An F-box protein contributes to the self-compatibility of sweet cherry ‘Lapins’.
Abstract
Recent studies have shown that loss of pollen-S function in S4′ pollen from sweet cherry (Prunus avium) is associated with a mutation in an S haplotype-specific F-box4 (SFB4) gene. However, how this mutation leads to self-compatibility is unclear. Here, we examined this mechanism by analyzing several self-compatible sweet cherry varieties. We determined that mutated SFB4 (SFB4ʹ) in S4′ pollen (pollen harboring the SFB4ʹ gene) is approximately 6 kD shorter than wild-type SFB4 due to a premature termination caused by a four-nucleotide deletion. SFB4′ did not interact with S-RNase. However, a protein in S4′ pollen ubiquitinated S-RNase, resulting in its degradation via the 26S proteasome pathway, indicating that factors in S4′ pollen other than SFB4 participate in S-RNase recognition and degradation. To identify these factors, we used S4-RNase as a bait to screen S4′ pollen proteins. Our screen identified the protein encoded by S4-SLFL2, a low-polymorphic gene that is closely linked to the S-locus. Further investigations indicate that SLFL2 ubiquitinates S-RNase, leading to its degradation. Subcellular localization analysis showed that SFB4 is primarily localized to the pollen tube tip, whereas SLFL2 is not. When S4-SLFL2 expression was suppressed by antisense oligonucleotide treatment in wild-type pollen tubes, pollen still had the capacity to ubiquitinate S-RNase; however, this ubiquitin-labeled S-RNase was not degraded via the 26S proteasome pathway, suggesting that SFB4 does not participate in the degradation of S-RNase. When SFB4 loses its function, S4-SLFL2 might mediate the ubiquitination and degradation of S-RNase, which is consistent with the self-compatibility of S4′ pollen.
In sweet cherry (Prunus avium), self-incompatibility is mainly controlled by the S-locus, which is located at the end of chromosome 6 (Akagi et al., 2016; Shirasawa et al., 2017). Although the vast majority of sweet cherry varieties show self-incompatibility, some self-compatible varieties have been identified, most of which resulted from the use of x-ray mutagenesis and continuous cross-breeding (Ushijima et al., 2004; Sonneveld et al., 2005). At present, naturally occurring self-compatible varieties are rare (Marchese et al., 2007; Wünsch et al., 2010; Ono et al., 2018). X-ray-induced mutations that have given rise to self-compatibility include a 4-bp deletion (TTAT) in the gene encoding an SFB4′ (S-locus F-box 4′) protein, located in the S-locus and regarded as the dominant pollen factor in self-incompatibility. This mutation is present in the first identified self-compatible sweet cherry variety, ‘Stellar’, as well as in a series of its self-compatible descendants, including ‘Lapins’, ‘Yanyang’, and ‘Sweet heart’ (Lapins, 1971; Ushijima et al., 2004). Deletion of SFB3 and a large fragment insertion in SFB5 have also been identified in other self-compatible sweet cherry varieties (Sonneveld et al., 2005; Marchese et al., 2007). Additionally, a mutation not linked to the S-locus (linked instead to the M-locus) could also cause self-compatibility in sweet cherry and closely related species such as apricot (Prunus armeniaca; Wünsch et al., 2010; Zuriaga et al., 2013; Muñoz-Sanz et al., 2017; Ono et al., 2018). Much of the self-compatibility in Prunus species seems to be closely linked to mutation of SFB in the S-locus (Zhu et al., 2004; Muñoz-Espinoza et al., 2017); however, the mechanism of how this mutation of SFB causes self-compatibility is unknown.
The gene composition of the S-locus in sweet cherry differs from that of other gametophytic self-incompatible species, such as apple (Malus domestica), pear (Pyrus spp.), and petunia (Petunia spp.). In sweet cherry, in addition to a single S-RNase gene, the S-locus contains one SFB gene, which has a high level of allelic polymorphism, and three SLFL (S-locus F-box-like) genes with low levels of, or no, allelic polymorphism (Ushijima et al., 2004; Matsumoto et al., 2008). By contrast, the apple, pear, and petunia S-locus usually contains one S-RNase and 16 to 20 F-box genes (Kakui et al., 2011; Okada et al., 2011, 2013; Minamikawa et al., 2014; Williams et al., 2014a; Yuan et al., 2014; Kubo et al., 2015; Pratas et al., 2018). The F-box gene, named SFBB (S-locus F-box brother) in apple and pear and SLF (S-locus F-box) in petunia, exhibits higher sequence similarity with SLFL than with SFB from sweet cherry (Matsumoto et al., 2008; Tao and Iezzoni, 2010). The protein encoded by SLF in the petunia S-locus is thought to be part of an SCF (Skp, Cullin, F-box)-containing complex that recognizes nonself S-RNase and degrades it through the ubiquitin pathway (Kubo et al., 2010; Zhao et al., 2010; Chen et al., 2012; Entani et al., 2014; Li et al., 2014, 2016, 2017; Sun et al., 2018). In sweet cherry, a number of reports have described the expression and protein interactions of SFB, SLFL, Skp1, and Cullin (Ushijima et al., 2004; Matsumoto et al., 2012); however, only a few reports have examined the relationship between SFB/SLFL and S-RNase (Matsumoto and Tao, 2016, 2019), and none has investigated whether the SFB/SLFL proteins participate in the ubiquitin labeling of S-RNase.
Although the function of SFB4 and SLFL in self-compatibility is unknown, the observation that S4′ pollen tubes grow in sweet cherry pistils that harbor the same S alleles led us to speculate that S4′ pollen might inhibit the toxicity of self S-RNase. In petunia, the results of several studies have suggested that pollen tubes inhibit self S-RNase when an SLF gene from another S-locus haplotype is expressed (Sijacic et al., 2004; Kubo et al., 2010; Williams et al., 2014b; Sun et al., 2018). For example, when SLF2 from the S7 haplotype is heterologously expressed in pollen harboring the S9 or S11 haplotype, the S9 or S11 pollen acquire the capacity to inhibit self S-RNase and break down self-incompatibility (Kubo et al., 2010). The SLF2 protein in petunia has been proposed to ubiquitinate S9-RNase and S11-RNase and lead to its degradation through the 26S proteasome pathway (Entani et al., 2014). If SFB/SLFL in sweet cherry have a similar function, the S4′ pollen would not be expected to inhibit self S4-RNase, prompting the suggestion that the functions of SFB/SLFL in sweet cherry and SLF in petunia vary (Tao and Iezzoni, 2010; Matsumoto et al., 2012).
In this study, we used sweet cherry to investigate how S4′ pollen inhibits S-RNase and causes self-compatibility, focusing on the question of whether the SFB/SLFL protein can ubiquitinate S-RNase, resulting in its degradation.
RESULTS
SFB4 Mutation Leads to Self-Compatibility of ‘Lapins’
We performed self-pollination of sweet cherry cultivars ‘Lapins’ (S1S4′), ‘Rainer’ (S1S4), and ‘Hongdeng’ (S3S9) in April of 2013, 2014, and 2015, respectively. The self-fruiting rates of cv Lapins after self-pollination were 35.9%, 37.2%, and 42% in 2013, 2014, and 2015, respectively, exhibiting self-compatibility (Supplemental Table S1). By contrast, the self-fruiting rates of cv Rainer (S1S4) were 0%, 1%, and 1.8% and those of cv Hongdeng (S3S9) were 1.8%, 2.2%, and 1.6% during the same time periods, respectively, suggesting that these cultivars are self-incompatible (Supplemental Table S1). To investigate whether the self-compatibility of cv Lapins is caused by the style side or the pollen side, we performed reciprocal cross-pollination between cv Lapins (S1S4′) and cv Rainer (S1S4), which resulted in fruiting rates of 1.1%, 0%, and 2.8% in cv Lapins × cv Rainer in 2013, 2014, and 2015, respectively, while the fruiting rates of cv Rainer × cv Lapins were 39.6%, 43.4%, and 42.6%, indicating that the self-compatibility of cv Lapins is caused by mutations in pollen (Supplemental Table S1). The S genotype of the progeny from both cv Lapins self-pollination and the cv Rainer × cv Lapins cross fell into two classes, S1S4′ and S4S4′, with a ratio of almost 1:1 (Supplemental Table S2), suggesting that the self-compatibility of cv Lapins is caused by the presence of S4 alleles. In the cv Hongdeng × cv Lapins cross, the fruiting rate was 61.8%, 49.6%, and 60.2% in 2013, 2014, and 2015, respectively (Supplemental Table S1), and the S genotypes of progeny from the cv Hongdeng (S3S9) × cv Lapins (S1S4′) cross were S1S3, S1S9, S3S4′, and S4′S9, respectively (Supplemental Table S2), indicating that S4′ pollen could grow and develop normally in both self and nonself pistils.
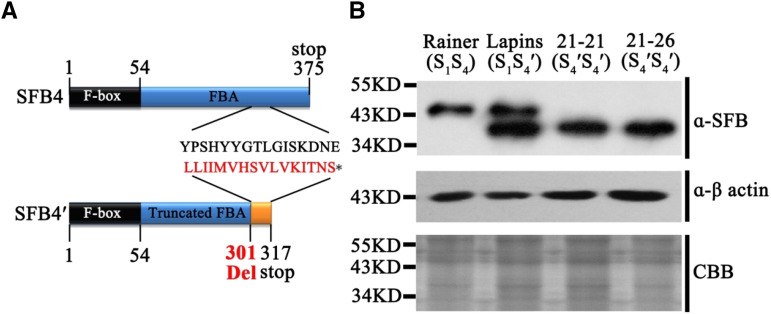
In cv Lapins S4′ pollen, a 4-bp deletion (TTAT) was detected in the SFB4 gene, which is predicted to lead to mistranslation at amino acid residue 301 and premature translation termination at amino acid residue 317 (Fig. 1A). To determine whether the prematurely terminated SFB4′ protein is present in S4′ pollen, we identified a highly specific SFB4′ protein fragment (Supplemental Fig. S1A), synthesized peptides corresponding to these sequences in vitro, and generated monoclonal antibodies to these peptides in rat. We subjected protein extracts from cv Rainer (S1S4), cv Lapins (S1S4′), cv 21-21 (S4′S4′), cv 21-26 (S4′S4′), cv Hongdeng (S3S9), cv Hongmi (S3S6), cv Yanyang (S3S4′), and cv Bin (S3S4) pollen tubes to immunoblot analysis with anti-SFB4 monoclonal antibodies. cv 21-21 and cv 21-26 are two progeny derived from the self-crossing of cv Lapins that harbor homozygous S4′ alleles. Immunoreactive signals were detected in cv Rainer (S1S4), cv Lapins (S1S4′), cv 21-21 (S4′S4′), cv 21-26 (S4′S4′), cv Yanyang (S3S4′), and cv Bin (S3S4), which contains SFB4 or SFB4′ protein. Notably, a 6-kD shorter protein band was detected in extracts from cv Lapins (S1S4′), cv 21-21 (S4′S4′), cv 21-26 (S4′S4′), and cv Yanyang (S3S4′), suggesting that the translation of SFB4′ protein is prematurely terminated in S4′ pollen (Fig. 1B; Supplemental Fig. S1B). A higher Mr band was also detected in cv Lapins, suggesting that the SFB4 antibody could also detect another SFB protein in the S1 haplotype.
Figure 1.
SFB4′ protein structure in extracts from pollen tubes. A, Schematic diagram of the SFB4′ protein from S4 pollen tubes showing mistranslation at amino acid 301 and a translation stop at amino acid 317. B, Immunoblot analysis of SFB4 and SFB4′ in pollen tube protein extracts from cv Rainer, cv Lapins, cv 21-21, and cv 21-26 using an SFB4-specific monoclonal antibody. Actin protein was used as an equal loading control. Coomassie Brilliant Blue (CBB) staining shows crude pollen tube protein extracts from the indicated sweet cherry varieties; S1S4 refers to the S genotype.
Mutated SFB4 Fails to Interact with S4-RNase
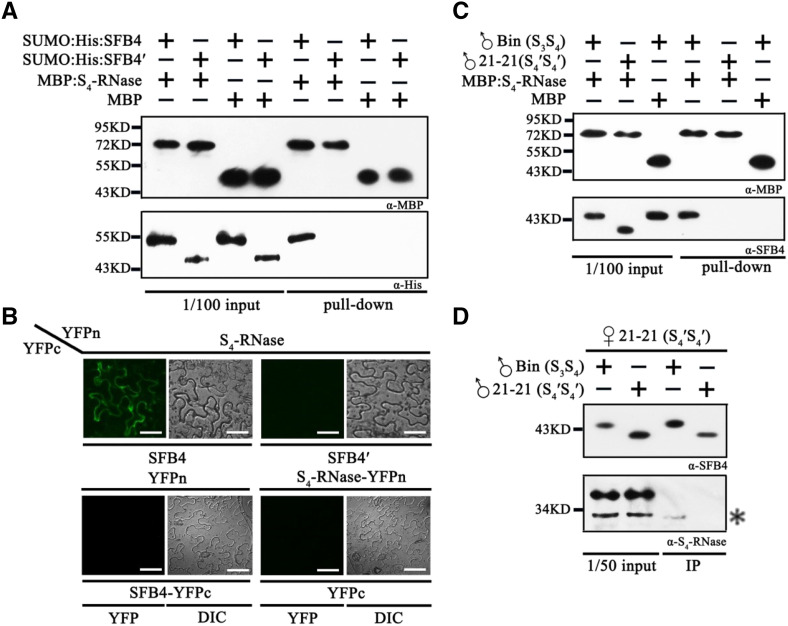
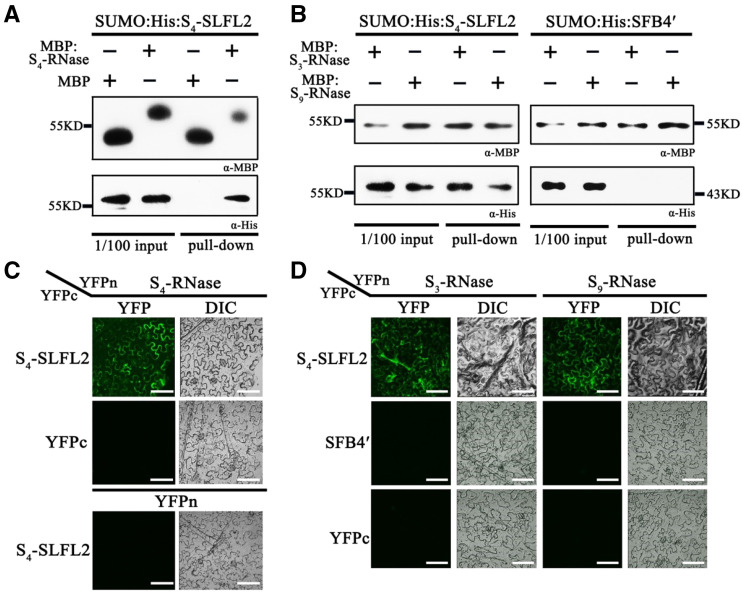
To investigate the mechanism of SFB4′-mediated self-compatibility, we examined the interaction between the SFB4/SFB4′ (truncated form of SFB4) protein and S4-RNase using a maltose-binding protein (MBP) pull-down assay. We purified recombinant MBP-S4-RNase, SUMO-His-SFB4, and SUMO-His-SFB4′ from Escherichia coli and subjected these fusion proteins to pull-down analysis. SUMO-His-SFB4 was detected with a His tag antibody following incubation with MBP-S4-RNase, whereas when we used the combination of SUMO-His-SFB4 and MBP, SUMO-His-SFB4 was not detected. In the other two combinations, SUMO-His-SFB4′/MBP-S4-RNase and SUMO-His-SFB4′/MBP, SUMO-His-SFB4′ was not detected. These results demonstrate that SUMO-His-SFB4 could bind to MBP-S4-RNase, whereas SUMO-His-SFB4′ and the negative control could not (Fig. 2A), indicating that SFB4′ lost its ability to interact with S4-RNase. This in vitro interaction was further confirmed using a bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana leaves. In particular, S4-RNase and SFB4/SFB4′ were fused to the N or C terminus of yellow fluorescent protein, generating YFPn and YFPc, respectively. We expressed these vectors in pairs (S4-RNase-YFPn/SFB4-YFPc and S4-RNase-YFPn/SFB4′-YFPc) in N. benthamiana leaves. Five days after infiltration, strong YFP fluorescence was observed in the cytosol when S4-RNase-YFPn was cotransformed with SFB4-YFPc, whereas no YFP signal was detected when S4-RNase-YFPn was cotransformed with SFB4′-YFPc. Moreover, when the pairs S4-RNase-YFPn/YFPc and YFPn/SFB4-YFPc were cotransformed into N. benthamiana leaves, no YFP signal was detected (Fig. 2B). These results indicate that SFB4 and S4-RNase physically interact with each other, while SFB4′ and S4-RNase do not.
Figure 2.
SFB4′ protein does not interact with S4-RNase. A, Pull-down assay to detect the interaction between S4-RNase and SFB4/SFB4′. SFB4 and SFB4′ were fused with SUMO and His tags, and the molecular masses were almost 55 and 45 kD, respectively. S4-RNase was fused with an MBP tag, and the molecular mass reached around 72 kD. SFB4 and SFB4′ were detected by His tag monoclonal antibody, while S4-RNase and MBP were detected by MBP monoclonal antibody. B, BiFC assay to detect the interaction between S4-RNase and SFB4/SFB4′. S4-RNase was fused with the N-terminal end of YFP, SFB4 and SFB4′ were fused with the C-terminal end of YFP, and the combinations S4-RNase/SFB4, S4-RNase/SFB4′, S4-RNase/YFPc, and YFPn/SFB4 were expressed in N. benthamiana leaves. DIC, Differential interference contrast. Bars = 20 μm. C, Semi-in vivo pull-down assay to further detect the interaction between S4-RNase and SFB4/SFB4′. Pollen tube proteins from cv Bin and cv 21-21 were incubated with MBP protein and MBP-tagged S4-RNase. The molecular masses of SFB4 and SFB4′ were almost 45 and 39 kD in pollen tubes, respectively. D, Semi-in vivo immunoprecipitation (IP) assay to confirm the interaction. Pistil proteins from cv 21-21 were incubated with pollen tube proteins from cv Bin and cv 21-21. The molecular mass of S4-RNase was almost 30 kD. The asterisk represents the signal of S4-RNase. The higher molecular mass band is a nonspecific band. α-His, His tag antibody; α-MBP, MBP tag antibody; α-S4-RNase, S4-RNase antibody; α-SFB4, SFB4 antibody.
To further confirm the relationship between SFB4/SFB4′ and S4-RNase, we performed a semi-in vivo pull-down assay by using purified MBP-S4-RNase as a bait to incubate pollen tube proteins extracted from cv 21-21 (S4′S4′) and cv Bin (S3S4) and used anti-SFB4 antibody to detect SFB4 proteins. SFB4 was only detected with the combination of MBP-S4-RNase/cv Bin pollen tube proteins, whereas no SFB4 signal was detected when we used the combination of MBP/cv Bin (S3S4) and MBP-S4-RNase/cv 21-21 (S4′S4′) pollen tube proteins. These results indicate that MBP-S4-RNase combines with SFB4 other than SFB4′ in pollen tubes (Fig. 2C). We separately incubated cv 21-21 (S4′S4′) style proteins with pollen tube proteins from cv 21-21 (S4′S4′) and cv Bin (S3S4) and performed semi-in vivo immunoprecipitation using anti-SFB4 antibody. Anti-S4-RNase polyclonal antibody generated using MBP-S4-RNase (Supplemental Fig. S2) was used to detect the S4-RNase signal. Two immunoreactive bands were detected in one lane. The higher Mr band is nonspecific, whereas the lower band is S4-RNase, which was only detected when cv 21-21 (S4′S4′) style proteins were incubated with cv Bin pollen tube proteins following immunoprecipitation (Fig. 2D), further confirming the interaction between SFB4 and S4-RNases and the loss of function of SFB4′.
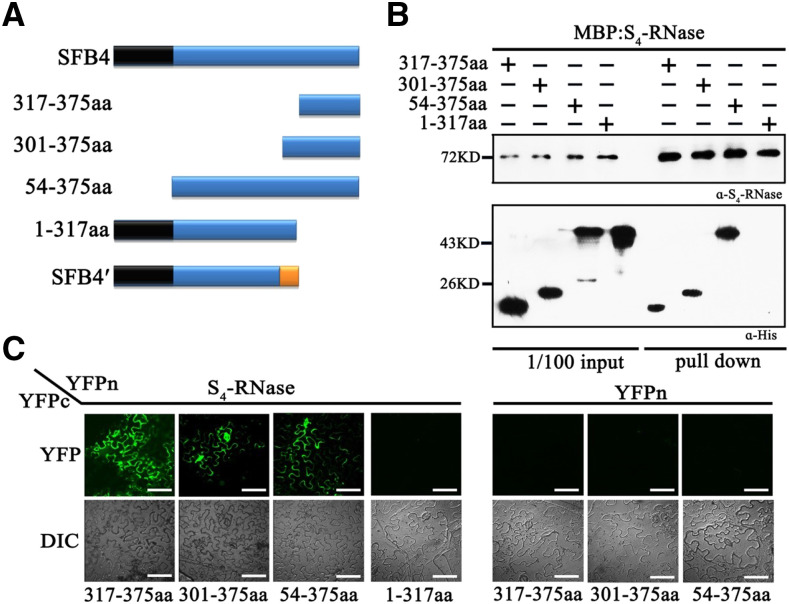
In SFB4′, 16 amino acids (amino acids 301 to 317) were predicted to be mistranslated, followed by 58 deleted amino acids. To determine whether loss of the SFB4′/S4-RNase interaction was caused by the mistranslation or loss of the 58 amino acids, we generated four types of truncated SFB4 proteins: amino acids 317 to 375 (SFB4-317-375aa), amino acids 301 to 375 (SFB4-301-375aa), amino acids 54 to 375 (SFB4-54-375aa), and amino acids 1 to 317 (SFB4-1-317aa; Fig. 3A). We fused the truncated proteins with SUMO and His tags in E. coli and conducted pull-down assays with MBP-S4-RNase. The SFB4-317-375aa, SFB4-301-375aa, and SFB4-54-375aa signals were detected by anti-His antibody using the combinations MBP-S4-RNase/SFB4-317-375aa, MBP-S4-RNase/SFB4-301-375aa, and MBP-S4-RNase/SFB4-54-375aa, whereas no signal was detected using MBP-S4-RNase and SFB4-1-317aa (Fig. 3B), suggesting that the deficiency of these 58 amino acids in SFB4′ was responsible for the absence of interaction between S4-RNase and SFB4′. To further confirm these interactions, we performed BiFC assays in N. benthamiana leaves in which S4-RNase and SFB4-317-375aa/301-375aa/54-375aa/1-317aa were fused to the N or C terminus of YFP, giving rise to YFPn and YFPc, respectively. Strong YFP fluorescence was observed on day 5 after infiltration in S4-RNase-YFPn/SFB4-317-375aa-YFPc, S4-RNase-YFPn/SFB4-301-375aa-YFPc, and S4-RNase-YFPn/SFB4-54-375aa-YFPc cotransformation (Fig. 3C), whereas no YFP signal was detected when S4-RNase-YFPn was cotransformed with SFB4-1-317aa-YFPc (Fig. 3C). These results further confirmed that the interaction between SFB4′ and S4-RNase is blocked by the lack of the C-terminal 58-amino acid region.
Figure 3.
The SFB4 C-terminal region is important for its ability to interact with S4-RNase. A, Schematic diagram of the four SFB4 fragments (317-375aa, 301-375aa, 54-375aa, and 1-317aa). B, Pull-down assay to detect the interaction between S4-RNase and different SFB4 fragments. S4-RNase was fused to the MBP tag and detected by S4-RNase antibody. The SFB4 fragments were fused to two tags of SUMO and His and detected by His tag antibody. The molecular masses of 317-375aa, 301-375aa, 54-375aa, and 1-317aa fragments were almost 21, 24, 50, and 50 kD, respectively. α-His, His tag antibody; α-S4-RNase, S4-RNase antibody. C, BiFC assay to further confirm the interaction. S4-RNase was fused with the N-terminal end of YFP, the SFB4 fragments were fused with the C-terminal end of YFP, and the combinations S4-RNase/SFB4-317-375aa, S4-RNase/SFB4-301-375aa, S4-RNase/SFB4-54-375aa, S4-RNase/SFB4-1-317aa, YFPn/SFB4-317-375aa YFPn/SFB4-301-375aa, and YFPn/SFB4-54-375aa were coexpressed in N. benthamiana leaves. DIC, Differential interference contrast. Bars = 20 μm.
S4′ Pollen Protein Degrades Both Self and Nonself S-RNase through the Ubiquitin Pathway
Although SFB4′ did not interact with S4-RNase, pollen tubes harboring the S4′ alleles showed self-compatibility, suggesting that they are able to resist the toxicity of S4-RNase. To investigate this possibility, cv 21-21 (S4′S4′) and cv 21-26 (S4′S4′) were used with self-fruiting rates of 46.1%, 47.4%, and 37.2% (cv 21-21) and 40.5%, 35.3%, and 50.5% (cv 21-26) in 2014, 2015, and 2016, respectively (Supplemental Table S3). When pollinated with cv Rainer (S1S4) pollen, the S genotype of all the progeny was S1S4, indicating that cv 21-21 and cv 21-26 did not lose their S function in pistils.
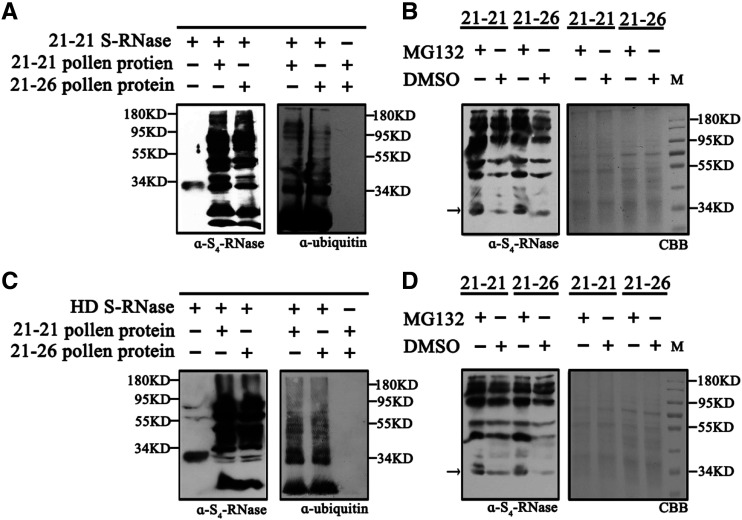
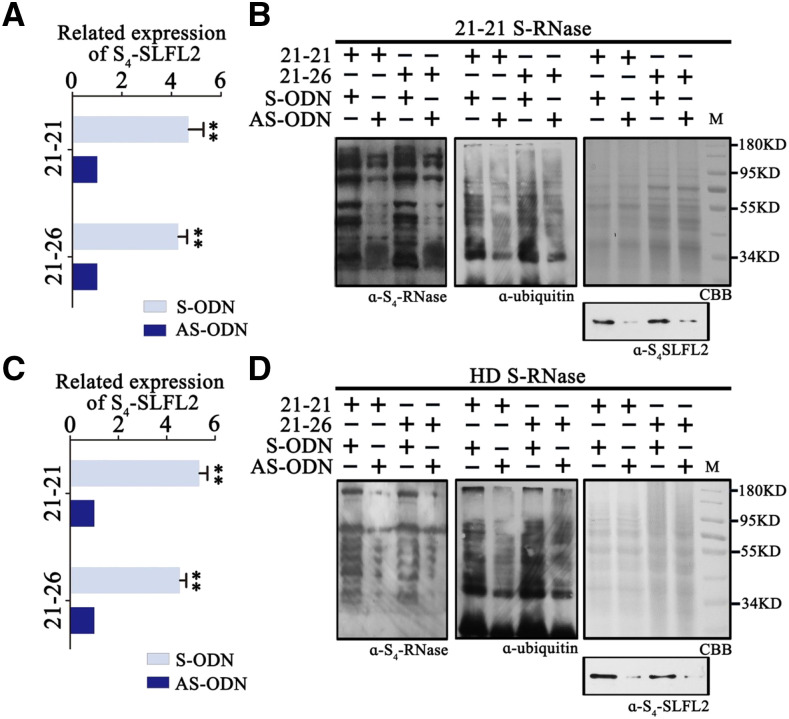
We used a cation-exchange column to purify S4-RNase from cv 21-21 pistils (Supplemental Fig. S3) as well as S-RNase from cv Hongdeng (Supplemental Fig. S3). After incubating S4-RNase with cv 21-21 or cv 21-26 pollen tube proteins overnight, we used S4-RNase and ubiquitin antibodies for immunoblot analyses of the protein extracts. When S4-RNase was incubated with either cv 21-21 or cv 21-26 proteins, the ubiquitin-labeled S4-RNase band was detected above the S4-RNase band (Fig. 4A), indicating that S4-RNase was labeled with ubiquitin by both cv 21-21 and cv 21-26 pollen tube proteins. To verify that the labeled S4-RNase was degraded through the 26S proteasome pathway, we added MG132 (an inhibitor of the 26S proteasome) to the S4-RNase/cv 21-21 and cv 21-26 protein mixtures and used dimethyl sulfoxide (DMSO) as a control. After overnight incubation, S4-RNase degraded more slowly in MG132-treated samples than the control (Fig. 4B), suggesting that the pollen tube proteins degrade S4-RNase through the 26S proteasome pathway.
Figure 4.
S4′ pollen tubes ubiquitinate self and nonself S-RNase. A, S-RNase from cv 21-21 pistils was incubated with cv 21-21 and cv 21-26 pollen tube proteins, and S4-RNase and ubiquitin antibodies were used to detect the proteins. B, MG132 and DMSO were used when S4-RNase was mixed with cv 21-21or cv 21-26 pollen tube proteins. The labeled proteins were detected by S4-RNase antibody. The protein content of each sample is shown on the SDS-PAGE gel stained by Coomassie Brilliant Blue (CBB). C, S-RNase from cv Hongdeng (HD) pistils was incubated with cv 21-21 and cv 21-26 pollen tube proteins, and S4-RNase and ubiquitin antibodies were used to detect the proteins. D, MG132 and DMSO were used when cv Hongdeng S-RNase was mixed with cv 21-21 or cv 21-26 pollen tube proteins. The labeled proteins were detected by S4-RNase antibody. The protein content of each sample is shown on the SDS-PAGE gel stained by Coomassie Brilliant Blue. α-S4-RNase, S4-RNase antibody; α-ubiquitin, ubiquitin antibody.
To better understand why S4′ pollen tubes grew in S3S9 pistils (Supplemental Table S3), we investigated the relationship between SFB4′ and both S3-RNase and S9-RNase. We purified recombinant MBP-S3-RNase, MBP-S9-RNase, and SUMO-His-SFB4′ from E. coli and subjected these samples to pull-down assays. SUMO-His-SFB4′ failed to be detected by His tag antibody using both combinations, SUMO-His-SFB4′/MBP-S3-RNase and SUMO-His-SFB4′/MBP-S9-RNase (Fig. 5B), indicating that SFB4′ cannot physically interact with S3-RNase or S9-RNase. This result was further confirmed by BiFC, where S3-RNase/S9-RNase and SFB4′ were fused to the N or C terminus of YFP (YFPn or YFPc). These vectors were overexpressed in pairs (S3-RNase-YFPn/SFB4′-YFPc and S9-RNase-YFPn/SFB4′-YFPc) in N. benthamiana leaves via Agrobacterium tumefaciens-mediated infiltration. YFP signal was not detected using any of these combinations, indicating that SFB4′ could not interact with S3-RNase or S9-RNase in vitro (Fig. 5D). However, when we incubated pollen tube protein extracts from cv 21-21 or cv 21-26 with purified pistil proteins from cv Hongdeng, the S-RNase was labeled with ubiquitin and degraded (Fig. 4, C and D). Together, these results suggest that S4′ pollen tubes can degrade self and nonself S-RNase through the 26S proteasome pathway.
Figure 5.
S-RNase interacts with S4-SLFL2 but not with SFB4′. A, Pull-down assay to detect the interaction between S4-RNase and S4-SLFL2. S4-RNase was fused with an MBP tag and detected by MBP antibody. S4-SLFL2 was fused with SUMO and His tags and detected by His antibody. The molecular mass of SUMO-His-S4-SLFL2 was almost 58 kD. B, Pull-down assay to detect the interaction of S3-RNase/S4-SLFL2, S9-RNase/S4-SLFL2, S3-RNase/SFB4′, and S9-RNase/SFB4′. S3-RNase and S9-RNase were fused with MBP tag and detected by MBP antibody. S4-SLFL2 and SFB4′ were both fused with SUMO and His tags, which was detected by His antibody. α-His, His tag antibody; α-MBP, MBP tag antibody. C, BiFC assay to further confirm the interaction between S4-RNase and S4-SLFL2. S4-RNase was fused with the N-terminal end of YFP, while S4-SLFL2 was fused with the C-terminal end of YFP. The S4-RNase/SLFL2 combination was expressed in N. benthamiana leaves. DIC, Differential interference contrast. Bars = 20 μm. D, BiFC assay to further confirm the interaction of S3-RNase/S4-SLFL2, S9-RNase/S4-SLFL2, S3-RNase/SFB4′, and S9-RNase/SFB4′. S3-RNase and S9-RNase proteins were fused to the YFP N-terminal end, while SFB4′ and S4-SLFL2 were fused with the YFP C-terminal end. Six combinations, S3-RNase/S4-SLFL2, S9-RNase/S4-SLFL2, S3-RNase/SFB4′, S9-RNase/SFB4′, S3-RNase/YFPc, and S9-RNase/YFPc, were expressed in N. benthamiana leaves. Bars = 20 μm.
Some pollen tube protein may also be labeled with ubiquitin and degraded via the 26S proteasome pathway. To further confirm that the labeled band detected by anti-ubiquitin antibody was S-RNase, we incubated pollen tube proteins from cv 21-21 or cv 21-26 without S-RNase overnight at 37°C, boiled the samples, and subsequently subjected them to immunoblot analysis using anti-ubiquitin antibody. When S-RNase was incubated with pollen tube proteins, the immune signal from labeled S-RNase was usually detected after 60 to 90 s of exposure time, whereas even when the exposure time was prolonged to 270 s, the protein band could not be detected without S-RNase application (Supplemental Fig. S4A). These results suggest that the ubiquitin-labeled protein from pollen tubes could not be detected as a protein band in our ubiquitin assay system and that the protein band was indeed S-RNase. Additionally, we examined the ubiquitination activity of the combination of cv Hongdeng S-RNase/cv Rainer pollen proteins and cv 21-21 S-RNase/cv Rainer pollen proteins. Both cv Hongdeng S-RNase and cv 21-21 S-RNase were ubiquitinated by cv Rainer pollen proteins, whereas no protein band was detected in the absence of S-RNase, further confirming that ubiquitin-labeled protein from pollen tubes could not be detected (Supplemental Fig. S4, B and C).
As S4′ pollen tubes are capable of degrading self and nonself S-RNase, to explore whether this process is specific or not, we incubated another protein purified from cv 21-21 pistils with a pollen tube protein extracted from cv 21-21 and cv 21-26. This protein (∼50 kD) was obtained from another eluting peak after the S-RNase peak in our S-RNase purification system (Supplemental Fig. S5A). Peptide mass fingerprinting analysis showed that this protein shares high sequence homology with hypothetical protein PRUPE_1G435600 (Hyp protein) from peach (Prunus persica). After incubating Hyp with pollen tube protein, only one protein band was detected by the antibody against Hyp protein (Supplemental Fig. S5B). Therefore, the Hyp protein could not be labeled with ubiquitin after incubating with cv 21-21 or cv 26-26 pollen protein, indicating that the process of S-RNase ubiquitination is specific. In all, the S4′ pollen tube has the ability to specifically degrade self and nonself S-RNase.
S4-SLFL2 Interacts with S-RNases
To screen proteins that are responsible for ubiquitination and degradation of S4-RNase among total pollen proteins, we purified recombinant MBP-S4-RNase and MBP from E. coli. Then they were incubated with proteins extracted from cv 21-21 pollen tubes. We identified potential S4-RNase-interacting proteins from these semi-in vivo pull-down assays using liquid chromatography-mass spectrometry (LC-MS) analysis. To make the results more accurate, prior to LC-MS analysis, we performed long-read transcriptome sequencing on the PacBio sequencing platform (Wang et al., 2016) to obtain full-length transcripts from sweet cherry pollen tubes. From the transcriptome data, we totally identified 16,409 transcripts, of which 292 were transposable elements. A total of 2,043 alternative splicing events were predicted, and 284 long noncoding RNAs were identified (Li et al., 2019). Then we used the translated sequences to construct a pollen tube protein database for LC-MS analysis to identify candidate proteins that interact with S4-RNase. Using the combination of MBP-S4-RNase and cv 21-21 pollen tube proteins, 82 proteins were identified by LC-MS analysis after the MBP pull-down, 39 of which were also found when cv 21-21 pollen tube proteins were pulled down by MBP, suggesting that the other 43 proteins in pollen tubes interact with S4-RNase.
In an effort to identify a factor involved in the recognition and ubiquitination of S4-RNase, we found a unique F-box protein among the 43 proteins, namely, S4-SLFL2, which is encoded by S4-SLFL2 in the S-locus (Supplemental Fig. S6). We used pull-down and BiFC assays to further confirm the interaction between S4-SLFL2 and S4-RNase. First, we incubated SUMO-His-S4-SLFL2 with MBP-S4-RNase or MBP. After the MBP pull-down, SUMO-His-S4-SLFL2 was only detected in the SUMO-His-S4-SLFL2 and MBP-S4-RNase combination, indicating that S4-RNase physically interacts with S4-SLFL2 (Fig. 5A). In addition, we transiently expressed the combinations S4-RNase-YFPn/S4-SLFL2-YFPc, S4-RNase-YFPn/YFPc, and YFPn/S4-SLFL2-YFPc in N. benthamiana leaves; the Skp1-like gene was also expressed together with the three groups, respectively, to maintain the stability of S4-SLFL2 (Matsumoto and Tao, 2016). Strong fluorescent signals were only detected when we used the S4-RNase-YFPn/S4-SLFL2-YFPc pair, indicating that S4-SLFL2 interacts with S4-RNase (Fig. 5C).
We also investigated the interaction between S4-SLFL2 and other S haplotype RNases. When purified MBP-S3-RNase and MBP-S9-RNase were used as bait to pull down purified SUMO-His-S4-SLFL2 (Fig. 5B), S4-SLFL2 signals were detected using the combinations MBP-S3-RNase/SUMO-His-S4-SLFL2 and MBP-S9-RNase/SUMO-His-S4-SLFL2, respectively, indicating that S4-SLFL2 physically interacts with both S3- and S9-RNase. Finally, to verify the interaction between S3-RNase/S9-RNase and S4-SLFL2 in vivo, we conducted a BiFC assay in N. benthamiana leaves. S4-SLFL2-YFPc accompanied by Skp1 was transiently expressed with S3-RNase-YFPn or S9-RNase-YFPn, respectively. As expected, epidermal cells expressing S3-RNase-YFPn/S4-SLFL2-YFPc and S9-RNase-YFPn/S4-SLFL2-YFPc showed strong fluorescence (Fig. 5D). Thus, in S4′ pollen tubes, S4-SLFL2 interacts with S-RNases, whereas SFB4′ does not.
S4-SLFL2 Helps Ubiquitinate S-RNase
We next investigated whether S4-SLFL2 is involved in the ubiquitination of S-RNase using an antisense oligonucleotide experiment in pollen tubes. Before the antisense oligonucleotide was designed, the sequences obtained from the long-read transcriptome were annotated based on National Center for Biotechnology Information (NCBI) BLASTN (https://blast.ncbi.nlm.nih.gov/). We identified 17 F-box genes that are expressed in pollen tubes and determined their chromosomal locations based on the sweet cherry genome (Shirasawa et al., 2017; Supplemental Fig. S7). Based on these F-box sequences, we synthesized a phosphorothioate antisense oligodeoxynucleotide (AS-ODN) of S4-SLFL2 and its sense control (S-ODN) and transfected them into cultured cv 21-21 and cv 21-26 pollen tubes. S4-SLFL2 expression was significantly suppressed after transfection with AS-ODN versus S-ODN, as determined by reverse transcription quantitative PCR (RT-qPCR; Fig. 6A), whereas the expression of the 16 other F-box genes had not changed under either AS-ODN or S-ODN treatment (Supplemental Fig. S8). We also detected the relative expression levels of SLFL2 in different varieties, and there was no significant difference among the varieties (Supplemental Fig. S9).
Figure 6.
S4-SLFL2 can ubiquitinate self and nonself S-RNase. A, An S4-SLFL2 antisense oligonucleotide gene was transfected into cv 21-21 and cv 21-26 pollen tubes for ubiquitin assays with self S-RNase. Error bars represent sd calculated from two biological replicates (separate biological material). Asterisks indicate significance by Student’s t test (**P < 0.01). B, The self S-RNase purified from cv 21-21 pistils was incubated with cv 21-21 and cv 21-26 pollen tube proteins and MG132 was added. The protein samples were stained by Coomassie Brilliant Blue (CBB), and S4-SLFL2 was detected by S4-SLFL2 antibody. C, An antisense S4-SLFL2 oligonucleotide gene was transfected into cv 21-21 and cv 21-26 pollen tubes for ubiquitin assays with nonself S-RNase. Error bars represent sd calculated from two biological replicates (separate biological material). Asterisks indicate significance by Student’s t test (**P < 0.01). D, The nonself S-RNase purified from cv Hongdeng (HD) pistils was incubated with cv 21-21 and cv 21-26 pollen tube proteins and MG132 was added. The protein samples were stained by Coomassie Brilliant Blue, and S4-SLFL2 was detected by S4-SLFL2 antibody. α-S4-RNase, S4-RNase antibody; α-S4-SLFL2, S4-SLFL2 antibody; α-ubiquitin, ubiquitin antibody.
We extracted proteins from S4-SLFL2-suppressed pollen tubes from cv 21-21 and cv 21-26 and incubated them overnight with purified S4-RNase from cv 21-21 and MG132. To confirm that the translation level of S4-SLFL2 was also suppressed, we generated anti-S4-SLFL2 monoclonal antibodies to detect the levels of S4-SLFL2 in pollen tube proteins. As expected, S4-SLFL2 levels were reduced in S4-SLFL2-suppressed pollen tubes from both cv 21-21 and cv 21-26 (Fig. 6B). We also used anti-S4-RNase and anti-ubiquitin antibody to detect the degree of ubiquitination of proteins from S4-SLFL2-suppressed pollen tubes. The degree of ubiquitination of S4-RNase was substantially reduced (Fig. 6B), indicating that S4-SLFL2 is involved in the ubiquitination of S4-RNase. As S4-SLFL2 also interacts with S3-RNase and S9-RNase, we investigated whether S4-SLFL2 might participate in the ubiquitination of S3-RNase and S9-RNase. We purified S-RNase from cv Hongdeng (containing S3-RNase and S9-RNase) and incubated it overnight with S4-SLFL2-suppressed cv 21-21 and cv 21-26 pollen tube proteins plus MG132. After confirming that the transcript level of S4-SLFL2 was suppressed (Fig. 6C) and the content of S4-SLFL2 was reduced (Fig. 6D), we detected S-RNases using anti-S4-RNase and anti-ubiquitin antibodies. The level of ubiquitin-labeled S-RNase was also reduced in the samples, suggesting that S4-SLFL2 is involved in the ubiquitination of S3-RNase and S9-RNase.
To further confirm the notion that S4-SLFL2 participates in the ubiquitination of S-RNase, we expressed S4-RNase, S3-RNase, S9-RNase, and S4-SLFL2 fused with SUMO and His tag as well as Cullin 1, Skp1, and Rbx1 fused with GST tag in E. coli and subjected these proteins to an in vitro ubiquitination assay using anti-S4-RNase, His, and GST antibodies for detection. In the absence of ubiquitin or S4-SLFL2, ubiquitin-labeled S-RNase was not detected. However, when both ubiquitin and S4-SLFL2 were applied to the reaction, self S4-RNase, nonself S3-RNase, and S9-RNase were labeled (Supplemental Fig. S10A), suggesting that S4-SLFL2 is required for the ubiquitination of S-RNase. Together, these findings indicate that S4-SLFL2 participates in the ubiquitination of both self and nonself S-RNase.
Pollen Tip-Localized SFB4 Ubiquitinates S4-RNase and Exhibits Different Activity from S4-SLFL2
In S4′ pollen tubes, SFB4′ has lost its recognition capability, whereas in S4 pollen tubes, both SFB4 and S4-SLFL2 interact with S4-RNase. We demonstrated that S4-SLFL2 participates in the ubiquitination of S4-RNase. To explore the function of SFB4, we purified S4-RNase and SFB4/SFB4′ fused with SUMO and His tag from E. coli and incubated the proteins with Cullin 1, Skp1, and Rbx1 in an in vitro ubiquitination assay. Labeled S4-RNase was not detected when ubiquitin was absent or SFB4 was replaced by SFB4′. However, when ubiquitin and SFB4 were added to the reaction mixture, labeled S4-RNase was observed (Supplemental Fig. S10B). These results suggest that SFB4 also ubiquitinates S4-RNase.
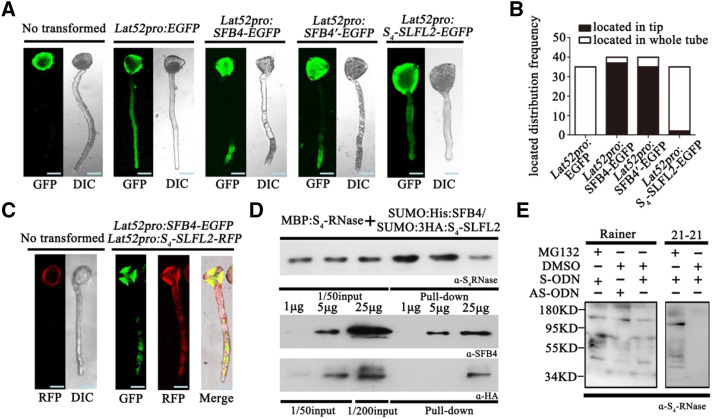
As both SFB4 and S4-SLFL2 ubiquitinated S4-RNase in vitro, we explored the functions of these proteins in S4 pollen tubes. We used the pollen-specific Lat52 promoter to drive the expression of SFB4, SFB4′, and S4-SLFL2 in sweet cherry pollen tubes. We fused the full-length SFB4, SFB4′, and S4-SLFL2 sequences to enhanced GFP (EGFP) in the pEZS vector and transformed these constructs into cv Rainer and cv 21-21 pollen tubes by particle bombardment. Transformation with empty vector containing Lat52-driven GFP and nontransformed pollen was used as the negative controls. After 3 h of culture, fluorescence from EGFP was captured by confocal microscopy. In nontransformed cv Rainer and cv 21-21 pollen tubes, fluorescence was observed in pollen grains but not pollen tubes, showing that there was autofluorescence in the pollen grains (Fig. 7A; Supplemental Fig. S11A). By contrast, in transformed pollen tubes, fluorescence was detected in pollen tubes, suggesting that this fluorescence was from EGFP. In pollen tubes transformed with EGFP and S4-SLFL2-EGFP, fluorescence was observed throughout the pollen tubes, indicating that GFP and S4-SLFL2 were evenly distributed in the pollen tube cytoplasm, whereas in pollen tubes transformed with SFB4-EGFP and SFB4′-EGFP, fluorescence was mainly observed at the tip of the pollen tube (Fig. 7A; Supplemental Fig. S10A).
Figure 7.
The functions of SFB4 and S4-SLFL2 are distinctly different. A, SFB4, SFB4′, and S4-SLFL2 GFP fusions expressed in cv Rainer pollen tubes. Nontransformed and pollen tubes expressing empty vector were used as the control. DIC, Differential interference contrast. Bars = 10 μm. B, Number of cv Rainer pollen tubes where the proteins were located in the tip (black bars) or in the whole tube (white bars). C, Coexpression of SFB4 fused to GFP and S4-SLFL2 fused to RFP in cv Rainer pollen tubes. Bars = 10 μm. D, MBP pull-down assay to detect the interaction between MBP-S4-RNase and mixed SUMO-His-SFB4/SUMO-3HA-S4-SLFL2. Recombinant MBP-S4-RNase, SUMO-His-SFB4, and SUMO-3HA-S4-SLFL2 were purified from E. coli. Equal amounts of MBP-S4-RNase with 1, 5, and 25 μg of SUMO-His-SFB4 and SUMO-3HA-S4-SLFL2S4-RNase were detected by S4-RNase antibody. SFB4 was detected by SFB4 antibody, and S4-SLFL2 was detected by HA tag antibody. E, cv Rainer and cv 21-21 pollen tubes were cultured in pollen tube medium with S4-RNase in the presence and absence of MG132. After overnight incubation, pollen tube proteins were extracted, and S4-RNase antibody was used to detect the proteins. α-HA, HA tag antibody; α-S4-RNase, S4-RNase antibody; α-SFB4, SFB4 antibody.
The different distribution patterns of SFB4 and S4-SLF2 in these pollen tubes were confirmed by quantification. In SFB4-transformed pollen tubes, in 37 out of 40 cv Rainer pollen tubes and 30 out of 34 cv 21-21 pollen tubes, fluorescent signals were observed in the pollen tube tip. A similar pattern was observed for SFB4′-transformed pollen tubes, where 35 out of 40 cv Rainer pollen tubes and 31 out of 36 cv 21-21 pollen tubes showed fluorescence in the pollen tube tip. By contrast, in S4-SLFL2-transformed pollen tubes, only two out of 35 cv Rainer pollen tubes and zero out of 32 cv 21-21 pollen tubes showed fluorescent signals in the pollen tube tip (Fig. 7B; Supplemental Fig. S11B). As SFB4, SFB4′, and S4-SLFL2 were all driven by the same promoter and the intensity of stimulated luminescence was also the same, these findings indicate that SFB4/SFB4′ and S4-SLFL2 have distinct distribution patterns and that SFB4/SFB4′ mainly localizes to the tip of the pollen tube.
To further investigate the locations of SFB4 and S4-SLFL2 in the pollen tube tip, we cotransformed S4-SLFL2 fused with RFP and SFB4 fused with EGFP into cv Rainer pollen tubes; nontransformed pollen tubes were used as the negative control. In nontransformed pollen tubes, RFP fluorescence was only detected in pollen grains, whereas in cotransformed pollen tubes, fluorescent signals from both RFP and EGFP were detected at the tip of pollen tubes. These results indicate that SFB4 and S4-SLFL2 only colocalize to the tip of pollen tubes (Fig. 7C). To explore the recognition between S4-RNase and SFB4/S4-SLFL2 in pollen tube tips, we performed an in vitro pull-down assay. After purifying recombinant MBP-S4-RNase, SUMO-His-SFB4, and SUMO-3HA-S4-SLFL2 from E. coli, we coincubated equal amounts of MBP-S4-RNase with 1, 5, and 25 μg of SUMO-His-SFB4 and SUMO-3HA-S4-SLFL2. After MBP pull-down analysis, both SFB4 and S4-SLFL2 were detected using the combination of MBP-S4-RNase and SUMO-His-SFB4 (25 μg)/SUMO-3HA-S4-SLFL2 (25 μg), whereas when we used the combination of MBP-S4-RNase and SUMO-His-SFB4 (5 μg)/SUMO-3HA-S4-SLFL2 (5 μg), only SFB4 was detected. These results suggest that S4-RNase more readily interacts with SFB4 than with S4-SLFL2 (Fig. 7D).
To confirm this notion, we performed a BiFC assay. S4-RNase and SFB4/S4-SLFL2 were fused to the N or C terminus of YFP (YFPn and YFPc), and the full-length SFB4 and S4-SLFL2 sequences were inserted into the same vector but with the YFP sequence eliminated. A. tumefaciens containing S4-RNase-YFPn was coinfiltrated into 10 N. benthamiana leaves with two equal parts of A. tumefaciens containing SFB4-YFPc and S4-SLFL2 at the same concentrations; the Skp1-like gene was also infiltrated into the N. benthamiana leaves. After 5 d of infiltration, YFP fluorescence was observed in all 10 leaves. When S4-RNase-YFPn was coexpressed with SFB4 and S4-SLFL2-YFPc in 10 leaves in the same manner, YFP fluorescence was also observed in the 10 leaves. When SFB4-YFPc and 10 times the amount of S4-SLFL2 were coexpressed with S4-RNase-YFPn in 10 leaves, YFP fluorescence was observed in six leaves, and when SFB4 and 10 times the amount of S4-SLFL2-YFPc were coexpressed with S4-RNase-YFPn in 10 leaves, fluorescence was not detected in any of the leaves (Supplemental Fig. S12A). These results further support the notion that S4-RNase more readily interacts with SFB4 in pollen tube compared with S4-SLFL2.
Finally, we explored the functional difference between SFB4 and S4-SLFL2 in S4 pollen tubes. When cv 21-21 pollen tubes were cultured with S4-RNase in the absence of MG132, S4-RNase was degraded in the pollen tubes. However, in cv Rainer pollen tubes, regardless of whether S4-SLFL2 was silenced, S4-RNase was not degraded (Fig. 7E). When MG132 combined with cv 21-21 S-RNase was present in cv Rainer pollen tubes, an increased amount of ubiquitinated S4-RNase was detected. Perhaps in cv Rainer S1 pollen tubes, S4-RNase is ubiquitinated and degraded (due to SLFL2 activity), whereas in cv Rainer S4 pollen tubes, S4-RNase is ubiquitinated but not degraded (due to SFB4 activity; Fig. 7E). We also examined the degradation of cv 21-21 S-RNase in extracted cv Rainer pollen tube proteins, finding that the S-RNase was ubiquitinated but not degraded regardless of the presence of MG132, perhaps due to SFB4 activity in mixed pollen extracts from S1 and S4 pollen tubes (Supplemental Fig. S12B). These results suggest that SFB4 does not promote the degradation of S4-RNase.
DISCUSSION
The sweet cherry cultivar ‘Lapins’ (S1S4′) is a progeny of ‘Stella’ (S3S4′). Both cultivars show self-compatibility. cv Stella was obtained by hybridizing cv Lambert (S3S4) with the self-compatible cv JI2420 (S4S4′) variety, whereas cv JI2420 was obtained from a cross between cv Emperor Francis (S3S4) and x-ray-induced cv Napoleon (S3S4) pollen (Lapins, 1971). A common attribute of all these self-compatible varieties is a 4-bp deletion in the SFB4 gene (Ushijima et al., 2004). This deletion has been shown to be closely linked to self-compatibility (Zhu et al., 2004; Muñoz‐Espinoza et al., 2017), but the mechanistic relationship between this mutation and the self-compatibility of S4′ pollen is unclear. In this study, we investigated the mechanism behind the self-compatibility of cv Lapins. We demonstrated that SFB4′ lost the ability to interact with S4-RNase and that S4′ pollen tubes are able to degrade self S-RNase via the 26S proteasome pathway. S4-SLFL2, which was identified from S4′, interacts with and ubiquitinates S-RNase, resulting in its degradation. Based on a comparison of the interactions of SFB4 and S4-SLFL2 with S4-RNase in pollen harboring the S4 haplotype, we propose that the self-compatibility mechanism of S4′ pollen involves the ubiquitin-mediated degradation of S-RNase by S4-SLFL2 when it is not recognized by SFB4.
Although the SFB4′ mutation in S4′ pollen tubes may lead to the early termination of the protein during translation, it was not clear whether SFB4′ protein was still present in S4′ pollen tubes. We determined that SFB4′ is ∼6 kD smaller than the wild-type SFB4. In general, changes in amino acid sequences have one of two outcomes: the gain of a new function or a loss of function (Boyle, 2005). If SFB4′ gained a new function, it would have inhibited self S4-RNase; however, in our analysis, SFB4′ did not physically interact with S4-RNase, indicating that SFB4′ protein might lose its specific function against S-RNase (Matsumoto et al., 2012). Until now, the molecular mechanism underlying how S4′ pollen avoids S4-RNase-mediated arrest has remained unclear. Petunia, like sweet cherry, exhibits S-RNase-based self-incompatibility (Fujii et al., 2016). Petunia pollen usually inhibits S-RNase activity via ubiquitin modification and degradation through the 26S proteasome pathway (Entani et al., 2014). We hypothesized that in sweet cherry, S4′ pollen inhibits S-RNase via the ubiquitin-proteasome pathway. Indeed, S4′ homozygous pollen tube protein could induce S-RNase degradation through the 26S proteasome pathway, which is consistent with our hypothesis. However, it is not clear which protein in S4′ pollen tubes plays a role in S-RNase inhibition. The petunia SLF protein from nonself S-loci participates in the degradation of self S-RNase (Kubo et al., 2010; Entani et al., 2014; Sun et al., 2018). In sweet cherry S4′ pollen, SFB4′ protein has lost its function, but some studies have shown that the S-locus SLFL protein interacts with S-RNase (Matsumoto and Tao, 2016; Chen et al., 2018), and the SLFL sequence shares higher levels of sequence similarity with SLF than with SFB in petunia (Akagi et al., 2016). It was previously proposed that SLFL protein in sweet cherry is also able to inhibit S-RNase activity (Matsumoto and Tao, 2016). Indeed, our pull-down assays showed that SLFL2 from S4′ pollen tubes interacts with S4-RNase, and a semi-in vivo ubiquitination assay demonstrated that S-RNase is ubiquitinated and degraded.
The single-copy SFB gene from the S-locus is a typical F-box gene based on sequence and protein structure (Ushijima et al., 2003; Yamane et al., 2003). SFB interacts with Skp1 and Cullin to form SCF complexes (Matsumoto et al., 2012), which is consistent with its proposed role in ubiquitin labeling of its substrates. However, the finding that S4′ pollen shows self-compatibility indicates that S4-RNase plays a role in inhibiting self-pollen growth when the SFB4 gene is wild type. Consequently, it has been proposed that SFB protein recognizes self S-RNase and protects its ability to inhibit self-pollen tube formation (Meng et al., 2011; Matsumoto et al., 2012).
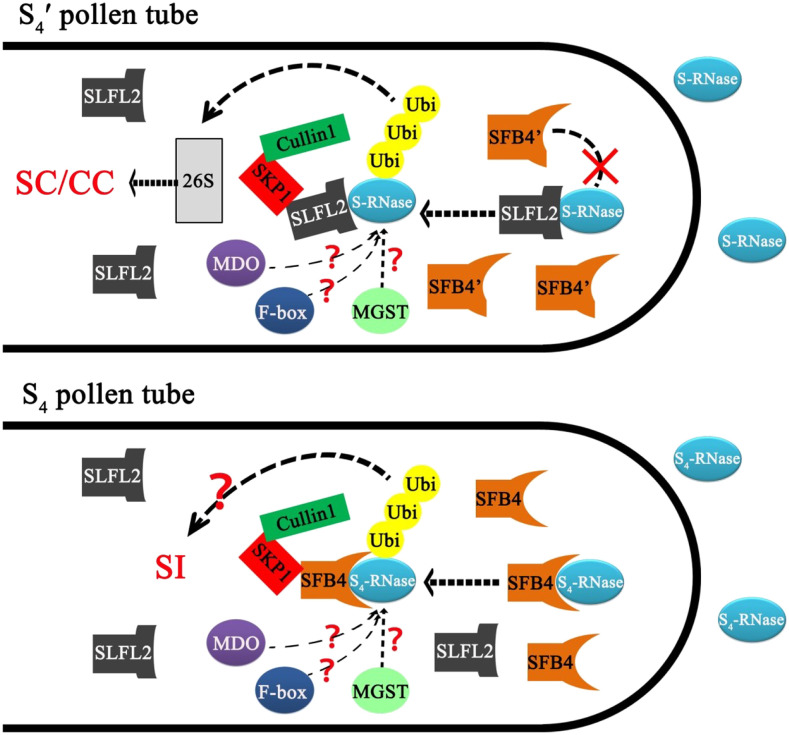
Here, we established that SFB4 protein is distributed in pollen tube tips, unlike SLFL2, which is localized to cytoplasm and primarily functions in the ubiquitin labeling of S-RNase for degradation. Ubiquitination is not only correlated to protein degradation but also for regulating protein endocytosis and other processes (Dubeaux and Vert, 2017). Thus, we speculate that the function of SFB protein is different from that of SLFL2, in that it may not contribute to the degradation of S-RNase. Rather, it recognizes S-RNase via an unknown mechanism. Whereas in S4′ pollen tubes, SFB4′ lost its ability to recognize S4-RNase, S4-SLFL2 participates in the ubiquitination and degradation of S4-RNase, which contributes to the self-compatibility of S4′ pollen tubes (Fig. 8).
Figure 8.
Schematic diagram showing the model of the self-incompatibility mechanism in S4′ pollen tubes in sweet cherry. When the S4′ pollen tube grows in pistils harboring the S4 allele or other S alleles, S-RNase enters into pollen tubes and may be regulated by some proteins like M-locus-encoded GST (MGST) or M-locus-encoded disulfide bond A-like oxidoreductase (MDO). The SFB4′ protein is located at the tip of the pollen tube, but it does not interact with S-RNase. In contrast, the S4-SLFL protein interacts with S-RNase and ubiquitinates it, the labeled S-RNase is degraded via the 26S proteasome pathway, and so it cannot inhibit the growth of the pollen tube, resulting in compatibility regardless of self- or cross-pollination. Other F-box proteins may also bind to S-RNase and be involved in self-incompatibility, but the mechanism is unclear. In S4 pollen tubes, SFB4 protein could recognize S4-RNase when it grows in pistils with the S4 haplotype. S4-RNase was labeled by ubiquitin, which led to self-incompatibility via an unknown pathway. CC, Cross-compatibility; SC, self-compatibility; SI, self-incompatibility.
Various proteins such as M-locus-encoded GST and M-locus-encoded disulfide bond A-like oxidoreductase also help maintain self-incompatibility in sweet cherry and related species such as apricot (Wünsch et al., 2010; Zuriaga et al., 2013; Muñoz-Sanz et al., 2017; Ono et al., 2018). An F-box gene other than SLF also encodes a protein that binds to S-RNase (Matsumoto and Tao, 2019), suggesting that it might play a role in self-incompatibility. However, the roles of these proteins in S4 or S4′ pollen tubes is unclear (Fig. 8). Indeed, the mechanism of self-incompatibility in sweet cherry remains largely unknown and requires further investigation.
MATERIALS AND METHODS
Plant Materials, Pollination, and Determination of S Genotype
Ten-year-old sweet cherry (Prunus avium) ‘Rainer’, ‘Lapins’, ‘Hongdeng’, ‘21-21’, and ‘21-26’ were grown at the Beijing Academy of Agriculture and Forestry. The cv 21-21 and cv 21-26 varieties are offspring of cv Lapins, following self-pollination 8 years ago. The anthers and pistils of each variety were collected in the spring, when the flowers were just opening. Pistils were separated from the flowers and stored at –80°C. Anthers were dissected from the filaments, and they were in a shady and cool condition until the pollen grains were released. The pollen grains were stored at –20°C. Self- and cross-pollination experiments were performed on the above-mentioned sweet cherry trees, and the progeny were cultured in a greenhouse until the leaves were sampled. Leaf DNA was extracted for S genotype determination, as previously described (Chen et al., 2004; Wang et al., 2010); the primers for S genotype detection are listed in Supplemental Table S4.
Extraction of Pollen Tube Proteins
Pollen grains were suspended in liquid germination medium (10% [w/v] Suc, 0.01% [w/v] H3BO3, and 0.015% [w/v] CaCl2). After 3 h at room temperature, the growth of pollen tubes was observed by microscopy and germinated pollen tubes were counted to determine the pollen germination rate. The liquid germination medium was removed using a pipette, and the pollen was washed three times with phosphate-buffered saline (PBS; pH 7). Pollen tube proteins were extracted using a plant protein extraction solution kit (Huaxing Bio, HX18612). Approximately 200 mg of pollen tubes was mixed with 0.5 mL of precooled extraction solution, ground in a pestle and mortar on ice, incubated at 4°C for 30 min, and then centrifuged (12,000g, 15 min, and 4°C). The supernatant was collected and stored at –80°C.
Purification of the S-RNase Fraction from Pistils
Sixty pistils of cv 21-21 or cv Hongdeng were homogenized in precooled acetone using a glass mortar and a 1.5-mL centrifuge tube. After centrifugation at 20,000g for 15 min, the supernatants were discarded. The sediment was air dried and then suspended in extraction buffer (50 mm PBS, pH 7, 1 mm dithiothreitol [DTT], 1% [v/v] Nonidet P-40, and protease inhibitor cocktail (EDTA-free; Roche). After centrifugation at 20,000g for 15 min, the supernatant was filtered through a 0.22-μm filter (Millipore, SLGP033RB) and the S-RNase fraction was purified using an AKTA FPLC system with a mono S 5/50 GL column (GE Healthcare). The AKTA FPLC system including the cation-exchange column was first washed with buffer A (50 mm PBS, pH 7), and then the filtered supernatant was loaded. Gradient-raised buffer B (50 mm PBS, pH 7, and 1 m NaCl) was used to elute the S-RNase fraction. S-RNase usually eluted when the salt conductivity was almost 13. The S-RNase sample volume was reduced using an Amicon Ultra-0.5 (Millipore) spin column.
Antibody Preparation
SFB4/SFB4′ Rat Monoclonal Antibody Preparation
A highly specific SFB4/SFB4′ protein fragment was identified (Supplemental Fig. S1A), peptides were synthesized in vitro corresponding to these sequences, and then rat monoclonal antibodies were generated to these peptides by the Beijing Genomics Institute.
S4-RNase Rabbit Polyclonal Antibody Preparation
The S4-RNase cDNA sequence from pistils of ‘Rainer’ sweet cherry varieties was inserted into the pMal-c2x (http://www.addgene.org) vector and transformed into Transetta (DE3) chemically competent cells (TransGen Biotech, CD801-01) after the signal peptide sequence was removed. Culture growth was induced at 16°C for 16 h, and the cells were then suspended in cell buffer (10 mm Tris-HCl, pH 7.4, and 30 mm NaCl). The Escherichia coli suspensions were sonicated for 20 min and centrifuged (12,000g, 1 h, and 4°C), and the supernatants were collected and incubated with MBP Amylose Beads (NEB, E8021V) at 4°C for 1 h. After discarding the supernatant, the remaining beads were washed three times with wash buffer (20 mm Tris-HCl, pH 7.4, 1 mm EDTA, 200 mm NaCl, and 1 mm DTT) and eluted with elution buffer (20 mm Tris-HCl, pH 7.4, 1 mm EDTA, 200 mm NaCl, 1 mm DTT, and 10 mm maltose). The eluted protein was detected by Coomassie Brilliant Blue staining and peptide mass fingerprinting (Supplemental Fig. S2).
S4-SLFL2 Mouse Monoclonal Antibody Preparation
The full-length S4-SLFL2 sequence was inserted into a modified pET28a vector containing a highly soluble and monomeric tag named SUMO (Liu and Gao, 2018) between the NheI and BamHI sites. The vector was transformed into BL21(DE3)pLysS chemically competent cells (TransGen Biotech, CD701-01), which were induced at 16°C for 16 h and resuspended in cell buffer (10 mm Tris-HCl, pH 7.4, and 30 mm NaCl). The E. coli suspensions were sonicated for 20 min, centrifuged (12,000g, 1 h, and 4°C), and the supernatants were incubated with Ni-NTA resin (Thermo Scientific, 88221) at 4°C for 1 h. After discarding the supernatants, the Ni-NTA resin was washed three times with 10 column volumes of wash buffer (20 mm Tris-HCl, pH 7.4, 20 mm imidazole, and 500 mm NaCl), then eluted with elution buffer (20 mm Tris-HCl, pH 7.4, 500 mm imidazole, and 500 mm NaCl). The eluted S4-SLFL2 was used to generate rat monoclonal antibodies by the Beijing Genomics Institute.
Hyp Rabbit Polyclonal Antibody Preparation
In our S-RNase purification system, after the S-RNase was eluted, another protein could be eluted, the mass of which was almost 53 kD (Supplemental Fig. S5A). This protein was used to generate rabbit polyclonal antibody by BaiTaiLong.
In Vitro Pull-Down and Immunoprecipitation Assays
The cDNA sequences of S3-RNase, S4-RNase, and S9-RNase from pistils of ‘Rainer’ and ‘Hongdeng’ sweet cherry varieties were inserted into the pMal-c2x (http://www.addgene.org) vector and transformed into Transetta (DE3) chemically competent cells (TransGen Biotech, CD801-01) after the signal peptide sequence was removed. Culture growth was induced at 16°C for 16 h, and the cells were then suspended in cell buffer (10 mm Tris-HCl, pH 7.4, and 30 mm NaCl). The E. coli suspensions were sonicated for 20 min, centrifuged (12,000g, 1 h, and 4°C), and the supernatants were collected and incubated with MBP Amylose Beads (NEB, E8035) at 4°C for 1 h. After discarding the supernatant, the remaining beads were washed and eluted, as described above. Full-length SFB4, SFB4′, and S4-SLFL2 and truncated SFB4 sequences were inserted into a modified pET28a vector containing a highly soluble and monomeric tag named SUMO. 3×HA tag was then inserted before the 5′ region of S4-SLFL2 to construct the SUMO-3HA-S4-SLFL2 vector. The resulting vectors were transformed into BL21(DE3)pLysS chemically competent cells (TransGen Biotech, CD701-01). The proteins were purified as described above.
For the MBP-tagged pull-down analysis, purified MBP-tagged S-RNase proteins were incubated with SFB, S4-SLFL2, or the proteins extracted from the pollen .tube of ‘21-21’ and ‘Bin’ sweet cherry varieties for 2 h at 4°C. After discarding the supernatant, MBP Amylose Beads (NEB, E8035) were rinsed three times with 500 μL of balance buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, and 1 mm DTT). The beads were then incubated with balance buffer plus 10 mm maltose, and the elution product was boiled with SDS-PAGE loading buffer (CWBIO, cw0028s) for 10 min and subjected to immunoblot analysis with His tag or SFB4 antibodies.
For semi-in vivo immunoprecipitation of S-RNase and SFB4, equal amounts of total pollen tube and stylar proteins (3–5 µg) were added to 1 mL of lysis buffer (50 mm PBS, pH 7.4, 1 mm DTT, 1% [v/v] Nonidet P-40, and protease inhibitor cocktail [EDTA-free; Roche]) and incubated for 2 h at 4°C. Twenty microliters of SFB4 monoclonal antibody was incubated with 20 µL (bed volume) of protein G-Sepharose beads (Roche Healthcare) for 2 h at 4°C, incubated with the protein mixture, and then centrifuged. The immunoprecipitates were washed three times with lysis buffer, and the concentrates were then resuspended in SDS-PAGE loading buffer (CWBIO, cw0028s), boiled for 10 min, and subjected to immunoblot analysis with S4-RNase antibodies.
For the MBP-binding assay (Fig. 7D) between MBP-S4-RNase and SUMO-His-SFB4/SUMO-3HA-S4-SLFL2, MBP-S4-RNase was coincubated with 1 μg of SUMO-His-SFB4 and 1 μg of SUMO-3HA-S4-SLFL2, 5 μg of SUMO-His-SFB4, 5 μg of SUMO-3HA-S4-SLFL2, 25 μg of SUMO-His-SFB4, and 25 μg of SUMO-3HA-S4-SLFL2 for 2 h at 4°C. After discarding the supernatant, MBP Amylose Beads (NEB, E8035) were rinsed three times with 500 μL of balance buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, and 1 mm DTT). The beads were then incubated with balance buffer plus 10 mm maltose, after which the elution product was boiled with SDS-PAGE loading buffer (CWBIO, cw0028s) for 10 min and then subjected to immunoblot analysis with HA tag or SFB4 antibodies.
BiFC Assays
Full-length or truncated S-RNase, SFB4, SFB4′, and S4-SLFL2 sequences were cloned into pCambia1300 35S-CYFPn-c vector (Meng et al., 2014) harboring a YFP coding sequence to generate either N-terminal or C-terminal fusion proteins. The full-length SFB4, S4-SLFL2, and Skp1 were also cloned into pCambia1300 vector with the YFP sequence missing. The resulting constructs were transformed into Agrobacterium tumefaciens strain EHA105. The vector-containing A. tumefaciens strains were infiltrated into 2-month-old Nicotiana benthamiana leaves for transient expression (Meng et al., 2014). When S4-SLFL2 was expressed in N. benthamiana leaves, a Skp1-like gene was also expressed together to maintain the stability of S4-SLFL2 (Matsumoto and Tao, 2016). YFP fluorescence was imaged 5 d after transformation using an Olympus BX61 confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, the light intensity was 250, and emission fluorescence was detected at 500 to 542 nm.
Ubiquitin Assay
For semi-in vivo protein ubiquitin assays, 25 μg of the S-RNase extracts was mixed with pollen tube protein and DMSO/MG132 (MedchemExpress, 133407-82-6), and ubiquitination assays were carried out overnight at 37°C. For the detection of S-RNases and ubiquitin by immunoblotting, rabbit S-RNase antisera and anti-ubiquitin antisera (Biyuntian Technology) were used as primary antibodies at 1:500 and 1:1,000 dilutions, respectively. The rabbit S-RNase antibody was produced as previously described.
For in vitro protein ubiquitin assays, full-length S3-RNase, S4-RNase, and S9-RNase were inserted into a modified pET28a vector containing a highly soluble and monomeric tag named SUMO, and the vectors were transformed into BL21(DE3)pLysS chemically competent cells (TransGen Biotech, CD701-01). These proteins were purified as described above. Different S-RNases were mixed with E1, E2, and ubiquitin (Enzo Life Science, BML-UW9410-0050, BML-UW9020-0100, and BML-UW8610-0001, respectively), ATP (Thermo Fisher Scientific, PV3227), and three subunits (GST-Skp1, GST-Cullin, and GST-Rbx1) of SCF complex purified from an E. coli system as previous described (Chen et al., 2018). SFB4, SFB4′, and S4-SLFL2 that contained SUMO and His tag were purified from E. coli and also added into the mixture, which was incubated overnight at 37°C. Rabbit S4-RNase antibody was used to detect the signal.
For in vivo protein ubiquitin assays, the pollen tubes of different varieties were first cultured in germination medium (10% [w/v] Suc, 0.01% [w/v] H3BO3, and 0.015% [w/v] CaCl2) for 30 min, the expression of S4-SLFL2 was down-regulated by antisense oligonucleotide, and 200 μg of S4-RNase protein purified from cv 21-21 was added into germination medium overnight. The pollen tubes were then collected and washed in 50 mm PBS. Pollen tube proteins were extracted as previously described, and western-blot analysis by rabbit S4-RNase antibody was used to confirm the presence of S4-RNase protein.
Antisense Oligonucleotide Experiments
Antisense oligonucleotide experiments were performed as previously described (Moutinho et al., 2001; Li et al., 2018). Briefly, phosphorothioated AS-ODN and S-ODN were synthesized (Taihe Biotechnology) in order to down-regulate the expression of SLFL2. The antisense and sense sequences were 5′-ctgAATTCCTCGAGTGCTGGATtgc-3′ and 5′-gcaATCCAGCACTCGAGGAATAcag-3′, respectively (where lowercase letters indicate the modified bases). For transfection, 20 μL (10 pm) of oligonucleotides (AS-ODN or S-ODN), 28 μL of cytofectin buffer, and 4 μL of cytofectin were premixed and added immediately to 200 μL of germination medium including pollen tubes. After 2 h, pollen tubes were collected for RNA and protein extraction.
PacBio Single-Molecule Sequencing of Pollen Tubes and Identification of F-Box Genes
Pollen grains from ‘Rainer’, ‘Lapins’, and ‘21-21’ sweet cherry varieties were cultured as described above. Then the liquid germination medium was removed with a pipette and the pollen samples were washed three times with PBS buffer. RNA was extracted from the pollen using an EASY Spin Plus Plant RNA Kit (Aidlab, RN40) and stored at –80°C. One microgram of pollen tube RNA of each variety was mixed as the sample for library construction. RNA samples were reverse transcribed using the Clontech SMARTer cDNA synthesis kit (TaKaRa) in separate PCR tubes. To generate barcoded full-length cDNA, three RT reactions were run in parallel. PCR optimization was used to determine the optimal amplification cycle number for the downstream large-scale PCR. A single primer (primer IIA from the Clontech SMARTer kit; 5′-AAGCAGTGGTATCAACGCAGAGTAC-3′) was used for all PCRs following RT. Large-scale PCR products were purified with AMPure PB beads, and quality control was performed on a 2100 BioAnalyzer (Agilent). Size fractions eluted from the run were subjected to quality control and pooled in equimolar ratios for subsequent reamplification to yield three libraries (1–2, 2–3, and 3–6). A total of six SMRT cells were sequenced on the PacBio RS II platform using P6-C4 chemistry with 3- to 4-h movies by the Tianjin Biological Chip Technology.
The SMRT analysis software (version smrtanalysis_2.3.0.140936.p4.150482) was used to identify final isoforms from raw reads. First, the raw reads were processed into error-corrected reads of inserts with minFullPass = 0 and minPredictedAccuracy = 0.8. Then, the full-length nonchimeric transcripts were determined by searching for the poly(A) tail signal and the 5′ and 3′ cDNA primers. Iterative Clustering for Error Correction was used to obtain consensus isoforms. Non-full-length sequences were used to filter the consensus isoform sequences by Quiver software to obtain the polished isoforms, with the parameter set at 0.99. Full-length and non-full-length reads were aligned to the sweet cherry genome using the Genome Mapping and Alignment Program (Wu and Watanabe, 2005). Next, insertions/deletions and mismatches were corrected using the reference sweet cherry genome (http://cherry.kazusa.or.jp/).
The final sequence was annotated by NCBI BLASTN (https://blast.ncbi.nlm.nih.gov/) with a cutoff e-value of 1e-10. F-box genes were selected according to the annotation results. We obtained a total of 17 F-box genes, all of which were successfully amplified in the three cultivars. SLFL2 was present in all three cultivars, and the sequence was identical across them. In fact, except for SFB (including SFB1 and SFB4, the sequence homology is 86.83%) and SLFL1 (including S1-SLFL1 and S4-SLFL1, the sequence homology is 92.28%), the remaining genes were identical across the cultivars. To better understand the 17 F-box genes, they were compared with the sweet cherry genome (http://cherry.kazusa.or.jp/). The genome was released in 2017, after a sweet cherry variety named ‘Satonishiki’ was sequenced. Supplemental Figure S7 was drawn when the location of each F-box gene from the sweet cherry genome was obtained.
LC-MS Analysis to Identify S4-RNase-Interacting Proteins in Pollen Tubes
After MBP pull-down of the two combinations (MBP-S4-RNase/cv 21-21 pollen tube protein and MBP/cv 21-21 pollen tube protein), the pH of the two eluted protein samples was adjusted to 8.5 with 1 m ammonium bicarbonate, followed by chemical reduction for 1 h at 60°C by adjusting the solution to 10 mm DTT, and then carboxyamidomethylating the samples in a final concentration of 55 mm iodoacetamide for 45 min at room temperature in the dark. Trypsin Gold (Promega) was added to a final substrate:enzyme ratio of 30:1 (w/w). The trypsin digest was performed at 37°C for 16 h. After digestion, the peptide mixture was acidified with 10 μL of formic acid and subjected to MS analysis. After protein digestion, each peptide sample was desalted using a Strata X column (Phenomenex), vacuum dried, and then resuspended in 200 μL of buffer A (2% [v/v] acetonitrile and 0.1% [v/v] formic acid). After centrifugation (20,000g, 4°C, and 10 min), the supernatant was recovered to obtain a peptide solution with a final concentration of approximately 0.5 μg μL−1. Approximately 10 μL of supernatant was loaded onto a 2-cm C18 trap column (Waters, 186002808) in an LC-20AD nano-HPLC system (Shimadzu) using an autosampler. The peptides were eluted onto a 10-cm analytical C18 column (i.d. 75 μm). The samples were loaded at 8 μL min−1 for 4 min, and a 44-min gradient was run at 300 nL min−1, starting with 2% to 80% buffer B (98% [v/v] acetonitrile and 0.1% [v/v] formic acid), followed by a 2-min linear gradient to 80%, maintenance at 80% buffer B for 4 min, and finally a return to 5% over 1 min. The peptides were subjected to nanoelectrospray ionization followed by tandem mass spectrometry in a Q EXACTIVE (Thermo Fisher Scientific) at Beijing Qinglian Biotech.
Before analyzing the LC-MS data, the obtained nucleotide sequences from PacBio sequencing were first translated to amino acid sequences, and they were assembled as the pollen tube protein database. The resulting tandem mass spectrometry data were processed using Proteome Discoverer 1.4, with the pollen tube protein database as the reference. The search parameters were as follows: Enzyme, trypsin; Static modification, C carboxyamidomethylation (57.021 D)/M oxidation (15.995 D); Precursor ion mass tolerance, ±15 ppm; Fragment ion mass tolerance, ±20 amu; Max missed cleavages, 2. After comparing the identified proteins in the two samples, the proteins that only existed in the sample of MBP-S4-RNase/cv 21-21 pollen tube protein were considered as the candidate proteins that interacted with S4-RNase.
RT-qPCR
The first cDNA strand of total RNA from pollen tubes of different varieties of sweet cherry was synthesized using oligo(dT) primers, and RT-qPCR was performed using the SuperReal PreMix Plus kit (SYBR Green; Tiangen, FP205-01). The relative RNA abundance was calculated using the 2−ΔΔCT method as previously described (Zhang et al., 2018), and the sweet cherry ubiquitin gene was used as a reference gene. Primers used for RT-qPCR are listed in Supplemental Table S4.
Subcellular Localization in Pollen Tubes
To determine the subcellular localization of SFB4, SFB4′, and S4-SLFL2 in sweet cherry pollen tubes, the SFB4, SFB4′, and S4-SLFL2 coding sequences were cloned into the pEZS-NL vector containing EGFP (Li et al., 2018) driven by the Lat52 promoter. In the Lat52-S4-SLFL2-EGFP vector, the sequence of EGFP was replaced by the sequence of RFP. For single-vector transformation, 1 µg of recombinant plasmid was mixed with 4 μL of spermidine (0.1 m), 10 μL of CaCl2 (2.5 m), and 6 μL of standard concentration powdered gold (Meng et al., 2014). A 5-min vortex oscillation was used to mix each ingredient. Particle bombardment (Bio-Rad, PDS-1000) was used to deliver the recombinant plasmid into sweet cherry pollen grains of ‘21-21’ and ‘Rainer’. Before the transformation, 1 mg of pollen grains was hydrated in pollen culture medium (10% [w/v] Suc, 0.01% [w/v] H3BO3, and 0.015% [w/v] CaCl2) for 30 min, then the pollen grains were transferred onto a piece of nitrocellulose filter membrane and rapidly dried in laminar flow. The membrane was put on the Target Shelf, and the mixture containing vector was loaded on the Microcarrier Launch Assembly. Then the pollen grains were transformed with a rupture disk pressure of 1,100 p.s.i. For the coexpression assay, 1 µg of plasmid was divided into 0.5 µg of Lat52-SFB4-EGFP vector and 0.5 µg of Lat52-S4-SLFL2-EGFP vector. After particle bombardment, pollen samples were cultured for approximately 3 h and imaged for GFP/RFP fluorescence using an Olympus BX61 confocal laser scanning microscope. Fluorescence was detected at 493 to 542 nm for GFP and at 578 to 625 nm for RFP. The intensity of the excitation light was 400.
Accession Numbers
Sequence data from this article can be found in the GenBank/NCBI data libraries under the following accession numbers: XM_021978640.1 (sweet cherry polyubiquitin), AJ298312.1 (sweet cherry S3-RNase), AB028154.1 (sweet cherry S4-RNase), AJ635270.1 (sweet cherry S9-RNase), JQ322646.1 (Skp1-like protein1), JQ322649.1 (Cullin1-like protein A), and AB280954.1 (S4-SLFL2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The SFB4-specific monoclonal antibody was prepared and tested.
Supplemental Figure S2. Coomassie Brilliant Blue staining of S4-RNase fused with an MBP tag.
Supplemental Figure S3. S-RNase purified from cv 21-21 (S4′S4′) and cv Hongdeng (S3S9) pistils were tested by Coomassie Brilliant Blue staining and immunoblot analysis.
Supplemental Figure S4. The ubiquitin-labeled pollen tube proteins could not be detected by immunoblot analysis.
Supplemental Figure S5. Hyp protein purified from pistils could not react with pollen tube protein.
Supplemental Figure S6. The SLFL2 protein was identified by LC-MS analysis using S4-RNase as a bait to pull down interacting cv 21-21 pollen tube proteins.
Supplemental Figure S7. Distribution of pollen tube-expressed F-box genes on different chromosomes according to the long-read sequencing data and the sweet cherry genome sequence.
Supplemental Figure S8. Relative expression levels of F-box genes, other than S4-SLFL2, in cv 21-21 and cv 21-26 pollen tubes after S4-SLFL2 silencing by AS-ODN and S-ODN.
Supplemental Figure S9. Relative expression levels (RT-qPCR) of SLFL2 in different varieties.
Supplemental Figure S10. In vitro ubiquitination assay for self S4-RNase and nonself S3-RNase/S9-RNase.
Supplemental Figure S11. SFB4 and S4-SLFL2 subcellular localization are distinctly different.
Supplemental Figure S12. S4-RNase more readily interacts with SFB4 compared with S4-SLFL2, and the ubiquitinated S-RNase labeled by cv Rainer pollen tube protein was not degraded.
Supplemental Table S1. Self-fruiting rates and cross-fruiting rates in sweet cherry cultivars.
Supplemental Table S2. S genotype of sweet cherry progeny from different pollination.
Supplemental Table S3. Self-fruiting rates or cross-fruiting rates of sweet cherry harboring homozygous S alleles.
Supplemental Table S4. Primers used in this study.
Acknowledgments
We thank Tao Ryutaro (Kyoto University) and Yongbiao Xue (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with in vivo S-RNase purification.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31630066 and 31272123) and The Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (grant no. CEFF–PXM2019_014207_000032).
References
- Akagi T, Henry IM, Morimoto T, Tao R(2016) Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S Locus-related F-box genes. Plant Cell Physiol 57: 1281–1294 [DOI] [PubMed] [Google Scholar]
- Boyle J.(2005) Lehninger Principles of Biochemistry, (4th ed.): Nelson, D., and Cox, M. Biochem Mol Biol Educ 33: 74–75 [Google Scholar]
- Chen G, Zhang B, Liu L, Li Q, Zhang Y, Xie Q, Xue Y(2012) Identification of a ubiquitin-binding structure in the S-locus F-box protein controlling S-RNase-based self-incompatibility. J Genet Genomics 39: 93–102 [DOI] [PubMed] [Google Scholar]
- Chen Q, Meng D, Gu Z, Li W, Yuan H, Duan X, Yang Q, Li Y, Li T(2018) SLFL genes participate in the ubiquitination and degradation reaction of S-RNase in self-compatible peach. Front Plant Sci 9: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XL, Chen XS, Shu HR(2004) Identifying the S genotypes of sweet cherry (Prunus avium L.) cultivars. Yi Chuan Xue Bao 31: 1142–1148 [PubMed] [Google Scholar]
- Dubeaux G, Vert G(2017) Zooming into plant ubiquitin-mediated endocytosis. Curr Opin Plant Biol 40: 56–62 [DOI] [PubMed] [Google Scholar]
- Entani T, Kubo K, Isogai S, Fukao Y, Shirakawa M, Isogai A, Takayama S(2014) Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction. Plant J 78: 1014–1021 [DOI] [PubMed] [Google Scholar]
- Fujii S, Kubo K, Takayama S(2016) Non-self- and self-recognition models in plant self-incompatibility. Nat Plants 2: 16130. [DOI] [PubMed] [Google Scholar]
- Kakui H, Kato M, Ushijima K, Kitaguchi M, Kato S, Sassa H(2011) Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J 68: 1028–1038 [DOI] [PubMed] [Google Scholar]
- Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, et al. (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799 [DOI] [PubMed] [Google Scholar]
- Kubo K, Paape T, Hatakeyama M, Entani T, Takara A, Kajihara K, Tsukahara M, Shimizu-Inatsugi R, Shimizu KK, Takayama S(2015) Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat Plants 1: 14005. [DOI] [PubMed] [Google Scholar]
- Lapins KO.(1971) Stella, a self-fruitful sweet cherry. Can J Plant Sci 51: 252–253 [Google Scholar]
- Li J, Zhang Y, Song Y, Zhang H, Fan J, Li Q, Zhang D, Xue Y(2017) Electrostatic potentials of the S-locus F-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J 89: 45–57 [DOI] [PubMed] [Google Scholar]
- Li S, Sun P, Williams JS, Kao TH(2014) Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflata. Plant Reprod 27: 31–45 [DOI] [PubMed] [Google Scholar]
- Li S, Williams JS, Sun P, Kao TH(2016) All 17 S-locus F-box proteins of the S2- and S3-haplotypes of Petunia inflata are assembled into similar SCF complexes with a specific function in self-incompatibility. Plant J 87: 606–616 [DOI] [PubMed] [Google Scholar]
- Li W, Meng D, Gu Z, Yang Q, Yuan H, Li Y, Chen Q, Yu J, Liu C, Li T(2018) Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol 218: 579–593 [DOI] [PubMed] [Google Scholar]
- Li Y, Wu C, Liu C, Yu J, Duan X, Fan W, Wang J, Zhang X, Yan G, Li T, et al. (2019) Functional identification of lncRNAs in sweet cherry (Prunus avium) pollen tubes via transcriptome analysis using single-molecule long-read sequencing. Hortic Res 6: 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Gao FH(2018) SUMO1/sentrin/SMT3 specific peptidase 2 modulates target molecules and its corresponding functions. Biochimie 152: 6–13 [DOI] [PubMed] [Google Scholar]
- Marchese A, Bosković RI, Caruso T, Raimondo A, Cutuli M, Tobutt KR(2007) A new self-compatibility haplotype in the sweet cherry ‘Kronio’, S5′, attributable to a pollen-part mutation in the SFB gene. J Exp Bot 58: 4347–4356 [DOI] [PubMed] [Google Scholar]
- Matsumoto D, Tao R(2016) Recognition of a wide-range of S-RNases by S locus F-box like 2, a general-inhibitor candidate in the Prunus-specific S-RNase-based self-incompatibility system. Plant Mol Biol 91: 459–469 [DOI] [PubMed] [Google Scholar]
- Matsumoto D, Tao R(2019) Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system. Plant Mol Biol 100: 367–378 [DOI] [PubMed] [Google Scholar]
- Matsumoto D, Yamane H, Abe K, Tao R(2012) Identification of a Skp1-like protein interacting with SFB, the pollen S determinant of the gametophytic self-incompatibility in Prunus. Plant Physiol 159: 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto D, Yamane H, Tao R(2008) Characterization of SLFL1, a pollen-expressed F-box gene located in the Prunus S locus. Sex Plant Reprod 21: 113–121 [Google Scholar]
- Meng D, Gu Z, Li W, Wang A, Yuan H, Yang Q, Li T(2014) Apple MdABCF assists in the transportation of S-RNase into pollen tubes. Plant J 78: 990–1002 [DOI] [PubMed] [Google Scholar]
- Meng X, Sun P, Kao TH(2011) S-RNase-based self-incompatibility in Petunia inflata. Ann Bot 108: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamikawa MF, Koyano R, Kikuchi S, Koba T, Sassa H(2014) Identification of SFBB-containing canonical and noncanonical SCF complexes in pollen of apple (Malus × domestica). PLoS ONE 9: e97642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho A, Camacho L, Haley A, Pais MS, Trewavas A, Malho R(2001) Antisense perturbation of protein function in living pollen tubes. Sex Plant Reprod 14: 101–104 [Google Scholar]
- Muñoz‐Espinoza C, Espinosa E, Bascuñán R, Tapia S, Meneses C, Almeida AM(2017) Development of a molecular marker for self-compatible S4′ haplotype in sweet cherry (Prunus avium L.) using high-resolution melting. Plant Breed 136: 6 [Google Scholar]
- Muñoz-Sanz JV, Zuriaga E, Badenes ML, Romero C(2017) A disulfide bond A-like oxidoreductase is a strong candidate gene for self-incompatibility in apricot (Prunus armeniaca) pollen. J Exp Bot 68: 5069–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Moriya S, Haji T, Abe K(2013) Isolation and characterization of multiple F-box genes linked to the S9- and S10-RNase in apple (Malus × domestica Borkh.). Plant Reprod 26: 101–111 [DOI] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Taguchi T, Ichikawa T, Sawamura Y, Nakanishi T, Takasaki-Yasuda T(2011) Related polymorphic F-box protein genes between haplotypes clustering in the BAC contig sequences around the S-RNase of Japanese pear. J Exp Bot 62: 1887–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Akagi T, Morimoto T, Wünsch A, Tao R(2018) Genome re-sequencing of diverse sweet cherry (Prunus avium) individuals reveals a modifier gene mutation conferring pollen-part self-compatibility. Plant Cell Physiol 59: 1265–1275 [DOI] [PubMed] [Google Scholar]
- Pratas MI, Aguiar B, Vieira J, Nunes V, Teixeira V, Fonseca NA, Iezzoni A, van Nocker S, Vieira CP(2018) Inferences on specificity recognition at the Malus × domestica gametophytic self-incompatibility system. Sci Rep 8: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa K, Isuzugawa K, Ikenaga M, Saito Y, Yamamoto T, Hirakawa H, Isobe S(2017) The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res 24: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH(2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305 [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP(2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Williams JS, Li S, Wu L, Khatri WA, Stone PG, Keebaugh MD, Kao TH(2018) S-Locus F-box proteins are solely responsible for S-RNase-based self-incompatibility of petunia pollen. Plant Cell 30: 2959–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Iezzoni AF(2010) The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci Hortic (Amsterdam) 124: 423–433 [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H(2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao R(2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39: 573–586 [DOI] [PubMed] [Google Scholar]
- Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D(2016) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7: 11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang K, Su H(2010) Identification of the S-genotypes of several sweet cherry (Prunus avium L.) cultivars by AS-PCR and pollination. Afr J Agric Res 5: 250–256 [Google Scholar]
- Williams JS, Der JP, dePamphilis CW, Kao TH(2014a) Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata. Plant Cell 26: 2873–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Natale CA, Wang N, Li S, Brubaker TR, Sun P, Kao TH(2014b) Four previously identified Petunia inflata S-locus F-box genes are involved in pollen specificity in self-incompatibility. Mol Plant 7: 567–569 [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK(2005) GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Wünsch A, Tao R, Hormaza JI(2010) Self-compatibility in ‘Cristobalina’ sweet cherry is not associated with duplications or modified transcription levels of S-locus genes. Plant Cell Rep 29: 715–721 [DOI] [PubMed] [Google Scholar]
- Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R(2003) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44: 764–769 [DOI] [PubMed] [Google Scholar]
- Yuan H, Meng D, Gu Z, Li W, Wang A, Yang Q, Zhu Y, Li T(2014) A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. J Exp Bot 65: 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ma C, Zhang Y, Gu Z, Li W, Duan X, Wang S, Hao L, Wang Y, Wang S, et al. (2018) A single-nucleotide polymorphism in the promoter of a hairpin RNA contributes to Alternaria alternata leaf spot resistance in apple (Malus × domestica.). Plant Cell 30: 1924–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y(2010) The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J 62: 52–63 [DOI] [PubMed] [Google Scholar]
- Zhu M, Zhang X, Zhang K, Jiang L, Zhang L(2004) Development of a simple molecular marker specific for detecting the self-compatible S4′ haplotype in sweet cherry (Prunus avium L.). Plant Mol Biol Rep 22: 387–398 [Google Scholar]
- Zuriaga E, Muñoz-Sanz JV, Molina L, Gisbert AD, Badenes ML, Romero C(2013) An S-locus independent pollen factor confers self-compatibility in ‘Katy’ apricot. PLoS ONE 8: e53947. [DOI] [PMC free article] [PubMed] [Google Scholar]