Abstract
Preeclampsia (PE) is a pregnancy-specific complication characterized by hypertension and proteinuria, and it is one of the primary global causes of maternal and perinatal mortality. Poor remodeling of placental arteries and endothelial dysfunction serve important roles in the pathogenesis of PE. Peptide derived from complement C4 A chain (PDCC4) was identified in our previous peptidome analysis of serum from patients with PE. The present study aimed to investigate the effect of PDCC4 on endothelial dysfunction in PE. TNF-α stimulated HUVECs were employed to mimic endothelial dysfunction in PE, and Cell Counting Kit 8 assay, wound healing assay, tube formation assay, RNA-sequencing (seq) and western blot analysis were performed using HUVECs. Moreover, an in vivo model of PE was established using pregnant rats treated with lipopolysaccharide (LPS), and blood pressure monitoring, histopathological examination, ELISA and immunohistochemistry were performed on rats. It was found that TNF-α impaired proliferation, migration and tube formation of HUVECs, but pretreatment with PDCC4 moderated these effects. RNA-seq and western blotting demonstrated that the PI3K/mTOR/HIF1α signaling pathway was activated by PDCC4, and a selective PI3K inhibitor reversed the protective function of PDCC4 on TNF-α stimulated HUVECs. Additionally, PDCC4 alleviated hypertension, histopathological changes of placenta and kidney and the expression levels of endothelial injury markers and inflammatory cytokines induced by LPS in rats. These results suggested that PDCC4 relieved endothelial dysfunction in PE via PI3K/mTOR/HIF1α signaling pathway and may be a potential therapy for PE.
Keywords: novel peptide, endothelial dysfunction, preeclampsia, PI3K signaling pathway
Introduction
Preeclampsia (PE), a specific complication of pregnancy characterized by hypertension and proteinuria (1), is a threat to maternal and pediatric health and affects 5-7% of all pregnant women worldwide every year (2-4). In severe cases, PE can lead to multiorgan failure and even mortality (5,6). Low-dose aspirin treatment is reported to decrease the prevalence of hypertension in pregnancy (7-9). Moreover, trophoblast complement inhibitory therapy with heparin reduced the incidence of primary outcomes (such as severe preeclampsia, newborn weight ≤5th percentile or major abruptio placentae) in women with adverse pregnancy outcomes in antecedent pregnancy (10). As PE is life-threatening and lacks effective treatment, the development of specific therapies targeting the pathogenesis of PE is urgently required.
Although the precise mechanisms underlying the occurrence of PE remain unknown, the leading hypotheses suggest the involvement of inadequate uterine trophoblast invasion and poor remodeling of placental spiral arteries (11-13). Importantly, the widespread endothelial dysfunction caused by impaired remodeling of spiral arteries accelerates the onset of symptomatic PE via the release of anti-angiogenic factors, such as soluble fms-like tyrosine kinase 1 (sFLT-1) and soluble endoglin (sENG) (14-16). Metformin is an oral hypoglycaemic agent that is widely accepted for the treatment of gestational diabetes. Recently, Brownfoot et al (17) revealed that metformin decreased sFLT-1 and sENG secretion, as well as and improved endothelial dysfunction in patients with PE. Hence, a medication that alleviates endothelial dysfunction may demonstrate promise for the prevention and treatment of PE.
Numerous peptides have been reported to be biologically active and have been researched for therapeutic applications in various diseases, including cancer, hypertension and diabetes (18-21). Apelin has been observed to function in regulating fluid balance, local regulation of blood vessels and cardiac contractility. For instance, Ho et al (22) revealed that, in rodents, apelin prevented the development of systemic hypertension and preserved the cellular architecture of the kidney that is often impaired in PE. Currently, increasing numbers of peptides have been approved for clinical research, such as liraglutide and abaloparatide (23,24), but specific peptides available for PE treatment are limited. Our previous study systemically evaluated the peptide profile in the serum from patients with PE (25). However, additional efforts are required to examine the peptides with unknown functions.
Based on previous findings, the present study aimed to investigated the biological functions of a novel peptide derived from the complement C4 A chain (PDCC4), which has a high stability and a low molecular weight (25). Subsequently, the potential mechanism underlying the protective effects provided by PDCC4 on endothelial dysfunction were examined.
Materials and methods
Peptide synthesis and administration
The amino acid sequence of PDCC4 is NGFKSHALQLNNRQIR. PDCC4 and N-terminal fluorescein isothiocyanate-labelled PDCC4 (FITC-PDCC4) were chemically synthesized with >95% purity by Shanghai Science Peptide Biological Technology Co., Ltd. The peptide was dissolved in sterile water to a concentration of 10 mM for storage and diluted to the indicated concentration before use.
Cell culture and treatment
Human umbilical vein endothelial cells (HUVECs) were purchased from ScienCell Research Laboratories, Inc. HUVECs were cultured using DMEM (ScienCell Research Laboratories, Inc.) supplemented with 10% FBS (ScienCell Research Laboratories, Inc.) in a 37°C humidified incubator with 5% CO2. Cells were randomly divided into four groups for treatment: Control group, PDCC4 group, TNF-α group and TNF-α + PDCC4 group. The TNF-α and TNF-α + PDCC4 groups were administered 20 ng/ml TNF-α (23-25), while the control group received an equal volume of sterile water. A total of 30 min before TNF-α treatment, the PDCC4 and TNF-α + PDCC4 groups were administered 50 µM PDCC4, while the TNF-α group received an equal volume of sterile water.
HUVECs were administrated with 50 µM FITC-PDCC4 and incubated for 1 h at 37°C in the dark, then imaged with a laser confocal fluorescence microscopy (Zeiss AG; magnification, ×400) to observe the cell penetrating capacity of PDCC4. To investigate the function of PDCC4, various concentrations (10, 20, 50 and 100 µM) of PDCC4 were administrated to HUVECs and then cells were cultured for 30 min at 37°C. Subsequently, HUVECs were incubated with 20 ng/ml TNF-α (Sigma-Aldrich; Merck KGaA) for 24 h at 37°C to induce endothelial dysfunction, as previously described (26-28). Additionally, to investigate the effect of inhibiting PI3K on function of PDCC4, HUVECs were preconditioned with 2 µM LY294002 (Sigma-Aldrich; Merck KGaA) for 1 h at 37°C before administration of PDCC4.
5-Ethynyl-2′-deoxyuridine (EdU) assay
HUVECs were seeded in 24-well plates at a concentration of 5,000 cells/well, and were then incubated for 12 h at 37°C. After cells attached, HUVECs were treated as aforementioned for 24 h. Then, the proliferation of the HUVECs was measured with an EdU assay kit (Beyotime Institute of Biotechnology) as previously described (29). EdU was added to the culture medium at a concentration of 10 µM. After incubation at 37°C for 2 h, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were washed twice with PBS and treated with 0.3% Triton X-100 for 10 min at room temperature. Finally, Alexa Fluor 488 azide and DAPI were used successively to stain the cells for 30 and 10 min in the dark at room temperature. Images were acquired using a fluorescence microscope (magnification, ×200) (30,31).
Wound healing assays
HUVECs were seeded in 6-well plates at a concentration of 5×105 cells per well and cultured until they reached confluency. After the cells were treated as aforementioned, a scratch was made down the center of the layers of cells in each well using a sterile 1,000 µl micropipette tip. Cells were then washed with PBS twice and cultured in medium without FBS. Images of three random visual fields of each group were captured using a bright-field microscope (magnification, ×40) at 0 and 24 h after scratching. The width of the healed wound was quantified using ImageJ software (v1.52a; National Institutes of Health), and the migratory rate was calculated (32,33).
Tube formation assay
A total of 100 µl liquefied Matrigel (Becton, Dickinson and Company) was added to the 24-well plate. The plate was incubated at 37°C until the Matrigel solidified. Subsequently, 200 µl HUVECs (1×105 cells/ml) that were treated as aforementioned were seeded in a 24-well plate. The preliminary experiment indicated that untreated HUVECs completely formed tubular structures on the Matrigel after 6-8 h of incubation (data not shown). Thus, the tube formation of the HUVECs was observed using a bright-field microscope (magnification, ×200) at 6-8 h, as also previously described (34-36). The branch number and length of the tubes were quantified using ImageJ software (v1.52a; National Institutes of Health).
Mitochondrial membrane potential assay
HUVECs were seeded in 6-well plates at a concentration of 5×105 cells per well for treatment as aforementioned. Then, a 5,5′,6,6′,tetra-chloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) staining kit (Beyotime Institute of Biotechnology) was used according to the manufacturer's instructions to detect the mitochondrial membrane potential of the HUVECs (37,38). Briefly, the culture medium in each well was replaced with JC-1 staining medium (5 µg/ml). After a 20 min incubation in the dark at 37°C, HUVECs were washed twice with PBS and a fluorescence microscope (magnification, ×400) was used to capture images of the mitochondria.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay kit (cat. no. CK04-13; Dojindo Molecular Technologies, Inc.) was applied to evaluate the effect of different concentrations of PDCC4 on the proliferation of HUVECs according to the manufacturer's instructions. The cells were seeded in 96-well plates at a concentration of 3,000 cells/well. After the cells were attached, they were pretreated with PDCC4 (10, 20, 50 and 100 µM) for 30 min at 37°C and then cotreated with 20 ng/ml TNF-α in a 37°C humidified incubator with 5% CO2. At 0, 24, 48 and 72 h, 10 µl CCK-8 solution was added to the medium at 37°C (39,40). The absorbance at 450 nm was measured using a multifunctional microplate reader (Hybrid Technology; BioTek Instruments, Inc.).
Animal protocol
All animal studies were approved by the Animal Care and Ethical Committee of Nanjing Medical University. Animal experiments were performed according to the Guide for the Care and Use of Laboratory Animals (41). A total of 72 Sprague-Dawley (SD) rats (48 female, 24 male; age, 8 weeks; weight, 280-320 g) were purchased from the Animal Center of Nanjing Medical University. The rats were housed with 60% humidity and a 12-h light/dark cycle at 26°C and free to standard rat chow and water. Female rats were mated with males after 7 days of acclimatization. Gestational day (GD) 0 was defined via a vaginal smear analysis of sperm. On GD14, the female SD rats were randomly divided into four groups (n=12 rats in each): Saline group, Saline + PDCC4 group, LPS group and LPS + PDCC4 group. On GD14, the rats in the LPS group and LPS + PDCC4 group were intravenously injected with LPS (Sigma-Aldrich; Merck KGaA; 1 µg/kg dissolved in 2 ml sterile saline) via an infusion pump (infusion rate, 2 ml/h) as previously described (42), while the rats in the Saline group and Saline + PDCC4 group were injected with an equal volume of saline via an infusion pump (infusion rate, 2 ml/h). Rats in the Saline + PDCC4 group and LPS + PDCC4 group were intravenously injected with PDCC4 (10 mg/kg) once per day on GD16, 17 and 18, while the rats in the Saline group and LPS group were injected with an equal volume of saline.
Measurement of blood pressure, urinary protein and creatinine
On GD7, 11, 14, 16 and 18, a non-invasive volume-pressure recording blood pressure monitoring system (Visitech BP2000; VisiTech International Ltd.) was employed to measure the systolic blood pressure (SBP) of the rats.
Beginning on GD19, 5 ml of 24-h urine was collected from the rats using individual metabolic cages. Then, the concentration of urinary protein was determined using the pyrogallol red method as previously described (43), and the concentration of creatinine was assayed using a commercial kit (cat. no. C011-2; Nanjing Jiancheng Bioengineering Institute), according to the manufacturer's instructions.
Pathological examination
On GD20, the rats were anaesthetized and sacrificed. Then, the fetuses, placentas and kidneys were isolated. Living and dead fetuses were counted, and the live fetuses were weighed. The placentas and kidneys were fixed with 4% paraformaldehyde overnight at room temperature, embedded in paraffin and sliced into 3-µm thick sections. Then, tissue sections were subjected to hematoxylin and eosin (H&E) staining, which was performed as previously described (44,45), and sections were imaged using light microscopy (magnification, ×100).
Immunohistochemistry
The protein expression levels of proinflammatory and anti-inflammatory factors in rat placentas were estimated via immunohistochemistry analysis. Paraffin-embedded sections of placentas were washed with xylene and ascending alcohol, for air-drying and hydration, and were then incubated in Tris EDTA buffer for 15 min at 95°C for antigen retrieval. The paraffin sections were incubated at room temperature in 3% hydrogen peroxide solution for 20 min to eliminate endogenous peroxidase activity. The sections were incubated in PBS containing 3% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 15 min at room temperature to block non-specific binding. The sections were incubated overnight in a 4°C humidified incubator with primary antibodies against IL-4 (1:200; cat. no. bs-20686R; BIOSS) and IL-6 (1:200; cat. no. bs-0782R; BIOSS). Then, tissue sections were washed three times with PBS, and incubated with a secondary antibody (1:100; cat. no. bs-0295G-Bio; BIOSS) and a tertiary antibody (1:500; cat. no. bs-0437P-HRP; BIOSS) in a 37°C incubator for 30 min. Finally, the labelled antibody was visualized with a 3,3′-diaminobenzidine kit (46). The stained sections were imaged using light microscopy (magnification, ×100), and images were loaded to ImageJ software (v1.52a; National Institutes of Health) to quantify the expression of proteins.
ELISA
A total of 500 µl blood was drawn via the tail vein before sacrifice. The supernatant serum was collected after the blood samples centrifuged at 1,500 × g for 15 min at 4°C. The sFlt-1 and sEng levels were determined according to the manufacture's instruction of the ELISA kits (cat. nos. CSB-E07350r and CSB-EL007667RA; Cusabio Technology LLC). The concentrations were measured according to the absorbance of the samples and standards at the 450 nm wavelength (47).
RNA sequencing (seq)
HUVECs were lysed using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and total RNA was extracted using RNAprep pure Cell/Bacteria kit (cat. no. DP430; Tiangen Biotech Co., Ltd.), and then the integrity of total RNA was inspected via a Bioanalyzer 2100 (Agilent Technologies, Inc.). Qualified total RNA was further purified using RNA Clean XP kit (Beckman Coulter, Inc.) and RNase-Free DNase Set (Qiagen AB) according to the manu-facturer's instructions. Ribosomal RNA (rRNA) was removed from total RNA using the Ribo-Zero™ Gold kit (Epicentre; Illumina, Inc.), and fragmented rRNA-depleted RNA was converted to double-stranded cDNAs using ScriptSeq™ v2 RNA-Seq Library Preparation kit (Epicentre; Illumina, Inc.) according to the manufacturer's instructions. cDNAs were used for end repair, 3′ adenylation, adapter ligation and cDNA enrichment using a VAHTS Stranded mRNA-seq Library Prep kit (Illumina, Inc.). The quality of libraries and quantification were inspected using Bioanalyzer 2100 and qPCR with a Library Quantification kit (KAPA Biosystems; Roche Diagnostics, Inc.). Then, 4 nM cDNA libraries were run on HiSeq. 2500 platform (Illumina, Inc.) in rapid run mode for 2X 100 bp paired-end reads using a TruSeq PE Cluster kit and a TruSeq SBS kit (cat. nos. PE-401-3001 and FC-401-3001; Illumina, Inc.) and data were summarized, normalized and quality controlled using HiSeq Control software (v 2.2.68; Illumina, Inc.).
To analyze the differential expression of genes, the raw reads were normalized to Fragments per Kilobase of exon per Million mapped reads. The t-test results and P-values were adjusted for the False Discovery Rate (FDR) using the Benjamini-Hochberg procedure, and a FDR-adjusted P-value<0.05 was considered the threshold for statistical significance. Differentially expressed genes were uploaded to Database for Annotation, Visualization and Integrated Discovery (v6.8; https://david.ncifcrf.gov/tools.jsp) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Reverse transcription-quantitative PCR (RT-qPCR)
Cells and placenta tissues of rats were lysed using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was extracted with a Total RNA Isolation kit (Tiangen Biotech Co., Ltd.) and then subjected to reverse transcription using a SuperScript IV kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. cDNA was synthesized at 50°C for 10 min and 80°C for 10 min. RT-qPCR was performed using PowerUp™ SYBR™ Green Master mix (Thermo Fisher Scientific, Inc.) and an ABI ViiA 7 System (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The following thermocycling conditions were used: Initial denaturation at 95°C for 120 sec, denaturing at 95°C for 15 sec and extension at 60°C for 60 sec for 40 cycles. The primer sequences are listed in Table SI. Expression values of targeted genes were normalized to the values of GAPDH using the 2−ΔΔCq method (48).
Western blotting
HUVECs were lysed with RIPA buffer (Beyotime Institute of Biotechnology), and total protein was extracted. The protein concentration was measured using the BCA method (Beyotime Institute of Biotechnology). After processing with loading buffer, proteins (30 µg) were subjected to electrophoresis with a 10% polyacrylamide gel. Subsequently, the proteins were transferred to PVDF (EMD Millipore) membranes. After blocking with 5% fat-free milk for 1 h at room temperature, the membranes were incubated with primary antibodies at 4°C overnight. The primary antibodies used in this study were phosphorylated (p)-PI3K (1:2,000; cat. no. bs-3332R; BIOSS), PI3K (1:2,000; cat. no. 4249; Cell Signaling Technology, Inc.), p-mTOR (1:2,000; Ser2481; cat. no. ab137133; Abcam), mTOR (1:2,000; cat. no. 2983; Cell Signaling Technology, Inc.), HIF1α (1:2,000; cat. no. 36169; Cell Signaling Technology, Inc.) and β-actin (1:5,000; cat. no. 20536-1-AP; ProteinTech Group, Inc.) (49). After washing three times with TBST containing 0.1% Tween-20 (10 min every time) at room temperature, the membranes were incubated with goat anti-rabbit IgG horseradish peroxidase (1:2,000; cat. no. ab205718; Abcam) for 1 h at room temperature. After washing again with TBST, the immunoblots were visualized using chemiluminescence reagents (EMD Millipore) and grey value of immunoblots were semi-quantified using ImageJ software (v1.52a; National Institutes of Health).
Statistical analysis
GraphPad Prism 7 software (GraphPad Software, Inc.) was used for statistical analysis and data plot-ting. Quantitative data are presented as the mean ± SD. Data with two groups were compared using the two-tailed Student's t-test, and multiple group comparisons were determined via one-way ANOVA and Bonferroni's multiple comparison test. χ2 tests were used to compare survival rate of fetal rats. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed for three independent times with six duplications each time.
Results
PDCC4 at a concentration of 50 µM has a significant effect on the expression of biomarkers of endothelial dysfunction
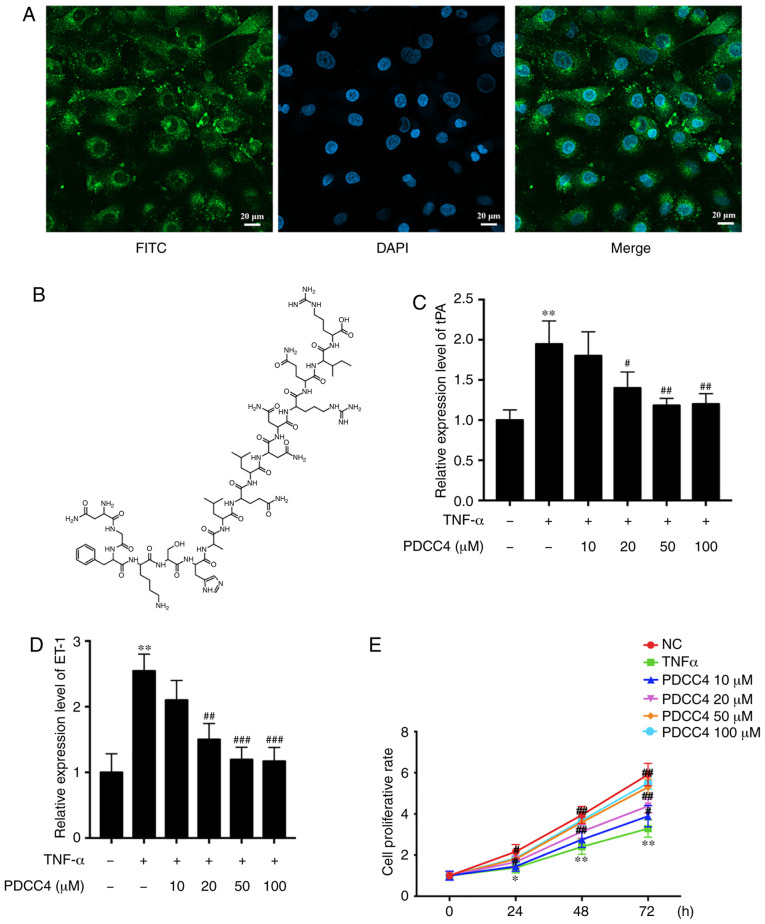
The ability of PDCC4 to penetrate cells was first evaluated, before analysis of its function in HUVECs. To track the delivery of the peptide, PDCC4 was conjugated to FITC. HUVECs were treated with 50 µM FITC-labelled PDCC4 for 1 h. The fluorescent images demonstrated that PDCC4 crossed the membrane, and the majority was located in the cytoplasm, suggesting a favorable cell-penetrating ability of PDCC4 (Fig. 1A). The chemical formula of PDCC4 is presented in Fig. 1B.
Figure 1.
PDCC4 is located in the cytoplasm and reduces the expression of markers of endothelial dysfunction induced by TNF-α. (A) Representative fluores-cent image of HUVECs after incubation with FITC-labelled PDCC4 for 1 h and staining with DAPI indicating the cell-penetrating and cytoplasm-localized properties of PDCC4. Scale bar, 20 µm. (B) Chemical formula of PDCC4. Reverse transcription-quantitative PCR indicated that PDCC4 moderated the increase in (C) tPA and (D) ET-1 expression levels induced by TNF-α in a dose-dependent manner. (E) HUVECs were stimulated with different concentrations of PDCC4 (10, 20, 50 and 100 µM) and assessed using a Cell Counting Kit-8 assay (n=3 per group). *P<0.05 and **P<0.01 vs. NC group; #P<0.05, ##P<0.01 and ###P<0.001 vs. TNF-α (+) group. HUVEC, human umbilical vein endothelial cells; PDCC4, peptide derived from complement C4 A chain; ET-1, endothelin-1; tPA, tissue plasminogen activator; NC, negative control.
To investigate the biological function of PDCC4 in HUVECs, biomarkers of endothelial dysfunction were detected via RT-qPCR following treatment of cells with different concentrations of PDCC4. A dose-dependent decline in the mRNA expression of relevant markers, including tissue plasminogen activator (tPA) and endothelin-1 (ET-1), was observed after PDCC4 treatment (Fig. 1C and D). Compared with the TNF-α treatment alone group, the group treated with 50 µM PDCC4 had the most significant effect on the mRNA expression levels of tPA and ET-1.
PDCC4 moderates the suppressed proliferation of TNF-α-induced HUVECs
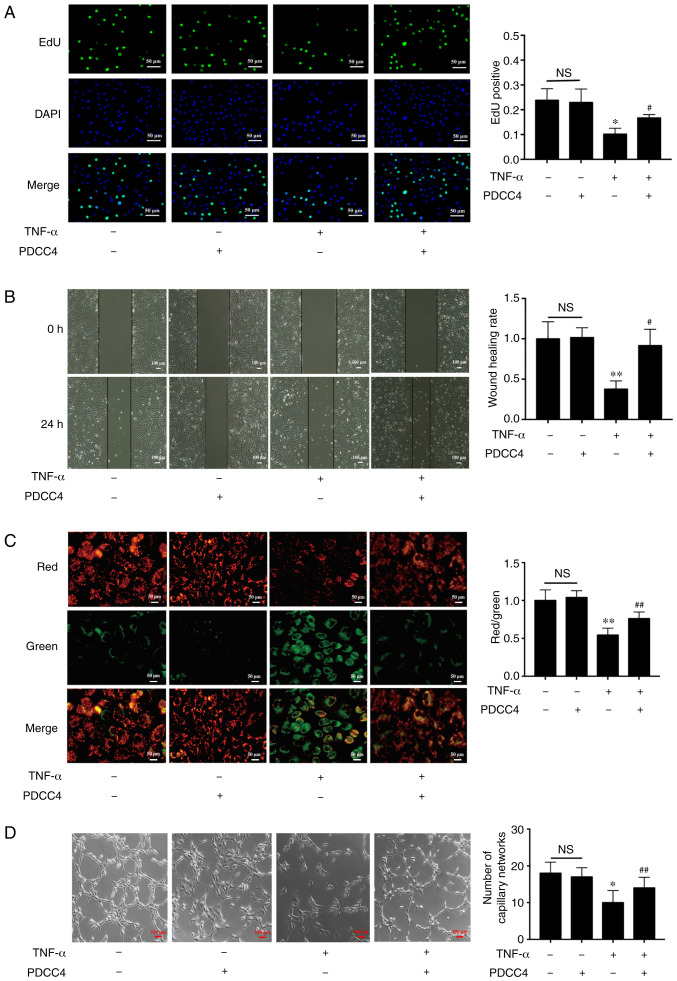
A CCK-8 assay was conducted to determine whether PDCC4 (10, 20, 50 and 100 µM) reversed the decreased proliferation capacity of HUVECs. The results demonstrated that PDCC4 moderated proliferation inhibition induced by TNF-α in a dose-dependent manner (Fig. 1E). PDCC4 had the most significant effect on the proliferation capacity of HUVECs at 50 µM. Therefore, a concentration of 50 µM was selected as the appropriate concentration for further study. An EdU assay was performed to measure the effect of PDCC4 on HUVEC proliferation. EdU was used to label newly synthesized DNA with green fluorescence. The total DNA was determined via staining with DAPI (blue fluorescence). Proliferation is indicated as the green/blue ratio. It was found that the proliferation of HUVECs was significantly inhibited in the TNF-α group, but that this inhibition was reversed in the TNF-α + PDCC4 group (Fig. 2A).
Figure 2.
PDCC4 rescues the impaired proliferation, migration, mitochondrial function and tube formation of HUVECs treated with TNF-α. (A) Proliferative HUVECs were treated with EdU, representative fluorescent images were captured and the rate of EdU labelling was calculated. The proliferation of HUVECs was inhibited by TNF-α, and this change was rescued by PDCC4 (n=3 per group). Scale bar, 50 µm. (B) Representative images of wound healing were captured at 0 and 24 h after scratching the cells, and the wound healing rate was calculated. PDCC4 prevented the migration of HUVECs from inhibition by TNF-α (n=3 per group). Scale bar, 100 µm. (C) Representative fluorescent images of HUVECs after staining with JC-1; the mitochondrial membrane potential is presented as the ratio of red to green fluorescent density. PDCC4 increased the mitochondrial membrane potential of HUVECs exposed to TNF-α (n=3 per group). Scale bar, 50 µm. (D) Representative images of the tube formation assays indicated that PDCC4 preserved the HUVEC tube formation capacity, which was impaired by TNF-α (n=3 per group). Scale bar, 100 µm. *P<0.05 and **P<0.01 vs. negative control group; #P<0.05 and ##P<0.01 vs. TNF-α (+) group. HUVEC, human umbilical vein endothelial cells; PDCC4, peptide derived from complement C4 A chain; EdU, 5-ethynyl-2′-deoxyuridine; NS, not significant.
PDCC4 alleviates the damage to HUVEC migration and tube formation
The results suggested that the migratory capacity of TNF-α-induced HUVECs was suppressed compared with that of the control group. After treatment with PDCC4, the impaired migratory function was significantly reversed (Fig. 2B). Representative images indicated that the number of branch points of the tube-like structures of the TNF-α + PDCC4 group was greater compared with that of the TNF-α group, suggesting that tube formation was enhanced by PDCC4 treatment (Fig. 2D).
PDCC4 relieves the mitochondrial depolarization induced by TNF-α
By examining mitochondrial depolarization, the protective effect of PDCC4 on TNF-α-induced cell death in HUVECs were further determined (Fig. 2C). Control HUVECs stained with JC-1 emitted mitochondrial orange-red fluorescence and exhibited little green fluorescence. The fluorescence images in the TNF-α group demonstrate obvious green fluorescence and little orange-red fluorescence. Compared with the TNF-α group, the TNF-α + PDCC4 group had decreased mitochondrial depolarization and notable orange-red fluorescence.
PI3K/mTOR/HIF1α pathway may be involved in the PDCC4-mediated rescue of endothelial dysfunction
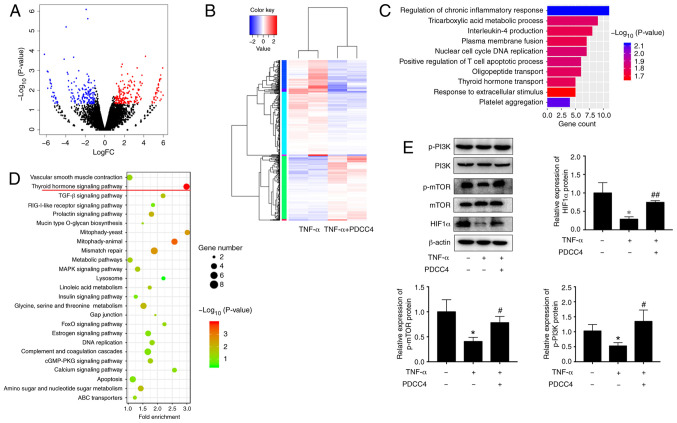
To determine the underlying mechanism of PDCC4 function, the transcriptome after PDCC4 treatment was examine via RNA-seq. A total of 442 genes, including 178 downregulated genes and 264 upregulated genes, had significant differences when comparing the group treated with PDCC4 and the control group (Fig. 3A and B). GO analysis identified enrichment in important biological processes, which included the 'regulation of chronic inflammatory response' and 'thyroid hormone transport', indicating that the relevant genes may be involved in these processes (Fig. 3C). KEGG pathway analysis was also applied to analyze the dysregulated genes involved in various signaling pathways, and it was observed that the 'thyroid hormone signaling pathway' had the highest enrichment (Fig. 3D).
Figure 3.
PDCC4 reverses the dysregulation of the PI3K/mTOR/HIF1α pathway in response to TNF-α treatment. (A) HUVECs in the TNF-α group and TNF-α + PDCC4 group were sent for RNA-sequencing analysis. Differentially expressed genes were identified with cut-off values of P<0.05 and |logFC|>1. (B) A heat map of the differentially expressed genes is presented, in which red represents high expression and blue represents low expression. (C) Gene Ontology and (D) Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed with the differentially expressed genes. (E) Immune blotting indicated that the dysregulation of the PI3K/mTOR/HIF1α pathway induced by TNF-α was reversed by PDCC4 treatment (n=3 per group). *P<0.05 vs. negative control group; #P<0.05 and ##P<0.01 vs. TNF-α (+) group. HUVEC, human umbilical vein endothelial cells; PDCC4, peptide derived from complement C4 A chain; HIF1α, hypoxia inducible factor-1α; p-, phosphorylated.
Previous studies have revealed that key proteins of the thyroid hormone signaling pathway, including PI3K, mTOR and HIF1α, can be involved in the pathological process of PE development (50,51). Therefore, it was hypothesized that PDCC4 may function via the PI3K/mTOR/HIF1α signaling pathway. Western blot analyses were conducted to assess the key proteins involved in these pathways. After treatment with PDCC4, increase in protein expression levels were observed for p-PI3K, mTOR and total HIF1α (Fig. 3E). These results suggested that the activation of the PI3K/mTOR/HIF1α signaling pathway may be associated with the functions of PDCC4.
Relationship between the PI3K/mTOR/HIF1α pathway and the role of PDCC4 in relieving endothelial dysfunction
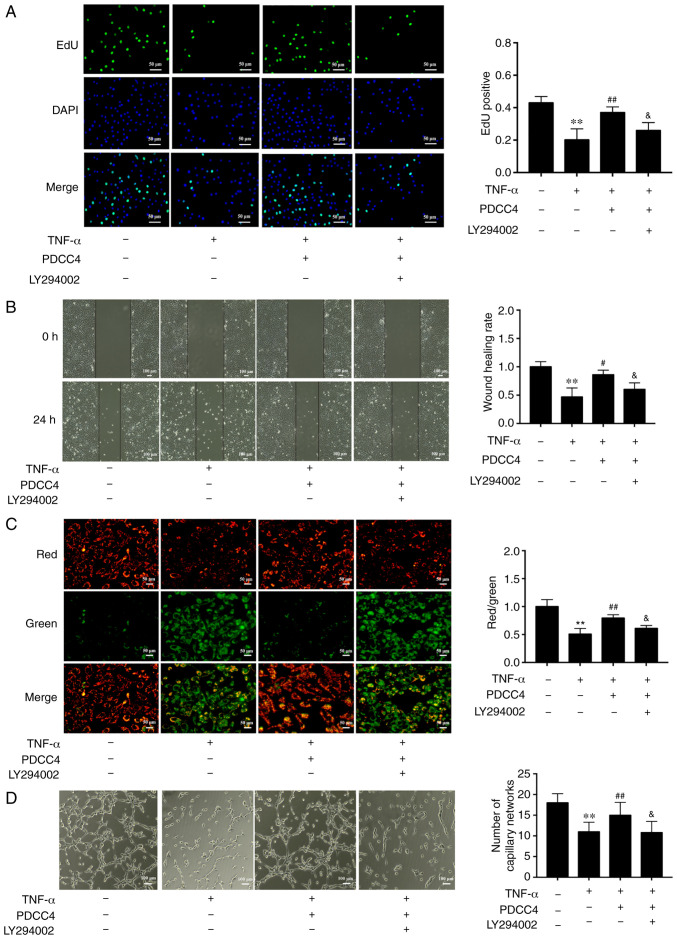
To further determine the relationship between the PI3K/mTOR/HIF1α pathway and PDCC4 functions, the highly selective PI3K inhibitor LY294002 was used to treat HUVECs. The results demonstrated that administration of 2 µM LY294002 inhibited the proliferation and migration promoted by PDCC4 (Fig. 4A and B). Moreover, LY294002 dampened the reversal effect of PDCC4 on impaired mitochondrial membrane potential induced by TNF-α (Fig. 4C). However, treatment of LY294002 alone had no effect on mitochondrial membrane potential of HUVECs (data not shown). The number of branch points of tube-like structures was also diminished following treatment with the inhibitor LY294002 in comparison with the TNF-α + PDCC4 group (Fig. 4D).
Figure 4.
PDCC4 attenuates the endothelial dysfunction induced by TNF-α via the PI3K/mTOR/HIF1α pathway. (A) Proliferative HUVECs were treated with EdU, representative fluorescent images were captured, and the rate of EdU labelling calculated. PDCC4 attenuated the antiproliferative effect of TNF-α, which was inhibited by LY294002 (n=3 per group). Scale bar, 50 µm. (B) Representative images of wound healing were captured at 0 and 24 h after scratching, and the wound healing rate was calculated. PDCC4 attenuated the antimigration effect of TNF-α, which was inhibited by LY294002 (n=3 per group). Scale bar, 100 µm. (C) Representative fluorescent images of HUVECs after staining with JC-1 are illustrated, and the mitochondrial membrane potential is presented as the ratio of red to green fluorescent density. LY294002 inhibited PDCC4′s ability to increase the mitochondrial membrane potential of HUVECs exposed to TNF-α (n=3 per group). Scale bar, 50 µm. (D) Representative images of the tube formation assay indicated that PDCC4 prevented HUVEC tube formation from being impaired by TNF-α, which was inhibited by LY294002 (n=3 per group). Scale bar, 100 µm. **P<0.01 vs. negative control group; #P<0.05 and ##P<0.01 vs. TNF-α (+) group; &P<0.05 vs. TNF-α (+) + PDCC4 group. HUVEC, human umbilical vein endothelial cells; PDCC4, peptide derived from complement C4 A chain; EdU, 5-ethynyl-2′-deoxyuridine.
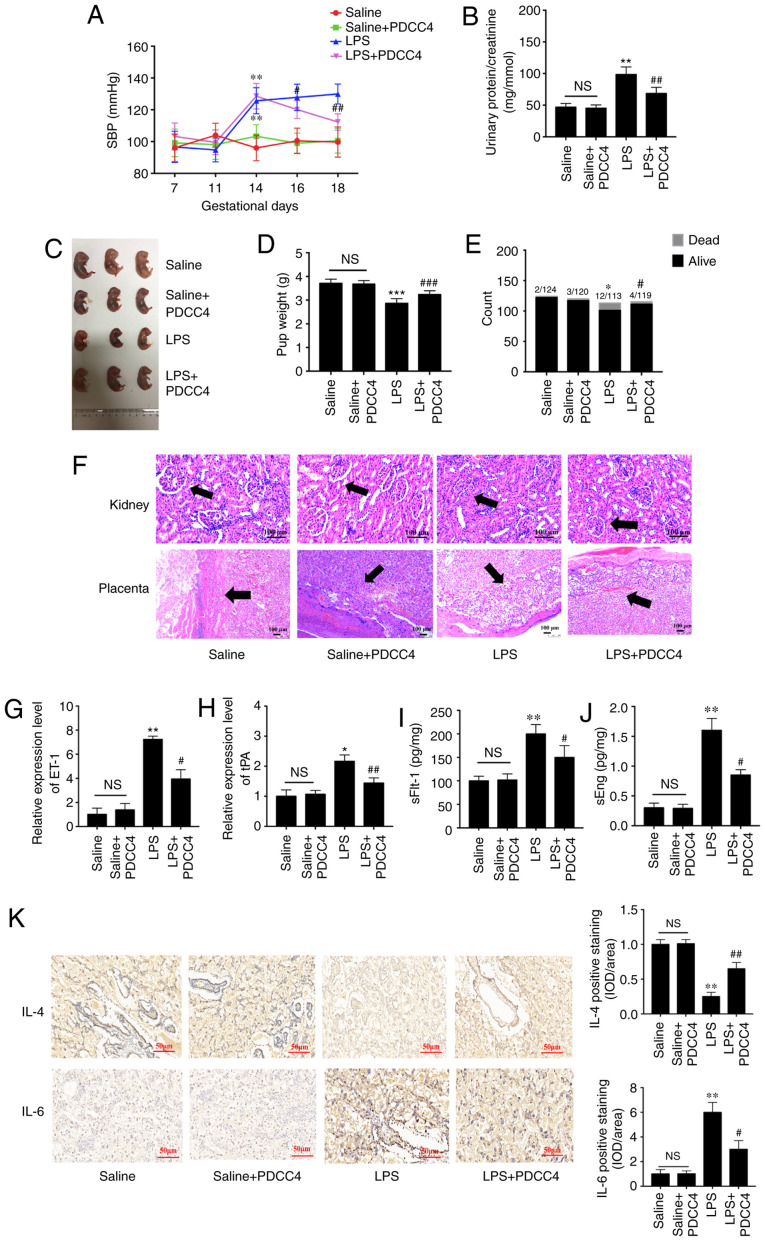
PDCC4 moderates the pathological process of LPS-induced PE in a rat model
To evaluate the effects of PDCC4 on LPS-stimulated rat, a three treatment protocol (5, 10 and 20 mg/kg) was established in the preliminary experiment. It was found that 5 mg/kg had no effect on SBP and the urinary protein of rat, while 10 and 20 mg/kg significantly attenuated LPS induced hypertension and proteinuria. However, no obvious difference between the effect of 10 and 20 mg/kg was observed (data not shown). Thus, 10 mg/kg PDCC4 was used in the following experiments. There were significant differences in the SBP values among the four groups on GD16 after treatment with LPS. The high SBP values in the LPS group persisted after treatment with only LPS during pregnancy. Treatment with 10 mg/kg PDCC4 significantly alleviated the SBP values compared with the LPS group (Fig. 5A). The ratio of urinary protein and creatinine in the LPS group was increased, while PDCC4 treatment decreased the urinary protein level (Fig. 5B). There were more live fetuses in the LPS + PDCC4 group compared with in the LPS group (Fig. 5C). In addition, the LPS + PDCC4 group had an increase in the birth weight of survivors compared with the LPS group (Fig. 5D). The numbers of live fetuses and resorptions were significantly different among the groups (Fig. 5E). Stillbirth was induced by LPS, resulting in an increased fetal mortality rate, and the LPS + PDCC4 group had a decreased rate of stillbirth (Fig. 5E).
Figure 5.
PDCC4 relieves the pathological manifestations of preeclampsia in a rat model. (A) SBP of the four groups was evaluated on GD7, 11, 14, 16 and 18. The SBP of the rats increased significantly after treatment with LPS, and this change was rescued by PDCC4 (n=18 per group). (B) Ratio of urinary protein to creatinine in the four groups was analyzed (n=12 per group). (C) Representative pictures of fetal appearance were captured for the four groups (n=12 per group). (D) Fetal weight and the (E) numbers of live and dead fetuses in each group were determined (n=12 per group). (F) Pathological alterations of the kidneys and placentas were observed after hematoxylin and eosin staining. Black arrows indicated glomerulus in kidney (upper) and basal zone in placenta (lower). The PDCC4-treated group exhibited mild inflammation compared with the LPS-treated group in a microscopic analysis of morphology. Scale bar, 100 µm. PDCC4 decreased the expression levels of markers of endothelial dysfunction, (G) ET-1 and (H) tPA, in the rat placentas (n=12 per group). Increasing serum levels of (I) sFlt-1 and (J) sEng in each group were declined by PDCC4 (n=12 per group). (K) Immunohistochemical staining demonstrated that PDCC4 decreased proinflammatory cytokines (IL-6) and increased anti-inflammatory cytokines (IL-4) in PE rats (n=12 per group). Scale bar, 50 µm. *P<0.05, **P<0.01 and ***P<0.001 vs. negative control group; #P<0.05, ##P<0.01 and ###P<0.001 vs. PE group. SBP, systolic blood pressure; GD, gestational day; PDCC4, peptide derived from complement C4 A chain; tPA, tissue plasminogen activator; ET-1, endothelin; sFlt-1, soluble fms-like tyrosine kinase 1; sEng, soluble endoglin; NS, not significant; LPS, lipopolysaccharide; OD, optical density.
Changes in kidney structure and function in the pregnant rats injected with LPS exhibited an increased glomerular area, capillary structure disorder and abnormal protein production. After treatment with PDCC4, the renal histology of pregnant rats was significantly relieved compared with that of the LPS group (Fig. 5F). For instance, the placentas of the LPS-treated rats mostly displayed severe inflammation, whereas those of the LPS + PDCC4 group generally exhibited less severe inflammation (Fig. 5F).
The expression levels of markers of endothelial dysfunction, including tPA and ET-1, in the placentas were detected. The results demonstrated that LPS increased the expression levels of these marker, while PDCC4 decreased the mRNA expression levels (Fig. 5G and H). ELISA was conducted to investigate the effect of different treatments on circulating angiogenic factors, such as sFlt-1 and sEng. The LPS group had a significant increase in the sFlt-1 and sEng levels compared with the Saline group, and this increase was attenuated by PDCC4 (Fig. 5I and J). The expression of the proinflammatory cytokine IL-6 in the placentas was increased in the LPS-treated group compared with the Saline group. By constant, this expression level was significantly decreased by PDCC4 (Fig. 5K). The anti-inflammatory cytokine IL-4 was significantly downregulated in the LPS group but upregulated in the LPS + PDCC4 group (Fig. 5K).
Discussion
PE is unique to human pregnancy, and the most effective treatment for this complication is delivery (52). It is widely accepted that disturbances in placental blood vessel function and in placental development contribute to the pathophysiology of PE (53-55). However, the etiology of PE is yet to be fully elucidated. In the present study, PDCC4 was identified as a novel peptide that moderated endothelial dysfunction by regulating the PI3K/mTOR/HIF1α pathway. The present findings indicated that PDCC4 was a promising candidate for PE treatment.
Various peptides have been researched for potential applications in the diagnosis and treatment of PE. For example, Szabo et al (56) reported that increased B-type natriuretic peptide (BNP) levels serve as biomarkers for severe PE. Additionally, BNP levels in patients with early-onset PE are significantly higher compared with those in patients late-onset PE (57). VG1177, a competitive antagonist peptide for MHC class II invariant chain peptide, could also both prevent and treat PE-like features in mice (58).
It has been reported that immune system activation serves a major role in the pathophysiology of PE, characterized by imbalanced proinflammatory and anti-inflammatory cytokines (49). In patients with PE, circulating proinflammatory cytokines, such as TNF-α, are increased in maternal and cord blood, resulting in endothelial dysfunction via diverse mechanisms, such as oxidative stress (59). Other proinflammatory cytokines, such as IL-6, IL-8 and monocyte chemotactic protein 1, are secreted by endothelial cells in response to TNF-α (60). Additionally, there is an increase in the expression levels of tPA and ET-1, two biomarkers of endothelial dysfunction (61,62). In the present study, treatment with PDCC4 resulted in a decline in the expression of IL-6 in placental tissue. The peptide PDCC4 also attenuated endothelial dysfunction, as demonstrated by a decrease in the expression levels of tPA and ET-1 both in vitro and in vivo. Several clinical studies have revealed that serum and placental anti-inflammatory cytokines, IL-4 and IL-10, are decreased in patients with PE. Consistently, the present results indicated that the expression of IL-4 in the placentas of PE-like rats was decreased, but it was upregulated in the PDCC4-treated group. Therefore, PDCC4 may participate in the regulation of immune responses and moderate endothelial dysfunction.
PE is a complicated pathological process involving various molecules and signaling pathways. For instance, PI3Ks modulate cell metabolism, proliferation and survival by phosphorylating the 3-OH group of inositol membrane lipids to activate or deactivate intracellular proteins (63,64). mTOR, one of the conserved PI3Ks, integrates various biochemical and growth factor signals, such as glucose, insulin, amino acids and ATP (65). Previous studies have revealed a decline in the phosphorylation of PI3K and mTOR in response to oxidative stress in PE (47,50,66). Consistently, the present findings indicated that the expression levels of p-PI3K and p-mTOR were downregulated in response to endothelial dysfunction induced by TNF-α, which was attenuated by PDCC4. The expression of HIF1α is shown to be upregulated in response to activation of the PI3K/mTOR signaling pathway, which contributes to neovascularization in benign and malignant diseases (51,67,68). In the present study, the expression of HIF1α was downregulated in response to TNF-α treatment and was partially rescued by PDCC4 in vivo or in vitro, which suggested that neovascularization promotes PDCC4. Additionally, inhibition of PI3K decreased the proliferation, migration and tube formation of HUVECs. It was also identified that the mitochondrial potential of HUVECs increased after PI3K inhibition. Therefore, the current results suggested that PDCC4 relieved endothelial dysfunction by influencing the PI3K/mTOR/HIF1α signaling pathway.
To investigate the anti-PE effect of PDCC4 in vivo, SD rats were injected with LPS on GD14 to establish a model of PE. To the best of our knowledge, PDCC4 is a novel peptide discovered for the first time by our previous study, and its pharmacokinetic parameters mostly remain unknown. PDCC4 treatment decreased SBP and urinary protein levels in rats with PE, which was similar to the major clinical characteristics observed in patients with PE (4). As later clinical manifestations include poor placental vessel remodeling and placental ischemia, there is an increased prevalence of fetal growth restriction and low birth weight (69). In the present study, the low fetal weight and fetal length in rats with PE were attenuated by PDCC4 treatment. Aberrant immune responses during pregnancy are considered a major pathogenic characteristic of PE (70). In the current study, not only were the relative expression levels of inflammatory cytokines and chemokines elevated, as described previously (71), but the placenta and kidney tissues from the rats exhibited inflammatory cell infiltration and tissue necrosis, which were relieved by PDCC4 treatment.
In the current research, the anti-PE effect of PDCC4 was verified both in vitro and in vivo, and an underlying mechanism was proposed. However, due to the unavailability of liquid chromatography and mass spectrum equipment, it was not possible to detect the exact amounts of PDCC4 in rats. Moreover, short duration between first PDCC4 intervention and SBP detection may partially interfere the evaluation of the effects of PDCC on LPS induced hypertension. In a future investigation, the effects of PDCC on SBP will be investigated for a longer duration. Additionally, the direct target of PDCC4 remains unknown and requires further research.
In conclusion, the present study provides insight into a newly identified peptide that moderates vascular endothelial cell injury by activating the PI3K/mTOR/HIF1α signaling pathway, which is promising for PE treatment.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant nos. 81771604, 81571444 and 81971393), the Nanjing Science and Technology Development Project (grant no. 201715052) and The Project of Invigorating Health Care through Science, Technology and Education-Jiangsu Provincial Medical Youth Talent (grant no. QNRC2016112).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LX, RJ and HD designed the present study, which was performed by LX, KX, LW and XY. LX, KX, LW and XY made substantial contributions to acquisition and analysis of data. WL and CL also made contributions to interpretation of data. LX wrote the initial draft of the manuscript. KX revised it critically for important intellectual content. RJ and HD gave final approval of the version to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal studies were approved by the Animal Care and Ethical Committee of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mol BWJ, Roberts CT, Thangaratinam S, Magee LA, de Groot CJM, Hofmeyr GJ. Preeclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 2.Khan KS, Wojdyla D, Say L, Gulmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: A systematic review. Lancet. 2006;367:1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 3.Duley L. The global impact of preeclampsia and eclampsia. Semin Perinatol. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Rana S, Lemoine E, Granger JP, Karumanchi SA. Preeclampsia: Pathophysiology, challenges, and perspectives. Circ Res. 2019;124:1094–1112. doi: 10.1161/CIRCRESAHA.118.313276. [DOI] [PubMed] [Google Scholar]
- 5.Wirka RC, Quertermous T. Circulating peptide prevents preeclampsia. Science. 2017;357:643–644. doi: 10.1126/science.aao2642. [DOI] [PubMed] [Google Scholar]
- 6.Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semin Perinatol. 2012;36:56–59. doi: 10.1053/j.semperi.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Rolnik DL, Wright D, Poon LC, O'Gorman N, Syngelaki A, de Paco Matallana C, Akolekar R, Cicero S, Janga D, Singh M, et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. N Engl J Med. 2017;377:613–622. doi: 10.1056/NEJMoa1704559. [DOI] [PubMed] [Google Scholar]
- 8.LeFevre ML, U.S. Preventive Services Task Force Low-dose aspirin use for the prevention of morbidity and mortality from preeclampsia: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;161:819–826. doi: 10.7326/M14-1884. [DOI] [PubMed] [Google Scholar]
- 9.Duley L, Meher S, Abalos E. Management of preeclampsia. BMJ. 2006;332:463–468. doi: 10.1136/bmj.332.7539.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain N, Schatz F, Paidas MJ. Heparin and maternal fetal interface: Why should it work to prevent pregnancy complications? Thromb Res. 2009;124:653–655. doi: 10.1016/j.thromres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Lowe SA, Brown MA, Dekker GA, Gatt S, McLintock CK, McMahon LP, Mangos G, Moore MP, Muller P, Paech M, et al. Guidelines for the management of hypertensive disorders of pregnancy 2008. Aust N Z J Obstet Gynaecol. 2009;49:242–246. doi: 10.1111/j.1479-828X.2009.01003.x. [DOI] [PubMed] [Google Scholar]
- 12.Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta. 2006;27:939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Jeyabalan A. Epidemiology of preeclampsia: Impact of obesity. Nutr Rev. 2013;71(Suppl 1):S18–S25. doi: 10.1111/nure.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powe CE, Levine RJ, Karumanchi SA. Preeclampsia, a disease of the maternal endothelium: The role of antiangiogenic factors and implications for later cardiovascular disease. Circulation. 2011;123:2856–2869. doi: 10.1161/CIRCULATIONAHA.109.853127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young BC, Levine RJ, Karumanchi SA. Pathogenesis of preeclampsia. Annu Rev Pathol. 2010;5:173–192. doi: 10.1146/annurev-pathol-121808-102149. [DOI] [PubMed] [Google Scholar]
- 16.Roberts JM, Taylor RN, Musci TJ, Rodgers GM, Hubel CA, McLaughlin MK. Preeclampsia: An endothelial cell disorder. Am J Obstet Gynecol. 1989;161:1200–1204. doi: 10.1016/0002-9378(89)90665-0. [DOI] [PubMed] [Google Scholar]
- 17.Brownfoot FC, Hastie R, Hannan NJ, Cannon P, Tuohey L, Parry LJ, Senadheera S, Illanes SE, Kaitu'u-Lino TJ, Tong S. Metformin as a prevention and treatment for preeclampsia: Effects on soluble fms-like tyrosine kinase 1 and soluble endoglin secretion and endothelial dysfunction. Am J Obstet Gynecol. 2016;214:356.e1–356.e15. doi: 10.1016/j.ajog.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell Mol Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PubMed] [Google Scholar]
- 19.Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Rio GD, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Xiao J, Wan J, Cohen P, Yen K. Mitochondrially derived peptides as novel regulators of metabolism. J Physiol. 2017;595:6613–6621. doi: 10.1113/JP274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkov V, Mosgoeller W, Ziesche R, Raderer M, Stiebellehner L, Vonbank K, Funk GC, Hamilton G, Novotny C, Burian B, Block LH. Vasoactive intestinal peptide as a new drug for treatment of primary pulmonary hypertension. J Clin Invest. 2003;111:1339–1346. doi: 10.1172/JCI17500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho L, van Dijk M, Chye STJ, Messerschmidt DM, Chng SC, Ong S, Yi LK, Boussata S, Goh GH, Afink GB, et al. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science. 2017;357:707–713. doi: 10.1126/science.aam6607. [DOI] [PubMed] [Google Scholar]
- 23.Jacobsen LV, Flint A, Olsen AK, Ingwersen SH. Liraglutide in type 2 diabetes mellitus: Clinical pharmacokinetics and pharmacodynamics. Clin Pharmacokinet. 2016;55:657–672. doi: 10.1007/s40262-015-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleeman A, Clements JN. Abaloparatide: A new pharmacological option for osteoporosis. Am J Health Syst Pharm. 2019;76:130–135. doi: 10.1093/ajhp/zxy022. [DOI] [PubMed] [Google Scholar]
- 25.Dai X, Song X, Rui C, Meng L, Xue X, Ding H, Shen R, Li J, Li J, Lu Y, Long W. Peptidome analysis of human serum from normal and preeclamptic pregnancies. J Cell Biochem. 2017;118:4341–4348. doi: 10.1002/jcb.26087. [DOI] [PubMed] [Google Scholar]
- 26.Chen TG, Zhong ZY, Sun GF, Zhou YX, Zhao Y. Effects of tumour necrosis factor-alpha on activity and nitric oxide synthase of endothelial progenitor cells from peripheral blood. Cell Prolif. 2011;44:352–359. doi: 10.1111/j.1365-2184.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerhardt S, König V, Doll M, Hailemariam-Jahn T, Hrgovic I, Zöller N, Kaufmann R, Kippenberger S, Meissner M. Dimethylfumarate protects against TNF-α-induced secretion of inflammatory cytokines in human endothelial cells. J Inflamm (Lond) 2015;12:49. doi: 10.1186/s12950-015-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fratantonio D, Speciale A, Molonia MS, Bashllari R, Palumbo M, Saija A, Cimino F, Monastra G, Virgili F. Alpha-lipoic acid, but not di-hydrolipoic acid, activates Nrf2 response in primary human umbilical-vein endothelial cells and protects against TNF-α induced endothelium dysfunction. Arch Biochem Biophys. 2018;655:18–25. doi: 10.1016/j.abb.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Zeng RJ, Zheng CW, Gu JE, Zhang HX, Xie L, Xu LY, Li EM. RAC1 inhibition reverses cisplatin resistance in esophageal squamous cell carcinoma and induces downregulation of glycolytic enzymes. Mol Oncol. 2019;13:2010–2030. doi: 10.1002/1878-0261.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai S, Zhu Q, Guo C, Yuan R, Zhang X, Nie Y, Chen L, Fang Y, Chen K, Zhang J, et al. MLL1 promotes myogenesis by epigenetically regulating Myf5. Cell Prolif. 2020;53:e12744. doi: 10.1111/cpr.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Q, Li Q, Xie F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther. 2019;12:6297–6307. doi: 10.2147/OTT.S214689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv Y, Lu X, Li C, Fan Y, Ji X, Long W, Meng L, Wu L, Wang L, Lv M, Ding H. miR-145-5p promotes trophoblast cell growth and invasion by targeting FLT1. Life Sci. 2019;239:117008. doi: 10.1016/j.lfs.2019.117008. [DOI] [PubMed] [Google Scholar]
- 33.Lip SV, Boekschoten MV, Hooiveld GJ, van Pampus MG, Scherjon SA, Plösch T, Faas MM. Early-onset preeclampsia, plasma microRNAs, and endothelial cell function. Am J Obstet Gynecol. 2020;222:497.e1–497.e12. doi: 10.1016/j.ajog.2019.11.1286. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Bi N, Wu L, Ding X, Men Y, Zhou W, Li L, Zhang W, Shi S, Song Y, Wang L. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16:50. doi: 10.1186/s12943-017-0620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Tao W, Wang Y, Bikow J, Lu B, Keating A, Verma S, Parker TG, Han R, Wen XY. Rosuvastatin, identified from a zebrafish chemical genetic screen for antiangiogenic compounds, suppresses the growth of prostate cancer. Eur Urol. 2010;58:418–426. doi: 10.1016/j.eururo.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Zhang W, Xu Q, Zhang J, Chen W, Xu Z, Li C, Wang Z, Zhang Y, Zhen Y, et al. Ang-(17) protects HUVECs from high glucose-induced injury and inflammation via inhibition of the JAK2/STAT3 pathway. Int J Mol Med. 2018;41:2865–2878. doi: 10.3892/ijmm.2018.3507. [DOI] [PubMed] [Google Scholar]
- 37.Jin J, Qiu S, Wang P, Liang X, Huang F, Wu H, Zhang B, Zhang W, Tian X, Xu R, et al. Cardamonin inhibits breast cancer growth by repressing HIF-1α-dependent metabolic reprogramming. J Exp Clin Cancer Res. 2019;38:377. doi: 10.1186/s13046-019-1351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi L, Zhang Y, Xu Y, Zheng X, Zhang X, Wang Z, Xue M, Jin X. HMGB1/RAGE pro-inflammatory axis promotes vascular endothelial cell apoptosis in limb ischemia/reperfusion injury. Biomed Pharmacother. 2019;116:109005. doi: 10.1016/j.biopha.2019.109005. [DOI] [PubMed] [Google Scholar]
- 39.Xiang Y, Yao X, Wang X, Zhao H, Zou H, Wang L, Zhang QX. Houshiheisan promotes angiogenesis via HIF-1α/VEGF and SDF-1/CXCR4 pathways: In vivo and in vitro. Biosci Rep. 2019;39:BSR20191006. doi: 10.1042/BSR20191006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Yang J, Bao J, Li X, Ye A, Zhang G, Liu H. Activation of the cholinergic anti-inflammatory pathway by nicotine ameliorates lipopolysaccharide-induced preeclampsia-like symptoms in pregnant rats. Placenta. 2017;49:23–32. doi: 10.1016/j.placenta.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 41.National Research Council Committee for the Update of the Guide for the Care and use of Laboratory Animals, editor. Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press; Washington, DC: 2011. The National Academies Collection: Reports funded by National Institutes of Health. [Google Scholar]
- 42.Shu W, Li H, Gong H, Zhang M, Niu X, Ma Y, Zhang X, Cai W, Yang G, Wei M, et al. Evaluation of blood vessel injury, oxidative stress and circulating inflammatory factors in an L-NAME-induced preeclampsia-like rat model. Exp Ther Med. 2018;16:585–594. doi: 10.3892/etm.2018.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yalamati P, Bhongir AV, Karra M, Beedu SR. comparative analysis of urinary total proteins by bicinchoninic acid and pyrogallol red molybdate methods. J Clin Diagn Res. 2015;9:Bc01–04. doi: 10.7860/JCDR/2015/13543.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai WS, Ding YL. GNG7 silencing promotes the proliferation and differentiation of placental cytotrophoblasts in preeclampsia rats through activation of the mTOR signaling pathway. Int J Mol Med. 2019;43:1939–1950. doi: 10.3892/ijmm.2019.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014;1180:31–43. doi: 10.1007/978-1-4939-1050-2_3. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto K, Arao T, Tanaka K, Kaneda H, Kudo K, Fujita Y, Tamura D, Aomatsu K, Tamura T, Yamada Y, et al. mTOR signal and hypoxia-inducible factor-1 alpha regulate CD133 expression in cancer cells. Cancer Res. 2009;69:7160–7164. doi: 10.1158/0008-5472.CAN-09-1289. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Y, Shan N, Tan B, Deng Q, Liu Y, Wang H, Luo X, He C, Luo X, Zhang H, et al. SRC-3 plays a critical role in human umbilical vein endothelial cells by regulating the PI3K/Akt/mTOR pathway in preeclampsia. Reprod Sci. 2018;25:748–758. doi: 10.1177/1933719117725818. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Jonsson Y, Rubèr M, Matthiesen L, Berg G, Nieminen K, Sharma S, Ernerudh J, Ekerfelt C. Cytokine mapping of sera from women with preeclampsia and normal pregnancies. J Reprod Immunol. 2006;70:83–91. doi: 10.1016/j.jri.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Yue C, Xu J, Zhan Y, Zhao H, Li Y, Ye Y. Downregulation of receptor tyrosine kinase-like orphan receptor 1 in preeclampsia placenta inhibits human trophoblast cell proliferation, migration, and invasion by PI3K/AKT/mTOR pathway accommodation. Placenta. 2019;82:17–24. doi: 10.1016/j.placenta.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Harati-Sadegh M, Kohan L, Teimoori B, Mehrabani M, Salimi S. The association of the placental Hypoxia-inducible factor1-α polymorphisms and HIF1-α mRNA expression with preeclampsia. Placenta. 2018;67:31–37. doi: 10.1016/j.placenta.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Grotegut CA. Prevention of preeclampsia. J Clin Invest. 2016;126:4396–4398. doi: 10.1172/JCI91300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen JM, Kramer MS, Platt RW, Basso O, Evans RW, Kahn SR. The association between maternal antioxidant levels in midpregnancy and preeclampsia. Am J Obstet Gynecol. 2015;213:695.e1–613. doi: 10.1016/j.ajog.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 54.Bahado-Singh RO, Syngelaki A, Akolekar R, Mandal R, Bjondahl TC, Han B, Dong E, Bauer S, Alpay-Savasan Z, Graham S, et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am J Obstet Gynecol. 2015;213:530.e1–530.e10. doi: 10.1016/j.ajog.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 55.Ray JG, Booth GL, Alter DA, Vermeulen MJ. Prognosis after maternal placental events and revascularization: PAMPER study. Am J Obstet Gynecol. 2016;214:106.e1–106.e14. doi: 10.1016/j.ajog.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Szabo G, Molvarec A, Nagy B, Rigo J., Jr Increased B-type natriuretic peptide levels in early-onset versus late-onset preeclampsia. Clin Chem Lab Med. 2014;52:281–288. doi: 10.1515/cclm-2013-0307. [DOI] [PubMed] [Google Scholar]
- 57.Borges VTM, Zanati SG, Peraçoli MTS, Poiati JR, Romão-Veiga M, Peraçoli JC, Thilaganathan B. Maternal left ventricular hypertrophy and diastolic dysfunction and brain natriuretic peptide concentration in early- and late-onset pre-eclampsia. Ultrasound Obstet Gynecol. 2018;51:519–523. doi: 10.1002/uog.17495. [DOI] [PubMed] [Google Scholar]
- 58.Chatterjee P, Chiasson VL, Seerangan G, De Guzman E, Milad M, Bounds KR, Gasheva O, Tobin RP, Hatahet M, Kopriva S, et al. Depletion of MHC class II invariant chain peptide or γ-δ T-cells ameliorates experimental preeclampsia. Clin Sci (Lond) 2017;131:2047–2058. doi: 10.1042/CS20171008. [DOI] [PubMed] [Google Scholar]
- 59.Kim J, Lee KS, Kim JH, Lee DK, Park M, Choi S, Park W, Kim S, Choi YK, Hwang JY, et al. Aspirin prevents TNF-α-induced endothelial cell dysfunction by regulating the NF-κB-dependent miR-155/eNOS pathway: Role of a miR-155/eNOS axis in preeclampsia. Free Radic Biol Med. 2017;104:185–198. doi: 10.1016/j.freeradbiomed.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Shaw J, Tang Z, Schneider H, Salje K, Hansson SR, Guller S. Inflammatory processes are specifically enhanced in endothelial cells by placental-derived TNF-α: Implications in preeclampsia (PE) Placenta. 2016;43:1–8. doi: 10.1016/j.placenta.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 61.Barone Gibbs B, Dobrosielski DA, Bonekamp S, Stewart KJ, Clark JM. A randomized trial of exercise for blood pressure reduction in type 2 diabetes: Effect on flow-mediated dilation and circulating biomarkers of endothelial function. Atherosclerosis. 2012;224:446–453. doi: 10.1016/j.atherosclerosis.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Onda K, Tong S, Nakahara A, Kondo M, Monchusho H, Hirano T, Kaitu'u-Lino T, Beard S, Binder N, Tuohey L, et al. Sofalcone upregulates the nuclear factor (erythroid-derived 2)-like 2/heme oxygenase-1 pathway, reduces soluble fms-like tyrosine kinase-1, and quenches endothelial dysfunction: Potential therapeutic for preeclampsia. Hypertension. 2015;65:855–862. doi: 10.1161/HYPERTENSIONAHA.114.04781. [DOI] [PubMed] [Google Scholar]
- 63.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: The path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 64.Bartholomeusz C, Gonzalez-Angulo AM. Targeting the PI3K signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:121–130. doi: 10.1517/14728222.2011.644788. [DOI] [PubMed] [Google Scholar]
- 65.Kawahara T, Asthana S, Kneteman NM. m-TOR inhibitors: What role in liver transplantation? J Hepatol. 2011;55:1441–1451. doi: 10.1016/j.jhep.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Li Q, Jiang W, Xiong X, Li H, Zhao J, Qi H. LAMA5 promotes human umbilical vein endothelial cells migration, proliferation, and angiogenesis and is decreased in preeclampsia. J Matern Fetal Neonatal Med. 2020;33:1114–1124. doi: 10.1080/14767058.2018.1514597. [DOI] [PubMed] [Google Scholar]
- 67.Oladipupo S, Hu S, Kovalski J, Yao J, Santeford A, Sohn RE, Shohet R, Maslov K, Wang LV, Arbeit JM. VEGF is essential for hypoxia-inducible factor-mediated neovascularization but dispensable for endothelial sprouting. Proc Natl Acad Sci USA. 2011;108:13264–13269. doi: 10.1073/pnas.1101321108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukushima K, Murata M, Hachisuga M, Tsukimori K, Seki H, Takeda S, Asanoma K, Wake N. Hypoxia inducible factor 1 alpha regulates matrigel-induced endovascular differentiation under normoxia in a human extravillous trophoblast cell line. Placenta. 2008;29:324–331. doi: 10.1016/j.placenta.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 69.Kovo M, Schreiber L, Elyashiv O, Ben-Haroush A, Abraham G, Bar J. Pregnancy outcome and placental findings in pregnancies complicated by fetal growth restriction with and without preeclampsia. Reprod Sci. 2015;22:316–321. doi: 10.1177/1933719114542024. [DOI] [PubMed] [Google Scholar]
- 70.Cotechini T, Komisarenko M, Sperou A, Macdonald-Goodfellow S, Adams MA, Graham CH. Inflammation in rat pregnancy inhibits spiral artery remodeling leading to fetal growth restriction and features of preeclampsia. J Exp Med. 2014;211:165–179. doi: 10.1084/jem.20130295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szarka A, Rigó J, Jr, Lázár L, Beko G, Molvarec A. Circulating cytokines, chemokines and adhesion molecules in normal pregnancy and preeclampsia determined by multiplex suspension array. BMC immunol. 2010;11:59. doi: 10.1186/1471-2172-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.