasmids pGL3-CRY2-3′UTR-WT and pGL3-CRY2-3′UTR-MUT were respectively co-transfected with miR-214-5p mimics or NC in SKOV3 cells (Fig. 4A). The luciferase activity of the WT reporter was inhibited; however, the MUT reporter group remained unchanged (Fig. 4B). Hence, CRY2 may be a potential target gene of miR-214-5p. RT-qPCR analysis demonstrated that CRY2 expression significantly declined in the ovarian cancer cell lines and tissues (Fig. 4C and D). Pearson's correlation analysis revealed that SNHG9 expression was positively correlated with CRY2 expression (r=0.327; P=0.021; Fig. 4E). Subsequently, SKOV3 cells were treated with sh-NC and sh-SNHG9, or co-transfected with sh-SNHG9 and anti-miR-214-5p. RT-qPCR analysis indicated that low SNHG9 expression significantly decreased CRY2 expression compared with the sh-NC group; however, this effect was partially reversed by miR-214-5p inhibitors in SKOV3 cells (P<0.05; Fig. 4F). Taken together, these results suggest that SNHG9 regulates CRY2 expression via interaction with miR-214-5p in ovarian cancer cells.
Figure 4.
SNHG9 regulates CRY2 expression via miR-214-5p. (A) Mutant and binding sites between miR-214-5p and CRY2. (B) Luciferase activity in SKOV3 cells co-transfected with miR-214-5p/miR-NC mimics and CRY2-WT or CRY2-MUT reporter plasmids. CRY2 mRNA expression levels in (C) ovarian cancer tissues and paracancerous normal tissues (n=25) and (D) ovarian cancer and normal ovarian cell lines. (E) Pearson's correlation analysis demonstrated a positive correlation between the mRNA expression levels of SNHG9 and CRY2 in ovarian cancer tissues. (F) Relative expression of CRY2 in SKOV3 cells treated with sh-NC or sh-SNHG9, with or without anti-miR-214-5p. All experiments were performed in triplicate and data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001 vs. control group or ISOE80 group. SNHG9, small nucleolar RNA host gene 9; CRY2, cryptochrome circadian regulator 2; miR, microRNA; WT, wild type; MUT, mutant; sh, short hairpin; NC, negative control.
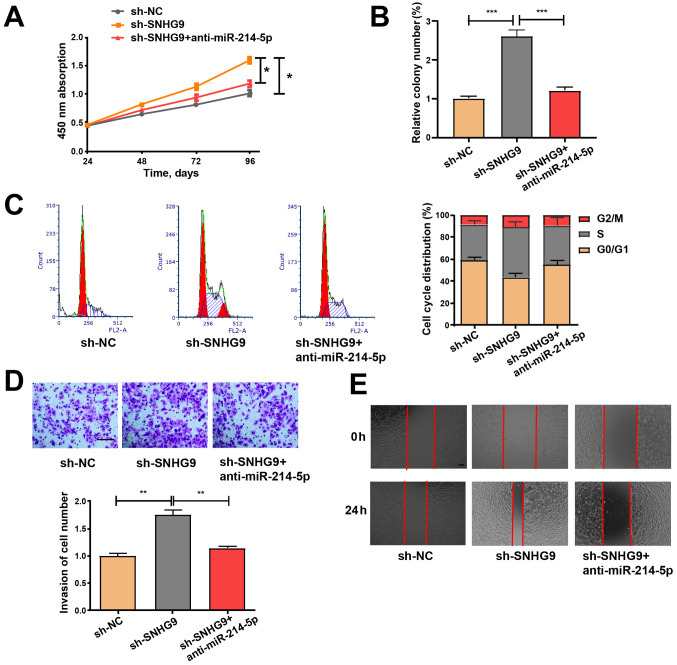
SNHG9/miR-214-5p axis regulates the properties of ovarian cancer cells
SKOV3 cells were treated with sh-NC and sh-SNHG9, or co-transfected with sh-SNHG9 and anti-miR-214-5p to determine whether the inhibitory role of low SNHG9 expression in ovarian cancer is mediated by miR-214-5p. Cell viability was detected via a CCK-8 assay, while colony formation was assessed via a colony formation assay. Subsequently, flow cytometry was performed to assess cell cycle progression, and wound healing and Transwell assays were performed to examine cell migration and invasion abilities. The effects of low SNHG9 expression on cell migration, colony formation, proliferation, cell cycle progression and invasiveness were partially reversed by transfection with anti-miR-214-5p (Fig. 5A-E), thus suggesting that anti-miR-214-5p may regulate the roles of decreased SNHG9 in ovarian cancer cells to a certain extent.
Figure 5.
miR-214-5p inhibitor partially reverses the promoting effect of sh-SNHG9 on cell function. (A) Cell Counting Kit-8, (B) colony formation, (C) cell cycle progression, (D) Matrigel invasion (scale bar, 100 µm) and (E) wound healing (scale bar, 100 µm) assays were performed on SKOV3 cells to assess cell proliferation, cycle, invasiveness and migration. All experiments were performed in triplicate and data are presented as the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001. miR, microRNA; sh, short hairpin; SNHG9, small nucleolar RNA host gene 9; NC, negative control.
Discussion
Aberrant regulation of lncRNAs is associated with tumor development and carcinogenesis, and lncRNAs may be applied as diagnostic, prognostic or therapeutic biomarkers for several types of cancer, including gastric cancer (29), osteosarcoma (30) and ovarian cancer (31–33). For example, Zhao et al (34) reported that the lncRNA LINC00092 induces glycolysis in fibroblasts associated with cancer, and influences the progression of ovarian cancer. Furthermore, Zhang et al (35) demonstrated that the lncRNA, MIR4697HG induces cell growth and metastasis in ovarian cancer. In the present study, SNHG9 expression levels were detected in three ovarian cancer cell lines (SKOV3, OVCAR-3 and A2780) and 25 patients with ovarian cancer. The results demonstrated that SNHG9 expression was notably downregulated in ovarian cancer cell lines and tissues compared with the normal cell line and tissue samples, respectively. Furthermore, decreased SNHG9 expression was indicated to be associated with low overall survival time in patients with ovarian cancer. In vitro analysis demonstrated that transfection with sh-SNHG9 induced the migration, colony formation, proliferation and invasion of ovarian cancer cells.
Subsequent experimentations were performed to determine the underlying molecular mechanisms of SNHG9. Dual-luciferase reporter assays, bioinformatics analyses and the RIP assay verified that miR-214-5p is a direct target of SNHG9 in ovarian cancer cells, and that CRY2 may be an underlying target of miR-214-5p. Consistent with these results, miR-214-5p expression increased in ovarian cancer tissues compared with paracancerous normal tissues; however, CRY2 expression decreased in ovarian cancer tissues compared with paracancerous normal tissues. Notably, Pearson's correlation analysis demonstrated a negative association between miR-214-5p and SNHG9, while a positive association was indicated between CRY2 and SNHG9 expression levels in ovarian cancer tissues, and transfection with a miR-214-5p inhibitor partially reversed the effects of sh-SNHG9 on the proliferation, cell cycle, migration and invasiveness of ovarian cancer cells. Taken together, the present results suggest that SNHG9 may regulate the expression of CRY2 by competitively binding to miR-214-5p, thereby affecting the pathophysiological process of ovarian cancer.
The current study presents a number of limitations. First, a larger population size is required to further investigate the clinical value of SNHG9. Secondly, the biological role of SNHG9 in ovarian cancer should be investigated using more ovarian cancer cell lines. Thirdly, more target genes or miRNAs should be investigated to evaluate their interaction with SNHG9. Furthermore, the function of SNHG9 in the human body should be verified, and the molecular mechanism of SNHG9 in ovarian cancer should be investigated via further assays. For example, whether SNHG9 regulates the progression of ovarian cancer via the PI3K/AKT, NF-kB or Wnt signaling pathways should be assessed.
In summary, SNHG9 was demonstrated to be downregulated in human ovarian cancer cell lines and tissues, and its expression was positively associated with overall survival in patients with ovarian cancer. Decreased SNHG9 expression in ovarian cancer cells was demonstrated to promote cell migration, colony formation, proliferation, and invasion in vitro. Taken together, the results suggest that SNHG9 may participate in ovarian cancer development and progression by regulating the miR-214-5p/CRY2 axis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YL made substantial contributions to the conception and design of the present study. GC, ZL, YC and YL contributed to the acquisition, analysis and interpretation of data, drafted the initial manuscript and critically revised it for important intellectual content. All authors read and approved the final manuscript. and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved (according to the ICMJE).
Ethics approval and consent to participate
The present study was approved by the Institutional Ethics Committee of The Second People's Hospital of Taizhou City (Taizhou, China). All population-associated experiments were performed in accordance with The Declaration of Helsinki, and written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tew WP. Ovarian cancer in the older woman. J Geriatr Oncol. 2016;7:354–361. doi: 10.1016/j.jgo.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Dong X, Men X, Zhang W, Lei P. Advances in tumor markers of ovarian cancer for early diagnosis. Indian J Cancer. 2014;51(Suppl 3):e72–e76. doi: 10.4103/0019-509X.154049. [DOI] [PubMed] [Google Scholar]
- 4.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497–2505. doi: 10.3892/ijo.2015.2943. [DOI] [PubMed] [Google Scholar]
- 6.Lai XJ, Cheng HF. lncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol Sci. 2018;22:322–328. doi: 10.26355/eurrev_201801_14175. [DOI] [PubMed] [Google Scholar]
- 7.Liu SP, Yang JX, Cao DY, Shen K. Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol Med. 2013;10:138–141. doi: 10.7497/j.issn.2095-3941.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017;21:3–15. doi: 10.18869/acadpub.ibj.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 10.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Bunch H. Gene regulation of mammalian long non-coding RNA. Mol Genet Genomics. 2018;293:1–15. doi: 10.1007/s00438-017-1370-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: Potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Kirby JE, Sunwoo H, Lee JT. Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 2016;30:1747–1760. doi: 10.1101/gad.281162.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanduri C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim Biophys Acta. 2016;1859:102–111. doi: 10.1016/j.bbagrm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Malakootian M, Mirzadeh Azad F, Fouani Y, Taheri Bajgan E, Saberi H, Mowla SJ. Anti-differentiation non-coding RNA, ANCR, is differentially expressed in different types of brain tumors. J Neurooncol. 2018;138:261–270. doi: 10.1007/s11060-018-2809-5. [DOI] [PubMed] [Google Scholar]
- 16.Ranzani V, Arrigoni A, Rossetti G, Panzeri I, Abrignani S, Bonnal RJP, Pagani M. Next-generation sequencing analysis of long noncoding RNAs in CD4+ T cell differentiation. Methods Mol Biol. 2017;1514:173–185. doi: 10.1007/978-1-4939-6548-9_14. [DOI] [PubMed] [Google Scholar]
- 17.Dong J, Xu J, Wang X, Jin B. Influence of the interaction between long noncoding RNAs and hypoxia on tumorigenesis. Tumour Biol. 2016;37:1379–1385. doi: 10.1007/s13277-015-4457-0. [DOI] [PubMed] [Google Scholar]
- 18.Lou Y, Jiang H, Cui Z, Wang L, Wang X, Tian T. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/β-catenin signaling. Oncotarget. 2017;8:69983–69994. doi: 10.18632/oncotarget.19545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng ZG, Xu H, Suo SS, Xu XL, Ni MW, Gu LH, Chen W, Wang LY, Zhao Y, Tian B, Hua YJ. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. 2016;6:26093. doi: 10.1038/srep26093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao C, Zhao D, Zhao Q, Dong D, Mu L, Zhao X, Guo M, Xu A, Fang L, Liu Q, Che J. Microarray profiling and co-expression network analysis of lncRNAs and mRNAs in ovarian cancer. Cell Death Discov. 2019;5:93. doi: 10.1038/s41420-019-0173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10:2648–2658. [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Li Y, Zhou L, Yang G, Wang M, Hong Y. Cryptochrome 2 (CRY2) suppresses proliferation and migration and regulates clock gene network in osteosarcoma cells. Med Sci Monit. 2018;24:3856–3862. doi: 10.12659/MSM.908596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang L, Yang Z, Zhou J, Tung JY, Hsiao CD, Wang L, Deng Y, Wang P, Wang J, Lee MH. Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Mol Cancer Ther. 2015;14:1476–1487. doi: 10.1158/1535-7163.MCT-15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira JI, Higashimoto M, Mashiko M, Ito K, Niikura H, Takenoshita SI, Yaegashi N. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87:1060–1070. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Helder-Woolderink JM, Blok EA, Vasen HF, Hollema H, Mourits MJ, De Bock GH. Ovarian cancer in Lynch syndrome; a systematic review. Eur J Cancer. 2016;55:65–73. doi: 10.1016/j.ejca.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Miao H, Wang L, Zhan H, Dai J, Chang Y, Wu F, Liu T, Liu Z, Gao C, Li L, Song X. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019;15:e1008144. doi: 10.1371/journal.pgen.1008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Shen H, Wang Y. CRY2 is suppressed by FOXM1 mediated promoter hypermethylation in breast cancer. Biochem Biophys Res Commun. 2017;490:44–50. doi: 10.1016/j.bbrc.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Li P, Tong L, Song Y, Sun J, Shi J, Wu Z, Diao Y, Li Y, Wang Z. Long noncoding RNA H19 participates in metformin-mediated inhibition of gastric cancer cell invasion. J Cell Physiol. 2019;234:4515–4527. doi: 10.1002/jcp.27269. [DOI] [PubMed] [Google Scholar]
- 30.Yao P, Ni Y, Liu C. Long non-coding RNA 691 regulated PTEN/PI3K/AKT signaling pathway in osteosarcoma through miRNA-9-5p. Onco Targets Ther. 2020;13:4597–4606. doi: 10.2147/OTT.S249827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu F, Zhou G, Huang K, Fan X, Li G, Chen B, Dong P, Zheng J. Serum lincRNA-p21 as a potential biomarker of liver fibrosis in chronic hepatitis B patients. J Viral Hepat. 2017;24:580–588. doi: 10.1111/jvh.12680. [DOI] [PubMed] [Google Scholar]
- 32.Birgani MT, Hajjari M, Shahrisa A, Khoshnevisan A, Shoja Z, Motahari P, Farhangi B. Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol Oncol Res. 2018;24:329–337. doi: 10.1007/s12253-017-0241-3. [DOI] [PubMed] [Google Scholar]
- 33.Fu LL, Li CJ, Xu Y, Li LY, Zhou X, Li DD, Chen SX, Wang FG, Zhang XY, Zheng LW. Role of lncRNAs as novel biomarkers and therapeutic targets in ovarian cancer. Crit Rev Eukaryot Gene Expr. 2017;27:183–195. doi: 10.1615/CritRevEukaryotGeneExpr.2017019244. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, Zhang Y, Yang H, Xuan Y, Yang Y, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]
- 35.Zhang LQ, Yang SQ, Wang Y, Fang Q, Chen XJ, Lu HS, Zhao LP. Long noncoding RNA MIR4697HG promotes cell growth and metastasis in human ovarian cancer. Anal Cell Pathol (Amst) 2017;2017:8267863. doi: 10.1155/2017/8267863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.