Abstract
The development of chemotherapeutic dug resistance hinders the clinical treatment of cancer. MicroRNAs (miRNAs/miRs) have been revealed to serve essential roles in the drug resistance of numerous types of cancer. miR-139-5p was previously reported to be associated with cisplatin (DDP) sensitivity in human nasopharyngeal carcinoma cells and colorectal cancer cells. However, the effect and underlying mechanism of miR-139-5p in DDP sensitivity in non-small cell lung cancer (NSCLC) cells has not yet been fully elucidated. In the present study, the expression of miR-139-5p and Homeobox protein Hox-B2 (HOXB2) in NSCLC tissues was examined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting. Subsequently, the effect of miR-139-5p on the DDP sensitivity of NSCLC cells in vitro was investigated. Cell proliferation was examined using a Cell Counting Kit-8 assay. Western blotting was used to evaluate the protein expression of HOXB2, phosphorylated (p)-PI3K, p-AKT, caspase-3 and cleaved-caspase-3, and RT-qPCR was used to evaluate the expression of miR-139-5p, and the mRNA expression levels of HOXB2, PI3K, AKT and caspase-3. The apoptotic rate of the cells was detected using flow cytometry. miR-139-5p expression in NSCLC tissues was shown to be significantly lower compared with that in adjacent tissues. Additionally, miR-139-5p increased cell apoptosis and inhibited NSCLC cell proliferation induced by DDP in vitro via modulating the PI3K/AKT/caspase-3 signaling pathway. Furthermore, HOXB2 was identified to be a target of miR-139-5p, and miR-139-5p was revealed to sensitize NSCLC cells to DDP via the targeting of HOXB2. Taken together, the results of the present study demonstrated that regulating the expression of miR-139-5p could provide a novel approach to reverse DDP resistance and increase chemosensitivity in the treatment of NSCLC.
Keywords: microRNA-139-5p, non-small cell lung cancer, cisplatin sensitivity, PI3K/AKT, cleaved-caspase-3
Introduction
Lung cancer is one of the most common cancers leading to mortality, and its incidence is increasing on a yearly basis (1,2). It is reported that there were >2.09 million new lung cancer cases in 2018 worldwide, ranking first among all cancers (3). Lung cancer is the second most common cancer in USA and the most common cancer in China (4). In some developing countries, due to the high smoking prevalence, lung cancer incidence is still rising (5). In relation to sex differences, lung cancer is more common and prevalent in males globally (5). According to epidemiological data, the incidence of lung cancer in China was around 69% among men and 31% among women in 2015; meanwhile, the incidence in United States was 52% in males and 48% in females (6). Non-small cell lung cancer (NSCLC) is a relatively common type of lung cancer. For instance, >0.23 million new NSCLC cases, far more than breast and colon cases, were reported in the USA in 2018 (7). Chemotherapy, which promotes tumor cell apoptosis, is a commonly used method to treat NSCLC, however, NSCLC cells exhibit high drug tolerance (8–11). Therefore, exploring novel molecular targets that have the ability to increase lung cancer drug-sensitivity is of great importance for the treatment of lung cancer.
MicroRNAs (miRNAs), a class of endogenous non-coding RNA molecules of ~20 nucleotides (nts) in length, have been revealed to have an essential role in the drug resistance of multiple types of cancer, such as colorectal cancer and epithelial ovarian cancer (12–14). The role of miRNAs in a variety of intracellular signaling processes, including those associated with tumors, has been verified (15,16). They are also involved in the regulation of gene expression at the transcriptional and post-transcriptional levels (17), and in the regulation of the cell cycle (18), cell proliferation (19,20), cell differentiation (21) and apoptosis (22,23). A previous study claimed that miRNAs are abnormally expressed in lung cancer tissues, and are associated with tumor invasion, metastasis and prognosis (24). miRNAs have been suggested as potential biomarkers in lung cancer diagnosis and adenocarcinoma (25,26). For instance, miR-139-5p was significantly reduced in NSCLC tissues, acting as a tumor suppressor gene in the occurrence and development of NSCLC (27). Meanwhile, miR-139-5p was also reported to suppress proliferation or induce apoptosis by targeting tyrosine kinase receptor cellular-mesenchymal to epithelial transition factor in lung cancer (28). In terms of drug resistance, miR-139-5p was previously reported to be associated with cisplatin (DDP) sensitivity in human nasopharyngeal carcinoma cells (29) and colorectal cancer cells (30). In addition, miR-139-5p was effective in inhibiting NSCLC cell proliferation and invasion (31). However, the effect of miR-139-5p on DDP sensitivity in NSCLC cells and the underlying mechanisms have not yet been fully elucidated.
The HOX genes are master transcription factors that are expressed in coordinated spatiotemporal patterns in order to ensure normal development (32). Moreover, HOX genes are reported to be involved in multiple cellular processes, including cell differentiation, proliferation and survival. Homeobox protein Hox-B2 (HOXB2) is a key HOX gene, which also serves as an oncogene in several cancers, such as bladder cancer (33) and osteosarcoma (34). Moreover, HOXB2 was reported to regulate the PI3K/AKT signaling pathway to modulate the pathogenesis of different tumors, such as osteosarcoma (34) and acute myeloid leukemia (35). In terms of lung cancer, HOXB2 was reported to promote the invasion of lung cancer cells (36), functioning as a novel prognostic biomarker for lung adenocarcinomas (37). However, the association between HOXB2 and miR-139-5p in the regulation of DDP sensitivity in NSCLC still remains unclear.
Therefore, the present study primarily focused on investigating the effect of miR-139-5p, combined with DDP, on cell proliferation and apoptosis of NSCLC cells. The results obtained could provide a novel approach in the reversal of DDP resistance and increasing chemosensitivity in the treatment of NSCLC.
Materials and methods
Patients and tissues
Tissue samples were taken from a total of 60 patients (60–75 years, 69.5±5.2 years, 39 males and 21 females) with NSCLC between July 2016 and July 2017 from Weifang Yidu Central Hospital (Weifang, China). The inclusion criteria included: i) Patients diagnosed with NSCLC via biopsy; ii) >18 years of age; iii) histopathological stage confirmed as early or metastatic, adenocarcinoma or squamous cell carcinoma. Exclusion criteria included: i) Currently suffering from any other types of cancer; and ii) currently taking medications or any adjuvant treatments.
The tumor and paracarcinoma tissues (5 cm away from the tumor tissues) were removed by surgical section, and immediately placed in liquid nitrogen or 10% formalin. All samples were confirmed by pathological examination, and no radiotherapy or chemotherapy was performed prior to surgery. In addition, the present study was approved by the Ethics Committee of Weifang Yidu Central Hospital, and written informed consent was obtained from each participant prior to the study.
Cell culture and cell transfection
The A549 lung adenocarcinoma cell line was purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China) and cultured in Hyclone™ Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 80 U/ml penicillin and 0.08 mg/ml streptomycin (all from Gibco, Thermo Fisher Scientific, Inc.). Cells were cultured in an incubator at 37°C with an atmosphere of 5% CO2. The medium was replaced every other day. Cells in the exponential phase of growth were used for subsequent experiments.
Cells were seeded in 6-well plates at a density of 2×104 cells/well, and subsequently divided into three groups: i) Control group (Control); ii) negative control group (NC); and iii) miR-139-5p mimics group (mimics). The miR-139-5p mimics and control plasmids were obtained from New England Biolabs, Inc. The sequences used were as follows: miR-NC, 5′-GUGUAUUCUACAGUGCACGUGUCUCCAGUGU-3′; miR-139-5p mimics, 5′-GGCUCGGAGGCUGGAGACGCGGCCCUGUUGGAGUAAC-3′. Cells were transfected with 500 ng plasmids or miRusing 2.5 µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following cell incubation for 6 h, the medium was replaced with fresh medium containing 10% FBS. Experiments were performed in triplicate 48 h post-transfection.
In a separate experiment, 1×105 cells/well cells were seeded into 24-well plates and divided into four groups: i) Control group; ii) DDP group; iii) DDP + miR-NC mimics; and iv) mimics group (DDP + miR-139-5p mimics). Prior to transfection, all cells were treated with 1.0 µM DDP (cat. no. R00313; Rechemscience Co., Ltd.) and cultured for 2 h at 37°C, 5% CO2, as described previously (38). Transfections were then performed at 37°C with 5% CO2 for 24 h.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to determine the differences in miR-139-5p and HOXB2 expression in tumor tissues and paracarcinoma tissues, and to investigate the mRNA expression levels of PI3K, AKT and caspase-3 in cells. Total RNA was extracted with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and the concentration of RNA was determined using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Subsequently, the RNA was reversed-transcribed into complementary DNA using a PrimeScript™ RT reagent kit (Takara Bio, Inc.) at 42°C for 10 min. To detect mRNA expression, RNA was reverse-transcribed into cDNA using a Mir-X™ miRNA First Strand Synthesis kit, and RT-qPCR was performed with a Mir-X™ miRNA RT-qPCR SYBR® kit (both from Takara Bio, Inc.). SYBR-Green (10 µl), molecular grade water (6 ml), 1 µl each of the forward and reverse primers and 2 µl cDNA were mixed in each reaction on the PCR plate. Amplification was performed using the following thermocycling conditions: 10 min at 95°C, 50 cycles at 95°C for 15 sec, and 60°C for 60 sec. Each reaction was performed three times. Primer sequences used in the present study are shown in Table I. The comparative cycle quantification (Cq) method was used to analyze the RT-qPCR data, and 2−ΔΔCq values were chosen to reflect the mRNA difference (39). The U6 gene was selected as an internal control for miR-139-5p, and GAPDH was used as an internal control for HOXB2, PI3K, AKT and caspase-3. All experiments were performed in triplicate. All primers were displayed in Table I.
Table I.
Oligonucleotide primers used for reverse transcription-quantitative PCR.
| Gene | Primer sequences (5→3) |
|---|---|
| miR-139-5p | F: GCCTCTACAGTGCACGTGTCTC |
| R: CGCTGTTCTCATCTGTCTCGC | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| HOXB2 | F: TCCTCCTTTCGAGCAAACCTTCC |
| R: AGTGGAATTCCTTCTCCAGTTCC | |
| PI3K | F: TCAATGTCCATCTCCATTCTCCT |
| R: GATTGCCTCCAGTTGCTTCC | |
| AKT | F: CATCCTCATGGAAGAGATCCGC |
| R: GAGGAAGAACCTGTGCTCCATG | |
| Caspase-3 | F: CATGGAAGCGAATCAATGGACT |
| R: CTGTACCAGACCGAGATGTCA | |
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT |
| R: GGCTGTTGTCATACTTCTCATGG |
F, forward; R, reverse; miR, microRNA; HOXB2, Homeobox protein Hox-B2.
Luciferase reporter assay
The binding sites of miR-139-5p and HOXB2 were determined using the TargetScan online tool (40). To observe the binding of miR-139-5p to HOXB2 mRNA, the 3′-untranslated region (3′-UTR) segment of HOXB2 mRNA was amplified by PCR as aforementioned and inserted into the pGL3/luciferase vector (Promega Corporation). The wild-type 3′-UTR of HOXB2 mRNA was obtained by PCR amplification as aforementioned and cloned and inserted into pGL3/luciferase, as described for the wild-type 3′-UTR plasmid. Co-transfections of the HOXB2 3′-UTR or mutated HOXB2 3′-UTR plasmid with miR-139-5p mimics into the cells were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was measured 48 h after transfection using the Dual-Luciferase Reporter assay system (Promega Corporation) and normalized to Renilla luciferase. Experiments were performed in triplicate.
Cell proliferation assay
Cell proliferation was measured using a Cell Counting Kit-8 (CCK-8; ApexBio Technology LLC) assay according to the manufacturer's protocol. Transfected cells were seeded into 96-well plates at a density of 5,000 cells/well, and the cells were cultured for 6 h. Aliquots of 10 µl CCK-8 were added into each well, and the cell mixture was incubated at 37°C for 4 h. Optical density (OD) was detected at a wavelength of 450 nm.
Cell apoptosis assay
Apoptosis of transfected A549 cells (early and late apoptosis) was measured by flow cytometry using Annexin V/propidium iodide (PI) double staining. Cells were seeded into 600 mm-diameter culture dishes at a density of 4×104 cells/well. Cells were harvested, and washed twice with ice-cold PBS. Subsequently, the cells were resuspended in 300 ml binding buffer containing Annexin V (3 µl) and PI (3 µl) for 15 min at room temperature in the dark. Finally, cell apoptosis was quantified using an FACScan flow cytometer (Becton-Dickinson and Company) and analyzed using CellQuest software version 5.1 (BD Biosciences). During the course of these experiments, at least 10,000 cells for each sample were analyzed.
Western blot analysis
Total protein of the tissues and transfected cells was extracted using a protein extraction kit (Beyotime Institute of Biotechnology), and protein concentration was determined using a bicinchoninic acid kit (Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were separated via SDS-PAGE on 10% gel, and subsequently transferred to a polyvinylidene difluoride membrane. After blocking for 2 h in 5% skimmed milk at room temperature, the membranes were incubated with the following primary antibodies overnight at 4°C: Anti-HOXB2 (cat. no. ab220390; Abcam), anti-phosphorylated (p)-PI3K (cat. no. ab32089; Abcam) anti-p-AKT (cat. no. 4058; Cell Signaling Technology, Inc.), anti-caspase-3 (cat. no. 9662; Cell Signaling Technology, Inc.), anti-cleaved-caspase-3 (cat. no. 9664; Cell Signaling Technology, Inc.) and anti-GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.). The primary antibodies were used at 1:1,000 dilution. After washing three times with TBS with 0.3% Tween-20, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.) at 37°C for 30 min. Protein expression was visualized using ECL-Plus reagent (Santa Cruz Biotechnology, Inc.) and analyzed with the ChemiDoc™ XRS imaging system (Bio-Rad Laboratories, Inc.). GAPDH was used as the loading control.
Statistical analysis
All data are expressed as the mean ± standard error of the mean. Student's t-test was performed to test differences between two groups, whereas one-way analysis of variance, followed by Tukey's post hoc analysis, was used to examine differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-139-5p expression is downregulated, whereas HOXB2 expression is upregulated, in human NSCLC tissues
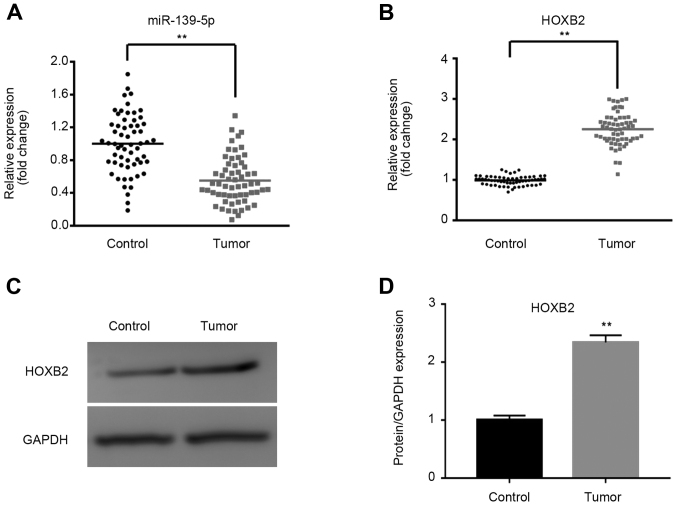
The mRNA and protein expression levels of miR-139-5p and HOXB2 in tissues were determined using RT-qPCR and western blotting. As shown in Fig. 1A and B, miR-139-5p was significantly downregulated, whereas HOXB2 mRNA was significantly upregulated, in NSCLC tissues compared with paracarcinoma tissues (P<0.01). Furthermore, the expression of HOXB2 protein was significantly upregulated in tumor tissues, as shown in Fig. 1C and D.
Figure 1.
Expression of miR-139-5p and HOXB2 in lung cancer tissues. The relative expression levels of miR-139-5p and HOXB2 in tissues were determined by reverse transcription-quantitative PCR and western blotting. Compared with the control tissues, miR-139-5p was significantly downregulated and HOXB2 was significantly upregulated in lung cancer tissues. Relative mRNA expression of (A) miR-139-5p and (B) HOXB2 in tumor and paracarcinoma tissues. (C) Relative protein expression of HOXB2 in tumor and paracarcinoma tissues. (D) Semi-quantitative analysis of western blotting. **P<0.01, tumor vs. control group. miR, microRNA; HOXB2, Homeobox protein Hox-B2.
miR-139-5p expression in cells is upregulated following transfection with mimics
As shown in Fig. 2, miR-139-5p was upregulated in the mimics group compared with the Control and NC groups (P<0.01) after transfection with miR-139-5p mimics.
Figure 2.
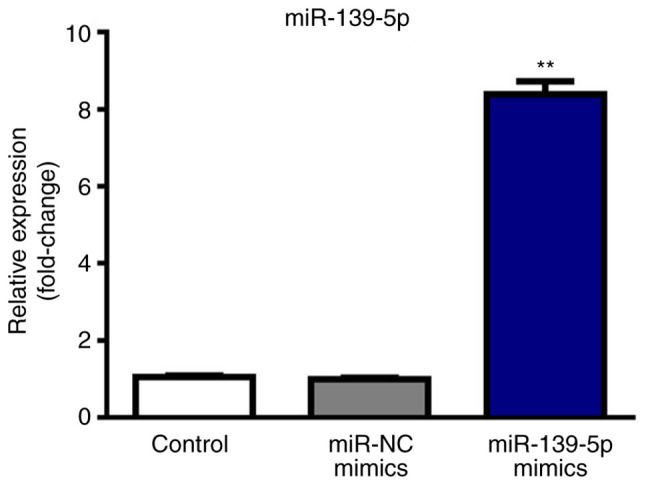
Expression of miR-139-5p after transfection with miR-139-5p mimics. The relative expression of miR-139-5p in tissues was determined by reverse transcription-quantitative PCR after transfection with mimics followed by incubation at 37°C with 5% CO2. Compared with the miR-NC mimics group, transfection with miR-139-5p mimics induced a significant increase in the expression of miR-139-5p. The three groups used were the Control group (non-treated group), the NC group (cells transfected with control plasmids) and the mimics group (cells transfected with miR-139-5p mimics). **P<0.01, miR-139-5p mimics group vs. miR-NC mimics group. miR, microRNA; NC, negative control.
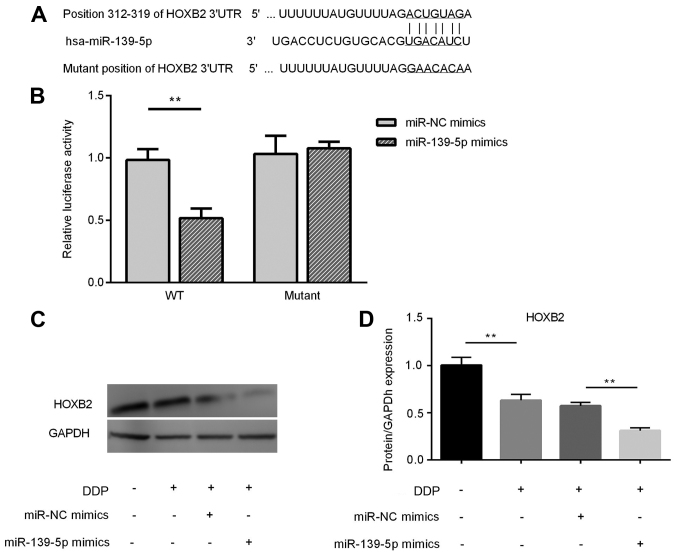
HOXB2 is a direct target of miR-139-5p and its expression is downregulated in DDP-treated A549 cells following miR-139-5p overexpression
Based on putative target sequences at nt positions 312–319 of the HOXB2 3′-UTR (Fig. 3A), HOXB2 was selected as a potential target of miR-139-5p. A luciferase reporter assay was used to confirm HOXB2 as a direct target of miR-139-5p. As shown in Fig. 3B, miR-139-5p led to a significant decrease in the luciferase activity of the wild-type HOXB2 3′-UTR, but not that of the mutant 3′-UTR, in A549 cells. Compared with the Control group, the expression of HOXB2 was significantly decreased in the cisplatin group (P<0.01). The protein expression of HOXB2 in DDP-treated A549 cells transfected with miR-139-5p mimics was significantly downregulated compared with the DDP + miR-NC group (P<0.01) (Fig. 3C and D).
Figure 3.
Target verification via a luciferase reporter assay and HOXB2 protein expression in A549 cells. HOXB2 was found to be a direct target of miR-139-5p and the expression of HOXB2 was downregulated significantly in DDP-treated A549 cells after transfection with miR-139-5p mimics. (A) TargetScan was used for target prediction. (B) Relative luciferase activity after transfection. (C) Relative protein expression of HOXB2 in A549 cells and (D) semi-quantitative analysis of western blotting. **P<0.01. miR, microRNA; HOXB2, Homeobox protein Hox-B2; NC, negative control; WT, wild-type; UTR, untranslated region; DDP, cisplatin.
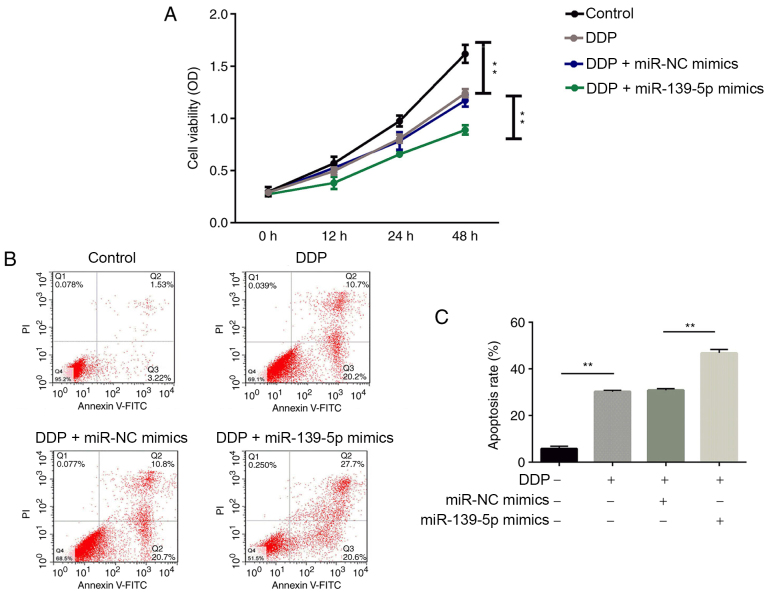
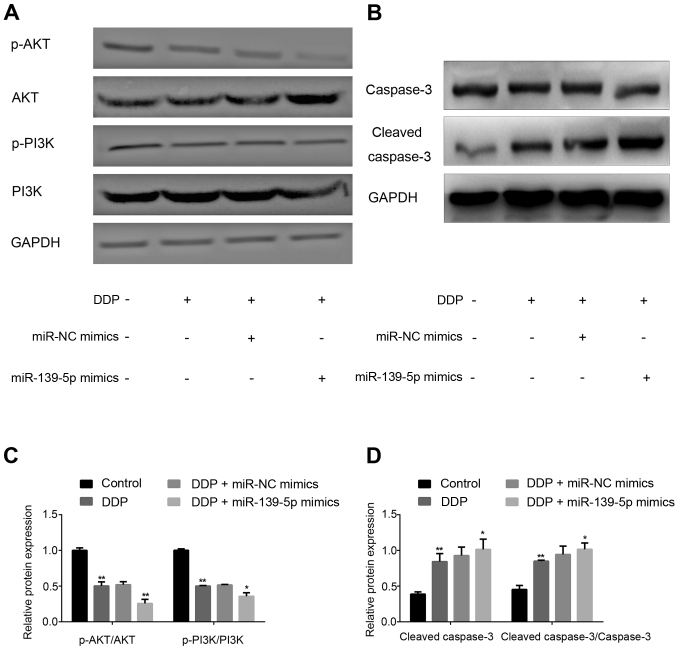
Transfection with miR-139-5p mimics in DDP-treated A549 cells inhibits NSCLC cell proliferation and promotes apoptosis. As shown in Fig. 4A, cell proliferation was significantly suppressed by DDP treatment compared with the Control group. Furthermore, cell proliferation in the DDP + miR-139-5p group was significantly decreased compared with the DDP + miR-NC mimics group (P<0.01).
Figure 4.
Effects of miR-139-5p on cell proliferation and apoptosis in DDP-treated cells. Non-small cell lung cancer cell proliferation was inhibited and apoptosis was increased significantly in DDP-treated A549 cells after transfection with miR-139-5p mimics. (A) Cell proliferation was measured by a Cell Counting Kit-8 assay. (B) Cell apoptosis was measured by flow cytometry. (C) The rate of apoptosis. **P<0.01. miR, microRNA; NC, negative control; DDP, cisplatin.
The results of the flow cytometry analysis are shown in Fig. 4B and C. Significantly increased rates of cell apoptosis were observed in the DDP group compared with the Control group. The apoptosis rate of A549 cells treated with miR-139-5p mimics combined with DDP was significantly elevated compared with the DDP + miR-NC mimics group (P<0.01).
p-AKT and p-PI3K expression is downregulated, whereas caspase-3 expression is upregulated, in DDP-treated A549 cells following miR-139-5p overexpression
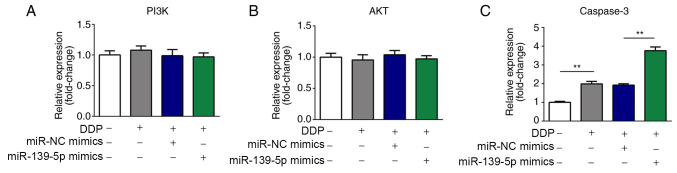
The relative expression levels of PI3K, AKT and caspase-3 in cells were determined by RT-qPCR. As shown in Fig. 5A-C, the mRNA expression levels of AKT and PI3K were not altered, whereas caspase-3 mRNA expression was significantly upregulated in DDP-treated A549 cells following miR-139-5p overexpression.
Figure 5.
Effects of miR-139-5p on the expression of AKT, PI3K and caspase-3 mRNA in DDP-treated cells. The relative expression levels of PI3K, AKT and caspase-3 in cells were determined by reverse transcription-quantitative PCR. mRNA expression of (A) PI3K and (B) AKT were not significantly altered, whereas (C) caspase-3 was upregulated significantly in DDP-treated A549 cells after transfection with miR-139-5p mimics. **P<0.01. miR, microRNA; NC, negative control; DDP, cisplatin.
As shown in Fig. 6A-D, the expression levels of p-AKT and p-PI3K were significantly decreased in the DDP group compared with the Control group (P<0.01). In addition, cells treated with DDP + miR-139-5p mimics revealed a significant decrease compared with the DDP + miR-NC mimics group (P<0.01). Furthermore, increased expression of cleaved-caspase-3 protein was also found. Therefore, miR-139-5p may be able to inhibit the expression of p-AKT and p-PI3K, while increasing cleaved-caspase-3 expression to enhance DDP sensitivity.
Figure 6.
Effects of miR-139-5p on the expression of p-AKT, p-PI3K and cleaved-caspase-3 protein in DDP-treated cells. Protein expression of p-AKT, p-PI3K were downregulated and cleaved-caspase-3 expression was upregulated significantly in DDP-treated A549 cells after transfection with miR-139-5p mimics. (A) The relative expression levels of p-AKT and p-PI3K in cells were determined by western blotting. (B) The relative expression levels of caspase-3 and cleaved-caspase-3 in cells were determined by western blotting. (C) Semi-quantification of p-AKT and p-PI3K expression. (D) Semi-quantification of caspase-3 and cleaved-caspase-3 expression. *P<0.05, **P<0.01, DDP + miR-139-5p mimics group vs. DDP + miR-NC mimics group, DDP group vs. control group. miR, microRNA; NC, negative control; p-, phosphorylated; DDP, cisplatin.
Discussion
DDP is generally considered to be the most suitable chemotherapeutic drug for the treatment of NSCLC (41). However, resistance to DDP is becoming increasingly problematic, and this is turning into a major impediment in the clinical treatment of patients with lung cancer (42). Alterations in drug uptake, drug metabolism and drug targets are the molecular mechanisms by which a cancer cell acquires resistance to a specific treatment (43).
miRNAs serve key roles in numerous biological processes, and are frequently dysregulated in tumor cells. An increasing number of miRNAs have been demonstrated to be involved in the drug-sensitivity of cancer cells in different tumor types via mechanisms such as inhibition of cell proliferation, apoptosis induction and cell cycle arrest (44–47). miR-139-5p, a potential biomarker, is often downregulated in numerous types of cancer (48–50). miR-139-5p suppresses cancer cell proliferation, migration, invasion and metastasis, suggesting that it may be a potential novel target for cancer treatment (51,52). At the same time, miR-139-5p has been reported to exert positive effects on drug resistance and radiotherapy resistance during the process of cancer treatment (53,54). The aim of the present study was to assess the effects of miR-139-5p on DDP-treated NSCLC cells, and to uncover the underlying mechanism. Tumor cell proliferation and dysregulation of apoptosis are important processes in tumorigenesis and development.
HOX genes have been implicated in numerous studies investigating prognosis and tumorigenesis in patients with cancer (55–57). The expression of Hox transcript antisense intergenic RNA was demonstrated to be higher in a DDP-resistant ovarian carcinoma cell line compared with the associated DDP-sensitive cell line (56). A previous study revealed that HOX gene homeobox protein BarH-like 2 could modulate DDP sensitivity in human epithelial ovarian cancer (57). As a member of the HOX gene family, HOXB2 was revealed to be overexpressed during cervical cancer progression (58). miR-139-5p was reported to target the HOXA10 transcript and to suppress endometrial cancer cell growth and migration during human endometrial cancer pathogenesis (59). In the present study, miR-139-5p was identified to have relevant binding sites in HOXB2, and HOXB2 was significantly upregulated in NSCLC tissues compared with paracarcinoma tissues. Compared with the Control group, the protein expression level of HOXB2 in A549 cells treated with DDP was significantly downregulated. In addition, the protein expression of HOXB2 in DDP-treated A549 cells transfected with miR-139-5p mimics was significantly decreased compared with the DDP + miR-NC mimics treatment group. These results indicated that miR-139-5p increased cell sensitivity to DDP via downregulation of HOXB2.
The PI3K/AKT signaling pathway, a crucial and extensively investigated intracellular signaling pathway, is instrumental in regulating cellular apoptosis, stimulating cell growth and increasing cell proliferation (60). caspase-3 is a member of the group of proteolytic enzymes that mediate apoptosis, and is an important effector molecule for apoptosis. miR-139-5p was demonstrated to increase cell proliferation by activating the PI3K/AKT signaling pathway in supratentorial pediatric low-grade gliomas (61). Inhibition of the PI3K/AKT/mTOR signaling pathway enhanced the sensitivity of the SKOV3/DDP ovarian cancer cell line to DDP in vitro (62). miRNAs were revealed to regulate DDP-induced gastric cancer cell death by modulating the PI3K/AKT/survivin pathway (63). The tyrosine kinase inhibitor, dasatinib, enhanced DDP sensitivity in human esophageal squamous cell carcinoma cells via suppression of the PI3K/AKT and STAT3 pathways (64). In the present study, the mRNA expression levels of PI3K and AKT in A549 cells were not affected by DDP treatment, whereas the mRNA expression of caspase-3 was increased upon treatment with DDP and DPP + miR-139-5p mimics. Overexpression of miR-139-5p further enhanced the DDP-induced decrease in the protein expression of p-AKT and p-PI3K, and further upregulated the DDP-induced increase in the expression of cleaved-caspase-3 protein. Therefore, miR-139-5p overexpression may lead to increased DDP sensitivity by modulating the PI3K/AKT/caspase-3 signaling pathway.
The present study aimed to investigate the effect of miR-139-5p expression on the modulation of DDP sensitivity in lung cancer, not the effect of DDP on the regulation of miR-139-5p expression. Hence, the expression of miR-139-5p after cisplatin treatment was not investigated. Moreover, the current study was limited as it only investigated the role of miR-139-5p in DDP sensitivity. Further studies will be conducted in the future to investigate whether miR-139-5p has a role in the sensitivity of cancer cells to other chemotherapy drugs.
In conclusion, the present study revealed that miR-139-5p could increase the efficacy of DDP on cell proliferation and apoptosis in vitro via modulation of the PI3K/AKT/caspase-3 pathway. The present study also demonstrated that miR-139-5p expression was significantly lower in NSCLC tissues compared with that in paracarcinoma tissues. Furthermore, HOXB2 was identified as a target of miR-139-5p, and miR-139-5p may function as a DDP sensitizer by targeting HOXB2 in NSCLC. The regulation of miR-139-5p expression could therefore provide a novel approach to reverse DDP resistance and increase chemosensitivity in the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HD, YB and CL collected, analyzed and interpreted the patient data. HD, AZ and YN contributed to the manuscript conception, experiment design, manuscript drafting and revising. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Weifang Yidu Central Hospital (approval no. 201601580), and written informed consent was obtained from each participant prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zha L, Sobue T, Kitamura T, Kitamura Y, Sawada N, Iwasaki M, Sasazuki S, Yamaji T, Shimazu T, Tsugane S. Changes in smoking Status and mortality from all causes and lung cancer: A longitudinal analysis of a population-based study in Japan. J Epidemiol. 2019;29:11–17. doi: 10.2188/jea.JE20170112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbimbo M, Vitali M, Fabbri A, Ottaviano M, Pasello G, Petrini I, Palmieri G, Berardi R, Zucali P, Ganzinelli M, et al. Relevent trial: Phase II trial of ramucirumab, carboplatin, and paclitaxel in previously untreated thymic carcinoma/B3 thymoma with area of carcinoma. Clin Lung Cancer. 2018;19:e811–e814. doi: 10.1016/j.cllc.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Yang D, Liu Y, Bai C, Wang X, Powell CA. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett. 2020;468:82–87. doi: 10.1016/j.canlet.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10:3–7. doi: 10.1111/1759-7714.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Neal RD, Sun F, Emery JD, Callister ME. Lung cancer. BMJ. 2019;365:l1725. doi: 10.1136/bmj.l1725. [DOI] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 8.Lee YG, Lee JH, Kim SH, Kim YJ, Lee H, Ahn S, Jang JS, Lee JS, Kim JH. Comparative analysis between combination and single-agent chemotherapy for elderly patients with advanced non-small cell lung cancer: A nationwide population-based outcome study. Lung Cancer. 2018;122:88–93. doi: 10.1016/j.lungcan.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Lee KH, Cho EK, Kim DW, Kim SW, Kim JH, Cho BC, Kang JH, Han JY, Min YJ, et al. Nivolumab in advanced non-small-cell lung cancer patients who failed prior platinum-based chemotherapy. Lung Cancer. 2018;122:234–242. doi: 10.1016/j.lungcan.2018.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Zhang L, Chen J. Effectiveness of treatment with endostatin in combination with emcitabine, carboplatin, and gemcitabine in patients with advanced non-small cell lung cancer: A retrospective study. Open Med (Wars) 2018;13:142–147. doi: 10.1515/med-2018-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino A, Miyazaki A, Tomoike A, Kimura H, Arimitsu K, Hirata M, Ohmomo Y, Nishii R, Okazawa H, Kiyono Y, et al. PET probe detecting non-small cell lung cancer susceptible to epidermal growth factor receptor tyrosine kinase inhibitor therapy. Bioorg Med Chem. 2018;26:1609–1613. doi: 10.1016/j.bmc.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 12.To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol. 2018;24:2949–2973. doi: 10.3748/wjg.v24.i27.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migliore C, Giordano S. Resistance to targeted therapies: A role for microRNAs? Trends Mol Med. 2013;19:633–642. doi: 10.1016/j.molmed.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Cao L, Wan Q, Li F, Tang CE. MiR-363 inhibits cisplatin chemoresistance of epithelial ovarian cancer by regulating snail-induced epithelial-mesenchymal transition. BMB Rep. 2018;51:456–461. doi: 10.5483/BMBRep.2018.51.9.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Zhao L, Zhang J, Meng Z, Zhou C, Wang G, Liu Y, Li M, Xi J, Niu W, et al. Chemotherapy-induced miR-141/MAP4K4 signaling suppresses progression of colorectal cancer. Biosci Rep. 2018;38:BSR20180978. doi: 10.1042/BSR20180978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Li B, Li Q, Wei S, He Z, Huang X, Wang L, Xia Y, Xu Z, Li Z, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9:854. doi: 10.1038/s41419-018-0928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aggarwal P, Challa KR, Rath M, Sunkara P, Nath U. Generation of inducible transgenic lines of arabidopsis transcription factors regulated by microRNAs. Methods Mol Biol. 2018;1830:61–79. doi: 10.1007/978-1-4939-8657-6_4. [DOI] [PubMed] [Google Scholar]
- 18.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Schneider C, Setty M, Holmes AB, Maute RL, Leslie CS, Mussolin L, Rosolen A, Dalla-Favera R, Basso K. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc Natl Acad Sci USA. 2014;111:8185–8190. doi: 10.1073/pnas.1322466111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukhari SI, Vasquez-Rifo A, Gagné D, Paquet ER, Zetka M, Robert C, Masson JY, Simard MJ. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res. 2012;22:1034–1045. doi: 10.1038/cr.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Zhang C, Li F, Zhang B, Zhang Y. Regulation of memory CD8+ T cell differentiation by microRNAs. Cell Physiol Biochem. 2018;47:2187–2198. doi: 10.1159/000491532. [DOI] [PubMed] [Google Scholar]
- 22.Fu X, He Y, Wang X, Peng D, Chen X, Li X, Wan Q. MicroRNA-16 promotes ovarian granulosa cell proliferation and suppresses apoptosis through targeting PDCD4 in polycystic ovarian syndrome. Cell Physiol Biochem. 2018;48:670–682. doi: 10.1159/000491894. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Zhou Q, Liang Y, Pan W, Bei Y, Zhang Y, Wang J, Jiao Z. miR-486 inhibits PM2.5-induced apoptosis and oxidative stress in human lung alveolar epithelial A549 cells. Ann Transl Med. 2018;6:209–218. doi: 10.21037/atm.2018.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Feng C, Shi S. miR-196b promotes lung cancer cell migration and invasion through the targeting of GATA6. Oncol Lett. 2018;16:247–252. doi: 10.3892/ol.2018.8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maemura K, Watanabe K, Ando T, Hiyama N, Sakatani T, Amano Y, Kage H, Nakajima J, Yatomi Y, Nagase T, et al. Altered editing level of microRNAs is a potential biomarker in lung adenocarcinoma. Cancer Sci. 2018;109:3326–3335. doi: 10.1111/cas.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulivi P, Zoli W. miRNAs as non-invasive biomarkers for lung cancer diagnosis. Molecules. 2014;19:8220–8237. doi: 10.3390/molecules19068220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yong-Hao Y, Xian-Guo W, Ming X, Jin-Ping Z. Expression and clinical significance of miR-139-5p in non-small cell lung cancer. J Int Med Res. 2019;47:867–874. doi: 10.1177/0300060518815379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C, Sang M, Li S, Sun X, Yang C, Xi Y, Wang L, Zhang F, Bi Y, Fu Y, et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6:39756–39792. doi: 10.18632/oncotarget.5476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. 2017;21:3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, Wang Z, Liu J, Cai G. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep. 2016;6:27157. doi: 10.1038/srep27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W, Hang M, Yuan CY, Wu FL, Chen SB, Xue K. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:3864–3870. [PMC free article] [PubMed] [Google Scholar]
- 32.Clemenceau A, Boucherat O, Landry-Truchon K, Lamontagne M, Biarde S, Joubert P, Gobeil S, Secco B, Laplante M, Morissette M, et al. Lung cancer susceptibility genetic variants modulate HOXB2 expression in the lung. Int J Dev Biol. 2018;62:857–864. doi: 10.1387/ijdb.180210yb. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Li S, Cheng X, Du P, Yang Y, Jiang WG. HOXB2 is a putative tumour promoter in human bladder cancer. Anticancer Res. 2019;39:6915–6921. doi: 10.21873/anticanres.13912. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. 2019;18:1281–1291. doi: 10.1080/15384101.2019.1618127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindblad O, Chougule RA, Moharram SA, Kabir NN, Sun J, Kazi JU, Rönnstrand L. The role of HOXB2 and HOXB3 in acute myeloid leukemia. Biochem Biophys Res Commun. 2015;467:742–747. doi: 10.1016/j.bbrc.2015.10.071. [DOI] [PubMed] [Google Scholar]
- 36.Inamura K, Togashi Y, Ninomiya H, Shimoji T, Noda T, Ishikawa Y. HOXB2, an adverse prognostic indicator for stage I lung adenocarcinomas, promotes invasion by transcriptional regulation of metastasis-related genes in HOP-62 non-small cell lung cancer cells. Anticancer Res. 2008;28:2121–2127. [PubMed] [Google Scholar]
- 37.Inamura K, Togashi Y, Okui M, Ninomiya H, Hiramatsu M, Satoh Y, Okumura S, Nakagawa K, Shimoji T, Noda T, et al. HOXB2 as a novel prognostic indicator for stage I lung adenocarcinomas. J Thorac Oncol. 2007;2:802–807. doi: 10.1097/JTO.0b013e3181461987. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H, Liu Y, Liang P, Wang B, Tan H, Zhang Y, Gao X, Gao J. TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis. Cell Biosci. 2018;8:23. doi: 10.1186/s13578-018-0221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo J, Jin D, Wu Y, Yang L, Du J, Gong K, Chen W, Dai J, Miao S, Xi S. The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells. EBioMedicine. 2018;35:204–221. doi: 10.1016/j.ebiom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L, Fang X. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One. 2014;9:e108133. doi: 10.1371/journal.pone.0108133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donzelli S, Mori F, Biagioni F, Bellissimo T, Pulito C, Muti P, Strano S, Blandino G. MicroRNAs: Short non-coding players in cancer chemoresistance. Mol Cell Ther. 2014;2:16. doi: 10.1186/2052-8426-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, Zhang J, Peng C, Lin Y, Chen J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ju J. Implications of miRNAs in colorectal cancer chemoresistance. Int Drug Discov. 2011;2011:2011. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang XH, Lu H, Li TS, Yu Y, Liu G, Peng X, Zhao JH. KLF8 induces breast stemness and chemoresistance via miRNAs associated with EMT. Mol Cancer Ther. 2013;12:A100–A100. [Google Scholar]
- 47.Carta A, Chetcuti R, Ayers D. An introspective update on the influence of miRNAs in breast carcinoma and neuroblastoma chemoresistance. Genet Res Int. 2014;2014:743050. doi: 10.1155/2014/743050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D, Le V, Yin P, Ni L, Liu J. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. FEBS J. 2014;281:3609–3624. doi: 10.1111/febs.12880. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Cao XY, Li YN, Qiu YY, Li YN, Li W, Wang H. Reversal of cisplatin resistance by microRNA-139-5p-independent RNF2 downregulation and MAPK inhibition in ovarian cancer. Am J Physiol Cell Physiol. 2018;315:C225–C235. doi: 10.1152/ajpcell.00283.2017. [DOI] [PubMed] [Google Scholar]
- 50.Ni H, Dai X, Leng X, Deng M, Qin Y, Ji Q, Xu C, Li J, Liu Y. Higher variety and quantity of microRNA-139-5p isoforms confer suppressive role in hepatocellular carcinoma. J Cell Biochem. 2018;119:6806–6813. doi: 10.1002/jcb.26874. [DOI] [PubMed] [Google Scholar]
- 51.Yue S, Wang L, Zhang H, Min Y, Lou Y, Sun H, Jiang Y, Zhang W, Liang A, Guo Y, et al. miR-139-5p suppresses cancer cell migration and invasion through targeting ZEB1 and ZEB2 in GBM. Tumour Biol. 2015;36:6741–6749. doi: 10.1007/s13277-015-3372-8. [DOI] [PubMed] [Google Scholar]
- 52.Hua S, Lei L, Deng L, Weng X, Liu C, Qi X, Wang S, Zhang D, Zou X, Cao C, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene. 2018;37:1624–1636. doi: 10.1038/s41388-017-0057-3. [DOI] [PubMed] [Google Scholar]
- 53.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212:643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 54.Pajic M, Froio D, Daly S, Doculara L, Millar E, Graham PH, Drury A, Steinmann A, de Bock CE, Boulghourjian A, et al. miR-139-5p modulates radiotherapy resistance in breast cancer by repressing multiple gene networks of DNA repair and ROS defense. Cancer Res. 2018;78:501–515. doi: 10.1158/0008-5472.CAN-16-3105. [DOI] [PubMed] [Google Scholar]
- 55.Boimel PJ, Cruz C, Segall JE. A functional in vivo screen for regulators of tumor progression identifies HOXB2 as a regulator of tumor growth in breast cancer. Genomics. 2011;98:164–172. doi: 10.1016/j.ygeno.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Wang H, Song T, Zou Y, Jiang J, Fang L, Li P. HOTAIR is a potential target for the treatment of cisplatin resistant ovarian cancer. Mol Med Rep. 2015;12:2211–2216. doi: 10.3892/mmr.2015.3562. [DOI] [PubMed] [Google Scholar]
- 57.Sellar GC, Watt KP, Li L, Nelkin BD, Rabiasz GJ, Porteous DJ, Smyth JF, Gabra H. The homeobox gene BARX2 can modulate cisplatin sensitivity in human epithelial ovarian cancer. Int J Oncol. 2002;21:929–933. [PubMed] [Google Scholar]
- 58.Gonzalez-Herrera A, Salgado-Bernabe M, Velazquez-Velazquez C, Salcedo-Vargas M, Andrade-Manzano A, Avila-Moreno F, Pina-Sanchez P. Increased expression of HOXB2 and HOXB13 proteins is associated with HPV infection and cervical cancer progression. Asian Pac J Cancer Prev. 2015;16:1349–1353. doi: 10.7314/APJCP.2015.16.4.1349. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Li C, Jiang Y, Wan Y, Zhou S, Cheng W. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int. 2018;18:51. doi: 10.1186/s12935-018-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015;1855:104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 61.Catanzaro G, Besharat ZM, Miele E, Chiacchiarini M, Po A, Carai A, Marras CE, Antonelli M, Badiali M, Raso A, et al. The miR-139-5p regulates proliferation of supratentorial paediatric low-grade gliomas by targeting the PI3K/AKT/mTORC1 signalling. Neuropathol Appl Neurobiol. 2018;44:687–706. doi: 10.1111/nan.12479. [DOI] [PubMed] [Google Scholar]
- 62.Cai Y, Tan X, Liu J, Shen Y, Wu D, Ren M, Huang P, Yu D. Inhibition of PI3K/Akt/mTOR signaling pathway enhances the sensitivity of the SKOV3/DDP ovarian cancer cell line to cisplatin in vitro. Chin J Cancer Res. 2014;26:564–572. doi: 10.3978/j.issn.1000-9604.2014.08.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, Jin Y, Wu Y. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35:1287–1295. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Lan T, Zhang W, Dong L, Kang N, Fu M, Liu B, Liu K, Zhang C, Hou J, et al. Dasatinib enhances cisplatin sensitivity in human esophageal squamous cell carcinoma (ESCC) cells via suppression of PI3K/AKT and Stat3 pathways. Arch Biochem Biophys. 2015;575:38–45. doi: 10.1016/j.abb.2014.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.