The capacity for rapid Na+-based signaling has evolved in ecologically important coccolithophore species via a novel class of voltage-gated Na+ channels, namely EukCatBs.
Abstract
The evolution of Na+-selective four-domain voltage-gated channels (4D-Navs) in animals allowed rapid Na+-dependent electrical excitability, and enabled the development of sophisticated systems for rapid and long-range signaling. While bacteria encode single-domain Na+-selective voltage-gated channels (BacNav), they typically exhibit much slower kinetics than 4D-Navs, and are not thought to have crossed the prokaryote–eukaryote boundary. As such, the capacity for rapid Na+-selective signaling is considered to be confined to certain animal taxa, and absent from photosynthetic eukaryotes. Certainly, in land plants, such as the Venus flytrap (Dionaea muscipula) where fast electrical excitability has been described, this is most likely based on fast anion channels. Here, we report a unique class of eukaryotic Na+-selective, single-domain channels (EukCatBs) that are present primarily in haptophyte algae, including the ecologically important calcifying coccolithophores, Emiliania huxleyi and Scyphosphaera apsteinii. The EukCatB channels exhibit very rapid voltage-dependent activation and inactivation kinetics, and isoform-specific sensitivity to the highly selective 4D-Nav blocker tetrodotoxin. The results demonstrate that the capacity for rapid Na+-based signaling in eukaryotes is not restricted to animals or to the presence of 4D-Navs. The EukCatB channels therefore represent an independent evolution of fast Na+-based electrical signaling in eukaryotes that likely contribute to sophisticated cellular control mechanisms operating on very short time scales in unicellular algae.
Electrical signals trigger rapid physiological events that underpin an array of fundamental processes in eukaryotes, from contractile amoeboid locomotion (Bingley and Thompson, 1962), to the action potentials of mammalian nerve and muscle cells (Hodgkin and Huxley, 1952). These events are mediated by voltage-gated ion channels (Brunet and Arendt, 2015). In excitable animal cells, Ca2+- or Na+-selective members of the four-domain voltage-gated cation channel family (4D-Cav/Nav) underpin well-characterized signaling processes (Catterall et al., 2017). The 4D-Cav/Nav family is broadly distributed across eukaryotes, contributing to signaling processes associated with motility in some unicellular protist and microalgal species (Fujiu et al., 2009; Lodh et al., 2016), although these channels are absent from land plants (Edel et al., 2017). It is likely that the ancestral 4D-Cav/Nav channel was Ca2+-permeable, with Na+-selective channels arising later within the animal lineage (Moran et al., 2015). This shift in ion selectivity represented an important innovation in animals, allowing rapid voltage-driven electrical excitability to be decoupled from intracellular Ca2+ signaling processes (Moran et al., 2015).
Na+-selective voltage-gated channels have not been described in other eukaryotes, although a large family of Na+-selective channels (BacNav) is present in prokaryotes (Ren et al., 2001; Koishi et al., 2004). BacNav are single-domain channels that form homotetramers, resembling the four-domain architecture of 4D-Cav/Nav. Studies of BacNav channels have provided considerable insight into the mechanisms of gating and selectivity in voltage-dependent ion channels (Payandeh et al., 2012; Zhang et al., 2012). The range of activation and inactivation kinetics of native BacNav are generally slower than observed for 4D-Nav, suggesting that the concatenation and subsequent differentiation of individual pore-forming subunits may have enabled 4D-Nav to develop specific properties such as fast inactivation, which is mediated by the conserved intracellular Ile–Phe–Met linker between domains III and IV (Fig. 1A; Irie et al., 2010; Catterall et al., 2017).
Figure 1.
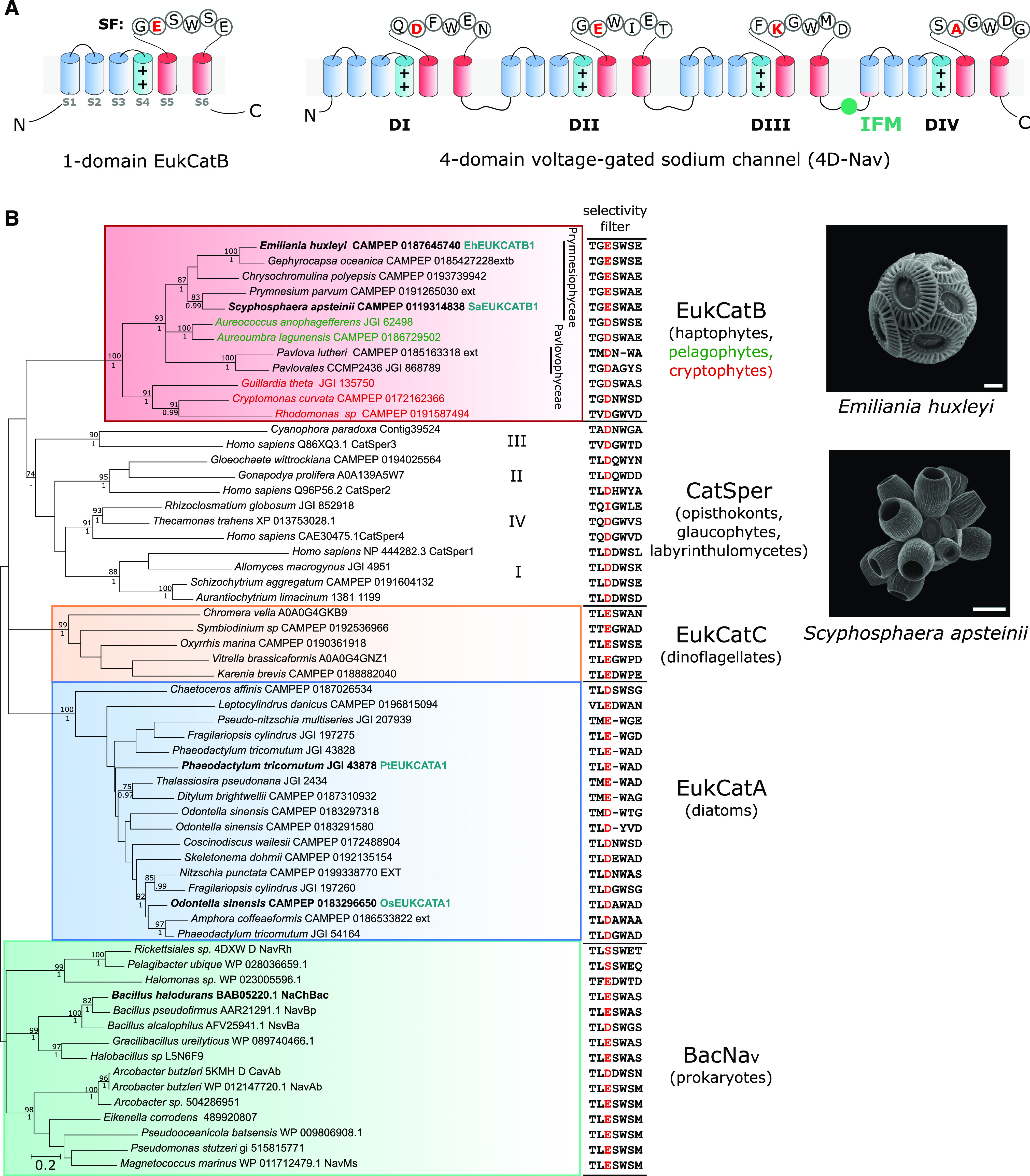
EukCatBs represent a novel class of single-domain channels. A, Schematic diagram of a single-domain EukCatB channel. The voltage-sensing module (S1–S4, blue), including conserved positively charged (++) residues of segment (S4) that responds to changes in membrane potential, is shown. The pore module (S5–S6, red) is also indicated, including the SF motif (Ren et al., 2001). The structure of a 4D-Nav (showing the SF of rat 4D-Nav1.4 with canonical “DEKA” locus of Na+-selective 4D-Nav1s) is also displayed (right). The Ile–Phe–Met motif of the fast inactivation gate is indicated (West et al., 1992) B, Maximum likelihood phylogenetic tree of single-domain, voltage-gated channels including BacNav and the three distinct classes of EukCat channels (EukCatA–C). Representatives of the specialized family of single-domain Ca2+ channels identified in mammalian sperm (CatSpers) are also included. SF for each sequence is shown (right). “Position 0” of the high-field–strength site that is known to be important in determining Na+ selectivity (Payandeh et al., 2011), is colored red. Channel sequences selected for functional characterization in this study are shown in bold. EukCatA sequences previously characterized (Helliwell et al., 2019) are also indicated, as is NaChBac channel from B. halodurans (Ren et al., 2001). Maximum likelihood bootstrap values (>70) and Bayesian posterior probabilities (>0.95) are above and below nodes, respectively. Scanning electron micrographs of coccolithophores E. huxleyi (scale bar = 2 μm) and S. apsteinii, (scale bar = 10 μm) are shown.
We recently identified several classes of ion channel (EukCats) bearing similarity to BacNav in the genomes of eukaryotic phytoplankton. Characterization of EukCatAs found in marine diatoms demonstrated that these voltage-gated channels are nonselective (exhibiting permeability to both Na+ and Ca2+) and play a role in depolarization-activated Ca2+ signaling (Helliwell et al., 2019). Two other distinct classes of single-domain channels (EukCatBs and EukCatCs) were also identified that remain uncharacterized. These channels are present in haptophytes, pelagophytes, and cryptophytes (EukCatBs), as well as dinoflagellates (EukCatCs; Helliwell et al., 2019). Although there is a degree of sequence similarity between the distinct EukCat clades, the relationships between clades are not well resolved, and there is not clear support for a monophyletic origin of EukCats. The diverse classes of EukCats may therefore exhibit significant functional differences. Characterization of these different classes of eukaryote single-domain channels is thus vital to our understanding of eukaryote ion channel structure, function, and evolution, and to our gaining insight into eukaryote membrane physiology more broadly.
Notably, EukCatB channels were found in ecologically important coccolithophores, a group of unicellular haptophyte algae that represent major primary producers in marine ecosystems. Coccolithophores are characterized by their ability to produce a cell covering of ornate calcium carbonate platelets (coccoliths; Fig. 1B; Taylor et al., 2017). The calcification process plays an important role in global carbon cycling, with the sinking of coccoliths representing a major flux of carbon to the deep ocean. Patch-clamp studies of coccolithophores indicate several unusual aspects of membrane physiology, such as an inwardly rectifying Cl− conductance and a large outward H+ conductance at positive membrane potentials, which may relate to the increased requirement for pH homeostasis associated with intracellular calcification. Here we report that EukCatB channels from two coccolithophore species (Emiliania huxleyi and Scyphosphaera apsteinii) act as very fast Na+-selective voltage-gated channels that exhibit many similarities to the 4D-Navs, which underpin neuronal signaling in animals. Thus, our findings demonstrate that the capacity for rapid Na+-based signaling has evolved in certain photosynthetic eukaryotes, contrary to previous widely held thinking.
RESULTS
Coccolithophore EukCatBs Are Fast Na+-Selective Voltage-Gated Channels
EukCatB sequences are present in haptophyte, cryptophyte, and pelagophyte taxa (Helliwell et al., 2019). They form a distinct phylogenetic group from the nonselective Ca2+- and Na+-permeable diatom EukCatAs and prokaryote BacNavs (Fig. 1B; Supplemental Table S1). Ion selectivity of voltage-gated channels is mediated by the conserved pore loop region between transmembrane segments S5 and S6, known as the selectivity filter (SF; Catterall et al., 2017). Six amino acids (LESWAS) make up this motif in the Na+-selective Bacillus halodurans BacNav channel (species highlighted in bold, Fig. 1B; Yue et al., 2002). Examination of the SFs of EukCat sequences indicates a clear diversification between the major EukCat clades. However, the SFs of coccolithophore (Prymnesiophyceae) EukCatB channels are strongly conserved (Fig. 1B) and exhibit high similarity to Na+-selective BacNav channels. In particular, the charged Glu residue occupying “position 0” in the high-field–strength site (highlighted red, Fig. 1B) of NaChBac and the adjacent Ser and Trp residues are highly conserved in EukCatB channels (Payandeh et al., 2011). In NaChBac, this conserved Glu plays a critical role in determining Na+-selectivity, as its mutation to Asp allows permeability to Ca2+ and K+ (DeCaen et al., 2014). By comparison, EukCatAs from the diatoms, which we showed previously to be permeable to Na+ and Ca2+ (Helliwell et al., 2019), differ significantly from the canonical Na+-selective SF of NaChBac. EukCatAs either have an Asp in position 0, or the conserved Glu, but not the adjacent Ser. Finally, dinoflagellate EukCatC sequences were highly diverse, ranging from SFs that were similar to the canonical Na+-selective motif (e.g. LESWSE, Oxyrrhis marina CAMPEP 0190361918) to SFs that resemble the Ca2+-selective NaChBac mutants (e.g. LEDWPE, Karenia brevis CAMPEP0188882040; DeCaen et al., 2014).
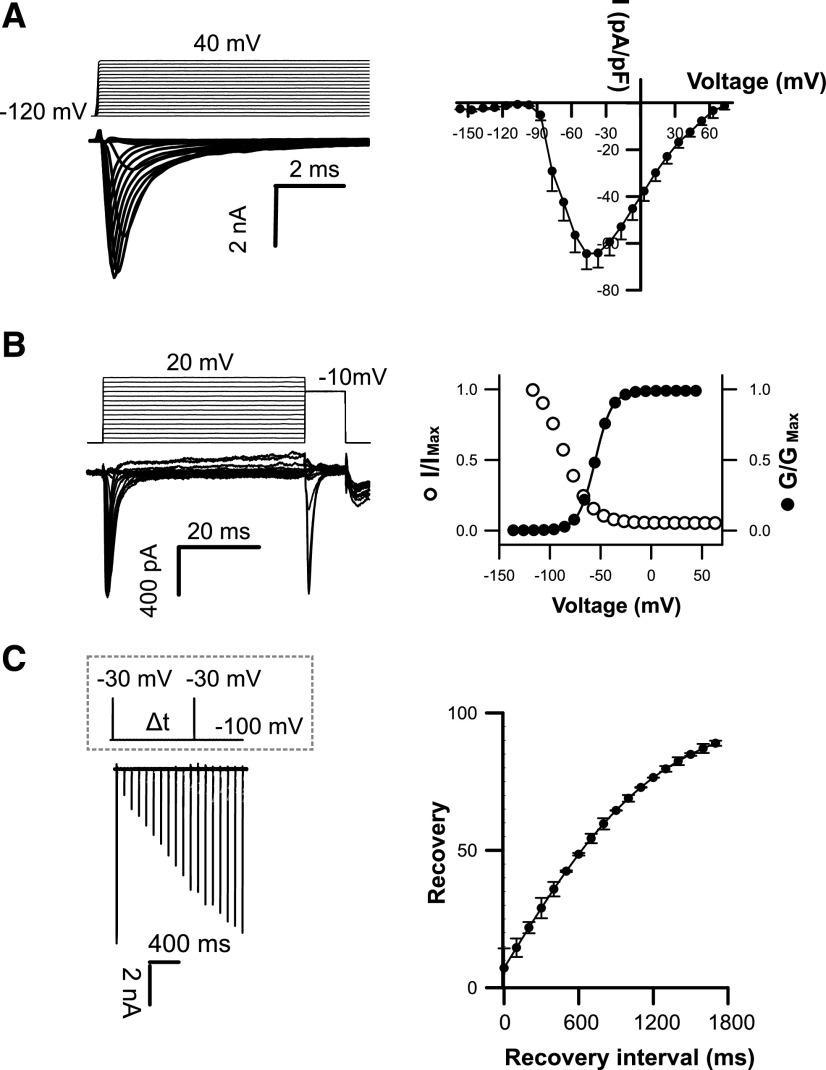
To determine the ion transport properties of coccolithophore EukCatB channels, we expressed representative channels from E. huxleyi (EhEUKCATB1, SF: TGESWSE) and S. apsteinii (SaEUKCATB1, SF: TGESWAE) in HEK293 cells (Supplemental Tables S2 and S3). In control conditions (external solution E1, Supplemental Table S4), patch-clamp approaches revealed that both proteins yielded robust voltage-dependent inward currents that were distinct from those recorded with nontransfected cells (Supplemental Fig. S1; He and Soderlund, 2010). EhEUKCATB1 and SaEUKCATB1 channel currents were activated by membrane depolarization, with extremely rapid activation and inactivation kinetics (τactivation < 0.6 ± 0.1 ms; τinactivation < 3.5 ± 0.9 ms; Fig. 2; Supplemental Fig. S2; Supplemental Table S3). The kinetics are faster than those previously reported for native BacNavs (Supplemental Table S3; Irie et al., 2010) and are remarkably similar to those observed in animal 4D-Nav1s (τactivation < 2 ms; τinactivation < 10 ms; Hille, 2001; Ren et al., 2001).
Figure 2.
Coccolithophore EukCatB channels expressed in HEK293 cells yield currents with very fast kinetics. A, Under control external solution (E1; Supplemental Table S4), typical current traces for cells expressing EhEUKCATB1 in response to membrane depolarization from a holding potential of –120 mV. Average peak current–voltage curves (right), where current was normalized to cell capacitance. Peak current = 64.45 ± 6.64 pA/pF (n = 57). B, Steady-state inactivation: representative examples of current traces (left) used to obtain steady-state inactivation curves (right) for EhEUKCATB1 (n = 14). Average normalized data were fitted using the Boltzmann equation (see “Materials and Methods”) for both activation (solid circles) and steady-state inactivation (open circles) curves (τactivation < 0.6 ± 0.1 ms; τinactivation < 3.5 ± 0.9 ms). Error bars: means ± se (see Supplemental Table S3) C, Recovery from inactivation; superimposed currents obtained by a double-pulse protocol using a varying interval (∆t) between the two voltage pulses (gray dashed box). Holding potential was –100 mV and test potential was −30 mV for 5 ms, followed by a recovery test pulse to –30 mV for 5 ms at 100 to 1,800 ms after the first test pulse. The peak currents elicited by the recovery pulse were normalized to construct the recovery curve. A single exponential was fitted to the averaged normalized recovery curve yielding τ for recovery (first-order exponential fit) of 632 ± 50.6 ms (n = 14).
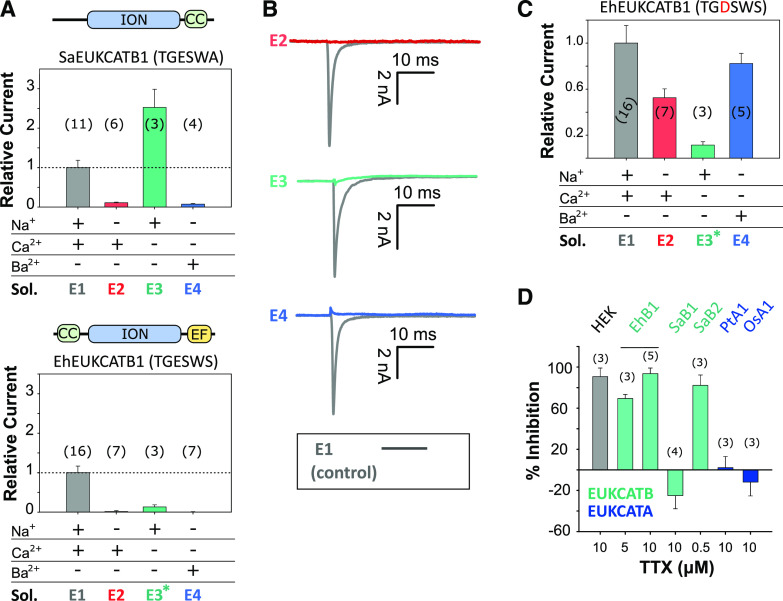
The currents produced by EhEUKCATB1 and SaEUKCATB1 channels exhibit reversal potentials of 73 and 60 mV, respectively (in control solution E1; Supplemental Table S4) that are similar to the equilibrium potential for Na+ (68 mV; Fig. 2A; Supplemental Fig. S2A), suggesting that the channels are Na+-selective. To characterize the ion selectivity of EukCatB channels further and to explore the functionality of the EF-hand domain located in the C terminus of EhEUKCATB1, we systematically replaced cations in the external solution. Omitting Na+ from the external solution (substituting with N-methyl-d-Gln (NMDG), a large synthetic cation) completely inhibited the peak inward currents of SaEUKCATB1, indicating that they have an absolute requirement for Na+ (Fig. 3A; Supplemental Table S4). By contrast, the removal of Ca2+ from the external solution significantly enhanced the channel conductance in SaEUKCATB1. These data suggest that SaEUKCATB1 represents a fast-activating Na+-selective channel that is entirely distinct from other Na+-selective channels previously described in eukaryotes (4D-Nav).
Figure 3.
Coccolithophore EukCatB channels are Na+-selective. A, Mean peak currents for coccolithophore EukCatB channels (SaEUKCATB1 and EhEUKCATB1) are shown after substitution of different cations in the external solution. Currents are shown relative to the standard control external solution (E1; Supplemental Table S4). Replacement of extracellular Na+ with NMDG abolished the current in both EukCatB channels. Removal of Ca2+ did not reduce currents for SaEUKCATB1, but greatly reduced those of EhEUKCATB1, which contains two Ca2+-binding EF-hand domains at the C terminus (asterisk indicates 100 μm of EGTA was added to EhEUKCATB1 experiments to ensure consistent removal of Ca2+). Schematic representation of domain architecture of each channel is shown above each graph. CC, Coiled-coil domain (Arrigoni et al., 2016); EF, EF-hand pair; ION, ion transport domain. Error bars: means ± se, n shown in parentheses. Extracellular and intracellular (pipette) solutions are given in Supplemental Table S4. B, Representative currents from A are shown for EhEUKCATB1. C, Mean peak currents after site-directed mutagenesis of the SF of EhEUKCATB1 from TGESWSE to TGDSWSE (E305D). Cation replacement experiments indicate that the EhEUKCATB1 E305D mutant conducts substantial currents in the absence of Na+ (i.e. when Na+ is replaced by NMDG in the external medium). The E305D mutation therefore causes the Na+-selective EhEUKCATB1 channel to become permeable to Ca2+ and Ba2+. Extracellular and intracellular (pipette) solutions are given in Supplemental Table S4, (asteriks indicates 100 μm of EGTA). Mean currents relative to control are shown (Supplemental Table S4). Error bars: means ± se. D, The effect of TTX, a selective Nav blocker, on the mean peak currents of EukCatB channels (E. huxleyi, EhB1; S. apsteinii, SaB1 and SaB2), compared to two EukCatA channels of diatoms (Phaeodactylum tricornutum, PtA1; Odontella sinensis, OsA1 (Helliwell et al., 2019). The effect of TTX on small native Na+ currents in HEK cells is also shown. Mean percent inhibition (relative to untreated control current) is shown, error bars: means ± se; n is given in parentheses.
The currents generated by EhEUKCATB1 were also strongly inhibited when Na+ was substituted in the external solution with NMDG (Fig. 3, A and B). The amplitude of EhEUKCATB1 channel currents were closely dependent on the concentration of external Na+ (Supplemental Fig. S3). This experiment was conducted without NMDG replacement of Na+, confirming that NMDG itself was not having an inhibitory effect. Furthermore, the EhEUKCATB1 channel current was inhibited by 98% in the presence of 105 mm of CaCl2 and 0 mm of Na+, compared to the control treatment (standard external solution E1, containing 140 mm of Na+ and 5 mm of Ca2+; Supplemental Fig. S4). However, EhEUKCATB1 channel currents were also absent in Ca2+-free media, suggesting that EhEUKCATB1 may represent a Ca2+-dependent Na+ channel (Fig. 3, A and B). EhEUKCATB1 possesses a C-terminal Ca2+-binding EF-hand pair that may contribute to the observed Ca2+-dependent activation. To confirm that the activity of EhEUKCATB1 is Na+-dependent, we used site-directed mutagenesis to modify its SF, changing the conserved Glu to Asp (E305D). The EhEUKCATB1-E305D channel was permeable to both Ca2+ and Ba2+ in the absence of Na+, indicating that it was no longer Na+-selective (Fig. 3C). This result indicates that the highly conserved Glu (0) in the SF of coccolithophore EukCatBs is a critical determinant of Na+-selectivity in a manner resembling BacNav channels (DeCaen et al., 2014).
The Na+-Selective Channel EhEUKCATB1 Is Tetrodoxin-Sensitive
Tetrodotoxin (TTX) is a highly selective and potent blocker of certain 4D-Nav1s (Ren et al., 2001; Klein et al., 2017), but single-domain BacNav are insensitive to TTX (Ren et al., 2001). We found that 5 and 10 μm of TTX substantially inhibited EhEUKCATB1 channel currents (Fig. 3D), lending strong support to this channel being a Ca2+-dependent Na+ channel. Although the sensitivity of EhEUKCATB1 to TTX is relatively low compared to the nanomolar sensitivity of some mammalian 4D-Navs (Klein et al., 2017), it seems likely that specific structural properties shared by these channels are targeted by TTX. A direct comparison of the residues responsible for binding TTX between 4D-Nav (pseudo-heterotetrameric) and single-domain EukCatBs (homotetrameric) awaits detailed structural information. Nevertheless, EukCatB channels possess a conserved Glu (E309 in EhEUKCATB1; black arrow, Supplemental Fig. S5), which is absent in TTX-insensitive BacNav and occupies a similar position to the “outer ring” of charged residues in the pore of 4D-Nav that have been implicated in TTX binding (Moczydlowski, 2013; Shen et al., 2018). Unlike EhEUKCATB1, SaEUKCATB1 was insensitive to TTX, despite being highly Na+-selective (Fig. 3D). By comparison, another highly similar EukCatB channel from S. apsteinii (SaEUKCATB2) was sensitive to 0.5 μm of TTX. Thus EukCatBs, like 4D-Navs (Klein et al., 2017), are not universally sensitive to TTX, and further studies will be required to identify the residues that contribute to the different TTX-sensitivities in EukCatB channels. It should be noted that both of the two previously characterized Ca+2- and Na+-permeable EukCatA channels from diatoms (Helliwell et al., 2019) were insensitive to TTX.
Coccolithophores Exhibit Fast Electrical Excitability
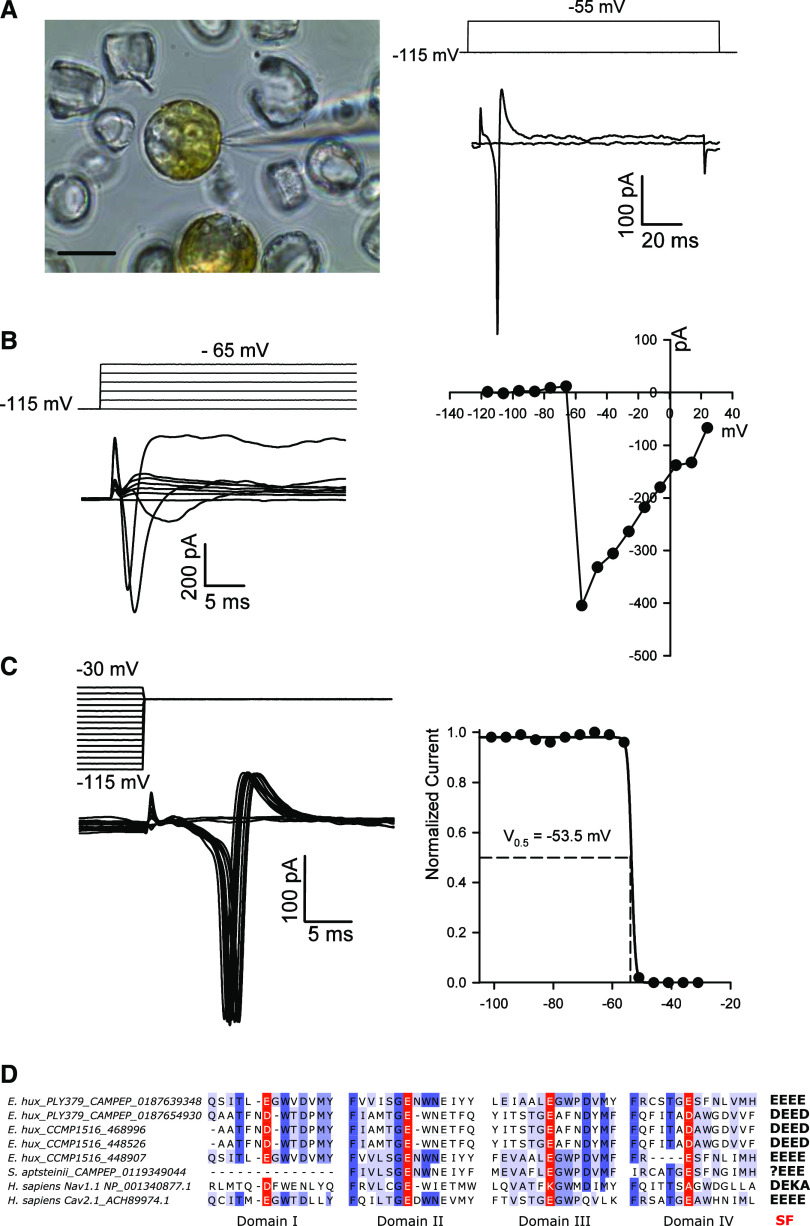
Given the presence of fast-activating Na+-selective, voltage-gated channels in coccolithophores, we examined the nature of membrane excitability in these cells. Application of patch-clamp approaches to decalcified S. apsteinii cells revealed fast-activating and fast-inactivating inward currents in response to membrane depolarization (Fig. 4). These extremely rapid kinetics (τactivation: 0.30 ± 0.07 ms and τinactivation: 0.73 ± 0.36 ms, n = 3) are similar to those that underlie metazoan Na+-based action potentials, and indicate that S. apsteinii is an excitable cell capable of rapid electrical signaling. The currents also exhibit similar kinetics to those produced by the heterologously expressed SaEUKCATB1 channel (Supplemental Fig. S2; Supplemental Table S3). However, both E. huxleyi and S. apsteinii also possess 4D-Cav/Nav genes in addition to EukCatBs (Supplemental Table S1) and so it is not clear which of these ion channels contribute to the observed membrane excitability. The coccolithophore 4D-Cav/Nav either exhibit the “EEEE” motif that is associated with Ca2+ selectivity in 4D-Cav1s, or a slight variant “DEED” (Fig. 4D; Verret et al., 2010). As the presence of a Lys in the SF appears to be important for Na+-selectivity, e.g. the DEKA of vertebrate 4D-Nav1s (Pozdnyakov et al., 2018), the coccolithophore 4D-Cav/Nav channels may not be Na+-selective, in contrast to the EukCatBs found in this lineage. Co-occurrence of both EukCatBs and 4D-Cav/Navs in coccolithophores may thus have driven functional diversification of voltage-gated ion channels. Investigation of the wider distribution and functional roles of EukCatBs and 4D-Nav/Cav in coccolithophores will provide insight into the selective pressures shaping the evolution of these different classes of ion channels.
Figure 4.
Fast voltage-activated inward currents in the coccolithophore S. apsteinii. As with C. pelagicus (Taylor and Brownlee, 2003), the coccolithophore S. apsteinii exhibits fast-activating and inactivating voltage-activated currents that likely underpin membrane excitability. The native currents observed in S. apsteinii cells have remarkably similar characteristics to the SaEUKCATA1 channel expressed in HEK cells. A, Image on the left shows a micrograph of patch-clamp pipette forming a seal on a decalcified S. apsteinii cell in normal seawater. Scale bar = 10 μm. The right shows fast-activating and inactivating inward current evoked by a voltage command from a holding potential of –115 mV to –55 mV (left). A second voltage stimulus to –45 mV evokes a short train of fast inward currents showing the recovery from inactivation is very fast (left). The cell was 19 pF and the seal was 5 GΩ. B, Family of depolarizing pulses from a holding potential of –115 mV in steps of + 10 mV (left) and corresponding current–voltage curve from the same cell recording (right) showing peak inward current at ∼–55 mV. The cell was 13 pF and the seal was 2 GΩ. C, Characteristic steady-state inactivation of native S. apsteinii voltage-activated currents. A prepulse protocol was used to depolarize the cell from a holding potential of −115 mV in + 5 mV increments for 100 ms before the test depolarization to –55 mV (left). Normalized peak inward current in response to the test pulse was plotted against prepulse voltage and fitted using a Boltzmann curve yielding a half-inactivation voltage of –53.5 mV (right). Cell was the same as in A. D, Multiple sequence alignment of the four pore domains of coccolithophore 4D-Cav/Navs. Sequences from coccolithophore genomes (E. huxleyi CCMP 1516) or transcriptomes (E. huxleyi PLY379 and S. apsteinii) are shown aligned to Na+-selective Nav1.1, and Ca2+-permeable Cav2.1 from humans. The coccolithophore sequences most closely resemble the EEEE motif found in metazoan Ca2+ channels and lack the conserved lysines found in Na+ channels. The 4D-Cav/Nav sequence found within the S. apsteinii transcriptome represents a partial sequence, as it lacks domain I.
DISCUSSION
Coccolithophore EukCatBs represent a novel class of highly Na+-selective single-domain voltage-gated channels in eukaryotes. The EukCatB channels are phylogenetically distinct from other classes of single-domain voltage-gated ion channels (including Kv channel subunits (Verret et al., 2010), but exhibit conserved features found in Na+-selective prokaryote BacNav channels. Their selectivity for Na+, together with their very rapid activation and inactivation kinetics, renders them distinct from the slower nonselective EukCatA channels recently identified in marine diatoms (Helliwell et al., 2019). Notably, many of the characteristics of the single-domain EukCatBs are highly similar to mammalian 4D-Navs, particularly in terms of their activation and inactivation kinetics. While the organization of the four pore-forming domains into a single protein has likely enabled development of specialized structures and functions within 4D-Navs (such as the intracellular linker connecting domains III and IV responsible for fast inactivation; West et al., 1992), our results indicate that concatenation is not a prerequisite for rapid gating kinetics, supporting observations from BacNav mutants (Irie et al., 2010; Shaya et al., 2014). Notably, certain coccolithophore EukCatBs also exhibited sensitivity to TTX, which is distinct from other single-domain channels such as BacNav and EukCatAs. However, as with mammalian 4D-Nav1s that are functionally divided into TTX-sensitive and TTX-resistant isoforms (Klein et al., 2017), we detected differences in sensitivity to TTX between EukCatBs. The EukCatBs may therefore prove to be useful models for studying Na+-channel gating and ion channel function more generally, as they are amenable to heterologous characterization and exhibit key functional differences from BacNav. The evolution of differing sensitivities of S. apsteinii EukCatBs also raises important ecological considerations. TTX is produced by a range of marine bacteria, and is also associated with harmful bloom-forming dinoflagellates (Turner et al., 2017). Environmental exposure of coccolithophores to TTX could therefore have detrimental effects on EukCatB channel function, and consequently drive the evolution of resistant isoforms.
Na+ selectivity has arisen independently in animal Navs at least twice (Gur Barzilai et al., 2012) and is thought to have been a critical step in the evolution of animal nervous systems, facilitating the rise of complex, fast-moving multicellular organisms. The discovery of EukCatBs indicates that eukaryotes have also evolved alternative mechanisms to the 4D-Navs for generating fast Na+-based action potentials. However, EukCatBs are not broadly distributed across eukaryotes and are unlikely to represent an ancestral channel in eukaryotes. While the Na+-based signaling systems in animals are typically associated with the rapid transmission of information between distant cells of a multicellular organism, the requirement for fast Na+-based signaling in a unicellular algal cell is likely to be very different. Haptophytes do exhibit very rapid responses to stimuli, such as the coiling of their haptonema, a unique flagella-like organelle that aids capture of bacterial prey for phagocytosis by rapidly coiling or bending toward the cell body (Parke et al., 1955; Kawachi et al., 1991). Retraction of the 100 µm of haptonema of Chrysochromulina sp. 21 NIES-4122 can occur within 5 to 10 ms (Nomura et al., 2019). However, given that the calcifying stages of coccolithophores from which action potentials have been recorded do not display a haptonema, broader roles are likely. Indeed, membrane excitability in coccolithophores may have a physiological role, as the major outward conductance at positive membrane potentials is H+, rather than K+ (Taylor et al., 2011). Repetitive action potentials in coccolithophores could therefore contribute to H+ efflux through Hv1 channels and maintain pH homeostasis (Taylor et al., 2011).
The precise role of the EukCatBs in coccolithophores will require further investigation, although techniques for genetic manipulation are currently lacking in coccolithophores, hindering functional characterization of individual genes in these organisms. Nevertheless, it is clear that the development of Na+-selective EukCatBs in haptophytes would have presented ancestral coccolithophores with the ability to uncouple electrical signaling from Ca2+ transport, as is the case in animal cells (Hille, 2001; Moran et al., 2015). Coccolithophores are unique among major calcifying organisms in that they produce their calcite structures (coccoliths) intracellularly, before they are secreted to the cell surface. While intracellular calcification allows precise control of the environment for calcite precipitation, it also necessitates massive fluxes of Ca2+ across cellular membranes and highly specialized mechanisms for Ca2+ homeostasis. The presence of Na+-selective channels, uncoupling electrical signaling from Ca2+ influx, may have allowed coccolithophores to develop the unique mechanisms for Ca2+ homeostasis required for intracellular calcification, without disrupting cellular signaling.
In summary, we show that coccolithophores possess a novel class of fast voltage-gated single-domain Na+ channels, indicating that the capacity for rapid Na+-based electrical signaling is not unique to animals. Further characterization of the structure and function of these unique Na+-channels is likely to inform us on the mechanisms underlying selectivity, gating, and pharmacology of the wider voltage-gated ion channel superfamily.
MATERIALS AND METHODS
Algal Cultures
Emiliania huxleyi (CCMP1516) was obtained from the Plymouth Culture Collection (Marine Biological Association). Scyphosphaera apsteinii (TW15) was from the Roscoff Culture Collection (Station Biologique de Roscoff, France). Cultures were maintained on a 12:12 light/dark cycle at 15°C with 100 and 50 µmol m−2 s−1 light. S. apsteinii was grown on medium that contained 90% filter-sterilized (Gulf Stream) seawater supplemented with Guillard’s f/2 nutrients and vitamins (Guillard and Ryther, 1962) and 10% (v/v) K media (Keller et al., 1987), whereas E. huxleyi was grown on natural seawater supplemented with Guillard’s f/2 nutrients and vitamins (Guillard and Ryther, 1962). Cultures were maintained in mid‐to‐late exponential growth by subculturing 1 mL into 40 or 250 mL of fresh media every 2 to 4 weeks.
Cell Lines
HEK293 cells (ATCC CRL-1573) were grown at 37°C in 5% (v/v) CO2 and 95% (v/v) O2 in a humidified incubator. High-Glc DMEM–Dulbecco’s Modified Eagle Medium growth medium with Antibiotic Antimycotic (Gibco) and 10% (v/v) FBS (Gibco) was used to culture cells, which were passaged every 3 to 4 d at 1:6 or 1:12 dilutions (cell/mm2).
Surveying for the Distribution of Single- and Four-Domain, Voltage-Gated Ion Channels
Sequence similarity searches were carried out as previously outlined by Helliwell et al. (2019) to survey haptophyte, pelagophyte, and cryptophyte genomes and transcriptomes for single- and four-domain, voltage-gated ion channels. Query sequences from Bacillus halodurans C-125 NaChBac (protein id: BAB05220.1) and BacNav-like sequences from the E. huxleyi CCMP1516 genome (protein id: 96075; Read et al., 2013), in addition to the 4D-Cav/Nav sequence of E. huxleyi (protein id: 468996) were used. Transcriptome databases surveyed were from the Marine Microbial Eukaryote Sequencing Project (MMETSP, https://www.imicrobe.us/#/projects/104; Keeling et al., 2014), alongside the reassembled sequence datasets (Johnson et al., 2019). In addition, eukaryote genomes including those from E. huxleyi (Read et al., 2013), Pavlovales sp. CCMP2436, Aureococcus anaphagefferens clone 1984, Pelagophyceae sp. CCMP2097, Guillardia theta CCMP2712, and Bigelowiella natans CCMP2755 were obtained from Joint Genome Institute http://genome.jgi.doe.gov/.
Databases were searched using BLASTP and TBLASTN (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with an E-value cutoff score of 1E−10. Each hit was inspected manually for relevant protein domains using Interpro (Apweiler et al., 2000), looking specifically for voltage-sensing domain (IPR005821), ion transport domain (IPR027359), and EF-hands (IPR011992). The presence of a minimum of three pore domains was used as a threshold for candidate 4D-Cav/Navs to distinguish them from other voltage-gated cation channels. IDs of all protein hits reported in this study are given in Supplemental Table S1.
Phylogenetic Analyses
The phylogenetic analyses of EukCat, Catsper, and BacNav sequences were performed using multiple sequence alignments generated with the program MUSCLE via the software Molecular Evolutionary Genetics Analysis (MEGA7; Kumar et al., 2016). After manual refinement, the tool GBLOCKS0.91B (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) was employed to remove poorly aligned residues, using the least stringent parameters (Castresana, 2000), resulting in an alignment of 172 and 211 amino acid residues for Figure 1, respectively. Maximum likelihood trees were generated using the software MEGA7 with 100 bootstraps. Model analysis was performed in MEGA7 to determine an appropriate substitution model (WAG+G+I). Bayesian posterior probabilities were additionally calculated using the program BEAST v1.8.4 (Drummond et al., 2012) running for 10,000,000 generations.
Synthesis of Heterologous Expression Plasmids for HEK293 Cells
Amino acid sequences of proteins used for heterologous expression are described in Supplemental Table S2. Coding sequences for EhEUKCATB1, SaEUKCATB1, and SaEUKCATB2 were obtained from MMETSP transcriptomic datasets: E. huxleyi CCMP 379 (CAMPEP_0187645740; MMETSP0994-7), and S. apsteinii RCC1455 (SaEUKCATB1: CAMPEP_0119314838, MMETSP1333; SaEUKCATB2: CAMPEP_0119345692, MMETSP1333). To confirm these sequences, we amplified the open reading frame from cDNA made from liquid cultures of S. apsteinii and E. huxleyi 1516 (using the primers: Sapsteinii_F1: ATGGTCGTCGCATCCTCAACG; Saptsteinii_R1: GCACCTCTGCACTCTGACATCTG; and Ehuxleyi_F1: ATGATCGCGGCGATACATAAC; Ehuxleyi_R1: TCACACACGCTGCGTCGT). cDNA was synthesized using SuperScript III reverse transcriptase from RNA extracted using ISOLATE II RNA Mini Kit (Bioline) following the manufacturer’s instructions. Codon-optimized versions of the transcripts were then synthesized (GenScript) for characterization in human expression systems, and subcloned into plasmid cloning DNA3.1-C-eGFP using HindIII and BamHI. A 6-bp Kozak sequence (GCCACC) was included upstream of the start codon (ATG), and the stop codon removed.
Site-Directed Mutagenesis of EhEUKCATB1
Site-directed mutagenesis of the SF region of EhEUKCATB1 was performed using a Q5 Site-Directed Mutagenesis Kit (New England BioLabs). Primer sequences (SDM1_F- TGACCGGAGAcTCCTGGTCTG; SDM1_R- GCACCTGAAACAGTGTGTAC) were designed (using the NEBase Changer Tool: https://nebasechanger.neb.com/), to replace a single Glu residue with Asp. Positive clones were screened by restriction analysis and confirmed by DNA sequencing of the entire gene.
Transfection of HEK293 Cells
HEK293 cells were plated for transfection onto glass-bottom (35-mm) petri-dishes coated with poly-l-Lys (ibidi) to help with cell adhesion. Transfections of HEK293 were performed with 4 μL of Lipofectamine 2000 (Invitrogen) and 1 to 2.5 μg of plasmid DNA per 35 mm2, each prepared separately with Opti-MEM (Gibco). The Lipofectamine and DNA were mixed and allowed to rest for 5 min before 200 μL of the mixture was added to each plate. After 12 to 30 h of incubation, cells were rinsed and maintained with fresh growth media and kept in the incubator at 37°C with 5% (v/v) CO2 and 95% (v/v) O2 until used for electrophysiological experiments. Cells exhibiting GFP fluorescence were subsequently selected for electrophysiological analysis.
HEK293 Whole Cell Patch-Clamp Electrophysiology
Electrophysiological recordings were carried out at room temperature with an AxoPatch 200B or Multiclamp 700B Amplifier (Molecular Devices) through a PC computer equipped with a model no. 1332 Analog-To-Digital Converter (Digidata) in conjunction with either the software pClamp 9.2 or pClamp10.1 (Molecular Devices). Patch electrodes were pulled from filamented borosilicate glass (1.5 mm outer diameter, 0.86 mm inner diameter) using a model no. P-97 Puller (Sutter Instruments) to resistances of 2 to 5 MΩ. In some experiments (analysis of SaEUKCATA1), unpolished electrode tips were coated with beeswax to minimize pipette capacitance. Pipette and extracellular solutions were as described in Supplemental Table S4. NMDG-Cl was prepared by titrating NMDG to neutrality with HCl. Data presented are leak-subtracted and adjusted for liquid junction potential. Whole-cell capacitance and seal resistance (leak) were periodically monitored during experiments. These corrected voltages were used to plot current-voltage curves and in all subsequent investigations. The amplitudes of the currents were measured from the baseline to the peak value and were normalized for cell capacitance as whole-cell current densities (pA/pF). Steady-state activation was studied by measuring the peak sodium conductance (GNa) during a 50-ms test pulse to various test potentials from a −120-mV holding voltage. GNa was calculated from the equation: GNa = INa/(V −Vrev), where INa is the peak sodium current during the test depolarization (V), and Vrev is the sodium reversal potential. Data were normalized to maximum peak conductance (Gmax) and fit to a two-state Boltzmann distribution: GNa/Gmax = (1 + exp[(V −V0.5)/k])−1, where V0.5 is the potential for half-maximal activation and k is the Boltzmann constant.
To study steady-state fast inactivation, cells were held at prepulse potentials ranging from −140 to +30 mV for 50 ms and then subjected to a –10-mV test pulse for 10 ms. Normalized peak currents were plotted versus prepulse potentials, and curves were fitted by the Boltzmann function: I/Imax = (1 + exp[(V −V0.5)/k])−1. Statistical analyses were performed with the software Sigma Plot 11.0 (Systat Software). Data are shown as the mean ± se (n, number of experiments).
Native Phytoplankton Cell Patch-Clamp Recording and Analysis
S. apsteinii cultures were maintained described in Drescher et al. (2012) and harvested in mid-exponential phase for decalcification using 0 mm of Ca2+ artificial seawater (ASW) supplemented with 20 mm of EGTA as described in Taylor and Brownlee (2003) and Taylor et al. (2011). After 20 to 30 min, cells were gently triturated with a transfer pipette to remove residual calcite and organic scales before plating onto a clean coverslip dish in full Ca2+ ASW. Whole-cell, patch-clamp recordings were obtained with a pipette solution comprising 200 mm of K-Glu, 5 mm of MgCl2, 5 mm of EGTA, and 100 mm of HEPES at pH 7.5. Osmolarity was balanced to 1,000 to 1,200 mOsmols using sorbitol. The extracellular ASW bathing solution contained 450 mm of NaCl, 8 mm of KCl, 30 mm of MgCl2, 16 mm of MgSO4, 10 mm of CaCl2, 2 mm of NaHCO3, and 20 mm of HEPES at pH 8.0. Data were acquired using an AxoPatch 200B Amplifier (Axon Instruments) controlled through a Digidata 1200 (Axon Instruments) with the acquisition software Clampex (Molecular Devices). Data presented are leak-subtracted and adjusted for liquid junction potential. Whole-cell capacitance and seal resistance (leak) were periodically monitored during experiments. Pipette resistance was 8 MΩ and whole-cell capacitance varied between 12 and 22 pF among cells.
Accession Numbers
Sequence data of EukCatBs characterized in this article can be found in Supplemental Table S2, alongside associated protein identifiers.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Typical current traces for native and heterologous channels in HEK293 cells.
Supplemental Figure S2. Coccolithophore EukCatB channel from S. apsteinii (SaEUKCATB1) expressed in HEK293 cells yield currents with fast kinetics resembling 4D-Nav1s.
Supplemental Figure S3. Ion selectivity of E. huxleyi EhEUKCATB1 currents.
Supplemental Figure S4. Weak permeability of EhEUKCATB1 currents to Ca2+.
Supplemental Figure S5. Comparison of the pore region of rat Nav1, EukCats, and NaChBac, indicating residues known to influence TTX binding affinity.
Supplemental Table S1. Distribution of 4D-Cav/Nav among representative eukaryotes that encode EukCatBs from transcriptome and genome data.
Supplemental Table S2. Amino acid sequences of EukCats used for heterologous characterization in this study.
Supplemental Table S3. Summary of kinetic properties and pharmacology of EhEUKCATB1 and SaEUKCATB1, compared to NaChBac and EukCatAs.
Supplemental Table S4. Composition of electrophysiology solutions (in millimolar).
Footnotes
This work was supported by the European Research Council (grant no. ERC–ADG–670390 to C.B.), the Natural Environment Research Council Independent Research Fellowship (grant no. NE/R015449/2 to K.E.H.), and the National Science Foundation (grant nos. 0949744 and 1638838 to A.R.T.).
Articles can be viewed without a subscription.
References
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. (2000) InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16: 1145–1150 [DOI] [PubMed] [Google Scholar]
- Arrigoni C, Rohaim A, Shaya D, Findeisen F, Stein RA, Nurva SR, Mishra S, Mchaourab HS, Minor DL Jr.(2016) Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell 164: 922–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingley MS, Thompson CM(1962) Bioelectric potentials in relation to movement in amoebae. J Theor Biol 2: 16–32 [Google Scholar]
- Brunet T, Arendt D(2015) From damage response to action potentials: Early evolution of neural and contractile modules in stem eukaryotes. Philos Trans R Soc B Biol Sci 371: 20150043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J.(2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Catterall WA, Wisedchaisri G, Zheng N(2017) The chemical basis for electrical signaling. Nat Chem Biol 13: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Takahashi Y, Krulwich TA, Ito M, Clapham DE(2014) Ionic selectivity and thermal adaptations within the voltage-gated sodium channel family of alkaliphilic Bacillus. eLife 3: e04387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher B, Dillaman RM, Taylor AR(2012) Coccolithogenesis in Scyphosphaera apsteinii (Prymnesiophyceae). J Phycol 48: 1343–1361 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A(2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel KH, Marchadier E, Brownlee C, Kudla J, Hetherington AM(2017) The evolution of calcium-based signalling in plants. Curr Biol 27: R667–R679 [DOI] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Yanagisawa A, Sokabe M, Yoshimura K(2009) Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr Biol 19: 133–139 [DOI] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH(1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8: 229–239 [DOI] [PubMed] [Google Scholar]
- Gur Barzilai M, Reitzel AM, Kraus JEM, Gordon D, Technau U, Gurevitz M, Moran Y(2012) Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep 2: 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Soderlund DM(2010) Human embryonic kidney (HEK293) cells express endogenous voltage-gated sodium currents and Nav 1.7 sodium channels. Neurosci Lett 469: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Chrachri A, Koester JA, Wharam S, Verret F, Taylor AR, Wheeler GL, Brownlee C(2019) Alternative mechanisms for fast Na+/Ca2+ signaling in eukaryotes via a novel class of single-domain voltage-gated channels. Curr Biol 29: 1503–1511.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B.(2001) Ion Channels of Excitable Membranes, 3rd ed Sinauer, Sunderland, MA [Google Scholar]
- Hodgkin AL, Huxley AF(1952) The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J Physiol 116: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Kitagawa K, Nagura H, Imai T, Shimomura T, Fujiyoshi Y(2010) Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J Biol Chem 285: 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LK, Alexander H, Brown CT(2019) Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. Gigascience 8: 323576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M, Inouye I, Maeda O, Chihara M(1991) The haptonema as a food-capturing device: Observations on Chrysochromulina hirta (Prymnesiophyceae). Phycologia 30: 563–573 [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. (2014) The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MD, Selvin RC, Claus W, Guillard RRL(1987) Media for the culture of oceanic ultraphytoplankton. J Phycol 23: 633–638 [Google Scholar]
- Klein AH, Vyshnevska A, Hartke TV, De Col R, Mankowski JL, Turnquist B, Bosmans F, Reeh PW, Schmelz M, Carr RW, et al. (2017) Sodium Channel Nav1.8 underlies TTX-resistant axonal action potential conduction in somatosensory C-fibers of distal cutaneous nerves. J Neurosci 37: 5204–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi R, Xu H, Ren D, Navarro B, Spiller BW, Shi Q, Clapham DE(2004) A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem 279: 9532–9538 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K(2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodh S, Yano J, Valentine MS, Van Houten JL(2016) Voltage-gated calcium channels of Paramecium cilia. J Exp Biol 219: 3028–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczydlowski EG.(2013) The molecular mystique of tetrodotoxin. Toxicon 63: 165–183 [DOI] [PubMed] [Google Scholar]
- Moran Y, Barzilai MG, Liebeskind BJ, Zakon HH(2015) Evolution of voltage-gated ion channels at the emergence of Metazoa. J Exp Biol 218: 515–525 [DOI] [PubMed] [Google Scholar]
- Nomura M, Atsuji K, Hirose K, Shiba K, Yanase R, Nakayama T, Ishida K-I, Inaba K(2019) Microtubule stabilizer reveals requirement of Ca2+-dependent conformational changes of microtubules for rapid coiling of haptonema in haptophyte algae. Biol Open 8: bio036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke M, Manton I, Clarke B(1955) Studies on marine flagellates II. Three new species of Chrysochromulina. J Mar Biol Assoc UK 34: 579–609 [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA(2012) Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486: 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA(2011) The crystal structure of a voltage-gated sodium channel. Nature 475: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozdnyakov I, Matantseva O, Skarlato S(2018) Diversity and evolution of four-domain voltage-gated cation channels of eukaryotes and their ancestral functional determinants. Sci Rep 8: 3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, Mayer C, Miller J, Monier A, Salamov A, et al. (2013) Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499: 209–213 [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE(2001) A prokaryotic voltage-gated sodium channel. Science 294: 2372–2375 [DOI] [PubMed] [Google Scholar]
- Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL Jr.(2014) Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol 426: 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li Z, Jiang Y, Pan X, Wu J, Cristofori-Armstrong B, Smith JJ, Chin YKY, Lei J, Zhou Q, et al. (2018) Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 362: eaau2596. [DOI] [PubMed] [Google Scholar]
- Taylor AR, Brownlee C(2003) A novel Cl- inward-rectifying current in the plasma membrane of the calcifying marine phytoplankton Coccolithus pelagicus. Plant Physiol 131: 1391–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AR, Brownlee C, Wheeler G(2017) Coccolithophore cell biology: Chalking up progress. Annu Rev Mar Sci 9: 283–310 [DOI] [PubMed] [Google Scholar]
- Taylor AR, Chrachri A, Wheeler G, Goddard H, Brownlee C(2011) A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol 9: e1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AD, Dhanji-Rapkova M, Coates L, Bickerstaff L, Milligan S, O’Neill A, Faulkner D, McEneny H, Baker-Austin C, Lees DN, et al. (2017) Detection of tetrodotoxin shellfish poisoning (TSP) toxins and causative factors in bivalve molluscs from the UK. Mar Drugs 15: E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C(2010) Calcium channels in photosynthetic eukaryotes: Implications for evolution of calcium-based signalling. New Phytol 187: 23–43 [DOI] [PubMed] [Google Scholar]
- West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA(1992) A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci USA 89: 10910–10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L, Navarro B, Ren D, Ramos A, Clapham DE(2002) The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol 120: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, et al. (2012) Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]