H3K27me3 demethylase REF6 suppresses seed dormancy and promotes seed germination by enhancing the catabolism of the plant hormone Abscisic acid.
Abstract
Seed dormancy is an adaptive trait that is crucial to plant survival. Abscisic acid (ABA) is the primary phytohormone that induces seed dormancy. However, little is known about how the level of ABA in seeds is determined. Here we show that the Arabidopsis (Arabidopsis thaliana) H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 (REF6) suppresses seed dormancy by inducing ABA catabolism in seeds. Seeds of the ref6 loss-of-function mutants displayed enhanced dormancy that was associated with increased endogenous ABA content. We further show that the transcripts of two genes key to ABA catabolism, CYP707A1 and CYP707A3, but not genes involved in ABA biosynthesis, were significantly reduced in ref6 mutants during seed development and germination. In developing siliques, REF6 bound directly to CYP707A1 and CYP707A3, and was responsible for reducing their H3K27me3 levels. Genetic analysis demonstrated that the enhanced seed dormancy and ABA concentration in ref6 depended mainly on the reduced expression of CYP707A1 and CYP707A3. Conversely, overexpression of CYP707A1 could offset the enhanced seed dormancy of ref6. Taken together, our results revealed an epigenetic regulation mechanism that is involved in the regulation of ABA content in seeds.
Seeds are vital for the survival and distribution of seed-plant species. Seed germination is the beginning of plant development, and the purpose of plant growth is to produce seeds. In mature seeds, the initiation of germination is determined by a period of quiescence, which is known as seed dormancy (Baskin and Baskin, 2004; Finch-Savage and Leubner-Metzger, 2006; Shu et al., 2013). Seed dormancy is an adaptive trait that ensures seeds germinate only when environmental conditions are favorable for seedling development and reproductive success, and dormant seeds do not germinate even under optimal conditions (Nonogaki, 2014; Shu et al., 2016; Chahtane et al., 2017; Yang et al., 2020). Under optimal conditions, nondormant Arabidopsis (Arabidopsis thaliana) seed sets usually finish germination within 48 h after imbibing, while dormant seed sets can germinate only partially or not at all. The speed of seed germination under suitable environments is widely used to evaluate seed dormancy of seed sets (Baskin and Baskin, 2004; Okamoto et al., 2006; Matakiadis et al., 2009; Shu et al., 2013). Seed dormancy is mainly induced by abscisic acid (ABA) generated from both zygotic tissues and the mother plant (Karssen et al., 1983; Kanno et al., 2010) and broken by GA during the imbibing stage of early germination (Yamauchi et al., 2004; Finch-Savage and Leubner-Metzger, 2006; Nonogaki, 2014; Shu et al., 2016). In developing Arabidopsis seeds, the endogenous ABA content reaches a maximum level in the middle stage of seed development, when embryo growth and seed filling occur, ∼9 to 10 d after flowering (Jo et al., 2019). An additional small peak of ABA accumulation takes place late in development at ∼15 to 16 d after flowering, when the seed-coat–browning process is initiated, and seeds (embryos) acquire desiccation tolerance and become quiescent (Okamoto et al., 2006; Kanno et al., 2010). On the other hand, the ABA in dry seeds decreases rapidly during seed imbibition and then maintains a basal level that is correlated with the germination potential of the seed (Okamoto et al., 2006). However, how the level of ABA during seed development and germination is determined is not well understood.
In Arabidopsis, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE2 (AtNCED2), AtNCED5, AtNCED6, AtNCED9, ABA DEFICIENT1, ABA DEFICIENT2, and ALDEHYDE OXIDASE3 (AtAAO3) are key enzymes involved in ABA biosynthesis. The loss of function of genes encoding these enzymes leads to decreased endogenous ABA content and thus reduced seed dormancy (González-Guzmán et al., 2002; Seo and Koshiba, 2002; Tan et al., 2003). Conversely, overexpression of these genes can increase ABA content in seed and enhance seed dormancy and thus delay seed germination (Tan et al., 2003; Kushiro et al., 2004; Chahtane et al., 2017).
ABA is hydroxylated at positions C-7′ and C-8′ in many plant species (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006), and a 9′-hydroxylation pathway has also been identified in several plants (Zhou et al., 2004). The ABA 8′-hydroxylation pathway is thought to be predominant in many physiological processes and catalyzed by three cytochrome P450 monooxygenases, CYP707A1, CYP707A2, and CYP707A3 (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006; Matakiadis et al., 2009). Loss-of-function mutations of ABA catabolism genes CYP707A1, CYP707A2, and CYP707A3 result in enhanced seed dormancy associated with increased endogenous ABA content in seeds (Kushiro et al., 2004; Okamoto et al., 2006). The CYP707A1 and CYP707A3 genes were found to be expressed predominantly during the middle stage of seed development, downregulated during the late maturation stage, and maintained at a low level in dry seed, while the CYP707A2 transcript levels increase from the late-stage to mature dry seed (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006; Matakiadis et al., 2009). The transcription of all three CYP707As is induced in the early imbibing stage.
Histone methylation and demethylation are essential for transcriptional regulation and play a fundamental role in regulating diverse developmental processes, such as flowering, architecture, and abiotic stress tolerance (for review, please see Liu et al., 2010a). Histone methylation is dynamically regulated by Lys (Lys, K) methyltransferases (“writers”), and demethylases (“erasers”). Methylation occurs mainly at Lys-4 (K4), Lys-9 (K9), Lys-27 (K27), and Lys-36 (K36) of histone H3 (Liu et al., 2010a). Those Lys residues can be augmented with different numbers (1 to 3) of methyl groups, which confer various biological functions. Generally, methylations at H3K9 and H3K27 are associated with gene silencing, whereas methylations at H3K4 and H3K36 are related to gene activation (Berger, 2007). As a repressive mark, H3K27me3 represses gene expression by a conserved mechanism in eukaryotes (Zhang et al., 2007; Hennig and Derkacheva, 2009). In Arabidopsis, H3K27me3 is catalyzed by histone methyltransferases, i.e. CURLY LEAF, MEDEA, or SWINGER, which are essential components of Polycomb Repressive Complex2 (Liu et al., 2010a). The demethylation of H3K27me3 in Arabidopsis is catalyzed by several Jumonji (JMJ) domain-containing proteins, including RELATIVE OF EARLY FLOWERING6 (REF6), EARLY FLOWERING6, and JMJ13, JMJ30, and JMJ32 (Liu et al., 2010a; Lu et al., 2011; Gan et al., 2014; Yan et al., 2018).
Arabidopsis REF6 was first reported as a repressor of FLOWERING LOCUS C to promote flowering (Noh et al., 2004; Lu et al., 2011; Gan et al., 2014). The loss-of-function ref6-1 mutant shows diverse phenotypes, including suppressed leaf senescence and lateral root formation (Wang et al., 2019a, 2019b), defects in cotyledon separation (Cui et al., 2016), and Brassinosteroid-related phenotype (Yu et al., 2008). The deletion of REF6 leads to the ectopic accumulation of H3K27me3 at hundreds of genes in seedlings (Lu et al., 2011; Li et al., 2016; Cui et al., 2016). Other groups and ours previously showed that REF6 zinc-finger domains could directly target genomic loci containing CTCTGYTY motifs (Y represents C or T; Li et al., 2016, 2018; Qiu et al., 2019; Wang et al., 2019a). In this study, we demonstrate that REF6 is a negative regulator of seed dormancy and is involved in the decrease of ABA content during seed germination. We show that REF6 suppresses seed dormancy via inducing ABA catabolism during seed development and germination. Our results revealed an epigenetic regulation mechanism involved in the regulation of ABA content in seeds.
RESULTS
Loss-of-Function of REF6 Results in Enhanced Seed Dormancy
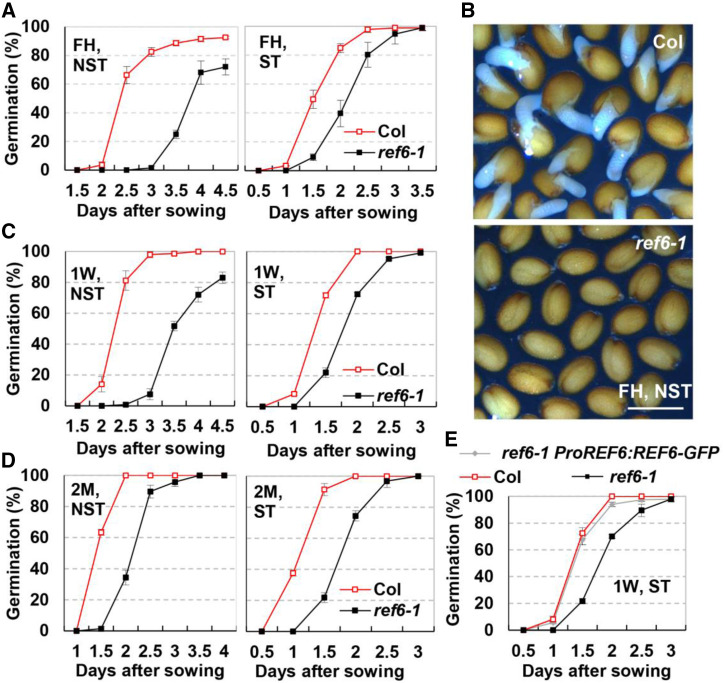
In a previous work on REF6 (Li et al., 2016), we occasionally observed that the seeds of ref6-1 germinated later than Columbia (Col) wild type. The expression of REF6 was observed during seed development, seed storage, and seed germination stages (Supplemental Fig. S1A). Furthermore, ABA treatment induced the expression of REF6 during seed germination (Supplemental Fig. S1B). These results imply that REF6 may be involved in the regulation of seed dormancy and germination. To test this hypothesis, we conducted detailed germination experiments using Col and ref6 mutant seeds that were stored for different time points. Without stratification treatment (ST), we observed that ∼70% of freshly harvested (FH) wild-type seeds germinated at 2.5 d after sowing, while no ref6-1 seeds germinated (Fig. 1A, left, and B). Although the difference in germination ratio between Col and ref6-1 seeds was reduced after ST compared with no stratification treatment (NST), the germination of ref6-1 seeds was still obviously delayed compared with that of wild-type Col (Fig. 1A, right). Seeds stored at room temperature for 1 week or 2 months (after-ripened) were further used in the germination assay. The results showed that, in both conditions, the germination of ref6-1 seeds was substantially slower than that of Col (Fig. 1, C and D). To confirm that the delay of germination was caused by the mutation in REF6, the seed germination rate of a complemented transgenic line, ref6-1 ProREF6:REF6-GFP (Li et al., 2016), was tested. The result showed that the seed germination rate of ref6-1 ProREF6:REF6-GFP was similar to Col (Fig. 1E). Taken together, these results demonstrate that REF6 is involved in suppressing seed dormancy.
Figure 1.
Loss-of-function mutant ref6-1 displayed enhanced seed dormancy. FH seeds were stored at room temperature for 0 weeks (FH), 1 week (1W), or 2 months (2M) before being subjected to analysis. Seeds were sterilized and stored at 4°C for 0 d (NST) or 4 d (ST) before sowing. A, The germination rate of FH seeds without (left) or with (right) ST. B, Representative pictures showing germinating seeds at 72 h after sowing. FH seeds without ST were sown on one-half strength Murashige and Skoog (1/2 MS). Scale bar = 0.6 mm. C and D, The germination rate of seeds stored for 1W (C), 2M (D) without (left), or with (right) ST on one-half strength MS medium. E, The germination rate of 1-week-stored seeds of Col, ref6-1, and ref6-1 ProREF6:REF6-GFP with ST on one-half strength MS medium. For A, C, D and E, values (mean ± sd) are from three biological replicates, each consisting of three technical replicates. At least 50 dry mature seeds were used in each technical replication.
The Endogenous ABA Content in Seeds of ref6-1 Is Significantly Increased
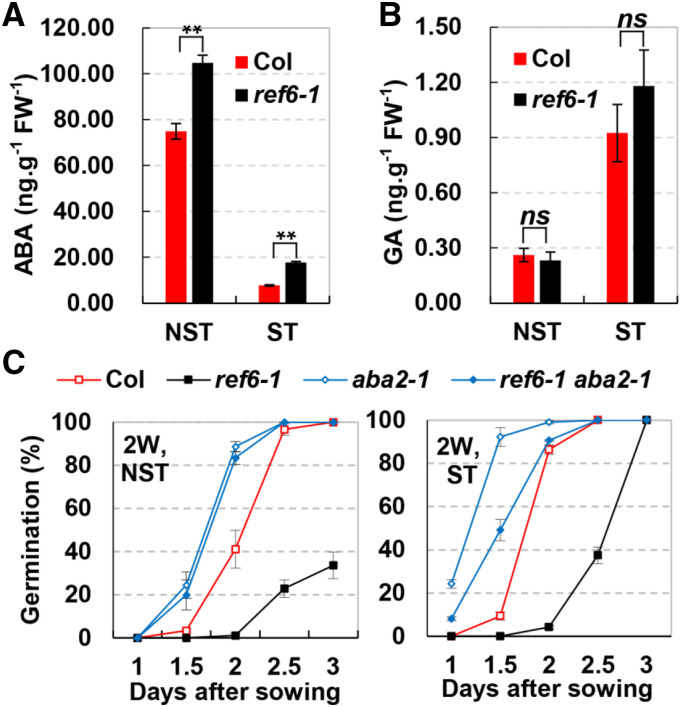
Seed dormancy is antagonistically regulated by ABA and GA (Bewley, 1997; Chahtane et al., 2017). The enhanced seed dormancy in ref6-1 could be due to reduced GA or increased ABA levels. Therefore, we quantified the endogenous ABA and GA contents in ref6-1 seeds using a liquid chromatography-tandem mass spectrometry system. In FH dry seeds with NST, the ABA level of ref6-1 was significantly higher than that of Col (Fig. 2A). After ST, although the ABA contents in seeds of both Col and ref6-1 were reduced, the ABA content in ref6-1 was still significantly higher than that in Col (Fig. 2A). In contrast to ABA, the GA level in the FH seed of ref6-1 was similar to that of Col with ST or NST (Fig. 2B). Thus, the loss of REF6 causes the increase of the ABA level in seeds.
Figure 2.
The enhanced seed dormancy of ref6 depends on the increased endogenous ABA content. A and B, The ABA (A) and GA (B) contents in FH seeds with ST or NST for 4 d. Values (mean ± sd) are from three biological replicates, each consisting of ∼100 mg of FH seeds. Asterisks indicate statistical significance of differences determined using Student’s t test (*P < 0.05 and **P < 0.01). ns, No significance. C, Loss-of-ABA2 reduced the enhanced seed dormancy of ref6-1. The germination rates of 2-week-stored (2W) seeds of various genotypes were indicated. Values are from three biological replicates (mean ± sd), each consisting of three technical replicates. At least 50 dry mature seeds were used in each technical replication.
To test if there is a causal relationship between the increased ABA level in ref6-1 and enhanced seed dormancy, we generated ref6-1 aba2-1 double mutants by crossing. aba2-1 is a mutant with substantially decreased ABA content due to the loss of function of ABA2, a gene encoding one of the key enzymes in ABA synthesis (González-Guzmán et al., 2002; Léon-Kloosterziel et al., 1996). Two-week-stored seeds were used in the germination assay. Without stratification, the germination rate of ref6-1 aba2-1 was similar to that of aba2-1 but substantially higher than that of ref6-1 (Fig. 2C, left). After stratification, the germination rate of ref6-1 aba2-1 was also dramatically higher than that of ref6-1 in the first 2.5 d (Fig. 2C, right). These results indicated that the reduction of ABA content by aba2 could reduce the seed dormancy of ref6-1. Together, these results support a notion that REF6 suppresses seed dormancy by reducing endogenous ABA levels in seeds.
The Transcripts of CYP707A1 and CYP707A3 Were Significantly Reduced in ref6-1 during Seed Development and Germination
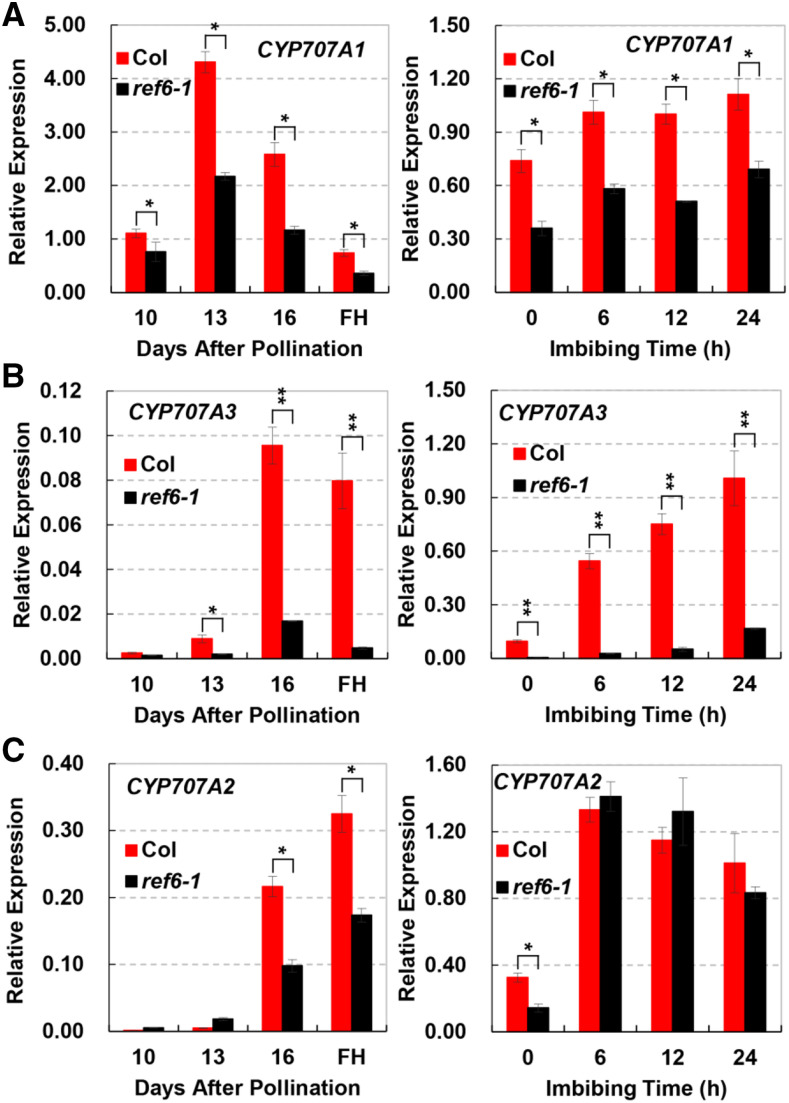
ABA content is dynamically regulated by enzymes involved in ABA biosynthesis and catabolism. To identify key factors that are responsible for the increased ABA content in seeds of ref6-1, we quantified the transcripts of genes related to ABA biosynthesis (ABA1, ABA2, AAO3, NCED2, NCED5, NCED6, and NCED9) and catabolism (CYP707A1, CYP707A2, and CYP707A3) during seed development and germination. The results indicated that, during seed development, there was no significant difference in the transcripts of ABA biosynthesis genes between Col and ref6 (Supplemental Fig. S2A). On the other hand, the expression levels of two ABA catabolism genes, CYP707A1 and CYP707A3, in ref6-1 were continuously and significantly lower throughout seed development and early germination stages than they were in Col (Fig. 3, A and B). The transcription of CYP707A2 in ref6-1 was substantially lower only at 16 d after pollination (DAP) and dry seed than in Col (Fig. 3C, left). During the early germination, the expression level of CYP707A2 was similar between ref6-1 and Col (Fig. 3C, right). It is worth noting that, during the seed development, the expression levels of CYP707A1 in both Col and ref6-1 were higher than those of CYP707A2 and CYP707A3, while during seed germination, the highest expressed gene was CYP707A2 (Supplemental Fig. S2B). These results indicate that the increased ABA concentration in ref6-1 is associated with the reduced expression of genes involved in the catabolism of ABA, but not with those involved in ABA biosynthesis.
Figure 3.
Suppressed ABA catabolism genes in seeds of ref6-1. A to C, The relative transcript levels of CYP707A1 (A), CYP707A3 (B), and CYP707A2 (C) in seeds of ref6-1 and Col during seed development and germination. The developing siliques (10, 13, and 16 DAP) and FH seeds imbibed for different times (0, 6, 12, and 24 h) under dark conditions at 22°C were used in the assay. The FH seeds were the same as the seeds imbibed for 0 h. The gene expression was relative to ACTIN2. Values (mean ± sd) represent three biological replicates, each consisting of three technical replicates. Asterisks indicate statistical significance of differences determined using Student’s t test (*P < 0.05 and **P < 0.01).
REF6 Directly Binds to The CYP707A1 and CYP707A3 Genes during Seed Development
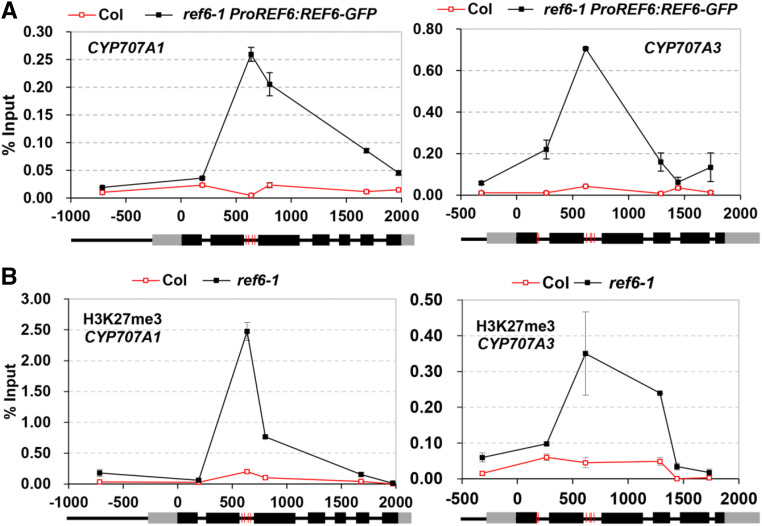
We investigated whether the involvement of REF6 in CYP707A1 and CYP707A3 expression during seed development is direct. So, we used chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) to detect the binding of REF6 to CYP707A1 and CYP707A3 in developing siliques (11 to 15 DAP) of ref6-1 ProREF6:REF6-GFP. The results displayed that REF6 was substantially enriched at the second intron of CYP707A1 and CYP707A3 (Fig. 4A; Supplemental Fig. S3A). No REF6 binding signal was found in CYP707A2 (Supplemental Fig. S3B). The REF6 binding peak was centered on the four CTCTGYTY motifs in CYP707A1 and CYP707A3, respectively, whereas CYP707A2 had no detectable CTCTGYTY motif. These results suggest that REF6 directly binds to the CYP707A1 and CYP707A3 but not CYP707A2 in developing siliques. We further examined the levels of H3K27me3 at the CYP707A1, CYP707A2, and CYP707A3 genes in siliques of Col and ref6-1 by ChIP-qPCR. Consistent with the results above, the H3K27me3 levels at the CYP707A1 and CYP707A3 genes in ref6 were remarkably higher than those in Col (Fig. 4B; Supplemental Fig. S3B), whereas there was no noticeable increase of H3K27me3 at CYP707A2 (Supplemental Fig. S3D). Together, these data support the idea that REF6 directly promotes the expression of CYP707A1 and CYP707A3 genes during seed development by reducing their H3K27me3 levels.
Figure 4.
REF6 binds to CYP707A1 and CYP707A3 to reduce the level of H3K27me3 in developing seeds. A, ChIP-qPCR showing that REF6 binds to the CYP707A1 and CYP707A3 genes during seed development. Developing siliques (10 to 15 DAP) of Col and ref6-1 ProREF6:REF6-GFP were used in this assay. B, ChIP-qPCR results showing the level of H3K27me3 at CYP707A1 and CYP707A3 in Col and ref6-1 during seed development. Developing siliques (13 DAP) of Col and ref6-1 were used in this assay. ChIP signals are displayed as the percentage of input DNA. Col plants were used as the negative control sample. Values (mean ± sd) represent three biological replicates, each consisting of three technical replicates. Schematic representation of the CYP707A1 and CYP707A3 genomic loci are shown underneath. Black and gray boxes represent exons and untranslated regions, respectively. The red vertical lines indicate the CTCTGYTY motifs in CYP707A1 and CYP707A3.
The Enhanced Seed Dormancy and Endogenous ABA Concentration of ref6-1 Depend on The Reduced Expression of CYP707A1 and CYP707A3
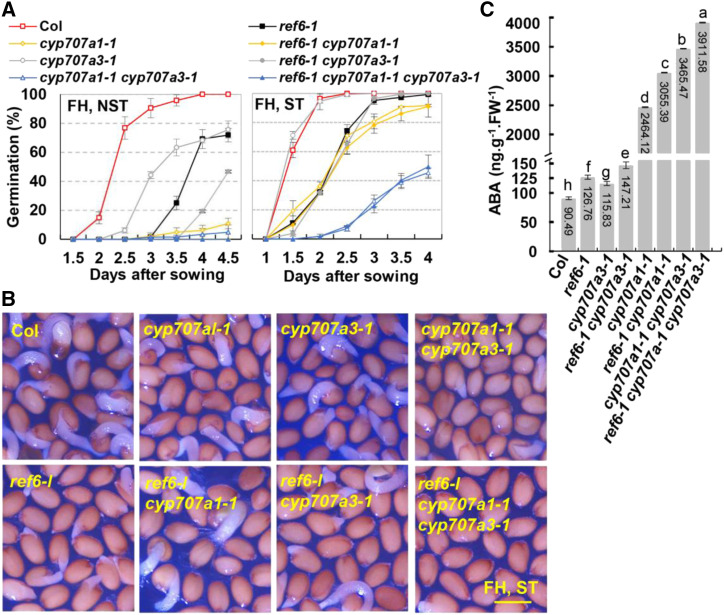
As described above, the transcription levels of the CYP707A1 and CYP707A3 in ref6-1 were significantly reduced during seed development and germination (Fig. 3, A and B). To explore the causal relationship between ABA catabolism genes and the enhanced seed dormancy of ref6-1, we generated double and triple mutants by crossing ref6-1 with loss-of-function mutants of CYP707A1 (cyp707a1-1), CYP707A2 (cyp707a2-1), and CYP707A3 (cyp707a3-1). FH harvested seeds were used in the seed germination assays. With NST, the seed germination rate of double mutant ref6-1 cyp707a3-1 was substantially lower than that of single mutant ref6-1 or cyp707a3-1 (Fig. 5A, left). After ST, the seed germination rate of ref6-1 cyp707a3-1 was lower than that of cyp707a3-1 in the first 2.5 d, but similar to that of ref6-1 (Fig. 5A, right, and B; Table 1). With NST, after 4 d, the seed germination rates of ref6-1 and cyp707a1-1 were ∼69% and 6%, respectively, while the seeds of ref6-1 cyp707a1-1 failed to germinate (Fig. 5A, left; Table 1). After ST, the seed germination rate of ref6-1 cyp707a1-1 was very close to that of cyp707a1-1 (Fig. 5A, right and B; Table 1). The results of germination assays of triple mutant ref6-1 cyp707a1-1 cyp707a3-1 demonstrated that, despite ST, the seed germination rate of ref6-1 cyp707a1-1 cyp707a3-1 were similar with that of double mutant cyp707a1-1 cyp707a3-1 (Fig. 5, A and B; Table 1). With regard to the cyp707a2-1, in contrast, we found that the seed germination percentages of double or triple mutants that contain ref6-1 were lower than that of single or double mutants that do not contain ref6-1, respectively (Table 1; Supplemental Fig. S4A). Furthermore, the quantification of ABA in FH seeds demonstrated that the increased endogenous ABA contents of various mutants well-corresponded with the seed germination results (Fig. 5C; Supplemental Fig. S4B). These results suggest that the enhanced seed dormancy of ref6-1 was predominantly dependent on the reduction of CYP707A1 and CYP707A3 but not dependent on CYP707A2.
Figure 5.
The enhanced seed dormancy and endogenous ABA concentration of ref6-1 depend on the reduced expression of CYP707A1 and CYP707A3. A, The seed germination rates of Col and various mutants. FH seeds with or without ST were used in the experiments. Values (mean ± sd) are from three biological replicates, each consisting of three technical replicates. At least 50 dry mature seeds were used in each technical replication. B, Representative pictures showing germinating seeds at 2.5 d after sowing. FH seeds with ST were sown on one-half strength MS. Scale bar = 0.6 mm. C, The endogenous ABA concentration in FH seeds of Col and various mutants. Values (mean ± sd) are from three biological replicates, each consisting of ∼100 mg of FH seeds. Lowercase letters indicate statistical differences determined using one-way ANOVA with Duncan’s multiple range test (for FH-NST-4D, the F value = 373,910 and degrees of freedom = 13).
Table 1. The seed germination percentages of each genotype at 4 or 2.5 d after sowing.
Lowercase letters indicate statistically significant differences, which were determined with the software SAS (https://welcome.oda.sas.com/login) using one-way ANOVA and Duncan’s multiple range test (for FH-NST-4 days, the F value = 536.51 and degrees of freedom = 13; for FH-ST-2.5 days, the F value = 260.08 and degrees of freedom = 13).
| Genotype | Germination (%) FH NST (4 d) | Germination (%) FH ST (2.5 d) |
|---|---|---|
| Col | 100 a | 100 a |
| ref6-1 | 69.20 ± 6.51 b | 74.37 ± 4.11 c |
| cyp707a1-1 | 6.22 ± 3.46 e | 70.91 ± 4.49 c,d |
| cyp707a2-1 | 29.14 ± 2.68 c | 89.39 ± 7.56 b |
| cyp707a3-1 | 68.22 ± 1.68 b | 99.21 ± 1.37 a |
| cyp707a1-1 cyp707a2-1 | 0 f | 3.86 ± 3.19 i,j |
| cyp707a1-1 cyp707a3-1 | 4.05 ± 3.3 e,f | 7.04 ± 1.75 h,i,j |
| cyp707a2-1 cyp707a3-1 | 5.80 ± 1.78 e | 36.52 ± 2.83 f |
| ref6-1 cyp707a1-1 | 0 f | 62.76 ± 4.27 e |
| ref6-1 cyp707a2-1 | 0 f | 19.95 ± 5.24 g |
| ref6-1 cyp707a3-1 | 19.39 ± 2.71 d | 65.18 ± 7.02 d,e |
| ref6-1 cyp707a1-1 cyp707a2-1 | 0 f | 0 j |
| ref6-1 cyp707a1-1 cyp707a3-1 | 0 f | 8.82 ± 0.49 h,i |
| ref6-1 cyp707a2-1 cyp707a3-1 | 0 f | 12.17 ± 4.01 h |
Overexpression of CYP707A1 Offset the Enhanced Seed Dormancy of ref6-1
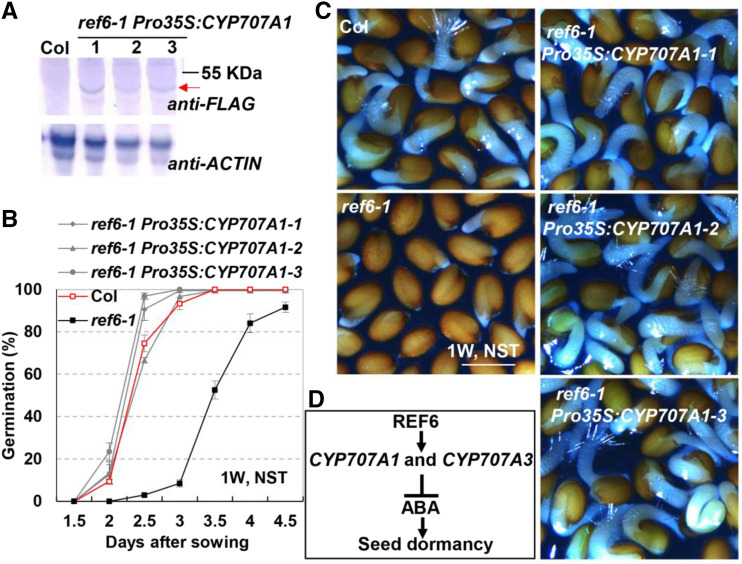
As described above, the decreased expression of CYP707A1 is mainly responsible for the enhanced seed dormancy of ref6-1. To further verify this, CYP707A1 was overexpressed in ref6-1 through generating transgenic plants ref6-1 Pro35S:CYP707A1. Three independent transgenic lines that showed overexpressed CYP707A1 were obtained (Fig. 6A). Seed germination analysis with the transgenic seeds exhibited that the germination rates of the three ref6-1 Pro35S:CYP707A1 lines were similar to that of Col, and were substantially higher than that of ref6-1 (Fig. 6, B and C). These results showed that overexpression of CYP707A1 could rescue the dormancy phenotype of ref6-1.
Figure 6.
Overexpression of CYP707A1 reduced the seed dormancy of ref6-1. A, Western-blotting analysis of three independent ref6-1 Pro35S:CYP707A1 transgenic lines. Seven-day-old seedlings were used to extract protein. ACTIN was used as the loading control. The red arrow indicates the recombinant protein CYP707A1-4XFLAG. Experiments were repeated twice, and the graph shows the results of one of the experiments. B, Seed germination rates of Col, ref6-1 and ref6-1 Pro35S:CYP707A1 lines. One-week-stored seeds without ST were subjected to analysis. Values (mean ± sd) are from three biological replicates, each consisting of three technical replicates. At least 50 dry mature seeds were used in each technical replication. C, Representative pictures showing germinated seeds at 72 h after sowing. One-week–stored seeds without ST were sown on one-half strength MS medium. Scale bar = 0.6 mm. D, A working model showing the molecular mechanism by which REF6 suppresses seed dormancy.
DISCUSSION
ABA is a crucial hormone for seed development, dormancy, germination, and abiotic stress responses (Chen et al., 2020b). However, how the level of ABA is regulated in seeds is far from clear. In this study, we found that histone H3K27 demethylase REF6 is involved in the reduction of ABA content during seed development and germination (Fig. 2A). Seed dormancy is conferred by ABA in dry seed (Finch-Savage and Leubner-Metzger, 2006; Shu et al., 2013) and released by GA generated during seed germination (Bewley, 1997; Chahtane et al., 2017). In the dry seeds, the ABA content of ref6-1 was significantly higher than that of Col (Fig. 2A), while no apparent difference in GA levels between them was observed (Fig. 2B), indicating that REF6 is involved in reducing ABA content in seeds. The loss of function of REF6 enhances seed dormancy and reduces seed germination (Fig. 1). Further genetic experiments support the notion that the increased ABA level is responsible for the enhanced seed dormancy of ref6-1 because aba2-1 ref6-1 double mutants reduced the seed dormancy of ref6-1 (Fig. 2C). Due to the reduced expression of CYP707A1 and CYP707A3 in developing ref6-1 seeds (Fig. 3, A and B), ABA content in the FH ref6-1 seeds was significantly higher than that in Col (Fig. 2A). During seed imbibing, the significantly reduced expression of CYP707A1 and CYP707A3 in ref6-1 will lead to reduced efficiency of ABA catabolism during the ST in ref6-1 seeds compared to Col. Together, the higher amount of ABA in FH ref6-1 seeds as well as the reduced catabolism of ABA during the stratification in ref6-1 seeds would result in higher ABA level in stratified ref6 seeds. Consistent with this, after ST, ABA concentration in ref6-1 seeds was still obviously higher than that in Col (Fig. 2A), while the seed germination rate of ref6-1 was lower than that of Col (Fig. 1A, right). After ST for 4 d, the ABA levels in both Col and ref6-1 were dramatically decreased, but the difference in ABA content between Col and ref6-1 became smaller (Fig. 2A). In addition, after ST, the GA content of ref6-1 was slightly higher than that of Col (Fig. 2B). These may be responsible for the smaller difference in seed germination rate between ref6 and Col after ST (Fig. 1A).
Both the biosynthesis and catabolic pathways of ABA in plants have been uncovered (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006; Matakiadis et al., 2009); however, how the key genes in the two pathways are transcriptionally regulated is still largely unknown. ABI4 has been found to inhibit the transcription of CYP707A1 and CYP707A2 and thus reduces the catabolism of ABA (Shu et al., 2013). Reactive oxygen species (ROS) were reported to promote the transcription of CYP707As and thus decrease the ABA content in seeds (Bailly and Bailly, 2004; Liu et al., 2010b; Chen et al., 2020a). We show here that REF6 is involved in the regulation of ABA catabolism genes (Fig. 3), but not of biosynthesis genes in seeds (Supplemental Fig. S2A). REF6 is an H3K27me3 demethylase that directly binds to CTCTGYTY motifs in its target loci (Lu et al., 2011, 2016, 2018; Cui et al., 2016; Qiu et al., 2019; Wang et al., 2019a). We found that REF6 directly bound to (Fig. 4A) and promoted the expression of CYP707A1 and CYP707A3 during seed development and germination (Fig. 3, A and B). Loss of REF6 led to the reduced transcript levels of CYP707A1 and CYP707A3 during seed development (Fig. 3A) that were accompanied by the increased H3K27me3 levels at the two genes (Fig. 4B). During seed development, the expression of REF6 was increased from 10 DAP to 13 DAP (Supplemental Fig. S1A). Therefore, it can be inferred that the H3K27me3 demethylation activity of REF6 should be stronger at 13 DAP than that at 10 DAP, and thus the expression of CYP707A1 and CYP707A3 at 13 DAP would be higher than that at 10 DAP. Consistent with this speculation, our results showed that the expression of CYP707A1 and CYP707A3 at 13 DAP was higher than that at 10 DAP (Fig. 3, A and B). The expression of REF6 also well corresponded to that of CYP707A1 and CYP707A3 during seed imbibing (Fig. 3, A and B; Supplemental Fig. S1A). These observations support the notion that the H3K27me3 demethylation activity of REF6 is likely coupled with the regulation of CYP707A1 and CYP707A3. It would be useful to generate a REF6 that only lacks the demethylation activity in ref6-1 mutant, and then measure whether the expression of CYP707A1 and CYP707A3 in this material could be recovered. Interestingly, REF6 did not bind to CYP707A2 (Supplemental Fig. S3B), indicating that the expression of CYP707A2 in the dry seed is likely indirectly regulated by REF6. The different regulation mechanisms between CYP707A1/CYP707A3 and CYP707A2 by REF6 may be responsible for the different spatiotemporal expression patterns observed for CYP707A2 and CYP707A1/CYP70A3 (Fig. 3; Kushiro et al., 2004; Okamoto et al., 2006).
All three CYP707As can be responsible for ABA catabolism; however, each CYP707 gene plays a different role during seed development and germination. CYP707A2 plays a major role in the rapid decrease of ABA levels during early seed imbibition (Kushiro et al., 2004; Matakiadis et al., 2009), whereas CYP707A1 and CYP707A3 are mainly involved in reducing the ABA content in developing seeds (Kushiro et al., 2004; Okamoto et al., 2006). A similar level of expression of CYP707A2 in imbibing seeds between Col and ref6-1 (Fig. 3C) argued against the involvement of CYP707A2 in REF6-regulated ABA catabolism (Fig. 2A). Consistent with this argument, the ABA content in the double mutant ref6-1 cyp707a2-1 was significantly higher than that of the single mutant ref6-1 or cyp707a2-1 (Supplemental Fig. S4B). Moreover, introduction of ref6-1 into cyp707a2-1 cyp707a3-1 and cyp707a1-1 cyp707a2-1 enhanced ABA content in both double mutants. As a result, the seed germination rate of double mutant ref6-1 cyp707a2-1 was substantially lower than that of single mutant ref6-1 or cyp707a2-1 (Table 1; Supplemental Fig. S4A). The seed germination rates of triple mutants ref6-1 cyp707a2-1 cyp707a3-1 and ref6-1 cyp707a1-1 cyp707a2-1 were also lower than those of cyp707a2-1 cyp707a3-1 and cyp707a1-1 cyp707a2-1, respectively (Supplemental Fig. S4A).
Previous studies have reported that CYP707A1 is the primary enzyme for ABA 8’-hydroxylation during the midmaturation stage of seed (Okamoto et al., 2006). We show here that the significantly reduced transcription of CYP707A1 and CYP707A3 (Fig. 3, A and B) in ref6-1 is responsible for the higher ABA content of ref6-1 (Fig. 2A). Consistently, the seed germination of triple mutant ref6-1 cyp707a1-1 cyp707a3-1 was similar to those of double mutant cyp707a1-1 cyp707a3-1 (Fig. 5A; Table 1). However, the germination rate of double mutant ref6-1 cyp707a1-1 was similar to that of cyp707a1-1, while the seed germination of double mutant ref6-1 cyp707a3-1 was similar to that of ref6-1 (Fig. 5A; Table 1). These results suggest that the enhanced seed dormancy of ref6-1 is predominantly dependent on the reduction of CYP707A1 and, to a lesser extent, of CYP707A3. Consistent with this, CYP707A1 had the highest expression level in developing seeds among the three CYP707As (Supplemental Fig. S2B). Moreover, overexpression of CYP707A1 in ref6-1 could completely offset the enhanced seed dormancy (Fig. 6).
In summary, we have revealed an epigenetic mechanism involved in the regulation of ABA concentration in seeds (Fig. 6D). We show that during seed development, REF6 directly upregulates the expression of CYP707A1 and CYP707A3 to promote the catabolism of ABA, leading to a relatively low level of ABA content, and thus a suppression of seed dormancy. As a result, the ref6 mutant seeds display an enhanced seed dormancy phenotype. Given the widespread conservation of the H3K27me3 demethylases in crops, including rice (Oryza sativa; Cheng et al., 2018) and maize (Zea mays; Qian et al., 2019), we anticipate our findings will prove informative for understanding the mechanisms governing the ABA level in diverse crops. Revealing how plants regulate the ABA level would represent a key step toward our understanding of the action of ABA in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0 [Col]) was used as wild type. Seeds were sterilized with 10% (v/v) NaClO and stratified at 4°C for 0 d (NST) or 4 d (ST) before being sown on one-half strength MS (Duchefa) medium. Seven-day-old seedlings were transferred to soil and grown in a greenhouse under 23°C ± 1°C with a 16-h photoperiod/8-h dark. For all experiments, plants were grown under identical conditions, and the seeds were harvested from different plants at the same time and pooled. The mutants ref6-1 and aba2-1 were obtained from the Arabidopsis Biological Resource Center at Ohio State University; https://abrc.osu.edu/). The mutants, cyp707a1-1, cyp707a2-1, and cyp707a3-1, were kindly provided by Eiji Nambara. The complemented line ref6-1 ProREF6:REF6-GFP was generated in our previous report (Li et al., 2016). Multiple mutants were generated by crossing the ref6-1 with aba2-1, cyp707a1-1, cyp707a2-1, and cyp707a3-1, respectively.
Seed Germination Assay
Plants of each genotype were grown under the same condition, and seeds were harvested at the same time. FH seeds stored for different times were used in seed germination assays. Radicle protrusion was regarded as the indicator of a germinated seed. The seed germination experiments were carried out three times, and each consists of three technical replicates. At least 50 dry mature seeds were used in each technical replication for each genotype.
Generation of Transgenic Plants
The open reading frame of CYP707A1 was amplified by PCR using the gene-specific primers listed in Supplemental Table S1, digested by KpnI and SalI, and subcloned into the pCAMBIA1306 vector, which drives the expression of CYP707A1-4×FLAG under the CaMV35S promoter. Resultant binary constructs were introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform the ref6-1 mutant plants using the floral dip method (Clough and Bent, 1998). The transformed seeds were selected by hygromycin to get transgenic plants ref6-1 Pro35S:CYP707A1, which can be detected by an anti-FLAG antibody in an immunoblotting analysis.
RNA Extraction and Gene Expression Analysis
Developing siliques and seeds of each genotype were used to extract total RNA. Total RNA was extracted and reverse-transcribed as described in Chen et al. (2020a). The relative gene expression was determined by quantitative real-time PCR, which was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using the SYBR Green Realtime PCR Master Mix (TOYOBO). Gene-specific primers are listed in Supplemental Table S1. Gene expression was relative to ACTIN2. Data represent three biological replicates, each consisting of three technical replicates.
Protein Extraction and Immunoblotting
One-hundred milligrams of 7-d-old seedlings were used to extract total protein. Seedlings were ground in liquid nitrogen and then incubated in 100 μL of protein extraction buffer (20 mm of Tris-HCl at pH7.5, 150 mm of NaCl, 0.5% [v/v] Tween 20, 1 mm of EDTA, and 1 mm of dithiothreitol) containing a protease inhibitor cocktail (“Complete,” catalog no. 04693116001; Roche). After gentle shaking for 30 min at 4°C, the supernatant was collected by two centrifugations (10 min, 16,000g, 4°C).
Immunoblotting analysis was performed with the specific antibodies anti-FLAG (catalog no. M20008; Abmart) and anti-ACTIN (catalog no. M20009; Abmart) according to Chen et al. (2018).
Quantification of ABA and GA
Plants of each genotype were grown under the same condition, and seeds were harvested from different plants at the same time. Approximately 100 mg of FH seeds were used to quantitate the endogenous ABA and GA concentrations. Seeds were frozen by liquid nitrogen, and then the ABA and GA contents were determined by liquid chromatography-tandem mass spectrometry (8030 Plus; Shimadzu) as previously described for ABA (Zhou et al., 2017) or GA (Yang et al., 2012). The experiments were repeated three times (three biological replicates, each consisting of three technical replicates), and similar results were obtained.
ChIP-qPCR
ChIP experiments were conducted as described in the literature (Li et al., 2016, 2018; Chen et al., 2017; Yu et al., 2020), with minor modifications. Briefly, 2 g of developing siliques (10 to 15 DAP) were collected and ground into fine powder in liquid nitrogen and then well homogenized with extraction buffer 1 (2 m of Suc, 1 m of Tris-HCl at pH 8.0, 1 m of MgCl2, 14.3 m of β-mercaptopropionate, and 0.1 m of phenylmethylsulfonyl fluoride [protease inhibitor cocktail]). After that, 37% (v/v) formaldehyde was added immediately to the homogenized buffer until the final concentration was 1%, and cross linked on a shaker for 15 min at 4°C and stopped by 0.125 m of Gly. Chromatin was isolated and sheared into 500- to 1,000-bp fragments by sonication. The sonicated chromatin was incubated with 4 μL of antibody to GFP (catalog no. ab290; Abcam) or H3K27me3 (catalog no. 07-449; Millipore) overnight at 4°C. Precipitated DNA was then dissolved with sterilized water. ChIP-qPCR was performed with three technical replicates, and results were calculated as the percentage of input DNA according to the Champion ChIP-qPCR user manual (SABioscience). Values are from three biological replicates. Sequences for the primers used in the ChIP-qPCR are listed in Supplemental Table S1.
Statistical Analysis
Statistical significance of differences was determined using Student’s t test and one-way ANOVA with Duncan’s multiple range test.
Accession Numbers
Genes referenced in this article can be found in The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: REF6 (At3g48430), CYP707A1 (At4g19230), CYP707A2 (At2g29090), CYP707A3 (At5g45340), NCED6 (At3g24220), NCED9 (At1g78390), NCED2 (At4g18350), NCED5 (At1g30100), ABA1 (At5g67030), ABA2 (At1g52340), and AAO3 (At2g27150).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The expression pattern of REF6 in seeds in different stages or treated by ABA.
Supplemental Figure S2. The transcription levels of ABA biosynthesis and catabolism genes in siliques or seeds of ref6-1 and Col.
Supplemental Figure S3. REF6 does not bind CYP707A2.
Supplemental Figure S4. CYP707A2 does not contribute to the enhanced seed dormancy of ref6-1.
Supplemental Table S1. Primer sequences used in this study.
Acknowledgments
We thank Eiji Nambara for cyp707a1-1, cyp707a2-1, and cyp707a3-1 seeds.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31870289 to C.L., 31700283 to H.C., 31570372 to L.X., and 31871716 to J.L.), the Fundamental Research Funds for the Central Universities (Sun Yat-sen University grant no. 18lgzd12 to C.L.), and the Natural Science and Engineering Research Council of Canada (grant no. RGPIN/04625–2017 to Y.C.).
Articles can be viewed without a subscription.
References
- Bailly C, Bailly C(2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14: 93–107 [Google Scholar]
- Baskin JM, Baskin CC(2004) A classification system for seed dormancy. Seed Sci Res 14: 1–16 [Google Scholar]
- Berger SL.(2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bewley JD.(1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahtane H, Kim W, Lopez-Molina L(2017) Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J Exp Bot 68: 857–869 [DOI] [PubMed] [Google Scholar]
- Chen C, Li C, Wang Y, Renaud J, Tian G, Kambhampati S, Saatian B, Nguyen V, Hannoufa A, Marsolais F, et al. (2017) Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3: 814–824 [DOI] [PubMed] [Google Scholar]
- Chen H, Ruan J, Chu P, Fu W, Liang Z, Li Y, Tong J, Xiao L, Liu J, Li C, et al. (2020a) AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J 101: 310–323 [DOI] [PubMed] [Google Scholar]
- Chen HH, Qu L, Xu ZH, Zhu JK, Xue HW(2018) EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Mol Plant 11: 706–719 [DOI] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y(2020b) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62: 25–54 [DOI] [PubMed] [Google Scholar]
- Cheng S, Tan F, Lu Y, Liu X, Li T, Yuan W, Zhao Y, Zhou DX(2018) WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res 46: 2356–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui X, Lu F, Qiu Q, Zhou B, Gu L, Zhang S, Kang Y, Cui X, Ma X, Yao Q, et al. (2016) REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat Genet 48: 694–699 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G(2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Gan ES, Xu Y, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T(2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5: 5098. [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL(2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M(2009) Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Jo L, Pelletier JM, Harada JJ(2019) Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J Integr Plant Biol 61: 564–580 [DOI] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M(2010) Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol 51: 1988–2001 [DOI] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DL, Breekland AE, Koornneef M(1983) Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E(2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M(1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Li C, Chen C, Chen H, Wang S, Chen X, Cui Y(2018) Verification of DNA motifs in Arabidopsis using CRISPR/Cas9-mediated mutagenesis. Plant Biotechnol J 16: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, et al. (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X(2010a) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye N, Liu R, Chen M, Zhang J(2010b) H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J Exp Bot 61: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X(2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou JP, Kamiya Y, Nambara E, Truong HN(2009) The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol 149: 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS(2004) Divergent roles of a pair of homologous Jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H.(2014) Seed dormancy and germination-emerging mechanisms and new hypotheses. Front Plant Sci 5: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E(2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Chen C, Jiang L, Zhang J, Ren Q(2019) Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genomics 20: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Mei H, Deng X, He K, Wu B, Yao Q, Zhang J, Lu F, Ma J, Cao X(2019) DNA methylation repels targeting of Arabidopsis REF6. Nat Commun 10: 2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M(2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koshiba T(2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH(2016) Two faces of one seed: Hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q(2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR(2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Wang X, Gao J, Gao S, Li Z, Kuai B, Ren G(2019a) REF6 promotes lateral root formation through de-repression of PIN1/3/7 genes. J Integr Plant Biol 61: 383–387 [DOI] [PubMed] [Google Scholar]
- Wang X, Gao J, Gao S, Song Y, Yang Z, Kuai B(2019b) The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet 15: e1008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S(2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Chen D, Smaczniak C, Engelhorn J, Liu H, Yang W, Graf A, Carles CC, Zhou DX, Kaufmann K(2018) Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat Plants 4: 681–689 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu S, Lin R(2020) The role of light in regulating seed dormancy and germination. J Integr Plant Biol 62: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y(2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liang Z, Song X, Fu W, Xu J, Lei Y, Yuan L, Ruan J, Chen C, Fu W, et al. (2020) BRAHMA-interacting proteins BRIP1 and BRIP2 are core subunits of Arabidopsis SWI/SNF complexes. Nat Plants 6: 996–1007 [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE(2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LJ, Xiao LT, Xue HW(2017) Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol 174: 1728–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Cutler AJ, Ambrose SJ, Galka MM, Nelson KM, Squires TM, Loewen MK, Jadhav AS, Ross AR, Taylor DC, et al. (2004) A new abscisic acid catabolic pathway. Plant Physiol 134: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]