Functional flavone synthase Is in liverworts are evolutionarily linked to seed plant flavanone 3β-hydroxylases, and bifunctional enzymes likely represent a transition between the two enzymes.
Abstract
During the course of evolution of land plants, different classes of flavonoids, including flavonols and anthocyanins, sequentially emerged, facilitating adaptation to the harsh terrestrial environment. Flavanone 3β-hydroxylase (F3H), an enzyme functioning in flavonol and anthocyanin biosynthesis and a member of the 2-oxoglutarate-dependent dioxygenase (2-ODD) family, catalyzes the hydroxylation of (2S)-flavanones to dihydroflavonols, but its origin and evolution remain elusive. Here, we demonstrate that functional flavone synthase Is (FNS Is) are widely distributed in the primitive land plants liverworts and evolutionarily connected to seed plant F3Hs. We identified and characterized a set of 2-ODD enzymes from several liverwort species and plants in various evolutionary clades of the plant kingdom. The bifunctional enzyme FNS I/F2H emerged in liverworts, and FNS I/F3H evolved in Physcomitrium (Physcomitrella) patens and Selaginella moellendorffii, suggesting that they represent the functional transition forms between canonical FNS Is and F3Hs. The functional transition from FNS Is to F3Hs provides a molecular basis for the chemical evolution of flavones to flavonols and anthocyanins, which contributes to the acquisition of a broader spectrum of flavonoids in seed plants and facilitates their adaptation to the terrestrial ecosystem.
The success of land plants in the colonization of and adaptation to terrestrial ecosystems has been particularly attributed to the emergence and evolution of a unique metabolic capacity that synthesizes diverse specialized metabolites, including flavonoids, a highly polymorphic class of polyphenols (Weng and Chapple, 2010). The flavonoid metabolites have been classified into several subgroups, namely flavanones, dihydroflavonols, flavones, flavonols, flavan-3,4-diols, flavan-3-ols, and anthocyanins, based on their oxidation status and substitution patterns of the core skeleton (Winkel-Shirley, 2001; Martens et al., 2010). Along with the evolution of land plants, different classes of flavonoids emerged (Koes et al., 1994). The basal land plants liverworts produce chalcones, flavanones, and flavones; whereas lycophytes gained the ability to produce proanthocyanidins (Markham, 1984; Koes et al., 1994). Furthermore, both pteridophyta and gymnosperms, while dominated with flavone production, began to produce flavonols (Markham, 1984; Koes et al., 1994). Finally, flavonols and anthocyanins are well represented in angiosperms. Flavonols, which bear a 3-hydroxyl group in the core structure, have been exploited as effective photoprotectants against UV-B radiation (Solovchenko and Schmitz-Eiberger, 2003), as signal providers to symbionts (Hungria et al., 1991), as regulators of the transport of phytohormones (Peer and Murphy, 2007), and as determinants of conditional male fertility (Muhlemann et al., 2018). Anthocyanins, derived from dihydroflavonol, are important for sexual reproduction, acting as attractants for insect pollinators and for animal dispersers of seed (Shimada et al., 2005). It is obvious that a clear chemical evolution trace from chalcones, flavanones, and flavones to flavonols and anthocyanins, occurs across plant phyla. However, the molecular basis for such a chemical evolution remains mysterious.
The biosynthesis of flavones and flavonols requires chemical conversion of a common precursor, (2S)-flavanone, and is catalyzed by flavone synthase I (FNS I) and flavanone 3β-hydroxylases (F3Hs), respectively. Both enzymes as well as flavonol synthase (FLS) and anthocyanidin synthase (ANS) belong to a larger enzyme family, the 2-oxoglutarate-dependent dioxygenases (2-ODDs; Farrow and Facchini, 2014). FNS I converts (2S)-flavanone to flavone via desaturation of carbon 2 and 3 of the heterologous ring of flavanone (Gebhardt et al., 2005, 2007), while F3H catalyzes the conversion of (2S)-flavanone to (2R,3R)-dihydroflavonol by hydroxylation of the C-3β position (Supplemental Fig. S1). Subsequently, FLS converts (2R,3R)-dihydroflavonols to their corresponding flavonols, and ANS catalyzes the nonpigmented leucoanthocyanidins (leucopelargonidin, leucocyanidin, and leucodelphinidin) to the pigmented anthocyanidins (pelargonidin, cyanidin, and delphinidin, respectively; Supplemental Fig. S1). These four classes of 2-ODD enzymes phylogenetically form two distinct subgroups, one consisting of F3H and FNS I and the other consisting of FLS and ANS. FNS I and F3H both use flavanone as substrate and exhibit, in general, a relatively narrow substrate specificity (Turnbull et al., 2000; Martens et al., 2003), whereas ANS and FLS display some degree of promiscuity in their substrate preferences and catalytic activities. For example, Arabidopsis (Arabidopsis thaliana) FLS1 is not only capable of converting dihydroflavonols to their corresponding flavonols but also mediates the oxidation of 2S-flavanone (naringenin) to both dihydrokaempferol enantiomers, an activity normally associated with F3H (Prescott et al., 2002). While F3Hs are ubiquitous in vascular plants, FNS Is appear to be confined to the Apiaceae family as well as a few non-Apiaceae species such as rice (Oryza sativa; Lee et al., 2008), maize (Zea mays), and Arabidopsis (Falcone Ferreyra et al., 2015). Prior to the discovery of FNS Is in those non-Apiaceae species, it was assumed that the gene encoding FNS I arose from duplication and mutation of F3H (Martens et al., 2001, 2003; Gebhardt et al., 2005, 2007). However, the FNS Is revealed in both Z. mays and Arabidopsis show very poor sequence similarity with those present in Apiaceae species, which suggests that the evolution of the FNS Is was not as clear-cut as was originally believed. It is likely that the evolution of FNS occurred several times independently. In several cereal crops, such as Z. mays, O. sativa, and wheat (Triticum aestivum), flavones are the major flavonoid substances, which protect the plants during pathogen attack and under biotic or abiotic stress conditions (Righini et al., 2019).
Previously, we found that the liverwort Plagiochasma appendiculatum FNS I (which should change to PaFNS I/F2H, according to the function) converted flavanone to 2-hydroxyflavanone and flavone (Han et al., 2014). The dual FNS I and F2H activities of PaFNS I/F2H, together with the fact that its amino acid sequence shares a higher identity with F3Hs than with FNS Is, implicates an evolutionary connection between liverwort FNS Is and seed plant F3Hs. On the other hand, previous in silico analysis failed in identifying any F3H sequences in either the bryophyte Physcomitrium (Physcomitrella) patens or the lycophyte Selaginella moellendorffii, even though both species produce dihydroflavonol-derived metabolites. To identify when and how F3H emerged and evolved to produce a vast variety of flavonoid metabolites, we systematically identified FNS I and F3H homologous sequences from species of different phyla, including liverworts, P. patens, S. moellendorffii, gymnosperms, and angiosperms. Subsequent biochemical characterization revealed that the functionally promiscuous FNS Is widely emerged in the liverworts, which evolved into a dual-function enzyme with both FNS I and F3H activities in both P. patens and S. moellendorffii. Further evolution led to the emergence of F3H with a minor level of FNS I activity in gymnosperm species, while those generated by angiosperm species showed a more specific F3H activity.
RESULTS
Identification of FNS I and F3H Homologous Sequences
When using the sequence of PaFNS I/F2H as a query to search the transcriptomic sequences of liverworts, six putative 2-ODD motif-containing genes were identified from different liverwort species. They were tentatively designated as Pa2ODD1, Mpa2ODD1, Me2ODD1, Cc2ODD2, Cj2ODD1, and Cj2ODD2 (Supplemental Table S1). To identify the potential FNS I and F3H homologous sequences in moss P. patens and lycophyte species S. moellendorffii, instead of a BLAST search we employed the 2OG-Fe (II) oxygenase superfamily signature PF03171 defined in Pfam (pfam.xfam.org/) to scan the genome sequences of P. patens and S. moellendorffii. Three sequences from P. patens and six from S. moellendorffii were recognized and tentatively designated as Pp2ODD1 to Pp2ODD3 and Sm2ODD1 to Sm2ODD6 (Supplemental Table S2).
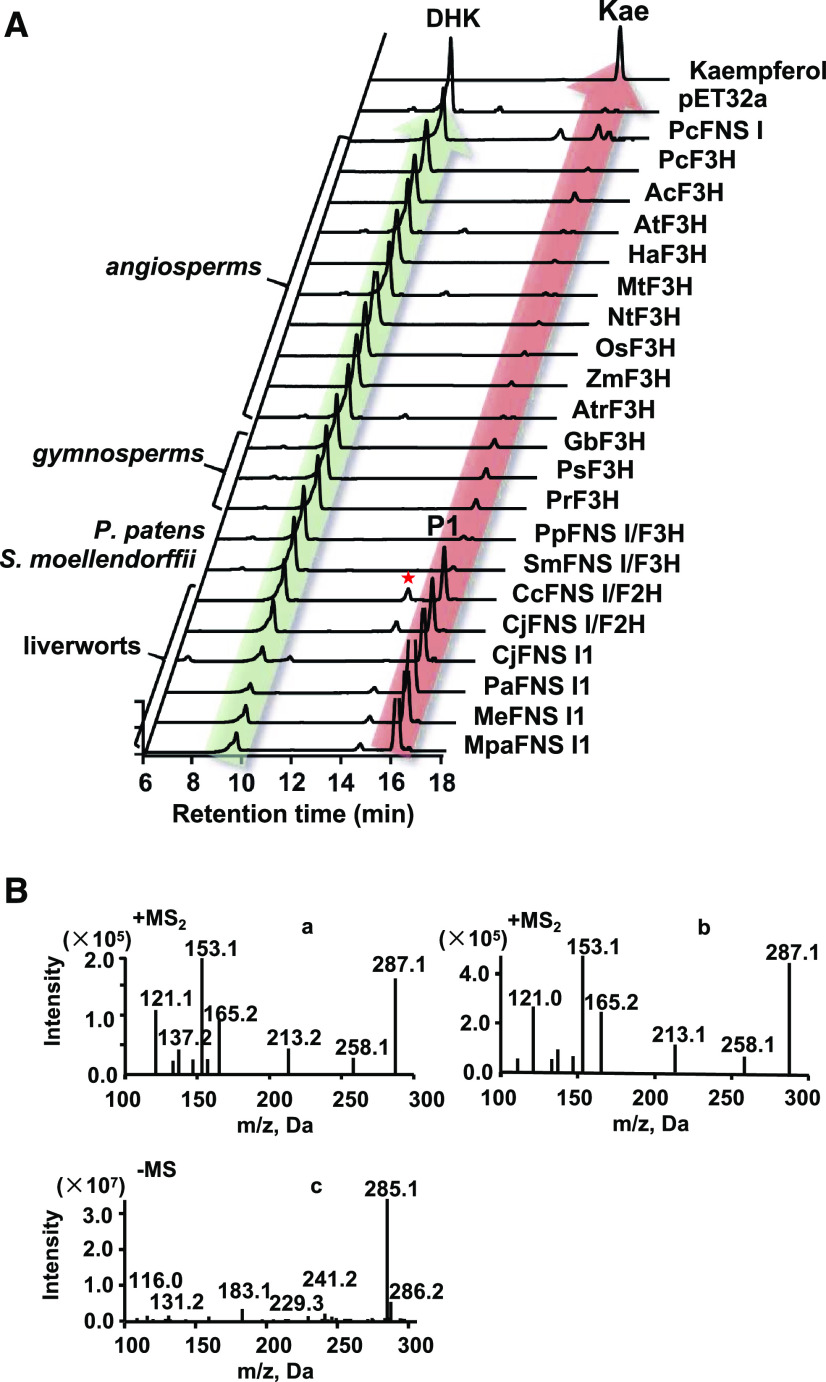
Gene Isolation, Protein Expression, and Functional Characterization
The full-length cDNAs of the identified putative 2-ODD genes from liverworts, P. patens, and S. moellendorffii were isolated from their respective species. In addition, an FNS I gene from parsley (Petroselinum crispum; AY817680), three F3Hs from gymnosperms Picea sitchensis (ABK25766), Ginkgo biloba (AAU93347), and Pinus radiata (AGY80772), and nine F3Hs from angiosperms O. sativa (XP_015634635.1), Z. mays (NM_001112225.1), Amborella trichopoda (XM_006851991), Nymphaea thermarum (JAANDH010000393.1), Helianthus annuus (XP_022015871.1), Arabidopsis (CAD37988), Medicago truncatula (XM_003629275), P. crispum (AY230248.1), and Aethusa cynapium (ABG78792.1) were also included for sequence alignment and functional characterization. The matrix with percentage of sequence identities for these proteins is included in Supplemental Table S3. The recombinant proteins were produced via heterologous expression in Escherichia coli BL21 and purified via affinity chromatography (Supplemental Fig. S2). The enzymatic assays were conducted with naringenin as substrate. The results showed that four of six liverwort enzymes (Pa2ODD1, Mpa2ODD1, Me2ODD1, and Cj2ODD1) and the Apiaceae PcFNS I converted naringenin into a single reaction product, P1 (Fig. 1A), that shares the same retention time as the authentic standard apigenin and has a mass-to-charge ratio (m/z) of 271 with daughter ions of m/z 153, 119, and 163, identical to that of apigenin (Fig. 1B, graphs a and b). These data suggest that these enzymes possess flavone synthase activity. In contrast, two liverwort enzymes, Cc2ODD2 and Cj2ODD2, catalyzed naringenin to apigenin and the additional product P2. Liquid chromatography-mass spectrometry (LC-MS) analysis revealed this additional product with m/z value of 289 (Fig. 1B, graph c), indicating the presence of an additional oxygen atom compared with the substrate naringenin. The MS/MS profile of this product showed fragmentations with m/z values of 153, 121, and 163, which is consistent with the characterized reaction product of PaFNS I/F2H, 2-hydroxynaringenin (Han et al., 2014).
Figure 1.
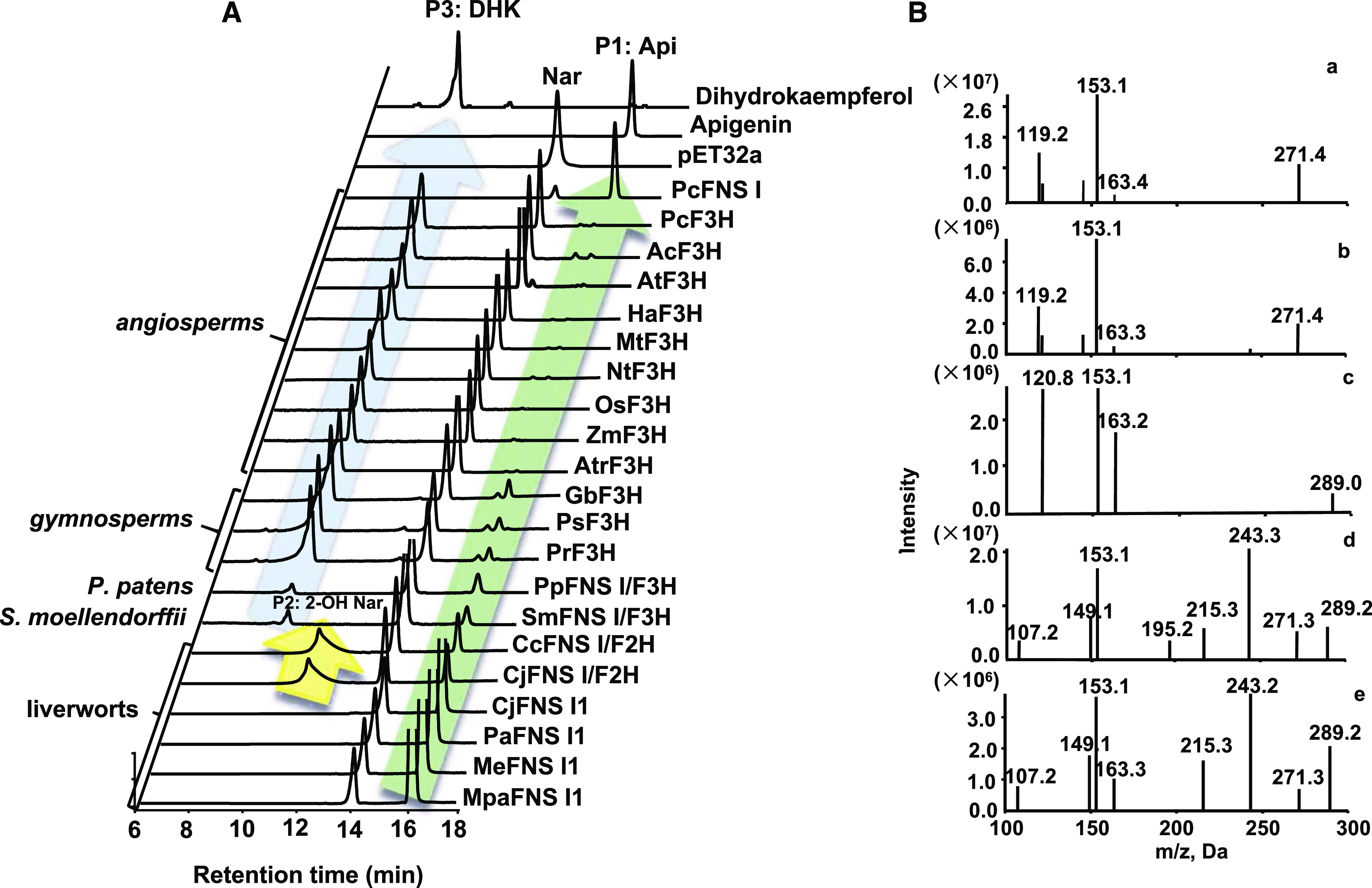
In vitro enzyme activity of purified recombinant proteins provided with naringenin as substrate. A, HPLC profiles of reaction products derived from various 2-ODD enzymes from different plant species, the empty vector control, and the standards. Api, Apigenin; DHK, dihydrokaempferol; Nar, Naringenin; 2-OHNar, 2-hydroxynaringenin. B, MS/MS profiles of the reactions P1 (a), P2 (c), and P3 (d), apigenin standard (b), and dihydrokaempferol standard (e).
This result suggests that two liverwort enzymes (Cc2ODD2 and Cj2ODD2) possess dual activities yielding both 2-hydroxyflavanone and flavone. Interestingly, both P. patens and S. moellendorffii enzymes (Pp2ODD1 and Sm2ODD1) also converted naringenin into two products, apigenin and a small amount of P3 that had an m/z value of 289 (Fig. 1B, graph d), and shared the same retention time and fragmentation pattern with the authentic dihydrokaempferol (Fig. 1B, graph e), suggesting that P. patens and S. moellendorffii enzymes possess both FNS I and F3H activities. When incubated with naringenin, the gymnosperm enzymes (GbF3H, PrF3H, and PsF3H) generated mostly dihydrokaempferol with minor apigenin, indicative of a catalytic activity shift to F3H (Fig. 1). The angiosperm enzymes (AtrF3H, AtF3H, MtF3H, HaF3H, NtF3H, PcF3H, AcF3H, OsF3H, and ZmF3H) produced exclusively dihydrokaempferol when they were incubated with naringenin (Fig. 1). The genes isolated from liverworts, P. patens, and S. moellendorffii were thereafter designated as CjFNS I1, MeFNS I1, MpaFNS I1, PaFNS I1, CjFNS I/F2H, CcFNS I/F2H, PpFNS I/F3H, and SmFNS I/F3H. The results demonstrated that while most liverwort enzymes exhibited flavone synthase activity to produce apigenin, as did the Apiaceae PcFNS I, CjFNS I/F2H and CcFNS I/F2H had dominant F2H activities in addition to their FNS I activities, enabling them to convert naringenin to apigenin and 2-hydroxynaringenin. P. patens and S. moellendorffii enzymes (PpFNS I/F3H and SmFNS I/F3H) acquired F3H activity in addition to acting as FNS I. Gymnosperm enzymes (GbF3H, PsF3H, and PrF3H) further elaborated their F3H function while retaining residual activity of FNS I. Finally, angiosperm enzymes (AtrF3H, AtF3H, MtF3H, HaF3H, NtF3H, PcF3H, AcF3H, OsF3H, and ZmF3H) evolved as authentic F3H, converting naringenin exclusively to dihydrokaempferol (Fig. 1).
The effects of pH on these 2-ODD enzyme activities were investigated using two buffers that spanned a pH range from 5 to 8.5 with naringenin as substrate. The optimum pH of PcFNS I, CjFNS I1, and CjFNS I/F2H activity was 6.5 in MES buffer (Supplemental Fig. S3). The enzymes of gymnosperms (PsF3H, PrF3H, and GbF3H) showed similar or comparable activities in different pH buffers (Supplemental Fig. S3). The F3H enzymes of angiosperms showed high enzyme activities at a broad pH range from 5.5 to 7 (Supplemental Fig. S4).
Under the optimal pH, the kinetic parameters of each 2-ODD enzyme were investigated (Table 1; Supplemental Figs. S5 and S6). In general, CjFNS I1, CjFNS I/F2H, and the F3Hs from spermatophytes showed high catalytic efficiency toward naringenin. PcFNS I kcat/Km values (8.7 × 104 m−1 s−1) were greater than the equivalents for CjFNS I1 and CjFNS I/F2H (2.6 × 104 and 1.2 × 104 m−1 s−1, respectively). The kcat/Km values of both PpFNS I/F3H (0.35 × 104 m−1 s−1) and SmFNS I/F3H (0.29 × 104 m−1 s−1) toward naringenin were much lower than those of enzymes from liverworts and spermatophytes. Except for AtrF3H (kcat/Km value of 1.7 × 104), the catalytic efficiency of the F3Hs from angiosperms (from 5.5 × 104 to 8.1 × 104 m−1 s−1) were higher than those of gymnosperms (from 3.6 × 104 to 4.5 × 104 m−1 s−1).
Table 1. Kinetic analysis of purified recombinant 2-ODD enzymes using naringenin as substrate.
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1 s−1 | |
| PcFNS I | 0.31 ± 0.064 | 0.027 ± 0.00026 | 8.7 × 104 |
| CjFNS I1 | 9.4 ± 1.6 | 0.25 ± 0.012 | 2.6 × 104 |
| CjFNS I/F2H | 14 ± 3.2 | 0.17 ± 0.0084 | 1.2 × 104 |
| SmFNS I/F3H | 91 ± 13 | 0.26 ± 0.014 | 0.29 × 104 |
| PpFNS I/F3H | 95 ± 11 | 0.33 ± 0.019 | 0.35 × 104 |
| PrF3H | 20 ± 3.5 | 0.91 ± 0.048 | 4.5 × 104 |
| PsF3H | 26 ± 5.2 | 0.95 ± 0.039 | 3.6 × 104 |
| GbF3H | 11 ± 2.2 | 0.44 ± 0.027 | 4.1 × 104 |
| AtrF3H | 22 ± 4.9 | 0.38 ± 0.016 | 1.7 × 104 |
| ZmF3H | 5.0 ± 1.1 | 0.27 ± 0.016 | 5.5 × 104 |
| OsF3H | 21 ± 4.2 | 1.2 ± 0.085 | 5.8 × 104 |
| NtF3H | 3.2 ± 0.48 | 0.18 ± 0.0052 | 5.7 × 104 |
| MtF3H | 12 ± 2.6 | 0.87 ± 0.026 | 7.4 × 104 |
| HaF3H | 2.6 ± 0.36 | 0.17 ± 0.0041 | 6.5 × 104 |
| AtF3H | 5.7 ± 0.72 | 0.44 ± 0.012 | 7.6 × 104 |
| AcF3H | 15 ± 2.1 | 0.83 ± 0.032 | 5.5 × 104 |
| PcF3H | 4.7 ± 1.1 | 0.38 ± 0.018 | 8.1 × 104 |
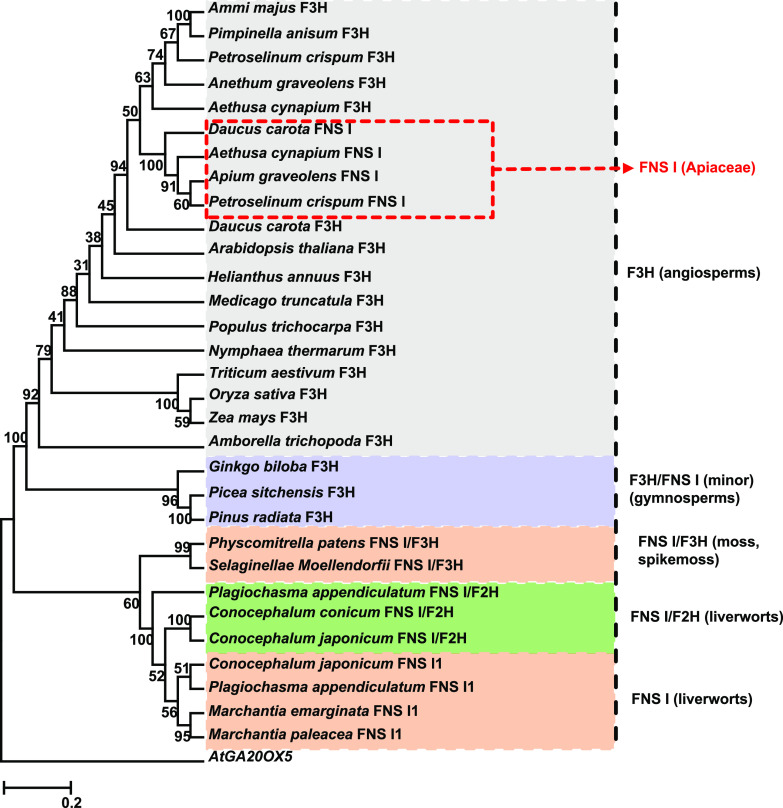
Phylogenetic analysis of the characterized F3Hs and FNS Is showed that the single-function FNS Is and the enzymes displaying both FNS I and F2H activities of liverworts are grouped in a single clade that is placed at the root of the phylogenetic tree; separated from this clade are the enzymes bearing both FNS I and F3H activity presented in P. patens and S. moellendorffii; the further extended third clade comprises both gymnosperm and angiosperm F3Hs. PcFNS I as a representative of the typical Apiaceae FNS I clustered with other Apiaceae FNS Is and F3Hs, away from the liverwort FNS Is (Fig. 2).
Figure 2.
Phylogeny of plant FNS Is and F3Hs. The length of each branch reflects the distance between nodes. AtGA20OX5 was used as an outlier. Accession numbers are as follows: Ammi majus F3H (AY817678), Pimpinella anisum F3H (AY817674), Petroselinum crispum F3H (AY230248), Anethum graveolens F3H (AY817679), Aethusa cynapium F3H (DQ683351), Daucus carota F3H (AF184270), D. carota FNS I (AY817675), P. crispum FNS I (AY817680), Aethusa cynapium FNS I (DQ683350), Apium graveolens FNS I (AY817676), Medicago truncatula F3H (XM_003629275), Populus trichocarpa F3H (XM_006383044), Arabidopsis thaliana F3H (NM_114983), Amborella trichopoda F3H (XM_006851991), Triticum aestivum F3H (EF463100), Helianthus annuus F3H (XP_022015871), Nymphaea thermarum F3H (JAANDH010000393), Zea mays F3H (NM_001136803), Oryza sativa F3H (XM_015779149), Ginkgo biloba F3H (AAU93347), Picea sitchensis F3H (ABK25766), Pinus radiata F3H (AGY80772), Physcomitrella patens FNS I/F3H (XP_001780809), Selaginella moellendorffii FNS I/F3H (XP_002985262), Plagiochasma appendiculatum FNS I/F2H (KJ439220), and Arabidopsis AtGA20OX5 (AT1G44090).
Expression of Liverworts and S. moellendorffii Genes in Arabidopsis
To further assess the in planta activity of the identified enzymes from primitive land plants, the genes encoding liverwort CjFNS I/F2H, which shows both FNS I and F2H activity, and CjFNS I1, the sole FNS I enzyme, as well as S. moellendorffii SmFNS I/F3H with both FNS I and F3H activities were constitutively expressed in Arabidopsis using the cauliflower mosaic virus 35S promoter. Reverse transcription (RT)-PCR detected transcripts from the 35S::CjFNS I1, 35S::CjFNS I/F2H, and 35S::SmFNS I/F3H transgenes in T2 plants, while this was not the case for control wild-type plants (Fig. 3A). Flavones are hardly detectable in wild-type Arabidopsis, although a native FNS I gene has recently been identified (Falcone Ferreyra et al., 2015). The methanolic extracts of T2 transgenic seedlings grown in a medium supplemented with 50 mm naringenin were analyzed using LC-MS (Fig. 3, B, E, and F). Plants expressing CjFNS I1 accumulated 15 to 30 μg apigenin g−1 dry weight, while the control plants accumulated approximately only 3 μg g−1. In plants expressing CjFNS I/F2H, the content of apigenin was about 40% higher than that detected in the control plants; 2-hydroxynaringenin, however, was not detected, presumably due to its spontaneous dehydration to apigenin in planta. Finally, the apigenin content of SmFNS I/F3H transgenic lines was about fourfold higher than that in control plants, while kaempferol content increased about 30% to 40% compared with the control, indicating that SmFNS I/F3H functions as both FNS I and F3H in planta. Mass spectrometry profiles of apigenin and kaempferol produced in Arabidopsis are shown in Figure 3, C and D.
Figure 3.
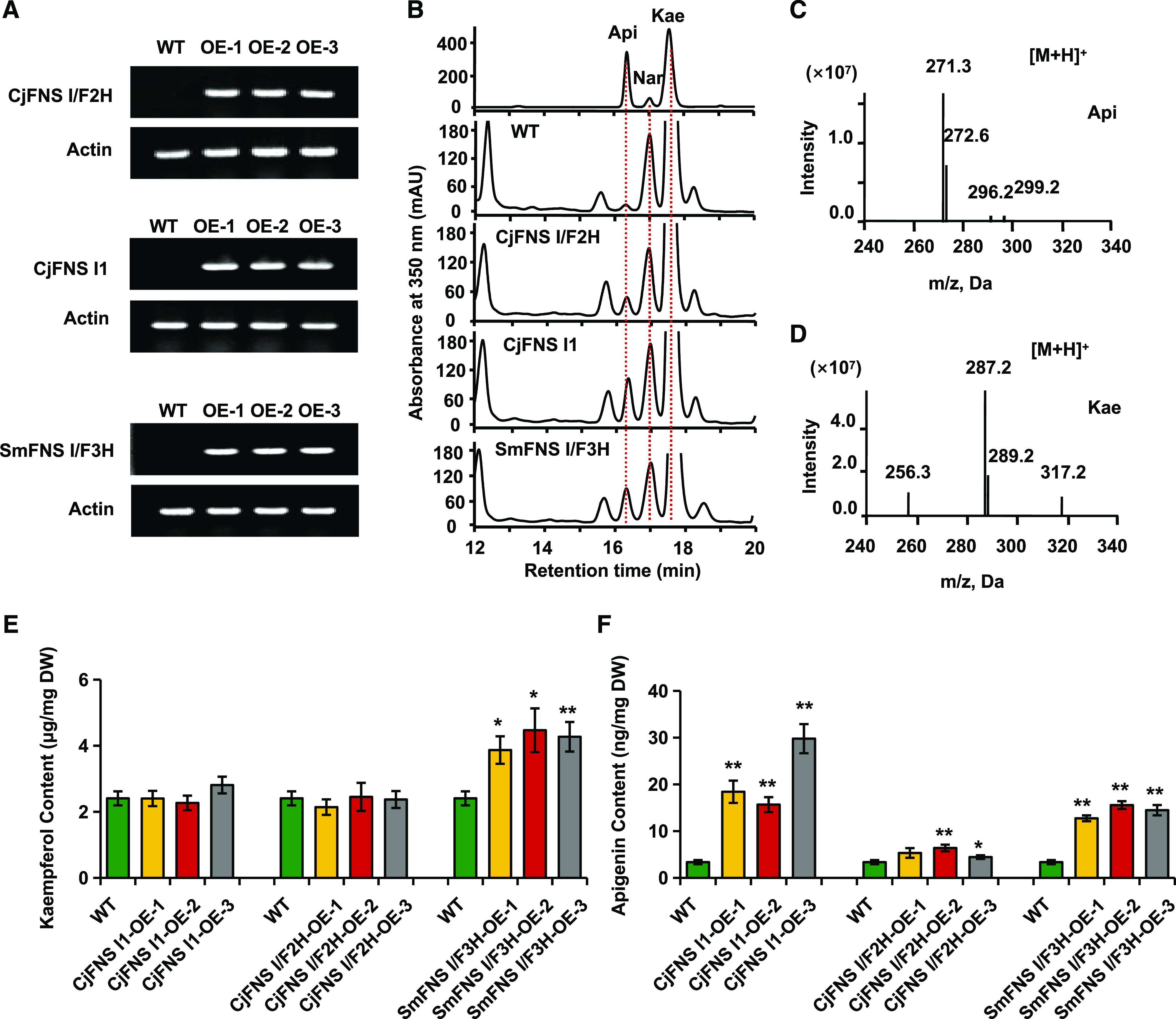
Constitutive expression of CjFNS I/F2H, CjFNS I1, and SmFNS I/F3H in Arabidopsis. A, Transgene transcript presence as determined by RT-PCR. B, Representative HPLC profiles of flavonoids produced by nontransgenic and transgenic Arabidopsis provided with naringenin as substrate. Api, Apigenin; Kae, kaempferol; Nar, naringenin. C and D, Mass spectrometry profiles of apigenin (C) and kaempferol (D) produced in Arabidopsis. E and F, Kaempferol (E) and apigenin (F) produced by nontransgenic Arabidopsis and three transgenic lines grown in the presence of naringenin. DW, Dry weight. Data are means ± sd of three biological replicates. Statistical analysis was performed with Student’s t test. Means differ from the wild type (WT) at P < 0.05 (*) and P < 0.01 (**).
Identification of Functional Residues
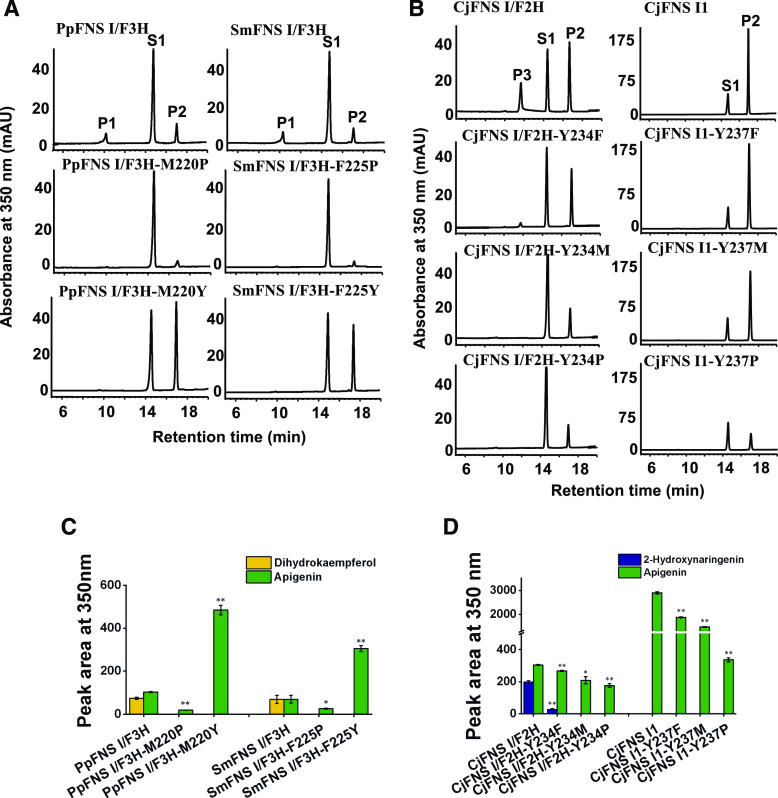
To probe the structural basis for functional evolution of the recognized flavonoid biosynthetic 2-ODD enzymes, we conducted protein homology modeling with the binding of naringenin and 2-oxoglutarate, based on the structure of ANS complexed with naringenin (Welford et al., 2005). The residues Y234, V142, L212, and V303 (numbering based on the liverwort CjFNS I/F2H sequence) formed a hydrophobic pocket that anchors the bound substrate (Supplemental Fig. S7A). Y240 in liverwort PaFNS I/F2H (i.e. Y234 in CjFNS I/F2H) is a key amino acid for the enzyme activity (Han et al., 2014). This key amino acid is necessary but not efficient for the naringenin 2-hydroxylase (F2H) activity. Alignment of the set of functional FNS Is and F3Hs revealed that this residue was replaced with P in both gymnosperm and angiosperm entries, with F in S. moellendorffii, and with M in P. patens (Supplemental Fig. S7B). We then conducted the following substitutions: (1) P of the gymnosperm and angiosperm F3Hs to either M, F, or Y; (2) Y of the liverwort FNS Is and FNS I/F2Hs to either M, F, or P; (3) M in the FNS I/F3H of P. patens to either P or Y; and (4) F in the FNS I/F3H of S. moellendorffii to either P or Y. When the P residue of seed plant F3Hs was altered to M, the mutant variants (GbF3H-P228M, PsF3H-P227M, PrF3H-P227M, AtF3H-P220M, MtF3H-P221M, AtrF3H-P221M, PcF3H-P221M, and AcF3H-P221M) all exhibited predominant F3H activity, while higher FNS I activity appeared than did the corresponding wild-type enzyme (Fig. 4). When the P residue of those enzymes was altered to F, the F3H activity of mutant variants (GbF3H-P228F, PsF3H-P227F, PrF3H-P227F, AtF3H-P220F, MtF3H-P221F, AtrF3H-P221F, PcF3H-P221F, and AcF3H-P221F) was reduced, while FNS I activity increased compared with the wild-type enzymes (Fig. 4). Notably, when P was substituted with Y, all the variants (GbF3H-P228Y, PsF3H-P227Y, PrF3H-P227Y, AtF3H-P220Y, MtF3H-P221Y, AtrF3H-P221Y, PcF3H-P221Y, and AcF3H-P221Y) exhibited dominant FNS I activity with dramatically reduced F3H activity (Fig. 4).
Figure 4.
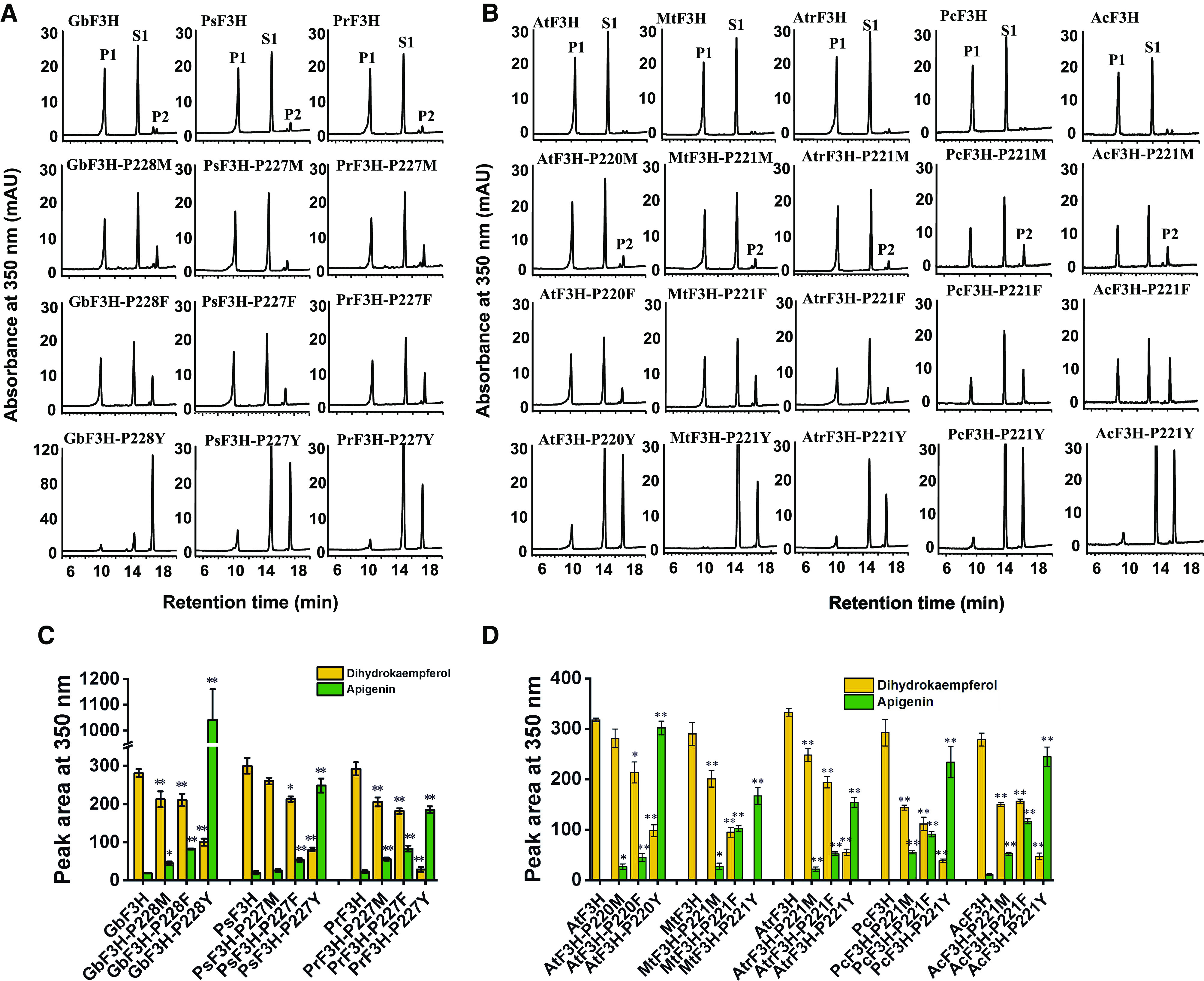
In vitro enzyme activity of mutant and wild-type GbF3H, PsF3H, PrF3H, AtF3H, MtF3H, AtrF3H, PcF3H, and AcF3H, based on naringenin as substrate. A and C, HPLC profiles of enzyme activity of mutant and wild-type GbF3H, PsF3H, and PrF3H in vitro (A) and their peak areas (C). B and D, HPLC profiles of enzyme activity of mutant and wild-type AtF3H, MtF3H, AtrF3H, PcF3H, and AcF3H in vitro (B) and their peak areas (D). P1, Dihydrokaempferol; P2, apigenin; S1, naringenin. Values shown are means ± sd (n = 3). Statistical analysis was performed with Student’s t test. Means differ from the wild-type enzyme at P < 0.05 (*) and P < 0.01 (**).
These data indicate that the P residue in the substrate-binding pocket of 2-ODD from gymnosperms or angiosperms is essential for acquiring and sustaining F3H activity and specificity. The variants harboring M or F at this position release the constraint of catalysis and make the enzymes versatile, exhibiting both F3H and FNS I activities, which is consistent with the native P. patens and S. moellendorffii enzymes PpFNS I/F3H and SmFNS I/F3H. Similarly, when M or F residues of the P. patens and S. moellendorffii enzymes were changed to Y (i.e. PpFNS I/F3H-M220Y and SmFNS I/F3H-F225Y), the original dual-functional enzymes were transformed into sole-functional ones, showing an enhanced FNS I activity and nearly complete loss of F3H activity, suggesting that the Y residue is critical for sustaining FNS I activity (Fig. 5, A and C). On the other hand, when the Y residue present in the liverwort CjFNS I/F2H and CjFNS I1 was mutated to either F, M, or P, the level of both FNS I and/or F2H activity was reduced, but F3H activity was not elevated (Fig. 5, B and D). When the M or F residue of P. patens and S. moellendorffii enzyme was changed to P, the activities of PpFNS I/F3H-M220P and SmFNS I/F3H-F225P in producing flavone and dihydrokaempferol were sharply decreased compared with those of the wild-type enzymes. The site-directed mutagenesis experiment indicated that single-residue substitution from P in gymnosperm and angiosperm F3Hs to Y that is conserved in liverwort FNS Is almost completely shifted the F3H to FNS I, further hinting toward the evolutionary relationship and functional connection of the liverwort FNS I with seed plant F3H.
Figure 5.
In vitro enzyme activity of wild-type and mutant PpFNS I/F3H, SmFNS I/F3H, CjFNS I/F2H, and CjFNS I1 provided with naringenin as substrate. A and C, HPLC profiles of enzyme activity of mutant and wild-type PpFNS I/F3H and SmFNS I/F3H in vitro (A) and their peak areas (C). B and D, HPLC profiles of enzyme activity of mutant and wild-type CjFNS I/F2H and CjFNS I1 in vitro (B) and their peak areas (D). P1, Dihydrokaempferol; P2, apigenin; P3, 2-hydroxynaringenin; S1, naringenin. Values shown are means ± sd (n = 3). Statistical analysis was performed with Student’s t test. Means differ from the wild-type enzyme at P < 0.05 (*) and P < 0.01 (**).
Promiscuity of Liverwort FNS Is and FNS I/F2Hs
When functional FNS Is and F3Hs were provided with dihydrokaempferol as substrate, the liverwort FNS I and FNS I/F2H enzymes all converted it to kaempferol (Fig. 6A), which was verified with the same retention time, m/z value of 287 under positive ion mode, and fragmentation pattern as the authentic standard (Fig. 6B). These results suggest that all the identified liverwort enzymes show FLS activity. In contrast, when P. patens, S. moellendorffii, and seed plant FNS Is and F3Hs were incubated with dihydrokaempferol, they were inactive or at best yielded trace amounts of kaempferol (Fig. 6A). The liverwort enzymes and PcFNS I also make another minor product with a retention time of about 14.5 min. LC-MS analysis revealed this additional product with m/z value of 285 under negative ion mode (Fig. 6B). These results indicated that this minor product showed the same molecular weight with kaempferol; however, it could not be identified due to the minor amounts. The effects of pH on CjFNS I1 and CjFNS I/F2H activities were investigated using two buffers that spanned a pH range from 5 to 8.5 with dihydrokaempferol as substrate. The optimum pH of CjFNS I1 and CjFNS I/F2H was 6.5 in MES buffer (Supplemental Fig. S8). Under the optimal pH, the kinetics parameters of CjFNS I1 and CjFNS I/F2H were investigated. The catalytic efficiency (kcat/Km value) of CjFNS I1 (2.3 × 104 m−1 s−1) was greater than that of CjFNS I/F2H (0.11 × 104 m−1 s−1) for dihydrokaempferol (Table 2; Supplemental Fig. S9). Moreover, the catalytic efficiency of CjFNS I1 for dihydrokaempferol (2.3 × 104 m−1 s−1) was similar to that for naringenin (2.6 × 104 m−1 s−1; Tables 1 and 2). However, CjFNS I/F2H preferred naringenin to dihydrokaempferol, according to the Km and kcat/Km values (Tables 1 and 2).
Figure 6.
Enzyme activity of purified recombinant proteins and the control (empty pET32a vector) measured in vitro with dihydrokaempferol as substrate. A, HPLC profiles of reaction products derived from various 2-ODD enzymes from different plant species, the empty vector control, and the standards. DHK, Dihydrokaempferol; Kae, kaempferol. B, MS/MS (MS2) profile of reaction product P1 (a), MS/MS profile of kaempferol (b), and MS profile of the unknown peak labeled with a red star in A (c).
Table 2. Kinetic analysis of recombinant CjFNS I1 and CjFNS I/F2H using dihydrokaempferol as substrate.
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1 s−1 | |
| CjFNS I1 | 13 ± 2.3 | 0.30 ± 0.0091 | 2.3 × 104 |
| CjFNS I/F2H | 38 ± 7.9 | 0.042 ± 0.0025 | 0.11 × 104 |
DISCUSSION
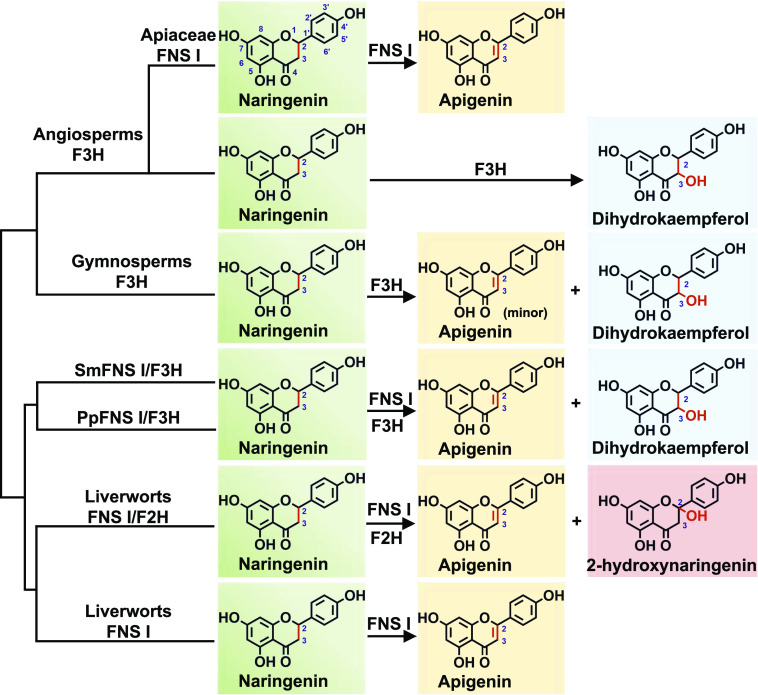
Flavonoids have been associated with a number of biological functions during plant development and growth. Their chemical diversity is thought to be responsible for the successful adaptation of plants to specific ecological niches (Falcone Ferreyra et al., 2015). Liverworts, a group of the most primitive land plants, produce the flavones apigenin and luteolin (Markham, 1988; Asakawa et al., 2013; Wang et al., 2013) but no appreciable quantities of flavonols. Consistently, our study reveals that each of five examined liverwort species possesses at least a functional FNS I gene but has no functional F3H detected. These data indicate that FNS I evolved in a much earlier period of land plant colonization, prior to the emergence of ancestral F3H, and also explain the lack of flavonols in liverworts, since F3H is required for dihydroflavonol production (otherwise flavonols would accumulate). Previously, it was documented that the genome of neither the bryophyte P. patens nor the lycophyte S. moellendorffii harbors true F3H genes, although both species do produce compounds derived from dihydroflavonol (Kawai et al., 2014). Our previous (Han et al., 2014) and current in vitro biochemical analyses demonstrate that liverworts have emerged with two kinds of FNS Is: some of the enzymes were able to convert flavanone to flavone directly by abstraction of protons associated with C-2 and C-3, while others showed additional oxygenation activity at the C-2 position in addition to their capacity for desaturation. This C-2 hydroxylation essentially resembles the F3H-catalzyed 3-hydroxylation reaction, although the positions of the addition of the oxygen atom differ from each other. More intriguingly, the homologs of both P. patens and S. moellendorffii displayed both FNS I and F3H activities. The latter activity was further strengthened in the homologous enzymes of gymnosperms (P. sitchensis, G. biloba, and P. radiata) and completely shifted in the angiosperm F3H. Combining with phylogenetic analysis, the data suggest that the FNS I in liverworts might serve as the ancestor of seed plant F3H. This early-evolved FNS I has gone through functional transition as FNS I/F2H and FNS I/F3H and eventually shifted to the bona fide F3H (Fig. 7).
Figure 7.
Summary of the reactions catalyzed by diverse plant 2-ODDs characterized in this investigation.
In most angiosperms, the capacity to express FNS I may have been lost once the physiological function of synthesizing flavones as UV-protecting pigments was carried out by the membrane-associated cytochrome P450 monooxygenase FNS II (Du et al., 2010a, 2010b; Fliegmann et al., 2010; Zhao et al., 2016). Genes encoding FNS II have been isolated and characterized from a range of angiosperm species (Du et al., 2016). FNS IIs belong to the CYP93B family in dicotyledons (Fliegmann et al., 2010; Zhao et al., 2016) and the CYP93G family in monocotyledons (Du et al., 2010a, 2010b). CYP93 family proteins are thought to be restricted to angiosperms (Du et al., 2016). Nevertheless, flavones are widely distributed among the gymnosperms, while gymnosperm F3Hs showed trace FNS I activity (Fig. 1), so it will be necessary and interesting to further dig out FNS II or FNS I enzymes to illustrate which kind of enzyme is involved in the flavone biosynthesis in gymnosperms.
FNS I might have evolved multiple times during land plant colonization and radiation. The FNS Is present in Apiaceae species show a high level of sequence identity and structural similarity with F3Hs, suggesting that they arose in ancestral Apiaceae plants following the duplication of F3H genes (Gebhardt et al., 2005, 2007). As the phylogeny indicated, FNS Is from liverworts and Apiaceae species are separated distantly, while the FNS Is and F3Hs in Apiaceae cluster as a particular branch. In angiosperm species, FNS Is were initially thought to be restricted to Apiaceae species (Britsch, 1990; Martens et al., 2001, 2003; Gebhardt et al., 2005), but they have also been identified in O. sativa (Lee et al., 2008), Equisetum arvense (Bredebach et al., 2011), and Z. mays and Arabidopsis (Falcone Ferreyra et al., 2015). The non-Apiaceae FNS Is share low sequence identity with either those from liverworts or those from Apiaceae species, implying that these proteins may have evolved through convergent or parallel evolution in different lineages. Replacement of seven amino acids of PcF3H into the PcFNS I sequence (M106T, I115T, V116I, I131F, D195E, L215V, and K216R) causes a nearly complete change in enzyme activity from F3H to FNS I (Gebhardt et al., 2007). Our previous results showing that the conversion of Y240 of PaFNS I/F2H into F reduces, and into P eliminates, the F2H activity are in agreement with the results shown for CjFNS I/F2H in Figure 5. This corresponding key amino acid residue in the F3Hs from gymnosperms and angiosperms was P. In this investigation, the site-directed mutagenesis experiments indicated that when the critical P residue present in gymnosperm and angiosperm F3Hs was altered to Y, the enzymes’ function moved from F3H to FNS I. Remarkably, the replacement is also effective in the F3H protein from P. crispum that has previously been compared with an Apiaceae-typical FNS from P. crispum (Gebhardt et al., 2007). However, this corresponding key amino acid residue in PcFNS I is P instead of Y.
These results indicate that FNS Is might originate from F3Hs by mutation of several amino acids as a fairly recent duplication event in Apiaceae (Gebhardt et al., 2007). Therefore, the path of FNS I and F3H enzyme evolution is likely to be the following: FNS Is emerged in liverwort species at a very early period of land plant colonization. Then those early-evolved FNS Is acquired additional oxygenase activity (F2H or F3H), and this function further elaborated in the evolution of vascular plants, eventually resulting in the completely F3H enzymes in the seed plants. Apiaceae might have gained FNS Is from F3Hs by gene duplication, which provides the capacity of flavone formation without losing their flavonols. Although the single-amino acid mutation from P to Y altered the F3Hs of gymnosperms and angiosperms almost to FNS Is, the reverse mutation in the liverwort FNS I sequence did not endow them with F3H activity (Fig. 5B). The implication is that the functional transition from FNS Is in the basal land plants to F3Hs in the more advanced taxa (such as the gymnosperm and angiosperm species) was possibly controlled by additional candidate residues.
It has been assumed that the substrate specificity and selectivity of both FNS Is and F3Hs are relatively narrow (Gebhardt et al., 2005). However, in stark contrast to the recombinant PcFNS I that showed high preference to 2S-naringenin but little activity with dihydrokaempferol, the liverwort FNS Is displayed significant FLS activity toward dihydrokaempferol. ANS and FLS mechanistically attack the substrate’s α-face, while FNS I and F3H target its β-face (Schofield and Zhang, 1999; Clifton et al., 2006). These data indicate that some of the liverwort FNS Is are functionally versatile, exhibiting FNS I, F2H, and FLS activity and able to dehydrogenate the carbons at both positions 2 and 3 of the flavanone substrate, no matter whether the carbon 3 is hydroxylated. It has been proposed that ancestral forms of enzymes are often functionally promiscuous and display broader substrate/product specificity than do the more evolved forms (Nam et al., 2012). The loose substrate and product specificity shown by the liverwort FNS Is reflects their prototype enzyme features.
In conclusion, this study has significantly contributed to our understanding of the evolution of F3Hs and FNS Is. Functional FNS Is emerged in the primitive land plant liverworts, which evolved into a dual-function enzyme with both FNS I and F3H activities in both P. patens and S. moellendorffii. Further evolution led to a higher specific F3H activity prevailing in gymnosperm and angiosperm plants. The bifunctional enzymes such as FNS I/F2H in the liverworts and FNS I/F3H in P. patens and S. moellendorffii likely represent a transition between canonical FNS Is and F3Hs. These findings provide a molecular basis for the chemical evolution from flavones to flavonols and anthocyanins, which are essential for land plants to flourish in terrestrial ecosystems.
MATERIALS AND METHODS
Plant Materials and Reagents
Marchantia paleacea, Plagiochasma appendiculatum, Marchantia emarginata, Conocephalum japonicum, Conocephalum conicum, Selaginella moellendorffii and Physcomitrium (Physcomitrella) patens samples were propagated via gemmal growth and sexual crossing in a greenhouse delivering a constant temperature of 25°C and a 12-h photoperiod. Medicago truncatula and Arabidopsis (Arabidopsis thaliana) plants were grown in soil at 22°C under a 12-h photoperiod. Naringenin, apigenin, and kaempferol were all purchased from Chengdu Must Bio-technology, and dihydrokaempferol was purchased from BioBioPha. All solvents used were of HPLC grade.
Sequence Alignment, Phylogenetic Analysis, and Homology Modeling
A scan of the transcriptomes of the five liverwort species P. appendiculatum (SRP073827), M. paleacea (SRP078650), M. emarginata (SRP078649), C. conicum (SRP076966), and C. japonicum (SRP078647) was carried out using PaFNS I/F2H (GenBank accession no. KJ439220) as the query sequence. This revealed six cDNAs, designated Pa2ODD1, Mpa2ODD1, Me2ODD1, Cc2ODD2, Cj2ODD1, and Cj2ODD2, the sequences of which have been deposited with GenBank under the accession numbers listed in Supplemental Table S1. Their deduced polypeptide sequences were aligned with that of PaFNS I/F2H using DNAMAN v7 software (Lynnon Biosoft). Phylogenetic trees were constructed using the neighbor-joining method implemented in MEGA v5.1 software, employing 1,000 bootstrap replicates. Homology models for CjFNS I/F2H, CjFNS I1, PpFNS I/F3H, SmFNS I/F3H, GbF3H, and AtF3H were obtained from the SWISS-MODEL server (Schwede et al., 2003). The crystal structure of Arabidopsis ANS complexed with 2-oxoglutarate and naringenin (Protein Data Bank entry 2brt) served as a template (Welford et al., 2005). The models were visualized using PyMOL (www.pymol.org/citing).
RNA Extraction and cDNA Synthesis
RNA was extracted from individual C. conicum, C. japonicum, P. appendiculatum, M. paleacea, M. emarginata, P. patens, and S. moellendorffii plants using a cetyl-trimethyl-ammonium bromide-based method (Porebski et al., 1997), while the RNA of Arabidopsis and M. truncatula was obtained using the TRIzol reagent (Invitrogen). Single-stranded cDNA was synthesized from a template of total RNA using a RevertAidFirst Strand cDNA Synthesis kit (MBI-Fermentas) primed by oligo(dT)18, following the manufacturer’s instructions.
Cloning and Construction of Expression Plasmids
Full-length cDNAs of Pa2ODD1, Mpa2ODD1, Me2ODD1, Cc2ODD2, Cj2ODD1, and Cj2ODD2 were amplified from the corresponding cDNA template with the primers whose sequences are given in Supplemental Table S4. The open reading frames of Pp2ODD1, Pp2ODD2, Pp2ODD3, Sm2ODD1, Sm2ODD2, Sm2ODD3, Sm2ODD4, Sm2ODD5, Sm2ODD6, AtF3H, and MtF3H were amplified from their appropriate cDNA template using the primer pairs listed in Supplemental Table S4. The amplicons were ligated into pMD19-T and transformed into the Escherichia coli DH5α strain for sequencing. The coding sequences were amplified from the recombinant plasmids and ligated into the appropriate cloning site of pET32a (Novagen) to allow for the translation of N-terminally His-tagged proteins (relevant primer sequences are given in Supplemental Table S5). The sequences corresponding to Petroselinum crispum FNS I, Picea sitchensis F3H, Ginkgo biloba F3H, Pinus radiata F3H, Nymphaea thermarum F3H, Helianthus annuus F3H, Amborella trichopoda F3H, Oryza sativa F3H, Zea mays F3H, P. crispum F3H, and Aethusa cynapium F3H were all synthesized by Genwiz Bio-technology and inserted into pET32a. After validation by sequencing, all constructs, including the empty vector as a negative control, were transformed into E. coli BL21 (DE3).
Expression and Purification of Recombinant Proteins
Proteins were heterologously expressed in E. coli BL21 (DE3; Novagen). Overnight cultures (2 mL) generated from a single transgenic colony were inoculated into 200 mL of selective Luria-Bertani medium containing 100 mg L−1 ampicillin, and the culture was held at 37°C with vigorous shaking until the OD600 had reached 0.6. Protein synthesis was then induced by the addition of 0.6 mm isopropyl-thio-β-d-galactopyranoside, and the culture was maintained for 20 h at 16°C. Cells were harvested by centrifugation (2,500g, 10 min, 4°C) and homogenized by sonication on ice. His-tagged proteins present in the supernatant were purified using an Ni-NTA purification system kit (Invitrogen) according to the manufacturer’s protocol. The concentration of purified recombinant protein was determined using the Bradford reagent (Beyotime) with BSA employed as the standard. Purified proteins were subjected to SDS-PAGE and visualized by Coomassie Blue R250 staining.
Enzyme Assays
A 30-μg aliquot of recombinant protein was incubated for 1 h at 37°C in 50 mm Tris-HCl (pH 6.5), 1 mm ascorbate, 100 μm FeSO4, 160 μm 2-oxoglutaric acid, and 50 μm substrate (naringenin or dihydrokaempferol) in a 0.5-mL reaction; as a negative control, protein was similarly prepared from cells harboring an empty pET32a plasmid. The reactions were extracted twice in 0.5 mL of ethyl acetate and centrifuged at 2,500g for 10 min. The samples were evaporated under vacuum, and the residue was dissolved in 0.1 mL of methanol. The reaction products were characterized by LC-MS, carried out using an Agilent 1100 device (Agilent Technologies) equipped with a multi-wavelength diode array detector and an electron spray ionization mass spectrometer. The samples were separated through a reverse-phase C18 column, and the products were detected at 350 nm. The two solvents were aqueous 0.1% (v/v) glacial acetic acid (A) and 100% (v/v) methanol (B). The separation program was as follows: 0 to 20 min, 30% to 80% (v/v) B; 20 to 25 min, 100% (v/v) B; and 25 to 30 min, 30% (v/v) B, with a flow rate of 0.8 mL min−1 and an injection volume of 20 μL.
To optimize the reaction pH values, the activities of the recombinant proteins were assessed with the substrate naringenin or dihydrokaempferol at serial pH values (5–8.5). The apparent Km values for different naringenin or dihydrokaempferol were determined by incubating 2 to 5 μg of recombinant enzyme with serial concentrations of naringenin (2, 4, 8, 10, 20, 50, 75, 100, 150, 200, 250, and 300 μm) in a final volume of 100 μL. The reactions were stopped with the addition of an equal volume of acetonitrile after 10 min of incubation at 37°C. Each reaction product was quantified using a standard curve generated from known concentrations of the corresponding flavone or flavonol. Apparent Km and Vmax values were calculated from Lineweaver-Burk plots using the GraphPad Prism 5 software. This experiment was carried out three times independently, and there were three technical replications used in each analysis to obtain the accurate apparent Km and Vmax values.
Site-Directed Mutagenesis
The variant proteins AtF3H-P220M, AtF3H-P220F, AtF3H-P220Y, AtrF3H-P221M, AtrF3H-P221F, AtrF3H-P221Y, MtF3H-P221M, MtF3H-P221F, MtF3H-P221Y, PcF3H-P221M, PcF3H-P221F, PcF3H-P221Y, AcF3H-P221M, AcF3H-P221F, AcF3H-P221Y, GbF3H-P228M, GbF3H-P228F, GbF3H-P228Y, PrF3H-P227M, PrF3H-P227F, PrF3H-P227Y, PsF3H-P227M, PsF3H-P227F, PsF3H-P227Y, CjFNS I/F2H-Y234F, CjFNS I/F2H-Y234M, CjFNS I/F2H-Y234P, CjFNS I1-Y237F, CjFNS I1-Y237M, CjFNS I1-Y237P, PpFNS I/F3H-M220P, PpFNS I/F3H-M220Y, SmFNS I/F3H-F225P, and SmFNS I/F3H-F225Y were generated using the Stratagene QuickChange site-directed mutagenesis method. The necessary primers were designed using PrimerX software (www.bioinformatics.org/primerx). The variant sequences were all validated by sequencing. The reactions were treated with DpnI (Thermo Fisher Scientific) to cleave methylated template DNA, and the products were then transformed into E. coli DH5α. In vitro enzyme assays were based on purified recombinant protein, as above. Catalytic efficiencies were estimated using a calibration curve. The primers used to generate the individual mutants are given in Supplemental Table S6.
Arabidopsis Transformation
The cDNAs of CjFNS I/F2H, CjFNS I1, and SmFNS I/F3H were amplified with the primer pairs listed in Supplemental Table S7 from the plasmids pET32a-CjFNS I/F2H, pET32a-CjFNS I1, and pET32a-SmFNS I/F3H. The PCR products were cloned into the Gateway Entry vector pDONR207 (Invitrogen). Positive clones were then used in an LR Clonase reaction for recombination into the pGWB5 destination vector (Curtis and Grossniklaus, 2003). The resulting pGWB5-CjFNS I/F2H, pGWB5-CjFNS I1, and pGWB5-SmFNS I/F3H constructs were transformed into Agrobacterium tumefaciens strain GV3101 using the freeze/thaw method (Thomzik, 1995). Arabidopsis ecotype Columbia-0 was transformed using the floral dip method (Clough and Bent, 1998). T1 transformants were selected on solidified Murashige and Skoog medium containing 30 mg L−1 hygromycin, and the gene expression was confirmed by RT-PCR (relevant primer sequences are given in Supplemental Table S7). Transgene homozygous T2 plants were used for characterization purposes. Seedlings were cultured in 100 mL of Murashige and Skoog liquid medium with added 50 mm naringenin and were exposed to 22°C under a 12-h photoperiod. After 2 weeks, the seedlings were rinsed in distilled water, freeze-dried, and powdered. Aliquots of 10 mg of the powder were extracted in 1 mL of 70% (v/v) methanol and sonicated at room temperature for 1 h. Following centrifugation at 5,000g for 20 min, the extracts were hydrolyzed with an equal amount of 2 n HCl at 70°C for 40 min. The resulting products were extracted twice with an equal volume of ethyl acetate. After evaporation in vacuo, the residue was redissolved in 100 μL of methanol, and a 20-μL aliquot was injected into an HPLC device. Samples were separated using a 5-μm reverse-phase XDB-C18 column (Phenomenex Luna) with a flow rate of 0.8 mL min–1. The eluents were water plus 0.1% (v/v) formic acid (A) and acetonitrile (B); a linear gradient of 95% (v/v) A and 5% (v/v) B to 55% (v/v) A and 45% (v/v) B was supplied over 40 min.
Accession Numbers
Sequence data of 2-ODDs from liverworts, P. patens, and S. moellendorffii can be found in the GenBank/EMBL data libraries under the accession numbers shown in Supplemental Tables S1 and S2. Sequence data of F3Hs or FNS Is from the gymnosperms and angiosperms can be obtained from the GenBank/EMBL data libraries under the following accession numbers: G. biloba F3H (AAU93347), P. sitchensis F3H (ABK25766), P. radiata F3H (AGY80772), P. crispum F3H (AY230248), M. truncatula F3H (XM_003629275), Arabidopsis F3H (NM_114983), A. trichopoda F3H (XM_006851991), H. annuus F3H (XP_022015871), N. thermarum F3H (JAANDH010000393), Z. mays F3H (NM_001136803), O. sativa F3H (XM_015779149), and P. crispum FNS I (AY817680).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The biosynthesis pathway of plant flavonoids.
Supplemental Figure S2. SDS-PAGE analysis of recombinant proteins.
Supplemental Figure S3. Effect of pH on enzyme activities of CjFNS I1, CjFNS I/F2H, PcFNS I, PpFNS I/F3H, SmFNS I/F3H, GbF3H, PsF3H, and PrF3H using naringenin as substrate.
Supplemental Figure S4. Effect of pH on enzyme activities of AtF3H, AcF3H, AtrF3H, MtF3H, OsF3H, PcF3H, HaF3H, NtF3H, and ZmF3H using naringenin as substrate.
Supplemental Figure S5. Michaelis-Menten kinetic characterization of PcFNS I, CjFNS I1, CjFNS I/F2H, PpFNS I/F3H, SmFNS I/F3H, GbF3H, PsF3H, and PrF3H using naringenin as substrate.
Supplemental Figure S6. Michaelis-Menten kinetic characterization of AtrF3H, ZmF3H, OsF3H, NtF3H, MtF3H, HaF3H, AtF3H, AcF3H, and PcF3H using naringenin as substrate.
Supplemental Figure S7. Three-dimensional crystal modeling and sequence analysis of 2-ODDs from different plants.
Supplemental Figure S8. Effect of pH on enzyme activities of CjFNS I1 and CjFNS I/F2H using dihydrokaempferol as substrate.
Supplemental Figure S9. Michaelis-Menten kinetic characterization of CjFNS I1 and CjFNS I/F2H using dihydrokaempferol as substrate.
Supplemental Table S1. The characteristics of 2-ODDs from liverworts.
Supplemental Table S2. The characteristics of 2-ODDs from P. patens and S. moellendorffii.
Supplemental Table S3. The matrix with percentage of sequence identities for 2-ODD proteins from different plant species.
Supplemental Table S4. The primers used to obtain the full-length of 2-ODD genes.
Supplemental Table S5. The primers used to construct prokaryotic expression vectors.
Supplemental Table S6. The primers used for site-directed mutagenesis vector construction.
Supplemental Table S7. The primers used to construct the overexpression vectors and conduct RT-PCR analysis in Arabidopsis.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31770330 to A.-X.C. and 81630093 to H.-X.L) and by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, specifically the Physical Biosciences program of the Chemical Sciences, Geosciences and Biosciences Division (grant no. SC0012704 to C.-J.L.).
References
- Asakawa Y, Ludwiczuk A, Nagashima F(2013) Chemical Constituents of Bryophytes: Bio- and Chemical Diversity, Biological Activity, and Chemosystematics. Springer, Vienna: [DOI] [PubMed] [Google Scholar]
- Bredebach M, Matern U, Martens S(2011) Three 2-oxoglutarate-dependent dioxygenase activities of Equisetum arvense L. forming flavone and flavonol from (2S)-naringenin. Phytochemistry 72: 557–563 [DOI] [PubMed] [Google Scholar]
- Britsch L.(1990) Purification and characterization of flavone synthase I, a 2-oxoglutarate-dependent desaturase. Arch Biochem Biophys 282: 152–160 [DOI] [PubMed] [Google Scholar]
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ(2006) Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem 100: 644–669 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U(2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Ran F, Dong HL, Wen J, Li JN, Liang Z(2016) Genome-wide analysis, classification, evolution, and expression analysis of the cytochrome P450 93 family in land plants. PLoS ONE 11: e0165020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chu H, Chu IK, Lo C(2010a) CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol 154: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chu H, Wang M, Chu IK, Lo C(2010b) Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene (SbFNSII) in sorghum. J Exp Bot 61: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Emiliani J, Rodriguez EJ, Campos-Bermudez VA, Grotewold E, Casati P(2015) The identification of maize and Arabidopsis type I FLAVONE SYNTHASEs links flavones with hormones and biotic interactions. Plant Physiol 169: 1090–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow SC, Facchini PJ(2014) Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci 5: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Furtwängler K, Malterer G, Cantarello C, Schüler G, Ebel J, Mithöfer A(2010) Flavone synthase II (CYP93B16) from soybean (Glycine max L.). Phytochemistry 71: 508–514 [DOI] [PubMed] [Google Scholar]
- Gebhardt Y, Witte S, Forkmann G, Lukacin R, Matern U, Martens S(2005) Molecular evolution of flavonoid dioxygenases in the family Apiaceae. Phytochemistry 66: 1273–1284 [DOI] [PubMed] [Google Scholar]
- Gebhardt YH, Witte S, Steuber H, Matern U, Martens S(2007) Evolution of flavone synthase I from parsley flavanone 3β-hydroxylase by site-directed mutagenesis. Plant Physiol 144: 1442–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han XJ, Wu YF, Gao S, Yu HN, Xu RX, Lou HX, Cheng AX(2014) Functional characterization of a Plagiochasma appendiculatum flavone synthase I showing flavanone 2-hydroxylase activity. FEBS Lett 588: 2307–2314 [DOI] [PubMed] [Google Scholar]
- Hungria M, Joseph CM, Phillips DA(1991) Anthocyanidins and flavonols, major nod gene inducers from seeds of a black-seeded common bean (Phaseolus vulgaris L.). Plant Physiol 97: 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Ono E, Mizutani M(2014) Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J 78: 328–343 [DOI] [PubMed] [Google Scholar]
- Koes RE, Quattrocchio F, Mol JNM(1994) The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 16: 123–132 [Google Scholar]
- Lee YJ, Kim JH, Kim BG, Lim Y, Ahn JH(2008) Characterization of flavone synthase I from rice. BMB Rep 41: 68–71 [DOI] [PubMed] [Google Scholar]
- Markham GD.(1984) Structure of the divalent metal ion activator binding site of S-adenosylmethionine synthetase studied by vanadyl(IV) electron paramagnetic resonance. Biochemistry 23: 470–478 [DOI] [PubMed] [Google Scholar]
- Markham KR.(1988) Distribution of flavonoids in the lower plants and its evolutionary significance In Harborne JB, ed, The Flavonoids. Springer, Boston, pp 427–468 [Google Scholar]
- Martens S, Forkmann G, Britsch L, Wellmann F, Matern U, Lukačin R(2003) Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley. FEBS Lett 544: 93–98 [DOI] [PubMed] [Google Scholar]
- Martens S, Forkmann G, Matern U, Lukacin R(2001) Cloning of parsley flavone synthase I. Phytochemistry 58: 43–46 [DOI] [PubMed] [Google Scholar]
- Martens S, Preuss A, Matern U(2010) Multifunctional flavonoid dioxygenases: Flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71: 1040–1049 [DOI] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK(2018) Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc Natl Acad Sci USA 115: E11188–E11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H, Lewis NE, Lerman JA, Lee DH, Chang RL, Kim D, Palsson BO(2012) Network context and selection in the evolution to enzyme specificity. Science 337: 1101–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS(2007) Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR(1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15: 8–15 [Google Scholar]
- Prescott AG, Stamford NP, Wheeler G, Firmin JL(2002) In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 60: 589–593 [DOI] [PubMed] [Google Scholar]
- Righini S, Rodriguez EJ, Berosich C, Grotewold E, Casati P, Falcone Ferreyra ML(2019) Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant Cell Environ 42: 495–508 [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Zhang Z(1999) Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr Opin Struct Biol 9: 722–731 [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC(2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Inoue YT, Sakuta M(2005) Anthocyanidin synthase in non-anthocyanin-producing Caryophyllales species. Plant J 44: 950–959 [DOI] [PubMed] [Google Scholar]
- Solovchenko A, Schmitz-Eiberger M(2003) Significance of skin flavonoids for UV-B-protection in apple fruits. J Exp Bot 54: 1977–1984 [DOI] [PubMed] [Google Scholar]
- Thomzik JE.(1995) Agrobacterium-mediated transformation of protoplasts from oilseed rape (Brassica napus L.). Methods Mol Biol 44: 71–78 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Sobey WJ, Aplin RT, Hassan A, Schofield CJ, Firmin JL, Prescott AG(2000) Are anthocyanidins the immediate products of anthocyanidin synthase? Chem Commun (Camb) 32: 2473–2474 [Google Scholar]
- Wang L, Wang LN, Zhao Y, Lou HX, Cheng AX(2013) Secondary metabolites from Marchantia paleacea calluses and their allelopathic effects on Arabidopsis seed growth. Nat Prod Res 27: 274–276 [DOI] [PubMed] [Google Scholar]
- Welford RW, Clifton IJ, Turnbull JJ, Wilson SC, Schofield CJ(2005) Structural and mechanistic studies on anthocyanidin synthase catalysed oxidation of flavanone substrates: The effect of C-2 stereochemistry on product selectivity and mechanism. Org Biomol Chem 3: 3117–3126 [DOI] [PubMed] [Google Scholar]
- Weng JK, Chapple C(2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B.(2001) Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Wang G, Hill L, Weng JK, Chen XY, Xue H, Martin C(2016) A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci Adv 2: e1501780. [DOI] [PMC free article] [PubMed] [Google Scholar]