The tomato membrane protein SlFERL regulates fruit ripening via modulating ethylene production.
Abstract
Fruit ripening is a complex and genetically programmed process modulated by transcription factors, hormones, and other regulators. However, the mechanism underlying the regulatory loop involving the membrane-protein targets of RIPENING-INHIBITOR (RIN) remains poorly understood. To unravel the function of tomato (Solanum lycopersicum) FERONIA Like (SlFERL), a putative MADS-box transcription factor target gene, we investigated and addressed the significance of SlFERL in fruit ripening by combining reverse genetics, biochemical, and cytological analyses. Here, we report that RIN and Tomato AGAMOUS-LIKE1 (TAGL1) directly bind to the promoter region of SlFERL and further activate its expression transcriptionally, suggesting a potential role of SlFERL in fruit ripening. Overexpression of SlFERL significantly accelerated the ripening process of tomato fruit, whereas RNA interference knockdown of SlFERL resulted in delayed fruit ripening. Moreover, a surface plasmon resonance assay coupled with tandem mass spectrometry and a protein interaction assay revealed that SlFERL interacts with the key enzyme S-adenosyl-Met synthetase 1 (SlSAMS1) in the ethylene biosynthesis pathway, leading to increased S-adenosyl-Met accumulation and elevated ethylene production. Thus, SlFERL serves as a positive regulator of ethylene production and fruit ripening. This study provides clues to the molecular regulatory networks underlying fruit ripening.
Fleshy fruits are important crops worldwide, accounting for a substantial fraction of the world’s agricultural output. Fruit ripening involves sophisticated biochemical and physiological changes in texture, pigmentation, aroma, and flavor during the ripening process, which directly determines the ultimate intrinsic quality and yield (Klee, 2004; Li et al., 2018b; Shinozaki et al., 2018). Therefore, comprehensive understandings of the mechanisms underlying fruit ripening may have both theoretical and practical values for the fruit industry. Based on the respiration pattern exhibited during ripening, fruit are classified into two groups; climacteric fruit, which are characterized by concomitant respiratory peak and ethylene burst upon initiation of ripening, and nonclimacteric fruit, which do not exhibit increased respiration and typically produce ethylene in trace amounts (McMurchie et al., 1972; Alexander and Grierson, 2002). Ethylene plays crucial roles in the ripening of climacteric fruit, for which great efforts have been made to identify components in the ethylene biosynthesis and signaling pathways (Klee, 2004; Ju and Chang, 2015; Cai et al., 2018).
In the transcription factor networks modulating fruit ripening, MADS-box transcription factors function as key regulators of ripening (Gapper et al., 2013). The MADS-box transcription factor RIPENING-INHIBITOR (RIN) is one of the major factors that regulate ripening, involving both ethylene-dependent and ethylene-independent processes (Vrebalov et al., 2002; Li et al., 2019). Originally, the rin mutant was found to exhibit a severe ripening-defective phenotype in which the fruit fail to soften and do not demonstrate a climacteric rise of respiration and ethylene production (Tigchelaar et al., 1978). However, more recent studies have suggested that rin mutation causes fusion of truncated RIN with adjacent MC genes (RIN-MC), which is a gain-of-function mutation producing a protein that represses ripening (Vrebalov et al., 2002; Ito et al., 2017; Li et al., 2018c). RIN mainly binds to the C-A/T-rich-G (the consensus CArG) motifs and interacts with the promoters of many ripening-related genes. Chromatin immunoprecipitation (ChIP) assays coupled with DNA microarray analysis (ChIP-chip) have been performed for genome-wide identification of direct RIN target genes in tomato (Solanum lycopersicum; Fujisawa et al., 2013). The first RIN ChIP-sequencing (ChIP-seq) analysis reported >4,000 genes harboring RIN binding sites, of which 292 are differentially expressed in the rin mutant when compared to wild-type fruit (Zhong et al., 2013). Further, RIN ChIP-seq at a higher sequencing depth found >10,000 RIN binding sites genome-wide, and most of them are co-occupied with Tomato AGAMOUS-LIKE1 (TAGL1; Lü et al., 2018). In addition, the in vivo transcriptional activity of RIN also required other MADS transcription factors, such as TAGL1 (Leseberg et al., 2008). Functional annotation of these targets may reveal functions of MADS-box transcription factors in a wide range of ripening-related processes, especially in ethylene production and signaling.
Originally identified as a membrane protein mediating male-female interaction (Huck et al., 2003), FERONIA (FER) belongs to the Catharanthus roseus receptor-like kinase1-like protein family (CrRLK1Ls) in Arabidopsis (Arabidopsis thaliana). Recently, it has also emerged as an important regulatory factor in many aspects of plant growth and development, including fertilization (Escobar-Restrepo et al., 2007; Duan et al., 2014, 2020), vegetative growth (Duan et al., 2010; Li et al., 2015), and responses to external biotic and abiotic stimuli (Keinath et al., 2010; Yu et al., 2012; Stegmann et al., 2017; Guo et al., 2018). FER also employs the conserved regulator ErbB3-binding protein1 (EBP1) and phosphorylates eIF4E1 to regulate cell growth (Li et al., 2018a; Zhu et al., 2020). Although FER is also implicated in ethylene production in Arabidopsis and apple (Malus domestica) fruit (Deslauriers and Larsen, 2010; Kessler et al., 2010; Mao et al., 2015; Jia et al., 2017), the relationship between FER and ripening-related transcription factors during autocatalytic ethylene production is still poorly understood. In this study, a homolog of Arabidopsis FERONIA, Solanum. lycopersicum FERONIA Like (SlFERL), was found to be involved in the regulation of fruit ripening in tomato. Its expression at both the mRNA and protein levels persistently increased during fruit ripening. Overexpression (OE) of SlFERL significantly accelerated the fruit ripening process, and the genes involved in various ripening-related processes were upregulated to different amplitudes, whereas the RNA interference (RNAi) lines displayed opposite phenotypes. A surface plasmon resonance assay coupled with tandem mass spectrometry (SPR-MS/MS) and a protein-protein interaction assay confirmed that SlFERL interacted with the key enzyme SlSAMS1 to modulate ethylene biosynthesis, in accordance with the results for ethylene production, lycopene accumulation, and other ripening-related phenotypes. These findings demonstrate that SlFERL positively regulates fruit ripening by modulating ethylene biosynthesis, and they add further insights to our understanding of the molecular network of fruit ripening regulation.
RESULTS
RIN and TAGL1 Bind to the Promoter Region of SlFERL and Activate Its Transcription
In a large-scale identification of direct targets of RIN in tomato fruit (Fujisawa et al., 2013), a total of 241 potential candidates for RIN binding were identified; however, it is unclear how these potential targets are elaborately regulated with regard to various traits related to the ripening process. In this study, a detailed examination of the expression profiles of these genes during fruit ripening was first performed using TomExpress database (http://tomexpress.toulouse.inra.fr; Zouine et al., 2017). Hierarchical clustering analysis revealed a total of 64 genes that showed significant 2-fold elevated or decreased transcript levels during fruit ripening (Supplemental Fig. S1A; Supplemental Dataset S1). In addition, we paid attention to the proteins in the plasma membrane (PM)-cell wall continuum under the cell component category following subcellular localization and Gene Ontology (GO) analysis, because these proteins may be closely related to fruit softening and responses of fruit to biotic/abiotic stresses at the cell-environment interface (Martin and Rose, 2014; Liu et al., 2015; Franck et al., 2018). A total of four nonredundant membrane proteins were identified in the 64 proteins (Supplemental Fig. S1, B and C; Supplemental Dataset S2). Among these proteins, a putative membrane protein (Solyc09g015830) was identified that showed characteristic variation in its expression level during fruit ripening.
As shown in protein sequence analysis using SMART (http://smart.embl.de/; Letunic et al., 2015), Solyc09g015830 possessed a conserved malectin domain, suggesting that it may belong to a previously identified family of C. roseus receptor-like kinase1-like proteins (CrRLK1Ls). In a previous study, a screen of the tomato genome revealed a CrRLK1L family consisting of 23 members (Sakamoto et al., 2012). Phylogenetic analysis of these CrRLK1L homologs with Arabidopsis CrRLK1Ls revealed that Solyc09g015830 was clustered in the same clade with AtFERONIA (AtFER) and had high amino acid sequence identity with it (Supplemental Fig. S1D). Therefore, it can be viewed as a homolog of AtFER and designated as SlFERL hereafter. Notably, in combination with the National Center for Biotechnology Information (NCBI) nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/), further sequencing for SlFERL showed that its sequence had one more malectin-like domain compared with the previously reported Solyc09g015830, and this was confirmed by immunoblot with the polyclonal antibody for SlFERL (anti-SlFERL). The open reading frame (ORF) of SlFERL was 2,670 bp in length and encoded a protein composed of 889 amino acids (GenBank: NC_015446; Supplemental Figs. S2 and S3). Similar to AtFER, SlFERL was composed of a signal peptide (1–27 amino acids), two malectin-like domains (38–171 and 223–374 amino acids), a transmembrane region (444–466 amino acids), and a typical kinase domain (538–803 amino acids; Supplemental Fig. S2, A and B).
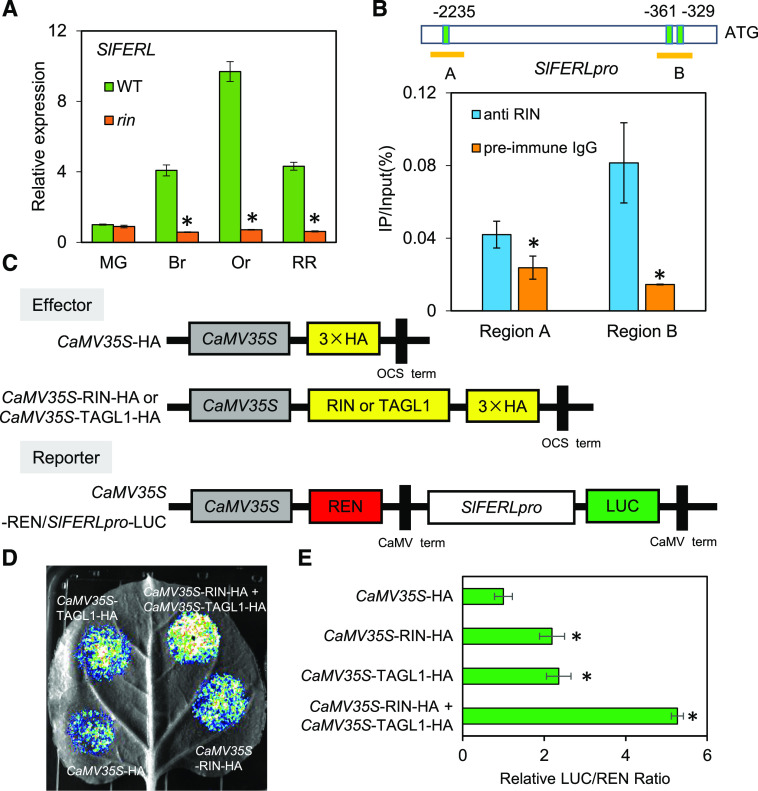
As shown in Figure 1A, its expression increased persistently in wild-type fruit during ripening, but did not vary significantly in the rin mutant, corresponding well with the RNAseq data in SOL Genomics Network (https://solgenomics.net/). Therefore, we were prompted to analyze the sequence of its promoter region across a 2,500-bp range upstream of the initiation codon ATG in order to ascertain its relationship with RIN. As a consequence, three C-A/T-rich-G (CArG box) motifs were detected at −2,235, −361, and −329 bp, implying that SlFERL expression may be substantially regulated by RIN binding at the transcriptional level on these three sites. To further confirm this hypothesis, ChIP with reverse transcription qPCR (ChIP-qPCR) analysis was performed to detect whether the three CArG box elements were enriched by RIN antibody. The results demonstrated that at least two CArG box elements were enriched by RIN antibody, as compared to the control with preimmune IgG (Fig. 1B). Moreover, as two previous RNA-seq analyses established that the SlFERL promoter harbors multiple RIN and TAGL1 binding sites (Zhong et al., 2013; Lü et al., 2018), to explore whether RIN and TAGL1 can regulate the activity of the SlFERL promoter in vivo, we further performed a dual-luciferase reporter assay in Nicotiana benthamiana leaves by coexpressing a reporter construct of firefly luciferase (LUC) driven by the SlFERL promoter and an effector construct expressing the RIN or TAGL1 protein (Fig. 1C). As shown in Figure 1D, CaMV35S:RIN-HA and CaMV35S:TAGL1-HA activated the LUC reporter gene, and the LUC/REN ratio of RIN and TAGL1 was significantly higher than that of the negative control (Fig. 1E). These data suggest that RIN and TAGL1 can bind to the promoter region and transcriptionally activate SlFERL.
Figure 1.
RIN and TAGL1 bind to the SlFERL promoter and activate its transcription. A, RT-qPCR analysis showing SlFERL expression in wild-type (WT) and rin. B, ChIP-qPCR assay showing enrichment of both CArG box elements by RIN antibody compared to the control with preimmune IgG. Three binding motifs of transcription factor RIN were found in the SlFERL promotor and divided in two regions (A and B) to carry out qPCR. Values are shown as the means ± sd. Asterisks indicate significant difference by Student’s t test (*P < 0.05). C to E, Transient expression of RIN and TAGL1 simultaneously enhances the promoter activity of SlFERL. The effector and reporter (C) were coexpressed in N. benthamiana leaves mediated by A. tumefaciens strain GV3101. After 24 h, the LUC image was captured (D). The activation of SlFERL promoter by RIN, TAGL1, and RIN+TAGL1 is shown by the LUC/REN ratio (E). Data are based on at least six replicates and represented as means ± sd. Asterisks indicate significant difference by Student’s t test (*P < 0.05).
SlFERL Is Ubiquitously Expressed in Tomato
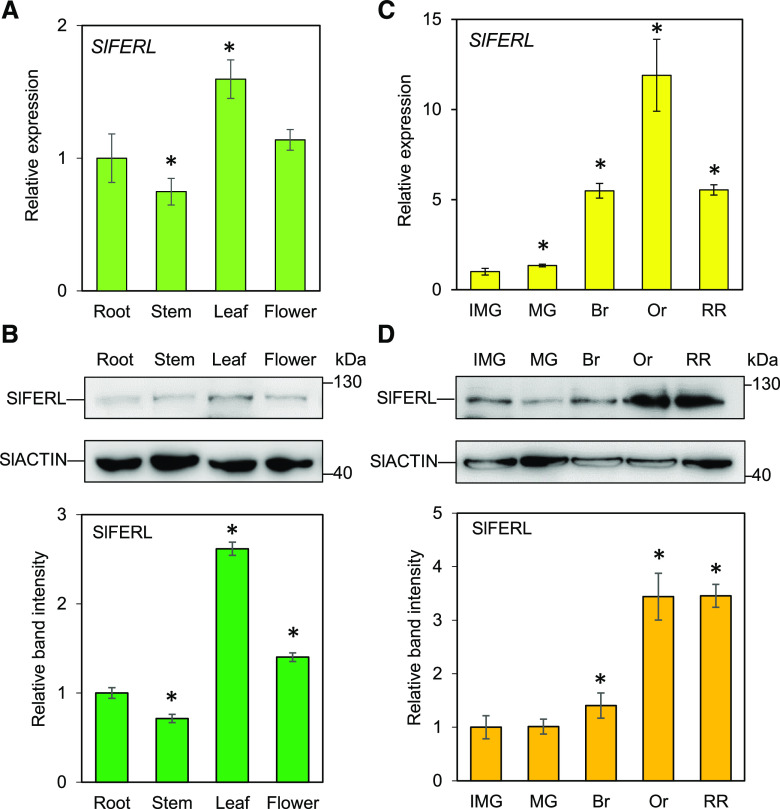
To ascertain the spatiotemporal expression pattern of SlFERL, RT-qPCR analysis was performed to evaluate the transcript abundance in various tissues, particularly in the process of fruit ripening. The results showed that SlFERL was ubiquitously expressed at relatively high levels in all the tissues examined, as indicated by RT-qPCR and immunoblot results (Fig. 2). Notably, the expression of SlFERL was almost persistently upregulated upon the onset of fruit ripening, and it reached its peak level at the orange stage (Or) and slightly decreased at red ripe (RR) stage (Fig. 2C), which displayed a similar pattern to RIN and coincided with the curve for autocatalytic ethylene burst (Shinozaki et al., 2018). SlFERL protein level began to increase at the breaker (Br) stage, then reached its peak level at the Or stage and was maintained at a stable level afterwards (Fig. 2D).
Figure 2.
SlFERL is ubiquitously expressed in various organs. A and B, Expression of SlFERL at mRNA (A) and protein (B) levels in root, stem, leaf, and flower. C and D, Expression of SlFERL at mRNA (C) and protein (D) levels during fruit ripening. SlACTIN was chosen as an internal control. Values are means ± sd of three replicates. The protein levels were quantified using the gray values for corresponding protein bands in at least three gels (represented as means ± sd) in ImageJ. Asterisks indicate significant difference by Student’s t test (*P < 0.05).
Subcellular Localization of SlFERL
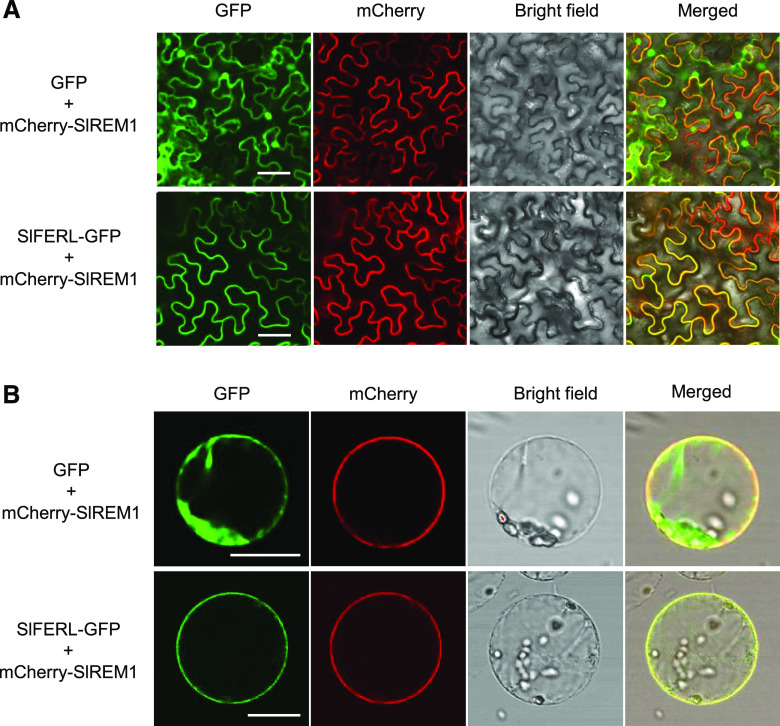
We were curious to know whether SlFERL may have subcellular localization identical to that of its counterpart AtFER in Arabidopsis (Duan et al., 2010). To check the subcellular localization of SlFERL, the full-length coding sequence (CDS) of SlFERL was fused to GFP and transiently expressed in epidermal cells of N. benthamiana leaves by infiltration with Agrobacterium tumefaciens. As expected, strong fluorescence signal of SlFERL-GFP was exclusively detected in the PM, whereas free GFP fluorescence of the control empty vector was observed in the cytoplasm and nucleus (Fig. 3A). To further confirm the PM localization of SlFERL, SlFERL-GFP was co-expressed with mCherry-SlREM1, a previously reported PM marker protein in leaf epidermal cells (Cai et al., 2018). We observed that SlFERL-GFP colocalized with mCherry-SlREM1 in the PM of protoplasts derived from N. benthamiana leaves (Fig. 3B).
Figure 3.
SlFERL localizes to the PM. SlFERL-GFP and mCherry-SlREM1 were coexpressed in epidermal cells of N. benthamiana leaves. Fluorescence images of epidermal cells of N. benthamiana (A) and protoplasts (B) were taken at 36 h after infiltration. The empty vector carrying GFP was chosen as a negative control for this assay. Scale bars = 25 μm.
SlFERL Is Involved in Tomato Fruit Ripening
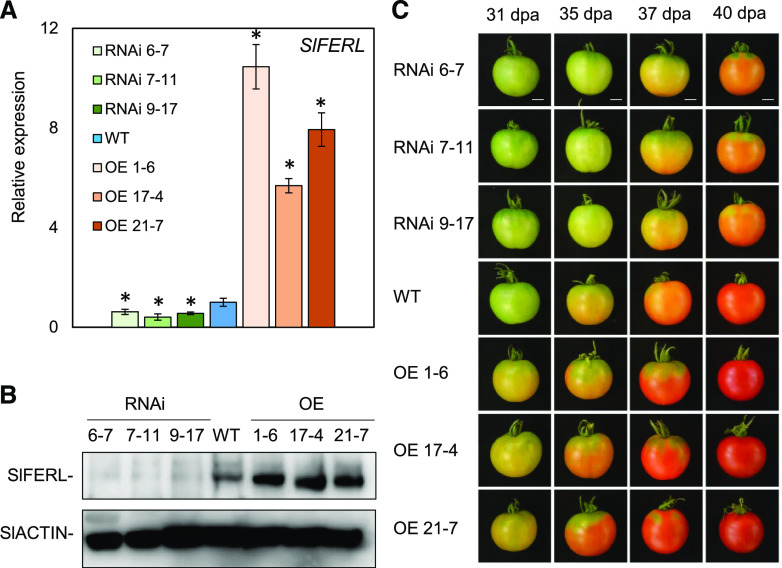
To further address the physiological functions of SlFERL during fruit ripening, stably transformed OE lines and RNAi lines for SlFERL were generated in the background of wild-type seedlings (cv Ailsa Craig) by A. tumefaciens-mediated genetic transformation. In total, 12 independent OE lines and 10 independent RNAi lines were obtained. Finally, three independent homozygous T2 transgenic OE and RNAi lines were identified by RT-qPCR analysis and used for further analysis.
As shown in Figure 4A, the transcriptional level of SlFERL was significantly higher in the OE lines and significantly downregulated in the RNAi lines. Expression for several close members with SlFERL was not significantly affected compared to the control, suggesting that SlFERL was specifically silenced (Supplemental Fig. S4). An immunoblotting assay using SlFERL antibody (anti-SlFERL) also showed that SlFERL was successfully overexpressed in the OE lines and downregulated in the RNAi lines (Fig. 4B). In terms of the fruit ripening process, obvious macroscopic changes in fruit color were observed at 35 d post anthesis (dpa). When wild-type fruit began to turn orange, RNAi fruit were still green, whereas the fruit for the SlFERL OE lines had attained an orange color (Fig. 4C). In comparison to wild-type fruit, the time span from anthesis to Br stage was delayed or accelerated by ∼3 to 4 d in SlFERL RNAi or OE fruit. These data indicate that SlFERL affects fruit ripening.
Figure 4.
RNAi or OE of SlFERL alters the fruit ripening process. A, SlFERL expression at the mRNA level in wild-type (WT) and transgenic lines. Total RNA was isolated from tomato fruit pericarp at 31 dpa from wild-type, OE (OE 1-6, OE 17-4, and OE 21-7), and RNAi (RNAi 6-7, RNAi 7-11, and RNAi 9-17) lines. SlACTIN was used as an internal control. Values are means ± sd of three replicates. Asterisks indicate significant difference by Student’s t test (*P < 0.05). B, SlFERL expression at protein level in wild-type and transgenic lines. Total protein was extracted from the fruit of wild-type and transgenic lines. SlACTIN was chosen as an internal control. C, Fruit ripening phenotype in wild-type and transgenic lines at 31, 35, 37, and 40 dpa. Scale bars = 1 cm.
SlFERL Affects Lycopene Accumulation and Ethylene Production of Fruit
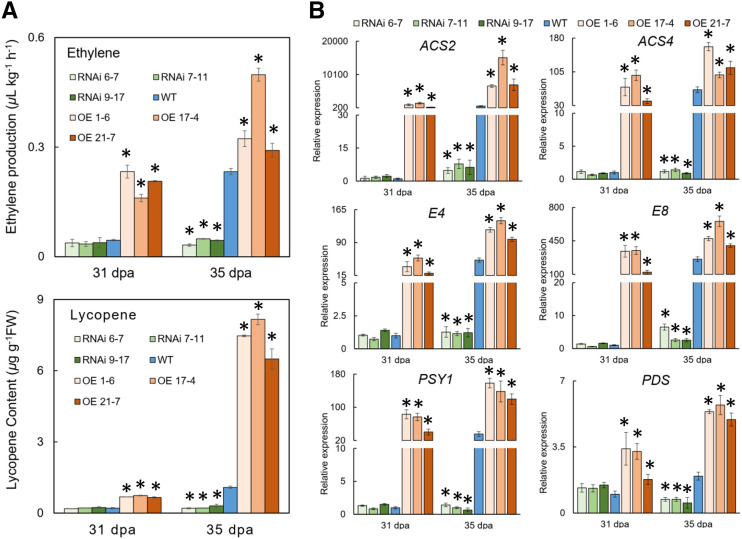
To further examine the underlying changes in other aspects, ethylene production and lycopene accumulation, two principal traits during the unripe-to-ripe phase transition, were investigated. As shown in Figure 5A, ethylene production was significantly higher in the OE fruit than in wild-type fruit, but it was much lower in the RNAi fruit. Moreover, the transcripts for several key enzymes involved in ethylene biosynthesis, namely ACS2 and ACS4, dramatically increased or decreased in the OE or RNAi fruit (Fig. 5B), which coincided well with altered ethylene production in the transgenic fruit (Fig. 5A). The ethylene-responsive and ripening-related genes E4 and E8 also demonstrated drastic increases or decreases in the OE or RNAi fruit compared with the wild type (Fig. 5B). Similarly, lycopene accumulation in the OE lines was significantly higher than in wild-type and RNAi lines at 31 and 35 dpa (Fig. 5A). Coincidently, the expression levels for the genes encoding phytoene synthase (PSY) and phytoene desaturase (PDS) were significantly upregulated at 31 and 35 dpa in the OE lines, whereas RNAi lines showed the opposite results (Fig. 5B).
Figure 5.
RNAi or OE of SlFERL changes ethylene production, lycopene content, and expression of ripening-related genes. A, Ethylene and lycopene content detection for wild type (WT) and transgenic lines. Fruit were harvested at 31 and 35 dpa for ethylene or lycopene measurement. Values are shown as the means ± sd of three replicates. B, RT-qPCR analyses for the genes related to ethylene production or lycopene biosynthesis. ACS2 and ACS4, aminocyclopropane-1-carboxylic acid synthases 2 and 4; E4 and E8, ethylene response genes 4 and 8; PSY1, phytoene synthase 1; PDS, phytoene desaturase. SlACTIN was used as an internal control. Error bars represent ± sd of three replicates. Aserisks indicate significant difference by Student’s t test (*P < 0.05).
Prokaryotic Expression of SlFERL-KD and SPR-MS/MS Analysis
To further explore substantial partners by which SlFERL was involved in fruit ripening, SPR-MS/MS was conducted to identify SlFERL-interacting proteins (Supplemental Figs. S5B and S6, A and B). As the SPR assay requires purified analytes, soluble SlFERL-KD (amino acids 467–889) was separated by affinity purification using nickel-nitrilotriacetic acid agarose resin. The protein was found to be stable during purification, as determined by SDS-PAGE (Supplemental Fig. S5A).
CM5 sensor chips of research grade (catalog no BR-1000-14, Biacore) were used for SPR experiments performed in a Biacore 3000 biosensor (GE Healthcare) at 4°C. SlFERL-KD was used as an immobilized ligand on the CM5 sensor chip, and tomato fruit at Br stage were used to prepare the interacting protein solution. Proteins bound to the immobilized SlFERL-KD were subjected to Nano-liquid chromatography MS/MS analysis, and the experiments were performed in duplicate (Supplemental Dataset S3). In total, 188 proteins in common were found in the two independent experiments, including SlFERL-KD (Supplemental Fig. S5C; Supplemental Dataset S4). GO analysis was performed to allocate these proteins to various functional categories, as shown in Supplemental Figure S5D and Supplemental Dataset S5. Interestingly, S-adenosyl-Met synthetase1 (SlSAMS1; Solyc01g101060) and SlSAMS2 (Solyc12g099000), two key enzymes in the ethylene biosynthesis pathway (Supplemental Dataset S5), were detected.
SlFERL Interacts with SlSAMS1
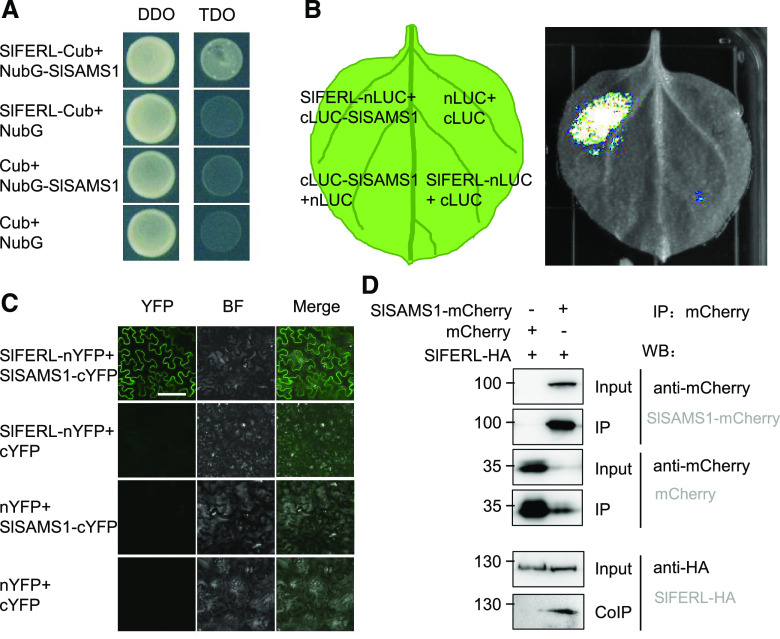
As SPR-MS/MS analysis detected SlSAMS1 and SlSAMS2 as potential interacting proteins for SlFERL, we hypothesized that SlFERL may interact with components in the ethylene biosynthesis pathway to modulate ethylene production. This hypothesis was tested by split ubiquitin membrane yeast two-hybrid (mY2H) analysis, which has been widely proven to be suitable for studying membrane protein interactions (Obrdlik et al., 2004). As a result, only the yeast cells co-transformed with SlFERL-Cub and NubG-SlSAMS1 could grow normally on the Triple dropout (TDO) medium (synthetic defined [SD] /–His/–Leu/–Trp), whereas none of the negative controls could grow on TDO (Fig. 6A), indicating that SlFERL could interact with SlSAMS1 in yeast cells. The same assay was performed to test the interaction between SlFERL and SlSAMS2, but as shown in Supplemental Figure S7, SlFERL did not interact with SlSAMS2.
Figure 6.
SlFERL interacts with SlSAMS1. A, Interaction between SlFERL and SlSAMS1 in yeast split-ubiquitin assay. Yeast AH109 cells cotransformed with NubG-SlSAMS1 with SlFERL-Cub resulted in growth of yeast cells on TDO medium. DDO, Double dropout medium (SD/–Leu/–Trp); TDO, triple dropout medium (SD/–His/–Leu/–Trp). B, Firefly luciferase complementation imaging (LCI) assay for interaction between SlFERL1-nLUC and cLUC-SlSAMS1 in N. benthamiana. C, Bimolecular fluorescence complementation (BiFC) detection of interaction between SlFERL1-nYFP and SlSAMS1-cYFP in N. benthamiana epidermal cells. Images were captured under a confocal microscope at 2 d postinfiltration. Bar = 100 μm, the scale bar for the top left image apply to all images. D, co-IP detection of interaction between SlFERL and SlSAMS1 in N. benthamiana leaves with antibodies against mCherry or HA at 2 d postinfiltration.
To further confirm the interaction between SlFERL and SlSAMS1, bimolecular fluorescence complementation (BiFC), split luciferase complementation, and coimmunoprecipitation assays were performed. The leaf areas cotransformed with SlFERL-nLUC and cLUC-SlSAMS1 displayed strong luciferase luminescence, whereas those cotransformed with SlFERL-nLUC/cLUC or nLUC/cLUC-SlSAMS1 exhibited no signal (Fig. 6B), indicating that SlFERL interacts with SlSAMS1. Similarly, after coinfiltration with the A. tumefaciens strains harboring constructs for BiFC assay, it was found that SlFERL can interact with SlSAMS1 on the PM (Fig. 6C). As shown in Figure 6D, the hemagglutinin-tagged SlFERL (SlFERL-HA) was coimmunoprecipitated by SlSAMS1-mCherry. Taken together, these results suggest that SlFERL interacts with SlSAMS1.
SlFERL and SlSAMS1 Synergistically Regulate Fruit Ripening
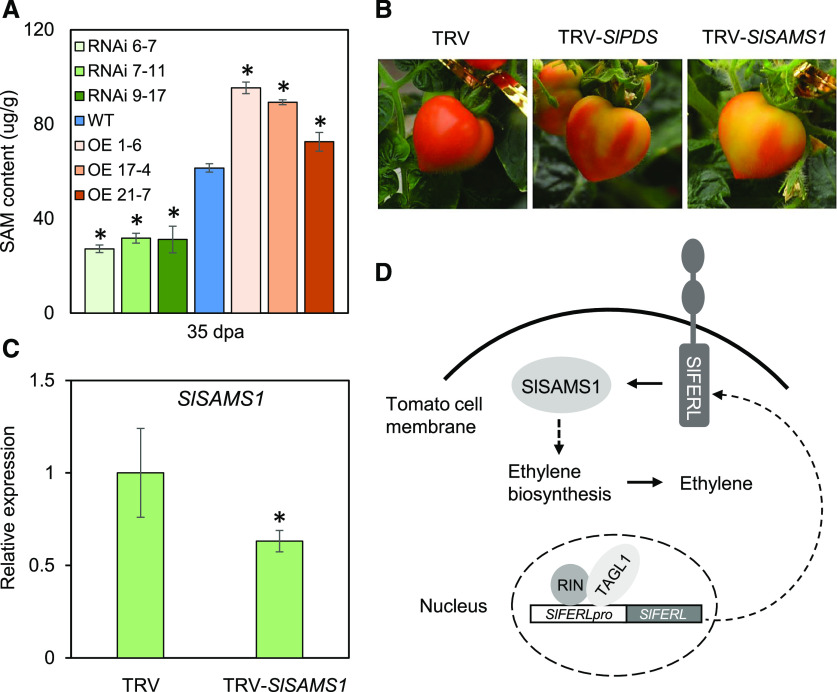
In a genome-wide screening for members of the SAMS family in tomato, four genes (SlSAMS1–SlSAMS4) were retrieved, although only SlSAMS1 showed a relatively high expression level during fruit ripening (Supplemental Fig. S8). As shown in Supplemental Figure S8, the expression of SlSAMS1 significantly decreased at the Br stage and was maintained at a stable level afterwards. To ascertain the interaction between SlFERL and SlSAMS1 in modulating ethylene production, we predicted that OE fruit would accumulate more SAM. To test this possibility, we examined the transcript levels of SlSAMSs and measured SAM content in OE and RNAi plants using HPLC (Van de Poel et al., 2010; Bulens et al., 2011). The OE fruit contained significantly higher levels of SAM compared to the wild-type control, whereas the RNAi fruit accumulated less SAM, corresponding well with the transcript level of SlSAMS1 (Fig. 7A; Supplemental Fig. S9). The SAM content measurements suggested that SlFERL may positively regulate SlSAMS1 activity. To investigate the potential role of SlSAMS1 in tomato fruit ripening, we performed a virus-induced gene silencing (VIGS) assay to downregulate the mRNA level of SlSAMS1 and further examined the phenotype related to fruit ripening. As shown in Figure 7B, the fruit showed an uneven coloration phenotype and the SlSAMS1 mRNA level was downregulated in the yellow part (Fig. 7C), suggesting that the fruit ripening process was delayed when SlSAMS1 was silenced. These results indicate that SlSAMS1 positively modulates fruit ripening.
Figure 7.
SlFERL and SlSAMS1 synergistically regulate fruit ripening. A, HPLC analysis for S-adenosyl-Met level in wild type (WT), OE, and RNAi fruit. Error bars represent means ± sd of three replicates. Asterisks indicate significant difference by Student’s t test (*P < 0.05). B, VIGS assay for SlSAMS1 in MicroTom fruit shows chimeric coloration in pericarp. C, RT-qPCR analysis of SlSAMS1 expression in VIGS fruit. Values are shown as the means ± sd of three replicates. Asterisks indicate significant difference by Student’s t test (*P < 0.05). D, A hypothetical working model proposed for SlFERL and SlSAMS1 in fruit ripening.
Collectively, a hypothetical model was proposed for the function of SlFERL in fruit ripening (Fig. 7D). After the transcription factors MADS-box RIN and TAGL1 bind to SlFERL promoter and activate its expression, SlFERL may interact with SlSAMS1 and positively affect SlSAMS1 activity, leading to SAM accumulation and elevated ethylene production in fruit, ultimately accelerating fruit ripening.
DISCUSSION
Fruit develop from carpels or adjacent floral tissues. They undergo sophisticated reprogramming of the gene expression network upon ripening, during which the bulk of genes may be activated or suppressed in a highly coordinated manner, eventually affecting fruit color, aroma, flavor, texture, nutritional contents, and other attributes (Klee and Giovannoni, 2011; Yang et al., 2019). This process is regulated by ethylene and major transcription factors with multiple gene targets (Wang et al., 2017; Cai et al., 2018; Zhou et al., 2019). Here, we report that a homologous gene to AtFER in tomato, SlFERL, was ubiquitously expressed in various tissues of tomato. The expression level of SlFERL was particularly high in fruit, which almost persistently increased during fruit ripening (Fig. 2).
The MADS-box transcription factor RIN has been reported as a crucial regulator of various aspects during the ripening process (Vrebalov et al., 2002; Giovannoni et al., 2017). Previous ChIP-Seq and ChIP-qPCR assays have revealed that RIN directly targets the promoter regions of ripening-related genes in tomato, including aroma formation genes ADH2 and LoxC, ubiquitin-proteasome related genes SlUBC32 and PSMD2), protease-coding gene VPE3, and other transcription factor genes CNR, NOR, and FUL1/2 (Qin et al., 2012; Fujisawa et al., 2013; Wang et al., 2014, 2017). When this MADS loop is activated in ripening fruit, a trace quantity of ethylene can rapidly drive the expression of downstream ripening genes (Lü et al., 2018). Although RIN binding sites have been identified previously using ChIP-chip and further microarray for ChIPed DNA samples (Fujisawa et al., 2011, 2013), a recent RIN ChIP-seq analysis identified more than 10,000 binding sites genome wide, most of which are also occupied with another MADS-box transcription factor, TAGL1 (Lü et al., 2018). In this study, the ChIP-qPCR assay demonstrated that RIN directly binds to the promoter region of SlFERL and further activates its transcription (Fig. 1), whereas TAGL1 also functions in activating its transcription, which corresponded well with the previously reported results identifying Solyc09g015830 (SlFERL in this study) as one of the potential targets of RIN and TAGL1 (Fujisawa et al., 2013; Zhong et al., 2013; Lü et al., 2018). Moreover, the expression level of SlFERL persistently increased during fruit ripening, showing an expression pattern similar to that of RIN during tomato fruit ripening.
Climacteric fruit, such as tomatoes and apples, display concurrent ethylene burst and respiratory peak at the commencement of ripening, further giving rise to ripening-related traits (Giovannoni, 2007). During this process, ethylene is utilized as a universal ripening signal for climacteric fruit, which are often harvested at low maturity and further treated to accomplish ripening (Gapper et al., 2013; Lü et al., 2018). However, excessive ethylene always results in rapid deterioration of fruit. Consequently, modulation of ethylene synthesis or signaling is of great practical importance during postharvest storage, shipping, and maintenance of intrinsic quality (Tian, 2013). In this study, the evidence from ChIP-qPCR analysis suggested that SlFERL may function in fruit ripening. Coupled SPR-MS/MS screening identified a key enzyme in the ethylene biosynthesis pathway, namely S-adenosyl-Met synthetase (SlSAMS1), as a coprecipitated protein with SlFERL. These results suggest that there is a substantial correlation between SlFERL and ethylene production. Further in vitro and in vivo experimental evidence confirmed the interaction between SlFERL and SlSAMS1 (Fig. 6).
S-adenosyl-l-Met synthetase (EC 2.5.1.6) catalyzes the conversion from l-Met to S-adenosyl-l-Met, which serves as an ethylene precursor (McMurchie et al., 1972). Although isoforms of ethylene biosynthesis genes that are specifically required for ethylene production in systems I and II have been identified in other studies, their regulation remains enigmatic, and results focusing on the correlation of SAM and fruit ripening are scarce (Wang et al., 2002; Li et al., 2019). Given that ethylene burst during climacteric ripening requires efficient recycling metabolism by the Yang cycle to deal with the high demand of Met consumption (Baur and Yang, 1972), SAM is a potential point of ethylene control as the intermediate metabolite between Met and 1-aminocyclopropane-1-carboxylic acid (ACC). Rice dwarf virus-encoded Pns11 increases the susceptibility of rice seedlings to Rice dwarf virus by interacting with OsSAMS1, thereby enhancing its enzymatic activity and leading to increasing production of SAM, ACC, and ethylene (Zhao et al., 2017). In this study, we found that SlFERL regulates tomato fruit ripening likely via mediating ethylene production by direct interaction with the ethylene biosynthesis enzyme SlSAMS1 (Fig. 6). As shown in previous reports (Van de Poel et al., 2013; Shinozaki et al., 2018) and in the expression pattern analysis in this study, SlSAMS1 is the only gene in the SlSAMS family that is involved in ethylene production in fruit. Its expression was observed to be stable at the immature green (IMG) and mature green (MG) stages, but began to decrease at the Br stage and was further maintained at a relatively low level afterward (Supplemental Fig. S8). Since high amounts of cellular SAM may inhibit ACS activity in vitro (Satoh and Yang, 1988), the SAM level may be stringently controlled during climacteric ripening (Van de Poel et al., 2013). At the IMG and MG stages, the expression of SlSAMS1 was maintained at a relatively high level, which may be appropriate for system I ethylene production. However, because trace quantities of ethylene could rapidly drive the expression of ripening-related genes, SlSAMS1 expression was suppressed at the commencement of fruit ripening, at least in response to autocatalytic ethylene production. All these results suggest that elaborate control of SAM levels is essential during high ethylene production rates. As SAM synthesis is an early step in ethylene production in plants (Yang and Hoffman 1984; Wang et al., 2002), higher ethylene levels may be a consequence of higher SAM levels, which control the initiation of changes in color, aromas, texture, flavor, and other biochemical and physiological attributes.
It was reported that AtFER interacts with S-adenosyl-Met synthetase and further negatively modulates SAM level and ethylene biosynthesis in Arabidopsis, which may be attributed to ethylene production, polyamine signaling, and methylation (Mao et al., 2015). Similarly, Jia et al. (2017) reported that MdFERL6 and MdFERL1 physically interact with MdSAMS, thereby negatively modulating ethylene production. MdFERL6 was expressed at a high level during early fruit development, but dramatically declined upon fruit ripening, implying that MdFERL6 may limit ethylene production prior to fruit development but induce ethylene production during fruit ripening (Jia et al., 2017). However, different from the expression patterns for MdFERL6 and MdFERL1, SlFERL was almost continuously upregulated upon the onset of tomato fruit ripening, peaking at the Or stage and slightly decreasing at the RR stage (Fig. 2C), which coincided with the expression pattern of RIN and the curve for system 2 ethylene production (Liu et al., 2015). In support of functional correlation between SlFERL and SAM in fruit ripening, it was demonstrated that the OE fruit had elevated levels of SAM and ethylene (Figs. 5A and 7A), suggesting that SlFERL may positively regulate SAM synthetase activity, thereby modulating SAM level and ethylene production. Since SlFERL possesses typical protein domains for a receptor-like kinase, it is initially anticipated that SlFERL may interact with SlSAMS1 by phosphorylation. In an attempt to verify this speculation, phosphorylation sites in SlSAMS1 were predicted using KinasePhos (http://kinasephos.mbc.nctu.edu.tw; Huang et al., 2005), which showed five potential phosphorylation sites (Supplemental Fig. S10A). Consequently, an IP-MS/MS assay was performed to identify SlSAMS1 phosphorylation sites in N. benthamiana leaves transiently coexpressing SlFERL-HA and SlSAMS1-mCherry. The result showed that phosphorylation was not detected at four of the five potential phosphorylation sites in SlSAMS1 at a sequence coverage of 72.8% (Supplemental Fig. S10B). The remaining potential phosphorylation site was not detected in the three biological replicates for N. benthamiana leaves, which may be attributed to the hydrophobicity of the peptide and the limits of MS identification, and a subsequent Phos-tag shift assay also did not show an obvious shift of the electrophoretic band for SlSAMS1. These findings suggest that SlFERL may interact with SlSAMS1 to modulate ethylene production, and that it may not be the result of phosphorylation, at least in the biological context of fruit ripening.
Alternatively, FER may also act as a scaffold protein to maintain proper localization of SAM1 in the PM and cytoplasm, as both the BiFC assay (Fig. 6C) and the colocalization analysis for SlFERL and SlSAM1 (Supplemental Fig. S11) indicated these two proteins interact at the PM. Similar cases have been reported for the assembly of the immune complex composed of RALF23-FLS2-BAK1-FER-LLG1/LLG2 (Stegmann et al., 2017; Xiao et al., 2019), FER may function in recruiting or stabilizing other signaling components. However, more experimental evidence is required to confirm this hypothesis. Haruta et al. (2014) demonstrated that specific regulatory mechanisms contribute differentially to FER downstream signaling, and these mechanisms were diversified among various cell types and tissues. Moreover, FER may work synergistically with other members of the CrRLK1L family, as demonstrated by Ge et al. (2017) in a study showing that Buddha’s Paper Seal1/2 can interact with two pollen-specific FER homologs (ANXUR1/2). This suggests that FER may act as a scaffolding component for the recruitment or assembly of signaling complexes (Keinath et al., 2010). Further identification of context-specific interacting proteins may facilitate our understanding of the diversified biological functions of SlFERL, including those implicated in fruit ripening. In addition, given that SAMSs/MET ADENOSYLTRANSFERASEs (MATs) are involved in both DNA and histone methylation (Zhong et al., 2013; Meng et al., 2018), accumulating evidence has confirmed that DNA methylation/demethylation and histone demethylation are closely related to fruit ripening (Lang et al., 2017; Li et al., 2020; Liu and Lang, 2020), which suggests a further role for the SlFERL-SAMS module in regulating substantial functions of DNA or histone methylation.
In summary, this study highlights an important role of SlFERL as a linker in fruit ripening by correlating RIN modulation upstream of ethylene biosynthesis. The results demonstrate that the MADS-box transcription factors RIN and TAGL1 bind to the promoter of SlFERL and activate its transcription, confirming that SlFERL is a target gene transcriptionally regulated by RIN and TAGL1. Moreover, SlFERL is involved in the regulation of fruit ripening by interacting with the key component in ethylene biosynthesis, SlSAMS1, further modulating ethylene production, lycopene synthesis, and the expression of crucial genes underlying fruit ripening. These results may provide more insight into the elaborate molecular regulatory network composed of key transcription factors, ethylene production, and signaling, as well as potential linker proteins.
MATERIALS AND METHODS
Plant Materials
Solanum lycopersicum ‘Ailsa Craig’ (AC; wild type), transgenic lines for SlFERL, and the rin mutant (cv AC background) were grown under controlled greenhouse conditions. Fruit were harvested at 21, 31, 35, 37, and 40 dpa, corresponding to the IMG, MG, Br, Or, and RR fruit-ripening stages. Pericarp tissues were sampled immediately after harvest, frozen in liquid nitrogen and stored at −80°C for further use.
Gene Cloning and Agrobacterium tumefaciens-Mediated Genetic Transformation
The full-length complementary DNA and protein sequence of SlFERL was obtained from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/) and Sol Genomics Network (https://solgenomics.net/). To generate the 35S:SlFERL construct, the ORF was cloned into pMDC83 between the 35S CaMV promoter and the NOS terminator. To construct the SlFERL RNAi vector, a 300-bp CDS (1,451–1,750 bp) was amplified, cloned into the pCR8/GW/TOPO vector, and ligated to pK7GWIWG2D using the Gateway strategy (Invitrogen). The destination vectors were confirmed by sequencing and respectively transformed into A. tumefaciens strain GV3101. Genetic transformation of tomato (‘AC’) was performed as described by Qin et al. (2016). The primers used are listed in Supplemental Table S1.
Phylogeny and Bioinformatics Analysis
The amino acid sequences were analyzed using ClustalW and a phylogenetic tree was constructed by the neighbor-joining method using MEGA X (Kumar et al., 2018). A bootstrap analysis with 1,000 replicates was performed to evaluate the statistical reliability of the tree topology. The AtFER and SlFERL protein sequences were aligned with Multalin, version 5.4.1 (Corpet, 1988).
Heml was used for hierarchical clustering (Deng et al., 2014) and Plant-mPLoc 2.0 for subcellular localization prediction (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; Chou and Shen, 2010). Gene cellular component and biological process classification used GO (http://geneontology.org/; Ashburner et al., 2000; The Gene Ontology Consortium, 2019). MS data analysis of SlSAMS1 phosphorylation sites used pFind (Chi et al., 2018).
ChIP-qPCR
ChIP was performed as described by Qin et al. (2012), using tomato fruit at the Or stage when the expression of RIN is strongly induced. The fruit pericarp was sliced, fixed with 1% (v/v) formaldehyde for 15 min under vacuum, ground to fine powder in liquid nitrogen, and then subjected to sonication to isolate nuclei. The nuclear pellets were sonicated until the average size of sheared DNA was ∼500 bp. The chromatin complexes were precleared with protein-A agarose and precipitated with affinity-purified polyclonal anti-RIN antibodies or preimmune IgG serum (negative control). The captured protein-DNA complexes were digested with proteinase K and reverse crosslinking. Finally, the immunoprecipitated DNA was purified using a PCR purification column (Qiagen) and analyzed by RT-qPCR.
Dual-LUC Reporter Assay
The double-reporter vector contains a LUC and a CaMV35S promoter-driven REN as the internal control. The SlFERL promoter was inserted into the pGreenII 0800-LUC double-reporter vector (Hellens et al., 2005), whereas HA, RIN-HA, and TAGL1-HA were cloned into the pCAMBIA2300 vector as effectors. All primers used are listed in Supplemental Table S1. The constructed effector and reporter plasmids were cotransformed into Nicotiana benthamiana leaves mediated by A. tumefaciens strain GV3101. After 24 h, the LUC image was captured by the 5200 Multi Chemiluminescent Imaging System (Tanon). LUC and REN luciferase activities were measured by GloMax 20/20 Luminometer (Promega) according to the Dual-Luciferase Reporter Assay System manual (Promega). The results are presented as the ratio of LUC to REN.
VIGS
pTRV1 and pTRV2 VIGS vectors have been described in previous work (Liu et al., 2002). A. tumefaciens strain GV3101 harboring pTRV1 or pTRV2 and its derivatives were used for the VIGS experiments. GV3101 harboring the TRV-VIGS vectors was grown at 28°C in Luria-Bertani medium containing 10 mm MES (pH 5.6) and 20 mm acetosyringone with appropriate antibiotics (gentamicin and rifampicin for GV3101 and kanamycin for pTRV1 or pTRV2). After cultivation overnight (28°C, 200 rpm), A. tumefaciens cells were harvested and resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES [pH 5.6], and 150 mm acetosyringone) to a final OD600 of 2.0 (for both pTRV1 or pTRV2 and its derivatives). A. tumefaciens strains carrying pTRV1 and pTRV2 or the recombinant vectors were mixed in a 1:1 ratio, and left for 2 h at room temperature before infiltration, as previously described (Fu et al., 2005). The ‘Micro Tom’ tomato inflorescence peduncles attached to the fruit were injected with cultures of A. tumefaciens harboring the vectors using a 1-mL syringe. To detect the accumulation of virus and silencing efficiency of specific genes in tomato fruit, RT-qPCR was performed. The primers are listed in Supplemental Tables S1 and S2.
Subcellular Localization
Subcellular localization was examined by fusing mCherry to the N terminus of SlREM1 and GFP to the C terminus of SlFERL. The A. tumefaciens strains GV3101 harboring the recombinant plasmids were infiltrated into the epidermal cells of N. benthamiana. For protoplast isolation, 1 g N. benthamiana leaves were incised with a razor blade into 1- to 3-mm stripes, and the stripes were incubated in isolation buffer (5 mm MES [pH 5.8], 1% [w/v] Cellulase R10, 0.5% [w/v] Macerozyme R10, 400 mm Mannitol, 0.2% [w/v] bovine serum albumin, and 20 mm KCl) for 4 h with gentle shaking (40 rpm) in the dark. Protoplasts were filtered using a 200-μm mesh griddle on ice. The leaves and protoplasts were observed at 48 h postinfiltration using an Olympus FV1000 MPE multiphoton laser scanning confocal microscope.
Protein Extraction
The protein extraction assay was performed according to the methods of Cai et al. (2018). Briefly, 100-mg tomato samples were ground and dissolved in 200 μL protein extraction buffer (100 mm Tris-HCl [pH 7.6] and 4% [w/v] SDS), incubated for 5 min at 95°C and then centrifuged at 16,000g for 15 min. The concentration of isolated protein was determined using the Pierce BCA protein assay kit (Thermo Scientific). The protein samples were analyzed by SDS-PAGE, followed by blotting with corresponding antibodies.
Microscopy
The A. tumefaciens strains carrying GFP/mCherry-fusion constructs were infiltrated into the epidermal cells of N. benthamiana following the procedures previously described (Chen et al., 2018). All transient expression assays were repeated at least three times. The fluorescence was detected under an Olympus FV1000 MPE multiphoton laser scanning confocal microscope. GFP was excited using a 488-nm laser, and the fluorescence signal was collected in the range 495 to 540 nm. mCherry were excited using a 543-nm laser, and the emission fluorescence of mCherry was collected in the range 600 to 650 nm.
Prokaryotic Expression and Recombinant Protein Purification
For expression and purification, a SlFERL fragment (1,399–2,667 bp, corresponding to the intracellular domain composed of amino acids 467–889) was cloned into pET-30a (Novagen) to generate pET-30a-SlFERL-KD, and the pET-30a-SlFERL-KD was transformed into Escherichia coli strain BL21 (DE3). The recombinant strain was induced at 16°C by supplementing 1 mm isopropyl β-d-1-thiogalactopyranoside for 12 h in Luria-Bertani medium. Cells were collected and lysed by sonication. SlFERL-KD was affinity-purified using nickel-nitrilotriacetic acid agarose resin (Novagen) according to the manufacturer’s instructions. The primers were listed in Supplemental Table S1.
Polyclonal Antibody Preparation
After sequence analysis for SlFERL, two highly conserved sequences (HTSGSAKTNTTGSYASSLP and KDLNESPGYDASMTDSRS; Abmart Shanghai) were selected for peptide synthesis and further used as the antigens for immunizing rabbits. Polyclonal antibody was affinity-purified from antisera using AminoLink Plus Coupling Resin according to the instructions for antibody purification (Thermo Scientific).
SPR-MS/MS Assay
The Biacore instrument (GE Healthcare) is controlled by BIA evaluation version 4.1. The fluidic system is washed with the running buffer (HEPES buffer [pH 7.4]), and then the sensor chip was activated by the addition of an equal volume of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride and N-hydroxysuccinimide, prepared according to the manufacturer’s instructions. The purified SlFERL-KD protein was injected at a flow rate of 10 mL min−1 before inactivation of the sensor chip by addition of 70 mL of 1 m ethanolamine and washing of the chip by 10 pulses of 5 mL 1% (v/v) acetic acid. The fluidic system was washed with the running buffer (HBS-N, 10 mm octyl β-D-glucopyranoside [OGP]). The fruit extracts were then injected into the immobilized ligand (peptides or protein) on the sensor chip with 10 mm OGP and 40 mL of the running buffer. The integrated m-fluidic fartridge (IFC) was washed with 50 mm NaOH, 50 mm OGP, and the running buffer. The flow cells were then washed with running buffer and deionized water. The recovery solution was injected and remained in the flow cells for an optional amount of time, separated by air segments. Finally, the recovery solution was eluted and transported to the recovery vial before a final wash of the IFC with 50 mm NaOH. The resulting proteins were lyophilized and used for MS identification. Calculation of data intersections was performed using Biovenn (Hulsen et al., 2008).
Ethylene Measurements
Three independent lines of SlFERL OE, RNAi, and wild-type tomato fruit were harvested at 31 and 35 dpa and ethylene production was measured as described by Cai et al. (2018).
Lycopene Measurements
Lycopene was detected as described by Fish et al. (2002), with modification. Briefly, 0.4 g fruit pericarp was suspended with 4 mL buffer containing hexane:acetone:ethanol (2:1:1 [v/v]) and shaken for 5 min. Afterward, 1.2 mL of deionized water was added and the samples were shaken for another 10 s. The vials were left on ice for 5 min to allow phase separation. The absorbance of the hexane was measured at 503 nm and subsequently used to calculate the lycopene concentration.
BiFC and LUC Complementation Imaging Assay
The plasmids used were previously described (Walter et al., 2004). The CDS of SlFERL without the stop codon was cloned into the 2YN-pBI vector, whereas SlSAMS1 was cloned into the 2YC-pBI vector, and they were respectively introduced into A. tumefaciens strain GV3101. The fluorescence was observed at 2 to 3 d after infiltration using protocols for A. tumefaciens-mediated transient expression in N. benthamiana leaves (Chen et al., 2018). The primers were listed in Supplemental Table S1.
LCI assays were carried out according to the method proposed by Cai et al. (2018). The ORFs of SlFERL and SlSAMS1 were cloned into pCAMBIA1300-cLUC/nLUC (kindly provided by Jianmin Zhou from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) to produce SlFERL-nLUC and cLUC-SlSAMS1, respectively. The primers are listed in Supplemental Table S1. The recombinant plasmids were transformed into A. tumefaciens strain GV3101 and infiltrated into N. benthamiana leaves as described above. At 2 d after infiltration, 1 mm luciferin containing 0.01% (v/v) Triton X-100 was sprayed onto leaves, kept in the dark for 3 min, and detached to observe the fluorescence. The LCI images were captured using a 5200 Multi Chemiluminescent Imaging System (Tanon).
Split-Ubiquitin yeast two hybrid analysis
Split-ubiquitin Y2H analysis was performed according to the directions provided for DUALmembrane kit 2 (Dualsystems Biotech). The ORFs of SlFERL and SlSAMS1 were amplified and subcloned into the pCCW-SUC bait vector and pDSL-Nx prey vector to generate SlFERL-Cub and NubG-SlSAMS1 respectively. The recombinant constructs were cotransformed into yeasts, screened on double dropout agar medium (SD/–Leu/–Trp) and TDO agar medium (SD/–His/–Leu/–Trp). Cub and NubG-SlSAMS1, SlFERL-Cub and NubG, and Cub and NubG were set as negative controls and cotransformed. The primers used are listed in Supplemental Table S1.
Coimmunoprecipitation Assay
Coimmunoprecipitation assays were performed as described previously (Kadota et al., 2016), with minor modifications. The vector pCAMBIA2300-35S:SlFERL-HA was used to generate the fusion protein SlFERL-HA, and the vector pCAMBIA2300-35S:SlSAMS1-mCherry was used to generate the fusion protein SlSAMS1-mCherry. The primers used are shown in Supplemental Table S1. SlFERL-HA and SlSAMS1-mCherry were cotransformed into N. benthamiana leaves, with cotransformation of SlFERL-HA and mCherry as controls. AntimCherry agarose beads (KT HEALTH) was used for purifying the protein complex. AntimCherry antibody (Solarbio) and anti-HA antibody (Abmart Shanghai) were used to detect tagged proteins.
Total RNA Isolation and RT-qPCR Analysis
Total RNA was isolated from fruit pericarp tissues according to the method of Wang et al. (2014). In order to remove DNA contamination, total RNA was treated with genomic DNA eraser and then reverse transcribed using the PrimeScript reagent kit (TaKaRa) according to the instructions. RT-qPCR reactions were carried out on the StepOne Plus Real-Time PCR System (Applied Biosystems) with SYBR Premix Ex Taq (Tli RNaseH Plus) ROX plus (TaKaRa) according to the manufacturer’s protocol. Gene-specific oligonucleotides (Supplemental Table S2) were designed using QuantPrime (Arvidsson et al., 2008). ACTIN was used as the internal control. Values reported represent the average of triplicate replicates.
Determination of SAM by HPLC
Cellular SAM level was determined using the HPLC method as previously described by Edwards and Cobb (1996) and Van de Poel et al. (2010). An Agilent 1100 HPLC system (Hewlett Packard) was combined with an Alltima C18 HP amide reverse-phase column (250 × 0.3 × 5 mm; Grace). Twenty milliliter samples were injected and further eluted with 0.1 m sodium acetate (pH 4.5) at 0.5 mL min−1. The separation was performed for 40 min and the corresponding SAM level was detected at 260 nm. The SAM standard is available as an iodide salt (with several impurities, 86% pure). To verify the SAM peak, the product was degraded by heating at 50°C as previously described (Van de Poel et al., 2010). The samples were spiked with 100 mm SAM-iodide standard to target the SAM peak. Each sample was measured in triplicate.
Accession Numbers
Sequence data from this article can be found in the SOL Genomics Network (https://solgenomics.net/) and NCBI under accession numbers: Solyc09g015830 (GenBank: NC_015446; SlFERL), Solyc03g025850 (SlREM1), Solyc01g101060 (SlSAMS1), Solyc12g099000 (SlSAMS2), Solyc09g008280 (SlSAMS3), Solyc10g083970 (SlSAMS4), Solyc01g095080 (SlACS2), Solyc05g050010 (SlACS4), Solyc03g111720 (SlE4), Solyc09g089580 (SlE8), Solyc03g123760 (SlPDS), Solyc03g031860 (SlPSY1)), and Solyc11g005330 (SlACTIN).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression pattern analyses of RIN target genes revealed that SlFERL is involved in fruit ripening.
Supplemental Figure S2. SlFERL shows high homology to AtFER.
Supplemental Figure S3. Gene cloning and immunoblot analysis for SlFERL.
Supplemental Figure S4. RT-qPCR analysis for transcript levels of other members of the CrRLK1L family in SlFERL-RNAi fruit.
Supplemental Figure S5. Identification of putative SlFERL-interacting proteins using SPR-MS/MS assay.
Supplemental Figure S6. Sensorgram of SPR experiment.
Supplemental Figure S7. SlFERL does not interact with SlSAMS2.
Supplemental Figure S8. Expression pattern analysis for the genes in SlSAMS family during tomato fruit ripening.
Supplemental Figure S9. Transcript levels of SlSAMS1 and its family members in SlFERL transgenic lines during fruit ripening.
Supplemental Figure S10. Potential phosphorylation site analysis of SlSAMS1.
Supplemental Figure S11. SlSAMS1 colocalizes with SlFERL to the PM.
Supplemental Table S1. List of primers for gene cloning and vector construction.
Supplemental Table S2. Primers for RT-qPCR.
Supplemental Dataset S1. Expression patterns of direct RIN target genes during fruit ripening.
Supplemental Dataset S2. Protein subcellular localization of RIN target genes.
Supplemental Dataset S3. MS identification of SlFERL-interacting proteins in SPR-MS/MS assay.
Supplemental Dataset S4. Intersection of replicates 1 and 2 in the SPR-MS/MS assay.
Supplemental Dataset S5. GO analysis for SlFERL-interacting proteins.
Acknowledgments
We thank Suhua Yang and Zhuang Lu (Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences), for their technical assistance in SPR-MS/MS analysis.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31930086, 31925035, and 32072637).
References
- Alexander L, Grierson D(2002) Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B(2008) QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur AH, Yang SF(1972) Methionine metabolism in apple tissue in relation to ethylene biosynthesis. Phytochemistry 11: 3207–3214 [Google Scholar]
- Bulens I, Van de Poel B, Hertog ML, De Proft MP, Geeraerd AH, Nicolaï BM(2011) Protocol: An updated integrated methodology for analysis of metabolites and enzyme activities of ethylene biosynthesis. Plant Methods 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Qin G, Chen T, Tian S(2018) The mode of action of remorin1 in regulating fruit ripening at transcriptional and post-transcriptional levels. New Phytol 219: 1406–1420 [DOI] [PubMed] [Google Scholar]
- Chen T, Ji D, Tian S(2018) Variable-angle epifluorescence microscopy characterizes protein dynamics in the vicinity of plasma membrane in plant cells. BMC Plant Biol 18: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, Liu C, Yang H, Zeng WF, Wu L, Zhou WJ, Wang RM, Niu XN, Ding YH, Zhang Y, et al. (2018) Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat Biotechnol 36: 1059–1061 [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB(2010) Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5: e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F.(1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Wang Y, Liu Z, Cheng H, Xue Y(2014) HemI: A toolkit for illustrating heatmaps. PLoS One 9: e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers SD, Larsen PB(2010) FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol Plant 3: 626–640 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY(2014) Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun 5: 3129. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM(2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Liu MJ, Kita D, Jordan SS, Yeh FJ, Yvon R, Carpenter H, Federico AN, Garcia-Valencia LE, Eyles SJ, et al. (2020) FERONIA controls pectin- and nitric oxide-mediated male-female interaction. Nature 579: 561–566 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Cobb AH(1996) Improved high-performance liquid chromatographic method for the analysis of potato (Solanum tuberosum) glycoalkaloids. J Agric Food Chem 44: 2705–2709 [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U(2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Fish WW, Perkins-Veazie P, Collins JK(2002) A quantitative assay for lycopene that utilizes reduced volumes of organic solvents. J Food Compost Anal 15: 309–317 [Google Scholar]
- Franck CM, Westermann J, Boisson-Dernier A(2018) Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69: 301–328 [DOI] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB(2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y(2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y(2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ(2013) Molecular and genetic regulation of fruit ripening. Plant Mol Biol 82: 575–591 [DOI] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z(2017) The epigenome and transcriptional dynamics of fruit ripening. Annu Rev Plant Biol 68: 61–84 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ.(2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Guo H, Nolan TM, Song G, Liu S, Xie Z, Chen J, Schnable PS, Walley JW, Yin Y(2018) FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr Biol 28: 3316–3324.e6 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR(2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HD, Lee TY, Tzeng SW, Horng JT(2005) KinasePhos: A web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res 33: W226–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U(2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W(2008) BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, Kotake-Nara E, Kawasaki S, Toki S(2017) Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat Plants 3: 866–874 [DOI] [PubMed] [Google Scholar]
- Jia M, Du P, Ding N, Zhang Q, Xing S, Wei L, Zhao Y, Mao W, Li J, Li B, et al. (2017) Two FERONIA-Like receptor kinases regulate apple fruit ripening by modulating ethylene production. Front Plant Sci 8: 1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Chang C(2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Macho AP, Zipfel C(2016) Immunoprecipitation of plasma membrane receptor-like kinases for identification of phosphorylation sites and associated proteins. Methods Mol Biol 1363: 133–144 [DOI] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R(2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U(2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ(2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Klee HJ.(2004) Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol 135: 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K(2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa AK, Zhu J-K(2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA 114: E4511–E4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L(2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P(2015) SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res 43: D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, Li X, Li X, Zhu S, Wang L, Wang Y, Liao H, Luan S, et al. (2018a) EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol 16: e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang X, Li C, Li H, Zhang J, Ye Z(2018b) Silencing GRAS2 reduces fruit weight in tomato. J Integr Plant Biol 60: 498–513 [DOI] [PubMed] [Google Scholar]
- Li S, Chen K, Grierson D(2019) A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol 221: 1724–1741 [DOI] [PubMed] [Google Scholar]
- Li S, Xu H, Ju Z, Cao D, Zhu H, Fu D, Grierson D, Qin G, Luo Y, Zhu B(2018c) The RIN-MC fusion of MADS-Box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol 176: 891–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang G, Liu X, Ding X, Zhang D, Wang X, Zhou Y, Yan H, Li T, Wu K, et al. (2020) Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening-related genes in tomato. New Phytol 227: 1138–1156 [DOI] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M(2015) Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol 169: 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lang Z(2020) The mechanism and function of active DNA demethylation in plants. J Integr Plant Biol 62: 148–159 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP(2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lü P, Yu S, Zhu N, Chen YR, Zhou B, Pan Y, Tzeng D, Fabi JP, Argyris J, Garcia-Mas J, et al. (2018) Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat Plants 4: 784–791 [DOI] [PubMed] [Google Scholar]
- Mao D, Yu F, Li J, Van de Poel B, Tan D, Li J, Liu Y, Li X, Dong M, Chen L, et al. (2015) FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ 38: 2566–2574 [DOI] [PubMed] [Google Scholar]
- Martin LB, Rose JK(2014) There’s more than one way to skin a fruit: Formation and functions of fruit cuticles. J Exp Bot 65: 4639–4651 [DOI] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL(1972) Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237: 235–236 [DOI] [PubMed] [Google Scholar]
- Meng J, Wang L, Wang J, Zhao X, Cheng J, Yu W, Jin D, Li Q, Gong Z(2018) METHIONINE ADENOSYLTRANSFERASE4 mediates DNA and histone methylation. Plant Physiol 177: 652–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S(2012) Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Qin G, Zhu Z, Wang W, Cai J, Chen Y, Li L, Tian S(2016) A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiol 172: 1596–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Deguchi M, Brustolini OJ, Santos AA, Silva FF, Fontes EP(2012) The tomato RLK superfamily: Phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol 12: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Yang SF(1988) S-adenosylmethionine-dependent inactivation and radiolabeling of 1-aminocyclopropane-1-carboxylate synthase isolated from tomato fruits. Plant Physiol 88: 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Nicolas P, Fernandez-Pozo N, Ma Q, Evanich DJ, Shi Y, Xu Y, Zheng Y, Snyder SI, Martin LBB, et al. (2018) High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat Commun 9: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C(2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47(D1): D330–D338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian SP.(2013) Molecular mechanisms of fruit ripening and senescence. Zhiwu Xuebao 48: 481–488 [Google Scholar]
- Tigchelaar EC, Mcglasson WB, Franklin MJ(1978) Natural and ethephon-stimulated ripening of F1 hybrids of ripening Inhibitor (rin) and non-ripening (nor) mutants of tomato (Lycopersicon esculentum Mill). Aust J Plant Physiol 5: 449–456 [Google Scholar]
- Van de Poel B, Bulens I, Lagrain P, Pollet J, Hertog ML, Lammertyn J, De Proft MP, Nicolaï BM, Geeraerd AH(2010) Determination of S-adenosyl-l-methionine in fruits by capillary electrophoresis. Phytochem Anal 21: 602–608 [DOI] [PubMed] [Google Scholar]
- Van de Poel B, Bulens I, Oppermann Y, Hertog ML, Nicolai BM, Sauter M, Geeraerd AH(2013) S-adenosyl-l-methionine usage during climacteric ripening of tomato in relation to ethylene and polyamine biosynthesis and transmethylation capacity. Physiol Plant 148: 176–188 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J(2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR(2002) Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cai J, Wang P, Tian S, Qin G(2017) Post-transcriptional regulation of fruit ripening and disease resistance in tomato by the vacuolar protease SlVPE3. Genome Biol 18: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S(2014) Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol 15: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J(2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE(1984) Ethylene biosynthesis and its regulation in higher-plants. Annu Rev Plant Physiol Plant Mol Biol 35: 155–189 [Google Scholar]
- Yang Y, Tang K, Datsenka TU, Liu W, Lv S, Lang Z, Wang X, Gao J, Wang W, Nie W, et al. (2019) Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J Integr Plant Biol 61: 1224–1242 [DOI] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, et al. (2012) FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA 109: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Hong W, Wu J, Wang Y, Ji S, Zhu S, Wei C, Zhang J, Li Y(2017) A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. eLife 6: e27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhou L, Tian S, Qin G(2019) RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol 20: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Estévez JM, Liao H, Zhu Y, Yang T, Li C, Wang Y, Li L, Liu X, Pacheco JM, et al. (2020) The RALF1-FERONIA complex phosphorylates eIF4E1 to promote protein synthesis and polar root hair growth. Mol Plant 13: 698–716 [DOI] [PubMed] [Google Scholar]
- Zouine M, Maza E, Djari A, Lauvernier M, Frasse P, Smouni A, Pirrello J, Bouzayen M(2017) TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J 92: 727–735 [DOI] [PubMed] [Google Scholar]