Transcriptomic and proteomic data from the basal angiosperm Amborella provide a resource to understand the evolution of male gametophyte functions in flowering plants.
Abstract
Flowering plants (angiosperms) are characterized by pollen tubes (PTs; male gametophytes) carrying two immobile sperm cells that grow over long distances through the carpel toward the ovules, where double fertilization is executed. It is not understood how these reproductive structures evolved, which genes occur de novo in male gametophytes of angiosperms, and to which extent PT functions are conserved among angiosperms. To contribute to a deeper understanding of the evolution of gametophyte functions, we generated RNA sequencing data from seven reproductive and two vegetative control tissues of the basal angiosperm Amborella trichopoda and complemented these with proteomic data of pollen grains (PGs) and PTs. The eudicot model plant Arabidopsis (Arabidopsis thaliana) served as a reference organism for data analysis, as more than 200 genes have been associated with male gametophyte functions in this species. We describe methods to collect bicellular A. trichopoda PGs, to induce their germination in vitro, and to monitor PT growth and germ cell division. Transcriptomic and proteomic analyses indicate that A. trichopoda PGs are prepared for germination requiring lipids, energy, but likely also reactive oxygen species, while PTs are especially characterized by catabolic/biosynthetic and transport processes including cell wall biosynthesis and gene regulation. Notably, a number of pollen-specific genes were lacking in Arabidopsis, and the number of genes involved in pollen signaling is significantly reduced in A. trichopoda. In conclusion, we provide insight into male gametophyte functions of the most basal angiosperm and establish a valuable resource for future studies on the evolution of flowering plants.
During the evolution of seed plants and the colonization of dry environments, male gametes (sperm cells) have lost their mobility and thus need a vehicle to be transported toward the major female gamete (egg cell). This is achieved by the development of a male gametophyte (pollen) capable of forming a tip-growing tube. In angiosperms (flowering plants), the pollen tube (PT) must grow over long distances through the maternal tissues of the carpel to reach the female gametophyte (embryo sac) inside an ovule that is sheltered within the ovary. Together with the double fertilization process involving two distinct female gametes (egg and central cell), these innovations represent main characteristics of angiosperms distinguishing them from all other plants, including gymnosperms. PTs carrying two immobile sperm cells over long distances thus not only play a key role during double fertilization, they are also highly interesting to understand the successful evolution of angiosperms, now representing about 90% of all living land plants (Sauquet et al., 2017). Notably, comparative studies of male gametophytes in gymnosperms and angiosperms also revealed remarkable differences in pollen grain (PG) morphology, PT walls, and growth rates, indicating that pollen–pollen played a central role in the rapid diversification of angiosperms (Williams, 2008, 2012).
In this context, analyses of the most basal living angiosperm, Amborella trichopoda, a sister of all other extant angiosperms, is especially interesting to understand the evolution of pollen–pollen function(s) and morphology that contributed to the success of flowering plants. A. trichopoda diverged very early on (about 130 million years ago) from all the other extant ∼300,000 species of angiosperms and is now considered as the most basal living angiosperm (Drew et al., 2014). It therefore has a unique position to shed light on the evolution of sexual organs and gametophyte development and the associated functions in angiosperms. A. trichopoda is a dioecious woody shrub endemic to New Caledonia. Once per year, male individuals produce thousands of small flowers (Fig. 1) that generate large numbers of PGs. Detailed morphological and histological studies have been performed to examine its PGs and to follow PT growth in vivo (Hesse, 2001; Williams, 2009). However, nothing is known about the underlying molecular and physiological processes. Although the A. trichopoda genome was published a few years ago (Amborella Genome Project, 2013), transcriptomic data are currently very limited, and comparisons with well-studied angiosperm model plants like Arabidopsis (Arabidopsis thaliana) are mostly restricted to predicted genes and gene families, respectively.
Figure 1.
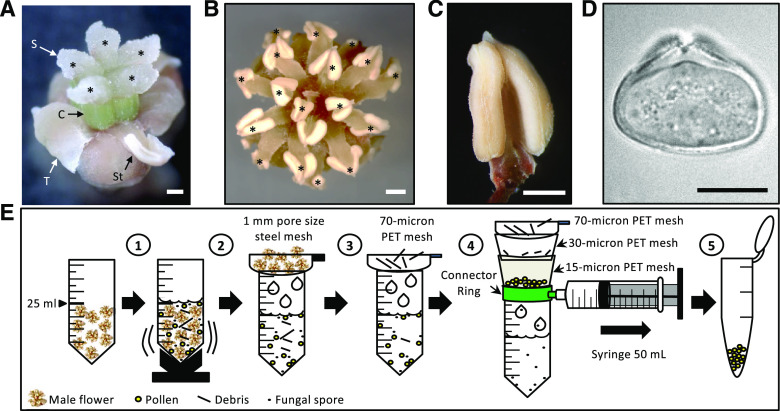
Reproductive organs of A. trichopoda and PG isolation procedure. A, Mature female flower consisting of five to six carpels (C) with a wet stigma (S; here the five stigmata are labeled with an asterisk), five to eight tepals (T), and two to three staminoids (St; here one is visible). B, Mature male flower showing 10 to 25 anthers (here 17 are labeled with an asterisk). C and D, Dissected anther (C) and mature PG showing the porus (D). E, Schematic diagram describing the process of PG isolation in five steps as indicated. See text for details. Scale bars = 1 mm (A–C) and 10 μm (D).
In contrast, a considerable amount of information has been accumulated during the past 30 years to understand male gametophyte development and function in Arabidopsis and to a lesser extent in other angiosperm species including crops like maize (Zea mays) and rice (Oryza sativa; Zhou and Dresselhaus, 2019). Among other discoveries, the research community has learned that Arabidopsis has lost genetic mechanisms to avoid self-fertilization (Nasrallah, 2019) and that rapid PT germination and proper navigation through the transmitting tract is critical for reproductive success (Johnson et al., 2019; Lopes et al., 2019). Furthermore, fundamental features and underlying molecular mechanisms of PT tip growth were elucidated, and mechanisms and players regulating cell wall rigidity of this fastest growing plant cell have been identified. Most progress has been made in the past years to understand the mechanisms how PTs exit the transmitting tract in Arabidopsis, how they are guided toward and inside the ovule, and how their burst is regulated to release their sperm cell cargo. Interactions of polymorphic peptide ligands with receptor-like kinases have been shown to play key roles during male-female interactions (Qu et al., 2015; Zhong and Qu, 2019). While mechanisms of polar PT tip growth appear conserved in angiosperms, different proteins are used, for example, for PT guidance and burst (Zhou and Dresselhaus, 2019). Studies involving the basal angiosperm A. trichopoda therefore have a high potential to reveal the evolution, conservation, and specificity of above-mentioned reproductive processes and their roles during reproductive isolation and thus speciation during angiosperm diversification.
As a first step to obtain a deeper understanding of the evolution of male gametophytes and their functions in angiosperms, we describe here methods to collect and purify mature bicellular PGs of A. trichopoda and how they can be germinated and monitored in vitro. PT growth was measured and timing of germ cell division determined. In addition, we generated transcriptomic data from nine different A. trichopoda tissues, including PGs as well as bicellular and tricellular PTs and studied transcriptome dynamics. Proteomic profiles were generated from PGs and tricellular PTs and compared with corresponding transcriptomic data. By using Arabidopsis as a reference angiosperm, we searched for putative orthologs of key proteins involved in male gametophyte function. In conclusion, we provide a valuable resource of transcriptomic and proteomic data of A. trichopoda for usage of evolutionary comparative studies of plant sexual reproduction. Furthermore, we identified putative conserved signaling pathways, many genes with yet-unknown functions as well as an the expansion of signaling components in Arabidopsis PTs, suggesting evolutionary diversification over the course of angiosperm evolution.
RESULTS
Bicellular A. trichopoda Pollen Generates Two Sperm Cells (Tricellular Stage) within 14 h during PT Growth In Vitro
It is difficult to cultivate A. trichopoda successfully outside their natural environment in New Caledonia. For many years, the Botanical Garden in Bonn (Germany) has optimized growth conditions for dioecious A. trichopoda plants (Grosse-Veldmann et al., 2011). We collected PG and ovules during the flowering period (September to early December) from male and female flowers born on male and female plants as well as vegetative plant material from a male plant through four successive years (2015–2018). To study A. trichopoda PGs and PTs (representing the male gametophyte), we first established protocols to isolate mature PGs in large quantities and to induce their germination in vitro. Despite the small size of A. trichopoda flowers (Fig. 1, A and B), bulk harvesting of mature PGs from anthers (Fig. 1, C and D) was successful using the procedure described in Figure 1E. In brief, this procedure consists of collecting 20 male flowers in a 50-mL tube, adding 25 mL of Camellia japonica pollen germination medium (CJ-PGM) and vortexing the tube for 45 s to release PGs into the solution. Afterward, the suspension is first filtered through a 1-mm pore size mesh to remove large debris and then subsequently filtered through strainers of 70-, 30-, and 15-micron pore size to remove small debris and to separate contaminating fungal spores that we found in all PG preparations. Finally, clean PGs with a diameter of about 25 µm (Fig. 1D) are retained on the 15-micron strainer, recovered from the surface, and transferred to a 1.5-mL collection tube for further analysis.
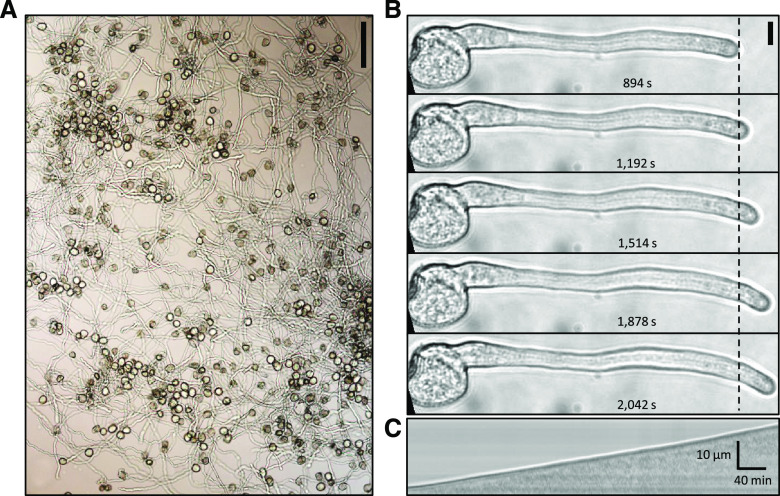
To induce in vitro pollen germination, five different PGMs were tested (Supplemental Table S1) and the germination ratio evaluated 10 h after incubation. The maximum germination rate (83.17%) was observed using liquid CJ-PGM (Fig. 2A; Nakamura et al., 1982). We next determined that A. trichopoda pollen tube growth rate (PTGR) on solid CJ-PGM was 100.46 ± 0.5 µm h−1 (Fig. 2B; Supplemental Table S2). Time-lapse imaging revealed that PT growth in A. trichopoda is linear in vitro and without recognizable oscillations as indicated in a x/t-projection (kymograph; Fig. 2C).
Figure 2.
A. trichopod in vitro pollen germination and PT growth. A, In vitro pollen germination on solidified CJ-PGM. Scale bar = 100 µm. B, Time series of a growing PT monitored 15 to 34 min after germination on solid medium. An average growth rate of 100.46 ± 0.5 µm h−1 was observed. Scale bar = 20 µm. C, Kymograph analysis of the PT shown in B. The growth profile appears linear lacking any signs of oscillation. Scale bars for time and distance are shown in the bottom right corner.
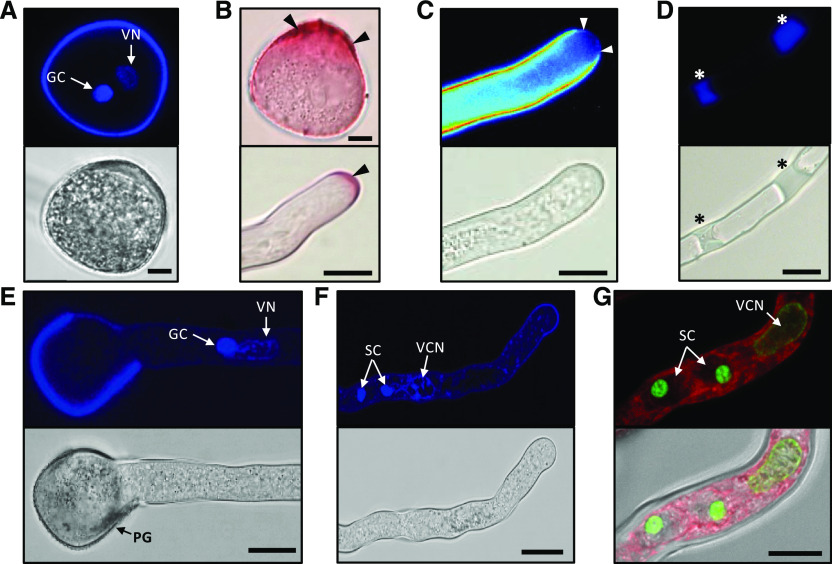
We then analyzed nuclei content and cell wall material of PGs and PTs. The 4′,6-diamino-phenylindole staining confirmed that A. trichopoda generates bicellular PGs including a single generative cell (GC) and a tube cell whose vegetative nucleus contains less compact chromatin compared with the GC (Fig. 3A). The presence and location of pectin in the cell wall of PGs and PTs was explored by staining with ruthenium red. The highest amounts of pectin were detectable at the aperture zone of the PG as well as in the tip of growing PTs but were also present at significant levels at the shank of the whole PT (Fig. 3B). Aniline blue staining showed the presence of callose deposition in the PT wall and the formation of extended callose plugs during progression of PT growth (Fig. 3, C and D). Notably, callose is fully absent from the PTs apex showing strong pectin staining. The vegetative nucleus and the GC remain associated during PG germination and PT growth, with the vegetative nucleus moving ahead (Fig. 3E). At 14 h after germination, the division of the GC generated two sperm cells (tricellular stage; Fig. 3F). SYBR GREEN I staining in combination with the membrane counter stain FM 4-64 revealed that on average the length of sperm cells in the PT is 10 μm. Compared with the chromatin of the GC and the vegetative tube cell, the chromatin of sperm cells is highly condensed (Fig. 3G). About 90% germinated PTs are tricellular 14 h after incubation on CJ-PGM. This time point was used to sample PTs at the tricellular stage for further experimentation.
Figure 3.
A. trichopoda PG and PT characteristics. A, 4′,6-diamino-phenylindole (DAPI) staining of a mature bicellular PG (top) and its brightfield image (bottom). B, Ruthenium red staining of a mature PG (top image) and a PT tip (bottom). Black arrowheads point at pectin-rich regions. C and D, Aniline blue staining of PTs (top) and respective brightfield images (bottom). The PT apex indicated by white arrowheads in C is devoid of callose-like substances, whereas callose plugs become visible in the shank of the PT marked by asterisks in D. E, PG 9 h after in vitro germination showing the PT with the vegetative nucleus followed by the GC. F, PT 14 h after in vitro germination showing the vegetative nucleus followed by the two sperm cells. G, SYBR GREEN I-stained PT 14 h after in vitro germination showing the male germ unit (the vegetative cell nucleus [VN] and two sperm cells [SC]) in more detail. FM 4-64 (red) served as counter stain for membranes. Note the highly compact sperm cell chromatin. The tip of the tube is to the right. Scale bars = 10 μm.
Transcriptomes of A. trichopoda Male Gametophytes at Different Stages Are Highly Similar and Show Low Dynamics
In order to visualize differences in gene expression patterns before and during PT germination, we investigated the A. trichopoda transcriptomes of mature pollen grains (PGs) as well as PTs at the bicellular (PT-Bi) and tricellular (PT-Tri) stages. After RNA extraction, library preparation, and Illumina sequencing, a total of 46.6 million reads were generated from PGs, 63.88 million reads from PT-Bi, and 58.01 million reads from PT-Tri stages and uniquely mapped to the A. trichopoda genome v1 (Amborella Genome Project, 2013), which was predicted to contain 27,313 genes. Additionally, RNA sequencing (RNA-seq) data were generated from A. trichopoda ovaries, leaves, male and female flowers, tepals, and roots, resulting in 8.91, 22.55, 80.25, 81.96, 50.17, and 18.60 million mapped reads, respectively. An overview of the RNA-seq data obtained for all these tissues is provided in Table 1. RNA-seq data were validated by RT-qPCR by selecting nine genes with high, medium, and low log2 fold change (log2FC) values among the tissue comparisons (Supplemental Fig. S1). A master table showing RNA-seq gene expression levels of all A. trichopoda genes in transcripts per million (TPM) values for each of the three biological replicates in the seven tissues is provided in Supplemental Table S3.
Table 1. Overview of RNA-seq data obtained from nine A. trichopoda tissues.
Mapped reads, number of expressed genes (TPM ≥ 1), and preferentially male gametophyte-expressed genes in PG and PT at the PT-Bi and PT-Tri stage. Male flowers (MALE) that contain male gametophytes were excluded from the analysis to identify preferentially expressed genes. Percentage refers to total number of protein-coding genes (27,313 genes; Amborella Genome Project, 2013).
| Tissue Type | Sample | Mapped Reads | No. of Expressed Genes | No. of Preferentially Expressed Genesa |
|---|---|---|---|---|
| Millions | % | % | ||
| Male Gametophytic Tissues | PG | 46.60 | 10,134 (37.10) | 470 (1.72) |
| PT-Bi | 63.88 | 11,384 (41.67) | 652 (2.38) | |
| PT-Tri | 58.01 | 11,002 (40.28) | 611 (2.23) | |
| Flower Tissues | MALE | 80.25 | 15,611 (57.15) | – |
| FEMALE | 81.96 | 16,802 (61.51) | – | |
| OVARY | 8.91 | 6,144 (22.49) | – | |
| TEPAL | 50.17 | 14,396 (52.70) | – | |
| Vegetative Tissues | LEAF | 22.55 | 15,003 (54.93) | – |
| ROOT | 18.60 | 16,174 (59.22) | – |
Genes with a TPM ≥ 1 in a given tissue and TPM ≤ 3 in sporophytic tissues (ovary, leaf, tepal and root; see Supplemental Table S4).
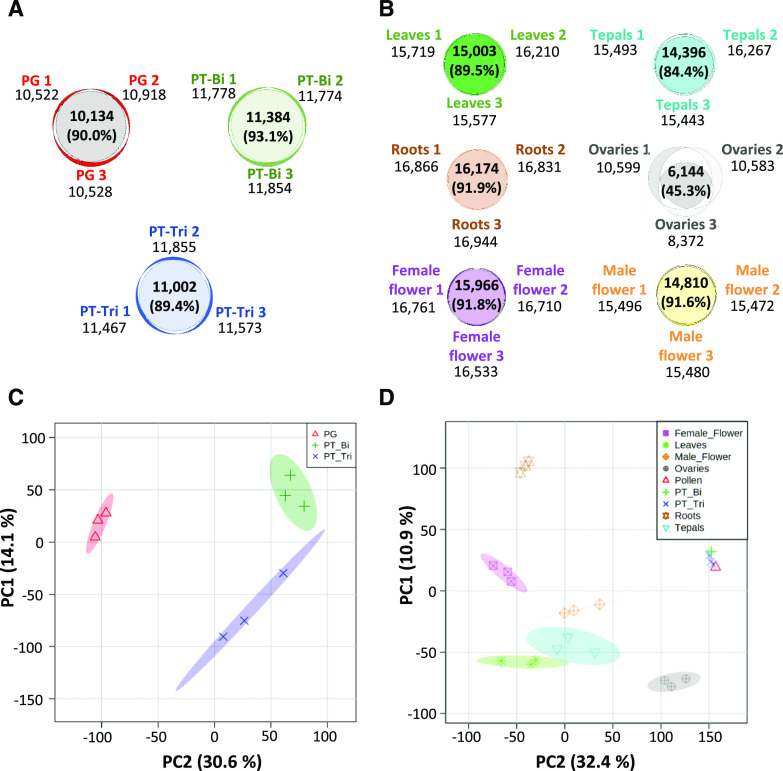
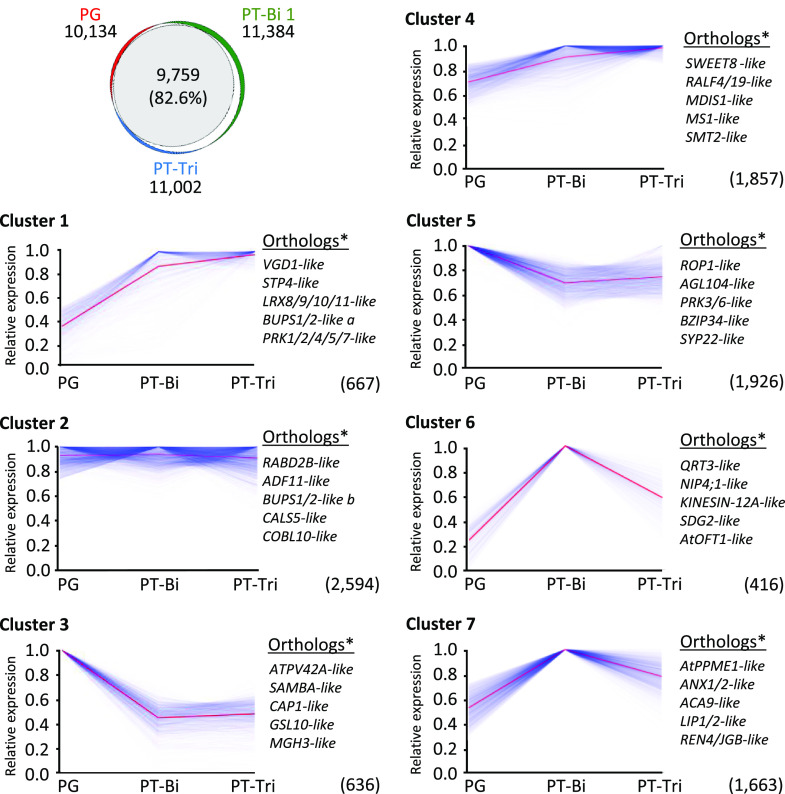
To determine the number of genes expressed in stages PG, PT-Bi, and PT-Tri, we considered a gene to be expressed at TPM values ≥ 1. A Venn diagram analysis revealed overlaps of 90.0%, 93.1%, and 89.4% within biological replicates from PG, PT-Bi, and PT-Tri samples, respectively (Fig. 4A). With the exception of ovule samples showing only an overlap of 45.3% between biological replicates and thus only 6,144 expressed genes in all samples, other control samples (leaves, roots, flowers, and tepals) showed an overlap between 84.4% to 91.9% and on average >15,000 expressed genes (Fig. 4B). We assume that the lower overlap between the three ovary samples that consequently resulted in a lower number of genes present in all samples results from tissue collection at slightly different developmental stages. There were 10,134 genes expressed in all PG samples, while PT-Bi showed 10,384 and PT-Tri 11,002 expressed genes. Additionally, principal component analyses (PCAs) were performed to analyze the similarity between the biological replicates and among the three pollen developmental stages and the control tissues. As shown in Figure 4C, with the exception of the PT-Tri stage, which still contained about 10% of PTs at stage PT-Bi, all biological replicates cluster closely together and the three male gametophyte stages differ considerably from each other. These findings support data validity and accurate sampling. A PCA of all nine samples, including leaves, roots, flowers, tepals, and ovaries, shows that the three male gametophyte stages group very closely together (Fig. 4D). In conclusion, the high overlap among biological replicates and the presence of different clearly separable subgroups in the PCAs underlines the high reproducibility among the different samples and indicates that transcriptomes of A. trichopoda male gametophytes at different developmental stages are highly similar.
Figure 4.
Venn diagrams and PCAs of transcriptomes from three male gametophyte stages and four sporophytic control tissues of A. trichopoda. A, Overlap of expressed genes (transcript per million [TPM] ≥ 1) in each of the three biological replicates generated from mature bicellular PGs and germinated PTs at the PT-Bi and PT-Tri stage, respectively. B, Overlap of expressed genes in sporophytic tissues indicated. For each, three biological replicates were generated. C, PCA plot of expressed genes within three biological replicates of PG, PT-Bi, and PT-Tri. D, PCA plot of samples shown in C compared with sporophytic tissues. For each tissue, three biological replicates are shown.
Next, we aimed to identify differentially expressed genes (DEGs) among PG, PT-Bi, and PT-Tri stages. For that purpose, we compared PG versus PT-Bi, PG versus PT-Tri, and PT-Bi versus PT-Tri by using the DESeq2 program (q < 0.05; Supplemental Table S4A). We consider those genes as DEGs whose TPM levels are ≥10 in all replicates of at least one of the compared tissues and a log2FC ≤ −2 and log2FC ≥ 2, respectively (Supplemental Table S4B). A total of only 301 DEGs were detected between PG and PT-Bi, 220 between PG and PT-Tri, and 26 between PT-Bi and PT-Tri. In order to obtain further insights into functional information during male gametophyte transition from PGs to tricellular PTs, we analyzed biological processes defined by MapMan bins (Usadel et al., 2005) in a pairwise manner with identified DEGs (Supplemental Table S4). Supplemental Figure S2 shows a summary bar chart representing the number of DEGs that are down- or upregulated in each comparison for a given bin. Despite the fact that many DEGs in each comparison are not assigned to any biological process, especially genes related to cell wall and cytoskeleton metabolism, protein degradation and synthesis as well as transcription regulation, signaling, and transport are up-regulated in PTs in PT-Bi and PT-Tri in comparison to PG. Only a few upregulated DEGs were identified in a PT-Bi-PT-Tri comparison, which are primarily associated to stress responses.
Additionally, we sought to explore the transcriptome dynamics between PG, PT-Bi, and PT-Tri stages and identify those genes with a similar expression pattern by a k-mean clustering analysis (Fig. 5; Supplemental Table S5A). The three male gametophyte stages show an overlap of 9,759 expressed genes and distribute into seven k-mean clusters. The largest cluster 2 containing 2,594 genes (26.58% of all overlapping genes) shows comparable expression levels in all samples. Stage-specific patterns are present in six out of the seven clusters, although, in general, transcript levels are only moderately changed (e.g. down-regulated to 50% in PTs compared to PGs in cluster 3). Cluster 1 and cluster 4 (together about 25% overlapping genes) show genes up-regulated during PT growth. These encode genes homologous to Arabidopsis genes coding for RALF, LRX, and BUPS, as well as other receptor kinases like PRKs (see next section and Supplemental Table S5B for details about gene identifiers). Cluster 6 and cluster 7 (together about 22% overlapping genes) display genes transiently upregulated at the PT-Bi stage. Among these genes there are a polygalacturonase QRT3 with a reported role in degrading the pollen mother cell wall during microspore development, a pollen-specific aquaporin (NIP4;1) required for pollen development, and a SET DOMAIN GROUP (SDG) gene reported to be involved in H3K4 methylation and important for gametophyte development. Additionally, these genes include a protein similar to the O-fucosyltransferase AtOFT1, whose mutation produces lower seed set and reduced fertility. Finally, cluster 3 and cluster 5 (together about 26% overlapping genes) show genes downregulated during PT growth. These include homologs of transcriptional regulators like AtPV42a and AGL104, members of MIKC MADS box genes, and other transcription factors like BZIP34. Genes associated with the cell cycle such as a regulator of the anaphase-promoting complex/cyclosome (SAMBA) or regulators of actin organization such as a cyclase associated protein1 (CAP1) are also downregulated, while homologs of signaling proteins like pollen-specific receptor kinases (PRK3 and PRK6) and pollen-specific RHO-RELATED PROTEIN FROM PLANTS1 GTPase (ROP1) appear only slightly downregulated.
Figure 5.
Cluster analysis to visualize transcriptome dynamics during transition from PGs to tricellular PTs in A. trichopoda. Top left, Venn diagram showing overlap between genes expressed in PGs, PT-Bi, and PT-Tri. Only genes with TPM values ≥ 1 in all replicates from a given tissue are considered. 9,759 genes expressed in all samples can be subdivided in seven k-mean clusters. y axis shows scaled gene expression values obtained for each gene by dividing its TPM value by the maximum value among the three samples. Total number of genes represented by each cluster are shown in brackets. Blue lines represent individual genes and average expression of genes in a given cluster is indicated with a red line. Orthologs* indicates the presence of the top five putative orthologs of Arabidopsis genes with known pollen function. A full list of genes in the k-mean clusters is shown in Supplemental Table S5.
We also explored which genes are enriched in male gametophytic tissues in comparison to sporophytic samples. To do so, A. trichopoda genes with an average expression of TPM ≥ 1 in a given tissue and TPM value ≤ 3 in all the sporophytic tissues (ovule, leaf, tepal, and root) were considered as enriched or preferentially expressed in that tissue. Despite the fact that this criterion could potentially identify genes with the same TPM value (e.g. TPM = 2) as enriched in a given tissue, in our data there are no genes that present this overlapping TPM. According to this criterion, the number of genes preferentially expressed in male gametophytic tissues are 470 in PG, 652 in PT-Bi, and 611 in PT-Tri (Table 1; see Supplemental Table S6A for details). These genes include pollen-specific genes described above and below, but again overall expression levels are similar in the three stages. In summary, the transcriptomes of the three different male gametophyte-specific stages are very similar and the vast majority of genes expressed at comparable levels.
Putative Orthologs of Essential Genes for Arabidopsis Male Gametophyte Functions Can Be Identified in A. trichopoda
To understand A. trichopoda-specific male gametophyte functions, we next explored the 30 most strongly expressed genes in the three developmental stages (Supplemental Table S6B). These genes are very highly expressed with TPM values up to 88,000. Expression values in the three stages were comparable. The six most highly expressed genes display TPM values between 25,000 to around 80,000. The corresponding predicted proteins do not have significant similarities with other described proteins and thus are prime candidates for functional studies. Notably, among the 30 most strongly expressed genes in PG, PT-Bi, and PT-Tri samples, we found 15, 13, and 14 genes lacking homology to known proteins, respectively (Supplemental Table S6B). Other highly expressed genes are homologous to genes coding for RAPID ALKALINIZATION FACTORS (RALFs), lipases/acylhydrolases with a Gly‐Asp‐Ser‐Leu‐like motif, allergen-like proteins, invertases, and pectin lyases. Because many genes essential for male gametophyte functions, including PT polarity, PT cell wall, and transport, physiology, growth, and guidance, as well as sperm release, have been characterized in the model flowering plant Arabidopsis, we next investigated the expression profiles of the respective homologous genes in A. trichopoda. Based on extensive literature data, we searched homologs for more than 200 genes with known functions in PGs and PTs in Arabidopsis. Following a BLASTp approach based on “best-hit,” many homologous genes were identified (Supplemental Table S7). Among the top 30 mostly strongly expressed A. trichopoda homologs, we identified genes encoding cell wall proteins like pectin methylesterases including AtPPME1 and VANGUARD1 (VGD1), a RALF4/19 homolog, and one gene encoding a LEU-RICH REPEAT/EXTENSIN (LRX) protein known to interact with RALFs (Mecchia et al., 2017). Most genes from the top 30 homology list encode proteins involved in signaling, including a number of receptor kinases like BUDDHAS PAPER SEAL1 (BUPS1) and BUPS/2 and small GTPases such as RHO-RELATED PROTEIN FROM PLANTS1 (ROP1) and ARABIDOPSIS RAB GTPASE HOMOLOG D2B (RABD2B) and RABD2C, but we also found MADS box keratin binding domain (MIK) C-type MADS box transcription factors. We could not identify homologs for 253 genes expressed in PG, 346 genes in PT-Bi, and 331 genes in PT-Tri by BLASTp searches, and about one-quarter of the 210 putative homologous genes listed in Supplemental Table S7 are lowly expressed in pollen with TPMs < 10.
As shown in Figure 3, the cell wall of A. trichopoda consists of pectin and callose, similar to PTs of Arabidopsis and other eudicots and monocots (Williams, 2008). It is thus not surprising that cell wall synthesis and modifying enzymes appear conserved, but so do general signaling components like small GTPases. To study conservation of key signaling pathways during pollen functions in A. trichopoda, we next focused on the presence and detailed expression pattern of different classes of receptor-like kinases (RLKs). To identify homologous and putative orthologous RLKs expressed in Arabidopsis and A. trichopoda pollen, we first compared annotated protein sequences phylogenetically and studied the expression pattern of the corresponding genes (Fig. 6). In Arabidopsis, especially members of the largest RLK subfamily, LRR-RLKs, have been shown to possess key functions in PT growth and guidance. These include pollen-specific receptor kinases (PRKs), MALE DISCOVERER1/2 (MDIS1/2), and MDIS-interacting RLKs (MIKs), as well as BAK1-ASSOCIATING RECEPTOR-LIKE KINASE1 (BARK1), STRUBBELIG-RECEPTOR FAMILY4/5 (SRF4/5), CYS-RICH RLK 1 (CRK1), and RECEPTOR-LIKE KINASE IN FLOWERS1 (RKF1). These were aligned with all 95 LRR-RLKs that were identified in the A. trichopoda genome (Fig. 6A; Liu et al., 2016). Based on the phylogenetic tree and expression pattern, we identified AMTR_s00130p00117400 as a putative ortholog of Arabidopsis MDIS1 and MDSI2. AMTR_00025p00206890 appears as a single homolog of PRK1, PRK2, PRK4, PRK5, and PRK7, while AMTR_00024p00252010 is the only homolog of PRK3, PRK6, and PRK8. This finding indicates an amplification of pollen-specific LRR-RLK genes in Arabidopsis. Notably, the genes coding for A. trichopoda proteins phylogenetically related to Arabidopsis MIK1 and MIK2 as well as to SRF4 and SRF5 do not show a remarkable expression pattern in PTs, similar to proteins in the same clade with Arabidopsis CRK1 and RFK1. The A. trichopoda LRR-RLK phylogenetically related to Arabidopsis BARK1 (AMTR_00117p00096300) shows only a weak expression level in PTs. Some A. trichopoda RLK genes such as AMTR_00010p00238570 and AMTR_00077p00153450, which are moderately expressed in PTs do not possess predominantly pollen-expressed LRR-RLK homologs in Arabidopsis. Most of the other A. trichopoda LRR-RLK genes are either not expressed in the tissues investigated or are expressed in vegetative tissues.
Figure 6.
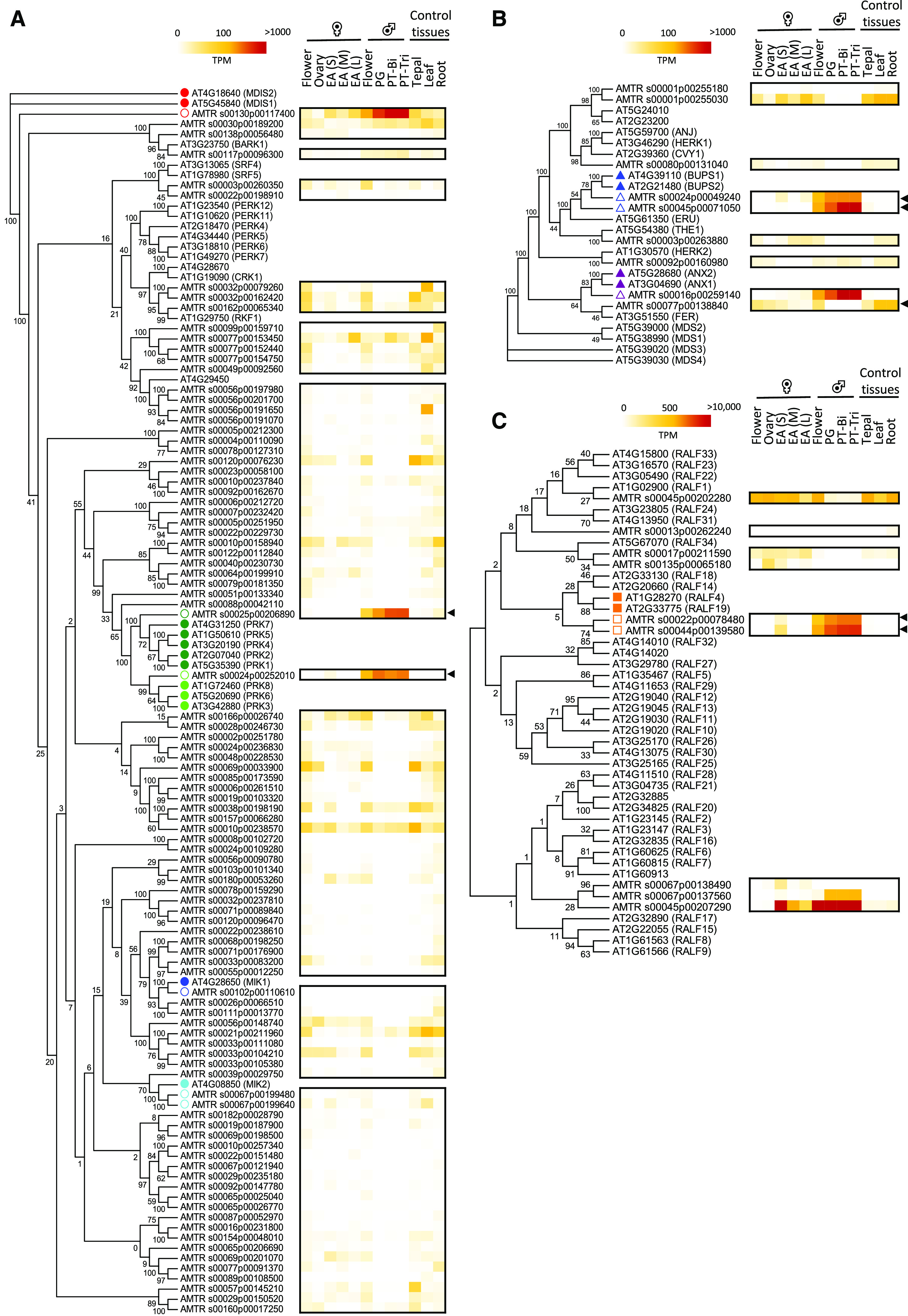
Phylogenetic tree and heatmap of A. trichopoda RLK and RALF genes to identify their putative Arabidopsis orthologs. A, LRR-RLKs predominantly expressed in PTs of Arabidopsis (24 genes) and all LRR-RLKs identified in A. trichopoda (95 genes). Arabidopsis genes involved in PT growth and guidance are indicated by filled dots, while their putative orthologs are shown by empty dots. B, CrRLK1L receptor genes identified in A. trichopoda (nine genes) and Arabidopsis (17 genes). Filled triangles indicate Arabidopsis CrRLK1Ls involved in maintenance of the PT integrity, and open triangles indicate putative A. trichopoda orthologs. The heatmap shows expression values (in TPM) of A. trichopoda CrRLK1Ls. C, RALFs identified in A. trichopoda (nine genes) and Arabidopsis (37 genes). Filled squares indicate Arabidopsis RALFs involved in maintenance of the PT integrity, and empty squares indicate putative A. trichopoda orthologs. Black arrowheads point toward highest expression in male gametophyte tissues. The phylogenetic tree was generated by using full-length protein sequences and the maximum likelihood method with 100 bootstraps. Female flower, EA(S), EA(M), and EA(L) represent small, medium, and large egg apparatus cells from A. trichopoda described in Flores-Tornero et al. (2019).
Another subclass of RLKs called Catharanthus roseus-RLK1-like kinases (CrRLK1Ls) is shown in Figure 6B. This subclass has 17 members in Arabidopsis and nine in A. trichopoda. Expression pattern and sequence similarity suggests that AMTR_00024p00049240 and AMTR_00045p00071050 are putative orthologs of BUPS1 and BUPS2 from Arabidopsis, while AMTR_s00016p00259140 appears to be the only putative ortholog of ANX1 and ANX2. The higher number of Arabidopsis RLK family members appears to be a general feature and suggests that gene duplication and diversification especially contributed to the expansion of RLK genes in evolutionary younger angiosperm species, but not A. trichopoda.
Genes encoding RALFs, which interact as mobile ligands with CrRLK1Ls, but also with cell-wall-located LRX proteins, confirm this finding. While the Arabidopsis genome encodes 37 RALF genes, A. trichopoda contains only nine genes, of which four are significantly expressed in male gametophytes (Fig. 6C). AMTR_s00022p00078480 and AMTR_s00044p00139580 are likely the putative RALF4/19 orthologs regulating cell wall stability during PT growth, while AMTR_s00045p00207290 and AMTR_s00067p00137560 are members of a unique clade and together with egg apparatus expressed AMTR_s00067p00138490. Function(s) for these RALFs have not been elucidated. In conclusion, the combination of phylogenetic and expression pattern analyses was successful in identifying candidate orthologs of Arabidopsis RLKs and their ligands with key functions in PT germination, growth, guidance, and burst in A. trichopoda.
Proteomic Studies Reveal That A. trichopoda PGs Are Well Prepared for Germination, While PTs Are Characterized by Catabolic, Biosynthetic, and Transport Processes
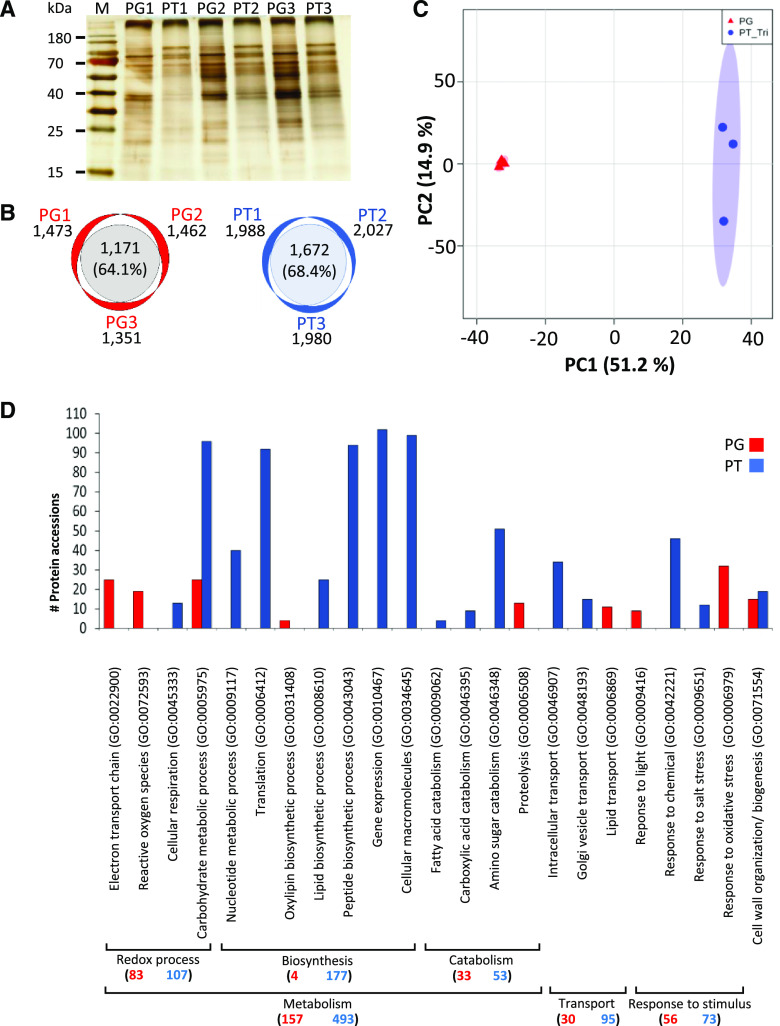
To obtain an additional level of insight into A. trichopoda male gametophyte functions, a proteomic analysis was conducted to compare mature A. trichopoda PGs and tricellular PTs. Three biological replicates each of PGs and PTs were processed and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Total protein extracts from three replicates for both samples were separated by SDS-PAGE and silver stained (Fig. 7A). PG and PT samples clearly displayed a different banding pattern pointing toward significant differences in protein levels. In total, 2,632 protein groups (Supplemental Table S8) were detected by LC-MS/MS. There were 1,667 protein groups were identified in PG and 2,288 protein groups in PT samples. A Venn diagram shows an overlap of 64.1% and 68.4% among the three PG and PT replicates, respectively (Fig. 7B). An additional PCA analysis groups the three biological replicates of PG and PT closely together and significantly apart from each other (Fig. 7C). These results indicate that the proteome of PG and PT are significantly different, while the biological replicates are very similar and thus reproducible.
Figure 7.
Proteomic analysis of PGs and tricellular PT from A. trichopoda. A, Characteristic protein pattern observed for PG and PT 17 h after germination. Prior to MS, protein samples of each of the three biological replicates were quality checked by silver staining after SDS-PAGE. B, Number of proteins detected by MS in three biological replicates of PGs and PTs. C, PCA plot of proteins detected within three biological replicates of PG and PT samples. D, Functional category distribution according to biological processes of identified proteins that were significantly upregulated in PG (red) and PT (blue; P < 0.05). x axis represents significantly enriched gene ontology (GO) terms (false discovery rate < 0.05), while y axis represents the number of protein accessions in each GO term. GO categories were made by using PANTHER web service version 14.1. Numbers in brackets indicate total number of proteins found in PG or PT for a given main biological process. Corresponding detailed information is listed in Supplemental Tables S8 and S9.
For further analysis, we considered only those proteins that were simultaneously detected in all three biological replicates from either PG (1,086 protein groups) or PT (1,581 protein groups). Among these, 270 protein groups were significantly enriched in PG and 926 in PT samples (q < 0.05, fold change > 2). However, only 11% and 15% expressed genes, respectively, have been detected as proteins, and thus it is difficult to compare transcriptome and proteome data. In order to elucidate the corresponding biological processes that these enriched proteins represent, a functional category analysis was performed by PANTHER v14.1 (Fig. 7D; Supplemental Table S9, A and B). The enriched functional categories were grouped into main biological processes. Within each biological process, the total number of protein accessions from PG (in red) and PT (in blue) is provided. Redox processes appear especially important for PG, which is enriched in proteins related to the electron transport chain, proteins involved in reactive oxygen species (ROS) biosynthesis and response to oxidative stress. Additionally, proteins related to oxylipin biosynthesis and lipid transport as well as carbohydrate metabolism are enriched. Altogether, this indicates that the PG is well prepared to start germination requiring a lot of energy, lipids, and sugars. Proteins involved in cell wall organization/biosynthesis further support this hypothesis. PT is especially characterized by processes related to biosynthesis, catabolism, and transport. Many proteins are required for carbohydrate, nucleotide, fatty acid, carboxylic acid, and amino sugar catabolism, while simultaneously biosynthetic processes play a major role to generate lipids, proteins, and macromolecules in general, including cell wall material. Intracellular transport and Golgi vesicle transport are further major functional categories. Finally, a large number of identified proteins are involved in transcription, indicating that gene expression is regulated during PT growth.
Notably, with exceptions like PG cell wall biosynthesis, many proteins involved in PG-/PT-specific functions such as signaling processes were not detected in the proteome data. This is not surprising, taking into consideration that only 11% and 15% of the expressed genes were represented in the proteomic data and that soluble proteins and highly abundant proteins, including those for housekeeping enzymes, are enriched. However, from the list of identified putative homologs of more than 200 genes with known functions in PGs and PTs in Arabidopsis (Supplemental Table S7), we have also detected peptides from 28 potential orthologs in the proteome data (Table 2). These include cell wall proteins like pectin methylesterase VGD1-like and AtPPME1-like, as well as a single homolog of LRR-expansin-like LRX-like9 to LRX-like11 proteins. The single putative A. trichopoda ortholog of PRK-like1, PRK-like2, PRK-like4, PRK-like5, and PRK-like7 RLKs was also detected. Peptides from other highly expressed signaling proteins like RALFs or BUPS/ANX RLKs as well as other PRKs and membrane transporters were not detected in the proteome data. These might be detectable in pollen membrane and cell wall fractions or might not have been detected due to low protein amounts despite high transcript levels. Finally, a few proteins were identified, like dynamin-like and actin filament bundling protein or small GTPases, which are lowly expressed, but which were detected in all samples of either PG or PT, indicating their high protein stability and low turnover rate. In summary, the proteomic analyses show that PGs seem to be well prepared for germination requiring lipids, energy, but likely also ROS molecules for signaling, while PTs are especially characterized by catabolic/biosynthesis and transport processes, including gene expression. Further experimentation involving fractionation of PG and PT samples is required to increase protein numbers and to detect proteins in specific subcompartments.
Table 2. Overview about A. trichopoda homologs of Arabidopsis genes reported to play a role in PG and PT functions, which were identified by using proteomic analyses.
TPM values represent the average expression of three biological replicates and are listed according to their expression level in PT-Tri. Significant enrichment of a given protein in PT or PG tissues is represented with a "x" (see Supplemental Tables S7 and S8 for details). Gene symbols according to putative Arabidopsis homologs.
| A. trichopoda Gene ID | PG | PT-Bi | PT-Tri | UniProt ID | PG | PT | Gene symbol | E-value | Description | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| AMTR_s00040p00188020 | 5,074 | 9,773 | 10,488 | W1PZU9 | – | x | VGD1 | 1.00E−132 | Pectin methylesterase | Jiang et al., 2005 |
| AMTR_s00007p00264370 | 4,953 | 8,603 | 8,447 | W1P6M7 | – | x | AtPPME1 | 5.00E−119 | Pectin methylesterase | Tian et al., 2006 |
| AMTR_s00029p00242570 | 829 | 2,137 | 2,718 | W1PRD0 | – | x | LRX10 | 4.00E−47 | Leu-rich repeat extensin-like protein |
Mecchia et al., 2017; Fabrice et al., 2018; Wang et al., 2018 |
| LRX11 | 7.00E−45 | |||||||||
| LRX8 | 3.00E−48 | |||||||||
| LRX9 | 2.00E−46 | |||||||||
| AMTR_s00025p00206890 | 263 | 522 | 531 | W1PWK6 | – | x | PRK1 | 0.00E+00 | Leu-rich repeat RLK |
Takeuchi and Higashiyama, 2016 |
| PRK2 | 0.00E+00 | |||||||||
| PRK4 | 0.00E+00 | |||||||||
| PRK5 | 0.00E+00 | |||||||||
| PRK7 | 4.00E−142 | |||||||||
| AMTR_s00066p00182490 | 291 | 300 | 289 | U5DFR5 | – | x | COBL10 | 0.00E+00 | GPI-anchored protein | Li et al., 2013 |
| AMTR_s00007p00221730 | 240 | 282 | 267 | W1P698 | – | x | ATAPY1 | 0.00E+00 | Apyrase | Wolf et al., 2007 |
| ATAPY2 | 0.00E+00 | |||||||||
| AMTR_s00099p00037820 | 200 | 161 | 179 | W1NYC9 | – | x | ATP | 3.00E−98 | Mitochondrial ATP synthase subunit | Geisler et al., 2012 |
| AMTR_s00047p00209120 | 106 | 98 | 100 | U5D8W3 | – | x | RGP1 | 0.00E+00 | UDP-l-arabinose mutase | Drakakaki et al., 2006 |
| RGP2 | 0.00E+00 | |||||||||
| AMTR_s00102p00131940 | 6 | 117 | 103 | W1P0U0 | – | x | QRT3 | 2.00E−155 | Polygalacturonase | Rhee et al., 2003 |
| AMTR_s00153p00086670 | 69 | 103 | 77 | W1PMJ7 | – | x | EXO70C2 | 5.00E−158 | Putative exocyst subunit | Synek et al., 2017 |
| AMTR_s00024p00207140 | 62 | 90 | 65 | W1PMG6 | – | x | PDAT1 | 0.00E+00 | Phosphodiacylglycerol acyltransferase | Zhang et al., 2009 |
| AMTR_s00055p00193060 | 43 | 71 | 61 | U5DD49 | – | x | FIM5 | 0.00E+00 | Actin bundling factor | Wu et al., 2010 |
| AMTR_s00044p00070480 | 29 | 32 | 54 | U5D9M4 | – | x | UGP1 | 0.00E+00 | UDP-Glc pyrophosphorylase | Park et al., 2010 |
| UGP2 | 0.00E+00 | |||||||||
| AMTR_s00010p00252880 | 37 | 47 | 35 | W1NFY8 | – | x | AtUTr1 | 0.00E+00 | UDP-Gal/ Glc transporter | Reyes et al., 2010 |
| AtUTr3 | 0.00E+00 | |||||||||
| AMTR_s00080p00116860 | 26 | 38 | 35 | W1PBE5 | – | x | DRP2A | 0.00E+00 | GTPase | Backues et al., 2010 |
| AMTR_s00010p00159850 | 27 | 22 | 31 | W1NED9 | – | x | PGM2 | 0.00E+00 | Phosphoglucomutase | Egli et al., 2010 |
| AMTR_s00020p00173350 | 18 | 22 | 24 | W1PVM3 | – | x | RGP5 | 1.00E−137 | UDP-l-arabinose mutase | Drakakaki et al., 2006 |
| AMTR_s00176p00051440 | 15 | 17 | 15 | W1PUK2 | – | x | DRP1C | 0.00E+00 | Dynamin-like protein | Kang et al., 2003 |
| AMTR_s00039p00217390 | 13 | 13 | 14 | U5D3A4 | – | x | GAPCp1 | 0.00E+00 | Phosphate dehydrogenase | Muñoz-Bertomeu et al., 2010 |
| GAPCp2 | 0.00E+00 | |||||||||
| AMTR_s00067p00130250 | 13 | 10 | 12 | U5DBN7 | – | x | MGP1 | 7.00E−116 | Mitochondrial F1F0-ATP synthase | Li et al., 2010 |
| AMTR_s00019p00240770 | 7 | 13 | 10 | W1PJV7 | – | x | VLN5 | 0.00E+00 | Actin filament bundling protein | Zhang et al., 2010 |
| AMTR_s00002p00227840 | 6 | 9 | 8 | W1P2M5 | – | x | ARGAH1 | 0.00E+00 | Arginase | Brownfield et al., 2008 |
| AMTR_s00013p00172160 | 5 | 5 | 5 | W1PIV7 | – | x | AtKDSA2 | 0.00E+00 | Phosphate synthetase | Delmas et al., 2008 |
| AMTR_s00003p00020850 | 2 | 4 | 5 | W1P5X2 | – | x | MIRO1 | 0.00E+00 | GTPases | Sørmo et al., 2011 |
| AMTR_s00019p00246290 | 1 | 1 | 1 | W1PHG5 | – | x | PUR4 | 0.00E+00 | Ribonucleotide synthase | Berthomé et al., 2008 |
| AMTR_s00155p00043060 | 1 | 2 | 1 | W1PJ58 | – | x | IPMDH2 | 0.00E+00 | 3-Isopropylmalate dehydrogenase | He et al., 2011 |
| IPMDH3 | 0.00E+00 | |||||||||
| AMTR_s00022p00227570 | 14 | 29 | 21 | W1PWI7 | x | – | RABA4D | 8.00E−138 | Rab GTPase |
Szumlanski and Nielsen, 2009 |
| AMTR_s00103p00075300 | 1 | 0 | 0 | W1NZP8 | x | – | STP6 | 0.00E+00 | Hexose transporter |
Scholz-Starke et al., 2003 |
DISCUSSION
The A. trichopoda Male Gametophyte Shows Typical Features of Other Extant Angiosperm Groups
In order to understand the evolution of male gametophyte functions in flowering plants, studies using A. trichopoda are unique, as this species diverged already about 130 million years ago from all other living angiosperms and thus may still contain, for example, ancient molecular mechanisms that may have been lost or have been amplified in sister clade angiosperms, for example, during reproductive isolation. Histochemical studies of A. trichopoda male gametophytes confirmed that PGs are bicellular at maturity as observed in A. trichopoda and other basal angiosperms from the Amborellales, Nymphaeales, Austrobaileyales clade like Trithuria bibracteata, Nuphar advena, or Austrobaileya scandens (Williams et al., 2014). Notably, bicellular PGs are also characteristic for a number of eudicots like Solanum lycopersicum (Chaturvedi et al., 2013) or Nicotiana tabacum (Bokvaj et al., 2015). We reported that division of the GC during in vitro PT growth is completed within 14 h after germination, while in S. lycopersicum and N. tabacum, for example, GCs produce sperm cells about 10 h after germination in vitro (Yang et al., 2005; Lu et al., 2015). Notably, it has been reported in N. tabacum that the GC does not produce two sperm cells unless reduced forms of nitrogen like specific amino acids or ammonium chloride are present in the medium (Read et al., 1993). CJ-PGM used for A. trichopoda PT growth does not contain reduced forms of nitrogen, and therefore it will be interesting to test in future studies whether its addition will accelerate mitosis in A. trichopoda GCs. Moreover, the vegetative nucleus always precedes the GC during the PT translocation phase and the two sperm cells during further PT growth, respectively, which is similar to many sister clade angiosperms (Heslop-Harrison and Heslop-Harrison, 1989; McCue et al., 2011; Palevitz, 1993).
According to a comparative study of PTGRs in different seed plants, PTs of A. trichopoda and other basal angiosperms grow much faster than those from gymnosperm species but slower than later-diverging angiosperms (Williams, 2008). Furthermore, when comparing studies of PTGRs in different seed plants both in vivo and in vitro systems, PTGRs tend to be significantly higher in vivo than in vitro (see Supplemental Table S2). However, we identified conditions allowing A. trichopoda PTs to grow in vitro even about 25% faster than in vivo, indicating that either support by the maternal tissues of the ovary is less important in A. trichopoda or that the germination medium selected mirrors quite well the natural environment of the ovary tissues. We further observed that PT growth appeared linear without noticeable oscillations that were reported in fast growing PTs like Lilium longiflorum and N. tabacum (Lassig et al., 2014). This does not exclude the existence of oscillation of secondary messengers like Ca2+ and ROS and the development of pH gradients, which have been reported to play a key role for polar tip growth in PTs of sister clade angiosperm species (Damineli et al., 2017; Mangano et al., 2018). Based on the above reported in vitro growth system, a transient transformation system for PTs could now be established to also address such questions in a basal angiosperm species.
A. trichopoda Male Gametophyte Transcriptomes Are Less Complex Compared with Those of Eudicots and Monocots
The richest resource to elucidate and compare the evolution of male gametophyte functions are the transcriptomic data reported above. Similar to other species like Arabidopsis (Becker et al., 2003; Honys and Twell, 2003), rice (Wei et al., 2010), or maize (Davidson et al., 2011), the transcriptome of PGs and PTs of A. trichopoda is less complex (about 40% of all protein encoding genes) compared with those of vegetative tissues such as roots and leaves (expression of almost 60% of all protein encoding genes). Around 2% expressed genes are enriched in male gametophytes, which is lower compared with reports for other species like Arabidopsis, rice, N. tabacum, or Glycine max showing 4% to 11% enriched genes (Rutley and Twell, 2015). There exists a very high overlap between the PG and PT transcriptomes of A. trichopoda and even gene expression levels appear quite similar: less than 40% variation in expression levels were observed for more than two-thirds of expressed genes. This is different in Arabidopsis, rice, or N. tabacum, showing a significant and progressive decrease in the diversity of transcripts during different stages of pollen development and germination (Rutley and Twell, 2015). In Arabidopsis, it was also shown that the PT transcriptome changes significantly depending on its growth in vitro or in vivo, as it was reported that more than 300 genes related to signaling, cell extension, and transcription were only found in semi-in vivo PTs in comparison to those germinated in vitro (Qin et al., 2009). Thus, it cannot be ruled out that such differences in expression will also occur in A. trichopoda semi-in vivo-grown PTs. All together, these findings suggest that the regulation of male gametophyte functions in sister clade angiosperms is likely more complex and imply that the A. trichopoda male gametophyte is more similar to that of the most recent common ancestor of all angiosperms. Likewise, the egg apparatus cells (egg and synergids) of A. trichopoda are morphologically highly similar and contain a rather large overlap of conjointly expressed genes (Flores-Tornero et al., 2019).
A detailed analysis by comparing the A. trichopoda male gametophyte transcriptomes with that of other angiosperms identified homologs of a large number of well-known and characterized proteins with male gametophyte functions. Many conserved genes encode proteins involved in cell wall metabolism and composition (Wallace and Williams, 2017; Fabrice et al., 2018), including pectin methylesterases, polygalacturonases, and LRR-extensin-like proteins that were also detected in the proteomic data. This finding confirms the histochemical data showing that the composition and arrangement of pectin and callose in the pollen cell wall in A. trichopoda, as well as the formation of extensive callose plugs, is typical for angiosperms, especially as described for C. japonica and L. longiflorum, respectively (Hasegawa et al., 2000). Notably, this is different from gymnosperm PT cell walls that show a different arrangement and composition, such as arabinogalactan protein abundancy and absence of pectins and callose depositions (Yatomi et al., 2002). As another remarkable finding, we detected high transcript levels of some genes that were not detected at the proteomic level and vice versa obtained enriched proteins in PGs or PTs, whose transcript levels were very low. Low correlation between protein abundance and mRNA expression levels has been reported before (e.g. Greenbaum et al., 2003; Walley et al., 2016) and can be explained as follows: on the one hand, the male gametophyte of plants shows a high level of translational regulation and thus transcripts are stored in the mature PG and are only translated during pollen germination in vivo (e.g. Fíla et al., 2017). Some proteins are very stable and thus are only represented by low transcript levels. Moreover, technical issues associated with proteomic studies still have a strong impact on detected proteins as a whole battery of protocols would be necessary to detect all proteins that are located at different membranes, organelles, cell wall, etc. Therefore, transmembrane proteins like BUPS1/2 and ANX1/2 or small secreted proteins (e.g. RALFs) can easily be missed or are underrepresented in proteomic studies. Usually, cytoplasmic proteins are particularly enriched in proteomic studies (Walley et al., 2016).
Identification of Genes Likely Essential for PT Functions
Both transcriptomic and proteomic data indicated that processes related to lipid and protein biosynthesis, as well as to transport mechanisms, are significantly upregulated during PT growth, which has also been shown in other angiosperms like rice (Dai et al., 2006), Arabidopsis (Zou et al., 2009), S. lycopersicum (Chaturvedi et al., 2013), or N. tabacum (Ischebeck et al., 2014). Moreover, the RALF-ANX/BUPS-LRR signaling pathway regulating PT cell wall integrity (Boisson-Dernier et al., 2009; Miyazaki et al., 2009; Ge et al., 2017, 2019) appears conserved, as homologs of all components could be clearly identified in the combined phylogenetic expression data. However, A. trichopoda additionally encodes a male/female gametophyte-specific RALF clade (Flores-Tornero et al., 2019) that does not contain homologs in other angiosperms (Galindo-Trigo et al., 2016) and may possess an ancient function or a function specific to basal angiosperms from the Amborellales, Nymphaeales, Austrobaileyales clade. Relatively few of the 94 LRR-RLKs encoded by the A. trichopoda genome (Liu et al., 2016) are expressed in male gametophytes. These include homologs of the Arabidopsis plasma membrane receptors PRK6 and MDIS1/2 that interact with female gametophyte-secreted PT attractant LURE1 peptides (Takeuchi and Higashiyama, 2016; Wang et al., 2016). Notably, putative homologs of further LURE1 interactors MIK1 and MIK2 (Wang et al., 2016) are not significantly expressed in the male gametophytes of A. trichopoda. It should also be emphasized that the number of PT-expressed PRKs including PRK1 and PRK3 that contribute to LURE1 binding is considerably expanded in Arabidopsis. The higher number of Arabidopsis RLK family members appears to be a general feature and suggests that gene duplication and diversification especially contributed to the expansion of RLK genes in evolutionary younger angiosperm species, but not A. trichopoda. Thus, taking into consideration that A. trichopoda lacks homologs of PT attractants like Arabidopsis LURE1 or maize EA1 peptides (Márton et al., 2005, 2012) and that Arabidopsis, for example, contains significantly more RLKs and candidate signaling peptides like CRPs expressed in male gametophytes, indicates that communication of PTs with the maternal tissues and the female gametophyte is less complex and more ancient in A. trichopoda and points to the possibility that amplification of these signaling mechanisms including polymorphic peptide ligands contributed significantly to reproductive isolation and thus angiosperm diversity.
Outlook
It will now be exciting to elucidate components and mechanisms regarding how and whether A. trichopoda can discriminate self from alien PGs, whether PT guidance mechanisms exist, and which are the roles of the highly and specifically expressed genes in A. trichopoda male gametophytes lacking homologs in sister clade angiosperms. The data generated in this study can be used for comparative gene expression studies, but also to select candidate genes and protein domains for investigations about protein evolution during angiosperm reproduction and diversification and can be used to complement, for example, corresponding mutants in Arabidopsis and other species. Functional studies of A. trichopoda male gametophyte-specific genes is still very difficult, as stable transformation systems are not available and cultivation of the species is quite challenging. However, overexpression studies of A. trichopoda male gametophyte-specific genes in other species, the development of a transient PT transformation system, and complementation approaches of reproductive mutants in sister clade angiosperms like Arabidopsis and maize with A. trichopoda genes could significantly help to better understand the evolution of reproductive gene families and mechanisms and thus could significantly help to better understand the evolution of the reproductive mechanism in this fascinating plant species and in flowering plants in general.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plant material was harvested from male and female flowers of Amborella trichopoda at the Botanical Garden in Bonn (Germany). Plants were grown in a shaded place inside a greenhouse under controlled conditions of 16°C to 18°C, constant humidity of 66%, and 12-h photoperiods. Tissues including male and female flowers, tepals, leaves, and roots were immediately frozen in liquid nitrogen after harvest and stored at −80°C. Fully opened male flowers were gathered in 50-mL Falcon conical tubes (Thermo Fisher), placed without lid in a hermetically sealed plastic box containing a bed of silica gel, and stored at 4°C until pollen isolation. Ovaries were isolated from fully opened female flowers, immediately frozen in liquid nitrogen, and stored at −80°C.
Pollen Isolation and Germination
As show in Figure 1, male flowers were collected in 50-mL Falcon conical tubes (Thermo Fisher) up to a volume of 25 mL. Next, tubes containing flowers were transferred without lid to a moisture chamber at room temperature for up to 30 min to allow rehydration. Afterward, 25 mL of fresh sterile-filtered liquid of CJ-PGM [5% (w/v) Suc, 300 µg mL−1 Ca(NO3)2, 200 µg mL−1 MgSO4, 100 µg mL−1 KNO3, 100 µg mL−1 H3BO3, pH 5.0] was added. The mixture was vortexed for 30 s at maximum speed to release PGs. Subsequently, the liquid germination medium containing PGs was filtered through a 1-mm pore size steel mesh to remove flowers tissues and debris. The pollen suspension was further filtered in a 50-mL Falcon conical tube containing a pluriStrainer 70-µm polyester mesh (PluriSelect) and a connector ring (PluriSelect) in which the sample is force-filtered using manual pressure from a syringe. After this filtration step, small debris were retained on the surface of the mesh and the pollen suspension was collected and refiltered through a stack of 70-30-15-micron pore size polyester strainers (PluriSelect) to remove cellular debris and separate bacterial and fungal spores from PGs. Finally, PGs were obtained as a beige-white clean film on the 15-micron strainer and removed from the membrane using a spatula, placed in a 1.5-mL collection tube and resuspended in 0.5-mL CJ-PGM. For germination in liquid medium, 1 mL of PG suspension of OD600 = 1 was resuspended in a final volume of 5 mL CJ-PGM, poured into 5.5-cm diameter glass petri dishes, covered with a lid, and incubated in a moisture chamber at room temperature for up to 20 h on the laboratory bench. To estimate PTGR in solidified medium, a small square plastic chamber (2 × 2 cm) was built on a glass slide, filled with 3 mL sterile CJ-PGM with 0.5% (w/v) low melting agarose (Carl Roth), and left to solidify. Alternatively, open flowers were dabbed on the surface of solid media to release PGs, and the small plastic chamber was immediately closed with its lid to keep humidity to allow PG germination under the microscope.
Histochemistry and Microscopy
Images of flowers were taken by using a Zeiss Discovery V8 stereo microscope. To calculate PTGRs, PGs were germinated on solid medium and recorded during 40 min under an inverted Zeiss LSM510 confocal laser scanning microscope using a 40×/1.3 NA oil immersion objective. From the movie generated, the length of 10 PTs was measured for 5 min every 30 s, and the average was considered as PTGR in vitro. Mature PGs were stained with DAPI (50 µg mL−1 4′,6-diamino-phenylindole, 0.2 m NaH2PO4, 10 mm EDTA, 0.5% [w/v] Triton X-100, pH 7.2). Liquid CJ-PGM was used to germinate mature PGs, and PTs were visualized with a Nikon Eclipse TE2000-S inverted microscope. To detect the presence of pectin in PGs and PTs, liquid CJ-PGM with 0.01% (w/v) ruthenium red (Sigma-Aldrich) was used. For callose plug staining, decolorized aniline blue solution (0.1% [w/v] aniline blue in 108 mm K3PO4, pH 11.0) was added to liquid media. A Zeiss Axioskop was used to visualize stained PGs and tubes. The presence of sperm cells and vegetative nucleus in PTs was determined by staining with SYBR Green I nucleic acid stain solution (Thermo Fisher) in a 1:1000 solution in combination with the membrane dye FM 4-64 (N-(3-triethylammoniumpropyl) 4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide; Thermo Fisher) and visualized using a Leica TCS SP8 confocal laser scanning microscope.
RNA Isolation, cDNA, and Library Preparation
Total RNA was isolated from three biological replicates of control tissues (tepals, leaves, and roots) as well as from PGs, PTs 10 h after germination (PT-Bi), and PTs 20 h after germination (PT-Tri) by using a Spectrum plant total RNA kit (Sigma-Aldrich) according to manufacturer’s instructions. Two hundred fifty nanograms of total RNA was used for each library construction. RNA-seq was carried out as described in the Illumina TruSeq stranded mRNA sample preparation guide for the Illumina HiSeq 1000 System (Illumina) and the KAPA library quantification kit (Kapa Biosystems). For ovary samples, three biological replicates containing 11 ovaries each were used to extract total RNA according to the “Purification of total RNA from animal and human cells” protocol of the RNeasy Plus micro kit (Qiagen). The SMARTer ultra low input RNA kit for sequencing v4 (Clontech Laboratories) was used to generate first strand cDNA from 0.5 ng total RNA. Double-stranded cDNA was amplified by LD PCR (15 cycles) and purified via magnetic bead cleanup. Library preparation was carried out as described in the Illumina Nextera XT Sample Preparation Guide (Illumina). One hundred fifty picograms of input cDNA were tagmented (tagged and fragmented) by the Nextera XT transposome. Products were purified and amplified via a limited-cycle PCR program to generate multiplexed sequencing libraries. For the PCR step, 1:5 dilutions of index 1 (i7) and index 2 (i5) primers were used. Libraries were quantified using the KAPA SYBR FAST ABI prism library quantification kit (Sigma-Aldrich). Equimolar amounts of each library were used for cluster generation on the cBot (TruSeq SR cluster kit v3; Illumina). Sequencing runs were performed on a HiSeq 1000 instrument using the indexed, 2 × 100 cycles paired-end protocol and the TruSeq SBS v3 kit. Image analysis and base calling resulted in “.bcl” files, which were converted into “.fastq” files using the CASAVA1.8.2 software (Illumina). Illumina deep sequencing was carried out at the genomics core facility of the University of Regensburg (Center for Fluorescent Bioanalytics KFB).
RNA-seq Data Analysis
RNA-seq data were processed using the LSTrAP-Lite pipeline (Tan and Mutwil, 2020), where Trimmomatic with default settings was used to trim sequence reads. All samples passed LSTrAP’s quality control. Reads were mapped to the Amborella genome v1.0 (Amborella Genome Project, 2013) obtained from Ensembl Plants (Kersey et al., 2016). Log2FC and Benjamini and Hochberg-corrected P values (Benjamini and Hochberg, 1995) for DEGs were calculated using DESeq2 (package version 1.10.1; Love et al., 2014). Significant DEGs (P < 0.05 after correction) were used to identify significantly enriched MapMan bins (MapMan v 3.6.0RC1; Thimm et al., 2004). Finally, gene expression levels were estimated using TPM values (Conesa et al., 2016) and clustered using Python Seaborn package with ‘Euclidean’ distance and ‘complete’ method. To estimate similarities between transcriptomes of the various samples, we used Jaccard index defined a: J(X,Y) = |X∩Y| / |X∪Y|, where X and Y are differentially regulated (e.g. up-regulated) genes in the two compared samples. K-means clustering and finding the optimal k with sum of squared distances was calculated using Python package scikit-learn (Pedregosa et al., 2011). Genes with TPM values ≥ 1 were considered as expressed. A comprehensive overview on normalized expression data are shown in Supplemental Table S3. RNA-seq data are deposited at the European Bioinformatics Institute under accession number E-MTAB-9190 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9190/).
RT-qPCR Validation of RNA-seq Data
For RNA-seq data validation, RT-qPCR was performed with nine randomly selected genes, including two genes with similar expression levels in all three tissues as reference genes and seven DEGs representing high, medium, and lowly expressed genes, respectively. RNA from PG, PT-Bi, and PT-Tri samples was isolated as previously described, and 100 ng of each sample used to prepare cDNA using SuperScript III reverse transcriptase (Thermo Fisher) according to the manufacturer’s instructions. Primers used for RT-qPCR are listed in Supplemental Figure S1. RT-qPCR was performed in 10-µL volumes with 1 µL cDNA by using KAPA SYBR FAST universal kit (Merck). Reactions were performed in three technical replicates per sample. RT-qPCR was performed using the Mastercycler Realplex2 (Eppendorf) in a 96-well reaction plate according to the manufacturer’s recommendations. Data were normalized to the expression levels of the two reference genes, and relative expression was calculated using the comparative Ct method (2_DΔCt). ΔCt values were compared to log2FC (TPM in tissue A versus tissue B) using Pearson’s correlation. The correlation of RNA-seq and RT-qPCR data are shown in Supplemental Figure S1.
Protein Extraction and LC-MS/MS Analysis
PG pellets were obtained from three independent experiments. Fifty milligrams of pellets from each isolation was immediately frozen in liquid nitrogen and stored at −80°C. The rest of the pellets from each isolation were resuspended in 1 mL liquid CJ-PGM until OD600 = 1, diluted to a final volume of 5 mL germination medium, and incubated at room temperature for 20 h in a moisture chamber. PTs were collected by filtration through a 40-µm strainer (PluriSelect) and washed twice with liquid germination medium to remove nongerminated PGs. Fifty milligrams of each isolation of PT pellet was collected from the surface of the strainer with a spatula, placed in a 1.5-mL collection tube, and immediately frozen in liquid nitrogen. Total proteins were extracted by grinding samples with liquid nitrogen in a mortar and subsequent addition of 250 µL of ice-cold extraction buffer (50 mm Tris/HCl, 150 mm NaCl, 0.1% [w/v] sodium deoxycholate, 0.1% [v/v] Triton-X100, 1 mm phenylmethylsulfonyl fluoride, pH 8.0). Pollen protein extracts were cleared twice in a bench centrifuge (13,000 rpm, 10 min, 4°C), distributed in aliquots of 50 µL and stored at −80°C. To analyze protein extracts, 10 µg of each biological replicate was loaded on a 12% SDS-PAGE using PageRuler prestained protein ladder as a marker (Thermo Fisher) and detected by silver staining according to Chevallet et al. (2006).
Proteomic analyses were done as previously described in Hafidh et al. (2018). In short, protein solutions were processed by filter-aided sample preparation using the Microcon device with molecular weight cut-off 30 kD (Merck Millipore) covering alkylation (iodoacetamide; Sigma-Aldrich) and digestion step (trypsin; Promega). LC-MS/MS analyses of peptide mixtures were done using the Ultimate 3000 RSLCnano system connected to an Orbitrap Elite hybrid mass spectrometer (Thermo Fisher). MS data were acquired in a data-dependent strategy, selecting up to the top 10 precursors for higher-energy collisional dissociation fragmentation. The analysis of mass spectrometric RAW data files was carried out using the Proteome Discoverer software (Thermo Fisher; v1.4) with in-house Mascot (Matrixscience; v2.6) and Sequest search engine utilization. MS/MS ion searches were done against modified cRAP database (based on http://www.thegpm.org/crap/) and UniProtKB protein database for A. trichopoda (http://www.uniprot.org/proteomes/UP000017836a; downloaded 18.4.2016, number of protein sequences 27,371). Percolator was used for postprocessing of the search results. Peptides with q value < 0.01, rank 1, and at least six amino acids long were considered only. Protein abundance was assessed using protein area calculated by Proteome Discoverer. Protein group reports from all individual samples were combined into a single supergroup (SG) report, where each SG is a list of proteins reported within a single protein group in at least one single sample report. Each SG was quantified as maximum of intensities of proteins in a protein group or as 90% of the minimal observed intensity in a sample. Then, log10-transformation was applied, and the data were linearly normalized to equalize the median intensities in all samples. The protein groups enriched in either PG or PT samples were detected by the Limma method (Smyth, 2004).
Statistical and Phylogenetic Analyses
Three biological replicates were generated and used for each tissue type. For statistical analysis, we calculated the average TPM values of the three replicates and used only those genes with TPM ≥ 1. PCA analysis was done by using MetaboAnalystR 3.0 online tool (Pang et al., 2020) with already normalized data. Colored regions from each tissue represent areas with 95% confidence to consider the replicates reproducible. GO term enrichment was performed online by using PANTHER v14.1 with a significance level of P < 0.05 and correction of false discovery rate < 0.05. See the “RNA-seq Data Analysis” section for statistical analysis related to DEGs. The LRR-RLK genes from Amborella and Arabidopsis (Arabidopsis thaliana) used for phylogenetic analysis were obtained from Liu et al. (2016) and Takeuchi and Higashiyama (2016) respectively, whereas the CrRLK1L genes were all obtained from Galindo-Trigo et al. (2016). Phylogenetic analyses were performed with MEGA7 software (Kumar et al., 2016) by using full-length protein sequences, aligning them by selecting the ClustalW option and using the maximum likelihood method with 500 bootstraps. Heatmaps were generated in Excel by using TPM values obtained from CoNekT online database (https://evorepro.sbs.ntu.edu.sg).
Accession Numbers
The raw sequencing data sets generated and analyzed during this study are available at the European Bioinformatics Institute under accession number E-MTAB-9190. Please see Supplemental Table S3 for detailed expression values of all annotated A. trichopoda genes.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Validation of RNA-seq data by RT-qPCR.
Supplemental Figure S2. DEG dendrogram of A. trichopoda PG and PT transcriptomes using MapMan ontology analysis.
Supplemental Table S1. PGM tested by using A. trichopoda PGs.
Supplemental Table S2. A. trichopoda PTGR observed in this study compared with another A. trichopoda study and those for other seed plants.
Supplemental Table S3. Gene expression values of all A. trichopoda tissues described in this report.
Supplemental Table S4. DEGs in male gametophyte tissues.
Supplemental Table S5. K-means clustering of all 9759 genes expressed in A. trichopoda PG, PT-Bi, and PT-Tri.
Supplemental Table S6. Most strongly expressed genes detected in male gametophytic tissues of A. trichopoda.
Supplemental Table S7. List of 210 Arabidopsis genes reported to play a role in PG and PT functions, their putative A. trichopoda orthologs, and average gene expression pattern.
Supplemental Table S8. Identification of proteins expressed in A. trichopoda PG and PT by mass spectrometry, and differentially expressed proteins (DEP) in pairwise comparisons between PG and PT.
Supplemental Table S9. List of gene ontology terms of significantly enriched proteins in A. trichopoda PG and PT.
Acknowledgments
The authors are grateful to Maximilian Weigend, Cornelia Löhne, and Bernhard Reinken (Botanical Garden of the University of Bonn, Germany) for providing A. trichopoda plant material.
Footnotes
This work was supported by the project Central European Institute of Technology 2020 (grant no. LQ1601), the Czech Infrastructure for Integrative Structural Biology research infrastructure project funded by Ministry of Education, Youth and Sport of the Czech Republic (grant no. LM2018127), and the German Research Council with financial support via European Research Area Network for Coordinating Action in Plant Sciences grant EVOREPRO (grant no. DR 334/12–1 to S.S. and T.D).
[OPEN]Articles can be viewed without a subscription.
References
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY(2010) The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Physiol 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA(2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Berthomé R, Thomasset M, Maene M, Bourgeois N, Froger N, Budar F(2008) pur4 mutations are lethal to the male, but not the female, gametophyte and affect sporophyte development in Arabidopsis. Plant Physiol 147: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U(2009) Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokvaj P, Hafidh S, Honys D(2015) Transcriptome profiling of male gametophyte development in Nicotiana tabacum. Genom Data 3: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield DL, Todd CD, Deyholos MK(2008) Analysis of Arabidopsis arginase gene transcription patterns indicates specific biological functions for recently diverged paralogs. Plant Mol Biol 67: 429–440 [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Ischebeck T, Egelhofer V, Lichtscheidl I, Weckwerth W(2013) Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J Proteome Res 12: 4892–4903 [DOI] [PubMed] [Google Scholar]
- Chevallet M, Luche S, Rabilloud T(2006) Silver staining of proteins in polyacrylamide gels. Nat Protoc 1: 1852–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Li L, Chen T, Chong K, Xue Y, Wang T(2006) Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 6: 2504–2529 [DOI] [PubMed] [Google Scholar]
- Damineli DS, Portes MT, Feijó JA(2017) One thousand and one oscillators at the pollen tube tip: The quest for a central pacemaker revisited In Obermeyer G, and Feijó J, eds, Pollen Tip Growth: From Biophysical Aspects to Systems Biology. Springer International Publishing, Cham, Switzerland, pp 391–413 [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. (2011) Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Delmas F, Séveno M, Northey JGB, Hernould M, Lerouge P, McCourt P, Chevalier C(2008) The synthesis of the rhamnogalacturonan II component 3-deoxy-D-manno-2-octulosonic acid (Kdo) is required for pollen tube growth and elongation. J Exp Bot 59: 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N(2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew BT, Ruhfel BR, Smith SA, Moore MJ, Briggs BG, Gitzendanner MA, Soltis PS, Soltis DE(2014) Another look at the root of the angiosperms reveals a familiar tale. Syst Biol 63: 368–382 [DOI] [PubMed] [Google Scholar]
- Egli B, Kölling K, Köhler C, Zeeman SC, Streb S(2010) Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol 154: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrice TN, Vogler H, Draeger C, Munglani G, Gupta S, Herger AG, Knox P, Grossniklaus U, Ringli C(2018) LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol 176: 1981–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fíla J, Záveská Drábková L, Gibalová A, Honys D(2017) When simple meets complex: Pollen and the -omics In Obermeyer G, and Feijó J, eds, Pollen Tip Growth: From Biophysical Aspects to Systems Biology. Springer International Publishing, Cham, Switzerland, pp 247–292 [Google Scholar]
- Flores-Tornero M, Proost S, Mutwil M, Scutt CP, Dresselhaus T, Sprunck S(2019) Transcriptomics of manually isolated Amborella trichopoda egg apparatus cells. Plant Reprod 32: 15–27 [DOI] [PubMed] [Google Scholar]
- Galindo-Trigo S, Gray JE, Smith LM(2016) Conserved roles of CrRLK1L receptor-like kinases in cell expansion and reproduction from algae to angiosperms. Front Plant Sci 7: 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu M-C, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Dresselhaus T, Qu LJ(2019) How CrRLK1L receptor complexes perceive RALF signals. Trends Plant Sci 24: 978–981 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Päpke C, Obata T, Nunes-Nesi A, Matthes A, Schneitz K, Maximova E, Araújo WL, Fernie AR, Persson S(2012) Downregulation of the δ-subunit reduces mitochondrial ATP synthase levels, alters respiration, and restricts growth and gametophyte development in Arabidopsis. Plant Cell 24: 2792–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M(2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Veldmann B, Korotkova N, Reinken B, Lobin W, Barthlott W(2011) Amborella trichopoda: Cultivation of the most ancestral angiosperm in botanic gardens. Sibbaldia: Int J Botanic Garden Hortic 9: 143–154 [Google Scholar]
- Hafidh S, Potěšil D, Müller K, Fíla J, Michailidis C, Herrmannová A, Feciková J, Ischebeck T, Valášek LS, Zdráhal Z, et al. (2018) Dynamics of the pollen sequestrome defined by subcellular coupled omics. Plant Physiol 178: 258–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Nakamura S, Uheda E, Nakamura N(2000) Immunolocalization and possible roles of pectins during pollen growth and callose plug formation in angiosperms. Grana 39: 46–55 [Google Scholar]
- He Y, Chen L, Zhou Y, Mawhinney TP, Chen B, Kang B-H, Hauser BA, Chen S(2011) Functional characterization of Arabidopsis thaliana isopropylmalate dehydrogenases reveals their important roles in gametophyte development. New Phytol 189: 160–175 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y(1989) Myosin associated with the surfaces of organelles, vegetative nuclei and generative cells in angiosperm pollen grains and tubes. J Cell Sci 94: 319–325 [Google Scholar]
- Hesse M.(2001) Pollen characters of Amborella trichopoda (Amborellaceae): A reinvestigation. Int J Plant Sci 162: 201–208 [Google Scholar]
- Honys D, Twell D(2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Valledor L, Lyon D, Gingl S, Nagler M, Meijon M, Egelhofer V, Weckwerth W(2014) Comprehensive cell-specific protein analysis in early and late pollen development from diploid microsporocytes to pollen tube growth. Mol Cell Proteomics 13: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang S-L, Xie L-F, Puah CS, Zhang X-Q, Yang W-C, Sundaresan V, Ye D(2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Harper JF, Palanivelu R(2019) A fruitful journey: Pollen tube navigation from germination to fertilization. Annu Rev Plant Biol 70: 809–837 [DOI] [PubMed] [Google Scholar]
- Kang B-H, Rancour DM, Bednarek SY(2003) The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J 35: 1–15 [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, et al. (2016) Ensembl Genomes 2016: More genomes, more complexity. Nucleic Acids Res 44(D1): D574–D580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K(2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassig R, Gutermuth T, Bey TD, Konrad KR, Romeis T(2014) Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J 78: 94–106 [DOI] [PubMed] [Google Scholar]
- Li S, Ge F-R, Xu M, Zhao X-Y, Huang G-Q, Zhou L-Z, Wang J-G, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Li W-Q, Zhang X-Q, Xia C, Deng Y, Ye D(2010) MALE GAMETOPHYTE DEFECTIVE 1, encoding the FAd subunit of mitochondrial F1F0-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant Cell Physiol 51: 923–935 [DOI] [PubMed] [Google Scholar]
- Liu PL, Xie LL, Li PW, Mao JF, Liu H, Gao SM, Shi PH, Gong JQ(2016) Duplication and divergence of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in basal angiosperm Amborella trichopoda. Front Plant Sci 7: 1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes AL, Moreira D, Ferreira MJ, Pereira AM, Coimbra S(2019) Insights into secrets along the pollen tube pathway in need to be discovered. J Exp Bot 70: 2979–2992 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wei L, Wang T(2015) Methods to isolate a large amount of generative cells, sperm cells and vegetative nuclei from tomato pollen for “omics” analysis. Front Plant Sci 6: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Martínez Pacheco J, Marino-Buslje C, Estevez JM(2018) How does pH fit in with oscillating polar growth? Trends Plant Sci 23: 479–489 [DOI] [PubMed] [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T(2005) Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- Márton ML, Fastner A, Uebler S, Dresselhaus T(2012) Overcoming hybridization barriers by the secretion of the maize pollen tube attractant ZmEA1 from Arabidopsis ovules. Curr Biol 22: 1194–1198 [DOI] [PubMed] [Google Scholar]
- McCue AD, Cresti M, Feijó JA, Slotkin RK(2011) Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: Potential roles of the male germ unit revisited. J Exp Bot 62: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603 [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M(2009) ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol 19: 1327–1331 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R(2010) The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Miki-Hirosige H, Iwanami Y(1982) Ultrastructural study of Camellia japonica pollen treated with myrmicacin, an ant-origin inhibitor. Am J Bot 69: 538–545 [Google Scholar]
- Nasrallah JB.(2019) Self-incompatibility in the Brassicaceae: Regulation and mechanism of self-recognition. Curr Top Dev Biol 131: 435–452 [DOI] [PubMed] [Google Scholar]
- Palevitz BA.(1993) Relationship between the generative cell and vegetative nucleus in pollen tubes of Nicotiana tabacum. Sex Plant Reprod 6: 1–10 [Google Scholar]
- Pang Z, Chong J, Li S, Xia J(2020) MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 10: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-I, Ishimizu T, Suwabe K, Sudo K, Masuko H, Hakozaki H, Nou I-S, Suzuki G, Watanabe Masao(2010) UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol 51: 981–996 [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V(2011) Scikit-learn: Machine learning in Python. J Mach Learn Res 12: 2825–2830 [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R(2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LJ, Li L, Lan Z, Dresselhaus T(2015) Peptide signalling during the pollen tube journey and double fertilization. J Exp Bot 66: 5139–5150 [DOI] [PubMed] [Google Scholar]
- Read SM, Clarke AE, Bacic A(1993) Requirements for division of the generative nucleus in cultured pollen tubes of Nicotiana. Protoplasma 174: 101–115 [Google Scholar]
- Reyes F, León G, Donoso M, Brandizzí F, Weber APM, Orellana A(2010) The nucleotide sugar transporters AtUTr1 and AtUTr3 are required for the incorporation of UDP-glucose into the endoplasmic reticulum, are essential for pollen development and are needed for embryo sac progress in Arabidopsis thaliana. Plant J 61: 423–435 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR(2003) Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol 133: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutley N, Twell D(2015) A decade of pollen transcriptomics. Plant Reprod 28: 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet H, von Balthazar M, Magallon S, Doyle JA, Endress PK, Bailes EJ, Barroso de Morais E, Bull-Herenu K, Carrive L, Chartier M, et al. (2017) The ancestral flower of angiosperms and its early diversification. Nat Commun 8: 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz-Starke J, Büttner M, Sauer N(2003) AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis. Plant Physiol 131: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK.(2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 [DOI] [PubMed] [Google Scholar]
- Sørmo CG, Brembu T, Winge P, Bones AM(2011) Arabidopsis thaliana MIRO1 and MIRO2 GTPases are unequally redundant in pollen tube growth and fusion of polar nuclei during female gametogenesis. PLoS One 6: e18530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L, Vukašinović N, Kulich I, Hála M, Aldorfová K, Fendrych M, Žárský V(2017) EXO70C2 is a key regulatory factor for optimal tip growth of pollen. Plant Physiol 174: 223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlanski AL, Nielsen E(2009) The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21: 526–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Higashiyama T(2016) Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531: 245–248 [DOI] [PubMed] [Google Scholar]
- Tan QW, Mutwil M(2020) Inferring biosynthetic and gene regulatory networks from Artemisia annua RNA sequencing data on a credit card-sized ARM computer. Biochim Biophys Acta Gene Regul Mech 1863: 194429. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M(2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tian G-W, Chen M-H, Zaltsman A, Citovsky V(2006) Pollen-specific pectin methylesterase involved in pollen tube growth. Dev Biol 294: 83–91 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S, Williams JH(2017) Evolutionary origins of pectin methylesterase genes associated with novel aspects of angiosperm pollen tube walls. Biochem Biophys Res Commun 487: 509–516 [DOI] [PubMed] [Google Scholar]
- Walley JW, Sartor RS, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, et al. (2016) Integration of omic networks in a developmental atlas of maize. Science 353: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Liang L, Xue Y, Jia PF, Chen W, Zhang MX, Wang YC, Li HJ, Yang WC(2016) A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang K, Yin G, Liu X, Liu M, Cao N, Duan Y, Gao H, Wang W, Ge W, et al. (2018) Pollen-expressed leucine-rich repeat extensins are essential for pollen germination and growth. Plant Physiol 176: 1993–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T(2010) Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH.(2009) Amborella trichopoda (Amborellaceae) and the evolutionary developmental origins of the angiosperm progamic phase. Am J Bot 96: 144–165 [DOI] [PubMed] [Google Scholar]
- Williams JH.(2008) Novelties of the flowering plant pollen tube underlie diversification of a key life history stage. Proc Natl Acad Sci USA 105: 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH.(2012) The evolution of pollen germination timing in flowering plants: Austrobaileya scandens (Austrobaileyaceae). AoB Plants 2012: pls010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Taylor ML, O’Meara BC(2014) Repeated evolution of tricellular (and bicellular) pollen. Am J Bot 101: 559–571 [DOI] [PubMed] [Google Scholar]
- Wolf C, Hennig M, Romanovicz D, Steinebrunner I(2007) Developmental defects and seedling lethality in apyrase AtAPY1 and AtAPY2 double knockout mutants. Plant Mol Biol 64: 657–672 [DOI] [PubMed] [Google Scholar]
- Wu Y, Yan J, Zhang R, Qu X, Ren S, Chen N, Huang S(2010) Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22: 3745–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Qiu YL, Xie CT, Tian HQ, Zhang Z, Russell SD(2005) Isolation of two populations of sperm cells and microelectrophoresis of pairs of sperm cells from pollen tubes of tobacco (Nicotiana tabacum). Sex Plant Reprod 18: 47–53 [Google Scholar]
- Yatomi R, Nakamura S, Nakamura N(2002) Immunochemical and cytochemical detection of wall components of germinated pollen of gymnosperms. Grana 41: 21–28 [Google Scholar]
- Zhang H, Qu X, Bao C, Khurana P, Wang Q, Xie Y, Zheng Y, Chen N, Blanchoin L, Staiger CJ, et al. (2010) Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB(2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Qu LJ(2019) Peptide/receptor-like kinase-mediated signaling involved in male-female interactions. Curr Opin Plant Biol 51: 7–14 [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Dresselhaus T(2019) Friend or foe: Signaling mechanisms during double fertilization in flowering seed plants. Curr Top Dev Biol 131: 453–496 [DOI] [PubMed] [Google Scholar]
- Zou J, Song L, Zhang W, Wang Y, Ruan S, Wu WH(2009) Comparative proteomic analysis of Arabidopsis mature pollen and germinated pollen. J Integr Plant Biol 51: 438–455 [DOI] [PubMed] [Google Scholar]