Abstract
The List of Prokaryotic names with Standing in Nomenclature (LPSN) was acquired in November 2019 by the DSMZ and was relaunched using an entirely new production system in February 2020. This article describes in detail the structure of the new site, navigation, page layout, search facilities and new features.
Keywords: DSMZ, database, List of Prokaryotic names with Standing in Nomenclature (LPSN), prokaryotic nomenclature
The List of Prokaryotic names with Standing in Nomenclature (LPSN) was established in 1997 by Professor Jean Euzéby as the List of Bacterial names with Standing in Nomenclature (LBSN). Launched on 28 March 1997 as anonymous ftp files and on the World Wide Web on 28 January 1998 [1], ‘Euzéby’s List’ rapidly became a key online resource for anyone interested in bacterial and archaeal nomenclature and classification; it changed ownership to Aidan C. Parte in July 2013 upon Euzéby’s retirement [2].
The LPSN has remained an invaluable resource for keeping abreast of the rapid changes in prokaryotic nomenclature and ever-increasing descriptions of novel taxa. The number of new prokaryotic names and new combinations validly published according to the International Code of Nomenclature of Prokaryotes (ICNP) [3] has exploded over the last three decades, and is set to continue increasing, irrespective of future modifications to the Code.
With the rise in actual and anticipated new prokaryotic names, the LPSN had become increasingly difficult to maintain, so a better technical and funding basis was sought. To that end, in November 2019, the LPSN was acquired by Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures GmbH, Germany. The DSMZ already had its Prokaryotic Nomenclature Up-to-date (PNU) service since 1993, and because there is significant overlap in the content of both sites, it was decided to merge the two services into a completely new one, still under the LPSN name, which has a higher profile in the community. On 17 February 2020, the new LPSN was launched at https://lpsn.dsmz.de/ under a modified Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license.
Technically, the new site is completely different. It uses a dedicated database and scripting to serve up pages on request as opposed to the old LPSN, which consisted of several thousand static HTML pages. The old site has been migrated to the archive in its entirety, where it will remain indefinitely. The new site maps between addresses to ensure that all addresses of the old site have a counterpart on the new site and are thereby redirected.
Responsive design
The new site has been designed to be ‘responsive’, so that it is fully functional on any screen size from smartphones up to large desktop monitors. Text flows and menus dynamically respond to screen orientation and window size, ensuring high usability and accessibility for all users. Additionally, users can manually display or hide selected content on most pages. The description of the site in this article is based on the desktop view of the LPSN.
Formatting of names in the LPSN
Correct names are given in bold/italic font in the taxon browser, in the search results and in other lists of taxa, such as child or sibling lists (e.g. Cavicella subterranea in Fig. 1); names not considered to be correct are given in italics only (e.g. Neisseria caviae in Fig. 1); names not considered to be validly published are given in italics and between quotation marks (e.g. ‘ Chlamydia caviae ’ in Fig. 1). Square brackets are used to indicate that the nomenclatural status is unreliable or uncertain, as would be the case for an artificial parent node of an incertae sedis taxon or a Candidatus species that lacks a genus description (this can occur when a paper proposes a Candidatus species and family, but not the genus that links them in the classification); the square brackets are also used where a preliminary entry in the LPSN lacks crucial information, for instance when a genus name for which we have full information can be assigned in the literature to a family for which we only have the name but not the authors, publication, etc. If the curators waited for the complete information for the family then the genus could not be found in the LPSN hierarchy.
Fig. 1.

Results from the Search taxonomy function using the search term ‘cavi’.
What is a ‘correct name’? Among a set of names in accordance with the rules of the ICNP [3], the LPSN curators select one taxon name as the correct name. This is based upon valid publication, legitimacy and priority of publication (Principle 6). Only correct names are to be used (Rule 23a Note 5). The situations in which the taxonomist has a choice between several names that could be regarded as the correct name are explained in an article freely available on the LPSN [4]. Whenever several options are available that are in accordance with the rules of the ICNP, the choice of a correct name in the LPSN reflects one of the taxonomic opinions expressed in the literature; this process is thus not fully automated as there are exceptions. Crucially, the ICNP does not specify that the most recent validly published name is to be regarded as the correct name. Other researchers may well express distinct taxonomic opinions and the Code permits them to do so. The LPSN indicates alternative taxonomic arrangements, if any, in addition to the preferred one.
Terminology of relationships between taxa
The terms ‘parent taxa’, ‘child taxa’ and ‘siblings’, or variants thereof, are used in the LPSN to define relationships between taxa immediately above, below or level in the taxonomic categories. So, the term ‘child taxon’ means that some taxon belongs to the category below a given taxon in terms of a hierarchical classification from domain down to subspecies. So, for a family, the child taxa would be its constituent genera; for a genus, the child taxa would be its constituent species, etc. Conversely, the ‘parent taxon’ of a family would be an order, the parent taxon of an order would by a class, etc. The ‘siblings’ of a species would be other species in the same genus, the siblings of a genus would be other genera in the same family, etc. The navigation menu at the top right of each taxon entry has ‘parent – siblings – children’ links, greyed-out as appropriate.
Navigation
Finding information about taxa is done through three channels – the Search taxonomy function, the Advanced search function or the Browse by menu. The Search taxonomy field is available at the top of the homepage and every other page in the LPSN – this search will find any phrase of four or more letters used in any taxon name. In the example given here, searching for ‘cavi’ returns 25 results, almost all of which are derived from the genus name of the guinea pig, Cavia porcellus. It also finds the genus name Micavibrio , where ‘cavi’ is split across two of the constituent words that form the genus name. Results are arranged by taxonomic category and sorted by the position of the search string within the taxon name, which is followed by an alphabetic sort.
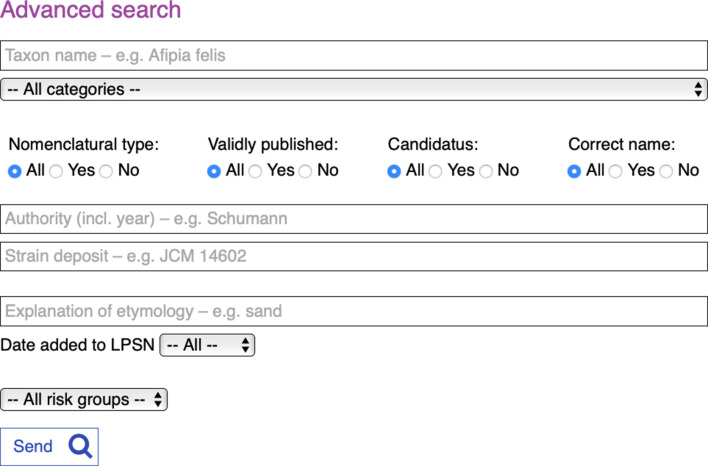
The Advanced search function (Fig. 2) enables searching of certain indexed fields in the database. In addition to the straight taxon name search described above, this search can be narrowed down using the All categories pull-down menu, which enables the user to select results by taxonomic category, for example class, family or species names. Further refinement of the results can be done using the radio buttons for Nomenclatural type, Validly published, Candidatus or Correct name. One of the benefits to the curators of such a search facility is that separate pages listing Candidatus names or other not validly published names need no longer be maintained; the benefit to users is that these names can now be found dynamically and will always be up to date.
Fig. 2.
New LPSN Advanced search page showing the default radio button settings.
The Authority field searches the defining publication entries, i.e. name and/or year. The Strain deposit field enables searching by culture collection abbreviation and/or deposit number, and the Explanation of etymology field allows searching by words or partial words that are translated into Greek or Latin for the taxon names. Results can also be filtered by Risk group classification (see Fig. 2). The curators hope that the etymology search will help authors of papers describing new taxa in the formation of new names. Additional search facilities will be added in the future depending on the growth of the database and specific user demands.
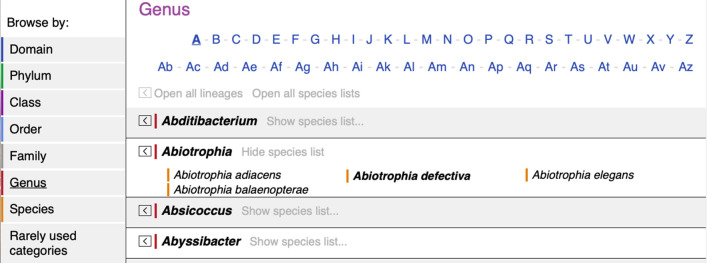
The Browse by menu (on the left of the page or in the drop-down menu, depending on responsive view) enables browsing of taxon names by category, from domain down to species. Under each category, the names are listed alphabetically and shortcuts are provided to initial letters to ease navigation of long lists of names. By default, the parental lineage and child taxa in each category are hidden in order to ease navigation and reduce information overload while browsing. The lineages and child taxon lists can be opened or closed en masse or individually as required. When browsing genera, clicking Open all lineages will expand to display from the family up to the domain, and clicking Open all species lists will display all species for all genera beginning with the letter A. Fig. 3 is an example showing where individual lineage and child lists are expanded for the genus Abiotrophia . Specific formatting is used to denote correct, non-correct and non-validly published names. Under Rarely used categories, taxonomic categories that are not in general use are listed. These categories include infrakingdom, tribe, subtribe, subphylum, superphylum, suborder and subgenus.
Fig. 3.
New Browse by function in the LPSN. Lineages and child lists can be expanded or closed en masse or individually by clicking on the appropriate arrow or text. Note the formatting used to denote correct, non-correct and non-validly published names.
Sample entry in the LPSN
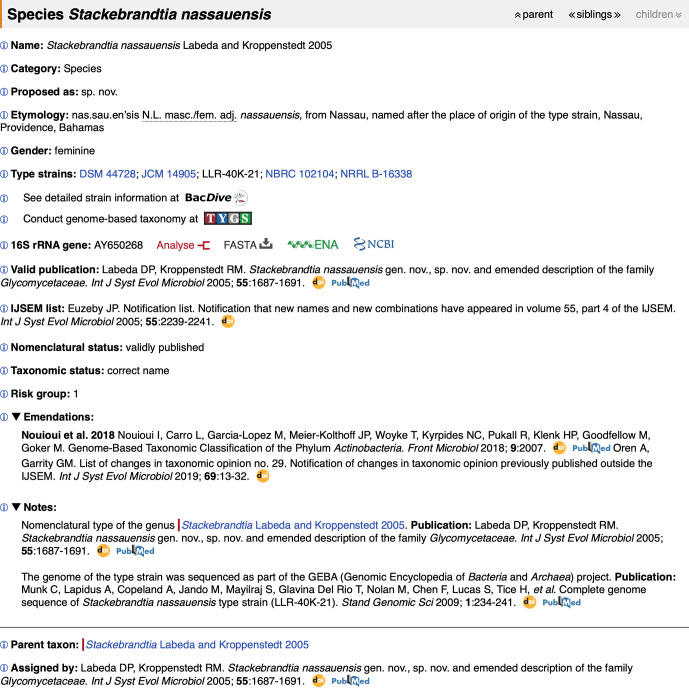
Fig. 4 shows the species entry in the LPSN for Stackebrandtia nassauensis . On the upper-right-hand-side of the page are the navigation buttons, for moving to the parent taxon (in this case the genus Stackebrandtia ), listing siblings (currently four) and children (greyed out in this case because there are no subspecies).
Fig. 4.
Sample taxon page in the LPSN showing information on the species Stackebrandtia nassauensis .
The first line gives the name and the authority, i.e. the author(s) and year of valid publication (year of original publication for not validly published names); the next lines give the taxonomic category, kind of taxonomic proposal made (in this case a new species, sp. nov.) and etymology of the name whenever possible. The gender of the name is also stated separately for clarity; in the example here, since the ending -ensis is used in either masculine or feminine adjectives to indicate geographical origin, so the Gender line clarifies that it is feminine, thus matching the gender of the genus name.
Type strain accession numbers are given with direct links to culture collection catalogue entries whenever the collections provide URLs that can be inferred from the strain accession number. The LPSN now links to the DSMZ’s BacDive service [5] for the type strains, which provides free access to a wide range of metadata about the strain, including morphology and physiology, images, culture and growth conditions, molecular biology, detailed strain location data, strain history and references. In addition, clicking on the TYGS button will preselect the respective whole-genome sequence in the submission form of the DSMZ’s Type Strain Genome Server (TYGS) [6], where genome-based taxonomic analysis can be performed. This type of analysis also works for entire genera and is relatively fast because precomputed data are used whenever available. TYGS analyses triggered by the LPSN page work even if the number of species in a particular genus exceeds the default TYGS upload cap for user-provided genome sequences.
For the 16S rRNA gene of the type strain, links to the DSMZ single-gene phylogeny server, direct sequence download in fasta format, the European Nucleotide Archive (ENA) sequence database and the NCBI sequence database are given. Triggering phylogenetic analysis and/or standardized sequence similarity calculation [7] as well as fasta download also work for entire general.
For taxon names published outside of the International Journal of Systematic and Evolutionary Microbiology (IJSEM), the effective publication is provided, followed by the reference to the Validation List under the IJSEM list subheading. In the example in Fig. 4, the taxon name was originally published in he IJSEM. The IJSEM list subheading thus shows a Notification List. Throughout the LPSN, whenever possible links to publications are provided using the Digital Object Identifier (DOI) and the PubMed ID.
The Nomenclatural status section states whether or not a name is validly published, or describes a special status, for example whether the name is an artificial parent node of Candidatus species lacking a genus description. The Taxonomic status gives the correctness of the name according to the criteria described above; if the name is regarded as a synonym the correct name is provided.
The Risk group section provides the assignment of the respective species or subspecies to a risk group as provided by Germany's official regulation, TRBA 466 (‘Classification of prokaryotes (bacteria and archaea) into risk groups’).
If the description of the taxon has been emended, the reference is provided to the paper and when available to the List of changes in taxonomic opinion that is published in the IJSEM. The Notes section provides information such as whether the taxon is a nomenclatural type, or other comments regarding errata, corrigenda, orthographical corrections to the name and to the etymology made on a Validation List, alternative classifications, etc.
The parent taxon name and defining publication are provided, and a reference to the article that assigned the taxon to its parent. The reference for the assignment of a species to a genus or a subspecies to a species is trivially identical to the reference that proposed the species or subspecies name because the genus name is part of a species name and the species name is part of a subspecies. This does not hold for the assignment of a genus to a family, of a family to an order, of an order to a class, etc. For this reason, the publication which assigned a certain child taxon above species rank to a certain parent taxon is not obvious from the name of the child taxon. The LPSN thus separately cites the publication in which the assignment of a child taxon to a parent taxon is found.
Updating schedule and data availability
The LPSN will be continuously updated with the addition of new taxon names as and when they become available in the IJSEM or the wider literature. At the time of writing (13 July 2020), 1878 entries have been added since the re-launch on 17 February 2020. The taxonomic status of added taxa will always be included and will be amended as appropriate over time.
The data are available under a modified CC BY-NC 4.0 license. The modifications can be found on the LPSN copyright page.
The Leibniz Institute DSMZ is a state-owned not-for-profit organisation and has one of the largest collections of micro-organisms and cell cultures worldwide. With the availability of 80 % of all bacterial species with a validly published name, the DSMZ hosts the majority of bacterial diversity. Under the auspices of the DSMZ, the future of the LPSN will remain secure both technically and financially, and will remain available without charge for researchers.
Funding information
This work received no specific grant from any funding agency.
Acknowledgements
A.C.P. would like to thank the DSMZ for securing the LPSN’s future and his co-authors for their tremendous work in developing the new service. The authors would like to thank the reviewers for their constructive comments, which have improved the paper and led to improvements to the LPSN itself.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: DSMZ, Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH/Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH; ICNP, International Code of Nomenclature of Prokaryotes; LPSN, List of Prokaryotic names with Standing in Nomenclature; PNU, Prokaryotic Nomenclature Up-to-date; TYGS, Type Strain Genome Server.
References
- 1.Euzéby JP. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 2.Oren A, Garrity G. Retirement of Professor Jean Paul Euzéby as list editor. Int J Syst Evol Microbiol. 2013;63:2373. doi: 10.1099/ijs.0.052316-0. [DOI] [Google Scholar]
- 3.Parker CT, Tindall BJ, Garrity GM. International Code of Nomenclature of Prokaryotes. Prokaryotic Code (2008 revision) Int J Syst Evol Microbiol. 2019;69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 4.Tindall BJ. Misunderstanding the Bacteriological Code. Int J Syst Bacteriol. 1999;49 Pt 3:1313–1316. doi: 10.1099/00207713-49-3-1313. [DOI] [PubMed] [Google Scholar]
- 5.Reimer LC, Vetcininova A, Carbasse JS, Söhngen C, Gleim D, et al. BacDive in 2019: bacterial phenotypic data for high-throughput biodiversity analysis. Nucleic Acids Res. 2019;47:D631–D636. doi: 10.1093/nar/gky879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]