Abstract
Combination antiretroviral therapy (cART) has transformed HIV-1 infection, once a fatal illness, into a manageable chronic condition. Drug resistance, severe side effects and treatment noncompliance bring challenges to the cART implementation in clinical settings and indicate the need for additional molecular targets. Here we have identified several small-molecule fusion inhibitors, guided by a neutralizing antibody, against an extensively studied vaccine target- the membrane proximal external region (MPER) of HIV-1 envelope (Env) spike. These compounds specifically inhibit the Env-mediated membrane fusion by blocking CD4-induced conformational changes. An NMR structure of one compound complexed with a trimeric MPER construct reveals that the compound partially inserts into a hydrophobic pocket formed exclusively by the MPER residues, thereby stabilizing its prefusion conformation. These results suggest that the MPER is a potential therapeutic target for developing fusion inhibitors and that strategies employing an antibody-guided search for novel therapeutics may be applied to other human diseases.
Introduction
Combination antiretroviral therapy (cART) has transformed HIV-1 infection from a once fatal illness into a manageable chronic condition1-3. The latest cART regimen uses several classes of antiviral therapeutics, including nucleoside/nucleotide reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, protease inhibitors, fusion inhibitors, coreceptor inhibitors and integrase inhibitors4,5. A typical therapy requires a combination of three or more drugs from at least two classes. Drug resistance, severe side effects and difficulties in patient compliance all call for additional drugs and drug targets. The first fusion inhibitor approved by FDA is Enfuvirtide, a 36-residue peptide derived from gp416,7. It has to be stored at low temperature, freshly reconstituted and injected subcutaneously twice a day. Moreover, injection site reactions, rapid emergence of resistant viruses and high cost of production have limited its long-term use8-10. The next-generation gp41 peptide-based fusion inhibitors, such as Sifuvirtide and Albuvirtide, may suffer similar disadvantages11-13. Many patients previously treated with Enfuvirtide have switched to oral CRIs14, thereby reducing the power of one of the potent weapons from the anti-HIV-1 arsenal. Developing small-molecule fusion inhibitors to overcome the limitations of peptide-based drugs is highly desirable.
HIV-1 envelope spike (Env) catalyzes the first critical step of infection - fusion of viral and target cell membranes15. The protein is first synthesized as a precursor, gp160, which trimerizes to (gp160)3 and then a furin-like protease cleaves it into two fragments: the receptor-binding surface subunit gp120 and the fusion-promoting transmembrane subunit gp41. Three copies of each form the mature viral spike (gp120/gp41)3. Gp120 binding to the primary receptor CD4 and a coreceptor (e.g., CCR5 or CXCR4) induces a series of refolding events in gp4116,17. The transmembrane subunit gp41 adopts a prefusion conformation when folded within the precursor gp16018-20. Cleavage between gp120 and gp41 primes the protein, making it metastable with respect to the postfusion conformation. When triggered, the gp41 fusion peptide at its N-terminus inserts into the target cell membrane, leading to formation of an extended conformation of gp41. This conformational state has the fusion peptide in the target cell membrane and the transmembrane segment in the viral membrane, and is referred to as the prehairpin intermediate21. This state is targeted by Enfuvirtide6, as well as by certain broadly neutralizing antibodies (bnAbs), including 2F5, 4E10 and 10E822,23. Subsequent rearrangements involve refolding of gp41 into a hairpin conformation, creating a six-helix bundle known as the postfusion conformation, which brings the two membranes together and leads to membrane fusion. Success of Enfuvirtide and Albuvirtide as effective therapeutics demonstrate that blocking gp41 refolding steps represents an effective antiviral strategy.
The MPER, a hydrophobic region of ~25 residues, adjacent to the viral membrane, is one of the most conserved regions in gp41 and is required for viral infectivity24. It is an extensively studied vaccine target recognized by a number of anti-gp41 bnAbs, including 2F5, 4E10, Z13e1 and 10E825-27. Its role in the mechanism of viral fusion is still unknown. These antibodies appear to block HIV-1 infection by a common mechanism - they bind the prehairpin intermediate state of gp41 with the help of their lipid binding activity22,23. To investigate whether small-molecule compounds can mimic these bnAbs to bind the MPER and block HIV-1 Env-mediated membrane fusion, we have identified several such small-molecule fusion inhibitors using a high throughput screen involving competition with 2F5. These compounds appear to be a promising lead series that can potentially be further optimized. Our studies show that the compounds target a hydrophobic pocket formed by the trimeric MPER and block CD4 binding to the intact, functional Env on the cell surfaces, suggesting that they block HIV-1 infection by preventing conformational changes in Env required for membrane fusion. Thus, the MPER, a long sought-after vaccine target, is also a potential therapeutic site for developing small-molecule fusion inhibitors. In addition, the antibody-guided search for novel therapeutics presented here should be a general strategy that may be applied to other human diseases.
Results
Small-molecule fusion inhibitors targeting the MPER
We previously designed a construct, designated gp41-inter, to capture the prehairpin intermediate conformation of gp41 using the following sequence: (HR2)-linker-[HR1-CCloop-HR2-MPER]-(trimerization foldon tag)22 (Supplementary Fig. 1). When the gp41-inter polypeptide chains trimerize, the N-terminal HR2 segments form a six-helix bundle with the HR1 segments, because the C-terminal HR2 segments, constrained by the foldon tag, will be unable to form a six-helix bundle. This construct can be pictured as the prehairpin intermediate captured by a covalently-linked HR2 peptide, such as Enfuvirtide (Supplementary Fig. 1). The purified gp41-inter protein is a stable and soluble trimer in solution. Extensive biochemical and antigenicity studies have confirmed that it indeed represents the prehairpin intermediate conformation of gp4122,23,28.
To screen small-molecule compounds that bind the MPER and may mimic the neutralizing antibodies to abort membrane fusion, we developed a sensitive, fluorescence polarization (FP) assay to detect binding of 2F5 to gp41-inter (Fig. 1a). Any compound that binds the antibody epitope with high enough affinity would likely disrupt this interaction and compete for binding to gp41-inter. Fluorescein isothiocyanate (FITC)-labeled 2F5 Fab bound with high affinity (Kd=12 nM) to gp41-inter as measured by fluorescence polarization (Fig. 1b). Unlabeled antibody effectively competed with the labeled Fab to diminish the fluorescence signal. The Z’ factor29 was 0.52 for this assay when we used the unlabeled Fab as a positive control and DMSO as a negative control, suggesting it was suitable for high throughput screening (HTS). We completed a screen with 162,106 compounds from libraries at the Harvard Medical School ICCB-Longwood Screening Facility (Supplementary Table 1). All screening was performed in duplicate, using the labeled 2F5 Fab. We eliminated any compounds that fluoresce or scatter light. We averaged duplicate values and selected those with a z-score of 5 or greater. We considered the 146 compounds meeting these criteria as “hits”, giving a hit rate of 0.09%. We further screened hits by eliminating those that fluoresce weakly but interfere with the FP assay, and selecting those that bind to gp41-inter but not to 2F5 using surface plasmon resonance (SPR) and those that inhibit cell-cell fusion mediated by HIV-1 Env but not by SIV Env at a high expression level.
Figure 1. Identification of dequalinium as a small-molecule fusion inhibitor targeting the MPER of HIV-1 Env.
(a) Diagram illustrating that fluoresceinated Fab fragment shown in orange can bind to gp41-inter, a construct, designed to capture the prehairpin intermediate conformation. F, FITC. (b) Binding curve for the association of 2F5 Fab with gp41-inter as measured by fluorescence polarization. The fraction bound, f, is plotted as a function of gp41-inter concentration (in nM), at a constant concentration of 2F5 (50 nM). KD is 12 nM. (c) Dequalinium structure. (d) and (e) SPR analysis of dequalinium binding to gp41-inter and 2F5 Fab, respectively. The immobilized protein was tested with various concentrations (2-20 μM) of dequalinium. Binding kinetics was evaluated using a 1:1 Langmuir binding model. The sensorgrams are shown in black and the fits in green. The experiments have been repeated independently twice with similar results. (f) Dequalinium was analyzed in the β-galactosidase-based cell-cell fusion assay using both HIV-1 92UG037.8 (red) and SIVmac251.30 (high expression level; green) Envs. Cytotoxicity (blue) was tested by a cell viability assay. Relative lum, relative luminescence. The experiments were performed in triplicates and repeated independently at least twice with similar results. The error bars represent the standard deviations calculated by the Excel STDEV function. (g) Inhibition of viral infectivity by dequalinium. BaL.26 and DJ263.8A are HIV-1 tier 1 isolates and X1254-3 is a tier 2 isolate; MuLV is a control. The experiments were performed using duplicate wells and performed at least twice with similar results. The error bars represent standard deviations as calculated using GraphPad Prism.
One hit compound, dequalinium (quinolinium,1,1’-(1,10-decanediyl)-bis(4-amino-2-methyl diiodide) (1); Fig. 1c), is an FDA-approved, antimicrobial drug30. It bound gp41-inter with an affinity of 11 μM, showed no binding to 2F5 (Fig. 1d and 1e), and effectively inhibited cell-cell fusion mediated by HIV-1 Env with an IC50 of 13.8 μM, but not by SIV Env when transfected at a high expression level (Fig. 1f). It showed a minimal level of cytotoxicity up to 50 μM within the assay time period (<3 hrs) by an assay measuring ATP concentration, which correlates with the amount of metabolically active cells. We further tested inhibition of HIV-1 infectivity using a luciferase-based virus neutralization assay with Env pseudoviruses in TZM.bl cells31,32. In this assay, which requires incubation for 48 hrs to allow for luciferase reporter gene expression, dequalinium showed more cytotoxicity than in the cell-cell fusion assay (see below). Nevertheless, it exhibited much greater inhibition to several HIV-1 isolates than to the murine leukemia virus (MuLV) negative control (Fig. 1g). Furthermore, dequalinium also specifically inhibited cell-cell fusion mediated by Envs of randomly selected, multiple primary HIV-1 isolates from different clades (Supplementary Fig. 2), suggesting that it recognizes a conserved binding site.
Structure–activity relationship studies of dequalinium
Dequalinium contains two aminoquinoline head groups connected by a 10 carbon linker. In a pilot structure–activity relationship (SAR) study using commercial analogs, we showed that two additional dequalinium-like compounds with different head groups were also active in blocking HIV-1 Env mediated cell-cell fusion, while the other two were not (Supplementary Fig. 3; compounds 2-5). The compound 4-aminoquinaldine (6) containing only the head group of dequalinium, also showed no activity. We noted that none of these compounds showed significant cytotoxicity within the tested concentration range. We next designed and synthesized 12 analog compounds, either varying the length of the carbon linker or modifying the head group of dequalinium (Fig. 2a and 2b). These compounds were tested for inhibition activity in the cell-cell fusion assay, as well as their cytotoxicity. Most compounds showed toxicity comparable to that of dequalinium, as indicated by the relative toxicity, with S1C5 (7) being the most toxic and S2C7 (8) the least toxic (Fig. 2c and 2d). Inhibition potency increased with the increasing linker length, but peaked at a length of 12 carbons. Smaller head groups such as S2C9 (9) and S2C11 (10) showed significant decreases in potency, as did the removal of the 2-methyl group (S2C1 (11)) and removal of 2-methyl and 4-amine groups (S2C10 (12)). Halogenated compounds S2C6 (13), S2C7 and S2C8 (14) showed modestly higher potency than dequalinium. A significant improvement was observed with compound S2C3 (15), which contains the addition of a cyclopentyl group at the 2,3 positions, suggesting that larger hydrophobic groups in these positions enhance the potency.
Figure 2. SAR study using synthesized dequalinium analogs.
(a) Design of five compounds with varying length of the linker (n) of 6, 8, 10, 12, 14 carbons. The 10-carbon-linker compound is dequalinium. (b) Design of eight dequalinium analogs with different head groups. (c) Synthesized compounds were analyzed in the cell-cell fusion assay. (d) Synthesized compounds were analyzed in the ATP-based cytotoxicity assay. DEQ, dequalinium. The experiment was performed in triplicates and repeated independently at least twice with similar results. The error bars represent the standard deviations calculated by the Excel STDEV function.
Binding of S2C3 to gp41-inter was further confirmed by SPR analysis. The compound interacted with gp41-inter with an affinity of 2.0 μM, but showed no binding to 2F5 Fab (Fig. 3a and Supplementary Fig. 4a). Three weaker compounds, S1C1 (16), and S2C10, S2C11, showed little or no binding to gp41-inter (Fig. 3b, 3c and Supplementary Fig. 4b), roughly correlating with their potency in blocking membrane fusion (Fig. 2c). Taken together with the binding data for dequalinium and S2C3, these results indicate that the inhibition efficiency of these compounds against HIV-1 Env mediated membrane fusion is primarily determined by their ability to bind their gp41 target. Furthermore, we confirmed by SPR that S2C3 competed with 2F5 and 4E10 for binding to gp41-inter, but not with an anti-gp41 cluster I antibody, 240D Fab, which recognizes an epitope in the C-C loop of gp4133 (Supplementary Fig. 4c-e), suggesting that the MPER remains the target of S2C3. Likewise, improvement in potency of S2C3 over dequalinium was also observed for inhibition of viral infectivity (Fig. 3d). The inhibitory potency of each selected compound in the virus inhibition assay correlated with that in the cell-cell fusion assay. A similar inhibition profile among selected compounds was also found against several other HIV-1 isolates albeit with reduced potencies; they showed significant cytotoxicity at high concentrations (Supplementary Fig. 5). Furthermore, S2C3 effectively blocks binding to the intact HIV-1 Env expressed on the cell surfaces by soluble CD4, but not by the CD4 binding site-directed neutralizing antibody VRC0134 or the prefusion conformation-specific neutralizing antibody PG1635 (Fig. 3e), suggesting that the compound specifically inhibits the Env function by interfering CD4-induced conformational changes required for membrane fusion.
Figure 3. Characterization of the most potent compound S2C3.
(a) SPR analysis of S2C3 binding to gp41-inter. The immobilized protein was tested with various concentrations (2-20 μM) of S2C3. Binding kinetics were evaluated using a 1:1 Langmuir binding model. The sensorgrams are shown in black and the fits in green. (b) and (c) Similar to (a), two weak compounds S1C1 and S2C10 were tested for binding to gp41-inter by SPR. In (a)-(c), the SPR binding experiments were repeated independently at least twice with similar results. (d) Comparison of inhibition of viral infectivity by dequalinium, S2C3, S2C7, S2C10 and S2C11. Buffer, sodium acetate, 50 mM, pH 4.5. The HIV-1 isolate used is the tier 2 virus X1254-3. The experiments were performed using duplicate wells and performed at least twice with similar results. The error bars represent standard deviations as calculated using GraphPad Prism. (e) Flow cytometry histograms of CD4, VRC01 and PG16 binding to the 92UG037.8 Env trimer on the cell surfaces in the absence (dotted line) or presence (solid line) of compound S2C3 (25 μM). Different ligand concentrations are shown in different colors as indicated. 293T cells were used as a negative control in black (dotted and solid lines). All the experiments have been repeated independently at least twice with similar results.
The MPER is highly conserved even among HIV-1, HIV-2 and SIV strains (Supplementary Fig. 6a). Our initial analysis showed the SIV Env was resistant to dequalinium when it was produced at a high expression level to match the fusion activity of HIV-1 Env (Fig. 1f), but further studies indicated that S2C3 could bind to a gp41-inter construct derived from the SIV Env sequence (Supplementary Fig. 6b). In our cell-cell fusion assay, the SIV Env-mediated membrane fusion was indeed inhibited by both dequalinium and S2C3 when expressed at a low, but fusion-active level (Supplementary Fig. 6c). In the pseudovirus assay, both SIV and HIV-2 Envs were sensitive to S2C3 inhibition while the control viruses pseudotyped by MuLV and VSV (vesicular stomatitis virus) envelope proteins are not (Supplementary Fig. 7a-f). These results support our conclusion that the observed inhibition of membrane fusion by these compounds is Env-dependent, not an off-target effect and that they are broad fusion inhibitors targeting a conserved site.
Additional evidence for the S2C3-MPER interaction
To gain further insights into the S2C3 binding site on gp41, we conducted a chemical shift perturbation study by titrating an MPER construct with increasing concentrations of S2C3 (Supplementary Fig. 8a). We recently reported an NMR structure of a gp41 fragment containing both the MPER and TMD (transmembrane domain) (residues 660-710) reconstituted in bicelles41. Using the same MPER-TMD/bicelle system, we recorded a series of 2D TROSY-HSQC spectra with increasing concentrations of either S2C3 or DMSO and the inactive compound S2C11 as negative controls. Titration of DMSO or S2C11 did not lead to any significant changes in either chemical shift or peak intensity, as expected (Supplementary Figs. 8 and 9). The most evident S2C3-dependent chemical shift changes were observed in residues of the MPER, including the backbones of L663, W672 and N677, as well as the side chains of W666, W670 and R683 (Supplementary Fig. 8b and 8c), suggesting that direct contacts of S2C3 to these residues led to changes of their chemical environment. In addition, the peak intensity of the MPER residues decreased by 40-60% after addition of S2C3, while the intensity of the N-terminal residues in the TMD (residues 683-698) was not affected (Supplementary Fig. 9). These observations imply that S2C3 binding may reduce the backbone dynamics of the MPER. Interestingly, the peak intensity of the C-terminal end of the TMD (residues 699-709) also decreased upon S2C3 addition, suggesting the conformational coupling between the MPER and the C-terminus of TMD.
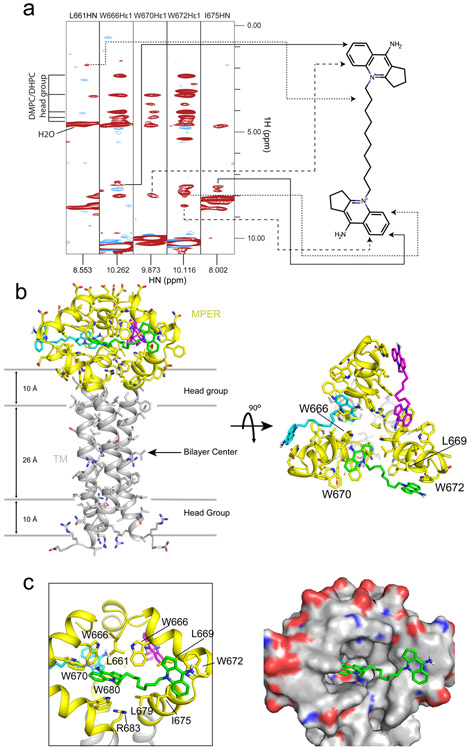
To further map residues that are in contact with S2C3, we acquired 3D 15N-edited-NOESY spectra from the bicelle-reconstituted (15N, 13C, 2H)-labeled MPER-TMD in the presence or absence of the compound. Under these conditions, only protons of S2C3 and labile protons of the MPER-TMD (backbone and side-chain amide protons) were detected, while non-labile protons of the MPER-TMD (attached to 13C) were replaced with deuterons and not detected in our NOE experiments. The acyl chains of detergent and lipid were also deuterated. To eliminate false positives due to incomplete deuteration of the 13C sites in the MPER-TMD, we performed JCH-modulated, 15N-edited NOESY36, which removes NOEs between 1H-15N and 1H-13C spectroscopically. The chemical shifts of S2C3 protons were assigned based on the 2D COSY experiment (Supplementary Fig. 10). The NOESY strips of the MPER-TMD/S2C3 showed similar patterns of the intra-protein NOE peaks as did the ones without the compound (Supplementary Fig. 11a and 11b), indicating S2C3 has little impact on those NOEs and the overall protein structure. We identified the NOE cross peaks between S2C3 protons and amide protons of residues L661, W666, W670, W672 and I675 in the MPER, but not with any residues in the TMD (Fig. 4a and Supplementary Fig. 11). The strongest NOE peaks, from the protons of S2C3 head groups, were observed in the strips of W666, W670 and W672 side chain amide proton (Hε1) and I675 backbone amide proton (HN). Additional NOEs indicated that the protons of the S2C3 carbon linker were in contact with L661HN of the MPER (Fig. 4a and Supplementary Fig. 11).
Figure 4. Structure of S2C3 in complex with the MPER-TMD.
(a) Strips from the 3D 15N-edited NOESY-TROSY-HSQC spectrum with J(13C-1H) modulation recorded using the 15N-, 13C-, 2H-labeled MPER-TMD protein (0.6 mM) in the presence of 2 mM S2C3. NOE peaks from the protons of S2C3 were observed in these strips and mapped to the S2C3 molecule on the right side as indicated by arrows. These NOE peaks were not observed in the spectrum of a control sample without S2C3 (Supplementary Fig. 11). The acyl chains of DHPC/DMPC in bicelles and the protein carbon side chains are deuterated. Solvent water shows a peak with a 1H chemical shift at ~4.7 ppm; protons of head groups of DMPC/DHPC bicelles give peaks with chemical shifts at 2.2-4.2 ppm. (b) Top and side views of the NMR structure of the S2C3-MPER complex. 15 structures with the lowest energies were selected for the final ensemble from 100 structures generated by Xplor-NIH software56. The average structure of the ensemble is shown with protein backbone in ribbon diagram and side chains in stick model. The lipid bilayer is indicated by gray lines schematically. The MPER is colored in yellow and the TMD in gray. Three S2C3 molecules occupying three binding pockets in an MPER trimer are shown in magenta, cyan and green. (c) Close-up views of the hydrophobic binding pocket of S2C3 formed by residues in the MPER in ribbon diagram (left) and electrostatic potential surface representation (right; blue: positively charged and red: negatively charged), respectively.
A small-molecule binding pocket formed by the MPER
We also initially observed similar but a smaller number of NOE peaks between dequalinium and the MPER in the 15N-edited NOESY spectrum, indicating direct contacts of the compound with residues L661, W666, W670 and W672 (Supplementary Fig. 12a and 12b). The cross peaks observed in the spectrum of MPER-TMD/dequalinium were consistent with those present in the spectrum of MPER-TMD/ S2C3 (Fig. 4a). We calculated a preliminary structure using these NOEs and found a binding pocket of dequalinium formed by the hydrophobic residues in the MPER (Supplementary Fig. 12c). Because dequalinium is less soluble in DMSO and has weaker affinity to gp41 than S2C3, we performed further structural studies using S2C3 only.
To define the binding site of S2C3 at the atomic level, we determined the structure of S2C3-bound MPER-TMD using NOE restraints between S2C3 and the MPER, as well as the intra-protein restraints reported previously36,37. The final ensemble of structures converged to RMSD of 1.187 Å and 1.729 Å for backbone and all heavy atoms, respectively (Supplementary Fig. 13 and Supplementary Table 2). The average structure of the ensemble is shown in Fig. 4b. The two head groups of S2C3 interact with a hydrophobic pocket formed by residues L661, W666, L669, W670, W672, I675, L679 and W680 from two neighboring MPER-TMD protomers (Fig. 4c). One head group projects outward, in contact with W672 and I675 of one MPER protomer. The other head group inserts into the hydrophobic core of the MPER formed by residues L661, W666, W670 and W680 of the other protomer. The S2C3 carbon linker also makes hydrophobic contacts with the side chains of L661, L669 and L679 and contributes to binding. In addition, the side chain of R683 projects toward the compound, explaining the S2C3 induced chemical shift changes of the side chain amide proton of R683 (Hε) (Supplementary Fig. 8).
S2C3 (also dequalinium) is a symmetrical molecule. Our NOE restraints cannot rule out the possibility that the two head groups of the compound may occupy each of two adjacent binding pockets instead of one. We therefore calculated the structure using the same NOE restraints but with an assumption that each of the two identical head groups of S2C3 makes contacts only with one MPER protomer. The resulted structures had much higher energy than the one shown in Fig. 4 because of the increased number of NOE violations, suggesting that one S2C3 molecule primarily, if not exclusively, occupies a single hydrophobic pocket formed by two neighboring MPER protomers. Indeed, the single-pocket binding mode is also consistent with the observation that there is an optimal length of the linker connecting the two head groups for its inhibitory activity (Fig. 2c).
MPER mutations affecting Env sensitivity to S2C3
To validate the NMR structure of the S2C3-MPER complex, we generated several mutants in the context of the full-length 92UG037.8 HIV-1 Env38, to alter the hydrophobicity of the binding pocket. S2C3 inhibition of these Env mutants were analyzed in the cell-cell fusion assay in comparison with the wild type Env. All mutants expressed comparable levels of Env with similar extents of cleavage between gp120 and gp41, and showed a readily detectable level of fusion activity ranging 20-100% of that of the wildtype Env (Supplementary Fig. 14a and 14b). In the presence of S2C3, the single mutant W666A showed an IC50 of 9.9 μM, as compared to 4.4 μM for the wild type Env (Supplementary Fig. 14c and Supplementary Table 3). A triple mutant W666S/L669S/I675S exhibited the greatest resistance to S2C3 with an IC50 of 16.7 μM. Interestingly, two other mutants, K683A and K683A/R696A, became more sensitive to S2C3 than the wildtype Env, suggesting the increased hydrophobicity of the binding pocket may lead to more effective recognition by the compound. As a comparison, a mutant (mTMD) containing multiple changes in the TMD even with reversed hydrophobicity in the region showed no significant difference in S2C3 inhibition from the wild type Env (Supplementary Fig. 14c and Supplementary Table 3). These results suggest that the hydrophobicity of the S2C3 binding pocket in the MPER is a key determinant critical for inhibition of HIV-1 Env-mediated membrane fusion.
Discussion
Modern drug discovery is a very time-consuming and increasingly expensive process. Most drug targets involve either an enzyme active site (such as those of HIV-1 reverse transcriptase and protease) or a ligand binding site (such as those of cell receptors)39,40. It has also been suggested that all the obvious human “druggable” targets may have been exhausted by conventional approaches41-43, and thus the pharmaceutical industry has begun to shift its focus towards protein-based biologics. There are several serious limitations of protein-based therapy, however, including high cost for production, inability to penetrate membranes to reach intracellular targets and unwanted immune responses. It is therefore still desirable, for treatment of most diseases, to develop small-molecule drugs.
In this study, we used a neutralizing monoclonal antibody targeting HIV-1 gp41 to guide the search for leads of novel therapeutics against a nonconventional site – the MPER. Monoclonal antibodies have been used as therapeutics to treat human diseases because they can specifically target functional sites of key proteins in disease-related pathways44,45. They too, however, may suffer from drawbacks similar to those of other biologics. We set out to turn a neutralizing antibody into small-molecule drug leads based on the following considerations. First, interactions between an antibody and its cognate antigen involve hydrophobic interactions, hydrogen bonds and salt bridges, similar to those between a small-molecule drug and its protein target. Second, protein-protein interactions often rely on a small set of contact residues (hot spot) for the majority of binding free energy despite large interfaces46, suggesting that a small-molecule compound may be sufficient not only to mimic how an inhibitory antibody binds its antigen, but also to compete with it for antigen binding. A small-molecule lead can thus be identified through competition with the antibody for antigen binding and it may mimic the action of the antibody to block or modulate physiological functions of the protein (antigen). Third, effective antibodies often target functionally critical sites (inhibitory or neutralizing epitopes) on a protein of interest, which may not necessarily be an active or ligand-binding site. This general strategy may expand our repertoire of druggable sites on disease-related proteins that are not accessible by conventional approaches.
As a proof of concept for the antibody-based screening strategy to search for promising drug leads or targets, we have identified dequalinium and its more potent analog S2C3 as small-molecule fusion inhibitors that effectively block HIV-1 infection. In particular, S2C3 binds a hydrophobic pocket formed exclusively by the residues in the MPER, as revealed by our NMR structure (Fig. 4). The MPER has long been considered a promising vaccine target because it contains linear epitopes recognized by several well-characterized (bnAbs)25-27. Previous structural studies have shown that it mainly adopts an α-helical conformation with or without a kink in the middle36. One such structure was determined by NMR using a monomeric MPER peptide reconstituted in detergent micelles, which folded into a kinked helix with many hydrophobic residues embedded in the micelles47, leading to a widely-held belief that the MPER should be buried in viral membrane. Nevertheless, none of these structures even hinted that the MPER could form a small-molecule binding site. Recently, our NMR structure of a gp41 construct containing both the MPER and TMD reconstituted in a lipid bilayer revealed that the MPER is not buried in membrane but instead forms a tightly packed trimeric cluster36. This new structure most likely represents a prefusion conformation of the MPER in a native Env spike, underscoring the important structural role of the lipid bilayer in maintaining physiologically relevant conformation of Env. Using the same system, we were able to confirm the binding pocket in the MPER formed by highly conserved hydrophobic residues and demonstrate how a small molecule interacts with this unexpected binding site in the prefusion Env, as further supported by the data showing that S2C3 blocks CD4-induced conformational changes (Fig. 3e). It is noteworthy that our HTS campaign started long before the structure determination of the MPER trimer, demonstrating the power of using a neutralizing antibody as a guide for searching novel small-molecule binding sites even in absence of any high-resolution structural information.
If S2C3 recognizes the prefusion conformation of the MPER, how does it compete with 2F5 for binding to the prehairpin intermediate state? Our previous studies indicate that the MPER-TMD in bicelles mainly adopts a conformation that is incompatible with 2F5 binding, but the MPER is conformationally dynamic and transiently samples various conformations, accessible up to ~10% of the time to 2F536. S2C3 can stabilize the prefusion conformation of the MPER, driving the conformational equilibrium towards the direction of disfavoring antibody binding and thus blocking 2F5 binding allosterically instead of by direct competition. Indeed, the decreased peak intensity of the MPER residues in the NMR titration experiment observed upon S2C3 addition suggested the reduced conformational dynamics of the MPER in the presence of the compound. Our data support a model of the mechanism by which S2C3-like compounds inhibit HIV-1 infection - by preventing conformational changes of Env from the prefusion state to the receptor-triggered fusion intermediate state, required for productive membrane fusion. We anticipate that these compounds may be useful reagents or probes to help dissect the functional roles of the MPER during HIV-1 entry in future investigations.
The discovery of a small-molecule binding site in the MPER drastically expands the potential medical relevance of this previously recognized vaccine target. Dequalinium is the active ingredient of several topical medications, such as Dequadin and Fluomizin, to treat bacterial infection48, but it has also been tested for treatment of cancer and malaria49,50. Because of their modest potency, dequalinium and its more potent derivatives, such as S2C3 and S2C7, are only first steps toward a useful anti-HIV-1 drug. The high-resolution structure of the trimeric 92UG024 MPER in complex with the hit compound S2C3 can motivate additional HTS campaigns and computational searches to identify more leads for drug candidates suitable for preclinical and clinical investigations. Finally, our antibody-based screening strategy for drug discovery should be applicable to many other human diseases.
Methods
Protein expression and purification
Gp41-inter proteins were produced as described previously22,28. Briefly, the proteins were overexpressed in Rosetta 2 codon plus cells (Novagen) as inclusion bodies after induction with 1 mM IPTG at 37 °C for 6 hours. The bacteria cells were lysed by freezing-thawing cycles and sonication; the gp41-inter proteins were purified by acid extraction and refolded by a rapid-dilution protocol as described22,51, and further purified by gel-filtration chromatography on a prep-grade Superdex 200 (GE Healthcare Life Sciences) in 25 mM Tris-HCl, pH 7.5 and 150 mM NaCl. Purified proteins were concentrated and stored at −80°C.
Anti-HIV-1 Env monoclonal antibodies and their Fab fragments were produced as described38,52. 2F5 Fab was labeled with fluorescein isothiocyanate (FITC). Briefly, 2F5 Fab was treated with a 10-fold molar excess of FITC in 50 mM borate, pH 8.5. The reaction was closely monitored by the 280nm/495nm absorbance ratio to avoid multiple labeling per Fab. When a single label was achieved (usually in 1 hr at room temperature), the reaction was quenched with sodium azide and free FITC molecules removed by dialysis. The labeled Fab was further purified using gel filtration chromatography.
Production of the MPER-TMD protein containing residues 660-710 from a clade D HIV-1 isolate 92UG024.2 Env was carried out as described36. Briefly, the protein was expressed as a trpLE fusion in Escherichia coli strain BL21 (DE3) cells using M9 minimal media supplemented with stable isotopes 15N, 13C or 2H according to the specific labeling requirement for each experiment. The protein was extracted from inclusion bodies, cleaved by cyanogen bromide, purified by Ni-NTA and HPLC, and then reconstituted in DHPC/DMPC bicelles following the previous protocols36.
High-throughput screening and chemical synthesis
All screening experiments were carried out at Harvard Medical School ICCB-Longwood Screening Facility. For the screening assay, 10 μl of the gp41-inter protein in phosphate buffered saline (PBS) at a concentration of 180 nM was added to each well of a Corning 384-well low volume microtitre plate using a Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific). 100 nl of each compound dissolved in DMSO with a concentration of ~10 mM was transferred to each well via pin transfer. Plates were gently vortexed for 5 seconds and then incubated for 1 hr at room temperature. After the incubation, 10 μl of FITC-labeled 2F5 Fab (100 nM in PBS) was added using the reagent dispenser, gently vortexed for 5 seconds and incubated for additional 30 min at room temperature. Plates were spun for 1 min prior to fluorescent measurements. For each screening plate, positive controls containing unlabeled 2F5 Fab and negative controls containing DMSO were included and Z’-factors were calculated as a quality control measure. Fluorescent polarization measurements were recorded on a PerkinElmer EnVision plate reader (excitation=480nm, emission=535nm, light=100%, number of flashes=50, detector gain=500). All screening was performed in duplicate. During data analysis, any compounds that fluoresce or scatter light, thus interfering with the FP calculation were eliminated. Duplicate values were averaged and those having a Z’ score of 5 or greater were selected for further analysis. Compound libraries at the ICCB-Longwood used for this project include the known bioactives collections (total of 9,659 compounds) and a number of commercial libraries (total of 160,127 compounds): Biomol1, 4 and Biomol ICCBL-2012 (Enzo Life Sciences International, Inc.), Microsource1, MS Discovery, NINDS Custom Collection (Discover Systems, Inc., Gaylordsville, CT), NIH clinical collection 1 and 2, Prestwick2 (Prestwick Chemical Inc), TocriScreen Mini Library (Tocris Bioscience), ActiMol TimTec 1 (Newark, DE), Asinex 1 (Winston-Salem, NC), Bionet (Ryan Scientific, Mount Pleasant, SC), CEREP (Redmond, WA), ChemDiv (San Diego, CA), and ChemBridge (San Diego, CA), ENAMINE (Ukraine), Life Chemicals (Burlington, ON, Canada), Maybridge (Trevillet, Tintagel, Cornwall, U.K.). We screened 162,106 compounds and 146 compounds met our criteria as “hits”, giving a hit rate of 0.09%. The synthesis of dequalinium analogs was conducted at Chemveda Life Sciences Pvt. Ltd., Plot #: B-11/1, IDA, Uppal, Hyderabad-500 039, Telangana, India. All compounds were purified by recrystallization and purity was confirmed by both mass spec and NMR analysis.
Cell-cell fusion assay and compound inhibition
The cell-cell fusion assay, based on the α-complementation of E. coli β-galactosidase, was conducted as described previously53, with minor modifications for analyzing inhibitory potency of small-molecule compounds. Briefly, 293T cells were cotransfected with expression constructs for either HIV-1 Env and the α fragment of β-galactosidase or CD4, CCR5 and the ω fragment of β-galactosidase. Env-expressing cells (1.0×106 cells/ml) were mixed with CD4- and CCR5-expressing cells (1.0×106 cells/ml). Cell-cell fusion was allowed to proceed at 37°C for 2 hr. Cell-cell fusion activity was quantified using a chemiluminescent assay system, Gal-Screen (Applied Biosystems, Foster City, CA). To analyze small-molecule compounds, Env-expressing cells were first incubated with each of them at various concentrations (10-100 μM) at 37°C for 20 min before mixing with CD4- and CCR5-expressing cells. Each compound was dissolved in DMSO to produce a 5 mM stock, which was subsequently diluted by 2-fold, 4-fold and 10-fold in DMSO, respectively. 1 μl of each of these compound solutions at different concentrations (0.5-5 mM) was mixed with 50 μl Env-expressing cells to give the final compound concentrations of 10-100 μM. The cells with equal amount of DMSO only were used as a negative control for compound inhibition and all fusion activity values were normalized by the readout of the DMSO control. For analyzing S2C3 inhibition with Env mutants, the final S2C3 concentrations after mixed with Env-expressing cells ranged between 2 and 25 μM, by addition of 1 μl S2C3 solutions at 0.1-1.25 mM to a well of 50 μl Env-expressing cells.
Cytotoxicity assay
The cytotoxicity assay was performed using the CellTiter-Glo 2.0 kit (Promega) to measure the cell viability (changes in the amount of ATP due to cell death) when exposed to different compounds. Another identical set of Env-expressing cells, CD4- and CCR5-expressing cells and compounds were mixed in parallel when the cell-cell fusion assay was performed, followed by incubation at 37°C for 2 hr. The cells were cooled to room temperature for 30 min before adding 100 μl of the CellTiter-Glo 2.0 reagent. The mixture was incubated in room temperature in dark for 10 min before recording luminescence using a Synergy Neo microplane reader (BioTek).
Viral infectivity assay and compound inhibition
Inhibition of HIV-1 infectivity was measured using a luciferase-based viral infectivity assay with Env pseudoviruses in TZM.bl cells according to a protocol described previously31,32. The assay measures the reduction in luciferase reporter gene expression in TZM.bl cells following a single round of virus infection. All the compounds were dissolved in a sodium acetate buffer (50 mM, pH 4.5), which showed less cytotoxicity than DMSO as a solvent, to produce stocks of 0.5 mM. Two-fold serial dilutions of compounds by 10% DMEM growth medium were performed in duplicate in a 96-well plate. The same dilution of the acetate buffer was performed as an empty control. Virus was added to each well, and the plate was incubated for 1 hr at 37°C. TZM.bl cells (1×104/well) in 10% DMEM growth medium containing DEAE-Dextran (Sigma) at a final concentration of 11 μg/ml were then added. Following a 48 hour incubation, luminescence was measured using Bright-Glo luciferase reagent (Promega). Murine leukemia virus (MuLV) and vesicular stomatitis virus (VSV) was used as a negative control. All HIV-1, HIV-2, and SIV Env pseudoviruses and negative control MuLV and VSV pseudoviruses were prepared via transfection of 293T cells as previously described36.
To determine cytotoxicity of the compounds in TZM.bl cells, they were diluted in the same manner as described above in the infectivity assay. The same dilution of the acetate buffer was also made as a sham control. TZM.bl cells (1×104/well) in 10% DMEM growth medium containing DEAE-Dextran (11 μg/ml) were added to the compounds without viruses. After a 48-hour incubation, excess medium was carefully removed carefully by aspirating and the cells were cooled to room temperature for 30 min before mixing with equal volume of the CellTiter-Glo 2.0 reagent. The mixture was incubated at room temperature in the dark for 10 minutes before recording luminescence.
SPR (Surface Plasmon Resonance) analysis
All experiments were performed with a Biacore 3000 system (GE Healthcare) at 25°C in HBS-E buffer (10 mM HEPES, pH 7.0, 150 mM NaCl, 3 mM EDTA) containing 0.5% DMSO. Protein immobilization to CM5 chips was performed following the standard amine coupling procedure as recommended by the manufacturer. The immobilization level was ~3,000 RU for small-molecule binding experiments unless specified. For S2C3 competition with antibodies for binding to gp41-inter, 2F5 Fab, 240D Fab was immobilized at a level of ~1,500 RU; 4E10 Fab at ~3,500 RU (4E10) to have a similar response of gp41-inter binding. Small molecule compounds were dissolved in DMSO and diluted in the HBS-E buffer by 200-fold, so that the final DMSO concentration matched that in the running buffer. Sensorgrams were recorded by passing various concentrations of an analyte over the immobilized ligand surface at a flow rate of 40 μl/min either with a 2-min association phase followed by a 10-min dissociation phase for binding to gp41-inter surfaces or with a 4-min association phase followed by a 10-min dissociation phase for binding to antibody surfaces. Identical injections over blank surfaces were subtracted from the data for kinetic analysis. Binding kinetics were analyzed by BiaEvaluation software using a 1:1 Langmuir binding model. All injections were carried out in duplicate and gave essentially identical results.
Flow cytometry
Flow cytometry was performed using a well-characterized stable 293T cell line expressing the wildtype HIV-1 92UG037.8 Env, as described previously38. Env-expressing cells were detached from plates using PBS, and washed with ice-cold PBS containing 1% BSA. 106 cells were incubated for 30~40 minutes on ice with either soluble 4 domain CD4 with a C-terminal histag, VRC01 Fab, or PG16 Fab at various concentrations in PBS containing 1% BSA in the presence of 25 μM S2C3 or the same volume of DMSO (control). The cells were then washed twice with PBS containing 1% BSA and stained with either by an Anti-His-PE antibody (Bergisch Gladbach, Germany) for the CD4 samples or by R-Phycoerythrin AffiniPure F(ab')2 fragment goat anti-human IgG, F(ab')2 Fragment specific secondary antibody (Jackson ImmunoResearch laboratories, West Grove, PA) for the Fab samples at 5 μg/ml. All the fluorescently labeled cells were washed twice with PBS containing 1% BSA and analyzed immediately using a BD LSRII instrument and program FACSDIVA (BD Biosciences, San Jose, CA). All data were analyzed by FlowJo (FlowJo, LLC, Ashland, OR).
Chemical shift perturbation upon S2C3 titration
NMR data of chemical shift perturbation were acquired on Bruker spectrometers operating at 1H frequency of 600 MHz and equipped with cryogenic probes at 35°C. A series of 2D 15N TROSY-HSQC spectra were acquired using 350 μl of the 15N-labeled MPER-TMD/bicelle (0.25 mM) after sequential addition of S2C3 to a final concentration of 0.5 mM, 1.5 mM and 2.5 mM, respectively. Specifically, a TROSY-HSQC spectrum was first acquired without S2C3 as a reference. S2C3 was dissolved in DMSO to make a 50 mM stock solution and it was added to the protein sample stepwise to give a final S2C3 concentration of 0.5 mM, 1.5 mM and 2.5 mM, respectively. At each step, a 2D TROSY-HSQC spectrum of the sample was acquired. As negative controls, TROSY-HSQC spectra were also acquired using the same batch of the MPER-TMD/bicelle sample (350 μl at 0.25 mM) after stepwise addition of equal amount of DMSO that was in the S2C3-added sample at each concentration. The pH was measured before and after adding S2C3 or DMSO, and no significant change was found. NMR data were processed with NMRpipe54. The spectra were analyzed using SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). All the parameters were identical for all data acquisition and processing. The chemical shift differences in 1H and 15N were averaged using the following equation to generate the averaged chemical shift difference (Supplementary Fig. 9):
The δN stands for the chemical shift difference in the 15N dimension. The δH stands for the chemical shift difference in the 1H dimension.
NMR structure determination
To obtain distance restraints between S2C3 and the MPER-TMD, a 13C-selected, 3D 15N-edited NOESY-TROSY-HSQC spectrum36 was acquired at 35°C on Bruker spectrometers operating at 1H frequency of 900 MHz using 0.6 mM 15N-, 13C- and 2H-labeled MPER-TMD reconstituted in perdeuterated bicelles in the presence of 2 mM S2C3. Perdeuteration was used to eliminate signals from carbon side chains of the MPER-TMD and acyl chains of DMPC/DHPC bicelles. 13C-1H J-coupling allowed us to remove any residual signals from protein carbon side chains in case deuteration was incomplete36. In the NOESY spectrum, only signals from S2C3, protein backbones, side-chain amide groups, head groups of DMPC and DHPC were detectable.
The NMR data were processed and analyzed using NMRpipe54, XEASY55 and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). The NOESY stripes of the MPER-TMD/S2C3 sample and its 2D TROSY-HSQC spectrum exhibited very similar patterns compared to those from an MPER-TMD sample without S2C3 (Supplementary Figs. 7a, 11a and 11b). Assignment of the amide group resonance was performed based on the assignments published previously36,53. Chemical shifts of most residues in the construct had no significant differences between the samples in the presence or absence of S2C3. To assign the proton peaks of S2C3, 2D 1H-1H COSY was acquired using 10mM S2C3 dissolved in DMSO-d6. The spectrum was analyzed in the software MestReNova (https://mestrelab.com/). Since S2C3 had little impact on NOESY and TROSY-HSQC spectra of the MPER-TMD, we used the inter- and intra-protomer NOE restraints and the dihedral angle restraints from the previously published MPER-TMD structure, together with distance restraints extracted from NOE peaks between S2C3 and the MPER-TMD for structure calculation of the MPER-TMD/S2C3 complex. We assumed that three S2C3 molecules bound with one MPER-TMD trimer and each compound molecule interacted with two neighboring protomers of the protein. 100 structures were generated in total by software XPLOR-NIH56, and 15 structures with the lowest energies were selected for the final ensemble (Supplementary Fig. 13).
For the MPER-TMD/dequalinium complex, 3D 15N-edited NOESY-TROSY-HSQC spectrum was acquired using 0.6 mM 15N, 2H-labeled perdeuterated MPER-TMD reconstituted in perdeuterated bicelles in the presence of 3 mM dequalinium at 35°C on Bruker spectrometers operating at 1H frequency of 800 MHz. Similar procedures of assignment and structure calculation were performed.
Western blot
293T cells were transiently transfected with 1 μg of the 92UG037.8 gp160 expression construct or its MPER mutants. Lysates of cells expressing Env or its mutants were prepared by resuspending the cells in PBS (phosphate buffered saline) at a density of 2×106 cells/ml, followed by treatment with Laemmli Sample Buffer (Bio-Rad, Hercules, CA) and boiling for 10 min. Env samples were resolved in 4-15% Mini-Protean TGX gel (Bio-Rad) and transferred onto PVDF membranes (Millipore, Billerica, MA) by an Iblot2 (Life Technologies). Membranes were blocked with 5% skimmed milk in PBS for 1 hour and incubated with anti-V3 loop antibody 3791 for another hour at room temperature. Alkaline phosphatase conjugated anti-human Fab IgG (1:5000) (Sigma-Aldrich) was used as a secondary antibody. Env proteins were visualized using one-step NBT/BCIP substrates (Thermo Scientific, Cambridge, MA).
Data availability
The atomic structure coordinates and NMR data are deposited in the ProteinDataBank under the accession number PDB ID 6V4T. All other related data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments:
We thank H. J. Ha, D. O’Neil Danis III, S. Rits-Volloch, H. Peng and Z. Liu for technical assistance, S.C. Harrison for critical reading of the manuscript. This work was supported by NIH grants AI129721 (to B.C.), AI112489 (to B.C.), AI127193 (to B.C. and J.J.C.), GM116898 (to J.J.C.), AI141002 (to B.C.) and AI106488 (to B.C.). The NMR data were collected on the 800 MHz at MIT-Harvard CMR (supported by NIH grant P41 EB-002026) and on the 900 MHz at National Facility for Protein Science in Shanghai, ZhangJiang Lab.
Footnotes
Competing Financial Interests:
Boston Children’s Hospital has filed a patent application based on this work with B. C., G. F. and T. X. listed as co-inventors. All other authors declare no competing interests.
References:
- 1.Hammer SM et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. The New England journal of medicine 337, 725–733, doi: 10.1056/NEJM199709113371101 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. The New England journal of medicine 337, 734–739, doi: 10.1056/NEJM199709113371102 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Palella FJ Jr. et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. The New England journal of medicine 338, 853–860, doi: 10.1056/NEJM199803263381301 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Grant M, Samuel R, Bettiker RL & Suh B Antiretroviral therapy 2010 update: current practices and controversies. Arch Pharm Res 34, 1045–1053, doi: 10.1007/s12272-011-0701-3 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Thompson MA et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. Jama 308, 387–402, doi: 10.1001/jama.2012.7961 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kilby JM & Eron JJ Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med 348, 2228–2238 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Robertson D US FDA approves new class of HIV therapeutics. Nat Biotechnol 21, 470–471, doi: 10.1038/nbt0503-470 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Poveda E et al. Dynamics of enfuvirtide resistance in HIV-infected patients during and after long-term enfuvirtide salvage therapy. J Clin Virol 34, 295–301, doi: 10.1016/j.jcv.2005.02.004 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Poveda E et al. Evolution of genotypic and phenotypic resistance to Enfuvirtide in HIV-infected patients experiencing prolonged virologic failure. Journal of medical virology 74, 21–28, doi: 10.1002/jmv.20141 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Sista PR et al. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. Aids 18, 1787–1794 (2004). [DOI] [PubMed] [Google Scholar]
- 11.He Y et al. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. The Journal of biological chemistry 283, 11126–11134, doi: 10.1074/jbc.M800200200 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Pan C, Cai L, Lu H, Qi Z & Jiang S Combinations of the first and next generations of human immunodeficiency virus (HIV) fusion inhibitors exhibit a highly potent synergistic effect against enfuvirtide- sensitive and -resistant HIV type 1 strains. Journal of virology 83, 7862–7872, doi: 10.1128/JVI.00168-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie D et al. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob Agents Chemother 54, 191–196, doi: 10.1128/AAC.00976-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos JR et al. Efficacy and safety of switching from enfuvirtide to raltegravir in patients with virological suppression. HIV Clin Trials 10, 432–438, doi: 10.1310/hct1006-432 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Chen B Molecular Mechanism of HIV-1 Entry. Trends Microbiol, doi: 10.1016/j.tim.2019.06.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ & Wiley DC Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Chan DC, Fass D, Berger JM & Kim PS Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Julien JP et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483, doi: 10.1126/science.1245625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyumkis D et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490, doi: 10.1126/science.1245627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pancera M et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461, doi: 10.1038/nature13808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan DC & Kim PS HIV entry and its inhibition. Cell 93, 681–684 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Frey G et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 105, 3739–3744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J et al. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. Journal of virology 88, 1249–1258, doi: 10.1128/JVI.02664-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montero M, van Houten NE, Wang X & Scott JK The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 72, 54–84, table of contents, doi:72/1/54 [pii] 10.1128/MMBR.00020-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster T et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67, 6642–6647 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiegler G et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 17, 1757–1765 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Huang J et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412, doi: 10.1038/nature11544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frey G et al. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nature structural & molecular biology 17, 1486–1491, doi: 10.1038/nsmb.1950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang JH, Chung TD & Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73, doi: 10.1177/108705719900400206 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Weissenbacher ER et al. A comparison of dequalinium chloride vaginal tablets (Fluomizin(R)) and clindamycin vaginal cream in the treatment of bacterial vaginosis: a single-blind, randomized clinical trial of efficacy and safety. Gynecologic and obstetric investigation 73, 8–15, doi: 10.1159/000332398 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Montefiori DC Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485, 395–405, doi: 10.1007/978-1-59745-170-3_26 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Sarzotti-Kelsoe M et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. Journal of immunological methods 409, 131–146, doi: 10.1016/j.jim.2013.11.022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson WE Jr. et al. Antibodies to the primary immunodominant domain of human immunodeficiency virus type 1 (HIV-1) glycoprotein gp41 enhance HIV-1 infection in vitro. J Virol 64, 5301–5305 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861, doi: 10.1126/science.1187659 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker LM et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289, doi:1178746 [pii] 10.1126/science.1178746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Q et al. Structure of the membrane proximal external region of HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A 115, E8892–E8899, doi: 10.1073/pnas.1807259115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Q, Piai A, Chen W, Xia K & Chou JJ Structure determination protocol for transmembrane domain oligomers. Nature protocols, doi: 10.1038/s41596-019-0188-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J et al. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349, 191–195, doi: 10.1126/science.aaa9804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imming P, Sinning C & Meyer A Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 5, 821–834, doi: 10.1038/nrd2132 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Overington JP, Al-Lazikani B & Hopkins AL How many drug targets are there? Nat Rev Drug Discov 5, 993–996, doi: 10.1038/nrd2199 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Kufareva I, Ilatovskiy AV & Abagyan R Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic acids research 40, D535–540, doi: 10.1093/nar/gkr825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos R et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16, 19–34, doi: 10.1038/nrd.2016.230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bojadzic D & Buchwald P Toward Small-Molecule Inhibition of Protein-Protein Interactions: General Aspects and Recent Progress in Targeting Costimulatory and Coinhibitory (Immune Checkpoint) Interactions. Curr Top Med Chem 18, 674–699, doi: 10.2174/1568026618666180531092503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiner GJ Building better monoclonal antibody-based therapeutics. Nat Rev Cancer 15, 361–370, doi: 10.1038/nrc3930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh S et al. Monoclonal Antibodies: A Review. Curr Clin Pharmacol 13, 85–99, doi: 10.2174/1574884712666170809124728 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Clackson T & Wells JA A hot spot of binding energy in a hormone-receptor interface. Science 267, 383–386, doi: 10.1126/science.7529940 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Sun ZY et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63, doi: 10.1016/j.immuni.2007.11.018 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Tischer M, Pradel G, Ohlsen K & Holzgrabe U Quaternary ammonium salts and their antimicrobial potential: targets or nonspecific interactions? ChemMedChem 7, 22–31, doi: 10.1002/cmdc.201100404 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Abeywickrama C, Rotenberg SA & Baker AD Inhibition of protein kinase C by dequalinium analogues: structure-activity studies on head group variations. Bioorg Med Chem 14, 7796–7803, doi: 10.1016/j.bmc.2006.07.067 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Berger O et al. Reverse-benzamidine antimalarial agents: design, synthesis, and biological evaluation. Bioorg Med Chem Lett 20, 5815–5817, doi: 10.1016/j.bmcl.2010.07.124 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Frey G et al. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci U S A 103, 13938–13943, doi:0601036103 [pii] 10.1073/pnas.0601036103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovacs JM et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proceedings of the National Academy of Sciences of the United States of America 109, 12111–12116, doi: 10.1073/pnas.1204533109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dev J et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 353, 172–175, doi: 10.1126/science.aaf7066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Bartels C, Xia TH, Billeter M, Guntert P & Wuthrich K The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. Journal of biomolecular NMR 6, 1–10, doi: 10.1007/BF00417486 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Schwieters CD, Kuszewski JJ, Tjandra N & Clore GM The Xplor-NIH NMR molecular structure determination package. Journal of magnetic resonance 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic structure coordinates and NMR data are deposited in the ProteinDataBank under the accession number PDB ID 6V4T. All other related data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.