A light signalling integrating factor improves the tolerance of Arabidopsis thaliana in UV-B stress by decreasing the expression level of a negative regulator.
Abstract
Understanding how the distinct cell types of the shoot apical meristem (SAM) withstand ultraviolet radiation (UVR) stress can improve cultivation of plants in high-UVR environments. Here, we show that UV-B irradiation selectively kills epidermal and niche cells in the shoot apex. Plants harboring a mutation in DECREASE WAX BIOSYNTHESIS (DEWAX) are tolerant to UV-B. Our data show that DEWAX negatively regulates genes involved in anthocyanin biosynthesis. ELONGATED HYPOCOTYL5 (HY5) binds to the DEWAX promoter elements and represses its expression to promote the anthocyanin biosynthesis. The HY5-DEWAX regulatory network regulates anthocyanin content in Arabidopsis (Arabidopsis thaliana) and influences the survivability of plants under UV-B irradiation stress. Our cell sorting-based study of the epidermal cell layer transcriptome confirms that core UV-B stress signaling pathway genes are conserved and upregulated in response to UV-B irradiation of the SAM. Furthermore, we show that UV-B induces genes involved in shoot development and organ patterning. We propose that the HY5-DEWAX regulatory relationship is conserved; however, changes in the expression levels of these genes can determine anthocyanin content in planta and, hence, fitness under UV-B irradiation stress.
The spectrum of light perceived by plants is broadly comprised of blue, red, far-red, and ultraviolet radiation (UVR; Heijde and Ulm, 2012). To effectively utilize the light signals and to adapt in diverse environments, plants have evolved multiple photoreceptors, which can detect an array of light wavelengths. In Arabidopsis (Arabidopsis thaliana), phytochrome genes encode photoreceptor proteins (PHYTOCHROME A to PHYTOCHROME E), which detect red/far-red light. CRYPTOCHROME1, CRYPTOCHROME2, PHOTOTROPHIN1, PHOTOTROPHIN2, ZEITLUPE/FLAVIN-BINDING, KELCH REPEAT, and F BOX1/LOV KELCH PROTEIN2 loci encode photoreceptors that perceive UV-A and blue light (Chen et al., 2004; Jenkins, 2009; Kami et al., 2010). UVB-RESISTANCE8 (UVR8) specifically detects UV-B light (Favory et al., 2009; Rizzini et al., 2011; Tilbrook et al., 2013). Despite showing diverse preferences for various wavelengths of light, all photoreceptors can promote photomorphogenesis by activating a common central integrator HY5 (Binkert et al., 2014; Gangappa and Botto, 2016). In response to UV-B irradiation, UVR8 moves to the nucleus, where it interacts with the chromatin containing HY5 (Cloix and Jenkins, 2008), and thus activates the transcription of target genes to impart UV-B tolerance (Kaiserli and Jenkins, 2007; Yin et al., 2016). Genetic studies also identified CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1), which encodes a RING-finger protein with WD-40 repeats. COP1 enables degradation of HY5 under dark conditions and promotes skotomorphogenesis (Ang et al., 1998). Interestingly, in UV-B conditions, COP1 interacts with HY5, and it further enhances gene expression responses to cope with UV-B irradiation stress in plants (Favory et al., 2009; Yin et al., 2016).
The shoot apical meristem (SAM) of angiosperms displays a complex organization consisting of cell layers and zones (Satina et al., 1940; Meyerowitz, 1997). The epidermal cell layer serves as an essential interface between the plant body and its surrounding environment. In the vegetative phase, SAMs are covered by leaves and do not come under radiation stress; however, upon the transition to flowering, they are exposed to direct sunlight. Transcriptome studies on the SAM epidermal cell layer have shown that genes involved in biotic and abiotic stresses are expressed mainly in the epidermis (Yadav et al., 2014). Despite the burgeoning field of light signaling, very little is known about how distinct cell types respond to UVR. Most of the UV-A (315–400 nm) and a modest amount of UV-B (280–315 nm) reaches earth’s surface, while UV-C is filtered out by the stratosphere ozone layer (Caldwell et al., 2007; McKenzie et al., 2007). UV-A has deeper penetration owing to its longer wavelength, and in humans (Homo sapiens), prolonged exposure can damage cells and tissues, increasing the likelihood of developing skin cancer and causing premature ageing (Noonan et al., 2012). UV-B has more energy compared to UV-A and can cause substantial damage to nuclear DNA as well as sunburns in animals and plants (Ichihashi et al., 2003). UV-C has the highest amount of energy and therefore is dangerous for all forms of life (Sagan, 1973; Diffey, 1991). Most organisms are well equipped to defend against UV-A and UV-B radiation stress; however, this regime would change in high altitudes, where atmospheric gases and water vapor are inadequate to prevent radiation from reaching plants.
The signaling cascade initiated by UVR8 promotes tolerance and acclimatization in response to UV-B in Arabidopsis. In the absence of UV-B, UVR8 remains a dimer, but when plants are exposed to UV-B, UVR8 monomerizes and interacts with COP1. UVR8-COP1 enters into the nucleus and prevents degradation of HY5 (Yin et al., 2016). As a result, HY5 further enhances the acclimatization and tolerance against UV-B by activating genes involved in DNA damage and repair, anthocyanin biosynthesis, and antioxidant production (Hideg et al., 2013). A number of HY5 target genes characterized in the past support this model. In addition, studies have shown that the cuticle also plays an important role in reflection of sunlight including UV-B and protects the plant from high-intensity light stress (Riederer and Schneider, 1990; Barnes et al., 1996). DEWAX expresses in the epidermis and is induced by dark (Go et al., 2014). Overexpression lines of DEWAX show a decrease in cuticular wax, suggesting that to improve the fitness of plants in high-intensity light and UV-B conditions, its expression needs to be curtailed in the daytime.
In this study, we investigated the effect of UV-B irradiation stress on Arabidopsis SAM. Plants having mutation in DEWAX, encoding an AP2/ERF transcription factor (TF), show enhanced tolerance to UV-B due to higher accumulation of UV-B sun screen compounds such as anthocyanin and flavonoids. DEWAX expression is restricted to the epidermis in most tissues. Further, we studied the role of HY5 in regulating DEWAX expression. Our results demonstrate that HY5 and DEWAX antagonistically regulate anthocyanin production. HY5 negatively regulates DEWAX to enhance the production of anthocyanin. Arabidopsis natural variants with changes in transcript levels of HY5 and DEWAX also display variation in anthocyanin content and UV-B tolerance.
RESULTS
The Epidermal Cell Layer Serves as an Interface between UV-B Radiation and the Plant Body
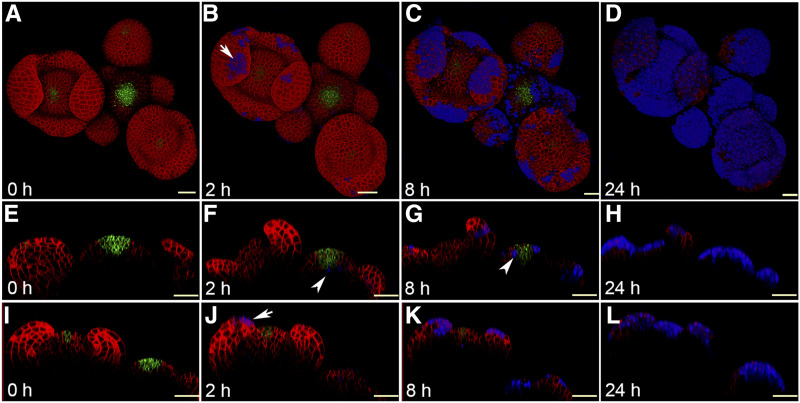
As in animals, the entire body of a plant is covered by an epidermal cell layer. To evaluate the impact of UV-B on epidermal cells, we exposed 4-week-old wild-type Arabidopsis ecotype Landsberg erecta (Ler) SAM expressing pCLV3::mGFP5-ER and 35::YFP29-1 to UV-B irradiation. YFP29-1 translation fusion was used in the past to visualize cell outlines, while pCLV3 marks the central zone (CZ)/stem cells in the SAM (Reddy et al., 2004). To treat the SAM with UV-B, older floral buds were removed carefully as described in Yadav et al. (2010), and plants were returned to the growth chamber for recovery overnight. Twelve hours later, plants were exposed to UV-B for 2 min using a UV-B transilluminator. To identify cells that had succumbed to UV-B stress in the SAM, we applied propidium iodide (PI) prior to each imaging session. PI stains walls of living cells but enters into dead cells or cells whose membrane integrity is compromised. To determine the effect of UV-B irradiation on SAM cell types, live imaging experiments were conducted at 0, 2, 8, 16, and 24 h after the initial exposure (Fig. 1, A–E and I; Supplemental Fig. S1, A–D). Cell death triggered by UV-B stress was spotted 2 h after exposure based on the PI staining in the epidermal cell layer and the stem cell niche (Fig. 1, B, F, and J). In stage 4 flowers, cell death was restricted to the epidermis in sepal primordia (Fig. 1, B and J). In the younger floral buttress and meristem, cell death did not occur, suggesting that young buds and meristematic cells are robust due to their developmental age or a hitherto unknown mechanism (Fig. 1B; Supplemental Fig. S1A). Still, cell death occurred in the stem cell niche, which is located four to five cell layers deep within the SAM, indicating that distinct cell types perhaps respond differently to UV-B irradiation stress. It is conceivable that the epidermal cells overlying the floral meristem showed more cell death due to their closeness to the UV-B radiation source as well as lesser vitality of this tissue compared to young floral buttress. Eight hours after UV-B exposure, cell death spreads within the meristematic cells in epidermis as well as in the subjacent subepidermis and corpus (Fig. 1, C, G, and K; Supplemental Fig. S1, B and F). In the floral meristem, most cells in the epidermis were killed after 16 h of UV-B exposure, indicating that UV-B stress affects lateral organs more than the SAM (Supplemental Fig. S1, C and G). Twenty-four hours after UV-B exposure, most cells had been killed in the SAM, including the stem cells (Fig. 1, D, H, and L; Supplemental Fig. S1, D and H). Taken together, these results show that UV-B induces cell death rapidly in the epidermis and stem cell niche. In UV-B-exposed plants, cell death occurs predominantly in the epidermis and spreads within and across the cell layers. Stem cells show tolerance to UV-B irradiation stress initially, but gradually succumb.
Figure 1.
UV-B irradiation causes cell death in the Arabidopsis SAM. A to D, Top views of the Arabidopsis inflorescence meristem expressing pCLV3::mGFP-ER (green) and 35S::LTI-YFP (red) imaged at 0 h (A), 2 h (B), 8 h (C) and 24 h (D) after 2 min of UV-B exposure, drawn from the confocal Z-section data. E to H, Reconstructed side views of the inflorescence meristem showing the layering organization of the SAM. I to L, Reconstructed side views of the floral meristem. Arrows in B and J indicate cell death in the epidermal cell layer in a stage 4 floral primordium visualized by PI staining (blue). PI marks the outline of cells in live tissue but enters into the dead cells when the membrane integrity is compromised. Arrowheads in F and G indicate cell death in the deeper cell layer of the SAM (blue). Scale bars = 40 μm.
SAM Epidermal Cell Layer-Enriched TFs Are Essential for UV-B Stress Response
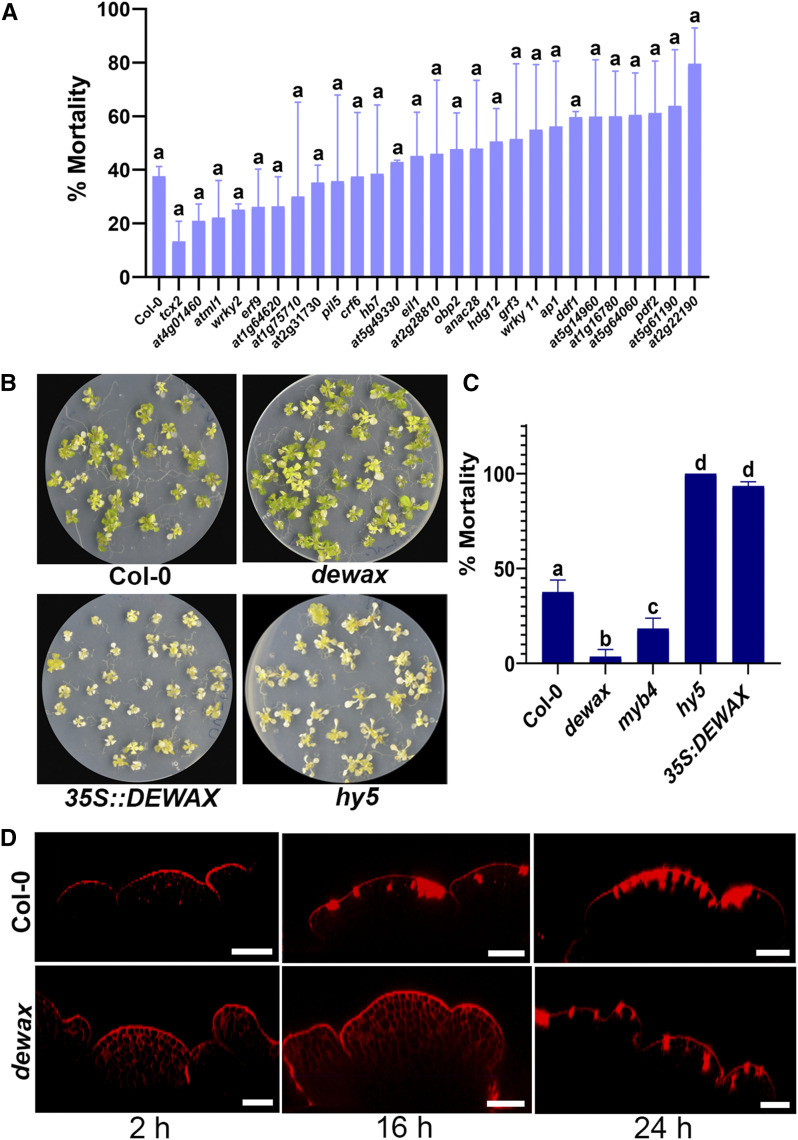
Our live imaging study revealed that the integrity of the epidermal cell layer and lateral organ growth are affected in the UV-B-exposed SAM. We asked how epidermal cells cope with UV-B irradiation stress. To identify the gene regulatory network that is involved in UV-B tolerance, we focused on TF genes enriched in the epidermal cell layer, because cells in the epidermal cell layer usually respond to environmental factors first in the plant body. Of the 44 TFs identified from epidermal cell layer in an earlier study (Yadav et al., 2014), we confirmed transfer DNA (T-DNA) insertion in 30 genes (Supplemental Table S1). We grew the T-DNA lines on Murashige and Skoog agar plates for 10 d and treated them to high-fluence UV-B for 10 min (312 nm, emitting ∼3.82 mW cm−2; Jin et al., 2000). The mutant lines were returned to the growth chamber and were scored for survival after 1 week of UV-B exposure (Fig. 2A). The dewax mutant plants survived UV-B irradiation stress and displayed the lowest mortality rate among the T-DNA lines screened (Fig. 2, B and C). hy5 was used in parallel as a control, since it is sensitive to UV-B (Fig. 2, B and C; Brown et al., 2005; Jenkins, 2009). Under UV-B, MYB4 negatively regulates CINNAMATE 4-HYDROXYLASE (C4H), which is involved in the production of cinnamate esters (Jin et al., 2000). In myb4 mutant, production of UV-B-absorbing cinnamate esters goes up, and as a result, plants show better survival than the wild type under UV-B irradiation stress. In our UV-B seedling assay, myb4 mutant seedlings showed less mortality compared to the control, confirming previous findings (Fig. 2C; Jin et al., 2000); nonetheless, dewax mutant plants showed extreme resilience in response to UV-B exposure. We also examined the overexpression line of DEWAX for its ability to withstand UV-B irradiation stress. In contrast to the loss of function mutant, 35S::DEWAX plants exposed to UV-B died faster than Arabidopsis ecotype Columbia (Col-0), suggesting that DEWAX has a negative influence on plant survival, particularly in UV-B conditions (Fig. 2, B and C). This observation motivated us to investigate the impact of UV-B on the SAM epidermal cell layer in the dewax mutant as well. For this, dewax mutant plants were raised until bolting stage, and older floral buds were removed before UV-B exposure to conduct the live imaging experiment. Both the dewax mutant and wild-type Col-0 plants were subjected to 2 min of UV-B exposure. As we observed in our UV-B seedling assay, plants lacking functional DEWAX in the SAM showed resilience to UV-B exposure and displayed an intact epidermal cell layer in the SAM without cell death in the first 16 h, whereas the control plants displayed signs of cell death in the epidermal cell layer and stem cell niche (Fig. 2D). This indicated that DEWAX affects the integrity of cells in UV-B conditions. Taken together; these observations show that functional DEWAX negatively affects cell viability both in vegetative and reproductive tissues in plants under high-fluence UV-B.
Figure 2.
UV-B induced cell death is delayed in the dewax mutant SAM. A, Mortality rate scored among the epidermal cell layer enriched TF mutant lines after UV-B exposure. B, Col-0, dewax, 35S::DEWAX, and hy5 seedlings were grown for 10 d and exposed to UV-B radiation. Images were taken 1 week after UV-B exposure to score the mortality rate. When exposed to UV-B, dewax mutant seedling showed more resilience compared to the Col-0 control. C, Mortality rate among the seedlings of Col-0, dewax, myb4, hy5, and 35S::DEWAX. Error bars represent the mean ± se of three biological replicates. In A and C, same lowercase letters indicate no statistical difference, while different letters denote within the group significant differences using ANOVA analysis followed by Uncorrected Fisher’s lsd multiple comparisons test (P < 0.01). D, Reconstructed side views of 4-week old dewax and Col-0 SAMs drawn from the confocal Z-series data. Plants were treated with UV-B for 2 min, and confocal images were acquired at the 2, 16, and 24 h time points. PI (red) was applied to stain the cell outlines and identify cell death in the growing SAM. Scale bars = 40 μm.
DEWAX Negatively Regulates Anthocyanin Biosynthesis
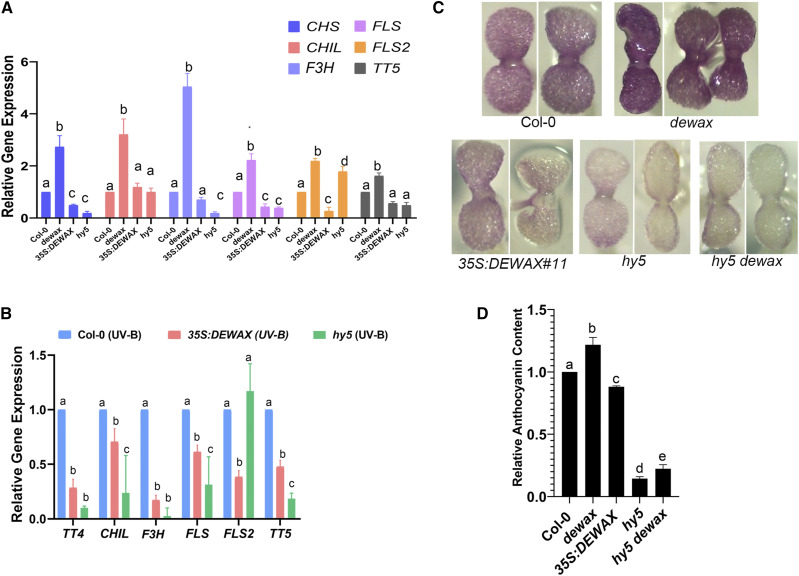
To identify the downstream transcriptional response genes of the DEWAX TF that contribute to survival under UV-B stress, we focused on the genome-wide gene expression responses of DEWAX reported by Go et al., (2014). In this study, DEWAX was ectopically expressed under the 35S promoter to identify its potential downstream target genes. In addition to genes involved in cuticle biosynthesis, we found that genes involved in anthocyanin and flavonoid biosynthesis are also negatively regulated by DEWAX. This includes CHALCONE SYNTHASE (CHS/TT4), CHALCONE ISOMERASE LIKE (CHIL), FLAVANONE 3-HYDROXYLASE (F3H/TT6), CINNAMATE 4-HYDROXYLASE (C4H), FLAVONOL SYNTHASE (FLS2), UDP GLC:FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UGT78D2), and FLAVONOL 3-O-GLUCOSYLTRANSFERASE (F3G; Supplemental Fig. S2; Go et al., 2014). Past studies have shown that genes involved in the production of flavonoids and anthocyanins are induced by UV-B in the epidermal cell layer (Hahlbrock and Scheel, 1989; Li et al., 1993). These phenolic compounds act as a UV-absorbing sun screen and thus provide protection against UV-B. To confirm the role of DEWAX independently in the regulation of anthocyanin biosynthesis, we studied the expression of CHS/TT4, CHIL, F3H, FLS, FLS2, and TT5 in the wild type, dewax, 35S::DEWAX, and hy5 mutant background by reverse transcription quantitative PCR (RT-qPCR). Expression of CHS/TT4, CHIL, F3H, FLS, FLS2, and TT5 was upregulated in the dewax mutant compared to the control. In contrast, plants overexpressing DEWAX showed a reduction in CHS/TT4, FLS, and FLS2 transcript levels, suggesting that DEWAX negatively regulates anthocyanin biosynthesis pathway genes (Fig. 3A; Supplemental Table S2). HY5 is involved in both photomorphogenesis and anthocyanin biosynthesis. HY5 induces CHS/TT4 and MYB12 in UV-B irradiation stress (Mehrtens et al., 2005; Stracke et al., 2010). In hy5 mutant, expression of CHS/TT4, F3H, and FLS was downregulated compared to control, confirming previous findings (Fig. 3A).
Figure 3.
The dewax mutant accumulates more UV-B sunscreen compounds. A, DEWAX negatively regulates genes involved in flavonoid and anthocyanin biosynthesis. CHS/TT4, CHIL, F3H, FLS, FLS2, and TT5 are upregulated in the dewax mutant. Plants overexpressing DEWAX show repression in CHS/TT4, FLS, and FLS2. B, Despite exposure to UV-B, a decrease in the transcript levels of CHS/TT4, CHIL, F3H, FLS, FLS2, and TT5 was observed in 35S::DEWAX and hy5. C, Five-day-old seedlings of Col-0, dewax, 35S::DEWAX, hy5, and hy5 dewax grown on Murashige and Skoog agar plates containing 5 μm norflurazon to assess anthocyanin content in planta. D, Anthocyanin content was measured by spectrophotometry. Error bars in A, B, and D represent the mean ± se of three replicates. Same lowercase letters indicate no statistical difference, whereas different letters denote significant differences between groups using ANOVA analysis followed by a linear step-up procedure (P < 0.01; Benjamini et al., 2006).
In the light of our findings that DEWAX negatively regulates genes involved in anthocyanin biosynthesis, we asked, how does DEWAX affect the survival of plants in UV-B stress at the molecular level? Past studies have shown that wild-type plants, when exposed to UV-B, show an increase in the expression of CHS/TT4, F3H/TT6, CHI, and FLS (Brosché et al., 2002; Ulm et al., 2004). To test the expression of anthocyanin biosynthesis genes in response to UV-B in our setting, we carried out RT-qPCR on UV-B-exposed Col-0 plants. RT-qPCR experiment revealed elevated expression of CHS/TT4, CHIL, F3H, FLS, FLS2, and TT5 in response to UV-B (Supplemental Fig. S3), suggesting that high-fluence UV-B induces the production of phenolics. In contrast, when exposed to UV-B, plants overexpressing DEWAX did not show an increase in the transcript levels of CHS/TT4, CHIL, F3H, FLS, FLS2,and TT5, supporting a role for DEWAX in negative regulation of anthocyanin biosynthesis (Fig. 3B). Consistent with this, we also did not see a change in the expression levels of CHS/TT4, CHIL, F3H, FLS, FLS2,and TT5 in a UV-B-exposed hy5 mutant, confirming that functional HY5 is required for elevated induction of anthocyanin production in response to high-fluence UV-B (Fig. 3B; Supplemental Table S2). Taken together, these data demonstrate that DEWAX negatively regulates expression of genes associated with the anthocyanin biosynthesis pathway. In contrast, HY5 promotes expression of genes involved in anthocyanin production and thus protects plants from UV-B.
To determine the significance of this regulatory network in anthocyanin production, we investigated the in vivo anthocyanin content in Col-0, dewax, 35S::DEWAX, and hy5 mutant plants by growing their seedlings on Murashige and Skoog agar plates containing 5 μm norflurazon (Fig. 3C). Norflurazon acts as a noncompetitive inhibitor for phytoene desaturase, a critical enzyme involved in carotenoid biosynthesis and chloroplast development. Norflurazon was applied to germinating seedlings to prevent the greening of cotyledons so that inherent accumulation of anthocyanin could be estimated visually. In parallel, we also analyzed anthocyanin accumulation in plants biochemically using spectrophotometry (Chory et al., 1989). In consistent with its role, plants lacking functional DEWAX accumulate higher amounts of anthocyanin content, whereas plants lacking HY5 activity showed significantly reduced anthocyanin levels, similar to the DEWAX overexpression lines (Fig. 3, C and D). To establish the role of functional HY5 in the higher accumulation of anthocyanin content in the dewax mutant background, we made the hy5 dewax double mutant and analyzed the anthocyanin content. As expected, hy5 dewax double mutant plants did not accumulate anthocyanin comparable to that in the wild type, suggesting that HY5 promotes anthocyanin biosynthesis, whereas DEWAX negatively regulates anthocyanin biosynthesis (Fig. 3, C and D).
Based on the anthocyanin content, we hypothesized that plants ectopically expressing DEWAX would show a reduction in the accumulation of critical intermediate metabolites compared to the dewax mutant. To quantify the intermediate metabolites involved in anthocyanin biosynthesis, and how their accumulation is affected in 35S::DEWAX and the dewax mutant, we carried out targeted metabolite analysis using gas chromatography-mass spectrometry (GC-MS; Kopka et al., 2004; Lisec et al., 2006; Masakapalli et al., 2014; Shree et al., 2019). Phe showed significant accumulation in the 35S::DEWAX sample compared to Col-0. In contrast, the dewax mutant accumulates significantly less Phe compared to the control (Supplemental Fig. S4). Phe is a major precursor of 4-coumarate, which is funneled into the anthocyanin biosynthesis pathway. Land plants subjected to biotic and abiotic stress conditions, including UV-B, usually show elevated levels of shikimic acid and its derivative. Interestingly, in our analysis, we found that the shikimic acid levels rose in 35S::DEWAX plants when exposed to UV-B (Supplemental Fig. S4), indicating that despite high UV-B, plants overexpressing DEWAX are unable to convert shikimic acid into UV-B-quenching sun screen compounds (Hahlbrock and Scheel, 1989). Taken together, the GC-MS data link DEWAX to the repression of anthocyanin biosynthesis in planta.
HY5 Directly Binds to the Promoter of DEWAX
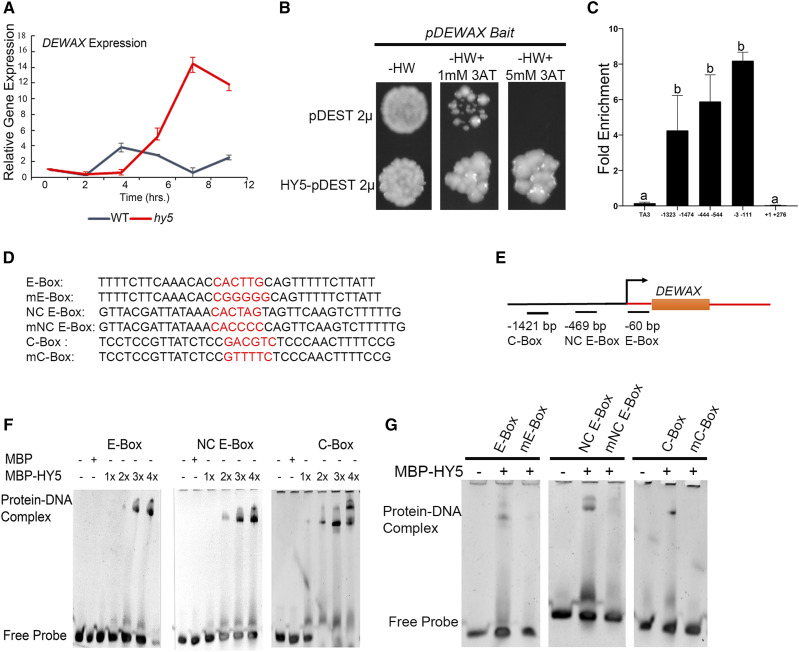
Two independent studies have shown that DEWAX is diurnally regulated (Go et al., 2014; Li et al., 2019). Presently it is not clear what regulates DEWAX. To test whether the negative regulation of DEWAX is linked to HY5 activity in the day time, we examined the expression of DEWAX in the hy5 mutant background using a time course experiment. For this, hy5 mutant and Col-0 plants were grown in the dark for 3 d, then shifted to white light (WL), and tissue was harvested for RNA isolation at 0, 2, 4, 6, 8, and 12 h after continuous light exposure. RT-qPCR experiments revealed that the expression of DEWAX plateaued in the control after 4 h of light exposure. Interestingly, in the hy5 mutant, we did not see repression of DEWAX even after 8 h of WL exposure (Fig. 4A). This finding suggests that the negative regulation of DEWAX in light might be dependent upon functional HY5.
Figure 4.
HY5 directly binds to the DEWAX promoter elements and regulates its expression. A, RT-qPCR experiment revealed the role of HY5 in transcriptional regulation of DEWAX. The red line indicates the expression profile of DEWAX in the hy5 mutant and the black line represents the corresponding expression in wild-type Col-0. B, HY5 binds to pDEWAX in the eY1H assay. C, ChIP-qPCR showing relative enrichment of the DEWAX regulatory regions bound by HY5. The ChIP assay was performed on pHY5::HY5-YFP hy5 and 35S::GFP in the wild type. DNA-protein complexes were immunoprecipitated using anti-GFP and IgG antibody as negative controls. Error bars represent the mean ± SE of two biological replicates. Same lowercase letters indicate no statistical difference, while different letters denote significant differences within the group using ANOVA analysis followed by uncorrected Fisher’s least significant difference multiple comparisons test (P < 0.01). D and E, E-Box (−60 bp), NC E-Box (−469 bp), and C-Box (−1,421 bp) elements present in the DEWAX promoters, as well as the corresponding mutated cis-elements (preceded by “m”), are highlighted (red letters; D) and their relative position depicted (E). F, EMSAs show binding of recombinant HY5 protein in increasing concentrations to oligonucleotides containing E-Box, NC E-Box, and C-Box regulatory elements. G, HY5 binding to E-Box, NC E-Box, and C-Box was abrogated upon introduction of mutation in the respective cis-elements.
To understand the molecular mechanism underlying DEWAX regulation, we carried out an enhanced yeast-one-hybrid (eY1H) assay to test the binding of HY5 to the DEWAX promoter. For this, a 3-kb DNA fragment upstream of the translational start site (TSS) of DEWAX was amplified (including the 5′ UTR from wild-type Ler genomic DNA) and cloned into the pMW2 vector as described in an earlier study (Taylor-Teeples et al., 2015). We confirmed the binding of HY5 with pDEWAX in the eY1H screen (Fig. 4B). To further investigate negative regulation by HY5, we looked at the ChIP-seq data from Burko et al. (2020) and found that HY5 preferentially binds closer to the TSS in the DEWAX promoter. We tested HY5 binding in the DEWAX regulatory region by chromatin immunoprecipitation (ChIP) coupled with qPCR. For this we used the pHY5::HY5-YFP hy5 lines and performed ChIP-qPCR by using anti-GFP antibodies (Supplemental Fig. S5). Plants expressing 35S::EGFP were used as controls. We obtained the immunoprecipitated DNA and found that the DEWAX promoter region closer to the TSS was highly enriched compared to the adjacent coding region (Fig. 4C). DEWAX regulatory regions away from the TSS also showed enrichment of HY5 binding in our ChIP-qPCR assay compared to control TA3, further confirming that HY5 binds to DEWAX in vivo.
To map HY5-binding within the DEWAX promoter region, we carried out electrophoretic mobility shift assays (EMSAs). We identified three putative binding sites for HY5 in the DEWAX promoter regions based on the ChIP enrichment and in silico analysis. Among the putative binding sites, E-Box (CACTTG, −60 bp), noncanonical (NC) E-Box (CACTAG, −469 bp), and C-Box (GACGTC, −1,421 bp) were selected for HY5 binding (Fig. 4, D and E). When the increasing concentration of HY5 was incubated with DNA containing the putative cis-element, we observed a shift in the HY5-DNA complex (Fig. 4F). To confirm whether the sites occupied by HY5 in the EMSA are indeed bona fide, we introduced a mutation in oligos containing the cis-elements and repeated EMSA experiments with the control (Fig. 4, E and G). The analysis of EMSA results revealed that HY5 binds to E-Box, NC E-Box, and C-Box in the DEWAX promoter.
HY5 Negatively Regulates DEWAX
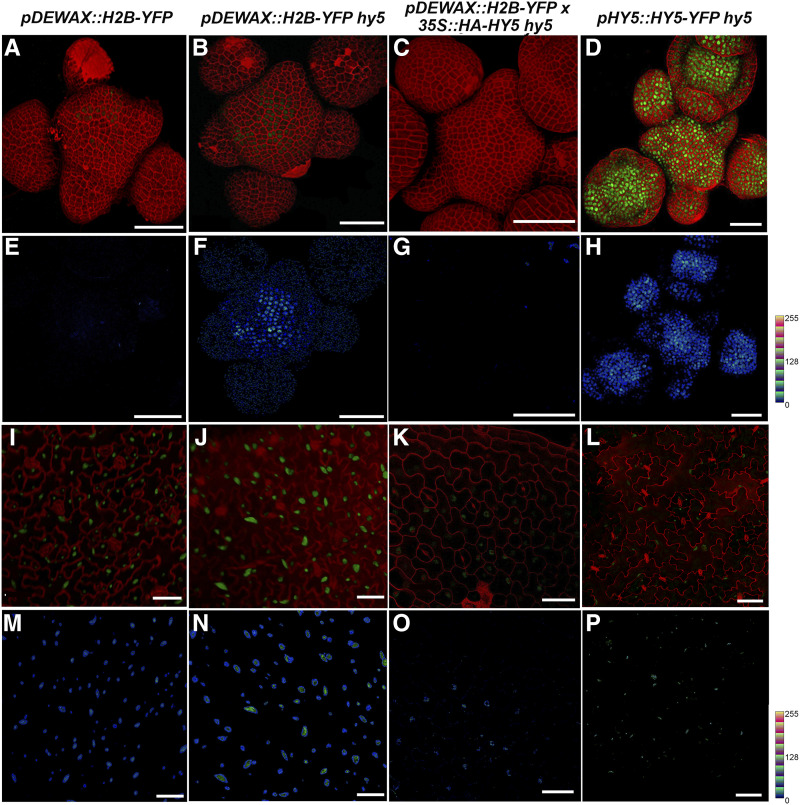
In our efforts to understand the regulation of DEWAX in planta, we tested the interaction of HY5 with DEWAX genetically. For this, we determined the spatiotemporal expression pattern of DEWAX by making transcriptional and translational fusion constructs. The 3-kb DNA fragment of DEWAX promoter used in eY1H was placed in front of the H2B-YFP sequence in pGreen for transcriptional fusion and adjacent to DEWAX-eGFP sequence in pCAMBIA for translational fusion. The resulting pDEWAX::H2B-YFP and pDEWAX::DEWAX-eGFP constructs were transformed by the floral dip method (Clough and Bent, 1998). We rescued 42 T1 lines for pDEWAX::H2B-YFP in wild-type Ler. Of these, we analyzed six lines in T2 that showed reporter activity in SAM and leaf (Fig. 5, A, E, I, and M). Interestingly, we noticed higher activity of the reporter gene in the leaf epidermal cells than in the SAM (Fig. 5, I and M). Similarly, we screened 101 T1 lines in the wild-type Ler background and isolated 14 lines positive for DEWAX-eGFP fusion protein in leaf pavement cells; however, we did not see the DEWAX-eGFP in the SAM epidermal cell layer (Supplemental Fig. S6, B and F). In Col-0, we screened 54 T1 lines and isolated 16 lines positive for DEWAX-eGFP in leaf tissue (Supplemental Fig. S6E). Of the 16 lines, five lines showed a weak expression of DEWAX-eGFP fusion protein restricted to the SAM epidermal cell layer (Supplemental Fig. S6A). One of the possible reasons for the apparent discrepancy in the Ler versus the Col-0 SAM could be the inflorescence architecture of these plants. Wild-type Ler SAM in the reproductive stage gets direct strikes of light, while Col-0 SAMs are always covered with floral buds.
Figure 5.
HY5 negatively regulates DEWAX. A to D, Reconstructed views of the L1-layer of SAM expressing pDEWAX::H2B-YFP (green) in wild-type Ler (A), the hy5 mutant (B), and 35S::HA-HY5 hy5 (C), and expressing HY5-YFP (green) in the hy5 mutant (D). Cell outlines were stained with PI (red). E to H, Quantification of YFP is depicted for the wild type (E), hy5 (F), 35S::HA-HY5 hy5 (G), and pHY5::HY5-YFP hy5 (H). Intensity plots were made using the LUT command in Fiji (0–256 scale represents gray values based on the LUT). I to L, Reconstructed views of the leaf epidermis showing expression of pDEWAX::H2B-YFP in wild-type Ler (I), hy5 (J), and 35S::HA-HY5 hy5 (K), and expression of pHY5::HY5-YFP hy5 in the mutant (L). M to P, Quantification of YFP was carried out in the wild type (M), hy5 (N), 35S::HA-HY5 hy5 (O), and pHY5::HY5-YFP hy5 (L) lines using Fiji. Quantification of HY5-YFP based on LUT in the SAM (H) and leaf epidermis (P) suggests relatively high activity of pHY5 in the SAM compared to leaf epidermis. Scale bars = 40 μm.
Next, we asked whether HY5 is elevated in the SAM and could this affect negatively the expression of DEWAX in the epidermal cell layer. For this, we used the pHY5::HY5-YFP hy5 line generated in Ler. pHY5::HY5-YFP fully rescued the hy5 phenotype (Binkert et al., 2016). As expected, we observed robust expression of HY5-YFP fusion protein across the clonal cell layers in the SAM (Fig. 5, D and H). In the leaf pavement cells, expression of pDEWAX::H2B-YFP was considerably higher than in SAM epidermis (Fig. 5, A, E, I, and M). By looking at the fluorescence intensity of YFP fluorochrome, we concluded that HY5 expression is higher in the SAM than in leaf pavement cells (Fig. 5, D, H, L, and P). Higher expression of HY5 could affect the expression of DEWAX. Thus, an elevated expression of HY5 can explain the reduced expression of DEWAX in the SAM, suggesting that HY5 could influence expression of the target gene depending upon the dosage.
To test the role of HY5 in transcriptional regulation of DEWAX in planta, we made use of the pDEWAX::H2B-YFP line established in wild-type Ler and set up crosses with 35S::HA-HY5 hy5. In F2, we identified plants that had 35S::HA-HY5 and the pDEWAX::H2B-YFP reporter. In this analysis, we also obtained plants segregating for hy5 based on the hypocotyl phenotype but carrying the pDEWAX::H2B-YFP transgene. Plants lacking HY5 activity showed an increase in pDEWAX::H2B-YFP expression in SAM, leaf, and hypocotyl tissues compared to the control (Fig. 5, B, F, J, and N; Supplemental Fig. S6, C, D, G, and H). In parallel, when plants carrying the 35S::HA-HY5 transgene were examined for pDEWAX::H2B-YFP expression in the SAM, we did not find reporter activity (Fig. 5, C and G), indicating that higher dosages of HY5 are affecting the expression of DEWAX in the SAM. Consistent with this observation, we also saw a decrease in DEWAX expression in leaf epidermis (Fig. 5, K and O). Taken together, these results show that HY5 negatively regulates DEWAX. This regulation clearly corresponds to the HY5 dosages in shoot and leaf tissue, respectively.
A High Level of DEWAX Expression Makes Plants Susceptible to UV-B-Mediated Damage
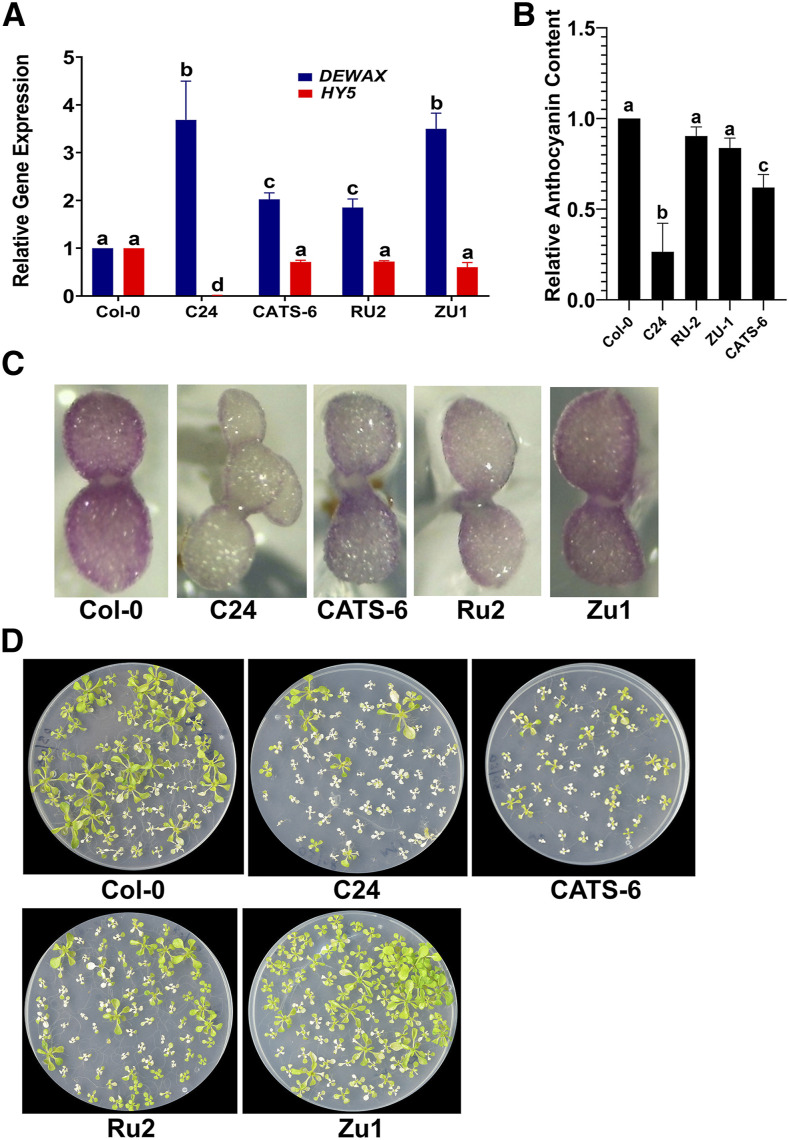
The quantity and quality of light received on planet earth are dependent on the altitude, latitude, and longitude of a place (McKenzie et al., 2011; Bais et al., 2015). To test the impact of light quality, specifically UV-B irradiation stress, in natural settings, we grew wild-type Col-0 at Leh (located at an altitude of 11,000 feet) in September 2018 when the conditions were conducive to grow in the open, and checked the expression of DEWAX. For this experiment, Col-0 seedling were grown on Murashige and Skoog agar plates, and lids of the plates were removed to expose the seedlings to natural sunlight for 3 to 4 h daily for 4 d. Plants grown at high altitude showed a 5-fold decrease in the DEWAX transcript levels compared to those grown in a growth chamber ( Supplemental Fig. S7). Based on this, we argued that the Arabidopsis accessions with higher DEWAX transcript levels would be susceptible to UV-B radiation stress. To test this hypothesis, we looked at the 1,001 genome database and checked the DEWAX transcript levels across 728 natural accessions of Arabidopsis using the newly developed ecotype-specific gene expression (EcoGEx) tool (https://github.com/sk-sahu/EcoGEx). DEWAX expression showed a high degree of variation in expression level, which ranged from 940 in Zu-1 to 17,872 in IP-Bra-0 (Gan et al., 2011). Based on the ecotype gene expression studies, we selected CATS-6, Cvi-0, LDV-18, PHW-3, Rakit-1, Rsch-4, Ru-2, Tuewa1-2, Zdrl-1, and Zu-1. C24 accession is reported to be highly sensitive to UV-B irradiation stress (Kalbina and Strid, 2006), although gene expression data were not available for this accession in EcoGEx, it was included in this study as well. We found elevated expression of DEWAX in C24, CATS-6, Ru-2, Tuewa1-2, and Zu-1 compared to the control under identical growth conditions (Fig. 6A; Supplemental Fig. S8B). Consistent with our findings, we found a correspondence between HY5 and DEWAX expression in C24, CATS-6, Ru-2, and Zu-1 accession. To further visualize the in vivo anthocyanin content, we grew Col-0, C24, CATS-6, Cvi-0, LDV-18, PHW-3, Rakit-1, Rsch-4, Ru-2, Tuewa1-2, Zdrl-1, and Zu-1 on Murashige and Skoog plates containing 5 μm norflurazon. We found significantly less anthocyanin content in C24 and Cvi-0 accessions than in other accessions (Fig. 6, B and C; Supplemental Fig. S8, A and C). However, we also found overall comparatively less anthocyanin content compared to the control in CATS-6, LDV-18, PHW-3, RAKIT-1, Ru-2, Tuewa1-2, and Zu-1 (Fig. 6, B and C; Supplemental Fig. S8, A and C). When subjected to high-fluence UV-B, C24, CATS-6, Ru2, and Zu1 succumbed to UV-B irradiation stress faster than the control (Fig. 6D), suggesting that DEWAX negatively affects survival in UV-B irradiation because plants with high expression of it tend to accumulate less anthocyanin and thus are susceptible to UV-B damage.
Figure 6.
DEWAX and HY5 antagonistically regulate fitness of Arabidopsis seedlings under UV-B stress. A and B, Relative expression of DEWAX and HY5 transcript levels (A) and anthocyanin content measured by spectrophotometry (B) in Col-0, C24, CATS-6, Ru-2, and Zu-1. Error bars represent the mean ± se of three replicates. Same lowercase letters indicate no statistical difference, while different letters indicate significant differences using ANOVA analysis followed by uncorrected Fisher’s least significant difference multiple comparisons test (P < 0.01). C, Five-day-old seedlings grown on Murashige and Skoog agar containing 5 μm norflurazon. Norflurazon was added to suppress the greening of cotyledons. Higher transcript levels of DEWAX negatively correlate with anthocyanin content in C24, CAT6, Ru2, and Zu-1 ecotypes (A–C). D, Ten-day-old seedlings of Col-0, C24, CATS-6, Ru-2, and Zu-1 exposed to UV-B for 10 min and scored for mortality 1 week later. The C24 ecotype displayed the highest mortality under UV-B.
In C24 and Cvi-0, we found significantly less HY5 expression intrinsically compared to Col-0 (Fig. 6A; Supplemental Fig. S8, A and C). Interestingly, the lower transcript levels of HY5 in these plants correlate with less anthocyanin, suggesting that HY5 is indeed required for stimulating anthocyanin production. CATS-6, Ru-2, Tuewa1-2, and Zu-1 have higher expression levels of HY5 but still accumulate less anthocyanin. The expression level of DEWAX is relatively high in these accessions, suggesting that despite modest expression of HY5, elevated levels of DEWAX could affect anthocyanin production. Taken together, the natural accession results show that the HY5-DEWAX regulatory network is critical for anthocyanin accumulation in plants, and this regulatory network fine-tunes the production of anthocyanins in plants.
UV-B Irradiation Stress Induces Genes Involved in Shoot Development and Organ Patterning
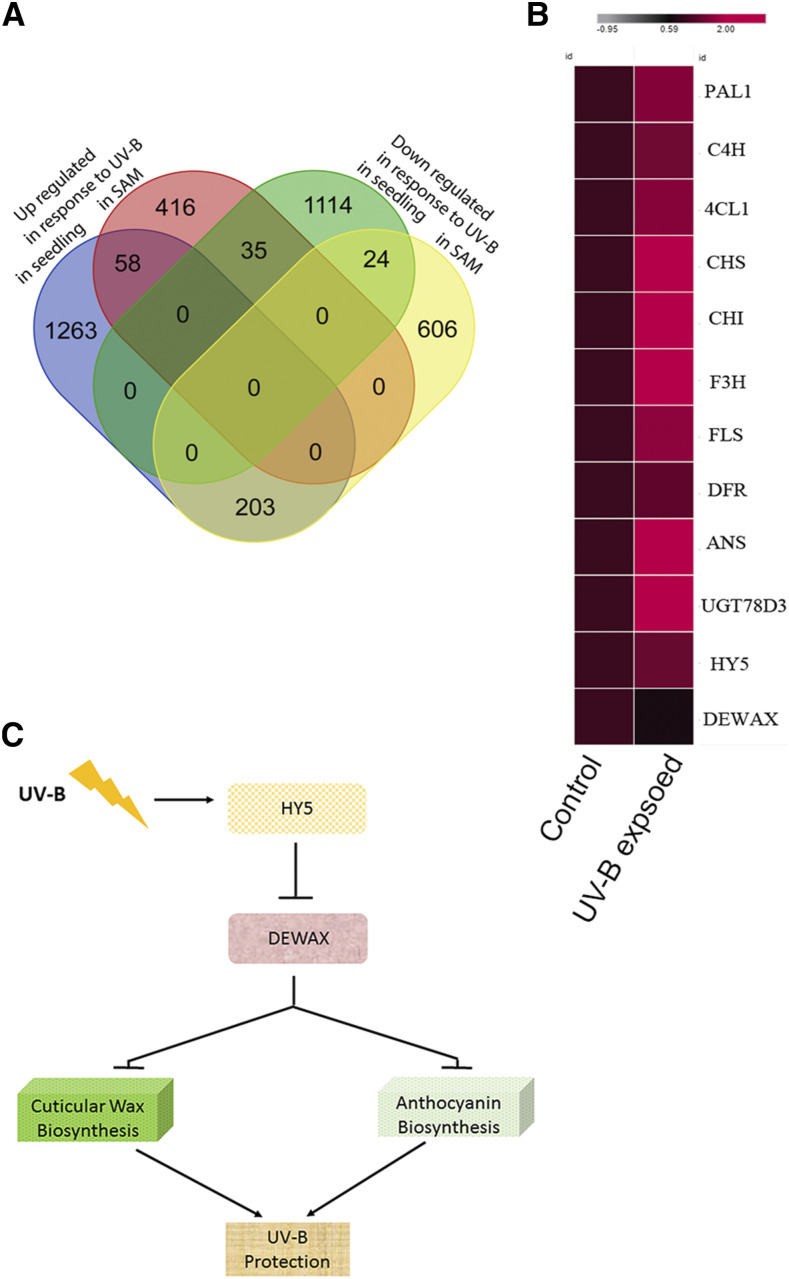
To find out genome wide responses to UV-B stress in shoot epidermal cell types, we made use of the pATHMGB15::H2B-YFP reporter in ap1-1;cal1-1. pATHMGB15 expresses in the meristematic cells of the SAM (Supplemental Fig. S9). Fluorescent-activated cell sorting of the epidermal cell layer was carried out as described previously to isolate SAM epidermal cells (Yadav et al., 2009, 2014). For this, plants were grown in normal WL for 4 weeks. One set of plants was treated with UV-B stress for 2 min, and returned to the plant growth chamber. Plants were harvested after 30 min for cell sorting. Control plants were processed without treatment for cell sorting, followed by RNA isolation. We compared the gene expression responses from sorted cells of UV-B-treated plants with that of the control, and found 1,342 differentially expressed genes (DEGs; upregulated ≥2-fold or downregulated ≤2-fold; P < 0.05). Of the 1,342 DEGs, 509 were upregulated while 833 were downregulated in response to UV-B (Supplemental Table S3). Next, we compared the overlap between genes that respond to UV-B in seedlings and SAMs. For seedling data, we analyzed the previously published microarray dataset and identified 2,697 DEGs (upregulated ≥2-fold or downregulated ≤2-fold; P < 0.05; Brown et al., 2005). A four-way analysis revealed 58 genes upregulated, whereas 24 genes were downregulated that were common to shoot and seedling samples (Fig. 7A). Among the 58 UV-B-induced genes listed (Fig. 7B), we found HY5, HY5-HOMOLOG (HYH), CRYPTOCHROME3 (CRY3), REPRESSOR OF UV-B PHOTOMORPHOGENESIS2 (RUP2), ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1 (ATBRCA1), PYRIDOXINE BIOSYNTHESIS 1 (PDX1), EARLY LIGHT INDUCIBLE PROTEIN1, (ELIP1), ELIP2, CHALCONE SYNTHASE (CHS), CHALCONE FLAVANONE ISOMERASE (CHI) FLAVONOL SYNTHASE1 (FLS1), and 4-COUMARATE: COA LIGASE3 (4CL3). The roles of HY5, HYH, and RUP2 are well established in UV-B signaling, whereas CHS, CHI, 4CL3, and FLS1 are involved in anthocyanin biosynthesis and are required in UV-B stress tolerance (Li et al., 1993; Mazza et al., 2000; Bieza and Lois, 2001; Brown and Jenkins, 2008). The downregulated genes are HISTONE H1.3 (HIS1-3), CBL-INTERACTING PROTEIN KINASE14 (CIPK14), and GA-STIMULATED ARABIDOPSIS6 (GASA6). Detection of HY5, HYH, RUP2, CHI, CHS, ELIP1, ELIP2, and PDX1 genes as UV-B induced in our dataset confirms earlier findings (Brown et al., 2005; Brown and Jenkins, 2008; Jenkins, 2009). MYB12, SUPEROXIDE DISMUTASE2 (SOD2), and ROOT UVB SENSITIVE1 (RUS1) were identified as differentially expressed in response to UV-B in the SAM epidermal cell layer. Interestingly, RUP1, UVR2, UVR3, MYB4, MYB111, F3H, and DAMAGED DNA BINDING2 (DDB2) were detected in response to UV-B exclusively in the seedling dataset (Brown et al., 2005). The differences observed in the transcriptome profile of seedling and SAM epidermal cell layers could be attributed to the tissue and cell types involved in the UV-B response. However, core UV-B signaling genes, as well as genes involved in UV-B tolerance, were activated in both tissue and cell population data, indicating that the core pathways of UV-B response and tolerance are conserved.
Figure 7.
Genes involved in anthocyanin biosynthesis are induced by UV-B irradiation stress in the SAM. A, A four-way analysis was carried out to identify a common set of genes induced and repressed by UV-B in seedlings and the SAM epidermal cell layer. B, Genes involved in anthocyanin biosynthesis were upregulated in response to UV-B, while DEWAX expression was downregulated compared to the unexposed control (comparison was made using normalized fragments per kilobase of transcript per million mapped reads values). C, Sketch of the HY5 and DEWAX gene regulatory network.
To further understand the role of the SAM epidermal cell layer in UV-B tolerance, we subjected the UV-B-responsive transcriptome to Gene Ontology (GO) enrichment analysis (Supplemental Table S4; Ge et al., 2019). As expected, genes involved in the UVR response, photosynthesis, flavonoid biosynthesis, and light intensity were overrepresented in the epidermal cell layer dataset. FIL, KAN2, PUCHI, ENHANCER OF SHOOT REGENERATION2, ANGUSTIFOLIA3 (AN3), GRF3, GRF4, GRF5, GRF7, SPL4, SPL9, REPLUMLESS, FD, TFL1, EPFL1, GIBBERELLIN 20 OXIDASE2, FOREVER YOUNG FLOWER, UNUSUAL FLORAL ORGANS, HAN, WUS, FLOWER PROMOTING FACTOR1 (FPF1), INFLORESCENCE DEFICIENT IN ABSCISSION LIKE-2 (IDL2), CYCD3;2, ROTUNDIFOLIA LIKE8, RGA2, TCP18/BRANCHED1 (BRC1), NGATHA-LIKE PROTEIN3 (NGAL3), KLUH, and BAM3 were identified in the shoot system category. Previous studies did not identify these genes as UV-B responsive in the seedling dataset. FIL, KAN2, and PUCHI are expressed in the peripheral zone of the SAM (Sawa et al., 1999; Kerstetter et al., 2001; Karim et al., 2009) and are involved in organ patterning. GRF3, GRF4, GRF5, and GRF7 belong to the GRF family of TFs, and are involved in cell proliferation in young emerging organs such as leaf (Kim et al., 2003; Omidbakhshfard et al., 2015). AN3 acts as a coactivator for GRFs (Horiguchi et al., 2005; Kawade et al., 2013). SPL4 and SPL9 belong to the SQUAMOSA PROMOTER BINDING PROTEIN family of TFs and are involved in the vegetative-to-reproductive phase transition (Xu et al., 2016). BAM3 is expressed in the shoot meristem and encodes receptor-like kinase, which is involved in shoot stem cell proliferation (DeYoung et al., 2006; Nimchuk et al., 2015). RGA2 is a member of the DELLA protein family and has been shown to reduce ROS accumulation by activating superoxide dismutase in addition to its role in inhibition of cell expansion (Achard et al., 2008). FPF1 is expressed in the shoot meristem in response to photoperiodic induction. Interestingly, FPF1 is involved in the integration of flowering time and floral organ development pathways (Kania et al., 1997). TCP18/BRC1 encodes a TCP TF, which is involved in axillary bud arrest outgrowth (Niwa et al., 2013). Taken together, the UV-B responsive genes belonging to the shoot system development category are primarily involved in lateral organ primordium patterning and meristem development. A subset of genes among them is also involved in cell proliferation. However, many critical negative regulators involved in bud outgrowth and inflorescence architecture are also activated, suggesting that UV-B acts as a growth stimulant at the cell level despite inhibiting the overall growth of floral buds and meristem. To cope with the irradiation stress, cells start activating pathways and genes that are part of ROS quenching. Thus, plants can use UV-B in many intriguing ways, which need to be explored to understand fully the impact of this type of radiation on overall growth and development.
DISCUSSION
UV-B fluence received by plants varies at different latitudes and longitudes on planet earth (McKenzie et al., 2011). It is crucial to understand how UV-B affects shoot meristem growth and development. Our live imaging study revealed that epidermal cells respond to UV-B by dying, but in dewax mutant plants, cell death in the epidermal cell layer appears late, indicating that higher dosages of DEWAX are detrimental to plant survival under UV-B. In the day, light and the UV-B signaling integrator HY5 negatively regulate DEWAX. The regulatory hierarchy between HY5 and DEWAX is consistent, given their roles in UV-B irradiation stress. These findings add to our understanding of how the central regulator of light signaling HY5 modulates expression of DEWAX and, as a result, influences the anthocyanin content. DEWAX inhibits anthocyanin accumulation and makes plants more sensitive to UV-B irradiation stress. However, in the nighttime, DEWAX inhibits cuticle and anthocyanin biosynthesis, which is vital for plant growth during the night (Fig. 7C; Go et al., 2014).
Past studies have shown the regulation of anthocyanin biosynthesis by HY5 (Hideg et al., 2013; Gangappa and Botto, 2016). However, these studies did not report negative regulation of anthocyanin biosynthesis by a diurnally regulated TF, nor how this switch functions in dark and light conditions. Since anthocyanin offers little value to the plant cell at night, its biosynthesis needs to be curtailed. To prevent the damage caused by high-intensity light and oxidative phosphorylation, plants used anthocyanin to quench the free radicals. Thus, light-dependent regulation of anthocyanin biosynthesis offers an excellent advantage for plants to efficiently utilize their resources under different growth conditions. Consistent with this hypothesis, we saw an increase in superoxide radical staining in 35S::DEWAX plant leaves compared to control leaves, as visualized by NBT staining (Supplemental Fig. S10).
Our natural variation data indicate that DEWAX is a critical component in UV-B irradiation tolerance. The quantity and quality of light received by plants on the planet vary depending upon latitude and longitude. Our studies on natural variation revealed an intricate link between HY5 and DEWAX in anthocyanin accumulation in planta. These studies led us to hypothesize that DEWAX could regulate anthocyanin contents under different environmental conditions. Overall, the HY5-DEWAX regulatory network imparts fitness to the plant under UV-B irradiation stress.
The SAM harbors distinct cell types, and they can respond to each environmental stimulus differently. Therefore, it is fundamental to understand their response at higher spatiotemporal resolution. We asked whether the genes that respond to UVR stress in seedlings are expressed in the SAM epidermal cell layer. For this, cells were sorted from UV-B-treated SAMs to study their gene expression responses. This analysis revealed that genes involved in UV-B irradiation stress tolerance are indeed induced in the SAM. The data also showed SAM development- and organ patterning-related functional categories. Among these, genes involved in organ patterning, flowering time, and cell proliferation were induced by UV-B. However, in this list, we also identified genes involved in axillary bud outgrowth arrest, organ abscission, and inhibition of flowering time. Some of these genes were induced by UV-B stress. However, genes involved in cell proliferation and organ patterning can be induced by ROS. Studies using seedlings were unable to identify shoot meristem development-related genes due to lack of relevant tissue in the sample. Further experiments are needed to establish the role of ROS signaling in the activation of these genes and how they regulate cell division and differentiation of stem cells into organs.
A role for DEWAX in wax biosynthesis has been established in previous studies (Go et al., 2014). It has been reported that cuticular wax content also varies depending upon the light conditions (Cameron et al., 2006). Interestingly, in the epidermal cell layer, KCS4 and KCS17 are induced by UV-B stress. However, GO enrichment analysis does not suggest an apparent increase in the expression of genes involved in wax biosynthesis, and thus it does not correspond directly to UV-B protection. It has been shown recently that SPL9 antagonizes DEWAX-mediated regulation of wax biosynthesis; consistent with this finding, we see upregulation of SPL9 in the epidermal cell layer in UV-B-exposed SAM (Li et al., 2019).
In summary, we identified DEWAX as a primary negative regulator of anthocyanin biosynthesis. HY5 negatively regulates DEWAX, and this regulation is further enhanced in UV-B irradiation stress. Diurnal regulation of wax and anthocyanin biosynthesis is vital for plants to cope with abiotic stresses such as light and drought.
MATERIAL AND METHODS
Plant Material and Growth Conditions
T-DNA insertion mutants for epidermal cell layer enriched TFs, including hy5 as well as Arabidopsis (Arabidopsis thaliana) natural accessions, were obtained from the Arabidopsis Biological Resource Center. Seeds were surface sterilized using 5% sodium hypochlorite and 70% ethanol and sown on 0.8% (w/v) agar plates containing Murashige and Skoog medium with 1% (w/v) Suc (pH 5.7). hy5 and dewax mutant plants were crossed to obtain the hy5 dewax double mutant. The plants were stratified at 4°C for 48 h and transferred to a Percival LED22C8 growth cabinet at 22°C, 70% humidity, and 120 μmol m−2 s−1 light in 16 h light and 8 h dark. For UV-B treatment, 10-d old seedlings were placed on a UV-B radiating (312 nm, 6×8w UV/white light transilluminators, fluence 3.82 mW cm−2) Genei UV transilluminator for 5 min, 10 min, and 15 min. The UV-B lamps radiate at a wavelength of >280 nm, with a peak at 312 nm. UV-B doses were measured with a UV-B radiometer where the filter transmits UV-B radiation with a peak near 313 nm and a half-bandwidth of 12 nm. The UV-B wavelength was measured using Compact CCD Spectrometers (CCS200, Thorlabs). After the treatment, seedlings were returned to the plant growth chamber for recovery in long-day conditions (16 h light and 8 h dark, PGC Flex, Conviron, and Percival Scientific).
For the high-altitude experiment, Col-0 plants were grown on a Murashige and Skoog agar plate for 4 d with the lid open for 3 to 4 h daily, then placed in the open. Ten-day-old plants were harvested, and tissue was stored in RNAlater solution. Environmental conditions were monitored for September, 2018, at Leh (India) using the CIRAS-3 portable photosynthesis system (PP Systems; Supplemental Table S5.
TF T-DNA Insertion Lines
T-DNA insertion lines for epidermal cell layer enriched TFs were obtained from the ABRC. For SALK, SAIL, and WISCONSIN lines, primers were designed using the iSect Primer design tool (http://signal.salk.edu/tdnaprimers.2.html). For GABI-KAT insertion lines, the primer sequence information was either obtained directly from the Web site (https://www.gabi-kat.de/db/primerdesign.php) or designed using the GK primer tool. Genomic DNA was isolated using a modified cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). Segregating insertion lines were confirmed for homozygosity by comparing the results of T-DNA lines and gene-specific amplification using Col-0 as wild type control. Plants having T-DNA insertion in both chromosomes did not show gene-specific amplification. We determined T-DNA insertion in the homozygous state in 30 TF-encoding genes. The complete list of T-DNA insertion mutants analyzed and primer sequence information related to T-DNA genotyping in this study is provided, along with the Arabidopsis gene identifier (AGI), in Supplemental Table S1.
Plasmid Constructs and Generation of Transgenic Lines
For ectopic expression, coding sequence of DEWAX was PCR amplified from a complementary DNA (cDNA) library of shoot apex. The forward primer, containing a CACC overhang, was designed to facilitate directional cloning of the inserted fragment into pENTR/D/TOPO. The resulting TOPO vectors were used to set up an LR reaction with the destination vector (pMDC32) to generate the 35S::DEWAX construct. The construct was sequence verified and transformed into wild-type Ler. To generate promoter::H2B-YFP transcriptional fusion constructs, a 3-kb fragment above the TSS of DEWAX was PCR amplified from the wild-type Ler genomic DNA template and cloned into pENTR/D/TOPO. The resulting clones were sequence verified and used to set up a LR reaction with the gateway-compatible pGreen0229 vector, as described by Yadav et al., (2014). Binary vectors with the promoter::H2B-YFP fusion were introduced into Agrobacterium tumefaciens GV3101 and were transformed into the wild-type Ler background by the floral dip method (Clough and Bent, 1998). T1 plants were selected on soil using BASTA (Bayer Crop Sciences) and screened using an upright epifluorescence microscope to isolate reporter lines. The pDEWAX::DEWAX-eGFP construct was assembled in a gateway-compatible pCAMBIA vector. First, a 3-kb promoter DNA fragment of the DEWAX promoter was PCR amplified from Ler genomic DNA and cloned into PspOMI and KpnI restriction sites in front of the gateway cassette containing attR1 and attR2 recombination sites. In the pENTR/D/TOPO eGFP vector, the DEWAX coding sequence was cloned at the N terminus of eGFP in SalI and StuI restriction sites by PCR amplification using DEWAX cDNA as a template. The resulting pENTR/D/TOPO DEWAX-eGFP vector was verified by sequencing to confirm the translation fusion, and was used to set up a LR reaction with the pDEWAX pCAMBIA vector to create pDEWAX::DEWAX-eGFP. The primer sequence information is given in Supplemental Table S6
To study the regulation of DEWAX in the gain- and loss-of-function HY5 background, the pDEWAX::H2B-YFP reporter line established in wild-type Ler was used to set up a cross with 35S::HA-HY5 hy5. In F2, hy5 mutant plants with elongated hypocotyls along with the pDEWAX::H2B-YFP reporter were isolated and analyzed for pDEWAX activity. 35S::HA-HY5 plants with the pDEWAX::H2B-YFP reporter were also identified from the segregating F2 plants based on the antibiotic resistance of the respective transgenes in the same plant.
Norflurazon Assay
Surface-sterilized seeds of the wild type, dewax, 35S::DEWAX, hy5, and hy5 dewax were directly plated on Murashige and Skoog agar plates containing 5 μm norflurazon. Similarly, Arabidopsis natural variants were grown to prevent the greening of seedlings on norflurazon.
Protoplasting of SAMs and RNA Sequencing
Approximately 200 SAMs of 28-d-old pATHMGB15:H2B-YFP ap1-1;cal1-1 plants were harvested within 15 min and placed in protoplasting solution (1.25% [w/v] Cellulase [Yakult], 0.3% [w/v] Macerozyme [Yakult], Hemicellulase [Sigma], 0.4 m d-mannitol, 20 mm MES, and 20 mm KCl, 0.1% [w/v] bovine serum albumin [BSA], 10 mm CaCl2, and 5 mm β-Mercaptoethanol for 1.25 h at 120 rpm in a 50-mL Falcon tube. Protoplasts were passed through a 40 μm cell strainer (BD Falcon) and collected in a 15-mL Falcon tube. The filtrate was centrifuged at 4°C for 15 min at 500g. The pellet was resuspended in 1 mL buffer A (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm MES, pH adjusted to 5.7 with KOH). Cells were sorted using fluorescence-activated cell sorter (BD FACSAria Fusion, BD Biosciences) fitted with a 100-µm nozzle at a rate of 2,000 to 5,000 events s−1 and sheath fluid pressure of 20 psi. Protoplasts from non-GFP-expressing ap1-1;cal1-1 were used as a negative control for establishing sorting criteria based on cell properties, as follows: (1) a cluster of live protoplasts with intact membranes was selected based on the forward/side scatter ratio; and (2) GFP-positive cells were selected by their emission intensity in the green channel (∼530 nm) above that of the negative control. Doublet exclusion was performed by plotting width versus area for forward and side scatter, and finally, gate was applied to identify the fluorescent cells by comparing it with nonfluorescent cells and sorting was performed. The purity of sorting events was independently confirmed through microscopic observation of sorted protoplasts. The number of fluorescently labeled protoplasts typically varied between 80,000 and 1,00,000 per experiment. Cells were sorted directly into lysis buffer (RLT buffer, Qiagen) for RNA extraction.
For RNA-seq, total RNA was isolated from the sorted cells using the QIAGEN RNeasy micro kit. The eluted total RNA from the column was precipitated with 0.5 volume 7.5 m ammonium acetate (Sigma-Aldrich), 2.5 volume ethanol (99.9%; Merck) and 0.5 μL Glycoblue (Invitrogen) overnight at −20°C. This was followed by centrifugation at 10,000 rpm at 4°C and the resulting pellet was washed with 70% ethanol, air dried and finally dissolved in 5 μL of RNAse free water. The samples showing an RNA integrity number ≥8.0 was used for library preparation. The TruSeq v2 RNA sample preparation kit (Illumina) was used to prepare cDNA libraries. From the total RNA, poly (A)+ RNA (mRNA) were purified using oligo(dT) magnetic beads and fragmented using divalent cations at elevated temperature. RNA fragments were reverse transcribed using random primers, followed by second-strand cDNA synthesis using RNase H/DNA Polymerase I. End repair and A-tailing of cDNA were followed by adapter ligation. The indexed cDNA products were purified and amplified by PCR (15 cycles) to create the final cDNA libraries. Library quality was checked on a 2200 TapeStation (Agilent Technologies) and individual libraries were quantified on the Qubit System (Invitrogen) prior to pooling. The pooled, indexed libraries were loaded and analyzed on an Illumina Hi-Seq 2500 platform. The library preparation and sequencing of libraries were carried out at Agrigenome Labs.
RNA Sequencing Data Analysis
RNA sequencing data (100-bp paired-end reads) were obtained using an Illumina Hi-Seq 2500. The FastQC tool was used for quality check of the raw reads. There were two to three biological replicates per sample. The reproducibility within the replicates was checked using the Spearman rank correlation coefficient. The Trimmomatic tool was used for adapter removal (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4103590/). The paired-end reads (2 × 100 bp) were mapped to The Arabidopsis Information Resource TAIR10 Arabidopsis genome (http//arabidopis.org) using STAR version 2.5.3 (https://academic.oup.com/bioinformatics/article/29/1/15/272537) and an annotation file from Araport (www.araport.org). On average, 85% of the reads were uniquely mapped to the genome. Gene expression was assessed for both read count and fragments per kilobase of transcript per million mapped reads using HTSeq-count (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4287950/) and Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/), respectively. DEG analysis was performed using the cuffdiff module from the Cufflinks package. For this, we used stringent criteria of adjusted P ≤ 0.05 and cutoffs of ≥2-fold upregulated and ≤2-fold downregulated.
GO Enrichment Analysis
GO enrichment analysis was carried out using web-based ShinyGO version 0.61 (http://bioinformatics.sdstate.edu/go/; Ge et al., 2019). The list of genes either upregulated or downregulated in response to UV-B were subjected to GO enrichment analysis. The top 50 categories were selected as output at P ≤ 0.05, as described in Supplemental Table S4.
eY1H Assay
For HY5 prey, the pENTR/D/TOPO HY5 clone was used to setup the LR reaction with the pDEST AD2μ vector. To create the bait, a 3-kb promoter fragment of DEWAX was first PCR amplified with primers containing attB4 and attB1R overhangs and cloned into the P4P1r vector using a BP reaction, then subcloned into the gateway-compatible pMW2 vector using an LR reaction as described in earlier studies (Taylor-Teeples et al., 2015). Primers used for making prey and bait are listed in Supplemental Table S6.
RNA Isolation and RT-qPCR
For RNA isolation, seedlings were grown on Murashige and Skoog agar plates for 10 d and exposed to UV-B for 4 min. Plates were returned to the plant growth chamber; after 30 min, seedlings were frozen and homogenized in liquid nitrogen. RNA extraction was performed using the ReliaPrep RNA miniprep kit from Promega according to the manufacturer’s instructions. Untreated plants were used as a control. Total RNA was used to set up the cDNA synthesis reaction using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was performed using SYBR Green (BioRad) with real-time PCR. Ubiquitin was used as a reference gene for normalization of Ct values.
Comparison of SAM and Seedling UV-B Responses
For SAM and seedling transcriptome comparison analysis, we downloaded the publicly available seedling UV-B response data from the National Center for Biotechnology Information's Gene Expression Omnibus database (accession number GSE3533; Brown et al., 2005). The raw data were normalized using the GCRMA package in R after downloading the Cel files (Wu and Irizarry, 2005). For DEG analysis, normalized expression values obtained by GCRMA were used as input in the LIMMA package, and the adjusted P-value statistic was examined for pairwise comparison including fold change values according to the methods of Benjamini and Hochberg (1995). Genes showing adjusted P ≤ 0.05 and ≥2-fold upregulation or ≤2-fold downregulation were considered differentially expressed. To identify the common genes that were either induced or suppressed in response to UV-B, a two-way Venn diagram analysis was performed for seedling and SAM cell type data using the AGIs for up- and downregulated genes.
EcoGEx
To obtain the ecotype-specific gene expression data, a source code was run in R (https://github.com/sk-sahu/EcoGEx). AGIs were used as input. Transcriptome data were retrieved from Gene Expression Omnibus dataset GSE80744 (Kawakatsu et al., 2016).
Anthocyanin Measurement
Quantitative estimation of anthocyanin content was performed as described by Chory et al. (1989). Wild-type and mutant seedlings, grown in parallel, were homogenized and incubated overnight in 0.3 mL 1% HCl in methanol at 4°C and extracted using an equal volume of chloroform after the addition of 0.2 mL of water. Total anthocyanin content was calculated using raw values from spectrophotometry with the formula (A530 − 0.25 × A657)/(tissue weight in milligrams). The data were plotted relative to wild-type.
Sample Preparation and Detection of Phe and Shikimic Acid
For detection of Phe and shikimic acid, 200 mg fresh Arabidopsis seedling tissue (quenched with liquid N2 and powdered) was extracted with 950 μL extraction solvent (methanol:water:chloroform [3:1:1 v/v/v]). Fifty microliters of ribitol (0.01 mg mL−1) was added to the extracts and used as an internal standard. The ribitol was further extracted at 70°C in a thermomixer for 10 min and then centrifuged at 13,000g for 15 min (Lisec et al., 2006). Aliquots of supernatant (50 μL) were dried using a speed vacuum. The dried samples were MeOX-TMS derivatized (using pyridine, methoxamine hydrochloride, and N-methyl-N-trimethylsilyl trifluoroacetamide [Sigma-Aldrich]) and subjected to GC-MS analysis (GC ALS-MS 5977B, Agilent Technologies) with 5% phenyl methyl siloxane (HP-5MS) column (19091S-433, Agilent; Masakapalli et al., 2014; Shree et al., 2019). Raw data were acquired, and files were processed using MassHunter software (Agilent). Phe and shikimic acid were identified based on the m/z fragments, retention time (RT) against standards, and probability of hits against the NIST 17 library (Kopka et al., 2004). The peak abundance of ribitol was used to normalize the relative abundances of Phe and shikimic acid levels among the samples. Basic statistical analysis was performed using GraphPad Prism 8.5.
Confocal Imaging and UV-B Treatment
For live imaging, surface-sterilized seeds of pCLV3::mGFP5-ER; 35S::YFP-PM29, wild-type Col-0, and dewax mutant lines were put on Murashige and Skoog plates and grown in plant growth chambers in long-day conditions (16 h light and 8 h dark) at 22°C for 7 d (120 μmol m−2 s−1 in a PGC Flex plant growth chamber (Conviron). Seedlings were transferred to round boxes containing Murashige and Skoog agar and grown for ∼2 to 3 weeks, until bolting, when the inflorescence became visible. Plants were allowed to recover overnight after dissection. Dissected plant SAMs showing active organogenesis were directly exposed to the UV-B source (Transilluminator; 312 nm, fluence 3.82 mW cm−2) for 2 min and returned to the growth chamber. Live-imaging of the exposed plants was carried out at 2, 8, 16, and 24 h after initial exposure. PI (100 mg mL−1) was applied to the SAM before each imaging session to stain the cell outline and identify UV-B-induced cell death. Confocal image stacks were obtained using a 63× long-distance water-dipping lens with hybrid detectors. Image acquisition was set to 514 × 514 or 1,024 × 1,024 pixels with 400 Hz scan speed. Confocal Z-sections were spaced 1.5 μm apart for live-imaging experiments, and a line average of 4 was applied. For mGFP excitation, a 488 nm laser line was used, whereas for YFP excitation, a 514 nm laser was used. PI was excited with 561 nm lasers. To quantify the H2B-YFP fluorescence signal in the hy5 mutant versus the wild type, 10% argon laser power was used to excite H2B-YFP with 16% laser gain. On top of this, a 450 digital gain was applied to collect the fluorescence spectra. PI was excited with 5% laser power with 8% laser gain. Fluorescence spectra for PI were collected after applying 120 digital gain. For live-imaging experiments, sequential imaging was performed. mGFP-ER was excited with 5.5% laser power with a 400 to 450 gain followed by YFP, with 7% laser power. Spectra were collected by adjusting the variable band-pass filter in the confocal image acquisition panel. Confocal image stacks were obtained using a SP8 upright confocal microscope (Leica).
ChIP-qPCR
ChIP was performed according to the method described by Gendrel et al., (2005). Ten-day-old seedlings of pHY5::HY5-YFP hy5 and 35S::EGFP in the wild type were fixed using 1% paraformaldehyde. Nuclei were isolated and then sheared using a Q700 sonicator (QSonica) with 15 cycles of 70% pulse amplitude for 5 s followed by 60 s pulse off time. Protein-DNA complexes were pulled down by protein-A magnetic beads (1614013, SureBeads) and anti-GFP antibody (a290, Abcam). Rabbit IgG serum was used as a negative control. The immunoprecipitated chromatin was washed and reverse crosslinked. qPCR was setup using SYBR Green (BioRad) according to the manufacturer’s instructions in a Touch Real-Time PCR Detection System (CFX96, BioRad). Fold enrichment was calculated by normalizing against the negative control, i.e. IgG. Data are presented as means ± se from two biological replicates. The primers used are listed in Supplemental Table S6.
EMSAs
Full-length coding sequences of HY5 were cloned into pENTR/D/TOPO and then subcloned into the gateway-compatible pMAL2C vector (for the N-terminal MBP tag) using LR clonase. Both the MBP-tagged HY5 and MBP protein (as control) were induced in the Rosetta strain of Escherichia coli using 0.1 mm isopropylthio-β-galactoside at 37°C and purified using amylose resin beads (New England Biolabs). Oligos containing the respective cis-elements were used to the synthesize double-strand DNA probe. 4% native polyacrylamide gel was prepared using acrylamide:bisacrylamide (29:1; Sigma Aldrich) in 0.5× Tris-borate/EDTA (44.5 mm Tris, 44.5 mm boric acid, and 1 mm EDTA). Fifty nanograms of probe DNA was incubated with 200 ng purified MBP-HY5 and MBP protein in binding buffer (0.2% glycerol, 50 mm NaCl, 10 mm dithiothreitol, 2.5 mm MgCl2, and 20 mm phenylmethylsulfonyl fluoride) for 0.5 h at 25°C. The gel was incubated at 4°C in 0.5× Tris-borate/EDTA for 1 h. Gels were run in a vertical electrophoresis unit (Mini PROTEAN Tetra cell, Bio-Rad) at 90 V for 3 h at 4°C. The gel was stained in ethidium bromide for 10 min and photographed using the Gel Documentation System (BioRad).
Statistical Analysis
Statistical analyses were performed using Prism 8.0.2 (GraphPad Software). Data were expressed as means ± se, and P-values were calculated using Student's t test or one-way/two-way ANOVA followed by uncorrected Fisher’s least significant difference multiple-comparisons test (P < 0.01) or the two-stage linear step-up procedure (P < 0.01; Benjamini et al., 2006). Significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and nonsignificant when P > 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank SRA data libraries under accession number PRJNA670063.
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Impact of UV-B radiation on Arabidopsis SAM.
Supplemental Figure S2. Flow chart of anthocyanin biosynthesis pathway.
Supplemental Figure S3. UV-B induces genes involved in anthocyanin biosynthesis.
Supplemental Figure S4. Metabolite profiling of UV-B treated plants with GC-MS.
Supplemental Figure S5. HY5-YFP fusion protein detected in 5-d-old seedlings.
Supplemental Figure S6. Expression of DEWAX in epidermis is regulated by HY5.
Supplemental Figure S7. Plant grown at high altitude show reduced expression of DEWAX.
Supplemental Figure S8. DEWAX antagonizes the HY5 mediated anthocyanin biosynthesis in Arabidopsis natural variants.
Supplemental Figure S9. Flow chart showing the protocol for sample preparation for cell-type specific transcriptomic study.
Supplemental Figure S10. NBT staining.
Supplemental Table S1. Primers used in the study to confirm the T-DNA insertion in the mutant lines.
Supplemental Table S2. Summary of RT-qPCR results.
Supplemental Table S3. Average and normalized fragments per kilobase of transcript per million mapped reads values of the epidermal cell population in control versus the UV-B-treated sample.
Supplemental Table S4. Enrichment analysis for genes showing up- and downregulation in response to UV-B light based on hypergeometric distribution.
Supplemental Table S5. Environmental conditions in Leh (India) at 11,000 feet.
Supplemental Table S6. List of primers used in the study.
Acknowledgments
We thank Roman Ulm for sharing pHY5::HY5-YFP hy5 seeds and Sourav Datta and Henrik Johannsson for 35S::HY5 and 35S::HA-HY5 hy5 lines. Live-imaging and cell sorting experiments were carried out at the confocal microscopy and FACS core facility, respectively, at the Indian Institutes of Science Education and Research Mohali. Confocal Microscopy and FACS facility are supported by Institute core grants. We thank Subham Agarwal for assisting in NBT staining. The Defence Research and Development Organization, Government of India, and the Defence Institute of High Altitude Research Leh supported this work.
Footnotes
This work was supported by the Department of Biotechnology, Ministry of Science and Technology (Ramalingaswami Fellowship and Innovative Young Biotechnologist Award to R.K.Y.), the Science and Engineering Research Board (Early Career Research grant no. ECR/2016/001176 to S.K.M.), and the University Grant Commission and Ministry of Education of India (PhD fellowship to P.S. and M.L.).
References
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P(2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW(1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Bais AF, McKenzie RL, Bernhard G, Aucamp PJ, Ilyas M, Madronich S, Tourpali K(2015) Ozone depletion and climate change: Impacts on UV radiation. Photochem Photobiol Sci 14: 19–52 [DOI] [PubMed] [Google Scholar]
- Barnes JD, Percy KE, Paul ND, Jones P, McLaughlin CK, Mullineaux PM, Creissen G, Wellburn AR(1996) The influence of UV-B radiation on the physicochemical nature of tobacco (Nicotiana tabacum L.) leaf surfaces. J Exp Bot 47: 99–109 [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D(2006) Adaptive linear step-up false discovery rate controlling procedures. Biometrika 93: 491–507 [Google Scholar]
- Bieza K, Lois R(2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Crocco CD, Ekundayo B, Lau K, Raffelberg S, Tilbrook K, Yin R, Chappuis R, Schalch T, Ulm R(2016) Revisiting chromatin binding of the Arabidopsis UV-B photoreceptor UVR8. BMC Plant Biol 16: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R(2014) UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosché M, Schuler MA, Kalbina I, Connor L, Strid A(2002) Gene regulation by low level UV-B radiation: Identification by DNA array analysis. Photochem Photobiol Sci 1: 656–664 [DOI] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI(2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI(2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J(2020) Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32: 967–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM, Bornman JF, Ballaré CL, Flint SD, Kulandaivelu G(2007) Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors. Photochem Photobiol Sci 6: 252–266 [DOI] [PubMed] [Google Scholar]
- Cameron KD, Teece MA, Smart LB(2006) Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol 140: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C(2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F(1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Cloix C, Jenkins GI(2008) Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE(2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45: 1–16 [DOI] [PubMed] [Google Scholar]
- Diffey BL.(1991) Solar ultraviolet radiation effects on biological systems. Phys Med Biol 36: 299–328 [DOI] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 10.1038/nature10414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF(2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Ge SX, Jung D, Yao R(2019) ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36: 2628–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V(2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Go YS, Kim H, Kim HJ, Suh MC(2014) Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26: 1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D(1989) Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol [Google Scholar]
- Heijde M, Ulm R(2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Hideg E, Jansen MAK, Strid A(2013) UV-B exposure, ROS, and stress: Inseparable companions or loosely linked associates? Trends Plant Sci 18: 107–115 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H(2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Ichihashi M, Ueda M, Budiyanto A, Bito T, Oka M, Fukunaga M, Tsuru K, Horikawa T(2003) UV-induced skin damage. Toxicology 189: 21–39 [DOI] [PubMed] [Google Scholar]
- Jenkins GI.(2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C(2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI(2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbina I, Strid A(2006) Supplementary ultraviolet-B irradiation reveals differences in stress responses between Arabidopsis thaliana ecotypes. Plant Cell Environ 29: 754–763 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C(2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S(1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MR, Hirota A, Kwiatkowska D, Tasaka M, Aida M(2009) A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H(2013) ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 23: 788–792 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS(2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H(2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL(1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RJ, Li LM, Liu XL, Kim JC, Jenks MA, Lü S(2019) Diurnal regulation of plant epidermal wax synthesis through antagonistic roles of the transcription factors SPL9 and DEWAX. Plant Cell 31: 2711–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR(2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Masakapalli SK, Bryant FM, Kruger NJ, Ratcliffe RG(2014) The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J 78: 964–977 [DOI] [PubMed] [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballaré CL(2000) Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol 122: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie RL, Aucamp PJ, Bais AF, Björn LO, Ilyas M(2007) Changes in biologically-active ultraviolet radiation reaching the Earth’s surface. Photochem Photobiol Sci 6: 218–231 [DOI] [PubMed] [Google Scholar]
- McKenzie RL, Aucamp PJ, Bais AF, Björn LO, Ilyas M, Madronich S(2011) Ozone depletion and climate change: Impacts on UV radiation. Photochem Photobiol Sci 10: 182–198 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B(2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM.(1997) Genetic control of cell division patterns in developing plants. Cell 88: 299–308 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF(1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM(2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, et al. (2013) BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25: 1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A, Cadet J, Douki T, Mouret S, Tucker MA, et al. (2012) Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun 3: 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B(2015) Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol Plant 8: 998–1010 [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM(2004) Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131: 4225–4237 [DOI] [PubMed] [Google Scholar]
- Riederer M, Schneider G(1990) The effect of the environment on the permeability and composition of Citrus leaf cuticles: II. Composition of soluble cuticular lipids and correlation with transport properties. Planta 180: 154–165 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Sagan C.(1973) Ultraviolet selection pressure on the earliest organisms. J Theor Biol 39: 195–200 [DOI] [PubMed] [Google Scholar]
- Satina S, Blakeslee AF, Avery AG(1940) Demonstration of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras on JSTOR. Am J Bot 27: 895–905 [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K(1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shree M, Lingwan M, Masakapalli SK(2019) Metabolite profiling and metabolomics of plant systems using 1H NMR and GC-MS In Banerjee R, Kumar GV, and Veejan Kumar SP, eds, OMICS-Based Approaches in Plant Biotechnology. Scrivener Publishing, Austin, TX, pp 129–144 [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R(2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al. (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R(2013) The UVR8 UV-B photoreceptor: Perception, signaling and response. The Arabidopsis Book 11: e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F(2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA(2005) Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol 12: 882–893 [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS(2016) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Reddy GV(2010) WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137(21): 3581–3589 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV(2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Xie M, Girke T, Venugopala RG(2014) A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 141: 2735–2744 [DOI] [PubMed] [Google Scholar]
- Yin R, Skvortsova MY, Loubéry S, Ulm R(2016) COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113: E4415–E4422 [DOI] [PMC free article] [PubMed] [Google Scholar]