Temperature-dependent shoot apical meristem development is conferred by temperature-gated physical interactions between ribonucleotide reductase subunits.
Abstract
Temperature is a major factor regulating plant growth. To reproduce at extreme temperatures, plants must develop normal reproductive organs when exposed to temperature changes. However, little is known about the underlying molecular mechanisms. Here, we identified the maize (Zea mays) mutant thermosensitive vanishing tassel1-R (tvt1-R), which lacks tassels at high (restrictive) temperatures due to shoot apical meristem (SAM) arrest, but forms normal tassels at moderate (permissive) temperatures. The critical stage for phenotypic conversion in tvt1-R mutants is V2 to V6 (Vn, where “n” is the number of leaves with collars visible). Positional cloning and allelism and complementation tests revealed that a G-to-A mutation causing a Arg277-to-His277 substitution in ZmRNRL1, a ribonucleotide reductase (RNR) large subunit (RNRL), confers the tvt1-R mutant phenotype. RNR regulates the rate of deoxyribonucleoside triphosphate (dNTP) production for DNA replication and damage repair. By expression, yeast two-hybrid, RNA sequencing, and flow cytometric analyses, we found that ZmRNRL1-tvt1-R failed to interact with all three RNR small subunits at 34°C due to the Arg277-to-His277 substitution, which could impede RNR holoenzyme (α2β2) formation, thereby decreasing the dNTP supply for DNA replication. Decreased dNTP supply may be especially severe for the SAM that requires a continuous, sufficient dNTP supply for rapid division, as demonstrated by the SAM arrest and tassel absence in tvt1-R mutants at restrictive temperatures. Our study reveals a novel mechanism of temperature-gated tassel formation in maize and provides insight into the role of RNRL in SAM maintenance.
Temperature is one of the most important environmental cues regulating plant growth and development (Heggie and Halliday, 2005; Franklin, 2009; Patel and Franklin, 2009; Li et al., 2016). Plants respond to temperature in two ways. First, temperature signals act as a stimulus that determines the timing of developmental transitions. For example, Arabidopsis (Arabidopsis thaliana) seed germination requires a period of cold for dormancy break (Heggie and Halliday, 2005; Penfield et al., 2005; Franklin, 2009). In addition, flowering is accelerated in many temperate species through prolonged exposure to cold, a process termed vernalization (Heggie and Halliday, 2005; Franklin, 2009; Li et al., 2016). Second, temperature sensing helps plants adjust their body plan to protect themselves from adverse temperatures. A typical example is the temperature-mediated regulation of Arabidopsis plant architecture. Arabidopsis plants grown at adverse temperatures (16°C and 28°C) exhibit substantially reduced leaf area and biomass compared to plants grown at favorable temperatures (22°C). Additionally, Arabidopsis plants display a heat-retaining, compact plant architecture at lower temperatures (16°C) and a heat-dissipating, loose plant architecture at higher temperatures (28°C; Heggie and Halliday, 2005; Franklin, 2009; Patel and Franklin, 2009).
To reproduce at extreme temperatures, plants depend on the normal formation and development of reproductive organs. Several genes required for pollen development at restrictive temperatures have been identified. thermosensitive genic male sterile5 (tms5) controls thermosensitive genic male sterility (TGMS) in rice (Oryza sativa). This gene encodes RNase ZS1 processing Ubiquitin fusion ribosomal protein L40 mRNA, whose excess accumulation leads to male sterility at restrictive temperatures (Zhou et al., 2014). photo- or thermo-sensitive genic male sterility locus on chromosome12 encodes a long noncoding RNA (Ding et al., 2012a; Zhou et al., 2012). A single nucleotide polymorphism (SNP) within this small RNA induces TGMS in the rice line ‘Peiai 64S’ through a complex mechanism, including transcriptional regulation mediated by small RNAs and DNA methylation (Ding et al., 2012b). UDP-Glc pyrophosphorylase1 encodes a UDP-Glc pyrophosphorylase that is essential for callose deposition in pollen. The co-suppression of this gene and its paralog UDP-Glc pyrophosphorylase2 using RNA interference results in a type of TGMS in which male sterility occurs at high temperatures but normal fertility occurs at low temperatures (Chen et al., 2007). Tms10, encoding a receptor-like kinase in rice, is essential for pollen development at high temperatures; its mutation confers TGMS in both the japonica and indica backgrounds (Yu et al., 2017). However, to date, no gene essential for floral organ formation at restrictive temperatures has been characterized.
The tassel, an important floral organ in maize (Zea mays), is formed from the inflorescence meristem (IM) and axillary meristem (AM), which are converted from the shoot apical meristem (SAM) and initiated from the IM, respectively (Bommert et al., 2005; Bortiri and Hake, 2007; Thompson and Hake, 2009; Tanaka et al., 2013; Zhang and Yuan, 2014). A deficiency in SAM maintenance or AM initiation leads to plants with no tassels or exhibiting similar phenotypes. Several genes controlling the early development of male inflorescences in maize have been identified, most of which are auxin-related genes required for AM initiation, including genes involved in auxin biosynthesis, such as Vanishing tassel2 (Vt2) and Sparse inflorescence1 (Gallavotti et al., 2008; Phillips et al., 2011); genes involved in auxin transport, such as Barren inflorescence2 (Bif2; McSteen et al., 2007); genes involved in auxin signaling, such as BIF1 and BIF4 (Galli et al., 2015); genes that function downstream of auxin, such as Barren stalk1 and Barren stalk2 (Gallavotti et al., 2004; Skirpan et al., 2009; Galli et al., 2015; Yao et al., 2019); and genes that interact with auxin, such as Needle1 (Ndl1; Liu et al., 2019). Little research has focused on the interplay of metabolites and early tassel development. The identification of Thiamine biosynthesis2, Tassel-less1, and Rotten ear demonstrated that metabolites such as thiamine and boron play important roles in regulating the early development of maize tassels (Woodward et al., 2010; Chatterjee et al., 2014; Durbak et al., 2014; Leonard et al., 2014). Thus, how metabolite sensing regulates early tassel development is worth exploring in more detail. Moreover, several mutants of the above-mentioned genes, such as vt2 and ndl1 (Phillips et al., 2011; Liu et al., 2019), are temperature-sensitive, but the underlying molecular mechanism is unclear.
Ribonucleotide reductase (RNR) is an enzyme that catalyzes the de novo synthesis of deoxyribonucleotide diphosphates from their corresponding ribonucleotide diphosphates. This process is the rate-limiting step in the biosynthesis of all four deoxyribonucleoside triphosphates (dNTPs) for DNA replication and damage repair in all living organisms (Reichard, 1988; Elledge et al., 1992; Mathews, 2006; Nordlund and Reichard, 2006). In eukaryotes, an RNR complex (α2β2) consists of two RNR large subunits (RNRLs) and two RNR small subunits (RNRSs; Jordan and Reichard, 1998). Several rnrl and/or rnrs mutants of flowering plants have been identified in Arabidopsis (rnrl: crinkled leaves8 [or defective in organelle DNA degradation2]; rnrs: tso2, rnr2a, and rnr2b; Wang and Liu, 2006; Garton et al., 2007; Tang et al., 2012), rice (rnrl: virescent3; rnrs: stripe1; Yoo et al., 2009; Chen et al., 2015; Qin et al., 2017), and Setaria italica (rnrl: sistl1; Tang et al., 2019). These rnr mutants are mainly characterized by defects in chloroplast biogenesis, and whether rnr mutations affect other biological functions in flowering plants deserves further exploration.
In this study, we identified a maize mutant thermosensitive vanishing tassel1-R (tvt1-R), which lacks tassels at high (restrictive) temperatures due to SAM arrest but has normal tassels at moderate (permissive) temperatures. Map-based cloning, an allelism test, and complementation tests revealed that a G-to-A mutation causing a Arg277-to-His277 substitution in the RNRL protein ZmRNRL1 underlies the tvt1-R mutant phenotypes. Yeast two-hybrid analysis at various temperatures indicated that ZmRNRL1-tvt1-R with the Arg277-to-His277 substitution failed to interact with all three RNRSs specifically at higher temperatures but not at low temperatures, thus leading to the no-tassel phenotype of tvt1-R mutants at high temperatures. Our results uncover a novel mechanism controlling tassel formation at restrictive temperatures in maize and provide a further perspective on the role of nucleotide homeostasis in SAM maintenance.
RESULTS
tvt1-R Is a Temperature-Sensitive Mutant Mainly Affected in Tassel Formation
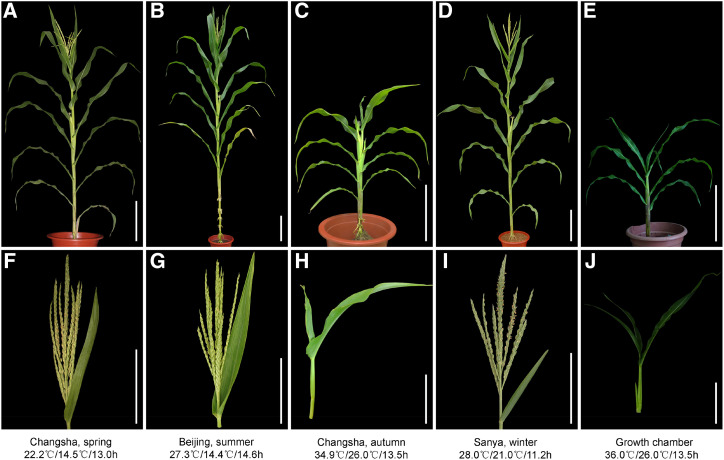
We obtained the tvt1-R mutant, which exhibited a lack of tassels, in Changsha (middle southern China; 28°N, 113°E) in the autumn (Fig. 1, C and H), from a mutant library generated by heavy-ion radiation. However, the tvt1-R mutant produced normal tassels when planted in Changsha in the spring (Fig. 1, A and F), in Beijing (northern China; 39°N, 116°E) in the summer (Fig. 1, B and G), and in Sanya (southern China; 18°N, 109°E) in the winter (Fig. 1, D and I). These phenotypes were observed consistently for 5 y (2014 to 2018) at the above-mentioned locations, indicating that tvt1-R is an environment-sensitive, no-tassel mutant.
Figure 1.
Phenotypes of tvt1-R mutants at the tasseling stage grown under different conditions. A to E, Plant morphology of the tvt1-R mutant grown in the indicated conditions. Scale bars = 30 cm. F to J, Shoot apex phenotypes of the plants shown in A to E. Scale bars = 15 cm. The plants were digitally extracted for comparison.
To determine which environmental factor regulates the no-tassel phenotype in tvt1-R mutants, we examined climatological data 10 to 30 d after sowing (roughly from after emergence to before SAM conversion into IM) in the three locations. The daily maximum average temperature, daily minimum average temperature, and average daylength were, respectively, 22.2°C, 14.5°C, and 13 h in Changsha in the spring; 27.3°C, 14.4°C, and 14.6 h in Beijing in the summer; 34.9°C, 26°C, and 13.5 h in Changsha in the autumn; and 28°C, 21°C, and 11.2 h in Sanya in the winter. These climatological data and the corresponding phenotypes of tvt1-R mutants indicate that this mutant is thermosensitive rather than photoperiod-sensitive, displaying the absence of tassels at high (restrictive) temperatures (above 35°C [daily maximum average temperature]) and the presence of tassels at moderate (permissive) temperatures (below 28°C [daily maximum average temperature]).
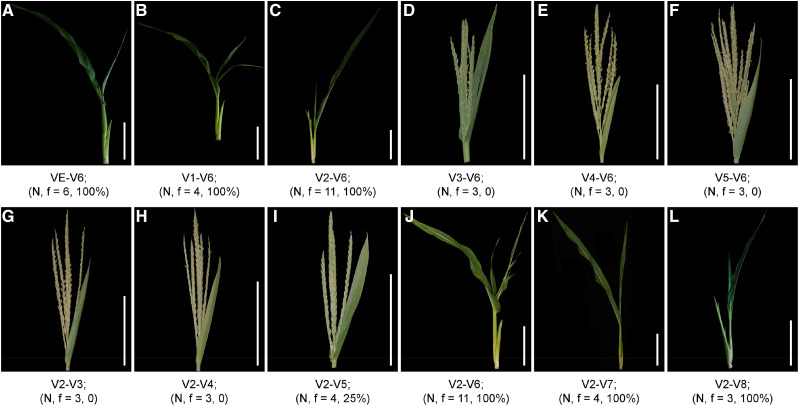
To eliminate the effects of natural temperature changes on the tvt1-R mutant phenotype, we simulated the light and temperature conditions of Changsha in the autumn using a growth chamber with an alternative light/dark cycle (13.5-h light at 36°C/10.5-h dark at 26°C [36°C/26°C/13.5 h]). When the tvt1-R mutant grew in this growth chamber, it had almost 100% penetrance of the no-tassel phenotype (Fig. 1, E and J). Hence, we used this condition to artificially create the restrictive temperatures required for the tvt1-R mutant phenotype. We conducted temperature-sensitivity–period trials in the growth chamber to investigate the critical stage (including the starting point and the end point) for the phenotypic conversion of the tvt1-R mutant. To detect the starting point, we first planted tvt1-R mutant seedlings outdoors (Beijing in the summer), then moved a group of seedlings into the growth chamber for treatment at each leaf stage from VE (emergence) to V5 (Vn, where “n” is number of leaves with collars visible; Ritchie et al., 1992), and finally moved all seedlings outdoors at V6. Seedlings that were moved into the growth chamber before or at V2 lacked tassels (Fig. 2, A–C), whereas seedlings that were moved into the growth chamber after V2 produced tassels (Fig. 2, D–F), indicating that the starting point of the critical stage is V2. Similarly, to detect the end point, we first planted tvt1-R mutant seedlings outdoors, and then moved all seedlings to the growth chamber for treatment at V2 (the starting point), and finally moved a group of seedlings outdoors at each leaf stage until V8. Seedlings that were moved outdoors during or after V6 lacked tassels (Fig. 2, J–L), whereas almost all seedlings that were moved outdoors before V6 produced tassels (Fig. 2, G–I), indicating that the end point of the critical stage is V6. Together, our results indicate that the no-tassel phenotype of the tvt1-R mutant requires continuous high temperatures from V2 to V6.
Figure 2.
Trials to detect the temperature-sensitive period. A to L, Shoot apex phenotypes of tvt1-R mutants grown in the growth chamber (36°C/26°C/13.5 h) at different growth stages. VE, V1, V2, etc. represent vegetative stages; VE represents emergence (the first vegetative stage); Vn represents where “n” is the number of leaves with collars visible. “N” represents the total number of treated plants and “f” represents the proportion of plants with no tassels. Scale bars = 15 cm. The plants were digitally extracted for comparison.
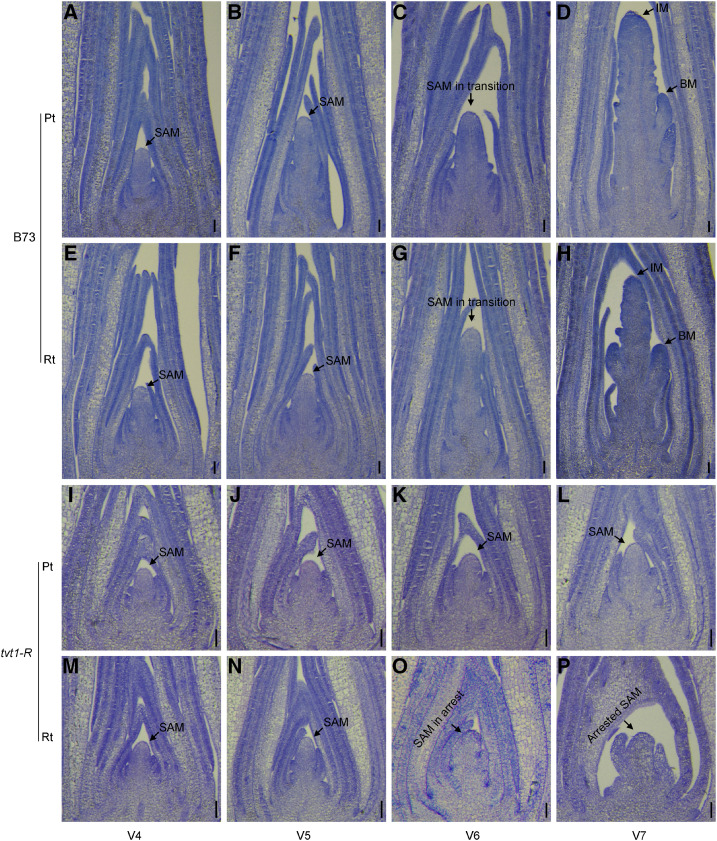
Phenotypic characterization showed that the tvt1-R mutant completely lacked tassels at restrictive temperatures, suggesting that its SAM might have been arrested. To further investigate the phenotype of tvt1-R mutant SAMs, we performed paraffin sectioning to create longitudinal sections of the shoot apices of maize inbred line B73 and the tvt1-R mutant seedlings from V4 to V7 grown at both permissive and restrictive temperatures. B73 SAMs were maintained, converted into IMs, and initiated branch meristems at both permissive (Fig. 3, A–D) and restrictive temperatures (Fig. 3, E–H). However, compared to the normal domed structure of tvt1-R mutant SAMs at permissive temperatures (Fig. 3, I–L), tvt1-R mutant SAMs at restrictive temperatures could not be maintained and began to exhibit an abnormal morphology at V6 (Fig. 3, M–P), which is in accordance with the end point of the critical stage for phenotypic conversion of this mutant. These results suggest that Tvt1 functions in SAM maintenance at restrictive temperatures.
Figure 3.
SAM phenotypes from V4 to V7 at permissive and restrictive temperatures. A to H, B73 SAMs were maintained, converted into IMs, and initiated branch meristems (BMs) at both permissive (A–D) and restrictive temperatures (E–H). I to L, The tvt1-R mutant SAMs were maintained and exhibited a normal domed structure at permissive temperatures. M to P, At restrictive temperatures, the tvt1-R mutant SAMs could not be maintained and began to exhibit an abnormal morphology at V6. Pt, permissive temperatures; Rt, restrictive temperatures. Scale bars = 100 μm.
To investigate other mutant phenotypes, we introgressed the tvt1-R mutant allele into the B73 genetic background for more than four generations. Unexpectedly, we observed an extremely low proportion of no-tassel tvt1-R-B73/tvt1-R-B73 plants at restrictive temperatures (6.9%, 2 of 29; Supplemental Fig. S1, A–J). However, in both the presence and absence of tassels, all tvt1-R-B73/tvt1-R-B73 plants at restrictive temperatures exhibited the striped-leaf phenotype (Supplemental Fig. S1, K–O). In addition, compared with Tvt1-B73/Tvt1-B73 plants, tvt1-R-B73/tvt1-R-B73 plants showed significantly reduced plant height, tassel branch number, and leaf number (Supplemental Fig. S1, P–R). These results indicate that the Tvt1 gene has pleiotropic effects on plant development.
Tvt1 Encodes a Large Subunit of RNR
To evaluate the number of loci controlling the thermosensitive missing tassel trait of tvt1-R, we separately crossed the tvt1-R mutant with inbred lines B73 and Mo17 to generate two F2 populations. Genetic analysis of the two F2 populations planted in Changsha in the autumn revealed that the tvt1-R × B73 F2 population segregated into 449 plants with tassels and 59 no-tassel plants (χ2 = 48.55), and the tvt1-R × Mo17 F2 population segregated into 1,250 plants with tassels and 42 no-tassel plants (χ2 = 325.95); both deviated from the 3:1 ratio expected for a single locus (χ20.05,1 = 3.84). Additionally, we carried out genetic analysis of the tvt1-R × B73 F2 population in the growth chamber. The population segregated into 90 plants with tassels and 10 no-tassel plants, which also deviated from the 3:1 ratio (χ2 = 12 > χ20.05,1 = 3.84). Together, these results suggest that the thermosensitive vanishing tassel trait of the tvt1-R mutant might be determined by a single major recessive locus and affected by other minor gene(s).
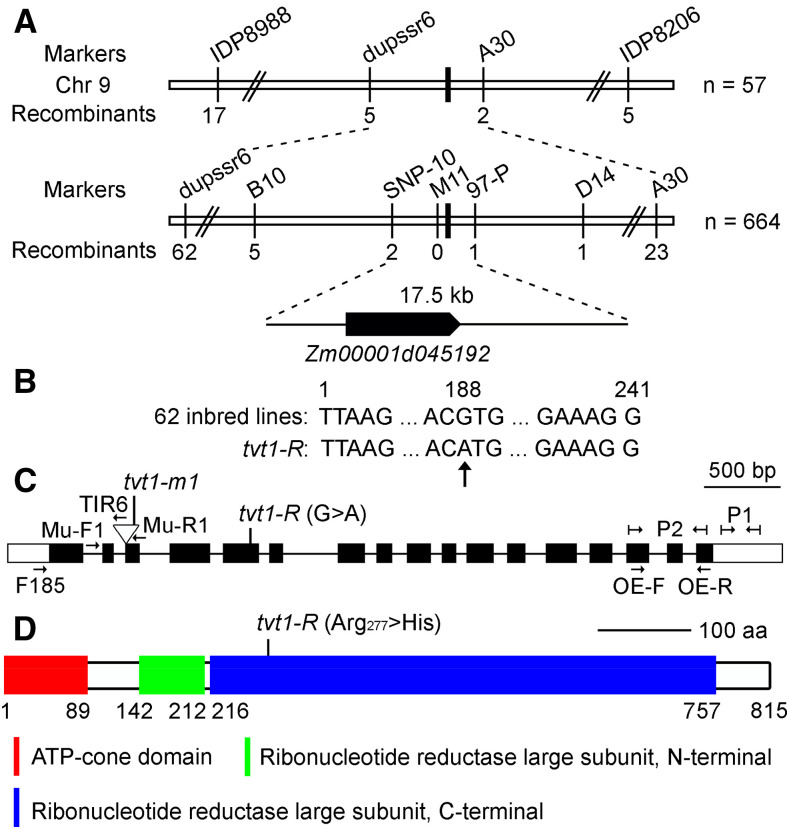
To map the Tvt1 gene, we performed linkage analysis of 57 no-tassel plants from the tvt1-R × B73 F2 population using 20 polymorphic markers distributed on 10 chromosomes. Two markers on chromosome 9, IDP8988 (χ2 = 58.45 > χ20.05,2 = 5.99) and IDP7884 (χ2 = 9.81 > χ20.05,2 = 5.99), were linked to the no-tassel phenotype. By developing additional markers around IDP8988, we initially mapped the Tvt1 gene between markers dupssr6 and A30 (Fig. 4A). Fine mapping was performed using 664 no-tassel plants, and the Tvt1 gene was ultimately narrowed down to a 17.5-kb interval between markers SNP-10 and 97-P, with two and one recombinants, respectively (Fig. 4A). There is only one protein-coding gene in this region, Zm00001d045192, according to the B73 reference genome version 4 (B73_RefGen_v4; Jiao et al., 2017). Using Sanger sequencing, we detected a G-to-A transition within the coding region of Zm00001d045192, which leads to an amino acid change from Arg277 (Arg; CGU) to His277 (His; CAU). To determine whether the G-to-A transition was present only in tvt1-R, we sequenced the variant site in 62 maize inbred lines and examined this site in maize haplotype version 3 (HapMap3; http://cbsusrv04.tc.cornell.edu/users/panzea/download.aspx?filegroupid=34) containing whole-genome sequencing data for 1,218 maize lines (Bukowski et al., 2018). The nucleotide “G” was absolutely conserved in the 62 maize inbred lines (Fig. 4B), and the G-to-A transition was not observed in HapMap3, suggesting that the transition is a mutation rather than an extant polymorphism. These results strongly suggest that Zm00001d045192 is the candidate gene.
Figure 4.
Mapping of Tvt1. A, Schematic diagram of the map-based cloning of the Tvt1 gene. The gene was localized to a 17.5-kb region on chromosome 9, which contains only one gene, Zm00001d045192. B, The G-to-A transition in exon 5 of Zm00001d045192 detected using Sanger sequencing. The nucleotide at the variation site is absolutely conserved in 62 maize inbred lines. The numbers showing the nucleotide position are equivalent to exon 5 of Zm00001d045192. C, Diagram showing the Zm00001d045192 gene structure and the position of the mutant alleles. Boxes represent exons (black boxes and white boxes indicate translated regions and untranslated regions, respectively), and lines represent introns. Primers in the diagram were designed for the following studies. Mu-F1, Mu-R1, and TIR6 were used to detect the Mu insertion type. P1 was designed for expression analysis of Tvt1, and P2 was designed to measure the relative expression level of Tvt1 in homozygous overexpression lines. A pair of specific primers (F185 + Mu-R1) was used to amplify the Mu transcript. A pair of intron-spanning primers (OE-F and OE-R) was designed to detect the overexpression of Tvt1 at the DNA level. D, Schematic diagram of ZmRNRL1 protein structure. aa, amino acids. The G-to-A point mutation in tvt1-R leads to a change in the 277th amino acid from Arg to His.
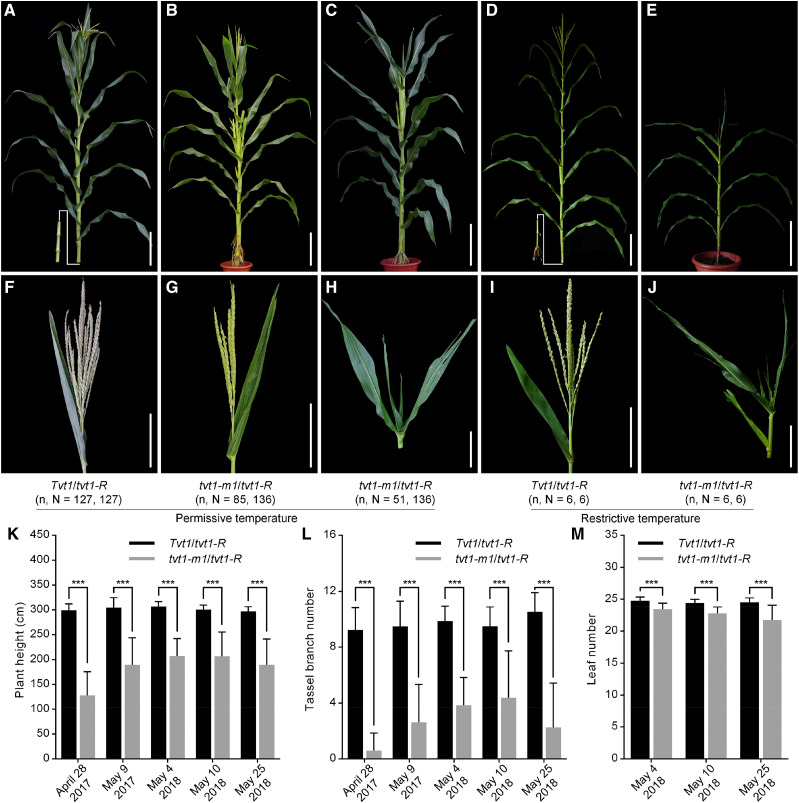
To confirm this candidate gene, we obtained the Mu insertion line tvt1-m1, which contains a 1,387-bp Mu1.4 insertion in the third exon of Zm00001d045192 (Fig. 4C; Supplemental Fig. S2, A, B, and E). The transcription of Zm00001d045192 was not completely blocked in the tvt1-m1 mutant (Supplemental Fig. S2C), likely due to the relatively short Mu1.4 insertion sequence. Combined PCR amplification and Sanger sequencing revealed two forms of Mu1.4 transcript simultaneously: the full 1,387-bp sequence resulting from complete transcription, and a 1,237-bp sequence resulting from selective transcription (Supplemental Fig. S2, D and E). Given that both forms of transcription cause the downstream frame-shift, we suggest that tvt1-m1 is a null allele of Zm00001d045192. tvt1-m1 mutants displayed serious developmental defects at both permissive and restrictive temperatures, with no tassels or striped leaves (Supplemental Fig. S2, F–I). These two mutant phenotypes resemble the phenotypes of tvt1-R mutants (Fig. 1, H and J) and tvt1-R-B73 homozygous plants at restrictive temperatures (Supplemental Fig. S1, N and O), respectively, implying that tvt1-m1 is allelic to tvt1-R. This was further confirmed by a subsequent allelism test in which heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) were crossed with tvt1-R mutants (Fig. 5, A–J). Approximately 6% to 72% tvt1-m1/tvt1-R plants displayed the no-tassel phenotype at permissive temperatures (Fig. 5, B and G; Table 1), and the proportion of no-tassel plants differed significantly among tvt1-m1/tvt1-R plants with different sowing dates (Table 1), further suggesting that the tvt1-R allele is environment-sensitive. Additionally, compared to Tvt1/tvt1-R plants, plant height, tassel branch number, and leaf number were significantly reduced in tvt1-m1/tvt1-R plants (Fig. 5, K–M). These phenotypes are consistent with the phenotypes of tvt1-R-B73/tvt1-R-B73 plants (Supplemental Fig. S1, P–R), highlighting the pleiotropic effects of Tvt1.
Figure 5.
Allelism test. A to E, Plant morphology of F1 progeny generated by crossing heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) with tvt1-R mutants. Scale bars = 30 cm. F to J, Shoot apex phenotypes of the plants shown in A to E. Scale bars = 15 cm. n represents the number of plants with the corresponding phenotype, and N represents the number of plants with the corresponding genotype. The plants were digitally extracted for comparison. K to M, Comparisons of plant height (K), tassel branch number (L), and leaf number (M) between Tvt1/tvt1-R and tvt1-m1/tvt1-R plants. April 28, 2017, May 9, 2017, May 4, 2018, May 10, 2018, and May 25, 2018 represent the five sowing dates in Beijing in the summer. Values are means ± sd. Two-tailed Student’s t test was used to detect significant differences (***P < 0.001).
Table 1. Statistical analysis of the proportion of plants without tassels in F1 progeny from an allelism test at five different sowing dates in Beijing during summer.
Statistical analysis of the percentage data between two samples was performed by χ2 test. The same letters indicate no significant difference (asymptotic significance [2-sided] > 0.05), and different letters indicate significant differences (asymptotic significance [2-sided] < 0.05).
| Sowing Date | Genotype | Total No. of Plants | No. of Plants with Tassels | No. of Plants without Tassels | Proportion of Plants without Tassels | α = 0.05 |
|---|---|---|---|---|---|---|
| April 28, 2017 | Tvt1/tvt1-R | 21 | 21 | 0 | ||
| tvt1-m1/tvt1-R | 22 | 6 | 16 | 72.73% | a | |
| May 9, 2017 | Tvt1/tvt1-R | 25 | 25 | 0 | ||
| tvt1-m1/tvt1-R | 21 | 13 | 8 | 38.10% | b | |
| May 4, 2018 | Tvt1/tvt1-R | 20 | 20 | 0 | ||
| tvt1-m1/tvt1-R | 32 | 30 | 2 | 6.25% | c | |
| May 10, 2018 | Tvt1/tvt1-R | 27 | 27 | 0 | ||
| tvt1-m1/tvt1-R | 29 | 22 | 7 | 24.14% | bc | |
| May 25, 2018 | Tvt1/tvt1-R | 34 | 34 | 0 | ||
| tvt1-m1/tvt1-R | 32 | 14 | 18 | 56.25% | ab |
We also developed a complementation construct with a 7,827-bp B73 genomic fragment containing the intact coding region and the 2.7-kb promoter region (Supplemental Fig. S3, A and B), and an overexpression construct with the coding sequence of Zm00001d045192 from B73 driven by the ubiquitin promoter (Supplemental Fig. S3, C and D). Using Agrobacterium tumefaciens-mediated transformation, two (GC-1 to GC-2) and five (OE-1 to OE-5) independent positive transgenic events for the complementation and overexpression construct, respectively, were obtained in the background of LH244. Each of the seven transgenic lines was crossed with tvt1-R mutants and each of the five overexpression lines was crossed with heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) and then selfed to generate the F2 progenies. All seven transgenic lines were capable of complementing the no-tassel phenotype of tvt1-R homozygous plants at restrictive temperatures (Fig. 6, A–H), and all five overexpression lines were capable of rescuing tvt1-m1 homozygous mutant phenotypes at permissive temperatures (Fig. 6, I–O), further supporting the notion that Tvt1 corresponds to Zm00001d045192. Two issues are worth noting. First, only four of the 24 tvt1-R/tvt1-R plants in the F2 population lacked tassels (Fig. 6A), which is consistent with the results of genetic analysis, suggesting that other loci likely control the no-tassel phenotype together with Tvt1. Second, 16 tvt1-m1/tvt1-m1;OE plants in the F2 progenies, including four tvt1-m1/tvt1-m1;OE-1 plants, one tvt1-m1/tvt1-m1;OE-2 plant, four tvt1-m1/tvt1-m1;OE-3 plants, and seven tvt1-m1/tvt1-m1;OE-4 plants displayed yellow-striped leaves that appeared in some homozygous overexpression lines (Supplemental Fig. S3, E and F). The proportion of this phenotype in homozygous overexpression lines OE-1, OE-2, OE-3, OE-4, and OE-5 was 94.4% (34 of 36), 15.4% (6 of 39), 22.5% (9 of 40), 0% (0 of 41), and 0% (0 of 47), respectively. The expression levels of Tvt1 in the five homozygous overexpression lines were measured by reverse transcription quantitative PCR (RT-qPCR) and were found to be highly correlated with the yellow-striped leaf phenotype (Supplemental Fig. S3G): yellow-striped leaves appeared in homozygous overexpression lines with relatively high-level Tvt1 expression and their corresponding F2 progenies (such as OE-1), but not in homozygous overexpression lines with relatively low expression levels of Tvt1 and their corresponding F2 offspring (such as OE-5). These results suggest that the yellow-striped leaf phenotype might be due to the excessive expression of Tvt1.
Figure 6.
Complementation tests. A to H, Shoot apex phenotypes of F2 progenies derived from crosses between the seven transgenic lines and the tvt1-R mutant at restrictive temperatures. I to O, Shoot apex or leaf phenotypes of F2 offsprings generated from crosses between the five overexpression lines and heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) at permissive temperatures. J1, Magnification of the white box in J. Scale bars = 15 cm. n represents the number of plants with the corresponding phenotype, and N represents the number of plants with the corresponding genotype. The plants were digitally extracted for comparison.
Tvt1 encodes a protein of 815 amino acid residues annotated as an RNRL (referred to as ZmRNRL1 hereafter). Phylogenetic analysis of ZmRNRL1 revealed that its homologs are present in all living organisms, which corresponds with its molecular function, and that another paralog (Zm00001d036322; designated as ZmRNRL2) is present in the maize genome (Supplemental Fig. S4). ZmRNRL1 shares 97%, 93%, 89%, 86%, 67%, and 56% sequence identity with ZmRNRL2, OsRNRL1, OsRNRL2 (Yoo et al., 2009), AtRNR1 (Garton et al., 2007; Tang et al., 2012), human (Homo sapiens) HsRNR1 (Pavloff et al., 1992), and yeast (Saccharomyces cerevisiae) ScRNR1 (Elledge and Davis, 1990), respectively (Supplemental Fig. S5). The high level of amino acid sequence similarity of RNRL-related proteins implies that these proteins share similar molecular functions in different species, as evidenced by a previous study wherein OsRNRL1 rescued the lethal phenotype of the yeast rnr1 rnr3 double mutant at 30°C (Yoo et al., 2009).
The Expression Patterns of ZmRNRL1 and ZmRNRL2
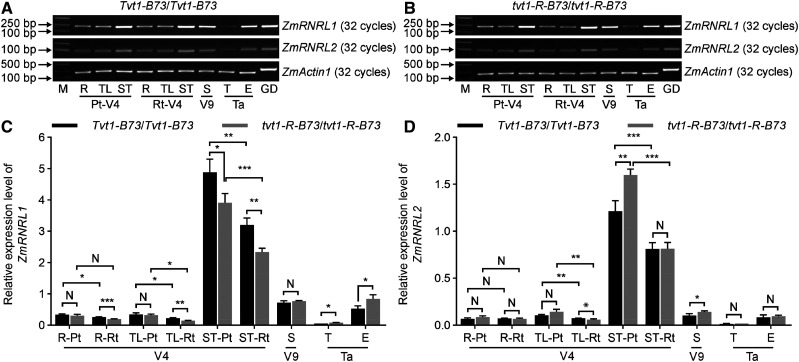
To decipher the tissue specificity and timing of ZmRNRL1 and ZmRNRL2 expression, we performed semiquantitative PCR (sq-PCR) and RT-qPCR analyses of complementary DNA (cDNA) derived from various tissues, including roots (V4), shoot tips (V4), topmost leaves (V4), stems wrapped by the ninth leaf (V9), tassels (VT, the last branch of the tassel completely visible), and ears (VT) of wild-type (Tvt1-B73/Tvt1-B73) plants. Based on the results of sq-PCR and RT-qPCR (Fig. 7), we obtained the following insights. First, ZmRNRL1 and ZmRNRL2 are expressed ubiquitously; second, ZmRNRL2 is expressed at a lower level than ZmRNRL1; and third, ZmRNRL1 and ZmRNRL2 are expressed at higher levels in developing tissues compared to mature tissues. Consistent results were also obtained when we examined publicly available RNA-sequencing (RNA-seq) data for 158 maize tissues in the qTeller platform (https://qteller.maizegdb.org/genes_by_name_B73v4.php; Kakumanu et al., 2012; Johnston et al., 2014; Forestan et al., 2016; Stelpflug et al., 2016; Walley et al., 2016; Waters et al., 2017).
Figure 7.
Expression levels of ZmRNRL1 and ZmRNRL2. A and B, Comparisons of ZmRNRL1 and ZmRNRL2 expression levels in various tissues of wild-type (A) and tvt1-R plants (B) in the B73 genetic background using sq-PCR. C and D, RT-qPCR analysis of ZmRNRL1 (C) and ZmRNRL2 (D) expression in various tissues of wild-type and tvt1-R plants in the B73 genetic background. Values are means ± sd (n = 3 biological replicates, each with three technical replicates). Two-tailed Student’s t test was used to determine significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001). N, No significant difference. E, Ears; GD, genomic DNA; M, DNA markers; Pt, permissive temperatures; R, roots; Rt, restrictive temperatures; S, ninth leaf-wrapped stems; ST, shoot tips; T, tassels; Ta, tasseling stage at permissive temperatures; TL, topmost visible leaves; V4, V4 stage; V9, V9 stage at permissive temperatures. ZmActin1 was used as the internal control.
To examine the effect of the point mutation on ZmRNRL1 and ZmRNRL2 expressions, we measured the spatial–temporal expression patterns of the two genes in tvt1-R (tvt1-R-B73/tvt1-R-B73) plants. Even if ZmRNRL1 and ZmRNRL2 are differentially expressed in some tissues of tvt1-R-B73/tvt1-R-B73 versus Tvt1-B73/Tvt1-B73 plants, the expression changes did not show consistent trends, suggesting that the point mutation had little effect on the transcription of the two genes (Fig. 7, C and D). Given that the tvt1-R mutant phenotype is temperature-sensitive, to investigate whether the expression levels of ZmRNRL1 and ZmRNRL2 are regulated by temperature, we analyzed their transcript levels in the roots (V4), shoot tips (V4), and topmost leaves (V4) of plants grown at restrictive temperatures. The transcript levels of ZmRNRL1 and ZmRNRL2 were significantly lower at restrictive versus permissive temperatures in most tissues of wild-type and tvt1-R plants (Fig. 7, C and D). Consistent with this result, the publicly available RNA-seq data showed that ZmRNRL1 and ZmRNRL2 were downregulated in 14-d–old B73 seedlings after heat treatment (50°C for 4 h; Fragments per kilobase of exon per million mapped reads [FPKM]ZmRNRL1, B73_control = 6.4, FPKMZmRNRL1, B73_heat = 0.7; FPKMZmRNRL2, B73_control = 2.2, FPKMZmRNRL2, B73_heat = 0.2; qTeller; Waters et al., 2017). Both results suggest that ZmRNRL1 and ZmRNRL2 expression is sensitive to restrictive temperatures. However, we believed that reduced expression levels of ZmRNRL1 and ZmRNRL2 is not the major factor leading to the no-tassel phenotype of tvt1-R mutants at restrictive temperatures. This is because downregulation of ZmRNRL1 expression at restrictive temperatures is less than half, whereas no mutant phenotype was observed in tvt1-m1 heterozygous plants, suggesting that half of ZmRNRL1 protein is sufficient to fully confer its function. Altogether, we reasoned that the tvt1-R mutation leading to the no-tassel phenotype at restrictive temperatures might affect the protein function of ZmRNRL1.
ZmRNRL1-tvt1-R Failed to Interact with the Three ZmRNRSs at High Temperatures Due to the Arg277-to-His277 Substitution
Multiple sequence alignment of RNRL-related proteins showed that Arg277 is absolutely conserved among various species (Supplemental Figs. S4 and S5). According to the Pfam database (http://pfam.xfam.org/), ZmRNRL1 contains three conserved domains: an ATP-cone domain (amino acid residues 1–89), an N-terminal (amino acid residues 142–212) and C-terminal RNRL domain (amino acid residues 216–757), and Arg277 in the third conserved domain (Fig. 4D). Arg277 of ZmRNRL1 was predicted to be a polypeptide-binding site/dimer interface according to the National Center for Biotechnology Information’s (NCBI’s) Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml; Marchler-Bauer et al., 2017). These results suggest that Arg277 is critical for ZmRNRL1 activity.
To examine the effect of the Arg277-to-His277 substitution on ZmRNRL1-tvt1-R, we separately predicted the three-dimensional structures of ZmRNRL1 proteins encoded by the wild-type and tvt1-R mutant allele using the three-dimensional structure of HsRNR1 as a template (Arnold et al., 2006; Benkert et al., 2011; Fairman et al., 2011; Biasini et al., 2014). Considering that ZmRNRL1-B73 and ZmRNRL1-tvt1-R protein sequences differ at three positions (amino acid residues 277, 320, and 385), we used ZmRNRL1 from an Chinese elite inbred HuangZao4 (designated as ZmRNRL1-HZ4) as an additional control, as ZmRNRL1-HZ4 only differs from ZmRNRL1-tvt1-R at a single position (amino acid residue 277; Supplemental Fig. S6A). The predicted three-dimensional structures of the three proteins were similar (Supplemental Fig. S6B), and the Arg277-to-His277 substitution did not alter the secondary structure of this protein (Supplemental Fig. S6C).
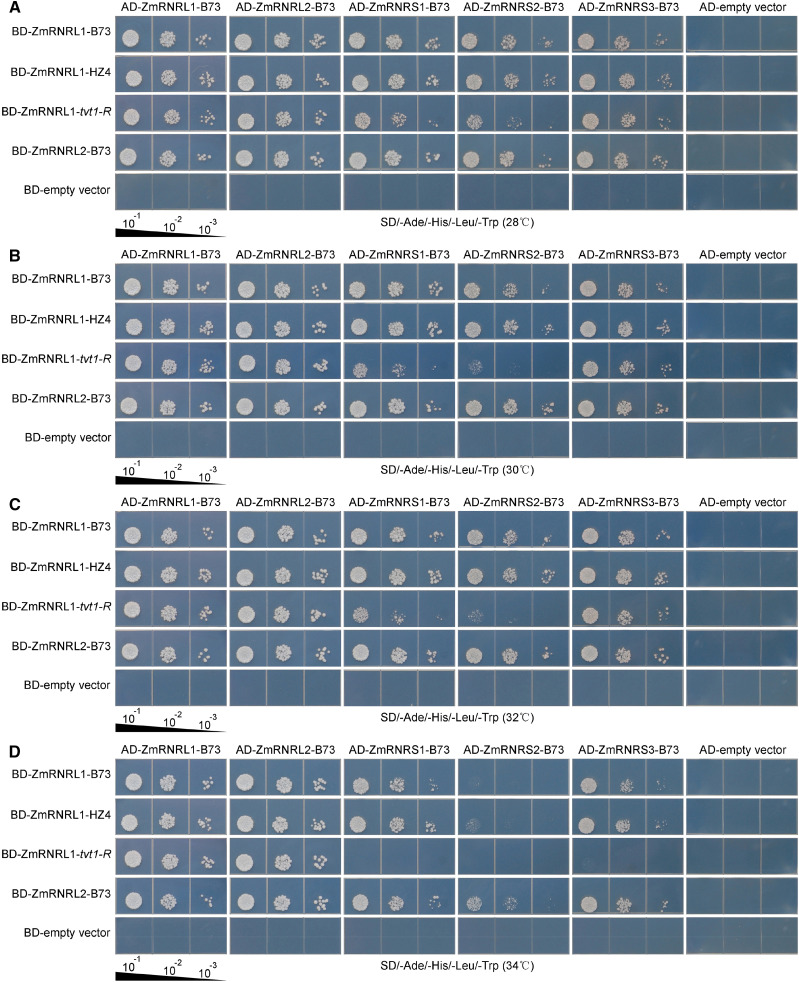
Based on the predictions that Arg277 is a polypeptide-binding site/dimer interface and that no changes in secondary structure occur among the proteins that differ in amino acid residue 277, we hypothesized that the Arg277-to-His277 substitution of ZmRNRL1-tvt1-R likely affects the interaction with its counterparts during the formation of RNR, especially at restrictive temperatures. To test this hypothesis, we performed yeast two-hybrid assays at five different temperatures: 28°C, 30°C, 32°C, 34°C, and 36°C. In maize genome, there were three genes encoding RNRS, Zm00001d045649, Zm00001d037337, and Zm00001d003164, designated as ZmRNRS1, ZmRNRS2, and ZmRNRS3, respectively. The coding sequences of ZmRNRL1-B73, ZmRNRL1-HZ4, ZmRNRL1-tvt1-R, and ZmRNRL2-B73 were separately cloned into pGBKT7 (GAL4 DNA-binding domain [BD]) vectors, and the coding sequences of ZmRNRL1-B73, ZmRNRL2-B73, ZmRNRS1-B73, ZmRNRS2-B73, and ZmRNRS3-B73 were individually cloned into pGADT7 (GAL4 activation domain [AD]) vectors. Using the empty vectors as control, each AD-related plasmid and BD-related plasmid pair was cotransformed into yeast strain AH109. Some AH109 co-transformants did not grow normally on double dropout (DDO; synthetic dextrose/−Leu/−Trp) agar plates at 36°C (Supplemental Fig. S7E), and we therefore analyzed the interactions between subunits at the remaining four different temperatures (28°C, 30°C, 32°C, and 34°C), at which the co-transformants grew normally on DDO agar plates (Supplemental Fig. S7, A–D). At lower temperatures (28°C, 30°C, and 32°C), wild-type proteins ZmRNRL1-B73 and ZmRNRL1-HZ4 interact with all RNR subunits, including two ZmRNRLs (ZmRNRL1-B73 and ZmRNRL2-B73) and three ZmRNRSs (ZmRNRS1-B73, ZmRNRS2-B73, and ZmRNRS3-B73; Fig. 8, A–C). The Arg277-to-His277 substitution in the mutant protein ZmRNRL1-tvt1-R had little effect on its protein interaction at these temperatures, although its interaction with ZmRNRS2-B73 was slightly affected (Fig. 8, A–C). At 34°C, wild-type proteins ZmRNRL1-B73 and ZmRNRL1-HZ4 could interact with ZmRNRS1-B73 and ZmRNRS3-B73; however, the mutant protein ZmRNRL1-tvt1-R failed to interact with all three ZmRNRSs (Fig. 8D). These results suggest that the formation of functional RNR holoenzyme (α2β2) in tvt1-R maize may be hindered at 34°C by the Arg277-to-His277 substitution, leading to the no-tassel phenotype in tvt1-R at 34°C.
Figure 8.
Spot assay on quadruple dropout (QDO) agar plates at four different temperatures. A to D, The indicated AH109 co-transformants were 10-fold serially diluted and spotted on QDO agar plates. Dilution factors are shown below the figures. The values 10−1, 10−2, and 10−3 indicate that the culture was diluted 10-fold, 100-fold, and 1,000-fold, respectively. The plates were incubated for 80 to 85 h at 28°C (A), 30°C (B), 32°C (C), and 34°C (D), respectively. BD, GAL4 DNA-binding domain fusion; AD, GAL4 activation domain fusion. Note that the mutant protein ZmRNRL1-tvt1-R (His277) failed to interact with all three ZmRNRSs (ZmRNRS1-B73, ZmRNRS2-B73, and ZmRNRS3-B73) at 34°C.
Putative Roles of ZmRNRL1 in DNA Replication at Restrictive Temperatures
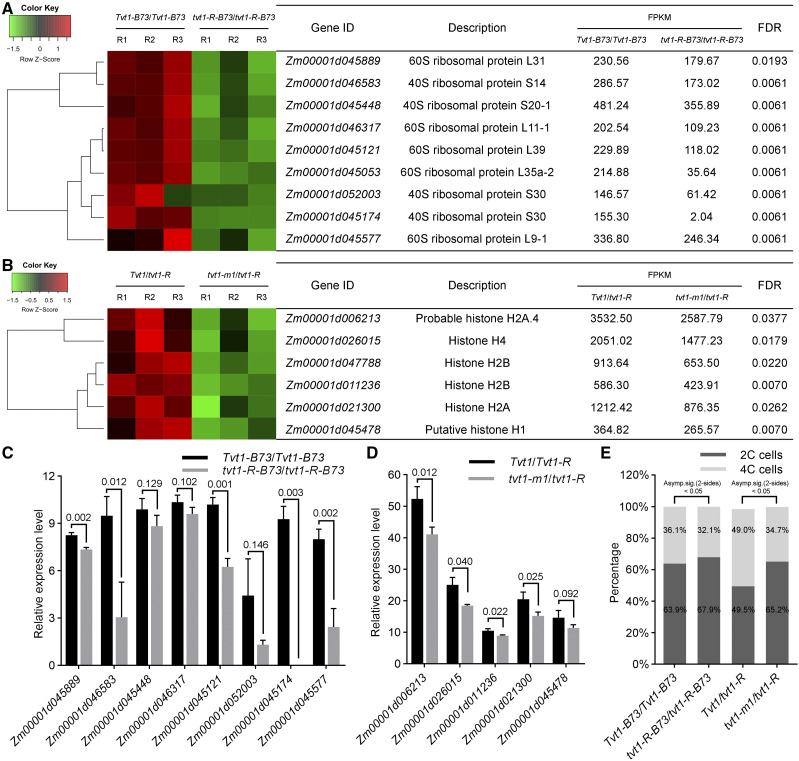
To better understand the molecular function of ZmRNRL1, we performed RNA-seq experiments using V4 shoot tips of wild-type plants and tvt1-R plants in the B73 genetic background grown at restrictive temperatures. We identified 264 differentially expressed genes (DEGs), including 204 downregulated DEGs and 60 upregulated DEGs in tvt1-R (tvt1-R-B73/tvt1-R-B73) versus wild-type (Tvt1-B73/Tvt1-B73) plants. Gene ontology (GO) analysis revealed that the 204 downregulated DEGs were only enriched in three significant GO terms belonging to the cellular component category, namely “cytosolic ribosome,” “cytosolic part,” and “ribosomal subunit” (Supplemental Fig. S8A). All three significant GO terms involved nine ribosomal protein genes (Fig. 9A). We selected eight ribosomal protein genes for RT-qPCR validation, and five of them could be verified to be significantly downregulated in tvt1-R-B73/tvt1-R-B73 plants (Fig. 9C). Ribosomal proteins play key roles in ribosome assembly and protein translation; the downregulation of the ribosomal protein genes usually leads to fewer ribosomes and reduced protein synthesis (de la Cruz et al., 2015; Graifer and Karpova, 2015; Zhou et al., 2015). Because RNR-synthesized dNTPs are used for DNA replication (Reichard, 1988; Elledge et al., 1992; Mathews, 2006; Nordlund and Reichard, 2006), and previous studies have uncovered links between DNA replication and protein synthesis (Abid et al., 1999; Berthon et al., 2008, 2009), we speculated that the downregulation of ribosomal protein genes might be caused by DNA replication stress in tvt1-R-B73/tvt1-R-B73 plants at restrictive temperatures.
Figure 9.
The rnrl1 mutations probably affect DNA replication at restrictive temperatures. A, Hierarchical clustering of nine downregulated ribosomal protein genes in tvt1-R-B73/tvt1-R-B73 plants compared to Tvt1-B73/Tvt1-B73 plants. B, Hierarchical clustering of six downregulated histone genes in tvt1-m1/tvt1-R plants compared to Tvt1/tvt1-R plants. In A and B, FPKM values of the three biological replicates are shown. C, RT-qPCR validation of eight downregulated ribosomal protein genes in tvt1-R-B73/tvt1-R-B73 plants compared to Tvt1-B73/Tvt1-B73 plants. D, RT-qPCR validation of five histone genes in tvt1-m1/tvt1-R plants compared to Tvt1/tvt1-R plants. In C and D, ZmActin1 was used as the internal control. Values are means ± sd (n = 3 biological replicates, each with three technical replicates). Two-tailed Student’s t test was used to determine significant differences. P values are indicated in the figure. E, Results of flow cytometric analysis of the second leaf tips at V2 in plants of the four different genotypes. The 2C cells, 4C cells, and all cycle events for each genotype are calculated from the sum of three biological replicates. All cycle events of Tvt1-B73/Tvt1-B73, tvt1-R-B73/tvt1-R-B73, Tvt1/tvt1-R, and tvt1-m1/tvt1-R plants are 16,734, 16,237, 17,700, and 17,776, respectively. Statistical analysis of the percentage data between two samples was performed by χ2 test.
Because an extremely low proportion of tvt1-R-B73/tvt1-R-B73 plants lacked tassels at restrictive temperatures, we also performed RNA-seq experiments using V6 shoot tips of Tvt1/tvt1-R plants and tvt1-m1/tvt1-R plants from an allelism test grown at restrictive temperatures. We identified 48 downregulated and 205 upregulated DEGs in tvt1-m1/tvt1-R versus Tvt1/tvt1-R plants. GO analysis of 48 downregulated DEGs in tvt1-m1/tvt1-R versus Tvt1/tvt1-R plants revealed 44 significantly enriched GO terms, including 10 terms belonging to the cellular component category, four terms belonging to the molecular function category, and 30 terms belonging to the biological process category (Supplemental Fig. S8B). A close scrutiny of the genes in these GO terms revealed several transcription factors and six histone genes (Fig. 9, B and D). During DNA replication, replicated DNA must be rapidly assembled into nucleosomes using both newly synthesized and parental histones to restore chromatin organization, a process termed DNA replication-coupled nucleosome assembly (MacAlpine and Almouzni, 2013; Prado and Maya, 2017; Serra-Cardona and Zhang, 2018). The downregulation of histone genes in tvt1-m1/tvt1-R plants suggests that DNA replication might be affected in this mutant at restrictive temperatures.
The two sets of RNA-seq data both suggest that the rnrl1 mutations probably affect DNA replication at restrictive temperatures. To verify this finding, we used flow cytometry to detect the proportion of 2C and 4C cells in the leaf tissues of plants of the four different genotypes at restrictive temperatures. The percentages of 4C cells were significantly lower in tvt1-R-B73/tvt1-R-B73 and tvt1-m1/tvt1-R plants than in Tvt1-B73/Tvt1-B73 and Tvt1/tvt1-R plants (Fig. 9E), respectively, further supporting that the rnrl1 mutations probably affect DNA replication at restrictive temperatures.
Given RNR-synthesized dNTPs are used for both DNA replication and DNA damage repair, we therefore tested whether DNA damage repair marker genes are upregulated in the mutants. Several DNA damage-repair-related marker genes have been identified in Arabidopsis, including ATRAD51 (Doutriaux et al., 1998), ATBRCA1 (Lafarge and Montané, 2003), ATGR1 (Uanschou et al., 2007), ATMRE11 (Bundock and Hooykaas, 2002), ATKU70 (Tamura et al., 2002), ATMSH2 and ATMSH6 (Lario et al., 2011), and ATPARP1 and ATPARP2 (Doucet-Chabeaud et al., 2001). According to the Gramene database (http://www.gramene.org/), there are 15 expressed genes (FPKM > 1) orthologous to the Arabidopsis DNA damage-repair-related marker genes in the maize genome. Of these 15 genes, 13 genes showed similar transcript levels in V4 shoot tips of Tvt1-B73/Tvt1-B73 versus tvt1-R-B73/tvt1-R-B73 individuals and in V6 shoot tips of Tvt1/tvt1-R versus tvt1-m1/tvt1-R individuals, and the remaining two genes were only slightly upregulated in V6 shoot tips of tvt1-m1/tvt1-R versus Tvt1/tvt1-R individuals (Supplemental Fig. S9A), indicating that the rnrl1 mutations had little effect on DNA damage repair at restrictive temperatures. Altogether, these results suggest that DNA replication is more sensitive to rnrl1 mutations compared to DNA damage repair. This finding is consistent with the observation that the Arabidopsis rnrs mutant tso2-1 exhibits defects in cell-cycle progression but does not show increased DNA damage compared to the wild type (Wang and Liu, 2006).
Both RNA-seq and flow cytometric analyses suggested that S phase for DNA replication might be affected by the rnrl1 mutations at restrictive temperatures. To analyze whether the rnrl1 mutations affect other cell-cycle stages at restrictive temperatures, we thus examined the expression levels of several representative cell-cycle-related genes in plants of the four different genotypes. Specifically, based on the Gramene database, we selected 21 expressed genes (FPKM > 1) that are orthologs of ATCYCD3;3, which is expressed in the G1 phase (Menges et al., 2005); ATCYCA3;1, which is highly expressed at the G1/S phase (Juraniec et al., 2016); ATCYCA2;1, whose transcripts peak at the end of the G2 phase (Shaul et al., 1996); ATCYCB1;2 and ATMAD2, which are markers for the G2/M phase (Menges et al., 2005; Juraniec et al., 2016); and ATCYCD4:1, which is expressed from the G2 to M phase (Kono et al., 2003). There was no significant difference in the expression levels of these cell-cycle–related genes between Tvt1-B73/Tvt1-B73 and tvt1-R-B73/tvt1-R-B73 individuals or between Tvt1/tvt1-R and tvt1-m1/tvt1-R individuals (Supplemental Fig. S9B), at least partially suggesting the rnrl1 mutations did not affect other cell-cycle stages at restrictive temperatures.
Previous studies have demonstrated that Knotted1 (Kn1; Zm00001d033859) is a useful meristem-maintenance marker gene (Jackson et al., 1994; Long et al., 1996), we also examined Kn1 transcript abundance in our RNA-seq data. No significant difference in Kn1 transcript abundance was detected between Tvt1-B73/Tvt1-B73 and tvt1-R-B73/tvt1-R-B73 individuals (FPKMTvt1-B73/Tvt1-B73 = 120.7; FPKMtvt1-R-B73/tvt1-R-B73 = 108.1; false discovery rate [FDR] = 0.999) or between Tvt1/tvt1-R and tvt1-m1/tvt1-R individuals (FPKMTvt1/tvt1-R = 110.9; FPKMtvt1-m1/tvt1-R = 119.0; FDR = 0.998), suggesting that SAM arrest in these two rnrl1 mutants was not due to differences in the expression levels of Kn1.
DISCUSSION
Our molecular genetic analyses of tvt1-R mutants led to the identification and characterization of ZmRNRL1, encoding a large subunit of RNR in maize. Expression, yeast two-hybrid, RNA-seq, and flow cytometric analyses suggested that ZmRNRL1-tvt1-R failed to interact with all three ZmRNRSs at a high temperature (34°C) due to the Arg277-to-His277 substitution, which could impede RNR heterodimer (α2β2) formation, thereby decreasing the dNTP supply for DNA replication; this effect may be especially severe for the SAMs that require a continuous, sufficient dNTP supply for rapid division, consequently resulting in SAM arrest and the no-tassel phenotype of tvt1-R mutants at restrictive temperatures. In addition to the terminal development of the SAM into tassels, the peripheral zone of the SAM is incorporated into leaf primordia and the rib zone to produces stem tissues (Jackson, 2009). Thus, SAM arrest not only leads to the absence of tassels, but it also affects leaf and stem development, as evidenced by our finding that the rnrl1 mutations significantly reduced leaf number and plant height (Fig. 5, K and M; Supplemental Fig. S1, P and R).
In Arabidopsis, several studies have provided a close link between cell-cycle regulation and meristem organization. For example, double-knockdown lines of the cyclin-dependent kinases ATCDKB2;1 and ATCDKB2;2 could result in severe SAM arrest phenotypes (Andersen et al., 2008). The gene jing he sheng1, a DNA replication helicase/nuclease2 mutant that indirectly affects cell-cycle progression, exhibited fasciated meristems (Jia et al., 2016). In these two studies, we noticed the S-phase–specific H4 expression, which was downregulated in ATCDKB2;1 and ATCDKB2;2 showing SAM arrest whereas it was upregulated in jing he sheng1 exhibiting fasciated SAM. It seems that the downregulation or upregulation of H4 expression in meristem mutants determines whether there is meristem arrest or over-proliferation. In this study, several histone genes were downregulated in tvt1-m1/tvt1-R plants displaying no tassels (SAM arrest) at restrictive temperatures (Fig. 9, B and D). This result coincides with previous studies, and supports our view.
The null allele, tvt1-m1, is the first frame-shift mutation of RNRL identified in flowering plants (Garton et al., 2007; Yoo et al., 2009; Tang et al., 2012, 2019). Homozygous tvt1-m1 mutants survived, although they exhibited severe developmental defects (Supplemental Fig. S2, F–I), suggesting the presence of a paralog to ZmRNRL1. An examination of the maize genome revealed that ZmRNRL1 indeed has a paralog, ZmRNRL2, which possesses a similar gene structure to ZmRNRL1 (the Gramene database) and whose coding protein shares 97% amino acid sequence identity with ZmRNRL1 (Supplemental Fig. S5). Moreover, ZmRNRL2 and ZmRNRL1 have similar spatial-temporal expression patterns (Fig. 7), and ZmRNRL2 and ZmRNRL1 proteins have same interaction partners at normal growth temperatures (Fig. 8, A–C). These results indicate that ZmRNRL2 performs molecular functions similar to ZmRNRL1, although they are expressed at different levels (Fig. 7, C and D).
Both the high amino acid sequence identity of RNRL-related proteins and previous study indicate that these proteins share similar molecular functions (Supplemental Fig. S5; Yoo et al., 2009). Previously identified rnr mutants were primarily characterized based on chloroplast deficiencies (Wang and Liu, 2006; Garton et al., 2007; Yoo et al., 2009; Tang et al., 2012, 2019; Chen et al., 2015; Qin et al., 2017), whereas the tvt1-R mutant was identified based on the absence of tassels, suggesting that ZmRNRL1 has different biological functions from the other RNRL-related proteins identified in flowering plants. The rnr mutants in flowering plants usually display decreased dNTP production (Wang and Liu, 2006; Garton et al., 2007; Yoo et al., 2009). Garton et al. (2007) and Tang et al. (2019) suggested that under a limited dNTP supply, inhibited chloroplast DNA replication might be a primary cause of chloroplast deficiency in rnr mutants. Based on our phenotypic analysis and RNA-seq results, it appears that SAM arrest leading to the lack of tassels at restrictive temperatures in tvt1-R mutants is affected by deficient nuclear DNA replication. We propose that whether the limited dNTP supply affects plastid DNA synthesis or nuclear DNA synthesis depends on two factors: the plant species and variable penetrance/expressivity.
Yoo et al. (2009) proposed that virescent3 (rice rnrl mutant) and stripe1 (rice rnrs mutant) exhibit chlorotic phenotypes due to a chloroplast sacrifice mechanism: the nuclear genome, which is critical for cell maintenance and cell division during growth and development, is the first priority, and the synthesis of plastid DNA for chloroplast biogenesis, which is less critical for plant survival, is the second priority. In contrast to rice, Arabidopsis, and S. italica, maize is a unisexual plant that produces tassels at the shoot apex and ears in the axils of vegetative leaves. Given that the ear, which directly produces offspring, develops from AMs, not the SAM, maize might lack this chloroplast sacrifice mechanism to ensure the establishment of the nuclear genome and SAM maintenance. Additionally, the number of RNRL and RNRS varies among species. For example, Arabidopsis contains one RNRL and three RNRSs (Wang and Liu, 2006; Garton et al., 2007; Tang et al., 2012), rice contains two RNRLs and two RNRSs (Yoo et al., 2009), maize contains two RNRLs and three RNRSs, and human contains one RNRL and two RNRSs (Qiu et al., 2006). More importantly, the interactions between RNRL and RNRS varies among species. Yeast two-hybrid assays showed that the maize RNRL could interact with all three RNRSs at normal growth temperatures (Fig. 8B). Similarly, the human RNRL hRRM1 could interact with two RNRSs, hRRM2 and p53R2 (Qiu et al., 2006). By contrast, rice RNRL only interacts with a specific RNRS (OsRNRL1:OsRNRS1 and OsRNRL2:OsRNRS2; Yoo et al., 2009). The differences in both the number of RNR subunits and the interactions between RNR subunits among species indirectly support the notion that the RNRL in different species has different biological functions.
In this study, when grown in the growth chamber, tvt1-R mutants had almost 100% penetrance of the no-tassel phenotype whereas tvt1-R-B73/tvt1-R-B73 plants only had 6.9% penetrance of that (Supplemental Fig. S1, I and J); tvt1-m1 homozygous plants exhibited either no tassels or striped leaves (Supplemental Fig. S2, F–I; Fig. 6, I and J). Both results suggest the variable penetrance/expressivity of mutant phenotypes. There are three possible explanations for this phenomenon:
The existence of modifier(s) for tvt1. Modifiers are nonallelic genetic variations that shape genotype–phenotype relationships (Meyer and Anderson, 2017). Maize contains abundant modifiers due to the high genetic variation among maize inbred lines, and previous studies have identified several modifiers in maize, such as the modifier for virescent yellow-like (Xing et al., 2014) and the modifier for Sympathy for the ligule (Anderson et al., 2019). In future research, it will be worth identifying the modifier(s) for tvt1 by a map-based cloning approach.
The nonuniform environment. It was clear that environmental factors can have a striking influence on penetrance/expressivity of the phenotype. A typical example in humans is phenotypic differences among monozygotic twins, such as monozygotic twins discordant for childhood leukemia and secondary thyroid carcinoma (Galetzka et al., 2012). Similarly, in plants, taking genic male fertility of rice as an example, it has been demonstrated that temperature (Chen et al., 2007; Ding et al., 2012a; Zhou et al., 2012, 2014; Yu et al., 2017), photoperiod (Ding et al., 2012a; Zhang et al., 2013; Fan et al., 2016), and humidity (Xue et al., 2018; Chen et al., 2020) can affect the penetrance of male sterility, corresponding to temperature-, photoperiod-, and humidity‐sensitive genic male sterility, respectively.
Transcriptional noise. This refers to how genetically identical individuals growing in the same environment display differences in expression, thereby showing different phenotypes (Cortijo and Locke, 2020). Based on the sources of noise in gene expression, transcriptional noise is often divided into intrinsic noise and extrinsic noise. Intrinsic noise is the inherent stochasticity that arises from the discrete nature of biochemical reactions, whereas extrinsic noise refers to random differences in expression regulated by the cell state, such as cell size, cell age, and cell-cycle position (Elowitz et al., 2002; Cortijo and Locke, 2020).
The overall activity of RNR is monitored by the ATP/dATP ratio. At high dATP concentrations, RNRL forms hexamers (α6) and translocates into the nucleus, allowing for binding to zinc-finger-RAN-binding-domain-containing-3, thereby impeding zinc-finger-RAN-binding-domain-containing-3–PCNA complexation that promotes DNA synthesis (Fu et al., 2018). Conversely, the high ATP/dATP ratio induces RNRL to form monomers (α) or dimers (α2), allowing for it to interact with RNRS to form the α2β2 quaternary structure to exert RNR activity (Fairman et al., 2011). Nucleotide-induced oligomerization of RNRL implies that it is regulated by complex allosteric mechanisms. Numerous studies have shown that temperature can induce the conformational changes of proteins (Zhang et al., 2015; Muneeswaran et al., 2018; Matt et al., 2019) and subsequently affect protein–protein interactions. Hence, we proposed that the failure in the interactions between the mutant protein ZmRNRL1-tvt1-R and the three RNRSs at 34°C might be due to alterations of its protein conformation at 34°C.
Interestingly, the homozygous ZmRNRL1-overexpression lines with relatively high ZmRNRL1 expression levels and their corresponding F2 progenies exhibited yellow-striped leaves (Supplemental Fig. S3, E and F). There are three possible explanations for this phenotype:
Excessive expression of ZmRNRL1 may lead to a high dNTP concentration in G1 phase, which arrests cell-cycle progression in the late G1 phase and prevents S-phase entry, thus affecting DNA synthesis and chloroplast biosynthesis. Naturally, RNRL gene expression peaks in the S phase and is negligible in the G0/G1 phase (Johansson et al., 1998). Correspondingly, dNTP concentrations are highest in the S phase and lowest in the G1 phase. Maintaining the low dNTP concentration in the G1 phase is necessary. In S. cerevisiae, the introduction of an additional GAL1-regulated RNR1 or rnr1-D57N allele led to continuously high dNTP concentrations, which inhibited cell-cycle progression in the late G1 phase and affected the activation of the replication origin (Chabes and Stillman, 2007).
Excessive expression of ZmRNRL1 may result in excessive ZmRNRL1 proteins, which interfere with RNR holoenzyme (α2β2) formation due to unequal ratio between RNR large and small subunits, thereby leading to an insufficient dNTP supply.
Overexpression of ZmRNRL1 may lead to homologous co-suppression and the degradation of ZmRNRL1 transcript from both the transgene and the endogenous ZmRNRL1 gene (Napoli et al., 1990). Further confirmation of this hypothesis would require phenotypic analysis of plants in which ZmRNRL1 is knocked-down.
Taken together, the yellow-striped leaves in ZmRNRL1-overexpression lines may be intrinsically similar with the striped leaves in rnrl1 mutants. Both phenotypes are probably caused by inhibited chloroplast DNA replication. Therefore, our study highlights the importance of nucleotide homeostasis for plant growth and development, especially for SAM maintenance and chloroplast biosynthesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The maize (Zea mays) tvt1-R mutant was obtained from the offspring of a breeder-selected, high-generation, inbred line N17 irradiated by heavy ions. No-tassel plant was first identified in the M3 generation of the mutant library sown in Changsha in the autumn of 2004. After several years of breeding, we obtained the no-tassel line, all plants of which exhibited the lack of tassels when planted in Changsha in the autumn of 2010, and we named it the tvt1-R mutant. Tvt1-B73 and tvt1-R-B73 homozygous plants were generated from BC4F2 or higher generation populations derived from a backcross between tvt1-R mutant (the donor parent) and inbred line B73 (the recurrent parent). The tvt1-m1 allele was isolated from Mu insertion stock mu-illumina-247673.4 (Williams-Carrier et al., 2010), which has been crossed with B73 three times before selfing. Seeds were selfed for one generation for allelism tests, and at least two generations for phenotypic analysis and other studies. The 62 maize inbred lines used for sequencing analysis are listed in Supplemental Dataset S1.
Field trials were carried out in Changsha (28°N, 113°E), Beijing (39°N, 116°E), and Sanya (18°N, 109°E), China from 2014 to 2018. Seeds were sown in Changsha in the spring, Beijing in summer, Changsha in autumn, and Sanya in winter on approximately April 1, May 1, July 11, and November 1, respectively. The climatological data for daily maximum average temperature, daily minimum average temperature, and average daylength were obtained from publicly available records (http://weather.zuzuche.com/ for daily average temperature and http://www.time.ac.cn/ for average daylength). Experiments in the growth chamber were carried out at 36°C/26°C/13.5 h and 70% relative humidity. Unless otherwise indicated, “permissive temperatures” and “restrictive temperatures” represent moderate temperatures of Beijing in the summer and high temperatures in the growth chamber, respectively.
Phenotyping and Statistical Analysis
Individuals lacking entire tassels (including the main axis, branches, and spikelets) at the tasseling stage were defined as no-tassel plants. Plant height, tassel branch number, and leaf number were measured at maturity. Plant height was measured from the shoot base to the shoot apex, and tassel branch number was counted using only primary branches. Analysis of significant differences in the average data was performed using the two-tailed Student’s t test (two samples) or Tukey’s multiple comparisons (three or more samples). The χ2 test was used for statistical analysis of differences in the percentage data between two samples.
Histological Analysis
Shoot tips at different leaf ages were sampled from B73 and tvt1-R mutants both at the permissive and restrictive temperatures and fixed in 3.7% (v/v) formalin-acetic acid-alcohol solution (10 volumes absolute ethanol, 1 volume glacial acetic acid, 2 volumes 37% [v/v] formaldehyde, and 7 volumes distilled water). The samples were dehydrated through a graded ethanol series, cleared in Histoclear (Sigma-Aldrich), embedded in paraffin (Thermo Fisher Scientific), and sliced into sections (8 to 12 μm) using a microtome. Wax strips that met the requirements were selected under a microscope, spread out in a water bath, adhered to poly-Lys slides, and dewaxed with xylene or Histoclear. The sections were stained with toluidine blue to reveal their histology.
Map-Based Cloning
An F2 mapping population was generated by crossing inbred line B73 with the tvt1-R mutant, and positional cloning was carried out using no-tassel plants obtained from the F2 population, which were planted in Changsha in the autumn. Polymorphic molecular markers (including simple sequence repeats, insertion–deletions, and SNPs for primary and fine mapping were derived from MaizeGDB (https://www.maizegdb.org/) or developed based on the B73 reference sequence (http://www.gramene.org/). The primers used for mapping are listed in Supplemental Dataset S1.
Allelism Test
Heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) were crossed with the tvt1-R mutant, and the progenies were planted at both permissive and restrictive temperatures. A pair of specific primers (Mu-F1 + TIR6) was used to identify tvt1-m1/tvt1-R plants and Tvt1/tvt1-R plants in the progeny.
Complementation Tests
To complement the tvt1-R mutant, seven transgenic lines (including the two complementation lines [GC-1 and GC-2] and five overexpression lines [OE-1 to OE-5]) were crossed with the tvt1-R mutant and then self-pollinated to generate the corresponding F2 progeny. Three pairs of primers (M11, GC-F + GC-R, and OE-F + OE-R) were used to identify tvt1-R/tvt1-R, tvt1-R/tvt1-R;GC, and tvt1-R/tvt1-R;OE plants among the offspring, which were planted at restrictive temperatures to observe their phenotypes.
To complement the tvt1-m1 mutant, F2 progenies derived from a cross between the five overexpression lines and heterozygous tvt1-m1 mutants (Tvt1/tvt1-m1) were generated and identified by PCR identification with three pairs of primers (Mu-F1 + Mu-R, Mu-F1 + TIR6, and OE-F + OE-R) to select tvt1-m1/tvt1-m1;OE and tvt1-m1/tvt1-m1 plants. The progenies were selected and planted at permissive temperatures to observe their phenotypes. The two complementation lines were not used to complement the tvt1-m1 mutant phenotype due to the difficulties in distinguishing between tvt1-m1/tvt1-m1;GC plants and Tvt1/tvt1-m1;GC plants in the F2 progenies.
The primers and restriction enzymes used for the complementation vector construction are listed in Supplemental Dataset S1. The ZmRNRL1-overexpression seeds were created by the Center for Crop Functional Genomics and Molecular Breeding of China Agricultural University.
Phylogenetic Analysis
Phylogenetic analysis was performed using MEGA7 (Kumar et al., 2016). Using the protein sequence of ZmRNRL1 as a query, a BLAST search for homologs in other species was performed against the Gramene (http://ensembl.gramene.org/Tools/Blast?db=core) and NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) databases. The proposed protein sequences were aligned using ClustalW with default parameters, and a neighbor-joining tree was constructed using the bootstrap method with 1,000 replications.
RACE, sq-PCR, and RT-qPCR
Total RNA was extracted from the samples using a Quick RNA isolation Kit (Huayueyang). Rapid amplification of cDNA ends (RACE) was performed with total RNA using a SMARTer RACE 5ʹ/3ʹ Kit (Clontech) following the manufacturer’s instructions. For sq-PCR and RT-qPCR, total RNA was subjected to genomic DNA removal and reverse transcription using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa). The synthesized cDNA was diluted 5- or 10-fold and used as a template for PCR. RT-qPCR was performed with TB Green Premix Ex Taq (TaKaRa). ZmActin1 (Zm00001d010159) was used as the internal control to normalize the gene expression data. Relative expression levels were calculated using the 2–ΔCT method.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). The primers and restriction enzymes used for AD-related and BD-related vector construction are listed in Supplemental Dataset S1. For the spot assay, more than 10 fresh colonies of each AH109 co-transformant were inoculated into 5 mL of DDO liquid medium and incubated at 30°C overnight (18 to 24 h) with shaking (250 rpm). The following day cultures were diluted to OD600 = 0.65, and then diluted 10‐, 100‐, and 1,000‐fold. Aliquots (8 μL) of 10-fold serial dilutions were spotted on DDO or QDO (synthetic dextrose/−Ade/−His/−Leu/−Trp) agar plates. Plates were incubated at five different temperatures (28°C, 30°C, 32°C, 34°C, and 36°C) for 80 to 85 h. Yeast two-hybrid analysis was repeated twice with consistent results.
RNA-Seq Analysis
Total RNA was extracted from V4 shoot tips from Tvt1-B73/Tvt1-B73 and tvt1-R-B73/tvt1-R-B73 plants and V6 shoot tips from Tvt1/tvt1-R and tvt1-m1/tvt1-R plants grown at restrictive temperatures. Each sample had three biological replicates. Paired-end libraries were constructed and sequenced using the Illumina NovaSeq 6000 platform. The generated raw reads were first trimmed using fastp with the default parameter (Chen et al., 2018). The trimmed reads were then mapped to the B73 AGPv4 genome with HISAT2 (Kim et al., 2015). Only reads with a mapping quality score ≥ 20 were retained for further analysis. Cufflinks v2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/) was used to estimate normalized gene expression values (FPKM; Trapnell et al., 2010). Differential expression analysis was performed with Cuffdiff v2.2.1 (Trapnell et al., 2013). Genes with FDR ≤ 0.05 were defined as differentially expressed. The Singular Enrichment Analysis tool in the program AGRIGO 2.0 was used for GO enrichment analysis of the DEGs (Tian et al., 2017).
Flow Cytometry
The tissues used for flow cytometric analysis were 2- to 4-cm samples of the second leaf tips (V2) of Tvt1-B73/Tvt1-B73, tvt1-R-B73/tvt1-R-B73, Tvt1/tvt1-R, and tvt1-m1/tvt1-R plants grown at restrictive temperatures. Each sample contained three biological replicates, each comprising three individual plants. Flow cytometry analysis was carried out at the Vegetable Research Center, Beijing Academy of Agricultural and Forestry Sciences.
Primers
The primers used in this study are listed in Supplemental Dataset S1.
Accession Numbers
The transcriptome data generated in this study have been deposited into NCBI under accession number SRP241128. Further sequence data from this article can be found in the Gramene (http://www.gramene.org/) or GenBank/EMBL databases under the following accession numbers: ZmRNRL1 (Zm00001d045192), ZmRNRL2 (Zm00001d036322), ZmRNRS1 (Zm00001d045649), ZmRNRS2 (Zm00001d037337), ZmRNRS3 (Zm00001d003164), OsRNRL1 (EU602344), OsRNRL2 (EU602346), AtRNR1 (At2g21790), HsRNR1 (NM_001033), ScRNR1 (NC_001137), ZmActin1 (Zm00001d010159), and Kn1 (Zm00001d033859).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes of wild-type and tvt1-R plants in the B73 genetic background at permissive and restrictive temperatures.
Supplemental Figure S2. Identification of the Mu insertion line.
Supplemental Figure S3. Identification of two complementation lines and five overexpression lines.
Supplemental Figure S4. Phylogenetic analysis of 42 RNRL-related homologs.
Supplemental Figure S5. Sequence alignment of RNRLs using DNAMAN (https://www.lynnon.com/) software.
Supplemental Figure S6. Prediction of protein structure.
Supplemental Figure S7. Spot assay on DDO agar plates at five different temperatures.
Supplemental Figure S8. GO analysis of DEGs.
Supplemental Figure S9. Expressions of genes associated with DNA damage repair and cell cycle.
Supplemental Dataset S1. The 62 maize inbred lines used for sequencing analysis and the primers used in this study.
Acknowledgments
We thank Dr. Alice Barkan and Susan Belcher for providing the mu-illumina-247673.4 seeds and communications about the genetic background, and the Center for Crop Functional Genomics and Molecular Breeding of China Agricultural University for creating the ZmRNRL1-overexpression seeds.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 91735305 and 31671709) and the National Key Research and Development Program (grant no. 2016YFD0101003).
References
- Abid MR, Li Y, Anthony C, De Benedetti A(1999) Translational regulation of ribonucleotide reductase by eukaryotic initiation factor 4E links protein synthesis to the control of DNA replication. J Biol Chem 274: 35991–35998 [DOI] [PubMed] [Google Scholar]
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, Schuster C, Lohmann JU(2008) Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, St Aubin B, Abraham-Juárez MJ, Leiboff S, Shen Z, Briggs S, Brunkard JO, Hake S(2019) The second site modifier, Sympathy for the ligule, encodes a homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4 and rescues the Liguleless narrow maize mutant. Plant Cell 31: 1829–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T(2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T(2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon J, Cortez D, Forterre P(2008) Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol 9: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon J, Fujikane R, Forterre P(2009) When DNA replication and protein synthesis come together. Trends Biochem Sci 34: 429–434 [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014) SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY(2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46: 69–78 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Hake S(2007) Flowering and determinacy in maize. J Exp Bot 58: 909–916 [DOI] [PubMed] [Google Scholar]
- Bukowski R, Guo X, Lu Y, Zou C, He B, Rong Z, Wang B, Xu D, Yang B, Xie C, et al. (2018) Construction of the third-generation Zea mays haplotype map. Gigascience 7: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P(2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14: 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Stillman B(2007) Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104: 1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Tabi Z, Galli M, Malcomber S, Buck A, Muszynski M, Gallavotti A(2014) The boron efflux transporter ROTTEN EAR is required for maize inflorescence development and fertility. Plant Cell 26: 2962–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang Z, Ni E, Lin J, Peng G, Huang J, Zhu L, Deng L, Yang F, Luo Q, et al. (2020) HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol 225: 2077–2093 [DOI] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G(2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J(2018) fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhu L, Xin L, Du K, Ran X, Cui X, Xiang Q, Zhang H, Xu P, Wu X(2015) Rice stripe1-2 and stripe1-3 mutants encoding the small subunit of ribonucleotide reductase are temperature sensitive and are required for chlorophyll biosynthesis. PLoS One 10: e0130172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo S, Locke JCW(2020) Does gene expression noise play a functional role in plants? Trends Plant Sci 25: 1041–1051 [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, Woolford JL Jr.(2015) Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84: 93–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q(2012a) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA 109: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Shen J, Mao H, Xie W, Li X, Zhang Q(2012b) RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol Plant 5: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M(2001) Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol Genet Genomics 265: 954–963 [DOI] [PubMed] [Google Scholar]
- Doutriaux MP, Couteau F, Bergounioux C, White C(1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Durbak AR, Phillips KA, Pike S, O’Neill MA, Mares J, Gallavotti A, Malcomber ST, Gassmann W, McSteen P(2014) Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell 26: 2978–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW(1990) Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 4: 740–751 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB(1992) Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci 17: 119–123 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS(2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, et al. (2011) Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol 18: 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yang J, Mathioni SM, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, et al. (2016) PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc Natl Acad Sci USA 113: 15144–15149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Aiese Cigliano R, Farinati S, Lunardon A, Sanseverino W, Varotto S(2016) Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci Rep 6: 30446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA.(2009) Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol 12: 63–68 [DOI] [PubMed] [Google Scholar]
- Fu Y, Long MJC, Wisitpitthaya S, Inayat H, Pierpont TM, Elsaid IM, Bloom JC, Ortega J, Weiss RS, Aye Y(2018) Nuclear RNR-α antagonizes cell proliferation by directly inhibiting ZRANB3. Nat Chem Biol 14: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetzka D, Hansmann T, El Hajj N, Weis E, Irmscher B, Ludwig M, Schneider-Rätzke B, Kohlschmidt N, Beyer V, Bartsch O, et al. (2012) Monozygotic twins discordant for constitutive BRCA1 promoter methylation, childhood cancer and secondary cancer. Epigenetics 7: 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P(2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ(2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al. (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112: 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ(2007) Crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase—leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50: 118–127 [DOI] [PubMed] [Google Scholar]
- Graifer D, Karpova G(2015) Roles of ribosomal proteins in the functioning of translational machinery of eukaryotes. Biochimie 109: 1–17 [DOI] [PubMed] [Google Scholar]
- Heggie L, Halliday KJ(2005) The highs and lows of plant life: Temperature and light interactions in development. Int J Dev Biol 49: 675–687 [DOI] [PubMed] [Google Scholar]
- Jackson D.(2009) Vegetative shoot meristems In Bennetzen JL, and Hake SC, eds, Handbook of Maize: Its Biology. Springer Science+Business Media, New York, pp 1–12 [Google Scholar]
- Jackson D, Veit B, Hake S(1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jia N, Liu X, Gao H(2016) A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol 171: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Hjortsberg K, Thelander L(1998) Two YY-1-binding proximal elements regulate the promoter strength of the TATA-less mouse ribonucleotide reductase R1 gene. J Biol Chem 273: 29816–29821 [DOI] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ(2014) Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26: 4718–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Reichard P(1998) Ribonucleotide reductases. Annu Rev Biochem 67: 71–98 [DOI] [PubMed] [Google Scholar]
- Juraniec M, Heyman J, Schubert V, Salis P, De Veylder L, Verbruggen N(2016) Arabidopsis COPPER MODIFIED RESISTANCE1/PATRONUS1 is essential for growth adaptation to stress and required for mitotic onset control. New Phytol 209: 177–191 [DOI] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MMR, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A(2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-seq. Plant Physiol 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL(2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Lee J, Ito M, Uchimiya H, Umeda M(2003) Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol 132: 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K(2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarge S, Montané MH(2003) Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res 31: 1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario LD, Ramirez-Parra E, Gutierrez C, Casati P, Spampinato CP(2011) Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J Exp Bot 62: 2925–2937 [DOI] [PubMed] [Google Scholar]
- Leonard A, Holloway B, Guo M, Rupe M, Yu G, Beatty M, Zastrow-Hayes G, Meeley R, Llaca V, Butler K, et al. (2014) Tassel-less1 encodes a boron channel protein required for inflorescence development in maize. Plant Cell Physiol 55: 1044–1054 [DOI] [PubMed] [Google Scholar]
- Li L, Li X, Liu Y, Liu H(2016) Flowering responses to light and temperature. Sci China Life Sci 59: 403–408 [DOI] [PubMed] [Google Scholar]
- Liu Q, Galli M, Liu X, Federici S, Buck A, Cody J, Labra M, Gallavotti A(2019) NEEDLE1 encodes a mitochondria localized ATP-dependent metalloprotease required for thermotolerant maize growth. Proc Natl Acad Sci USA 116: 19736–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK(1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Almouzni G(2013) Chromatin and DNA replication. Cold Spring Harb Perspect Biol 5: a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. (2017) CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1): D200–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews CK.(2006) DNA precursor metabolism and genomic stability. FASEB J 20: 1300–1314 [DOI] [PubMed] [Google Scholar]
- Matt A, Kuttich B, Grillo I, Weißheit S, Thiele CM, Stühn B(2019) Temperature induced conformational changes in the elastin-like peptide GVG(VPGVG)3. Soft Matter 15: 4192–4199 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S(2007) Barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH(2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression, and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Meyer KJ, Anderson MG(2017) Genetic modifiers as relevant biological variables of eye disorders. Hum Mol Genet 26(R1): R58–R67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneeswaran G, Kartheeswaran S, Muthukumar K, Karunakaran C(2018) Temperature-dependent conformational dynamics of cytochrome c: Implications in apoptosis. J Mol Graph Model 79: 140–148 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R(1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P(2006) Ribonucleotide reductases. Annu Rev Biochem 75: 681–706 [DOI] [PubMed] [Google Scholar]
- Patel D, Franklin KA(2009) Temperature-regulation of plant architecture. Plant Signal Behav 4: 577–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavloff N, Rivard D, Masson S, Shen SH, Mes-Masson AM(1992) Sequence analysis of the large and small subunits of human ribonucleotide reductase. DNA Seq 2: 227–234 [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA(2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P(2011) Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Maya D(2017) Regulation of replication fork advance and stability by nucleosome assembly. Genes (Basel) 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R, Zeng D, Liang R, Yang C, Akhter D, Alamin M, Jin X, Shi C(2017) Rice gene SDL/RNRS1, encoding the small subunit of ribonucleotide reductase, is required for chlorophyll synthesis and plant growth development. Gene 627: 351–362 [DOI] [PubMed] [Google Scholar]
- Qiu W, Zhou B, Darwish D, Shao J, Yen Y(2006) Characterization of enzymatic properties of human ribonucleotide reductase holoenzyme reconstituted in vitro from hRRM1, hRRM2, and p53R2 subunits. Biochem Biophys Res Commun 340: 428–434 [DOI] [PubMed] [Google Scholar]
- Reichard P.(1988) Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem 57: 349–374 [DOI] [PubMed] [Google Scholar]
- Ritchie SW, Hanway JJ, Benson GO(1992) How a Corn Plant Develops. Iowa State University Extension Special Report Iowa State University, Ames, IA [Google Scholar]
- Serra-Cardona A, Zhang Z(2018) Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem Sci 43: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O, Mironov V, Burssens S, Van Montagu M, Inze D(1996) Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc Natl Acad Sci USA 93: 4868–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A, Culler AH, Gallavotti A, Jackson D, Cohen JD, McSteen P(2009) BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol 50: 652–657 [DOI] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM(2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16 [DOI] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H(2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano HY(2013) Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol 54: 313–324 [DOI] [PubMed] [Google Scholar]
- Tang C, Tang S, Zhang S, Luo M, Jia G, Zhi H, Diao X(2019) SiSTL1, encoding a large subunit of ribonucleotide reductase, is crucial for plant growth, chloroplast biogenesis, and cell cycle progression in Setaria italica. J Exp Bot 70: 1167–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Matsushima R, Sakamoto W(2012) Mutations defective in ribonucleotide reductase activity interfere with pollen plastid DNA degradation mediated by DPD1 exonuclease. Plant J 70: 637–649 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Hake S(2009) Translational biology: From Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z(2017) agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L(2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L(2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlögelhofer P(2007) A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J 26: 5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, et al. (2016) Integration of omic networks in a developmental atlas of maize. Science 353: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z(2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Noshay J, Burghardt LT, Hirsch CN, Hirsch CD, Springer NM(2017) Natural variation for gene expression responses to abiotic stress in maize. Plant J 89: 706–717 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A(2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Woodward JB, Abeydeera ND, Paul D, Phillips K, Rapala-Kozik M, Freeling M, Begley TP, Ealick SE, McSteen P, Scanlon MJ(2010) A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell 22: 3305–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A, Williams ME, Bourett TM, Hu W, Hou Z, Meeley RB, Jaqueth J, Dam T, Li B(2014) A pair of homeolog ClpP5 genes underlies a virescent yellow-like mutant and its modifier in maize. Plant J 79: 192–205 [DOI] [PubMed] [Google Scholar]
- Xue Z, Xu X, Zhou Y, Wang X, Zhang Y, Liu D, Zhao B, Duan L, Qi X(2018) Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat Commun 9: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Skirpan A, Wardell B, Matthes MS, Best NB, McCubbin T, Durbak A, Smith T, Malcomber S, McSteen P(2019) The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant 12: 374–389 [DOI] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC(2009) Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150: 388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Han J, Kim YJ, Song M, Yang Z, He Y, Fu R, Luo Z, Hu J, Liang W, et al. (2017) Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc Natl Acad Sci USA 114: 12327–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yuan Z(2014) Molecular control of grass inflorescence development. Annu Rev Plant Biol 65: 553–578 [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu C, He Y, Zong J, Yang X, Si H, Sun Z, Hu J, Liang W, Zhang D(2013) Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc Natl Acad Sci USA 110: 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun L, Rossmann MG(2015) Temperature dependent conformational change of dengue virus. Curr Opin Virol 12: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu Q, Li J, Jiang D, Zhou L, Wu P, Lu S, Li F, Zhu L, Liu Z, et al. (2012) Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res 22: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L, et al. (2014) RNase Z(S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun 5: 4884–4892 [DOI] [PubMed] [Google Scholar]
- Zhou X, Liao WJ, Liao JM, Liao P, Lu H(2015) Ribosomal proteins: Functions beyond the ribosome. J Mol Cell Biol 7: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]