Abstract
The ongoing mutations in the structural proteins of SARS-CoV-2 are the major impediment for prevention and control of the COVID-19 disease. Presently we focused on evolution of the envelope (E) protein, one of the most enigmatic and less studied protein among the four structural proteins (S, E, M and N) associated with multitude of immunopathological functions of SARS-CoV-2. In the present study, we comprehensively analyzed 81,818 high quality E protein sequences of SARS-CoV-2 globally available in the GISAID database as of 20 August 2020. Compared to Wuhan reference strain, our mutational analysis explored only 1.2 % (982/81818) mutant strains undergoing a total of 115 unique amino acid (aa) substitutions in the E protein, highlighting the fact that most (98.8 %) of the E protein of SARS-CoV-2 strains are highly conserved. Moreover, we found 58.77 % (134 of 228) nucleotides (nt) positions of SARS-CoV-2 E gene encountering a total of 176 unique nt-level mutations globally, which may affect the efficacy of real time RT-PCR-based molecular detection of COVID-19. Importantly, higher aa variations observed in the C-terminal domain (CTD) of the E protein, particularly at Ser55-Phe56, Arg69 and the C-terminal end (DLLV: 72–75) may alter the binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 and thus could play a key role in COVID-19 pathogenesis. Furthermore, this study revealed the V25A mutation in the transmembrane domain which is a key factor for the homopentameric conformation of E protein. Our analysis also observed a triple cysteine motif harboring mutation (L39M, A41S, A41V, C43F, C43R, C43S, C44Y, N45R) which may hinder the binding of E protein with spike glycoprotein. These results therefore suggest the continuous monitoring of the structural proteins including the envelope protein of SARS-CoV-2 since the number of genome sequences from across the world are continuously increasing.
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus-2; E, envelope; S, spike; N, nucleocapsid; M, membrane; nt, nucleotide; aa, amino acid; TMD, transmembrane domain; CTD, C-terminal domain; NP, non-polar; PC, positively charged; NC, negatively charged
Keywords: SARS-CoV-2, Envelope protein, Mutations, Transmembrane domain, Triple cysteine motif
1. The study
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection, the COVID-19 disease has impacted the entire world, and created a public health emergency since December 2019 (Tang et al., 2020; Zhang and Holmes, 2020). The inherently higher mutations in the genome of SARS-CoV-2 have already produced many descendants from the original Wuhan strain, thereby escaping the host immune responses (DeDiego et al., 2008; Islam et al., 2020; Li et al., 2020; Rahman et al., 2020c). The genome of SARS-CoV-2 virus encodes for four major structural proteins such as spike (S) protein, nucleocapsid (N) protein, membrane (M) protein, and envelope (E) protein, all of which are required to complete a successful infectious event/replication cycle of the virus including entry, assembly, packaging and release of new virus particles within the human cells (McBride et al., 2014; Schoeman and Fielding, 2019; Hassan et al., 2020; Rahman et al., 2020a). The E protein is the smallest of the major structural proteins, and associated with viral assembly, budding, envelope formation, and pathogenesis (Hoque et al., 2020). During the replication cycle, SARS-CoV-2 virus expresses the E protein in a higher abundance inside the host cell, however, only a small portion is incorporated into the virion envelope. This protein carries out its functions by interacting with M and other accessory proteins viz. ORF3a, ORF7a, and host cell proteins (McBride et al., 2014; Schoeman and Fielding, 2019). Homologous assembling of the Coronavirus E protein forms pentameric channel with its transmembrane domain that directly affects virus replication. However, C-terminal domain of monomeric E protein affects the host's intracellular activities through interference with the endoplasmic reticulum, Golgi and the ER-Golgi intermediate compartment (Li et al., 2014). Interaction between the C-terminal domain binding motif (DLLV) of SARS Envelope and PALS1 disrupt tight junctions of epithelial cells promoting virus spread and thus play a key role in the SARS-CoV pathology and suspected to affect the integrity of the lung epithelia (De Maio et al., 2020; Toto et al., 2020).
The ongoing rapid transmission, and global spread of COVID-19 have raised intriguing questions whether the evolution and adaptation of SARS-CoV-2 is driven by synonymous mutations, deletions and/or replacements (Bal et al., 2020; Islam et al., 2020; Pachetti et al., 2020). Although, the mutational spectra of different structural proteins (S, M, and N) of SARS-CoV-2 has been reported by several research groups (Islam et al., 2020; Pachetti et al., 2020; Phan, 2020; Rahman et al., 2020b; Rahman et al., 2020c) over a short period of time, however, available literature on the nucleotide and aa-level mutations of E protein is till limited. However, the sequence identity between the E proteins of SARS-CoV and SARS-CoV-2 sturdily recommends the conservation of its functional features, thereby playing nearly identical roles in the pathogenesis of COVID-19 (Schoeman and Fielding, 2020). Indeed, a great discrepancy exists in the amino acid sequences of the E protein between the different coronavirus groups and, to an extent, within some of the groups.
To comprehensively analyze the mutational spectra of E protein of SARS-CoV-2 as a continuous part of the coronavirus genomic mutational research (Islam et al., 2020; Rahman et al., 2020b; Rahman et al., 2020c; Alam et al., 2020), we retrieved 83,607 complete or near-complete genome sequences of SARS-CoV-2 (human host) from the global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/) belonging to 159 countries or territories till 20 August 2020 (Supplementary Data 1). We obtained 81,818 cleaned sequences (97.86%) after removing the low-quality sequences and in frame stop codon. Multiple sequence alignment was performed through MAFFT using the Wuhan strain (NCBI accession no. NC_045512) (Katoh et al., 2019) as a reference, and nonsynonymous mutations were identified using the previously published methods (https://github.com/SShaminur/Mutation-Analysis) (Islam et al., 2020; Rahman et al., 2020b; Rahman et al., 2020c). Structure of E protein was predicted in Phyre2 at intensive modelling mode with 57 residues (77%) modelled at >90% accuracy (Kelley et al., 2015) and Gibbs free energy changes (ΔΔG) were predicted in FoldX (v5) integrated in YASARA with repaired mode (Delgado et al., 2019).
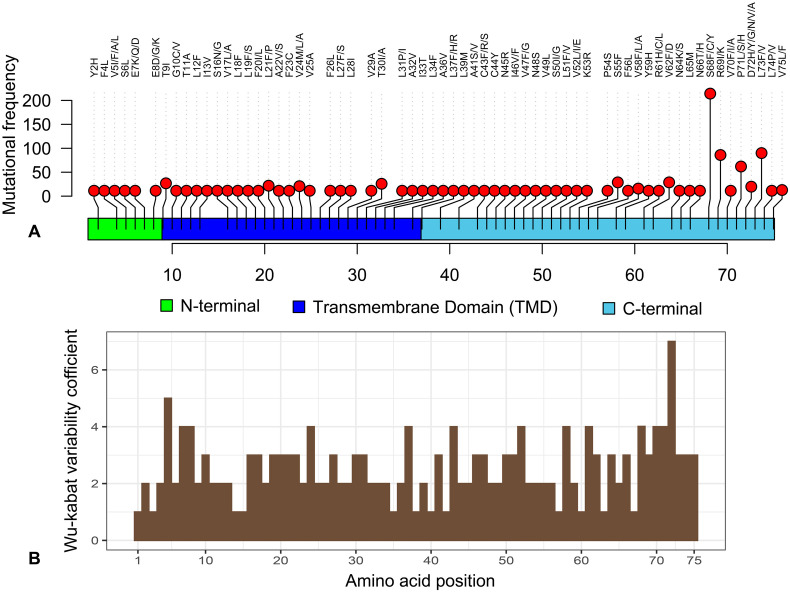
The mutational analysis of the present study revealed that only 1.2% (982/81818) strains possessed amino acid (aa) substitutions in 63 sites of the E protein. This result indicates the fact that most (98.8%) of the E protein of globally circulating SARS-CoV-2 strains are highly conserved. Moreover, compared to the ratios of mutant strains with respect to other structural proteins (S, M and N), the E protein evolved slowly (De Maio et al., 2020; Islam et al., 2020; Rahman et al., 2020b; Rahman et al., 2020c). Comparative conservancy of the E protein highlights the potentiality of the protein as target for vaccine and therapeutic interventions to prevent or control the COVID-19 pandemic. Highest aa mutations (n = 4) (C43R, C44Y, N45R, V47G) were found in a Moroccan strain of SARS-CoV-2 (EPI_ISL_467299) followed by two aa mutations in nine strains from England, Israel, Netherlands, Northern Ireland, Scotland, and Sierra Leone at different positions. Remarkably, rest of the strains (n = 972) possessed only one aa mutation in different positions of the E protein of the SARS-CoV-2 genome (Fig. 1, Supplementary Data 1).
Fig. 1.
Overview and variability coefficient of the envelope (E) protein of SARS-CoV-2. (A) Mapping and frequency distribution of mutations in the E protein of SARS-CoV-2 strains through Loliplot visualization. (B) Wu-Kabat variability coefficient of E protein of SARS-CoV-2. Here, variability coefficient 1 indicates the conservancy, whereas coefficients ˃ 1 indicate relative variability of the respective positions. The more the coefficient value the more the variability or diversity (Supplementary Data 1).
Overview and variability coefficient of the envelope (E) protein of SARS-CoV-2. (A) Mapping and frequency distribution of mutations in the E protein of SARS-CoV-2 strains through Loliplot visualization. (B) Wu-Kabat variability coefficient of E protein of SARS-CoV-2. Here, variability coefficient 1 indicates the conservancy, whereas coefficients ˃ 1 indicate relative variability of the respective positions. The more the coefficient value the more the variability or diversity (Supplementary Data 1).
SARS-CoV-2 E protein is 228 nucleotides long (with stop codon). The nucleotide (nt) level analysis of mutational spectra revealed 58.77% (134/228) mutated sites of the primary structure of the E protein. Among 134 positions nucleotide variation, 7 sites (9, 42, 114, 126, 189, 171, 180) had synonymous mutations whereas the remaining 127 nucleotide positions resulted in nonsynonymous aa substitutions at 63 aa positions (Supplementary Data 1). We found 7 nt variations in a Moroccan strain (EPI_ISL_467299), and 2 nt variations in 54 strains of SARS-CoV-2 from Australia, Austria, Canada, England, Guangdong, India, Iran, Israel, Netherlands, Northern_Ireland, Scotland, Sichuan, Sierra_Leone, Sweden, and Wales. The rest of the strains (n = 1514) showed only 1 nt mutation (Supplementary Data 1).
In this study, we also observed worldwide mutational variations within the primer-probes binding sites of SARS-CoV-2 E gene (Supplementary Table 1, Supplementary Data 1). We found a total of 74 nucleotide mutations that occupied the binding sites of primer-probes recommended by several research groups (Chu et al., 2020; D'Cruz et al., 2020; Park et al., 2020) for E gene targeted PCR-based detection of SARS-CoV-2 (Supplementary Table 1). The mutations occupy 5′ and/or 3′ ends of forward and/or reverse primers binding sites of virus strains from many countries (Supplementary Table 1). However, false-negative results may occur by mutations in the primer and probe target regions in the SARS-CoV-2 genome. Although it was attempted to design the real-time RT-PCR assay as precisely as possible based on the conserved structural regions of the SARS-CoV-2 genomes like the envelope (E) protein, variability causing incongruities between the primers and probes and the target sequences can lead to decrease in assay performance and potential false-negative results (Tahamtan and Ardebili, 2020). The results of the real-time RT-PCR using primers targeting different genes can be affected by the variation of viral RNA sequences. Genetic diversity and rapid evolution of the novel coronavirus (SARS-CoV-2) have been observed in different studies (Islam et al., 2020; Phan, 2020; Rahman et al., 2020b; Rahman et al., 2020c).
However, mutations in the primer binding sites, importantly at 3′ end of primers, may affect the RT-PCR-based COVID-19 diagnosis resulting in false negative interpretations (Nalla et al., 2020; Rana and Pokhrel, 2020). Besides primer mismatches, sequence variations in the probe recognition sites might also affect the efficiency of RT-PCR-based detection of COVID-19 providing false negative (unable to bind) or false positive (non-specific binding) results (Kamau et al., 2017; Nalla et al., 2020). Our study provides a global insight of E gene mutations, and its likely consequences on primer-based detection of SARS-CoV-2 by RT-PCR method. Overall, this study warrants continuous monitoring and to update the primer-probe sequences based on the regional viral genomic sequences for efficient and accurate detection of COVID-19.
The primary structure of the E protein contains 75 aa of which 84.0% (63/75) sites underwent to 115 unique aa mutations (Fig. 1). Our analysis showed that 35 sites in the E protein structure underwent to more than one aa mutation, and of them, aa position 5 and 72 had aa variation numbers of 4 and 6, respectively (Table 1). Comparing the individual strain level mutations, we found that the S68F mutation in 250 strains (highest frequency) followed by L73F, R69I, and P71L mutations noticed in 100, 88, and 59 strains, respectively. The N-terminal, transmembrane domain (TMD), and C-terminal domain (CTD) of the E protein had 7, 25, and 31 sites for aa substitutions, respectively (Fig. 1). Several earlier studies (Hassan et al., 2020; Islam et al., 2020) also reported aa mutations in 10 sites (aa positions: 26, 36, 37, 39, 46, 58, 68, 71, 72, 73) of the E protein corroborating our current findings. Importantly, CTD of the envelope (E) protein showed comparatively higher Wu-Kabat variability coefficient and mutational frequency reflecting the continuous evolution of this domain (Fig. 1, Supplementary Data 1). It is to be noted here, aa variations in this domain, particularly Ser55-Phe56 and Arg69 predicted to improve the interaction of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 than SARS-CoV (De Maio et al., 2020; Toto et al., 2020). Furthermore, aa at C-terminal end (DLLV: 72–75) directly interact with the PALS1 protein. More frequent aa variations with altered polarity of the side chains at these sites could play a critical role in COVID-19 pathogenesis leading to increased epithelial cell disruption and promoted virus spread (Table 1, Supplementary Data 1) (De Maio et al., 2020; Toto et al., 2020).
Table 1.
Amino acid (aa) variation in envelope (E) protein of SARS-CoV-2.
| Position | Number of aa variations | aa (Ref:position:strain) | Reference aa characteristics | Strains aa characteristics |
|---|---|---|---|---|
| 72 | 6 | D72Y,D72G,D72H,D72N,D72V,D72A | A | P,NP,B,P,NP,NP |
| 5 | 4 | V5I,V5F,V5A,V5L | NP | NP,NP,NP,NP |
| 7 | 3 | E7K,E7Q,E7D | A | B,P,A |
| 8 | 3 | E8G,E8D,E8K | A | NP,A,B |
| 24 | 3 | V24M,V24L,V24A | NP | NP,NP,NP |
| 37 | 3 | L37H,L37F,L37R | NP | B,NP,B |
| 43 | 3 | C43F,C43R,C43S | NP | NP,B,P |
| 52 | 3 | V52I,V52L,V52E | NP | NP,NP,A |
| 58 | 3 | V58F,V58L,V58A | NP | NP,NP,NP |
| 61 | 3 | R61H,R61C,R61L | B | B, P,NP |
| 68 | 3 | S68F,S68C,S68Y | P | NP,NP,P |
| 70 | 3 | V70F,V70I,V70A | NP | NP,NP,NP |
| 71 | 3 | P71L,P71S,P71H | NP | NP,P,B |
| 10 | 2 | G10C,G10V | NP | NP,NP |
| 16 | 2 | S16N,S16G | P | P,NP |
| 17 | 2 | V17L,V17A | NP | NP,NP |
| 19 | 2 | L19S,L19F | NP | P,NP |
| 20 | 2 | F20I,F20L | NP | NP,NP |
| 21 | 2 | L21F,L21P | NP | NP,NP |
| 22 | 2 | A22V,A22S | NP | NP,P |
| 27 | 2 | L27F,L27S | NP | NP,P |
| 30 | 2 | T30I,T30A | P | NP,NP |
| 31 | 2 | L31P,L31I | NP | NP,NP |
| 41 | 2 | A41S,A41V | NP | P,NP |
| 46 | 2 | I46V,I46F | NP | NP,NP |
| 47 | 2 | V47F,V47G | NP | NP,NP |
| 50 | 2 | S50I,S50G | P | NP,NP |
| 51 | 2 | L51F,L51V | NP | NP,NP |
| 62 | 2 | V62F,V62D | NP | NP,A |
| 64 | 2 | N64K,N64S | P | B,P |
| 66 | 2 | N66T,N66H | P | P,B |
| 69 | 2 | R69I,R69K | B | NP,B |
| 73 | 2 | L73F,L73V | NP | NP,NP |
| 74 | 2 | L74P,L74V | NP | NP,NP |
| 75 | 2 | V75L,V75F | NP | NP,NP |
NP = non-polar, P = polar, B = basic, A = acidic.
Identity, similarity, and the gap between SARS-CoV-2 (NC_045512.2) and SARS-CoV (NC_004718.3) E protein was 94.8%, 96.1%, and 1.3%, respectively. Two aa mutations, N15A and V25F were found in the TMD which may abolish the ion channeling capability of SARS-CoV E viroporin structure, a key factor of its homopentameric conformation (Torres et al., 2006; Torres et al., 2007; Verdiá-Báguena et al., 2012). We observed the V25A mutation in six strains from Spain, Canada, and England that may hamper the oligomerization of the E protein of SARS-CoV-2, at least to some extent. Recent evidences showed that recombination and DNA contamination can affect both synonymous and nonsynonymous mutation sites equally, and can be convincingly accepted for similar amino acid sequences between the two genomes (Chaw et al., 2020). Therefore, these two mutations may have some functional consequences and be worth investigating further. Moreover, a triple cysteine motif (38-NH2-LCAYCCN-COOH-44), and similar motif located in the C-terminus of S protein of SARS-CoV were predicted to interact with each other. This interaction can serve as a structural basis between E and S proteins which would be enhanced by the disulphide bonding to the corresponding cysteine residues (Wu et al., 2003; Schoeman and Fielding, 2019). Mutations (L39M, A41S, A41V, C43F, C43R, C43S, C44Y, N45R) in this interacting motif of the E protein were also evident in different strains (Supplementary Data 1). The C43F substitution was observed in six strains from England, Saudi Arabia, and the USA whereas C43R and C44Y mutations were noticed in two different strains of SARS-CoV-2 deposited to the GISAID from Morocco. We also found C43S mutation in one of the Australian strains (Supplementary Data 1). The mutations found in the E protein may change the structural conformation of this protein and subsequently alter the associated functions such as viral assembly, replication, propagation, and pathogenesis as also previously observed in SARS-CoV and MERS-CoV (Wu et al., 2003; DeDiego et al., 2014). Structural stability analyses categorized the top 13 frequent (frequency 20–205) aa substitutions into stabilizing (n = 3), Slightly stabilizing (n = 3), neutral (n = 5), destabilizing (n = 1) and highly destabilizing (n = 1) based on the changes of Gibbs free energy (ΔΔG = ΔGmutant − ΔGWT) in FoldX (Table 2). Stability changes of E protein may affect the viral conformation and functional process that ultimately affect the pathogenesis of SARS-CoV-2 although in vivo analyses needed for confirmation (Bromberg and Rost, 2009; Rahman et al., 2020b).
Table 2.
Effects of mutation on E protein structure stability using Gibbs free energy (ΔΔG = ΔGmutant − ΔGWT) in FoldX.
| SI no. | Mutation | Number of observed strains | ΔΔG (kcal/mol) | Stability changes |
|---|---|---|---|---|
| 1 | S68F | 205 | −0.43159 | Slightly stabilizing |
| 2 | L73F | 100 | 0.091489 | Neutral |
| 3 | R69I | 88 | −0.0100619 | Neutral |
| 4 | P71L | 59 | −0.528276 | Slightly stabilizing |
| 5 | S55F | 40 | −0.170826 | Neutral |
| 6 | V62F | 39 | 3.03747 | Highly destabilizing |
| 7 | T9I | 38 | 1.34748 | Destabilizing |
| 8 | L21F | 32 | 0.332089 | Neutral |
| 9 | T30I | 29 | −0.727664 | Stabilizing |
| 10 | V24M | 26 | −1.80842 | Stabilizing |
| 11 | V75L | 22 | −1.50965 | Stabilizing |
| 12 | V58F | 20 | −0.162121 | Slightly stabilizing |
| 13 | D72H | 20 | 0.164999 | Neutral |
Both synonymous and nonsynonymous mutations in the structural proteins of the SARS-CoV-2 genomes sequenced from different countries and ethnic groups have been reported (Islam et al., 2020; Rahman et al., 2020b) which is one of the limitations of this study. Moreover, many countries have not sequenced enough virus samples (such as African and Sub-Saharan countries), and some countries uploaded sequences collected from samples of single-source or zone of infection, hence the mutation pattern may be biased in specific country or continent. This study, therefore, opens up new perspectives to determine whether one of these frequent mutations will lead to biological differences, and their correlation with different geoclimatic conditions considering the Wuhan strains as a reference.
Therefore, highly conserved E protein can be a potential target for SARS-CoV-2 vaccine and therapeutic interventions to reduce of pathogenicity. However, these computational results shed the lights on the most enigmatic protein among the structural proteins of coronaviruses, the E protein, laying the foundations for a fundamental detailed “wet” experimental investigation of its interaction with host components and comparison with E protein of SARS-CoV in standardized infection model. The identification of the nucleotides and amino acids which are involved in virulence reduction should be investigated by further studies. The results of the present study should be interpreted cautiously given the existing uncertainty of SARS-CoV-2 genomic data to develop potential prophylaxis and mitigation for tackling the pandemic COVID-19 crisis.
The following are the supplementary data related to this article.
Mutation in primer probe binding sites of SARS-CoV-2 E gene.
Data availability
We used the SARS-CoV-2 genome sequences available in the open shared database (GISAID).
Ethical statements
We confirm that the ethical policies of the journal, as noted on the journal's authors guideline page, have been adhered to. No ethical approval was required since the study didn't include any animal or human sample.
CRediT authorship contribution statement
MSR conducted the overall study. MSR, MNH, MRI, II, and IDM interpreted the results and drafted the manuscript. MNH finally compiled and edited the manuscript. MMR, MS and MAH contributed intellectually to the interpretation and presentation of the results.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
The authors would like to acknowledge the frontliners who are working restlessly in this COVID-19 pandemic situation. The authors also appreciate the researchers worldwide who were kind enough to deposit and share the complete genomes of SARS-CoV-2 and other coronaviruses to the GISAID.
References
- Alam A.S.M.R.U., Islam M.R., Rahman M.S., Islam O.K., Hossain M.A. Understanding the possible origin and genotyping of the first Bangladeshi SARS-CoV-2 strain. J. Med. Virol. 2020:1–4. doi: 10.1002/jmv.26115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal A., Destras G., Gaymard A., Bouscambert-Duchamp M., Valette M., Escuret V., Frobert E., Billaud G., Trouillet-Assant S., Cheynet V. Molecular characterization of SARS-CoV-2 in the first COVID-19 cluster in France reveals an amino acid deletion in nsp2 (Asp268del) Clin. Microbiol. Infect. 2020;26(7):960–962. doi: 10.1016/j.cmi.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., Rost B. Correlating protein function and stability through the analysis of single amino acid substitutions. Bmc Bioinformatics. 2009;10(8):1–9. doi: 10.1186/1471-2105-10-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S.-M., Tai J.-H., Chen S.-L., Hsieh C.-H., Chang S.-Y., Yeh S.-H., Yang W.-S., Chen P.-J., Wang H.-Y. The origin and underlying driving forces of the SARS-CoV-2 outbreak. J. Biomed. Sci. 2020;27:1–12. doi: 10.1186/s12929-020-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Pan Y., Cheng S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Cruz R.J., Currier A.W., Sampson V.B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Frontiers in Cell and Developmental Biology. 2020;8(468):1–11. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio F., Cascio E.L., Babini G., Sali M., Della Longa S., Tilocca B., Roncada P., Arcovito A., Sanguinetti M., Scambia G. Improved binding of SARS-CoV-2 envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020:1–6. doi: 10.1016/j.micinf.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado J., Radusky L.G., Cianferoni D., Serrano L. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35:4168–4169. doi: 10.1093/bioinformatics/btz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Choudhury P.P., Roy B. SARS-CoV2 envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112:3890–3892. doi: 10.1016/j.ygeno.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M.N., Chaudhury A., Akanda M.A.M., Hossain M.A., Islam M.T. Genomic diversity and evolution, diagnosis, prevention, and therapeutics of the pandemic COVID-19 disease. PeerJ. 2020;8:1–35. doi: 10.7717/peerj.9689. e9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E., Agoti C.N., Lewa C.S., Oketch J., Owor B.E., Otieno G.P., Bett A., Cane P.A., Nokes D.J. Recent sequence variation in probe binding site affected detection of respiratory syncytial virus group B by real-time RT-PCR. J. Clin. Virol. 2017;88:21–25. doi: 10.1016/j.jcv.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Surya W., Claudine S., Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289:12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yang X., Wang N., Wang H., Yin B., Yang X., Jiang W. The divergence between SARS-CoV-2 and RaTG13 might be overestimated due to the extensive RNA modification. Futur. Virol. 2020;15(6):341–347. [Google Scholar]
- McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6):1–6. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Won J., Choi B.Y., Lee C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020;52:963–977. doi: 10.1038/s12276-020-0452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81(104260):1–3. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet-Ul-Alam A., Siddique M.A., Saha O., Rahaman M.M., Sultana M., Crandall K.A. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2, the etiologic agent of COVID-19 pandemic: an in silico approach. PeerJ. 2020;8:1–30. doi: 10.7717/peerj.9572. e9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.S., Islam M.R., Alam A.R.U., Islam I., Hoque M.N., Akter S., Rahaman M.M., Sultana M., Hossain M.A. Evolutionary dynamics of SARS‐CoV‐2 nucleocapsid protein and its consequences. J. Med. Virol. 2020:1–19. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- Rahman M.S., Islam M.R., Hoque M.N., Alam A.R.U., Akther M., Puspo J.A., Akter S., Anwar A., Sultana M., Hossain M.A. Comprehensive annotations of the mutational spectra of SARS-CoV-2 spike protein: a fast and accurate pipeline. Transbound. Emerg. Dis. 2020:1–13. doi: 10.1111/tbed.13834. (2020; 00) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D.R., Pokhrel N. Sequence mismatch in PCR probes may mask the COVID-19 detection in Nepal. Mol. Cell. Probes. 2020;53:1–3. doi: 10.1016/j.mcp.2020.101599. (101599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Is there a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front. Microbiol. 2020;11:2086. doi: 10.3389/fmicb.2020.02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. vol. 20. Taylor & Francis; 2020. Real-time RT-PCR in COVID-19 Detection: Issues Affecting the Results; pp. 453–454. (5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Parthasarathy K., Lin X., Saravanan R., Kukol A., Liu D.X. Model of a putative pore: the pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Maheswari U., Parthasarathy K., Ng L., Liu D.X., Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A., Ma S., Malagrinò F., Visconti L., Pagano L., Stromgaard K., Gianni S. Comparing the binding properties of peptides mimicking the envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020;29:2038–2042. doi: 10.1002/pro.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Zhang Y., Lü H., Wang J., He X., Liu Y., Ye C., Lin W., Hu J., Ji J. The E protein is a multifunctional membrane protein of SARS-CoV. Genomics, Proteomics & Bioinformatics. 2003;1:131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutation in primer probe binding sites of SARS-CoV-2 E gene.
Data Availability Statement
We used the SARS-CoV-2 genome sequences available in the open shared database (GISAID).